Corrigendum: Staphylococcus epidermidis Boosts Innate Immune Response by Activation of Gamma Delta T Cells and Induction of Perforin-2 in Human Skin
- 1Wound Healing and Regenerative Medicine Research Program, Dr. Phillip Frost Department of Dermatology and Cutaneous Surgery, University of Miami Miller School of Medicine, Miami, FL, United States
- 2Department of Microbiology and Immunology, University of Miami Miller School of Medicine, Miami, FL, United States
- 3Division of Plastic Surgery Dewitt Daughtry, Department of Surgery, University of Miami Miller School of Medicine, Miami, FL, United States
Perforin-2 (P-2) is an antimicrobial protein with unique properties to kill intracellular bacteria. Gamma delta (GD) T cells, as the major T cell population in epithelial tissues, play a central role in protective and pathogenic immune responses in the skin. However, the tissue-specific mechanisms that control the innate immune response and the effector functions of GD T cells, especially the cross-talk with commensal organisms, are not very well understood. We hypothesized that the most prevalent skin commensal microorganism, Staphylococcus epidermidis, may play a role in regulating GD T cell-mediated cutaneous responses. We analyzed antimicrobial protein P-2 expression in human skin at a single cell resolution using an amplified fluorescence in situ hybridization approach to detect P-2 mRNA in combination with immunophenotyping. We show that S. epidermidis activates GD T cells and upregulates P-2 in human skin ex vivo in a cell-specific manner. Furthermore, P-2 upregulation following S. epidermidis stimulation correlates with increased ability of skin cells to kill intracellular Staphylococcus aureus. Our findings are the first to reveal that skin commensal bacteria induce P-2 expression, which may be utilized beneficially to modulate host innate immune responses and protect from skin infections.
Introduction
Skin, in the same fashion as all other epithelial barrier sites (gastrointestinal, reproductive, and respiratory tracts) harbors a distinct community of commensal microbes that modulate the host immune system (1–3). One of the most common members of the healthy cutaneous microbiome is Staphylococcus epidermidis. S. epidermidis stimulates antimicrobial peptide production by skin cells (4–11), which may provide protection against pathogenic bacteria (4, 5, 10–12). Recent studies reported that colonization of mouse skin with S. epidermidis induced commensal-specific tissue (skin)-resident memory T cells that demonstrated immunoregulatory and tissue repair properties. This was proposed as a novel S. epidermidis mediated mechanism for rapid immune response and tissue protection from invasive pathogens (13–15).
Multiple lines of evidence have shown that gamma delta (GD) T cells display strong activities against bacteria (16–20), parasites (21), and viruses (22, 23). In marked contrast to αβ T lymphocytes (24–29), GD T cells recognize antigens independently of peptide processing and major histocompatibility complex (MHC)-restricted antigen presentation. They are activated by signs of tissue stress, including infected or transformed cells, and respond by deploying an immediate and efficient killing response or by regulating the immune response against them. Phosphoantigens and several other molecules of microbial origin have been proposed as GD T cell antigens accounting for the specific recognition of infected cells. These candidates include the Staphylococcus aureus superantigens Staphylococcal enterotoxin A (SEA) (and to a lesser extent staphylococcal enterotoxin E (SEE) (30, 31), which are recognized by the GD T cell receptor (TCR) independently from antigen processing and MHC presentation. Although GD T cells are one of the predominant lymphocyte subsets in mouse and human skin (32) that are essential for skin homeostatic and protective pathways against S. aureus (33), the contribution of commensal-derived antigens to the activation of GD T cells and their effector function, particularly their cytotoxic potential, has not been established. Furthermore, the extent to which GD T cells promote cutaneous tissue physiology remains to be determined.
Perforin-2 (P-2)/MPEG1 is a highly conserved member of the membrane attack complex (MAC)/perforin-like (PF)/cholesterol-dependent cytolysin (MACPF/CDC) superfamily (34–36). In contrast to all other MACPF/CDC members, P-2 is a type-1 transmembrane protein that traffics throughout the endosomal pathway to the late-endosome and phagosome (37–39). Therefore, P-2 can form pores in bacterial membranes and damage engulfed microbes within the phagolysosome (37, 40). In the absence of P-2, the other innate defense effectors including reactive oxygen species and nitric oxide, were unable to prevent the replication and systemic dissemination of intracellular pathogens (37, 41, 42). Dr. Eckhard Podack’s group was the first to report about major P-2 functions as an antibacterial effector protein of the innate immune system in phagocytic and in tissue forming cells (37, 41). Although we recently reported specific distribution of P-2 in normal human skin (43), the mechanisms involved in the regulation of P-2 expression have not been well established. Moreover, the effect of P-2 function within the complex system of host-microbe interactions has important implication for our understanding of skin immunity and diseases.
Here we established a human skin ex vivo model to study the effect of S. epidermidis on the skin innate immune response and on the novel antimicrobial protein P-2. We report that S. epidermidis activates skin GD T cells, specifically through P-2 induction, which has demonstrated antibacterial effects in other cell subsets (macrophages and fibroblasts) (37, 42). Importantly, S. epidermidis mediated induction of P-2 correlated with an enhanced ability of the skin cells to eliminate intracellular S. aureus.
Materials and Methods
Bacterial Strains and Culture Conditions
Staphylococcus epidermidis CCN021 and CCN0024, human commensal S. epidermidis strains, were obtained from GP (University of Miami). S. epidermidis ATCC 12228 was a gift from Prof. Davis (University of Miami). S. epidermidis CCN021 and CCN0024 were isolated from a healthy volunteer and characterized by phenotypic and qPCR identification techniques (44, 45). Staphylococci were routinely grown aerobically with agitation, at 37°C, in Luria-Bertani (LB) broth. For pre-treatment, bacteria were diluted in fresh LB and grown to mid-log growth phase. Before application on ex vivo human skin or single cell suspension, bacteria were harvested by centrifugation and washed with phosphate buffered saline (PBS). The bacterial density and the absence of contamination were controlled by numeration of colony forming units (CFU).
The GFP containing USA300 Methicillin resistant Staphylococcus aureus (MRSA) strain AH1726 [MRSA LAC (AH1263) + pCM29 (CmR)] (46) was obtained from GP (University of Miami). MRSA was grown aerobically with agitation overnight at 37°C in LB supplemented with 10 μg/mL chloramphenicol to retain the GFP plasmid.
Ex vivo Human Skin Explant System
Discarded human skin tissue was obtained from voluntary reduction surgeries (n = 6) at the University of Miami (UM) Hospital and as such were found to be exempt from human subject research under CFR46.101.2 by the Institutional Review Board at the UM Miller School of Medicine.
Skin samples were processed to remove subcutaneous fat and washed with PBS. Multiple 8 mm punch biopsies were obtained from each specimen and placed individually into 0.4 μm PET-membrane trans-wells (Millipore) in a 12 well plate containing 1 ml media per well (RPMI, 10% FBS, 1% HEPES). Skin specimens were maintained at the air-liquid interface as previously described (43, 47–50).
Human Skin Single Cell Suspension
Cells were isolated from healthy human skin using the MACS Whole Skin Dissociation Kit (Miltenyi 130-101-540). Briefly, subcutaneous fat was removed, and sterilization of human skin was optimized to remove any commensal or pathogenic microorganisms. Skin was washed with Gibco® Antibiotic-Antimycotic (ABAM) (Life Technologies) to prevent bacterial and fungal contamination. This solution contains 10,000 units/mL of penicillin, 10,000 μg/mL of streptomycin, and 25 μg/mL of Gibco Amphotericin B. After washings with ABAM, skin was washed in PBS (46). Three 4 mm diameter punches were digested overnight at 37°C using enzymes from a whole-skin dissociation kit (Miltenyi, Bergisch Gladbach, Germany). The resulting cell suspension was filtered through a 70 μm cell strainer and centrifuged at 1,500 r.p.m. for 10 min at 4°C. The supernatant was removed, and the pellet was washed once with PBS. Obtained cell suspensions were washed with IMDM (Gibco-Thermo Fisher Scientific) supplemented with 10% heat-inactivated FBS, 2 mM L-glutamine, 0.15% sodium hydrogencarbonate, 1 mM sodium pyruvate, and non-essential amino acids.
Ex vivo and in vitro Skin Stimulation With S. epidermidis
Staphylococcus epidermidis CCN021 was prepared for stimulation experiments as described above and 20 μL of the bacteria solution (approx. 6 Log CFU) was added centrally onto the epidermis while control samples were treated with PBS. After 24, 48, 72, and 96 h of incubation at 37°C in a 5% CO2 atmosphere, tissue samples were either digested with collagenase for further cell viability and FISH/Flow analysis (Supplementary Figure S2), preserved in RNA-later for RNA isolation, or used for CFU enumeration. CFU count was determined after overnight colony growth and expressed as CFU/ml.
Skin cell suspension obtained after whole-skin dissociation was used for in vitro stimulation with S. epidermidis CCN021, CCN0024, and ATCC 12228. Cells were plated on 24 well plates with 1 million cells per well and treated with S. epidermidis at a multiplicity of infection (MOI) of 20 for 24 h. The control cells were exposed to media only.
Intracellular MRSA Killing Assay
After 24 h stimulation with S. epidermidis, skin cells were washed twice with warmed plain IMDM and infected with MRSA at an MOI of 20 for 1 h. Cells were washed twice with IMDM after infection and fresh media containing gentamicin (50 μg/mL) was added for 30 min to eliminate extracellular bacteria. Samples were collected 30 and 90 min after intracellular infection for enumeration of intracellular colony forming units (CFU) and for FISH flow analysis as described before (43). To release intracellular bacterial load, cells were subjected to hypotonic lysis with 0.1% Triton X in PBS. Lysates were plated on agar plates containing 10 μg/mL chloramphenicol for CFU quantification (43).
FISH-Flow P-2 RNA Assay and Flow Cytometric Analysis
Single cell suspensions obtained from full thickness samples or after stimulation with S. epidermidis and MRSA were first labeled with live/dead detection kit (Yellow Amine, Thermo Fisher Scientific) and then with the following fluorescently labeled antibodies: CD45-Alexa Fluor 700, TCR GD-PE-Cy7, CD31-PacBlue, CD104-FITC, CD325-PerCPCy5.5, and CCRL1-PE (Biolegend, San Diego, CA, United States). We also stained cells with fluorescently labeled antibodies for TLR1-BV570, TLR2-PE, TLR6-BV605, and TCR GD 1 FITC (Biolegend, San Diego, CA, United States). P-2 mRNA was detected using an amplified signal FISH technique (PrimeFlow; Affymetrix/eBioscience-Thermo Fisher Scientific). For mRNA detection, target probe hybridization was performed using type 1 (AlexaFluor647) probes for P-2 as described (43). Approximately 20,000 cell events were acquired from each sample on flow cytometer equipped with 405 nm, 488 nm, 642 nm, and 785 nm (SSC) lasers (Fortessa X-50, BD Immunocytometry Systems, San Jose, CA, United States). Spectral compensation was completed using single color control samples and antibody capture beads (BD Biosciences). Data were analyzed using FlowJo version 10.2 (TreeStar).
GD T Cell Sorting and Real-Time PCR
Two-way sorting was performed to obtain purified GD T cells by sterile sorting on a SONY SH800S cell sorter (SONY Biotechnology, San Jose, CA, United States). Briefly, single cell suspensions were labeled with Live/Dead Violet, CD45, CD3, and TCR GD. GD T cells were sorted as Live/Dead-CD45+ CD3+ TCR GD+ cell population. 5,000–10,000 sorted cells were collected from three donors. Purity of sorted GD T population was >97%.
After sorting, cells were stimulated with S. epidermidis at an MOI of 20 for 1 h, washed with PBS, spun down, and kept on ice briefly prior to performing one-step reverse transcription and cDNA amplification of specific targets using a pool of TaqmanTM gene expression assays (Thermo Fisher Scientific). Resulting cDNA was loaded onto BioMark IFC 96 × 96 chip (Fluidigm) according to the manufacturer’s protocol. Raw data underwent “cellular detection rate” (CDR) filtering to remove outlier samples and genes based on dataset distribution (51, 52). CD74 (also known as HLADG) was used as a surrogate for the presence of a cell (i.e., loading control) due to its stable expression in lymphocytes. Cells that had low or absent CD74 expression exhibited reduced gene expression globally and were removed from analysis. Differential gene expression analysis was subsequently performed to contrast transcriptional profiles of GD T cells between unstimulated and S. epidermidis stimulated samples.
RT-PCR for Antimicrobial Peptides and Pro-inflammatory Cytokines
Total RNA from human skin was extracted using the miRNeasy kit (QIAGEN, Valencia, CA, United States) per manufacturer’s instructions as previously described (49). cDNA was made with qScriptTM Synthesis kit (Quanta BioSciences Inc.,Gaithersburg, MD, United States). ARPC2 was used as a reference gene for normalization, forward 5′-TCCGGGACTACCTGCACTAC-3′, reverse 5′-GGTTCAGCACCTTGAGGAAG-3′. All real-time PCR (qPCR) reactions were performed in triplicate using PerfeCTa® SYBR® Green SuperMix (Quanta BioSciences) and quantified using the ddCT method. The primer sequences were IL-1α forward 5′-AGATGCCTGAGATACCCAAAACC-3′ reverse 5′-CCAAGCACACCCAGTAGTCT-3′, defensin β4 (DefB4) forward 5′-GGTGGTATAGGCGATCCTGTT-3′ reverse 5′-AGGGCAAAAGACTGGATGACA-3′, and cathelicidin (LL37) forward 5′-GGGCAAAAGACTGGATGACA-3′ reverse 5′-TCTTGAAGTCACAATCCTCTGGT-3′.
Statistical Analysis
All experiments were conducted independently at least three times on different days. Comparisons of flow cytometry cell frequencies was measured by the two-way ANOVA test with Holm-Sidak multiple-comparison test, ∗p < 0.05, ∗∗p < 0.01, and ∗∗∗p < 0.001 or Student t-test using the Prism software (GraphPad software). Comparisons of PCR array data were performed using t-test (two tail distribution and equal variances between the two groups) based on the triplicate 2^(−ΔCT) values for each gene in the S. epidermidis treated group and control group. Error bars in all figures are reported as a SEM.
Results
S. epidermidis Contributes to Increased Number of Human GD T Cells
Staphylococcus epidermidis is an important skin commensal organism and modulator of cutaneous innate immune responses (9, 10). Here we established an ex vivo skin model of S. epidermidis colonization to study the effect of this commensal microorganism on skin innate immune responses including GD T cell activity. S. epidermidis was topically applied onto the epidermis. Tissue was collected at different time points during colonization and then dissociated into single cell suspensions (Figure 1). There were no statistical differences in viability of S. epidermidis treated and control tissue (Supplementary Figure S1). We analyzed the GD T cell subset in the control and S. epidermidis colonized human skin by flow cytometry and observed a statistically significant (p < 0.01) increase in the frequency as well as in the total number of GD T cells within live, CD45+ CD3+ skin cells after 72 h of S. epidermidis stimulation compared to control tissue (Figures 1A,B). We confirmed S. epidermidis colonization in human skin by CFU quantification (Figure 1C).
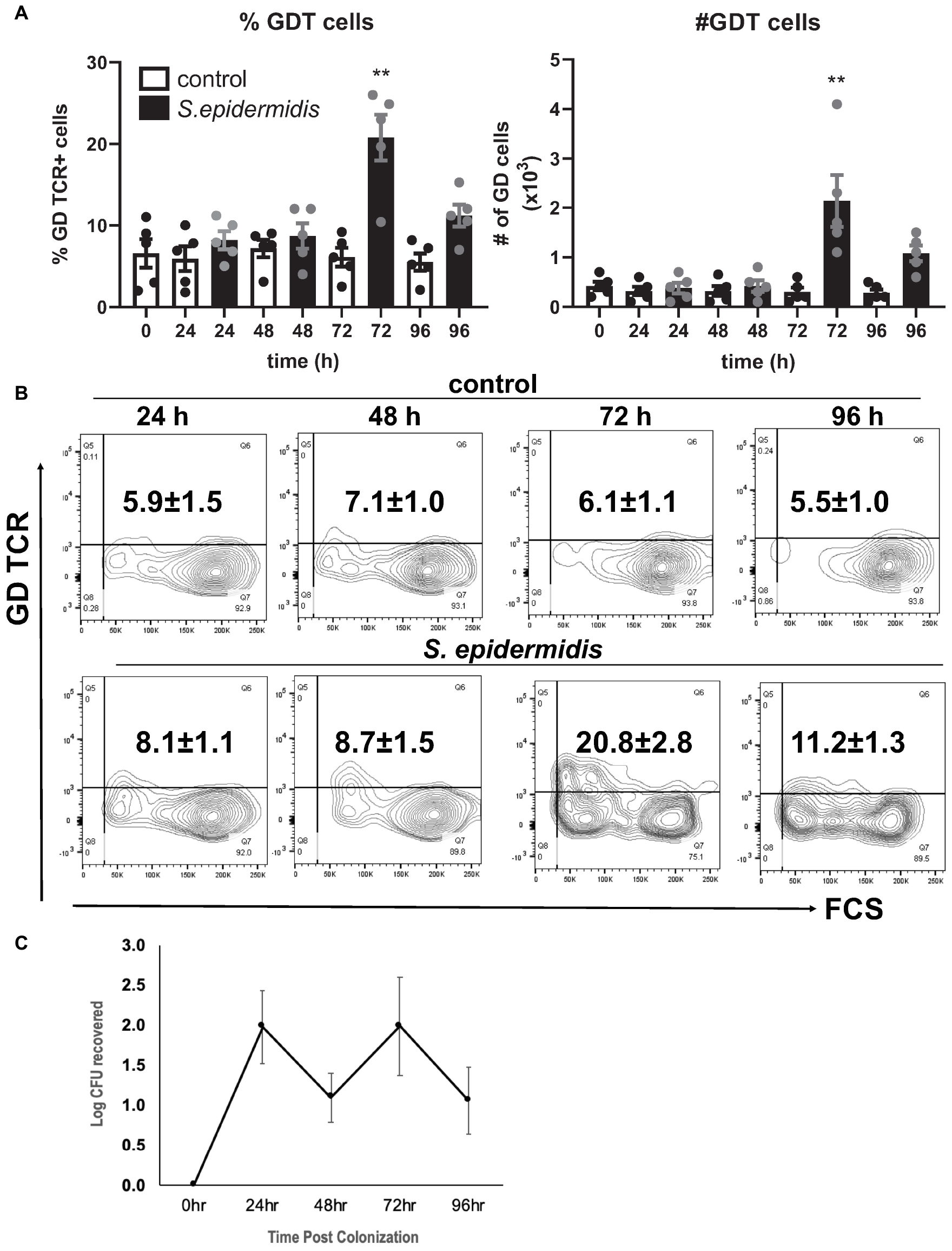
Figure 1. Staphylococcus epidermidis increases the number of GD T cells in human skin ex vivo. Control, uncolonized, and S. epidermidis colonized skin was maintained on air liquid interface and collected at indicated time points (0, 24, 48, 72, and 96 h). Single cell suspensions were obtained and labeled with live/dead stain, CD45, CD3, and GD TCR. (A) Cells were analyzed using flow cytometry and gated on the CD45+ CD3+ GDT+ population. Bar graphs show SEM frequency (%) and SEM number (#) of skin GD T cells (n = 5). (B) Representative contour plots showing frequency of GD TCR in control and S. epidermidis colonized skin. (C) Number of S. epidermidis colony forming units (CFU) recovered from ex vivo skin explants colonized with S. epidermidis CCN021 on day 0 through day 4. Data represent at least two technical replicates and five independent biological replicates per group. **p < 0.01 (two-way ANOVA with Holm-Sidak multiple-comparison test).
S. epidermidis Induces P-2 in Human GD T Cells, Keratinocytes, and Papillary Fibroblasts ex vivo
We have previously described an amplified fluorescence in situ hybridization (FISH) technique for detection of mRNA in combination with immune-phenotyping in human skin (43). We and others found that P-2 is an antimicrobial protein crucial for intracellular bacteria killing (37, 38, 40, 41). Here, we analyzed P-2 expression in different skin cell subsets after stimulation with S. epidermidis. First, we found that P-2 expression was significantly upregulated (p < 0.05 and p < 0.01) in GD T cells from human skin explants colonized with S. epidermidis at 24, 48, and 72 h compared to the uncolonized control (Figures 2A,B). Moreover, our analysis revealed that S. epidermidis stimulation for 96 h upregulated P-2 in the basal layer keratinocytes after an initial suppression observed at 24 h (CD45-CD31-CD104+ cells) (Figure 3). Two major human skin fibroblast subsets, papillary and reticular fibroblasts, based on their expression of CCRL1 and CD325, respectively (53, 54) were also tested. We found that only papillary fibroblasts, CCRL1+ cells, upregulate P-2 96 h post S. epidermidis colonization. P-2 expression in reticular fibroblasts was not affected at any time point and was lower overall compared to other cell subtypes (Figure 3).
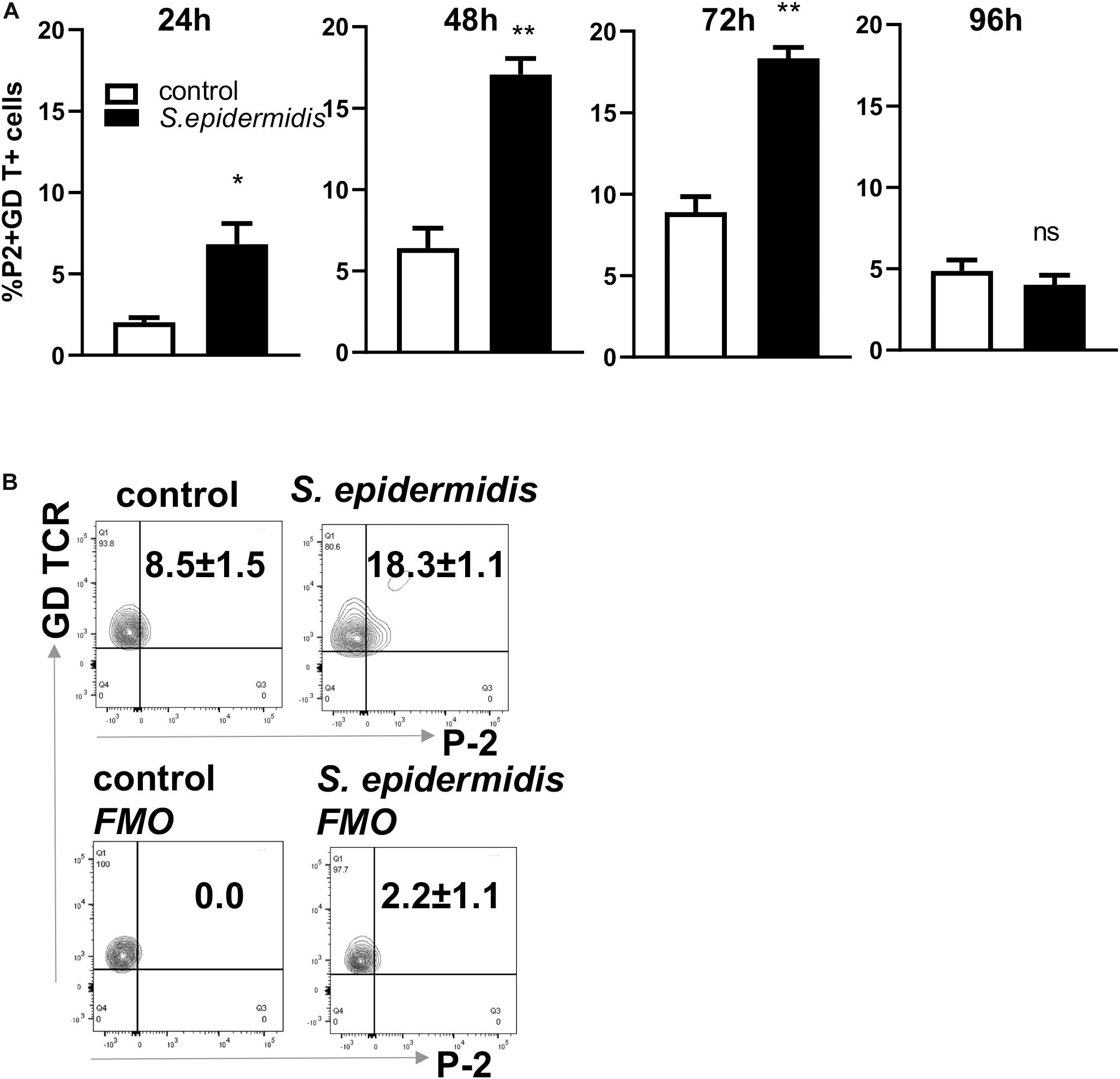
Figure 2. Colonization of human skin with Staphylococcus epidermis induces P-2 in GD T cells. (A) Control and S. epidermidis colonized human skin was collected at indicated time points (24, 48, 72, and 96 h) for FISH-Flow. Single cell suspensions were obtained using Collagenase D and labeled with live/dead stain. Using FISH-Flow RNA assay, P-2 RNA levels were analyzed in the CD45+, CD3+ GD TCR+ cell population. (B) Representative dot plot graph showing expression of mRNA P-2 in gated CD45+ CD3+ GD TCR+ T cells at 72 h in S. epidermidis colonized skin or non-colonized (control). FMO-fluorescence minus one. Bar graphs show SEM of P-2 mRNA positive cells within skin GD T cells (n = 3). Data represent at least two technical replicates with three independent biological replicates per group. *p < 0.05, **p < 0.01 as calculated using Student t-test.
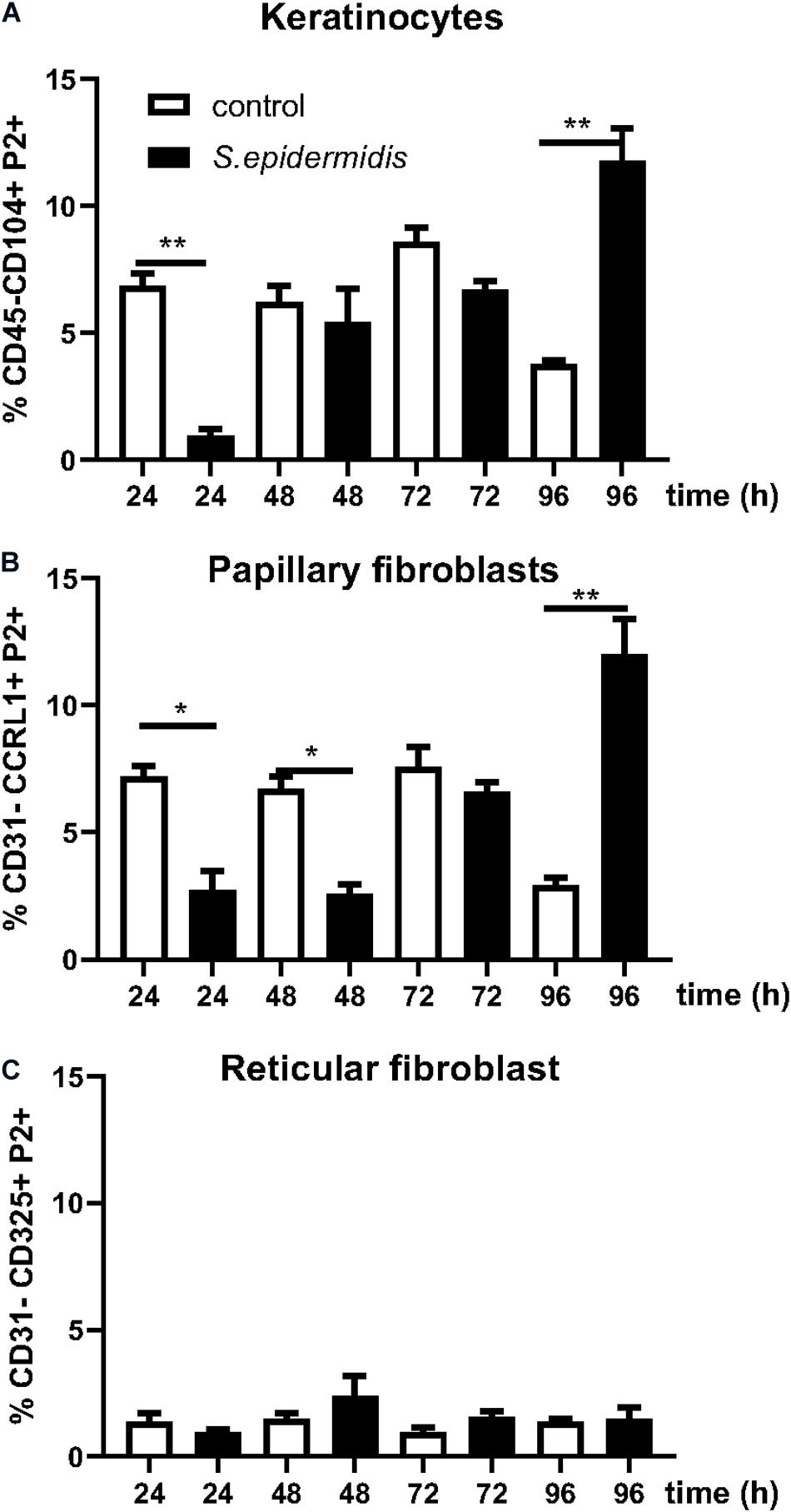
Figure 3. Staphylococcus epidermidis colonization induces P-2 in human keratinocytes and papillary fibroblasts. Control, non-colonized, and S. epidermidis colonized human skin was maintained at air liquid interface and collected at indicated time points (24, 48, 72, and 96 h) for FISH-Flow. Single cell suspensions were labeled with live/dead stain. Using FISH-Flow RNA assay, P-2 RNA levels were analyzed in (A) CD45-, CD31-, CD104+ cells (keratinocytes), or (B) CD45-, CD31-, CCRL1+ (papillary fibroblasts), and (C) CD45-, CD31-, CD325+ cells (reticular fibroblasts). Bar graphs show percentage of P-2 mRNA positive cells within each CD45-CD31-skin cell population. Data represent at least two experiments with three independent biological replicates per group. *p < 0.05, **p < 0.01 (two-way ANOVA with Holm-Sidak multiple-comparison test).
Antimicrobial Peptides Are Upregulated in Human Skin by S. epidermidis
Staphylococcus epidermidis isolates from healthy adults have been reported to show widespread production of bacteriocins (55) and in addition they can stimulate keratinocytes to produce antimicrobial peptides (4). We investigated if S. epidermidis triggers expression of antimicrobial peptides in our ex vivo skin model. We found that 24 h of S. epidermidis colonization significantly induced expression of defensin β4 (Defβ4) and cathelicidin (LL37) (p < 0.01) (Figure 4). Upregulation of LL37 was maintained 48 h post S. epidermidis colonization (p < 0.01) in contrast to defensin Defβ4 that was downregulated (p < 0.05). In addition, S. epidermidis colonization of human skin resulted in downregulation of pro-inflammatory IL-1α after 24 and 96 h (p < 0.05) (Figure 4).
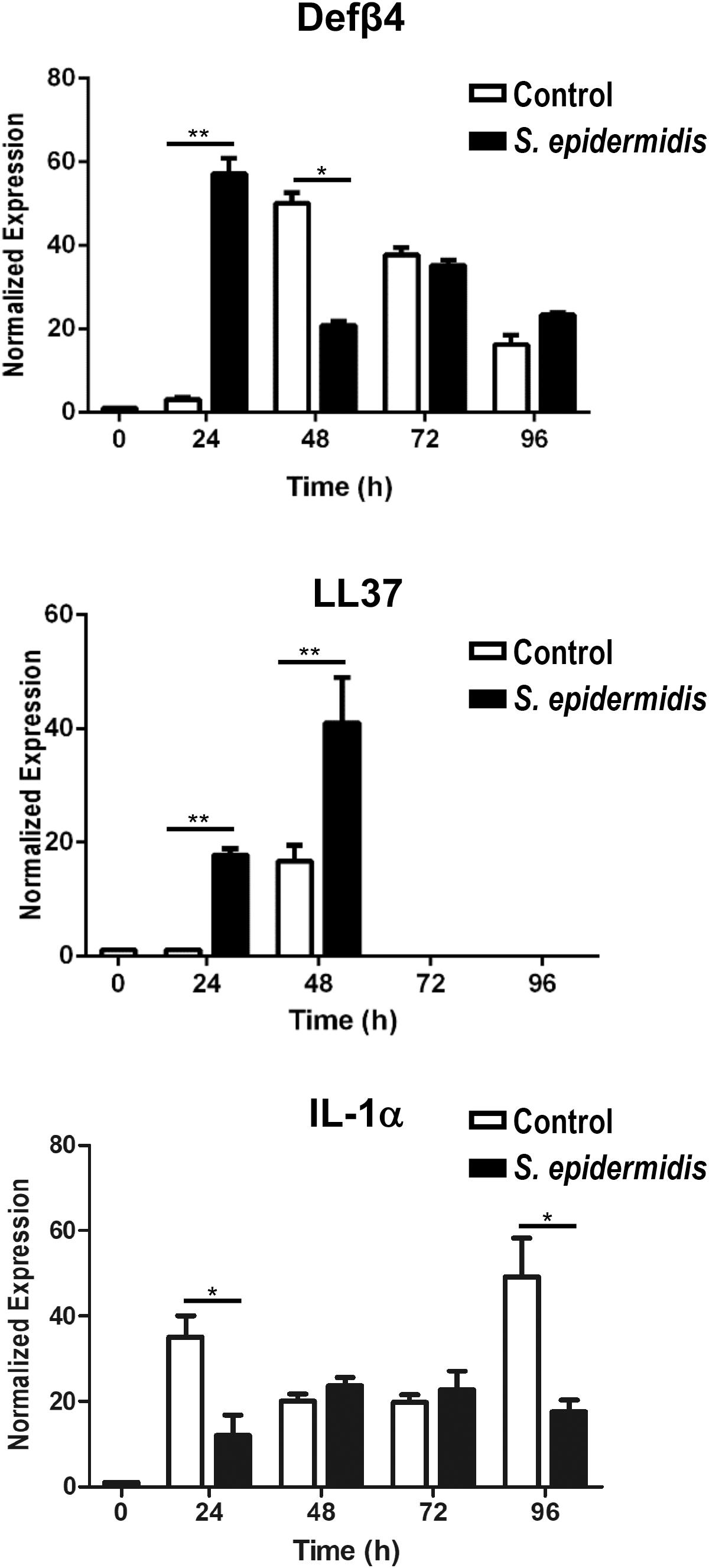
Figure 4. Antimicrobial and inflammatory responses mediated by Staphylococcus epidermidis in human ex vivo model. Expression levels of antimicrobial peptides Defβ4, LL-37, and pro-inflammatory cytokine IL-1α were evaluated by qPCR from non-colonized and S. epidermidis colonized skin at 24, 48, 72, and 96 h (n = 3 for each treatment group and time point). *p < 0.05, **p < 0.01 as calculated using Student t-test.
Early Regulation of GD T Cell Gene Expression by S. epidermidis
Human GD T cells in the skin exhibit both pro-inflammatory and regulatory functions (32). Deciphering the underlying mechanisms that contribute to induction of effector vs. regulatory GD T cell functions, including expression of cytotoxic molecules, is key to understanding skin homeostasis. To understand the effect of S. epidermidis on skin GD T cells during initial phases of colonization, we sorted GD T cells from normal skin (Figure 5A) and stimulated them with S. epidermidis for 1 h. We evaluated the expression of well-known genes previously described to play a role in GD T cell cytotoxic functions. We found a 6-7-fold induction in Fas ligand (FASLG) and Granulysin (GNLY) in S. epidermidis treated cells compared to control, untreated GD T cells (Figure 5B). Additionally, we observed increased expression of the transcription factor PLZF, which is responsible for selection of GD innate natural killer T cells (56), as well as CCL4, a monokine with inflammatory and chemokinetic properties (Figure 5B). Previous studies have observed increased CCL4 expression by GD T cells following engagement of the natural cytotoxicity receptor NKp30 on the GD T cell surface (57).
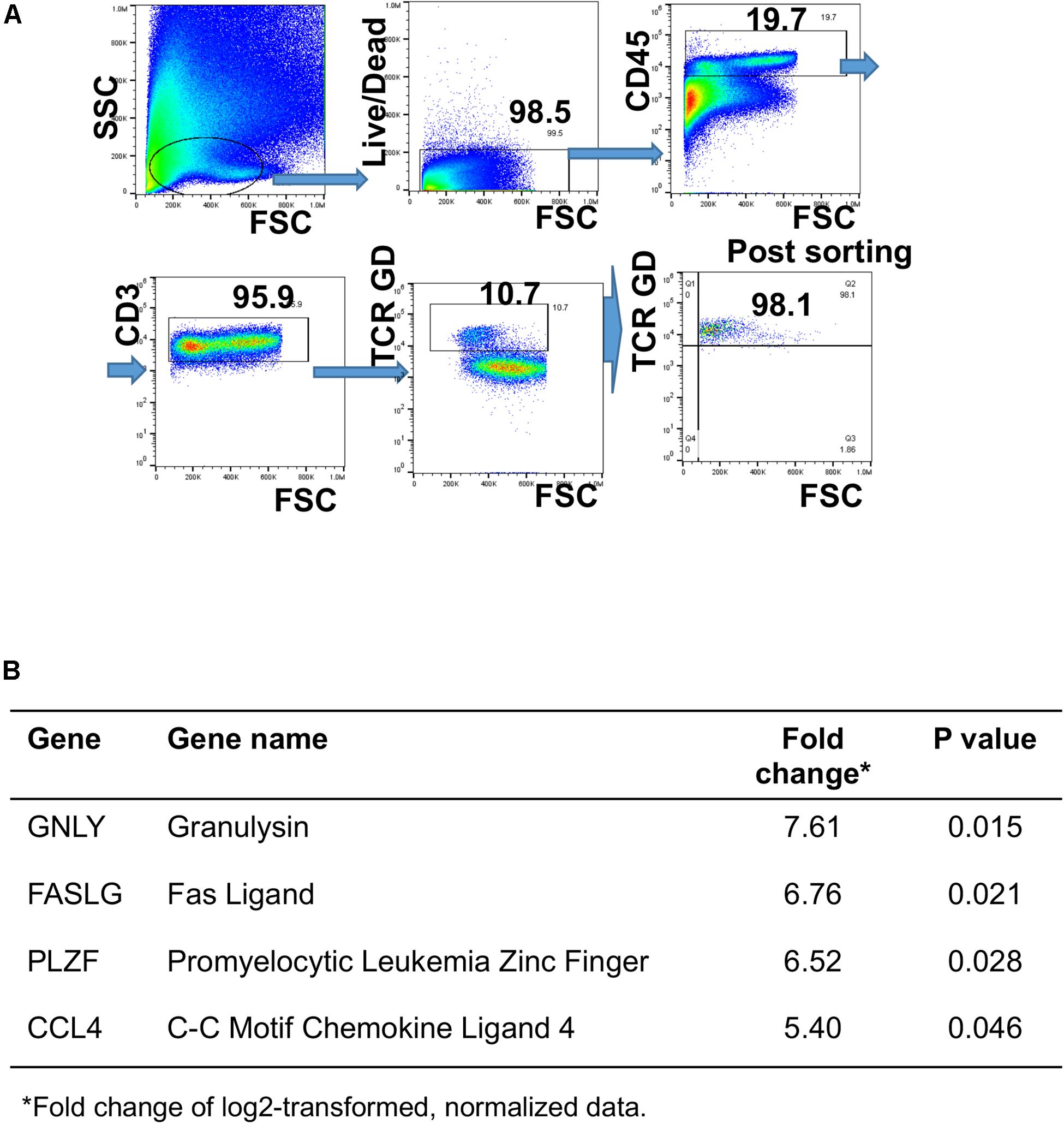
Figure 5. Differential gene expression in human GD T cells as an early response to Staphylococcus epidermidis. Human skin cells were isolated using the Miltenyi Whole Skin Dissociation kit (Miltenyi, Bergisch-Gladbach, Germany) and GD T cells were sorted from the skin cell suspension using fluorescence-activated cell sorting. (A) Gating strategy for GD T cell sorting. (B) Cells were stimulated with S. epidermidis for 1 h and changes in gene expression between uninfected and infected cells were measured using the BioMark IFC 96 × 96 chip (Fluidigm). Changes in gene expression are expressed as log2 of fold change (n = 3). P-values were calculated using Student t-test.
Intracellular MRSA Killing Is Enhanced After Exposure to S. epidermidis
We have previously reported that MRSA, the most common cutaneous pathogen, suppresses P-2 induction in skin cells (43), revealing a novel mechanism by which S. aureus may escape cutaneous immunity to cause persistent infections. Here, we report that in contrast to MRSA (43), S. epidermidis up-regulates P-2 expression in an ex vivo skin model (see Figure 1) and in the single cell suspension culture model (Figure 6). In order to further analyze S. epidermidis-mediated induction of P-2 in ex vivo human skin, we isolated skin cells and established a single cell type culture system. We found an increase in the frequency of GD T cells after 24 h stimulation with S. epidermidis (Figure 6A), which agrees with findings from the ex vivo human skin model (Figure 1). Furthermore, expression of P-2 was also increased in the GD T cells after 24 h of S. epidermidis stimulation (Figure 6B). Most importantly, we show that cells stimulated with S. epidermidis demonstrate an increased capability to kill intracellular MRSA (Figure 6C). We observed this same result after repeating stimulations with 2 additional S. epidermidis strains, S. epidermidis ATCC 12228 and commensal isolate S. epidermidis CCN0024 (Supplementary Figure S2). In addition, we found that S. epidermidis CCN021 stimulated GD T cells upregulate expression of TLR2 and TLR1, but not TLR6 (Figure 6D).
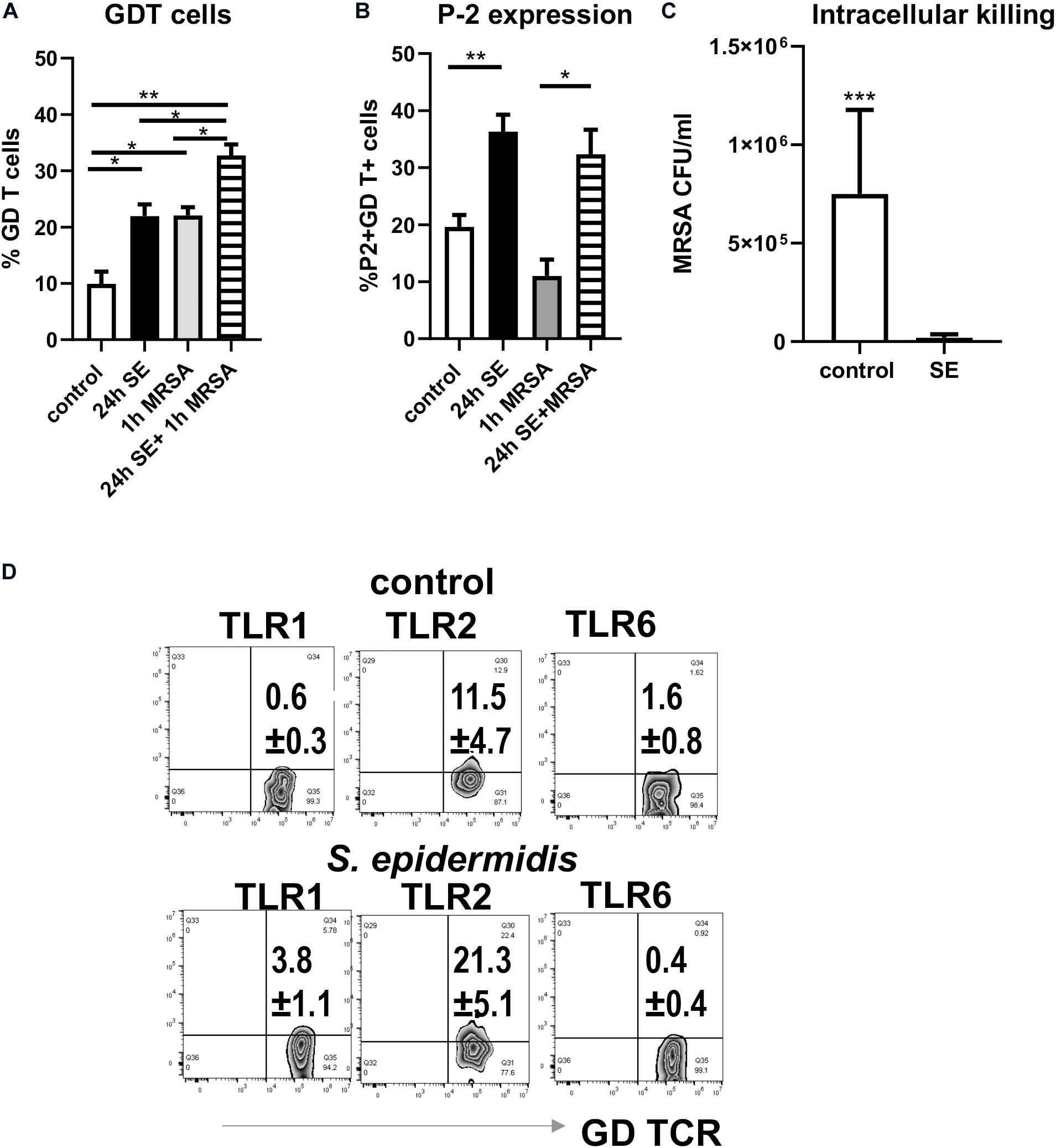
Figure 6. Pre-treatment of skin cells with Staphylococcus epidermidis increases frequency of GD T cells, stimulates P-2 expression, and limits survival of intracellular MRSA. Single skin cells were exposed to S. epidermidis at MOI 1:20 or media control for 24 h. After washing to remove S. epidermidis, cells were infected with MRSA (MOI 1:20) for 1 h to allow intracellular infection, and extracellular bacteria were subsequently removed by gentamicin treatment. (A) Frequency of GDT cells and (B) P2 mRNA expression in CD45+ CD3+ GD TCR+ cells as determined by FISH-Flow (n = 3 biological replicates). (C) Bar graph showing the number of intracellular MRSA (CFU/ml) upon hypotonic lysis of control and S. epidermidis pre-treated cells (n = 3 biological replicates). (D) Expression of TLR1, TLR2, and TLR6 on gated CD45+ CD3+ TCR GD+ T cells. Data represent at least two experiments with three independent biological replicates per group. *p < 0.05, **p < 0.01 as calculated using two-way ANOVA with Holm-Sidak multiple-comparison (A) and ***p < 0.001 as calculated using Student t-test (B).
Discussion
Pore-forming proteins permeabilize membranes of infected cells targeted for immune elimination and together with antimicrobial peptides represent the key effector molecules of the epithelial barriers. GD T cells, as surveillance cells in the skin, constitutively express mRNA for granzyme A and B and perforin and contain significant esterase activity (58). We provided the first evidence to show that human skin GD T cells constitutively express antimicrobial protein P-2 (43). In contrast to other secreted pore forming proteins, P-2 is a transmembrane protein that efficiently kills intracellular bacteria. IFNγ, type I interferons, and LPS have been implicated in the regulation of its expression (37, 38). We have previously reported that MRSA, the most common skin pathogen, suppresses P2-induction in skin cells (43), revealing a novel mechanism by which S. aureus may escape cutaneous immunity to cause persistent infections. Here we show that, in contrast to MRSA, S. epidermidis up-regulates P-2 expression in human skin and in single cell suspensions. We also demonstrate that S. epidermidis upregulates P-2 mRNA expression in multiple skin cell types including GD T cells, basal keratinocytes, and papillary fibroblasts. Most importantly, we observed a decrease in number of intracellular MRSA in skin stimulated by S. epidermidis, which correlates with S. epidermidis-mediated P-2 induction. These data provide new insights regarding mechanisms of P-2 expression and function and elucidate novel approaches to protect skin from infections caused by intracellular pathogens.
Gamma delta T cells represent a major T cell subset involved in the surveillance of epithelial surfaces (skin, gastrointestinal, reproductive, and respiratory tracts). It is well establish that GD T cells, upon recognition of pathogens, effectively proliferate, secrete pro-inflammatory cytokines, and activate their cytolytic machinery (perforin and granzymes) to kill the pathogen (59). Clonally expanded GD T cells can establish long-lasting immunity against recurrent S. aureus skin infections (33). In contrast, GD T cell deficient mice develop large skin lesions after infection with S. aureus (60). However, encounters with commensal microbes by skin GD T cells and how such interactions affect their response to pathogens remains poorly understood. Here we provide the first evidence that the common skin commensal, S. epidermidis, upregulates the frequency of GD T cells and induces the expression of P-2, which is associated with an increased capability to eliminate intracellular MRSA. Our data on increased frequency of skin GD T cells after colonization with S. epidermidis supports the hypothesis that under normal conditions the presence of S. epidermidis on the skin surface strengthens cutaneous innate defenses (9, 10).
We observed that during the early steps of colonization, prior to P-2 induction, S. epidermidis upregulates the GD T cell cytotoxic molecules Fas Ligand (FASLG) and granulysin (GNLY). This may contribute to the GD T cell mediated antimicrobial immune response, in addition to P-2 induction at later time points. We are currently expanding these studies to in vivo animal models. S. epidermidis, when topically applied to murine skin, induces specific IL-17 producing T cells that persist as tissue-resident memory T cells (13). However, to the best of our knowledge, our study is the first report that shows specific effect of S. epidermidis on induction of human skin GD T cell responses.
Previous reports indicate that a commensal strain of S. epidermidis and non-commensal strain S. carnosus have different modifications of the lipoprotein (Lpp) lipid moieties (61). The essential receptor for recognition of bacterial Lpp is TLR2. However, the degree of acylation at the lipid moiety can be discriminated by additional TLRs, such as TLR1 and TLR6, which form heterodimers with TLR2 (62–64). Importantly, these lipoprotein modifications were implicated in the differential immune responses where commensal staphylococcal species dampened IFNγ, TNFα, and IL-12 production compared to pathogenic staphylococcal species (61). We have found that a 24 h stimulation with S. epidermidis upregulates TLR2 and TLR1, but not TLR6 on GD T cells. We postulate that S. epidermidis, through recognition of TLR2/TLR1 heterodimers on cutaneous GD T cells, regulates not only Th1 responses but also cytotoxic mediators such as P-2. The recognition of TLR2/TLR1 heterodimers may even be strain specific (12) warranting further studies on the mechanisms of P-2 induction by S. epidermidis CCN021.
It has been shown that S. epidermidis colonization of skin induces AMP production by keratinocytes (5, 7, 8, 65). Our findings regarding induction of LL37 and Defβ4 during early phases of human skin colonization with S. epidermidis are in line with these results. Kinetics of P-2 induction upon S. epidermidis colonization shows dynamic control and cell specificity that integrates with the kinetics of AMP production. In GD T cells, the induction of P-2 is rapid and maintained, persisting from 24 to 72 h post colonization whereas in keratinocytes and papillary fibroblasts it shows complementary activation at 96 h. These data suggest that the initial protective response derives from GD T cells whereas in keratinocytes activation of P-2 follows initial activation of AMPs, cathelicidin, and β-defensin. The human skin ex vivo model is comprised of the epidermis and dermis with no circulation, thus limiting the studies on modulation of the immune response by S. epidermidis to resident innate immune cells, while the potential role of adaptive immunity would require in vivo models.
The initial findings presented here also provide a functional readout of S. epidermidis colonization and P-2 upregulation: decrease of the intracellular pathogen S. aureus. We observed suppression of the pro-inflammatory cytokine IL-1α after colonization with S. epidermidis. Previously, we showed that S. aureus induces IL-1α in non-healing diabetic foot ulcers (50), suggesting that S. epidermidis may have additional mechanisms to neutralize the damaging effects of pathogenic organisms. The limitation of our study was sequential stimulation of human skin and primary cells by S. epidermidis and S. aureus. Future in vivo studies are required to confirm antimicrobial effects of S. epidermidis in the presence of pathogenic S. aureus. Additionally, future studies that block P-2 expression will be necessary to confirm that enhanced S. aureus killing upon S. epidermidis treatment is solely due to P-2 upregulation. Despite these limitations, this work provides an intriguing possibility that colonization of S. epidermidis may prevent and/or protect from bacterial skin infections through modulation of P-2.
In summary, we confirmed that colonization with commensal S. epidermidis in human ex vivo skin modulates the innate immune system by activating GD T cells, promoting antimicrobial peptide production, and upregulating the antimicrobial protein P-2. Understanding how commensal bacteria regulate P-2 expression represents the first step toward identifying mechanisms by which P-2 contributes to cutaneous homeostasis and host defense mechanisms and may reveal new approaches for preventing and treating skin infections.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Ethics Statement
The studies involving human participants were reviewed and approved by Institutional Review Board at the UM Miller School of Medicine (exempt from human subject research under CFR46.101.2). The patients provided their written informed consent to participate in this study.
Author Contributions
MT-C and NS obtained funding, and conceived and coordinated the experiments. IP, KO’N, LP, CH, JB, VC, DG, OS, and SH performed the experiments and analyzed the data. KO’N, IP, and NS performed the statistical analyses. GP and ST provided reagents. NS, IP, and MT-C wrote the manuscript. All authors were involved in writing and had final approval of the submitted and published versions of the manuscript.
Funding
This work was supported by NIH NR015649 (MT-C and NS), DK119085 (MT-C), Dwoskin family gift to the Dr. Phillip Frost Department of Dermatology and Cutaneous Surgery (MT-C), and by Department of Microbiology and Immunology P2 funds (NS). VC was supported by Wound Healing Society Summer Research Fellowship.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We dedicate this work to late Dr. Eckhard Podack without whom studies of Perforin-2 would not be possible. We are grateful to all members of MT-C and NS laboratories for their overall support and Prof. Stephen C. Davis for the gift of S. epidermidis ATCC 12228.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2020.550946/full#supplementary-material
FIGURE S1 | Cell viability of ex vivo skin tissue with or without S. epidermidis colonization via flow cytometry (data represented as mean ± SEM, n = 3–4 skin samples).
FIGURE S2 | Pre-treatment of skin cells with S. epidermidis CCN0024 and ATCC 12228 strains limits intracellular MRSA survival. Single skin cells were exposed to single S. epidermidis strain at MOI 1:20 or media control for 24 h. After washing to remove S. epidermidis, cells were infected with MRSA (MOI 1:20) for 1 h to allow intracellular infection, and extracellular bacteria were subsequently removed by gentamicin treatment. Bar graph shows the number of intracellular MRSA (CFU/ml) upon hypotonic lysis of control and S. epidermidis pre-treated cells. ****p < 0.0001 as calculated using one-way ANOVA with Dunnett’s multiple comparisons test, which compared each S. epidermidis pre-treatment with non-pretreated control cells.
References
1. Belkaid Y, Harrison OJ. Homeostatic immunity and the microbiota. Immunity. (2017) 46:562–76. doi: 10.1016/j.immuni.2017.04.008
2. Stacy A, Belkaid Y. Microbial guardians of skin health. Science. (2019) 363:227–8. doi: 10.1126/science.aat4326
3. Lukic J, Chen V, Strahinic I, Begovic J, Lev-Tov H, Davis SC, et al. Probiotics or pro-healers: the role of beneficial bacteria in tissue repair. Wound Repair Regen. (2017) 25:912–22. doi: 10.1111/wrr.12607
4. Lai Y, Cogen AL, Radek KA, Park HJ, Macleod DT, Leichtle A, et al. Activation of TLR2 by a small molecule produced by Staphylococcus epidermidis increases antimicrobial defense against bacterial skin infections. J Invest Dermatol. (2010) 130:2211–21. doi: 10.1038/jid.2010.123
5. Ommori R, Ouji N, Mizuno F, Kita E, Ikada Y, Asada H. Selective induction of antimicrobial peptides from keratinocytes by staphylococcal bacteria. Microb Pathog. (2013) 56:35–9. doi: 10.1016/j.micpath.2012.11.005
6. Nakatsuji T, Chen TH, Narala S, Chun KA, Two AM, Yun T, et al. Antimicrobials from human skin commensal bacteria protect against Staphylococcus aureus and are deficient in atopic dermatitis. Sci Transl Med. (2017) 9:eaah4680. doi: 10.1126/scitranslmed.aah4680
7. Li D, Lei H, Li Z, Li H, Wang Y, Lai Y. A novel lipopeptide from skin commensal activates TLR2/CD36-p38 MAPK signaling to increase antibacterial defense against bacterial infection. PLoS One. (2013) 8:e58288. doi: 10.1371/journal.pone.0058288
8. Liu Q, Liu Q, Meng H, Lv H, Liu Y, Liu J, et al. Staphylococcus epidermidis contributes to healthy maturation of the nasal microbiome by stimulating antimicrobial peptide production. Cell Host Microbe. (2020) 27:68–78.e5. doi: 10.1016/j.chom.2019.11.003
9. Christensen GJ, Bruggemann H. Bacterial skin commensals and their role as host guardians. Benef Microbes. (2014) 5:201–15. doi: 10.3920/BM2012.0062
10. Gallo RL, Nakatsuji T. Microbial symbiosis with the innate immune defense system of the skin. J Invest Dermatol. (2011) 131:1974–80. doi: 10.1038/jid.2011.182
11. Cogen AL, Yamasaki K, Muto J, Sanchez KM, Crotty Alexander L, Tanios J, et al. Staphylococcus epidermidis antimicrobial delta-toxin (phenol-soluble modulin-gamma) cooperates with host antimicrobial peptides to kill group A Streptococcus. PLoS One. (2010) 5:e8557. doi: 10.1371/journal.pone.0008557
12. Zhou W, Spoto M, Hardy R, Guan C, Fleming E, Larson PJ, et al. Host-specific evolutionary and transmission dynamics shape the functional diversification of Staphylococcus epidermidis in human skin. Cell. (2020) 180:454–470.e18. doi: 10.1016/j.cell.2020.01.006
13. Naik S, Bouladoux N, Linehan JL, Han SJ, Harrison OJ, Wilhelm C, et al. Commensal-dendritic-cell interaction specifies a unique protective skin immune signature. Nature. (2015) 520:104–8. doi: 10.1038/nature14052
14. Linehan JL, Harrison OJ, Han SJ, Byrd AL, Vujkovic-Cvijin I, Villarino AV, et al. Non-classical immunity controls microbiota impact on skin immunity and tissue repair. Cell. (2018) 172:784–796.e18. doi: 10.1016/j.cell.2017.12.033
15. Scharschmidt TC, Vasquez KS, Truong HA, Gearty SV, Pauli ML, Nosbaum A, et al. A wave of regulatory T cells into neonatal skin mediates tolerance to commensal microbes. Immunity. (2015) 43:1011–21. doi: 10.1016/j.immuni.2015.10.016
16. Young JL, Goodall JC, Beacock-Sharp H, Gaston JS. Human gamma delta T-cell recognition of Yersinia enterocolitica. Immunology. (1997) 91:503–10. doi: 10.1046/j.1365-2567.1997.00289.x
17. Barisa M, Kramer AM, Majani Y, Moulding D, Saraiva L, Bajaj-Elliott M, et al. E. coli promotes human Vgamma9Vdelta2 T cell transition from cytokine-producing bactericidal effectors to professional phagocytic killers in a TCR-dependent manner. Sci Rep. (2017) 7:2805. doi: 10.1038/s41598-017-02886-8
18. Chen ZW. Protective immune responses of major Vgamma2Vdelta2 T-cell subset in M. tuberculosis infection. Curr Opin Immunol. (2016) 42:105–12. doi: 10.1016/j.coi.2016.06.005
19. Marx S, Wesch D, Kabelitz D. Activation of human gamma delta T cells by Mycobacterium tuberculosis and Daudi lymphoma cells: differential regulatory effect of IL-10 and IL-12. J Immunol. (1997) 158:2842–8.
20. Hara T, Mizuno Y, Takaki K, Takada H, Akeda H, Aoki T, et al. Predominant activation and expansion of V gamma 9-bearing gamma delta T cells in vivo as well as in vitro in Salmonella infection. J Clin Invest. (1992) 90:204–10. doi: 10.1172/JCI115837
21. Kurup SP, Harty JT. gammadelta T cells and immunity to human malaria in endemic regions. Ann Transl Med. (2015) 3(Suppl. 1):S22. doi: 10.3978/j.issn.2305-5839.2015.02.22
22. Poccia F, Agrati C, Martini F, Capobianchi MR, Wallace M, Malkovsky M. Antiviral reactivities of gammadelta T cells. Microbes Infect. (2005) 7:518–28. doi: 10.1016/j.micinf.2004.12.009
23. Knight A, Madrigal AJ, Grace S, Sivakumaran J, Kottaridis P, Mackinnon S, et al. The role of Vdelta2-negative gammadelta T cells during cytomegalovirus reactivation in recipients of allogeneic stem cell transplantation. Blood. (2010) 116:2164–72. doi: 10.1182/blood-2010-01-255166
24. Fisch P, Malkovsky M, Kovats S, Sturm E, Braakman E, Klein BS, et al. Recognition by human V gamma 9/V delta 2 T cells of a GroEL homolog on Daudi Burkitt’s lymphoma cells. Science. (1990) 250:1269–73. doi: 10.1126/science.1978758
25. Fisch P, Oettel K, Fudim N, Surfus JE, Malkovsky M, Sondel PM. MHC-unrestricted cytotoxic and proliferative responses of two distinct human gamma/delta T cell subsets to Daudi cells. J Immunol. (1992) 148:2315–23.
26. Havran WL, Allison JP. Developmentally ordered appearance of thymocytes expressing different T-cell antigen receptors. Nature. (1988) 335:443–5. doi: 10.1038/335443a0
27. Havran WL, Fitch FW. Characterization of murine cytolytic-helper hybrid T cell clones. Nature. (1987) 325:65–7. doi: 10.1038/325065a0
28. Havran WL, Grell S, Duwe G, Kimura J, Wilson A, Kruisbeek AM, et al. Limited diversity of T-cell receptor gamma-chain expression of murine Thy-1+ dendritic epidermal cells revealed by V gamma 3-specific monoclonal antibody. Proc Natl Acad Sci USA. (1989) 86:4185–9. doi: 10.1073/pnas.86.11.4185
29. Weintraub BC, Jackson MR, and Hedrick SM. Gamma delta T cells can recognize nonclassical MHC in the absence of conventional antigenic peptides. J Immunol. (1994) 153:3051–8.
30. Rust C, Orsini D, Kooy Y, Koning F. Reactivity of human gamma delta T cells to staphylococcal enterotoxins: a restricted reaction pattern mediated by two distinct recognition pathways. Scand J Immunol. (1993) 38:89–94. doi: 10.1111/j.1365-3083.1993.tb01698.x
31. Rust CJ, Koning F. Gamma delta T cell reactivity towards bacterial superantigens. Semin Immunol. (1993) 5:41–6. doi: 10.1006/smim.1993.1006
32. Nielsen MM, Witherden DA, Havran WL. gammadelta T cells in homeostasis and host defence of epithelial barrier tissues. Nat Rev Immunol. (2017) 17:733–45. doi: 10.1038/nri.2017.101
33. Dillen CA, Pinsker BL, Marusina AI, Merleev AA, Farber ON, Liu H, et al. Clonally expanded gammadelta T cells protect against Staphylococcus aureus skin reinfection. J Clin Invest. (2018) 128:1026–42. doi: 10.1172/JCI96481
34. He X, Zhang Y, Yu Z. An Mpeg (macrophage expressed gene) from the Pacific oyster Crassostrea gigas: molecular characterization and gene expression. Fish Shellfish Immunol. (2011) 30:870–6. doi: 10.1016/j.fsi.2011.01.009
35. Kemp IK, Coyne VE. Identification and characterisation of the Mpeg1 homologue in the South African abalone, Haliotis midae. Fish Shellfish Immunol. (2011) 31:754–64. doi: 10.1016/j.fsi.2011.07.010
36. McCormack R, Podack ER. Perforin-2/Mpeg1 and other pore-forming proteins throughout evolution. J Leukoc Biol. (2015) 98:761–8. doi: 10.1189/jlb.4MR1114-523RR
37. McCormack RM, de Armas LR, Shiratsuchi M, Fiorentino DG, Olsson ML, Lichtenheld MG, et al. Perforin-2 is essential for intracellular defense of parenchymal cells and phagocytes against pathogenic bacteria. eLife. (2015) 4:e06508. doi: 10.7554/eLife.06508
38. McCormack RM, Lyapichev K, Olsson ML, Podack ER, Munson GP. Enteric pathogens deploy cell cycle inhibiting factors to block the bactericidal activity of Perforin-2. eLife. (2015) 4:e06505. doi: 10.7554/eLife.06505
39. McCormack R, Bahnan W, Shrestha N, Boucher J, Barreto M, Barrera CM, et al. Perforin-2 protects host cells and mice by restricting the vacuole to cytosol transitioning of a bacterial pathogen. Infect Immun. (2016) 84:1083–91. doi: 10.1128/IAI.01434-15
40. Pang SS, Bayly-Jones C, Radjainia M, Spicer BA, Law RHP, Hodel AW, et al. The cryo-EM structure of the acid activatable pore-forming immune effector Macrophage-expressed gene 1. Nat Commun. (2019) 10:4288. doi: 10.1038/s41467-019-12279-2
41. McCormack R, de Armas L, Shiratsuchi M, Podack ER. Killing machines: three pore-forming proteins of the immune system. Immunol Res. (2013) 57:268–78. doi: 10.1007/s12026-013-8469-9
42. McCormack R, de Armas LR, Shiratsuchi M, Ramos JE, Podack ER. Inhibition of intracellular bacterial replication in fibroblasts is dependent on the perforin-like protein (perforin-2) encoded by macrophage-expressed gene 1. J Innate Immun. (2013) 5:185–94. doi: 10.1159/000345249
43. Strbo N, Pastar I, Romero L, Chen V, Vujanac M, Sawaya AP, et al. Single cell analyses reveal specific distribution of anti-bacterial molecule Perforin-2 in human skin and its modulation by wounding and Staphylococcus aureus infection. Exp Dermatol. (2019) 28:225–32. doi: 10.1111/exd.13870
44. Martineau F, Picard FJ, Ke D, Paradis S, Roy PH, Ouellette M, et al. Development of a PCR assay for identification of staphylococci at genus and species levels. J Clin Microbiol. (2001) 39:2541–7. doi: 10.1128/JCM.39.7.2541-2547.2001
45. Vandecasteele SJ, Peetermans WE, Merckx R, Van Eldere J. Quantification of expression of Staphylococcus epidermidis housekeeping genes with Taqman quantitative PCR during in vitro growth and under different conditions. J Bacteriol. (2001) 183:7094–101. doi: 10.1128/JB.183.24.7094-7101.2001
46. Pang YY, Schwartz J, Thoendel M, Ackermann LW, Horswill AR, Nauseef WM. agr-Dependent interactions of Staphylococcus aureus USA300 with human polymorphonuclear neutrophils. J Innate Immun. (2010) 2:546–59. doi: 10.1159/000319855
47. Yoon DJ, Fregoso DR, Nguyen D, Chen V, Strbo N, Fuentes JJ, et al. A tractable, simplified ex vivo human skin model of wound infection. Wound Repair Regen. (2019) 27:421–5. doi: 10.1111/wrr.12712
48. Pastar I, Stojadinovic O, Krzyzanowska A, Barrientos S, Stuelten C, Zimmerman K, et al. Attenuation of the transforming growth factor beta-signaling pathway in chronic venous ulcers. Mol Med. (2010) 16:92–101. doi: 10.2119/molmed.2009.00149
49. Ramirez HA, Pastar I, Jozic I, Stojadinovic O, Stone RC, Ojeh N, et al. Staphylococcus aureus triggers induction of miR-15B-5P to diminish DNA repair and deregulate inflammatory response in diabetic foot ulcers. J Invest Dermatol. (2018) 138:1187–96. doi: 10.1016/j.jid.2017.11.038
50. Stojadinovic O, Tomic-Canic M. Human ex vivo wound healing model. Methods Mol Biol. (2013) 1037:255–64. doi: 10.1007/978-1-62703-505-7_14
51. Finak G, McDavid A, Yajima M, Deng J, Gersuk V, Shalek AK, et al. MAST: a flexible statistical framework for assessing transcriptional changes and characterizing heterogeneity in single-cell RNA sequencing data. Genome Biol. (2015) 16:278. doi: 10.1186/s13059-015-0844-5
52. Bolton DL, McGinnis K, Finak G, Chattopadhyay P, Gottardo R, Roederer M. Combined single-cell quantitation of host and SIV genes and proteins ex vivo reveals host-pathogen interactions in individual cells. PLoS Pathog. (2017) 13:e1006445. doi: 10.1371/journal.ppat.1006445
53. Janson DG, Saintigny G, van Adrichem A, Mahe C, El Ghalbzouri A. Different gene expression patterns in human papillary and reticular fibroblasts. J Invest Dermatol. (2012) 132:2565–72. doi: 10.1038/jid.2012.192
54. Woodley DT. Distinct fibroblasts in the papillary and reticular dermis: implications for wound healing. Dermatol Clin. (2017) 35:95–100. doi: 10.1016/j.det.2016.07.004
55. Janek D, Zipperer A, Kulik A, Krismer B, Peschel A. High frequency and diversity of antimicrobial activities produced by nasal Staphylococcus strains against bacterial competitors. PLoS Pathog. (2016) 12:e1005812. doi: 10.1371/journal.ppat.1005812
56. Dimova T, Brouwer M, Gosselin F, Tassignon J, Leo O, Donner C, et al. Effector Vgamma9Vdelta2 T cells dominate the human fetal gammadelta T-cell repertoire. Proc Natl Acad Sci USA. (2015) 112:E556–65. doi: 10.1073/pnas.1412058112
57. Hudspeth K, Fogli M, Correia DV, Mikulak J, Roberto A, Della Bella S, et al. Engagement of NKp30 on Vdelta1 T cells induces the production of CCL3, CCL4, and CCL5 and suppresses HIV-1 replication. Blood. (2012) 119:4013–6. doi: 10.1182/blood-2011-11-390153
58. Mohamadzadeh M, McGuire MJ, Smith DJ, Gaspari AA, Bergstresser PR, Takashima A. Functional roles for granzymes in murine epidermal gamma(delta) T-cell-mediated killing of tumor targets. J Invest Dermatol. (1996) 107:738–42. doi: 10.1111/1523-1747.ep12365634
59. Bonneville M, O’Brien RL, Born WK. Gammadelta T cell effector functions: a blend of innate programming and acquired plasticity. Nat Rev Immunol. (2010) 10:467–78. doi: 10.1038/nri2781
60. Cho JS, Pietras EM, Garcia NC, Ramos RI, Farzam DM, Monroe HR, et al. IL-17 is essential for host defense against cutaneous Staphylococcus aureus infection in mice. J Clin Invest. (2010) 120:1762–73. doi: 10.1172/JCI40891
61. Nguyen MT, Uebele J, Kumari N, Nakayama H, Peter L, Ticha O, et al. Lipid moieties on lipoproteins of commensal and non-commensal staphylococci induce differential immune responses. Nat Commun. (2017) 8:2246. doi: 10.1038/s41467-017-02234-4
62. Kang JY, Nan X, Jin MS, Youn SJ, Ryu YH, Mah S, et al. Recognition of lipopeptide patterns by Toll-like receptor 2-Toll-like receptor 6 heterodimer. Immunity. (2009) 31:873–84. doi: 10.1016/j.immuni.2009.09.018
63. Takeda K, Takeuchi O, Akira S. Recognition of lipopeptides by Toll-like receptors. J Endotoxin Res. (2002) 8:459–63. doi: 10.1179/096805102125001073
64. Takeuchi O, Kawai T, Muhlradt PF, Morr M, Radolf JD, Zychlinsky A, et al. Discrimination of bacterial lipoproteins by Toll-like receptor 6. Int Immunol. (2001) 13:933–40. doi: 10.1093/intimm/13.7.933
Keywords: perforin-2/mpeg-1, human skin, innate immunity, Staphylococcus epidermidis, gamma delta T cells, cytotoxicity
Citation: Pastar I, O’Neill K, Padula L, Head CR, Burgess JL, Chen V, Garcia D, Stojadinovic O, Hower S, Plano GV, Thaller SR, Tomic-Canic M and Strbo N (2020) Staphylococcus epidermidis Boosts Innate Immune Response by Activation of Gamma Delta T Cells and Induction of Perforin-2 in Human Skin. Front. Immunol. 11:550946. doi: 10.3389/fimmu.2020.550946
Received: 11 April 2020; Accepted: 18 August 2020;
Published: 16 September 2020.
Edited by:
Gabriele Pradel, RWTH Aachen University, GermanyReviewed by:
Michael Otto, National Institutes of Health (NIH), United StatesJuan Carlos Cancino-Diaz, National Polytechnic Institute of Mexico (IPN), Mexico
Copyright © 2020 Pastar, O’Neill, Padula, Head, Burgess, Chen, Garcia, Stojadinovic, Hower, Plano, Thaller, Tomic-Canic and Strbo. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Marjana Tomic-Canic, TVRjYW5pY0BtZWQubWlhbWkuZWR1; Natasa Strbo, bnN0cmJvQG1lZC5taWFtaS5lZHU=
†These authors have contributed equally to this work and share first authorship
 Irena Pastar
Irena Pastar Katelyn O’Neill
Katelyn O’Neill Laura Padula
Laura Padula Cheyanne R. Head
Cheyanne R. Head Jamie L. Burgess
Jamie L. Burgess Vivien Chen
Vivien Chen Denisse Garcia
Denisse Garcia Olivera Stojadinovic
Olivera Stojadinovic Suzanne Hower
Suzanne Hower Gregory V. Plano
Gregory V. Plano Seth R. Thaller
Seth R. Thaller Marjana Tomic-Canic
Marjana Tomic-Canic Natasa Strbo
Natasa Strbo