- Department of Infectious Diseases, Frontier Science Center for Immunology and Metabolism, Medical Research Institute, Zhongnan Hospital of Wuhan University, Wuhan University, Wuhan, China
The membrane-associated RING-CH-type finger (MARCH) proteins of E3 ubiquitin ligases have emerged as critical regulators of immune responses. MARCH proteins target immune receptors, viral proteins as well as components in innate immune response for polyubiquitination and degradations via distinct routes. This review summarizes the current progress about MARCH proteins and their regulation on immune responses.
Introduction
Ubiquitination is one of the most common post-translational modifications, which plays a key role in regulating stability, localization, protein-protein interaction, and other properties of the substrates. Protein ubiquitination is carried in a multiple step process by the concerted action of E1 ubiquitin-activating enzyme, E2 conjugating enzyme, and E3 ubiquitin ligase. In the final step, the ubiquitin is ligated to a primary amine (i.e., that of lysine, cysteine, serine, threonine or the N-terminus) of substrate proteins by an E3 ligase. In most cases, E3 ubiquitin ligases dictate the specificity of substrates, therefore, are heavily studied.
Based on the structural properties, the E3 ubiquitin ligases are classified into three major types, including the really interesting new gene (RING), the homologous to the E6-AP carboxyl terminus (HECT), and the RING-between-RING (RBR) types of E3 ligases. The RING E3 ligases contain a characteristic RING finger domain, in which eight cysteine and histidine residues coordinate two Zn atoms in the interior of the protein (1).
Membrane-associated RING-CH-type finger (MARCH) proteins are a subfamily of the RING-type E3 ubiquitin ligases (2). MARCH proteins contain a C4HC3-type RING domain that has minor difference with the classic C3HC4-type RING domain in the identities of the fourth and fifth coordinating residues and the length of the peptide segments between the two (3). The MARCH proteins are originally identified as the mammalian structural homologs of the viral immunosuppressive membrane ubiquitin ligases K3 and K5 of Kaposi's sarcoma-associated herpesvirus (KSHV) (4, 5). K3 and K5 of gamma-2 herpesviruses and poxviruses, contained an N-terminal RING-CH domain followed by transmembrane domains, are found to down-regulate the surface expression of major histocompatibility complex I (MHC-I) (6–9). The first identified MARCH protein is c-MIR (now called MARCH8), which was identified by blast searches of the human genome databases (4). c-MIR is a functional homolog of herpesvirus proteins MIR1 and MIR2 and has similar substrate specificity (4). Further bioinformatics studies identified 10 more mammalian MARCH family members, which all possess RING-CH domains with E3 ubiquitin ligase activity.
Recent studies have demonstrated that MARCH proteins are critical regulators of immune responses, which act by catalyzing polyubiquitination of various immune receptors or certain organelle membrane-associated components involved in innate immune responses (3, 10). This review summarizes recent advances in our understanding of properties of MARCH proteins and their emerging roles in regulation of immune responses.
Properties of the MARCH E3 Ligase Family Members
Structures of MARCH Proteins
The most prominent properties of the MARCH family are the RING-CH domain and the transmembrane domains. Except for MARCH7 and MARCH10, the majority of 11 mammalian MARCH proteins share a similar structure, including an N-terminal RING-CH finger and two or more TM domains (Figure 1). MARCH7 and MARCH10 have no recognizable TM spans, with their RING-CH domains located at the C-terminus (3). Therefore, MARCH7 and MARCH10 represent two non-canonical members of the MARCH family. Phylogenetic analysis suggests that the TM-containing MARCH proteins can be classified into several sub-groups, including MARCH1/8, MARCH2/3, MARCH4/9/11, MARCH5, and MARCH6 sub-groups (Figure 1).
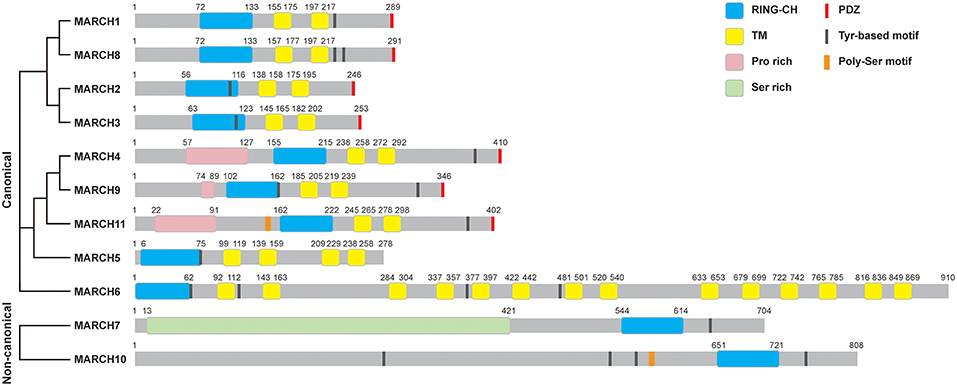
Figure 1. Structures of MARCH proteins. The domain organization for MARCH family members is shown. RING-CH, RING-CH finger domain; TM, transmembrane domain; PDZ, PDZ-binding domain; Pro rich, proline rich domain; Ser rich, serine rich domain; Tyr-based motif, tyrosine-based motif; Poly-Ser motif, poly-serine motif. The phylogenetic tree of MARCH proteins was generated with Clustal Omega alignment using the protein sequences in the Uniprot database.
Some MARCH proteins harbor other functional domains, such as C-terminal PDZ-binding domains that mediate protein interactions, and tyrosine-based YXXΦ (where X and Φ represent any amino acid and hydrophobic residues, respectively) motifs that are involved in endocytosis (Figure 1). MARCH11 mutant lack of the PDZ-binding domain does not interact with the PDZ-containing protein Veli3, suggesting that its PDZ-binding domain is crucial for its interaction with Veli3, which is important for its transport and subcellular localization (11). MARCH4, 9, and 11 possess an N-terminal proline rich domain that mediates protein interactions. MARCH7 contains an N-terminal disordered serine-rich region, which possesses high sequence similarity to serine/arginine-rich (SR) proteins and contains RNA recognition motifs. MARCH7 is localized on nuclear speckles, a site of the pre-mRNA processing and mRNA export, suggesting a potential role of MARCH7 in these processes (12, 13).
Subcellular Localizations of MARCH Proteins
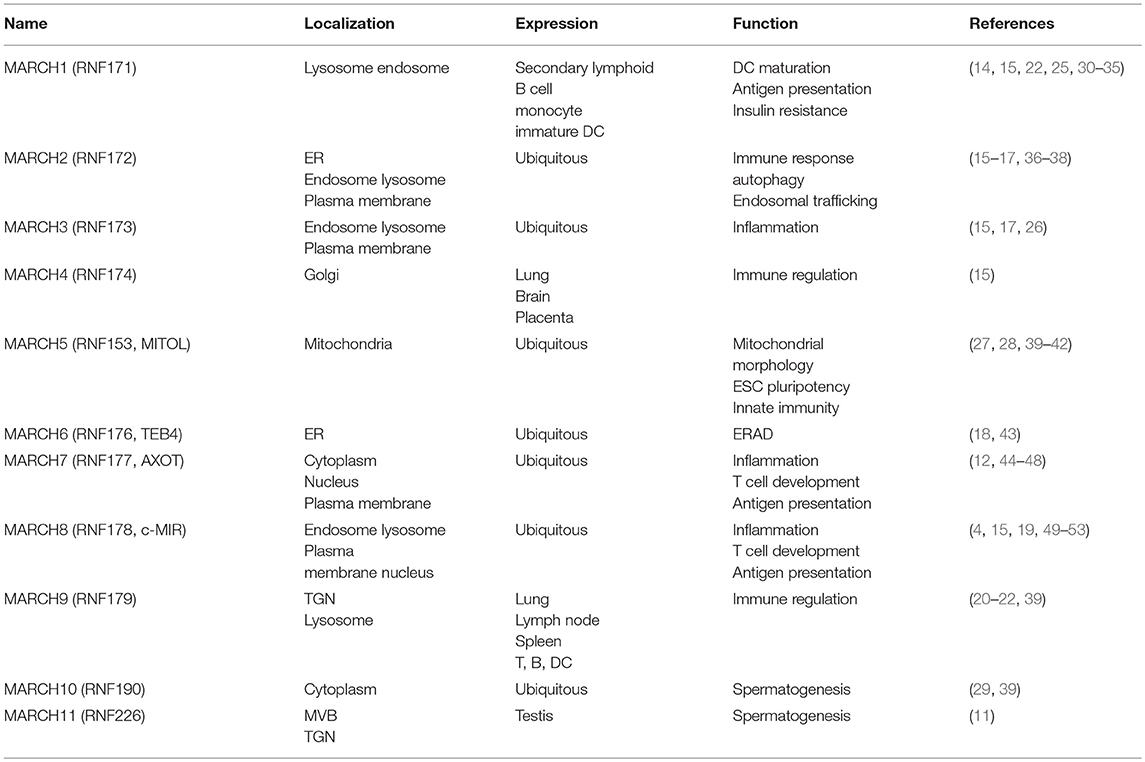
MARCH proteins are located in distinct cellular compartments, including the endosomes, lysosomes, endoplasmic reticulum (ER), Golgi apparatus, cytosol and plasma membrane. The localizations of MARCH proteins have direct effects on their functions. MARCH1, 2, 3, 8, and 9 are mostly located at the endosomes, lysosomes and plasma membrane (4, 14–22). Increasing evidences have shown that the localization of the MARCH proteins are important for their roles in regulation of immune receptors. For examples, a study using a chimeric CD86 protein has shown that CD86 is ubiquitinated by MARCH8 at the plasma membrane and followed by being internalized from the plasma membrane (23). MARCH1 is localized on LAMP-1-positive late endosomes/lysosomes, which is consistent with the finding that MARCH1 mediates ubiquitination and lysosome-dependent degradation of MHC class II molecules (24, 25). MARCH3 and MARCH8 regulate interleukin-1 (IL-1) receptor complex-mediated signaling (26). MARCH4 is located on the Golgi apparatus (15). MARCH5 is a mitochondrial outer membrane protein, which regulates mitochondrial morphology and immune response (27, 28). MARCH6 is an ER-resident ubiquitin ligase involved in ER-associated degradation (ERAD) (18). MARCH10 is localized in the cytosol (29). MARCH11 is localized on the multi-vesicular bodies (MVBs) and trans-Golgi network (TGN) (11). The PDZ-binding domain of the MARCH proteins plays a role in dictating their subcellular localizations. Mutations of the PDZ-binding domain of MARCH2 and 3 lead to their retention at the ER (16, 17).
Expression of MARCH Proteins
Most MARCH proteins are ubiquitously expressed in various tissues (Table 1), whereas MARCH1 is strictly expressed in the secondary lymphoid tissues and MARCH11 is mostly expressed in the testis (11, 15, 39). It has been reported that MARCH1-3, 5, 7–9 are highly expressed in immune cells, such as T cells, B cells, monocytes, macrophages and dentritic cells (DCs) (4, 25, 27, 30–32, 39, 44, 45, 49), implying for their potential roles in immune regulation.
Several studies have shown that the expression of MARCH proteins is regulated upon cellular stimulation. For examples, expression of MARCH1 is up-regulated following TNFα, IL-1β, and TGFβ stimulation (54). The transcription of MARCH1 gene is induced by IL-10 and LPS in monocytes or DCs, while its expression is decreased during maturation of DCs, indicating that MARCH1 plays important roles in regulating antigen presentation and DC maturation (22, 25, 31, 33, 34, 55). The transcription of MARCH9 gene is increased in human DCs upon Toll-like receptor (TLR) 3 or 4 activation (22). The transcription of MARCH2 gene is markedly induced upon human immunodeficiency virus 1 (HIV-1) infection, and MARCH2-deficiency increases HIV-1 infection in Jurkat and 293T cells (36). These studies suggest that MARCH2 plays unique inhibitory roles in HIV-1 infection. The transcription of MARCH3 gene is dramatically induced by LPS and TLR8 agonist in monocytes (56). Notably, it has been demonstrated that MARCH1 is highly expressed in human hepatocellular carcinoma (HCC) cells (55), MARCH5 is up-regulated in ovarian cancer tissues (28), and MARCH8 is highly expressed in esophageal tumors and associated with tumor aggression (50). These studies suggest that certain MARCH proteins may involve in tumorigenesis.
Post-translational Modifications (PTMs) of MARCH Proteins
Several studies have shown that MARCH proteins are tightly and delicately regulated by PTMs. The stability of several MARCH proteins is strictly regulated by ubiquitination. For examples, MARCH1 keeps a low protein level in antigen presentation cells (APCs) of human and mice by its TM-mediated dimerization, leading to autoubiquitination and degradation (57). However, it has been demonstrated that both wild-type and catalytically inactive MARCH1 are ubiquitinated in HeLa cells, suggesting that MARCH1 can be ubiquitinated by as yet unidentified E3 ligases (58). The stability of MARCH5-8, and 10 is tightly regulated by their RING-CH finger-mediated atutoubiquitination (4, 12, 15, 18, 27, 29). USP19 has been shown to remove K48-linked polyubiquitin moieties of MARCH6, which protects it from proteasome-dependent degradation (59). USP7 and USP9X deubiquitinate MARCH7, which promotes its stability (12).
The activity of MARCH ligases is regulated by phosphorylation. For example, MARCH3 is kept inactive by TYRO3-mediated phosphorylation in unstimulated cells. Upon IL-1β stimulation, CDC25A dephosphorylates MARCH3, which in turn activates MARCH3 and causes K48-linked polyubiquitination and degradation of IL-1 receptor type I (IL-1RI), leading to inhibition of IL-1β-triggered signaling (26).
Immune Regulation by MARCH Proteins
Regulation of MHCs by MARCH Proteins
MHCs are a set of cell-surface antigen-presenting proteins, which bind to pathogen-derived antigens and subsequently present them on the cell surface for recognition by the T cells (60). MHCs are important for the acquired immune system to recognize foreign molecules in vertebrates.
The expression and turnover of MHC-I on the surface of DCs is essential for their ability to activate CD8+ T cells (61). It has been demonstrated that several MARCH proteins can down-regulate MHC-I. Overexpression of MARCH9 causes increased endocytosis of MHC-I, and mediates polyubiquitination of MHC-I HLA-2.1 at its C-terminal lysine residues, leading to its lysosomal degradation (15, 22). Notably, knockdown of MARCH9 impairs the translocation of MHC-I from TGN to endosomes (22), indicating that MARCH9 plays a critical role in coordinating MHC-I access to endosomes and MHC-I-mediated antigen presentation. MARCH4 monoubiquitinates MHC-I, leading to endocytosis of MHC-I from cell surface and degradation (15).
MHC-II is essential for development and activation of CD4+ T cells (62). The expression of MHC-II is a prominent feature of professional APCs, and its cell surface expression is strictly regulated to efficiently control antigen presentation through endocytosis and subsequent lysosomal degradation (63). Several MARCH proteins regulate antigen presentation of MHC-II to CD4+ T cells and DC maturation (64). MARCH8-transgenic mice display impaired functions in antigen presentation and development of regulatory T cells (Tregs), and are resistant to the onset of experimental autoimmune encephalomyelitis (EAE) (53). MARCH8 mediates polyubiquitination of the β-chain of MHC-II at K225, leading to its lysosome-dependent degradation (53). Interestingly, it has been shown that Salmonella effector SteD inhibits antigen presentation and T cell activation by targeting MARCH8, which promotes MHC-II polyubiquitination and surface down-regulation (65). It has been reported that March8−/− mice display increased cell-surface MHC-II expression in thymic epithelial cells (TECs) and autoimmune regulator medullary (AIRE)− TECs. MARCH8-mediated polyubiquitination of MHC-II is regulated by CD83, which promotes MHC-II expression by impairing the interaction between MHC-II and MARCH1 (33, 52). Similar to MARCH8, MARCH1 has been identified as another physiological E3 ligase for MHC-II in mouse knockout studies (25, 30, 31, 66–68). In immature DCs, MHC-II undergoes sustaining polyubiquitination by MARCH1, leading to its down-regulation of cell-surface expression (25). During DC maturation, polyubiquitination of MHC-II is reduced and subsequently MHC-II accumulates at the cell surface (25). Further studies demonstrate that MHC-II is less polyubiquitinated and more stably expressed on the cell surface in the mature DCs and B cells in March1−/− compared with wild-type mice, resulting in enhanced antigen-presenting ability (68–70). Since MARCH1 plays crucial roles in regulating expression of cell-surface MHC-II, itself is tightly regulated at different DC maturation stages. These findings suggest that MARCH-mediated regulation of MHC molecules is important for immune regulation.
Regulation of the IL-1 Receptor Complex by MARCH3 and 8
The proinflammatory cytokine IL-1 is a central regulator in the initiation of inflammatory and immune responses. It also plays critical roles in the pathogenesis of different diseases, such as cancer, rheumatoid arthritis, neurodegenerative diseases, and atherosclerosis (71, 72). IL-1 consists of two separate ligands, IL-1α and IL-1β (73). Precursor IL-1α (pro-IL-1α) is biologically active, and is cleaved by calpain to generate mature IL-1α. Both forms of IL-1α are present mostly in the cell, unless released after cell death. In addition to binding to the cell-surface receptors, precursor IL-1α can translocate to the nucleus and affect transcription (74). By contrast, pro-IL-1β is biologically inactive and is cleaved by caspase-1 to produce an active protein (75). Although IL-1α and IL-1β share similar biological effects, IL-1β is more abundantly expressed during the early phase of inflammation response and is a major effector of inflammation (76). IL-1β signals through engagement of a membrane-bound receptor complex consisting of two subunits: IL-1RI and IL-1 receptor accessory protein (IL-1RAcP). Binding of IL-1β to IL-1RI initiates a ligand-induced conformational change of IL-1R1 that facilitates its recruitment of IL-1RAcP via Toll/interleukin-1 receptor (TIR)-TIR domain interaction, leading to the formation of an activated receptor complex. The receptor complex recruits the adaptor protein MyD88. MyD88 further recruits IRAK1, IRAK4, and TRAF6 to the receptor complex, where TRAF6 catalyzes K63-linked autoubiquitination to further recruit the TAK1-TAB2-TAB3 complex, leading to activation of the transcription factors NF-κB and AP-1, induction of downstream effector genes and inflammatory responses (77–85).
IL-1β-triggered signaling is tightly regulated to avoid excessive inflammatory response. Recent studies have shown that MARCH3 and 8 play important roles in terminating IL-1β-triggered inflammatory response (Figure 2). MARCH3-deficiency potentiates IL-1β-induced transcription of inflammatory genes in BMDMs, monocytes and primary mouse lung fibroblasts (MLFs). MARCH3-deficiency also increases the levels of serum inflammatory cytokines, as well as susceptibility to inflammatory death triggered by IL-1β injection or Listeria monocytogenes infection. Mechanistic studies indicate that MARCH3 is kept in inactive state by TYRO3-mediated phosphorylation. Upon IL-1β stimulation, MARCH3 is dephosphorylated by CDC25A, which in turn promotes its E3 ligase activity, leading to K48-linked polyubiquitination of IL-1RI at K409 and its lysosomal degradation (26). Another study has demonstrated that MARCH8 mediates K48-linked polyubiquitination of IL-1RAcP at K512 but not IL-1RI after IL-1β stimulation, leading to attenuation of IL-1β-triggered inflammatory response (51). These observations suggest that distinct members of the MARCH family mediate degradation of different components of the IL-1 receptor complex upon ligand stimulation, which ensures precise control of inflammatory response to avoid self-damage.
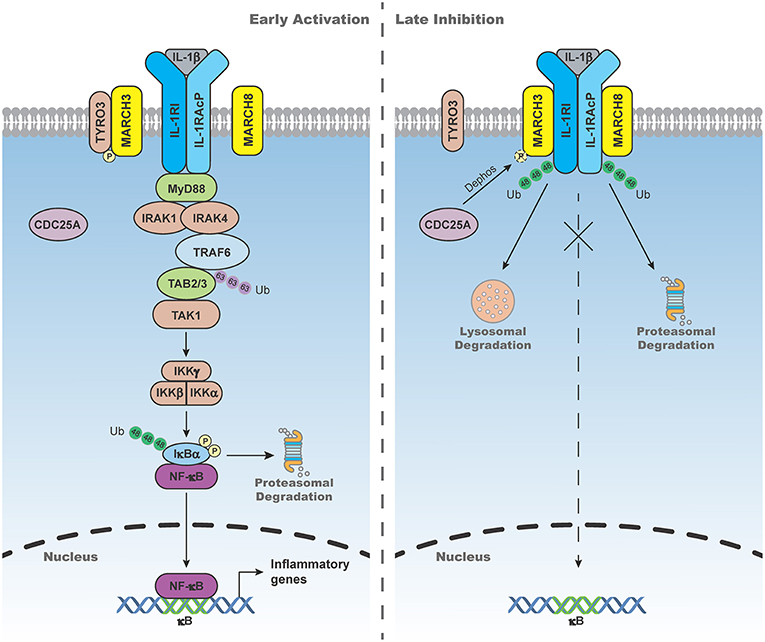
Figure 2. MARCH3- and 8-mediated negative regulation of IL-1β-triggered signaling. In the early phase of IL-1β stimulation, MARCH3 is kept in inactive state by the tyrosine-protein kinase receptor TYRO3-mediated phosphorylation. In the late phase of IL-1β stimulation, MARCH3 is dephosphorylated by cell division cycle 25A (CDC25A), which in turn promotes its E3 ligase activity, leading to K48-linked polyubiquitination of IL-1 receptor type I (IL-1RI) at K409 and its lysosomal degradation. MARCH8 mediates K48-linked polyubiquitination of IL-1 receptor accessory protein (IL-1RAcP) at K512 and its proteasomal degradation upon IL-1β stimulation, leading to attenuation of IL-1β-triggered inflammatory response.
Regulation of Other Immune Receptors by MARCH Proteins
MARCH proteins can also regulate stability of other cell-surface immune relevant receptors, such as cluster of differentiation 86 (CD86, also known as B7-2), TNF-related apoptosis inducing ligand receptor 1 (TRAIL-R1; also known as DR4), intercellular adhesion molecule 1 (ICAM-1, also known as CD54), Mult1, Fcγ Receptor IIb (FcγRIIb), and cystic fibrosis transmembrane conductance regulator (CFTR).
Regulation of CD86 by MARCH1 and 8
CD86 is an essential immune regulator expressed on APCs and provides costimulatory signals for T cell activation and survival (86, 87). It has been shown that MARCH8 mediates polyubiquitination of CD86 at its C-terminus, leading to its rapid endocytosis and lysosome-dependent degradation (4). It has also been shown that MARCH1 mediates polyubiquitination of CD86 at K267 and its degradation (34). Consequently, CD86 is more stably expressed on the cell surface of March1−/− DCs (34), indicating that MARCH1-mediated polyubiquitination of CD86 is a crucial mechanism in regulating antigen presentation by DCs.
Regulation of TRAIL-R1 by MARCH8
Binding of TRAIL to TRAIL-R1 induces apoptosis in cancer cells, which contributes to immunosurveillance (88, 89). MARCH8 targets TRAIL-R1 at K273 for polyubiquitination, which causes its lysosome-dependent degradation in breast cancer cells, leading to the inhibition of TRAIL-R1-mediated apoptosis signaling (90).
Regulation of ICAM-1 by MARCH9
ICAM-1 is an immune receptor that contains binding sites for a large amounts of immune-associated ligands. ICAM-1 plays important roles in facilitating transendothelial migration of activated lymphocytes across vascular endothelia in processes such as inflammatory responses (91, 92). It has been demonstrated that overexpression of MARCH9 mediates polyubiquitination of ICAM-1 for lysosome-dependent degradation (20). MARCH9 attenuates the oncogenic effects of ICAM-1, leading to the inhibition of migration and invasion in lung adenocarcinoma (LAC) cells (93). Interestingly, MARCH9 down-regulation in LAC correlates with poor clinical outcomes (93).
Regulation of Mult1 by MARCH4 and 9
Stimulation of immune cells through NKG2D receptor has been shown to involve in immune responses to infection and malignancy (94). Expression of the self-encoded ligands for NKG2D is tightly regulated to prevent autoimmune disorders. It has been reported that MARCH4 and 9 down-regulate cell-surface expression of Mult1 which is a ligand for NKG2D receptor, and prevents cells from being targeted for lysis by NK cells (95).
Regulation of FcγRIIb by MARCH3
FcγRIIb is an FcγR inhibitor that inhibits FcγR-mediated response to antibody-coated tumor cells. Regulation of FcγRIIb expression is crucial in tumor immunotherapy and autoimmune diseases (96, 97). It has been shown that MARCH3 is required for LPS-induced polyubiquitination and down-regulation of FcγRIIb (56). Notably, the ubiquitination of FcγRIIb precedes LPS-induced up-regulation of MARCH3 (56), suggesting that LPS had an earlier effect on MARCH3 activity.
Regulation of CFTR by MARCH2
CFTR is a plasma membrane cAMP-regulated chloride channel (98). Defective CFTR triggers aggresome formation and lung inflammation in cystic fibrosis (CF) by ROS-TG2-BECN1-mediated inhibition of autophagy (99, 100). CFTR also plays crucial roles in the progression and metastasis of cancer (101, 102). It has been reported that MARCH2 mediates polyubiquitination and degradation of CFTR, leading to attenuation of CFTR-mediated autophagy in tumor cells. MARCH2 interacts with CFTR via its PDZ-binding domain (37). These studies suggest that MARCH2 is a negative regulator of CFTR-mediated autophagy.
Regulation of Innate Immune Response by MARCH Proteins
It has been well-established that viral RNAs act as classic pathogen-associated molecular patterns (PAMPs), which are sensed by endosomal TLRs and cytosolic RIG-I-like receptors (RLRs) (103–106). Recognition of viral RNAs by these receptors links them to downstream adapter proteins, including TRIF, MyD88, and VISA (also called MAVS, IPS-1, and Cardif), leading to activation of the kinases TBK1 and IKKβ. These kinases phosphorylate and activate the transcription factors IRF3 and NF-κB, respectively, which cooperatively induce transcription of a set of antiviral genes including type I interferons (IFNs) (83, 107–119).
Several studies have demonstrated that MARCH proteins are involved in regulation of innate antiviral responses. MARCH5 catalyzes K63-linked polyubiquitination of TANK, which potentiates TLR7-mediated NF-κB activation (40). The protein level of VISA is tightly regulated to ensure its proper activation and timely termination of innate antiviral response. At the late phase of viral infection, VISA is phosphorylated at T54 by protein kinase A (PKA), which primes it for K48-linked polyubiquitination and degradation by MARCH5, leading to attenuation of innate immune response (120). The inactive rhomboid protease iRhom2 mediates proteasome-dependent degradation of MARCH5, resulting in the inhibition of virus-triggered degradation of VISA, which ensures proper level of VISA for innate antiviral response (121). These studies suggest that MARCH5 plays critical roles in temporal regulation of innate antiviral response.
MARCH proteins can regulate cellular antiviral response by targeting viral proteins. MARCH2 is induced upon HIV infection, which inhibits HIV-1 replication by mediating degradation of viral envelope proteins (36). It has also been shown that MARCH8 targets cell-surface envelope glycoproteins of HIV-1 for degradation, resulting in the inhibition of HIV-1 infection (49). MARCH8 catalyzes K63-linked polyubiquitination of hepatitis C virus (HCV) nonstructural 2 protein (NS2) and promotes viral assembly and envelopment (122). It has also been shown that MARCH8 is required for infection with Flaviviridae family members, such as HCV, dengue, and Zika viruses (122). These studies suggest that certain MARCH proteins can be potential host targets for antiviral strategies.
It has been shown that MARCH1-deficiency inhibits TLR3/4-mediated transcription of Tnfa gene in splenocytes (123). However, another study has shown that MARCH1-deficiency causes increased LPS-triggered production of proinflammatory cytokines, higher NK cell activation, as well as more susceptible to LPS-induced inflammatory death (124). Whether MARCH1 regulates TLR4-mediated inflammatory responses in a cell specific manner needs to be further investigated.
Functions of the Non-canonical MARCH Proteins
MARCH7 and MARCH10 are the two non-canonical MARCH family members. While it is unknown for the functions of MARCH10, studies have demonstrated that MARCH7 regulates leukemia inhibitory factor (LIF) secretion and inflammasome activation. In response to ConA, CD4+ T lymphocytes release LIF, which plays important roles in immune tolerance (125, 126). It has been shown that MARCH7-deficiency potentiates T cell proliferation and T cell-derived LIF secretion upon mitogen stimulation (46), suggesting that MARCH7 plays critical roles in immune tolerance. However, the mechanisms of MARCH7-mediated regulation of LIF secretion remains unclear.
NLRP3 inflammasome is composed of NLRP3, ASC and caspase-1, which is assembled in response to damage-associated molecular patterns (DAMPs) and PAMPs (127). It has been shown that MARCH7 mediates polyubiquitination of NLRP3 for degradation, leading to the inhibition of NLRP3-dependent inflammation triggered by dopamine D1 receptor (DRD1)-cAMP axis (47).
Concluding Remarks
The MARCH family of E3 ubiquitin ligases is unique in that they are mostly localized at plasma and/or organelle membranes. Most of the MARCH proteins are abundantly expressed in immune cells. This position them well in regulating immune receptors. In recent years, various studies have demonstrated that MARCH proteins target certain immune receptors for K48-linked polyubiquitination and degradation via distinct routes. In certain cases, MARCH proteins can also target the substrates for K63-linked polyubiquitination. In addition to immune receptors, certain MARCH proteins are also involved in regulation of viral proteins as well as components in innate immune responses. Therefore, the functions of MARCH proteins are not limited to immune receptors. Although considerable progress has been made for the functions and mechanisms of MARCH proteins, many outstanding questions remain. For examples, how are substrate specificities of MARCH proteins determined? Why are different subunits of a receptor complex targeted by distinct members of the MARCH family? One example is that the IL-1R complex components IL-1RI and IL-1RAcP are targeted by MARCH3 and 8 respectively. Whether do MARCH proteins play redundant functions in regulation of certain receptors? For examples, whether do MARCH1, 8 and 9 play redundant roles in down-regulation of MHC-II? Whether are other immune receptors regulated by MARCH proteins? How is the activity of MARCH proteins regulated by upstream signaling events? In addition to MARCH3, are other MARCH proteins regulated by phosphorylation or other post-translational modifications? Finally, we still know little about the pathological relevance of MARCH proteins in human. Further investigations into these outstanding questions would contribute to our understanding of the roles and mechanisms of MARCH proteins in physiological and pathological processes.
Author Contributions
H-BS and SL conceived and designed this review. HL performed manuscript preparation, literature search, and editing. H-BS, SL, and HL wrote the manuscript.
Funding
The work in the authors' laboratory was supported by grants from the Ministry of Science and Technology of China (2017YFA0505800 and 2016YFA0502102), the National Natural Science Foundation of China (31630045, 31830024, 31671465, and 31871415) and National Postdoctoral Program for Innovative Talents (BX20190255).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Deng Gao for help with the artwork of the figures and members of the Shu laboratory for helpful discussions.
References
1. Deshaies RJ, Joazeiro CA. RING domain E3 ubiquitin ligases. Annu Rev Biochem. (2009) 78:399–434. doi: 10.1146/annurev.biochem.78.101807.093809
2. Bauer J, Bakke O, Morth JP. Overview of the membrane-associated RING-CH (MARCH) E3 ligase family. N Biotechnol. (2017) 38(Pt A):7–15. doi: 10.1016/j.nbt.2016.12.002
3. Samji T, Hong S, Means RE. The membrane associated RING-CH proteins: a family of E3 ligases with diverse roles through the cell. Int Sch Res Notices. (2014) 2014:637295. doi: 10.1155/2014/637295
4. Goto E, Ishido S, Sato Y, Ohgimoto S, Ohgimoto K, Nagano-Fujii M, et al. c-MIR, a human E3 ubiquitin ligase, is a functional homolog of herpesvirus proteins MIR1 and MIR2 and has similar activity. J Biol Chem. (2003) 278:14657–68. doi: 10.1074/jbc.M211285200
5. Lehner PJ, Hoer S, Dodd R, Duncan LM. Downregulation of cell surface receptors by the K3 family of viral and cellular ubiquitin E3 ligases. Immunol Rev. (2005) 207:112–25. doi: 10.1111/j.0105-2896.2005.00314.x
6. Ishido S, Wang C, Lee BS, Cohen GB, Jung JU. Downregulation of major histocompatibility complex class I molecules by Kaposi's sarcoma-associated herpesvirus K3 and K5 proteins. J Virol. (2000) 74:5300–9. doi: 10.1128/JVI.74.11.5300-5309.2000
7. Stevenson PG, Efstathiou S, Doherty PC, Lehner PJ. Inhibition of MHC class I-restricted antigen presentation by gamma 2-herpesviruses. Proc Natl Acad Sci USA. (2000) 97:8455–60. doi: 10.1073/pnas.150240097
8. Coscoy L, Ganem D. Kaposi's sarcoma-associated herpesvirus encodes two proteins that block cell surface display of MHC class I chains by enhancing their endocytosis. Proc Natl Acad Sci USA. (2000) 97:8051–6. doi: 10.1073/pnas.140129797
9. Früh K, Bartee E, Gouveia K, Mansouri M. Immune evasion by a novel family of viral PHD/LAP-finger proteins of gamma-2 herpesviruses and poxviruses. Virus Res. (2002) 88:55–69. doi: 10.1016/S0168-1702(02)00120-X
10. Nakamura N. The role of the transmembrane RING finger proteins in cellular and organelle function. Membranes. (2011) 1:354–93. doi: 10.3390/membranes1040354
11. Morokuma Y, Nakamura N, Kato A, Notoya M, Yamamoto Y, Sakai Y, et al. MARCH-XI, a novel transmembrane ubiquitin ligase implicated in ubiquitin-dependent protein sorting in developing spermatids. J Biol Chem. (2007) 282:24806–15. doi: 10.1074/jbc.M700414200
12. Nathan JA, Sengupta S, Wood SA, Admon A, Markson G, Sanderson C, et al. The ubiquitin E3 ligase MARCH7 is differentially regulated by the deubiquitylating enzymes USP7 and USP9X. Traffic. (2008) 9:1130–45. doi: 10.1111/j.1600-0854.2008.00747.x
13. Flach K, Ramminger E, Hilbrich I, Arsalan-Werner A, Albrecht F, Herrmann L, et al. Axotrophin/MARCH7 acts as an E3 ubiquitin ligase and ubiquitinates tau protein in vitro impairing microtubule binding. Biochim Biophys Acta. (2014) 1842:1527–38. doi: 10.1016/j.bbadis.2014.05.029
14. Jabbour M, Campbell EM, Fares H, Lybarger L. Discrete domains of MARCH1 mediate its localization, functional interactions, and posttranscriptional control of expression. J Immunol. (2009) 183:6500–12. doi: 10.4049/jimmunol.0901521
15. Bartee E, Mansouri M, Hovey Nerenberg BT, Gouveia K, Früh K. Downregulation of major histocompatibility complex class I by human ubiquitin ligases related to viral immune evasion proteins. J Virol. (2004) 78:1109–20. doi: 10.1128/JVI.78.3.1109-1120.2004
16. Nakamura N, Fukuda H, Kato A, Hirose S. MARCH-II is a syntaxin-6-binding protein involved in endosomal trafficking. Mol Biol Cell. (2005) 16:1696–710. doi: 10.1091/mbc.e04-03-0216
17. Fukuda H, Nakamura N, Hirose S. MARCH-III Is a novel component of endosomes with properties similar to those of MARCH-II. J Biochem. (2006) 139:137–45. doi: 10.1093/jb/mvj012
18. Hassink G, Kikkert M, van Voorden S, Lee SJ, Spaapen R, van Laar T, et al. TEB4 is a C4HC3 RING finger-containing ubiquitin ligase of the endoplasmic reticulum. Biochem J. (2005) 388(Pt 2):647–55. doi: 10.1042/BJ20041241
19. Lapaque N, Jahnke M, Trowsdale J, Kelly AP. The HLA-DRalpha chain is modified by polyubiquitination. J Biol Chem. (2009) 284:7007–16. doi: 10.1074/jbc.M805736200
20. Hoer S, Smith L, Lehner PJ. MARCH-IX mediates ubiquitination and downregulation of ICAM-1. FEBS Lett. (2007) 581:45–51. doi: 10.1016/j.febslet.2006.11.075
21. Bartee E, Eyster CA, Viswanathan K, Mansouri M, Donaldson JG, Früh K. Membrane-Associated RING-CH proteins associate with Bap31 and target CD81 and CD44 to lysosomes. PLoS ONE. (2010) 5:e15132. doi: 10.1371/journal.pone.0015132
22. De Angelis Rigotti F, De Gassart A, Pforr C, Cano F, N'Guessan P, Combes A, et al. MARCH9-mediated ubiquitination regulates MHC I export from the TGN. Immunol Cell Biol. (2017) 95:753–64. doi: 10.1038/icb.2017.44
23. Goto E, Mito-Yoshida M, Uematsu M, Aoki M, Matsuki Y, Ohmura-Hoshino M, et al. An excellent monitoring system for surface ubiquitination-induced internalization in mammals. PLoS ONE. (2008) 3:e1490. doi: 10.1371/journal.pone.0001490
24. van Niel G, Wubbolts R, Ten Broeke T, Buschow SI, Ossendorp FA, Melief CJ, et al. Dendritic cells regulate exposure of MHC class II at their plasma membrane by oligoubiquitination. Immunity. (2006) 25:885–94. doi: 10.1016/j.immuni.2006.11.001
25. De Gassart A, Camosseto V, Thibodeau J, Ceppi M, Catalan N, Pierre P, et al. MHC class II stabilization at the surface of human dendritic cells is the result of maturation-dependent MARCH I down-regulation. Proc Natl Acad Sci USA. (2008) 105:3491–6. doi: 10.1073/pnas.0708874105
26. Lin H, Gao D, Hu MM, Zhang M, Wu XX, Feng L, et al. MARCH3 attenuates IL-1beta-triggered inflammation by mediating K48-linked polyubiquitination and degradation of IL-1RI. Proc Natl Acad Sci USA. (2018) 115:12483–8. doi: 10.1073/pnas.1806217115
27. Yonashiro R, Ishido S, Kyo S, Fukuda T, Goto E, Matsuki Y, et al. A novel mitochondrial ubiquitin ligase plays a critical role in mitochondrial dynamics. EMBO J. (2006) 25:3618–26. doi: 10.1038/sj.emboj.7601249
28. Hu J, Meng Y, Zhang Z, Yan Q, Jiang X, Lv Z, et al. MARCH5 RNA promotes autophagy, migration, and invasion of ovarian cancer cells. Autophagy. (2017) 13:333–44. doi: 10.1080/15548627.2016.1256520
29. Iyengar PV, Hirota T, Hirose S, Nakamura N. Membrane-associated RING-CH 10 (MARCH10 protein) is a microtubule-associated E3 ubiquitin ligase of the spermatid flagella. J Biol Chem. (2011) 286:39082–90. doi: 10.1074/jbc.M111.256875
30. Galbas T, Steimle V, Lapointe R, Ishido S, Thibodeau J. MARCH1 down-regulation in IL-10-activated B cells increases MHC class II expression. Cytokine. (2012) 59:27–30. doi: 10.1016/j.cyto.2012.03.015
31. Thibodeau J, Bourgeois-Daigneault MC, Huppé G, Tremblay J, Aumont A, Houde M, et al. Interleukin-10-induced MARCH1 mediates intracellular sequestration of MHC class II in monocytes. Eur J Immunol. (2008) 38:1225–30. doi: 10.1002/eji.200737902
32. Chattopadhyay G, Shevach EM. Antigen-specific induced T regulatory cells impair dendritic cell function via an IL-10/MARCH1-dependent mechanism. J Immunol. (2013) 191:5875–84. doi: 10.4049/jimmunol.1301693
33. Tze LE, Horikawa K, Domaschenz H, Howard DR, Roots CM, Rigby RJ, et al. CD83 increases MHC II and CD86 on dendritic cells by opposing IL-10-driven MARCH1-mediated ubiquitination and degradation. J Exp Med. (2011) 208:149–65. doi: 10.1084/jem.20092203
34. Baravalle G, Park H, McSweeney M, Ohmura-Hoshino M, Matsuki Y, Ishido S, et al. Ubiquitination of CD86 is a key mechanism in regulating antigen presentation by dendritic cells. J Immunol. (2011) 187:2966–73. doi: 10.4049/jimmunol.1101643
35. Nagarajan A, Petersen MC, Nasiri AR, Butrico G, Fung A, Ruan HB, et al. MARCH1 regulates insulin sensitivity by controlling cell surface insulin receptor levels. Nat Commun. (2016) 7:12639. doi: 10.1038/ncomms12639
36. Zhang Y, Lu J, Liu X. MARCH2 is upregulated in HIV-1 infection and inhibits HIV-1 production through envelope protein translocation or degradation. Virology. (2018) 518:293–300. doi: 10.1016/j.virol.2018.02.003
37. Xia D, Qu L, Li G, Hongdu B, Xu C, Lin X, et al. MARCH2 regulates autophagy by promoting CFTR ubiquitination and degradation and PIK3CA-AKT-MTOR signaling. Autophagy. (2016) 12:1614–30. doi: 10.1080/15548627.2016.1192752
38. Rhodes DR, Yu J, Shanker K, Deshpande N, Varambally R, Ghosh D, et al. ONCOMINE: a cancer microarray database and integrated data-mining platform. Neoplasia. (2004) 6:1–6. doi: 10.1016/S1476-5586(04)80047-2
39. Su AI, Cooke MP, Ching KA, Hakak Y, Walker JR, Wiltshire T, et al. Large-scale analysis of the human and mouse transcriptomes. Proc Natl Acad Sci USA. (2002) 99:4465–70. doi: 10.1073/pnas.012025199
40. Shi HX, Liu X, Wang Q, Tang PP, Liu XY, Shan YF, et al. Mitochondrial ubiquitin ligase MARCH5 promotes TLR7 signaling by attenuating TANK action. PLoS Pathog. (2011) 7:e1002057. doi: 10.1371/journal.ppat.1002057
41. Yoo YS, Park YY, Kim JH, Cho H, Kim SH, Lee HS, et al. The mitochondrial ubiquitin ligase MARCH5 resolves MAVS aggregates during antiviral signalling. Nat Commun. (2015) 6:7910. doi: 10.1038/ncomms8910
42. Gu H, Li Q, Huang S, Lu W, Cheng F, Gao P, et al. Mitochondrial E3 ligase March5 maintains stemness of mouse ES cells via suppression of ERK signalling. Nat Commun. (2015) 6:7112. doi: 10.1038/ncomms8112
43. Nagase T, Ishikawa K, Miyajima N, Tanaka A, Kotani H, Nomura N, et al. Prediction of the coding sequences of unidentified human genes. IX. The complete sequences of 100 new cDNA clones from brain which can code for large proteins in vitro. DNA Res. (1998) 5:31–9.
44. Ramalho-Santos M, Yoon S, Matsuzaki Y, Mulligan RC, Melton DA. “Stemness”: transcriptional profiling of embryonic and adult stem cells. Science. (2002) 298:597–600. doi: 10.1126/science.1072530
45. Szigyarto CA, Sibbons P, Williams G, Uhlen M, Metcalfe SM. The E3 ligase axotrophin/MARCH-7: protein expression profiling of human tissues reveals links to adult stem cells. J Histochem Cytochem. (2010) 58:301–8. doi: 10.1369/jhc.2009.954420
46. Metcalfe SM, Muthukumarana PA, Thompson HL, Haendel MA, Lyons GE. Leukaemia inhibitory factor (LIF) is functionally linked to axotrophin and both LIF and axotrophin are linked to regulatory immune tolerance. FEBS Lett. (2005) 579:609–14. doi: 10.1016/j.febslet.2004.12.027
47. Yan Y, Jiang W, Liu L, Wang X, Ding C, Tian Z, et al. Dopamine controls systemic inflammation through inhibition of NLRP3 inflammasome. Cell. (2015) 160:62–73. doi: 10.1016/j.cell.2014.11.047
48. Zhao K, Yang Y, Zhang G, Wang C, Wang D, Wu M, et al. Regulation of the Mdm2-p53 pathway by the ubiquitin E3 ligase MARCH7. EMBO Rep. (2018) 19:305–19. doi: 10.15252/embr.201744465
49. Tada T, Zhang Y, Koyama T, Tobiume M, Tsunetsugu-Yokota Y, Yamaoka S, et al. MARCH8 inhibits HIV-1 infection by reducing virion incorporation of envelope glycoproteins. Nat Med. (2015) 21:1502–7. doi: 10.1038/nm.3956
50. Singh S, Saraya A, Das P, Sharma R. Increased expression of MARCH8, an E3 ubiquitin ligase, is associated with growth of esophageal tumor. Cancer Cell Int. (2017) 17:116. doi: 10.1186/s12935-017-0490-y
51. Chen R, Li M, Zhang Y, Zhou Q, Shu HB. The E3 ubiquitin ligase MARCH8 negatively regulates IL-1beta-induced NF-kappaB activation by targeting the IL1RAP coreceptor for ubiquitination and degradation. Proc Natl Acad Sci USA. (2012) 109:14128–33. doi: 10.1073/pnas.1205246109
52. von Rohrscheidt J, Petrozziello E, Nedjic J, Federle C, Krzyzak L, Ploegh HL, et al. Thymic CD4 T cell selection requires attenuation of March8-mediated MHCII turnover in cortical epithelial cells through CD83. J Exp Med. (2016) 213:1685–94. doi: 10.1084/jem.20160316
53. Ohmura-Hoshino M, Matsuki Y, Aoki M, Goto E, Mito M, Uematsu M, et al. Inhibition of MHC class II expression and immune responses by c-MIR. J Immunol. (2006) 177:341–54. doi: 10.4049/jimmunol.177.5.3493-a
54. Meng Y, Hu J, Chen Y, Yu T, Hu L. Silencing MARCH1 suppresses proliferation, migration and invasion of ovarian cancer SKOV3 cells via downregulation of NF-kappaB and Wnt/beta-catenin pathways. Oncol Rep. (2016) 36:2463–70. doi: 10.3892/or.2016.5076
55. Xie L, Dai H, Li M, Yang W, Yu G, Wang X, et al. MARCH1 encourages tumour progression of hepatocellular carcinoma via regulation of PI3K-AKT-beta-catenin pathways. J Cell Mol Med. (2019) 23:3386–401. doi: 10.1111/jcmm.14235
56. Fatehchand K, Ren L, Elavazhagan S, Fang H, Mo X, Vasilakos JP, et al. Toll-like receptor 4 ligands down-regulate fcgamma receptor IIb (FcgammaRIIb) via MARCH3 protein-mediated ubiquitination. J Biol Chem. (2016) 291:3895–904. doi: 10.1074/jbc.M115.701151
57. Bourgeois-Daigneault MC, Thibodeau J. Autoregulation of MARCH1 expression by dimerization and autoubiquitination. J Immunol. (2012) 188:4959–70. doi: 10.4049/jimmunol.1102708
58. Lei L, Bandola-Simon J, Roche PA. Ubiquitin-conjugating enzyme E2 D1 (Ube2D1) mediates lysine-independent ubiquitination of the E3 ubiquitin ligase March-I. J Biol Chem. (2018) 293:3904–12. doi: 10.1074/jbc.RA117.001322
59. Nakamura N, Harada K, Kato M, Hirose S. Ubiquitin-specific protease 19 regulates the stability of the E3 ubiquitin ligase MARCH6. Exp Cell Res. (2014) 328:207–16. doi: 10.1016/j.yexcr.2014.07.025
60. Trowsdale J, Knight JC. Major histocompatibility complex genomics and human disease. Annu Rev Genomics Hum Genet. (2013) 14:301–23. doi: 10.1146/annurev-genom-091212-153455
61. Lehner PJ, Cresswell P. Recent developments in MHC-class-I-mediated antigen presentation. Curr Opin Immunol. (2004) 16:82–9. doi: 10.1016/j.coi.2003.11.012
62. Denzin LK, Cresswell P. HLA-DM induces CLIP dissociation from MHC class II alpha beta dimers and facilitates peptide loading. Cell. (1995) 82:155–65. doi: 10.1016/0092-8674(95)90061-6
63. Shin JS, Ebersold M, Pypaert M, Delamarre L, Hartley A, Mellman I. Surface expression of MHC class II in dendritic cells is controlled by regulated ubiquitination. Nature. (2006) 444:115–8. doi: 10.1038/nature05261
64. Ishido S, Kajikawa M. MHC class II fine tuning by ubiquitination: lesson from MARCHs. Immunogenetics. (2019) 71:197–201. doi: 10.1007/s00251-018-1094-y
65. Bayer-Santos E, Durkin CH, Rigano LA, Kupz A, Alix E, Cerny O, et al. The salmonella effector sted mediates MARCH8-dependent ubiquitination of MHC II molecules and inhibits T cell activation. Cell Host Microbe. (2016) 20:584–95. doi: 10.1016/j.chom.2016.10.007
66. Borges TJ, Murakami N, Machado FD, Murshid A, Lang BJ, Lopes RL, et al. March1-dependent modulation of donor MHC II on CD103(+) dendritic cells mitigates alloimmunity. Nat Commun. (2018) 9:3482. doi: 10.1038/s41467-018-05572-z
67. Oh J, Wu N, Baravalle G, Cohn B, Ma J, Lo B, et al. MARCH1-mediated MHCII ubiquitination promotes dendritic cell selection of natural regulatory T cells. J Exp Med. (2013) 210:1069–77. doi: 10.1084/jem.20122695
68. Matsuki Y, Ohmura-Hoshino M, Goto E, Aoki M, Mito-Yoshida M, Uematsu M, et al. Novel regulation of MHC class II function in B cells. EMBO J. (2007) 26:846–54. doi: 10.1038/sj.emboj.7601556
69. Ohmura-Hoshino M, Matsuki Y, Mito-Yoshida M, Goto E, Aoki-Kawasumi M, Nakayama M, et al. Cutting edge: requirement of MARCH-I-mediated MHC II ubiquitination for the maintenance of conventional dendritic cells. J Immunol. (2009) 183:6893–7. doi: 10.4049/jimmunol.0902178
70. Walseng E, Furuta K, Bosch B, Weih KA, Matsuki Y, Bakke O, et al. Ubiquitination regulates MHC class II-peptide complex retention and degradation in dendritic cells. Proc Natl Acad Sci USA. (2010) 107:20465–70. doi: 10.1073/pnas.1010990107
71. Dinarello CA. Anti-inflammatory agents: present and future. Cell. (2010) 140:935–50. doi: 10.1016/j.cell.2010.02.043
72. Dinarello CA. Immunological and inflammatory functions of the interleukin-1 family. Annu Rev Immunol. (2009) 27:519–50. doi: 10.1146/annurev.immunol.021908.132612
73. March CJ, Mosley B, Larsen A, Cerretti DP, Braedt G, Price V, et al. Cloning, sequence and expression of two distinct human interleukin-1 complementary DNAs. Nature. (1985) 315:641–7. doi: 10.1038/315641a0
74. Werman A, Werman-Venkert R, White R, Lee JK, Werman B, Krelin Y, et al. The precursor form of IL-1alpha is an intracrine proinflammatory activator of transcription. Proc Natl Acad Sci USA. (2004) 101:2434–9. doi: 10.1073/pnas.0308705101
75. Thornberry NA, Bull HG, Calaycay JR, Chapman KT, Howard AD, Kostura MJ, et al. A novel heterodimeric cysteine protease is required for interleukin-1 beta processing in monocytes. Nature. (1992) 356:768–74. doi: 10.1038/356768a0
76. Boraschi D, Tagliabue A. The interleukin-1 receptor family. Semin Immunol. (2013) 25:394–407. doi: 10.1016/j.smim.2013.10.023
77. Weber A, Wasiliew P, Kracht M. Interleukin-1 (IL-1) pathway. Sci Signal. (2010) 3:cm1. doi: 10.1126/scisignal.3105cm1
78. Chen ZJ. Ubiquitin signalling in the NF-kappaB pathway. Nat Cell Biol. (2005) 7:758–65. doi: 10.1038/ncb0805-758
79. Hu MM, Yang Q, Zhang J, Liu SM, Zhang Y, Lin H, et al. TRIM38 inhibits TNFalpha- and IL-1beta-triggered NF-kappaB activation by mediating lysosome-dependent degradation of TAB2/3. Proc Natl Acad Sci USA. (2014) 111:1509–14. doi: 10.1073/pnas.1318227111
80. Tian Y, Zhang Y, Zhong B, Wang YY, Diao FC, Wang RP, et al. RBCK1 negatively regulates tumor necrosis factor- and interleukin-1-triggered NF-kappaB activation by targeting TAB2/3 for degradation. J Biol Chem. (2007) 282:16776–82. doi: 10.1074/jbc.M701913200
81. Li Q, Yan J, Mao AP, Li C, Ran Y, Shu HB, et al. Tripartite motif 8 (TRIM8) modulates TNFalpha- and IL-1beta-triggered NF-kappaB activation by targeting TAK1 for K63-linked polyubiquitination. Proc Natl Acad Sci USA. (2011) 108:19341–6. doi: 10.1073/pnas.1110946108
82. Zheng H, Li Q, Chen R, Zhang J, Ran Y, He X, et al. The dual-specificity phosphatase DUSP14 negatively regulates tumor necrosis factor- and interleukin-1-induced nuclear factor-kappaB activation by dephosphorylating the protein kinase TAK1. J Biol Chem. (2013) 288:819–25. doi: 10.1074/jbc.M112.412643
83. Hu MM, Shu HB. Multifaceted roles of TRIM38 in innate immune and inflammatory responses. Cell Mol Immunol. (2017) 14:331–8. doi: 10.1038/cmi.2016.66
84. Liu LJ, Liu TT, Ran Y, Li Y, Zhang XD, Shu HB, et al. The E3 ubiquitin ligase MIB1 negatively regulates basal IkappaBalpha level and modulates NF-kappaB activation. Cell Res. (2012) 22:603–6. doi: 10.1038/cr.2011.199
85. He X, Li Y, Li C, Liu LJ, Zhang XD, Liu Y, et al. USP2a negatively regulates IL-1beta- and virus-induced NF-kappaB activation by deubiquitinating TRAF6. J Mol Cell Biol. (2013) 5:39–47. doi: 10.1093/jmcb/mjs024
86. Freeman GJ, Gribben JG, Boussiotis VA, Ng JW, Restivo VA, Lombard LA, et al. Cloning of B7–2: a CTLA-4 counter-receptor that costimulates human T cell proliferation. Science. (1993) 262:909–11. doi: 10.1126/science.7694363
87. Carreno BM, Collins M. The B7 family of ligands and its receptors: new pathways for costimulation and inhibition of immune responses. Annu Rev Immunol. (2002) 20:29–53.
88. Falschlehner C, Ganten TM, Koschny R, Schaefer U, Walczak H. TRAIL and other TRAIL receptor agonists as novel cancer therapeutics. Adv Exp Med Biol. (2009) 647:195–206. doi: 10.1007/978-0-387-89520-8_14
89. Baetu TM, Hiscott J. On the TRAIL to apoptosis. Cytokine Growth Factor Rev. (2002) 13:199–207. doi: 10.1016/S1359-6101(02)00006-0
90. van de Kooij B, Verbrugge I, de Vries E, Gijsen M, Montserrat V, Maas C, et al. Ubiquitination by the membrane-associated RING-CH-8 (MARCH-8) ligase controls steady-state cell surface expression of tumor necrosis factor-related apoptosis inducing ligand (TRAIL) receptor 1. J Biol Chem. (2013) 288:6617–28. doi: 10.1074/jbc.M112.448209
91. Rothlein R, Dustin ML, Marlin SD, Springer TA. A human intercellular adhesion molecule (ICAM-1) distinct from LFA-1. J Immunol. (1986) 137:1270–4.
92. Yang L, Froio RM, Sciuto TE, Dvorak AM, Alon R, Luscinskas FW. ICAM-1 regulates neutrophil adhesion and transcellular migration of TNF-alpha-activated vascular endothelium under flow. Blood. (2005) 106:584–92. doi: 10.1182/blood-2004-12-4942
93. Shen QM, Wang HY, Xu S. MARCH9 suppresses lung adenocarcinoma progression by downregulating ICAM-1. Cell Physiol Biochem. (2018) 50:92–107. doi: 10.1159/000493961
94. Guerra N, Tan YX, Joncker NT, Choy A, Gallardo F, Xiong N, et al. NKG2D-deficient mice are defective in tumor surveillance in models of spontaneous malignancy. Immunity. (2008) 28:571–80. doi: 10.1016/j.immuni.2008.02.016
95. Nice TJ, Deng W, Coscoy L, Raulet DH. Stress-regulated targeting of the NKG2D ligand Mult1 by a membrane-associated RING-CH family E3 ligase. J Immunol. (2010) 185:5369–76. doi: 10.4049/jimmunol.1000247
96. Muta T, Kurosaki T, Misulovin Z, Sanchez M, Nussenzweig MC, Ravetch JV. A 13-amino-acid motif in the cytoplasmic domain of Fc gamma RIIB modulates B-cell receptor signalling. Nature. (1994) 369:70–3.
97. Choquet D, Ku G, Cassard S, Malissen B, Korn H, Fridman WH, et al. Different patterns of calcium signaling triggered through two components of the B lymphocyte antigen receptor. J Biol Chem. (1994) 269:6491–7.
98. Li C, Naren AP. CFTR chloride channel in the apical compartments: spatiotemporal coupling to its interacting partners. Integr Biol. (2010) 2:161–77. doi: 10.1039/b924455g
99. Luciani A, Villella VR, Esposito S, Brunetti-Pierri N, Medina DL, Settembre C, et al. Cystic fibrosis: a disorder with defective autophagy. Autophagy. (2011) 7:104–6. doi: 10.4161/auto.7.1.13987
100. Luciani A, Villella VR, Esposito S, Brunetti-Pierri N, Medina D, Settembre C, et al. Defective CFTR induces aggresome formation and lung inflammation in cystic fibrosis through ROS-mediated autophagy inhibition. Nat Cell Biol. (2010) 12:863–75. doi: 10.1038/ncb2090
101. Xu J, Yong M, Li J, Dong X, Yu T, Fu X, et al. High level of CFTR expression is associated with tumor aggression and knockdown of CFTR suppresses proliferation of ovarian cancer in vitro and in vivo. Oncol Rep. (2015) 33:2227–34. doi: 10.3892/or.2015.3829
102. Peng X, Wu Z, Yu L, Li J, Xu W, Chan HC, et al. Overexpression of cystic fibrosis transmembrane conductance regulator (CFTR) is associated with human cervical cancer malignancy, progression and prognosis. Gynecol Oncol. (2012) 125:470–6. doi: 10.1016/j.ygyno.2012.02.015
103. Kawai T, Akira S. Toll-like receptors and their crosstalk with other innate receptors in infection and immunity. Immunity. (2011) 34:637–50. doi: 10.1016/j.immuni.2011.05.006
104. Yoneyama M, Fujita T. Structural mechanism of RNA recognition by the RIG-I-like receptors. Immunity. (2008) 29:178–81. doi: 10.1016/j.immuni.2008.07.009
105. Lian H, Zang R, Wei J, Ye W, Hu MM, Chen YD, et al. The zinc-finger protein ZCCHC3 binds RNA and facilitates viral RNA sensing and activation of the RIG-I-like receptors. Immunity. (2018) 49:438–48 e435. doi: 10.1016/j.immuni.2018.08.014
106. Luo WW, Shu HB. Delicate regulation of the cGAS-MITA-mediated innate immune response. Cell Mol Immunol. (2018) 15:666–75. doi: 10.1038/cmi.2016.51
107. Hu MM, Shu HB. Cytoplasmic mechanisms of recognition and defense of microbial nucleic acids. Annu Rev Cell Dev Biol. (2018) 34:357–79. doi: 10.1146/annurev-cellbio-100617-062903
108. Ran Y, Shu HB, Wang YY. MITA/STING: a central and multifaceted mediator in innate immune response. Cytokine Growth Factor Rev. (2014) 25:631–9. doi: 10.1016/j.cytogfr.2014.05.003
109. Chen LT, Hu MM, Xu ZS, Liu Y, Shu HB. MSX1 modulates RLR-mediated innate antiviral signaling by facilitating assembly of TBK1-associated complexes. J Immunol. (2016) 197:199–207. doi: 10.4049/jimmunol.1600039
110. Li Y, Li C, Xue P, Zhong B, Mao AP, Ran Y, et al. ISG56 is a negative-feedback regulator of virus-triggered signaling and cellular antiviral response. Proc Natl Acad Sci USA. (2009) 106:7945–50. doi: 10.1073/pnas.0900818106
111. Zhong B, Yang Y, Li S, Wang YY, Li Y, Diao F, et al. The adaptor protein MITA links virus-sensing receptors to IRF3 transcription factor activation. Immunity. (2008) 29:538–50. doi: 10.1016/j.immuni.2008.09.003
112. Hu MM, Liao CY, Yang Q, Xie XQ, Shu HB. Innate immunity to RNA virus is regulated by temporal and reversible sumoylation of RIG-I and MDA5. J Exp Med. (2017) 214:973–89. doi: 10.1084/jem.20161015
113. Zhong B, Zhang L, Lei C, Li Y, Mao AP, Yang Y, et al. The ubiquitin ligase RNF5 regulates antiviral responses by mediating degradation of the adaptor protein MITA. Immunity. (2009) 30:397–407. doi: 10.1016/j.immuni.2009.01.008
114. Wang YY, Liu LJ, Zhong B, Liu TT, Li Y, Yang Y, et al. WDR5 is essential for assembly of the VISA-associated signaling complex and virus-triggered IRF3 and NF-kappaB activation. Proc Natl Acad Sci USA. (2010) 107:815–20. doi: 10.1073/pnas.0908967107
115. Lei CQ, Zhong B, Zhang Y, Zhang J, Wang S, Shu HB. Glycogen synthase kinase 3beta regulates IRF3 transcription factor-mediated antiviral response via activation of the kinase TBK1. Immunity. (2010) 33:878–89. doi: 10.1016/j.immuni.2010.11.021
116. Li S, Zheng H, Mao AP, Zhong B, Li Y, Liu Y, et al. Regulation of virus-triggered signaling by OTUB1- and OTUB2-mediated deubiquitination of TRAF3 and TRAF6. J Biol Chem. (2010) 285:4291–7. doi: 10.1074/jbc.M109.074971
117. Yang Y, Liao B, Wang S, Yan B, Jin Y, Shu HB, et al. E3 ligase WWP2 negatively regulates TLR3-mediated innate immune response by targeting TRIF for ubiquitination and degradation. Proc Natl Acad Sci USA. (2013) 110:5115–20. doi: 10.1073/pnas.1220271110
118. Luo WW, Lian H, Zhong B, Shu HB, Li S. Kruppel-like factor 4 negatively regulates cellular antiviral immune response. Cell Mol Immunol. (2016) 13:65–72. doi: 10.1038/cmi.2014.125
119. Zhang J, Hu MM, Shu HB, Li S. Death-associated protein kinase 1 is an IRF3/7-interacting protein that is involved in the cellular antiviral immune response. Cell Mol Immunol. (2014) 11:245–52. doi: 10.1038/cmi.2013.65
120. Yan BR, Zhou L, Hu MM, Li M, Lin H, Yang Y, et al. PKACs attenuate innate antiviral response by phosphorylating VISA and priming it for MARCH5-mediated degradation. PLoS Pathog. (2017) 13:e1006648. doi: 10.1371/journal.ppat.1006648
121. Luo WW, Li S, Li C, Zheng ZQ, Cao P, Tong Z, et al. iRhom2 is essential for innate immunity to RNA virus by antagonizing ER- and mitochondria-associated degradation of VISA. PLoS Pathog. (2017) 13:e1006693. doi: 10.1371/journal.ppat.1006693
122. Kumar S, Barouch-Bentov R, Xiao F, Schor S, Pu S, Biquand E, et al. MARCH8 ubiquitinates the hepatitis C virus nonstructural 2 protein and mediates viral envelopment. Cell Rep. (2019) 26:1800–14 e1805. doi: 10.1016/j.celrep.2019.01.075
123. Bourgeois-Daigneault MC, Pezeshki AM, Galbas T, Houde M, Baril M, Früh K, et al. Tollip-induced down-regulation of MARCH1. Results Immunol. (2013) 3:17–25. doi: 10.1016/j.rinim.2013.02.002
124. Galbas T, Raymond M, Sabourin A, Bourgeois-Daigneault MC, Guimont-Desrochers F, Yun TJ, et al. MARCH1 E3 ubiquitin ligase dampens the innate inflammatory response by modulating monocyte functions in mice. J Immunol. (2017) 198:852–61. doi: 10.4049/jimmunol.1601168
125. Metcalfe SM, Watson TJ, Shurey S, Adams E, Green C. Leukemia inhibitory factor is linked to regulatory transplantation tolerance. Transplantation. (2005) 79:726–30. doi: 10.1097/01.TP.0000149324.42994.38
126. Heinrich PC, Behrmann I, Haan S, Hermanns HM, Müller-Newen G, Schaper F. Principles of interleukin (IL)-6-type cytokine signalling and its regulation. Biochem J. (2003) 374(Pt 1):1–20. doi: 10.1042/bj20030407
Keywords: MARCH proteins, E3 ligase, ubiquitination, immune regulation, immune receptors
Citation: Lin H, Li S and Shu H-B (2019) The Membrane-Associated MARCH E3 Ligase Family: Emerging Roles in Immune Regulation. Front. Immunol. 10:1751. doi: 10.3389/fimmu.2019.01751
Received: 06 May 2019; Accepted: 10 July 2019;
Published: 24 July 2019.
Edited by:
Liwu Li, Virginia Tech, United StatesReviewed by:
Kithiganahalli Narayanaswamy Balaji, Indian Institute of Science (IISc), IndiaJunji Xing, Houston Methodist Research Institute, United States
Copyright © 2019 Lin, Li and Shu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shu Li, c2h1bGkmI3gwMDA0MDt3aHUuZWR1LmNu; Hong-Bing Shu, c2h1aCYjeDAwMDQwO3dodS5lZHUuY24=
 Heng Lin
Heng Lin Shu Li*
Shu Li* Hong-Bing Shu
Hong-Bing Shu