
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Immunol. , 25 September 2018
Sec. T Cell Biology
Volume 9 - 2018 | https://doi.org/10.3389/fimmu.2018.02148
This article is part of the Research Topic Inhibitory Receptors and Pathways of Lymphocytes View all 15 articles
MiRNA targeting of key immunoregulatory molecules fine-tunes the immune response. This mechanism boosts or dampens immune functions to preserve homeostasis while supporting the full development of effector functions. MiRNA expression changes during T cell activation, highlighting that their function is constrained by a specific spatiotemporal frame related to the signals that induce T cell-based effector functions. Here, we update the state of the art regarding the miRNAs that are differentially expressed during T cell stimulation. We also revisit the existing data on miRNA function in T cell activation, with a special focus on the modulation of the most relevant immunoregulatory molecules.
MiRNAs are small (~19–24 nucleotides) single-stranded non-coding RNA species that act as post-transcriptional modulators; they control gene expression, either by promoting mRNAs degradation or repressing their translation (1). More than 2,500 human mature miRNA sequences have been already listed in MirBase (2) although the total amount of miRNAs is likely up to 10 times higher (3). Friedman et al. (4) estimated that miRNAs could modulate around 60% of protein-coding genes, indicating the relevance of these regulatory pathways in gene expression.
The miRNA repertoire changes upon T cell activation (5–11). Figure 1 summarizes miRNA species described to be either upregulated or downregulated upon T cell stimulation. Different studies have yielded data that may appear contradictory, likely due to T cell subset differences, the origin of the sample (murine or human) and the strategy of stimulation. Additional differences stem from the strategy used to evaluate miRNA expression, being arrays the most commonly employed technique, together with RT-qPCR and Northern Blot.
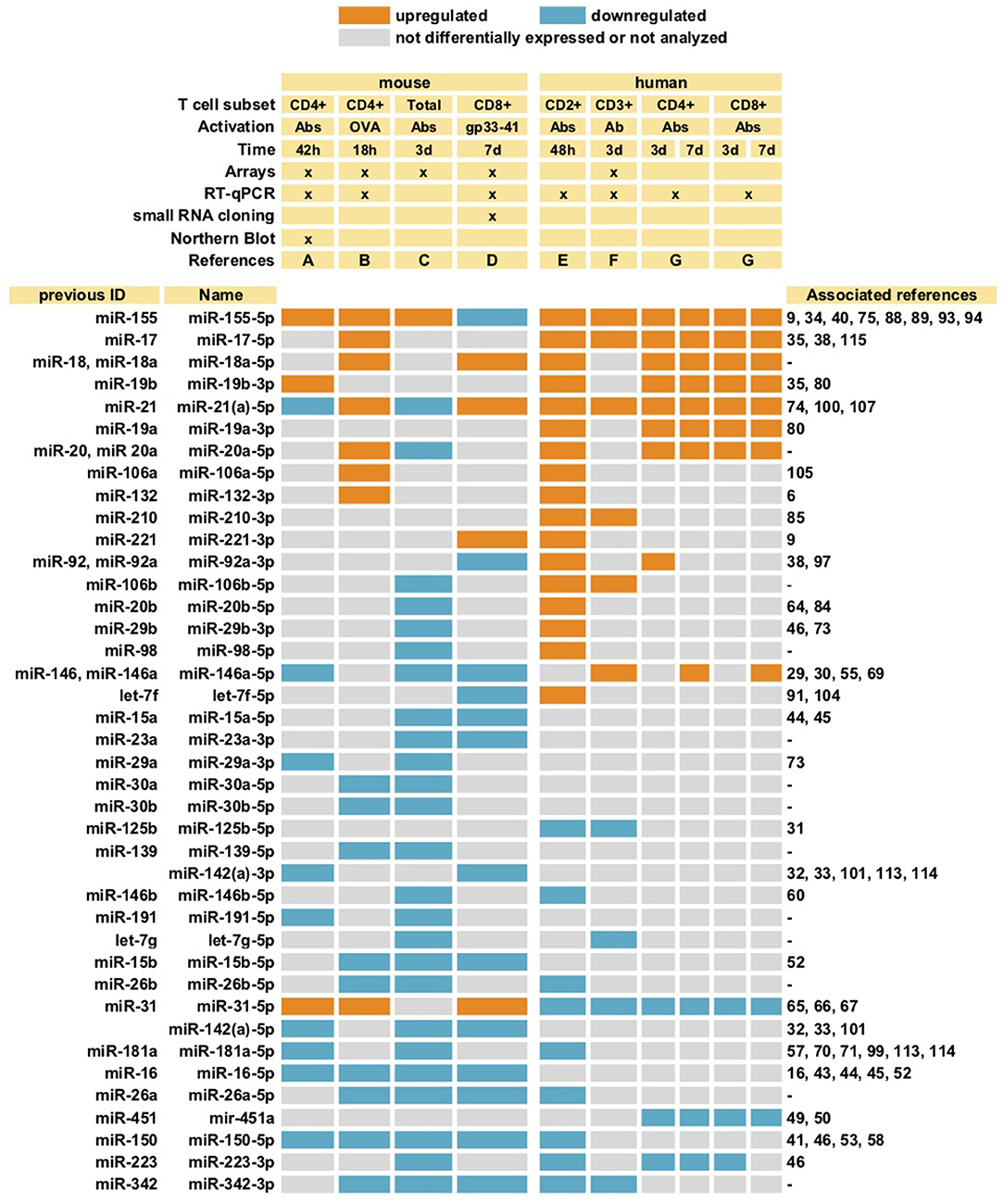
Figure 1. MiRNAs differentially expressed upon T cell stimulation. MiRNAs described in at least two different studies are summarized. Different subsets of T cells (both mouse and human) were activated with either antibodies against CD3 alone (Ab), or together with antibody against CD28 (Abs), or with specific peptides (OVA or gp33-41). Cells were stimulated during different lengths of time ranging from 18 h (18 h) to 7 days (7 d). The studies included in the table are: A (5), B (6), C (7), D (8), E (9), F (10), G (11). Whenever more than one detection method was used, only consistent data obtained with at least two techniques was selected (8). Most studies evaluated miRNA expression with miRNAs arrays, some together with RT-qPCR and Northern Blot, as indicated (x).
Despite variability, some trends are very consistent, including downregulation of miR-26a, miR-26b, miR-150, miR-181a, miR-223, and miR-342-3p; and upregulation of miR-155 and the miR-17~92 cluster (particularly miR-17-5p, miR-18a-5p, and miR-19b). MiR-146a was downregulated in mouse T cells, but upregulated in human upon activation, while miR-31 behaved in the opposite way, suggesting the existence of species-specific regulatory mechanisms.
In addition to variations in miRNA expression, it would be essential to consider the total abundance of each miRNA in the cell. Interestingly, only 7 miRNAs accounted for around 60% of the total sequencing reads in CD8+ T cells (8).
Beyond individual miRNA changes, it is important to highlight that miRNAs undergo a global downregulation upon stimulation. In this regard, almost three times higher total miRNA array hybridization signal has been detected in mouse CD8+ naive T cells compared to activated cells (8); similarly, an independent study found a significant downregulation of the total amount of miRNA in stimulated mouse and human CD4+ T cells compared to non-stimulated controls (5).
Dicer is an RNase III endonuclease that controls miRNA biogenesis. It processes precursor miRNA (pre-miRNA) into mature miRNA forms (12–14). Constitutive Dicer KO mice display embryonic lethality (15), indicating the relevance of this enzyme in development. Lineage-specific Dicer-deficient models were therefore required to study the consequences of reduced miRNA function in a tissue-specific manner.
Dicer-deficient CD4+ T cells were hyper-responsive to TCR stimulation and produced IL-2 in the absence of co-stimulation (16). After activation, CD4+ Dicer-deficient mice showed reduced proliferation, higher levels of apoptosis and a bias towards Th1 differentiation and IFN-γ release (17). In Th1 differentiation, IFN-γ production and a decline in IL-2 secretion occurred earlier in Dicer-deficient than in wild-type CD4+ T cells (17). Th2 cells presented reduced levels of GATA3 mRNA and failed to suppress IFN-γ expression (17). Consistently, similar phenotypes were observed in T cells lacking Drosha or its RNA-binding cofactor DGCR8, which form a complex responsible for primary miRNA transcript processing. Drosha-deficient naïve CD4+ T cells differentiated into Th1 and Th2, but expressed higher levels of IFN-γ than control cells (18). Similarly, DGCR8-deficient T lymphocytes showed reduced proliferation and an increase in IFN-γ secretion (19). A number of very comprehensive reports have addressed the role of miRNAs in T cell differentiation (20–24). In this review, immunoregulatory molecules responsible for differentiation have been discussed when closely related to T cell activation events.
CD4-specific Dicer deficiency also affects the regulatory T cell compartment, impairing Tregs development in the thymus and reducing their numbers in peripheral lymphoid organs (25). In addition, deficient naïve CD4+ T cells activated in the presence of TGF-β expressed significantly less FOXP3 than control cells (25). Besides, several studies have demonstrated that miRNA disruption in Treg cells leads to autoimmune diseases (18, 26, 27).
Dicer-deficient CD8+ T lymphocytes responded more rapidly to activation in vitro, as indicated by faster CD69 up-regulation and an earlier proliferative response, although their survival was reduced after 2 days (28). CD8+ Dicer KO cells also showed a delay in CD69 down-regulation after removal of the TCR-activating stimulus, suggesting a sustained activation of cytotoxic lymphocytes in the absence of miRNAs (28). Furthermore, CD8+ Dicer-deficient cells failed to produce an efficient in vivo effector response, including lower proliferation and impaired cytokine production (IFN-γ and TNF-α) (28).
Models with impaired miRNA synthesis machinery highlight the importance of miRNAs as positive (booster) and/or negative (brake) regulators of T cell development and function, which is a major focus of this review (Figure 2).
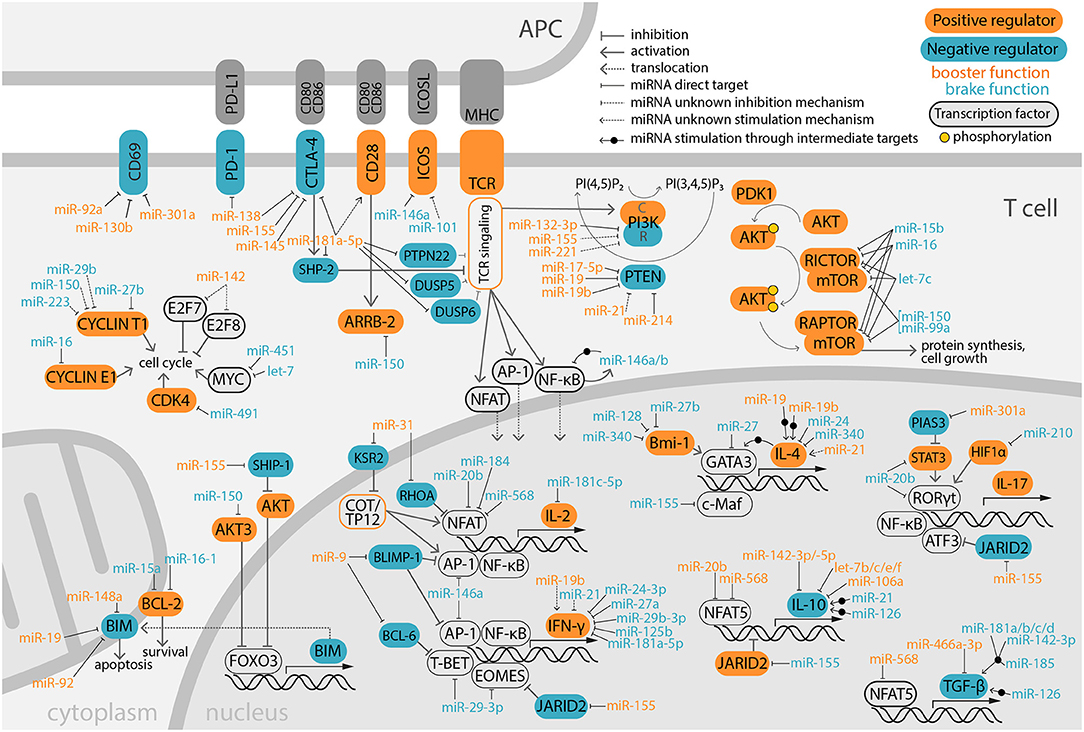
Figure 2. Overview of miRNA modulation on positive and negative immune-regulator molecules. Signaling coming from TCR and costimulatory molecules is integrated by the T lymphocyte promoting cell survival, proliferation and production of effector molecules, such as cytokines. This complex network is fine-tuned by miRNAs that target key immunoregulatory molecules, supporting either T cell activation (booster) or inhibition (brake). MiRNAs exert their function by targeting the mRNA 3′UTR in the cytoplasm, although for simplicity sake some have been depicted in the nucleus, close to their targeted immunoregulators. In PI3K, C and R designated the catalytic and regulatory subunits, respectively.
MiR-146a mainly acts as a “brake” miRNA, as miR-146a-deficient mice develop chronic inflammation and autoimmunity (29). CD4+ and CD8+ T cells from miR-146a deficient mice display less apoptosis and increased proliferation, expression of activation markers (CD25 and CD69) and effector cytokines (IL2, IFN-γ, and IL-17A) (30). Likewise, miR-125b is another negative regulator of T cell function, contributing to the maintenance of the naïve state in human CD4+ T cells, in which it appears at high levels (31). This effect is at least partly achieved via targeting key molecules for T cell activation, e.g., BLIMP-1, IL-2Rβ, IL-10Rα, and IFN-γ (31). Conversely, other miRNAs boost the immune response. For instance, miR-142-deficient mouse T cells showed reduced proliferation, deregulated cytokine expression and decreased secretion of pro-inflammatory cytokines such as IFN-γ, IL-17, and IL2 in response to activation (32, 33). Other examples of enhancer miRNAs are miR-155 and miR-17~92; miR-155-depleted mice are immunodeficient (34), whereas miR-17~92-deficient T cells exhibited reduced antitumoral responses (35).
T cell activation requires that the TCR recognizes a specific antigen bound to the MHC on the surface of an APC in the presence of co-stimulation. PI3K, AKT and mTOR are crucial mediators of T cell activation. Their positive signaling, downstream the TCR, is counter-balanced by negative regulators such as PTEN and BIM. Costimulatory signals are provided by surface receptors expressed on T lymphocytes that interact with specific ligands on APCs, and can be either activating (such as CD28 and ICOS) or inhibitory (like CTLA-4 and PD-1). These activating and inhibitory events are integrated into a net response that triggers the activation and/or repression of transcription factors (NFAT, AP-1, NF-κB, and others). Their nuclear localization promotes the synthesis of immune effector molecules, e.g., cytokines. MiRNAs also control the activation and integration of these pathways to support T cell effector functions while maintaining immune homeostasis. Herein, we review the miRNA-mediated regulation of key molecules involved in T cell activation.
The balance between BIM and BCL-2 molecules is essential for the fate of T lymphocytes, and their expression is tightly regulated by miRNAs, promoting either apoptosis or survival. BIM is a pro-apoptotic regulator and tumor suppressor downstream of AKT3, an important mediator of TCR signaling (36, 37). It destabilizes mitochondrial membrane, inducing CASPASE-9 activation and apoptosis. Within the miR-17~92 cluster, miR-19 and miR-92 target BIM 3′UTR mRNA (38). MiR-148a is upregulated in mouse Th1 cells after sustained activation (39). It also targets BIM, promoting cell survival (39). MiR-155 indirectly regulates BIM by targeting SHIP-1, which is a phosphatase that reduces AKT activity (40). In turn, AKT represses FOXO3, which is a transcription factor that promotes BIM expression, thus miR-155 limits BIM expression (40). Conversely, miR-150 promotes apoptosis by downregulating AKT3, which induces the accumulation of BIM (41). Human CD4+ T cells with high levels of miR-150 display reduced proliferation, increased apoptosis and lower T cell activation (41).
BCL-2 is an anti-apoptotic protein that antagonizes BIM, stabilizing the mitochondrial membrane and preventing its permeabilization (42). Treatment of mice with experimental autoimmune encephalomyelitis with 3,3′-Diindolylmethane (a plant-derived anti-inflammatory compound), induced the upregulation of miR-16 in brain CD4+ T cells and suppressed BCL-2; consistently, miR-16 overexpression in mouse CD4+ T cells downregulated BCL-2 (43). Interestingly, CD4+ T cells from relapsing-remitting multiple sclerosis patients (an autoimmune disease elicited by activated autoreactive T lymphocytes) displayed lower levels of miR-15a and miR-16, correlating with higher levels of their validated target BCL-2 mRNA (44, 45).
Molecules involved in cell cycle progression are essential mediators of T cell proliferation. miR-142-null T cells displayed gross cell cycle alterations, with cells differentially arrested in S and G2/M phases (32). Cell-cycle defects were associated to the transcription factors E2F7 and E2F8, which are putative targets for miR-142. MiR-142 is likely responsible of maintaining low levels of both molecules in resting T-cells and limiting their increase upon activation. Treatment of mice with miR-142 antagomir markedly increased survival and reduced clinical symptoms in a murine GVHD model, suggesting a potential new therapeutic strategy (32).
Cyclins are also directly targeted by miRNAs. Several miRNAs (miR-27b, miR-29b, miR-150, and miR-223) promote CYCLIN T1 downregulation in human resting CD4+ T cells. The levels of these miRNAs decrease upon activation, correlating with an upregulation of CYCLIN T1 (46). MiR-16 downregulates CYCLIN E1 in mouse CD4+ T cells (43). Another molecule involved in cell cycle progression is CDK4, a target of miR-491 in mouse CD8+ T cells (47). MYC is a transcription factor involved in cell cycle and proliferation, is targeted by let-7 in mouse CD8+ T cells (48) and by miR-451 in both mouse (49) and human (50) CD4+ T cells.
Mammalian Target Of Rapamycin (mTOR) is a metabolic regulator that promotes protein synthesis and cell growth during the onset of T lymphocyte function (51). mTOR kinase and Raptor are part of the complex mTORC1, while mTORC2 includes mTOR and Rictor. Both miR-16 and let-7c target the 3′UTR of mTOR and RICTOR (16). Elevated mTOR activity in Dicer-deficient CD4+ T cells and the subsequently increased AKT phosphorylation is associated with a lower activation threshold, overcoming the need of co-stimulation. MiRNA-mediated mTOR down-regulation contributes to the correct discrimination of activating and anergic stimuli and prevents co-stimulation independent IL-2, IFN-γ and TNF-α overproduction (16). mTOR signaling suppression is relevant for Treg induction. In this regard, miR-16 and miR-15b, which are abundantly expressed in Tregs, target RICTOR and mTOR mRNAs (52). Furthermore, miR-150 and miR-99a cooperatively target mTOR, promoting Treg induction (53).
Inducible co-stimulatory (ICOS) molecule and CD28 are surface receptors expressed on T cells that recognize specific ligands on APCs, acting as TCR signaling positive regulators (54). In germinal center responses, miR-146a upregulation in Tfh cells downregulates ICOS by interacting with its ligand on germinal center B cells, facilitating the termination of the immune response (55). MiR-101 is highly represented in human naïve CD4+ T cells and its transfection into the EL4 murine T cell line downregulates ICOS (56). Regarding CD28, miR-181a-5p overexpression in mouse T cells increases its levels (57), whereas miR-150 limits CD28 co-stimulation by targeting the arrestin β-2 protein (ARRB-2), with a subsequent increase in cAMP levels and inhibition of LCK, PI3K and AKT (58).
MiRNA regulation of cytokine expression can be due to direct cytokine mRNA targeting or targeting of transcription factors such as NF-κB, NFAT, or AP-1 or their regulators, often affecting multiple cytokines. For example, miR-146a is induced in mouse CD4+ and CD8+ T cells upon TCR engagement through NF-κB (30). This miRNA provides negative feedback regulation, downregulating NF-κB by targeting TRAF6 and IRAK1 (30, 59). Compared to wild-type cells, both CD4+ and CD8+ mouse T cells lacking miR-146a exhibited a higher induction of genes regulated by NF-κB, e.g., BCL-2, CD25, CD69, IL-2, IFN-γ, and IL-17A (30). TRAF6 is also targeted by miR-146b in mouse Tregs (60).
IL-2 is one of the main signatures of T cell activation. MiRNA-based IL-2 regulation relies on the inhibition of translation by miR-181c-5p (downregulated during T cell activation), which binds to the 3′UTR of IL-2 mRNA (61). It also depends on the miRNA-based downregulation of transcription factors such as NFAT or BLIMP-1. MiR-184 inhibits NFAT1 translation in human CD4+ T cells. This is particularly relevant in cells isolated from umbilical cord blood (62). MiR-568 transfection into human CD4+ T cells inhibited IL-2 expression after activation, through NFAT5 downregulation (63). MiR-20b also downregulated IL-2 through NFAT5 targeting (64). MiR-31 upregulates IL-2 by inhibiting RHOA, a small GTPase which suppresses NFAT (65, 66). It also targets the kinase suppressor of RAS2 (KSR2), which inhibits the COT/TPI2 signaling pathway (enhancer of IL-2 expression through NFAT and AP-1) (67). MiR-9 (upregulated in activated human CD4+ T cells) targets BLIMP-1, de-repressing IL-2 transcription (68). MiR-146a is upregulated around 8 days after stimulation in human CD4+ and CD8+ T cells, impairing IL-2 production, by targeting AP-1 (69).
IFN-γ release orchestrates Th1 immune responses by activating different cell lineages, e.g., dendritic cells, macrophages or NK cells. MiR-125b maintains T cell naïve state by targeting IFN-γ among other genes (31). Several miRNAs repress IFN-γ: miR-24-3p (70) and miR-181a-5p in human CD4+ T cells (70, 71); miR-24 and miR-27a in activated human CD8+ T cells (72); and miR-29 directly (73) and indirectly, by downregulating T-BET and EOMES, in mouse CD4+ T cells (19). On the other hand, miR-19b is required for normal IFN-γ production, restoring IFN-γ expression in miR-17~92-deficient mouse Th1 cells (35). MiR-9 suppresses BLIMP-1 and BCL-6 (repressors of AP-1 and T-BET, respectively), increasing IFN-γ secretion in activated human CD4+ T cells (68). Murine miR-21 KO CD4+ T cells re-stimulated in vitro produced more IFN-γ (74). Moreover, IFN-γ responsiveness is regulated by miR-155, which targets IFN-γRα in activated mouse CD4+ T cells, contributing to Th1 differentiation (75).
T cell activation stimulates the production of IL-4, leading to Th2 responses (76, 77). Its release is controlled directly by miR-24 [78] and miR-340 (78), or through the targeting of specific transcription factors and kinases/phosphatases. IL-4 triggers the upregulation of GATA3 dependent STAT6, repressing Th1 differentiation and inducing IL-4 production in a positive feedback loop. Conversely, MiR-27 targets the transcription factor GATA3 (79). BMI1 binds to GATA3, preventing its degradation. CD4+ T cells from MS patients display increased expression of miR-27b, miR-128 and miR-340 (78). These miRNAs inhibited Th2 development by targeting BMI1 (78). MiR-155 targets the 3′UTR of c-MAF mRNA, which is another transcription factor involved in IL-4 expression (34). MiR-21 contributes to IL-4 expression, since in vitro re-stimulated miR-21-null mouse CD4+ T cells produced less IL-4 than wild-type cells (74). Both miR-19a and miR-19b rescued IL-4 production in miR-17~92 cluster-deficient cells by targeting PTEN, SOCS1 and A20 (80).
TCR signaling promotes expression of the proinflammatory cytokine IL-17 (81–83). IL-17 expression depends on the transcription factor RORγt downstream of STAT3. miR-20b targets both molecules in mouse CD4+ T cells (84). RORγt transcription is promoted by HIF-1α, which is targeted by miR-210 (85). In turn, STAT3 is inhibited by the E3 SUMO-protein ligase PIAS3, a target of miR-301a that increases IL-17 secretion (86). MiR-212 targets BCL-6 3′UTR, which is a repressor of Th17 differentiation (87). JARID2, a chromatin-binding protein, recruits the polycomb repressive complex 2 (PRC2) and silences transcription of IL22, IL10, ATF3, TBX21, or EOMES through histone methylation (88). MiR-155 inhibits JARID2, releasing the repression of ATF3, which promotes IL-17 (88). ETS-1, a transcription factor that inhibits Th17 differentiation, is a target of miR-155 (89) and miR-326 (90). Li et al. (91) reported IL-17 downregulation due to IL-23R inhibition by let-7f.
CTLA-4 and PD-1 are both co-inhibitory receptors that repress TCR signaling via binding to co-stimulators expressed by APCs (54). CTLA-4 (a target of miR-145) is very abundant in human peripheral blood Tregs, in which miR-145 is downregulated (92). MiR-155 also targeted CTLA-4 in mouse (93) and human (94) CD4+ T cells. MiR-155 overexpression in human CD4+ T cells promoted proliferation, and could underlie chronic inflammation in atopic dermatitis, in which it is highly expressed also by CD4+ T cells present in skin lesions (94). MiR-138 targets CTLA-4 and PD-1, promoting tumor-regression by inhibiting tumor-infiltrating Tregs (95). MiR-181a-5p overexpression in mouse T cells decreased CTLA-4 expression, while increasing CD28 levels (57).
CD69 is an early surface marker of lymphocyte activation (96). Dicer KO CD8+ T cells up-regulated CD69 more rapidly upon stimulation and retained the expression longer after stimuli removal (28), indicating a potential miRNA-based repression of CD69 in naïve stages that restrains activation. MiR-130b and miR-301a increased their levels during CD8+ T cell activation and downregulated CD69 (28). MiR-92, which is downregulated in lamina propria leukocytes from rhesus macaques with chronic simian immunodeficiency virus infection, also targets the 3′UTR of CD69 mRNA (97).
TCR signaling is mediated by downstream kinases and phosphatases, which undergo a tight regulation that ensures functional activation while avoiding hyperreactivity.
Upon TCR and co-receptors engagement, PI3K phosphorylates PI(4,5)P2. PIK3R1 gene encodes the regulatory subunits p85, p50, and p55 (98). MiRNAs upregulated in CD4+ activated human T cells, e.g., miR-155 and miR-221 downregulate PIK3R1 (9). MiR-132-3p is upregulated in mouse dendritic cell-activated CD4+ T lymphocytes, targeting PIK3R1 mRNA (6).
Phosphatases downstream the TCR pathway counteract signaling by dephosphorylation. Downregulation of some of these phosphatases by miR-181a-5p generates high levels of phosphorylated intermediates in steady-state (57). MiR-181a-5p targets the phosphatases PTPN22, DUSP5 and DUSP6, which dephosphorylate LCK, ZAP70, and ERK1/2; and SHP-2, which mediates negative costimulatory signals from CTLA-4 (57). Therefore, the expression of this miRNA contributes to reduce the activation threshold, increasing the strength and sensitivity of the T cell to peptides with lower affinity (57). In elderly individuals, reduced expression of miR-181a in CD4+ naïve T cells is a cause of the declined T cell responsiveness associated with age (99).
PTEN dephosphorylates PI(3,4,5)P3, antagonizing PI3K. As such, PTEN curbs T cell activation, preserving self-tolerance. Transgenic mice overexpressing miR-17~92 cluster developed lymphoproliferative and autoimmune pathologies associated to the reduced expression of PTEN and BIM (38). PTEN is downregulated by several miRNAs that are increased upon T cell activation: miR-21 (100), miR-214 (7) and the miR-17~92 cluster [miR-17-5p (38), miR-19 (38), and miR-19b (35)]. Consistently, miR-21 and miR-214 expression increased T cell proliferation (7, 100).
IL-10 is an important anti-inflammatory cytokine mainly produced by Th2 and Tregs. It counteracts CD28 signaling and suppresses the expression of IFN-γ and IL-2. IL-10 is directly targeted by miR-142-3p, miR-142-5p (101), miR-let-7e (102), let-7c (103, 104), let-7b (104), let-7f (104), and miR-106a (105). MiRNAs further regulate IL-10 post-transcriptionally by modulating JARID2, NFAT5, p85-β or the programmed cell death protein 4 (PDCD4). JARID2 silences IL-10 and is a target of miR-155, which thus promotes IL-10 expression (88). MiR-568 (downregulated upon human CD4+ T cell activation) reduced IL-10 by targeting NFAT5 (63). NFAT5 was also targeted by miR-20b (64). MiR-126 is highly increased after Treg stimulation and promotes IL-10 expression (106), and miR-126 targeting of p85-β and PI3K/AKT pathway modulation is responsible of IL-10 release (106). MiR-21 is upregulated in CD4+ T cells from systemic lupus erythematosus patients, and its inhibition led to a decrease in IL-10 production (107). MiR-21 positive regulation of IL-10 secretion likely depends on its targeting of PDCD4, a translation inhibitor (107).
TGF-β is expressed in naïve T cells preventing T cell activation until sufficient TCR stimulation downregulates the TGF-β type 1 receptor (108–110). TGF-β induces FOXP3, a key transcription factor that promotes Treg differentiation (111). In addition to IL-10 modulation, miR-568 (63) and miR-126 (106) also regulate TGF-β release. In CD4+ mouse T cells from draining lymph nodes, miR-466a-3p (upregulated in mice after skin allograft) targets TGF-β2, limiting Treg generation (112). MiRNAs also regulate TGF-β function at different levels by targeting upstream molecules involved in cytokine production, TGF-β receptors and effector molecules of the TGF-β signaling pathway. GARP is a transmembrane protein specifically expressed in Tregs that cleaves the precursor form of TGF-β1 (113). GARP is targeted by miRNAs which are less abundant in human Tregs than in T helper subsets, e.g., miR-142-3p, miR-185, and miR-181a/b/c/d (113, 114). MiR-17 targets TGFBR2 (TGF-β receptor II) in mouse and human CD4+ T cells (35, 115). In addition, it has been found that a set of miRNAs upregulated in naïve CD4+ T cells from multiple sclerosis patients target TGFBR1 and/or SMAD4 (both involved in the TGF-β signaling pathway) limiting differentiation into Tregs (116).
MiRNA-mediated modulation of molecules involved in T cell activation remains far from being fully understood, although strides have been made in recent years. There is a need to advance towards a “network study” of miRNA function. Considering more than one miRNA in experimental designs increases its technical complication, but also enables models that simulate the complexity of the physiological scenarios, in which individual miRNAs interact with a set of targets and each target in turn can be regulated by several miRNAs, at different levels, either directly targeting the molecule or indirectly regulating its expression via targeting its receptor and/or transcription factors.
Finally, integrating basic and clinical research (e.g., cancer, autoimmunity, and GVHD) could help to achieve a better understanding of T cell immune-regulation to design new strategies for therapy in T cell related malignancies.
AR-G wrote the draft manuscript and designed the Figures. LF-M corrected and edited the manuscript. FS-M edited the manuscript. AR-G, LF-M, and FS-M discussed all the items in the manuscript.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We thank Dr M. Vicente-Manzanares for critical reading of the manuscript and for assistance with English editing. This study was supported by the following grants from the Spanish Ministry of Economy and Competitiveness, (grant SAF2017-82886-R to FSM), CIBER CARDIOVASCULAR and PIE 13.0004-BIOIMID from the Instituto de Salud Carlos III (Fondo de Investigación Sanitaria del Instituto de Salud Carlos III with co-funding from the Fondo Europeo de Desarrollo Regional; FEDER), Programa de Actividades en Biomedicina de la Comunidad de Madrid-B2017/BMD-3671-INFLAMUNE to FS-M, and ERC-2011-AdG294340-GENTRIS to FS-M. The Centro Nacional de Investigaciones Cardiovasculares (CNIC) is supported by the Spanish Ministry of Economy and Competitiveness (MINECO) and the Pro-CNIC Foundation and is a Severo Ochoa Center of Excellence (MINECO award SEV-2015-0505). AR-G is supported by the FPU program (Spanish Ministry of Education). LF-M is funded by the CIBER CARDIOVASCULAR.
AKT3, v-akt murine thymoma viral oncogene homolog 3; APC, antigen-presenting cell; BIM, B-cell lymphoma 2 (Bcl-2) interacting mediator of cell death; CTLA-4, Cytotoxic T lymphocyte-associated antigen 4; GVHD, Graft versus host disease; IL, Interleukin; PD-1, Programmed Death 1; PI(3, 4, 5)P3, phosphatidylinositol-(3,4,5)-triphosphate; PI(4, 5)P2, phosphatidylinositol-(4,5)-biphosphate; PTEN, phosphatase and tensin homolog; TCR, T-cell receptor; Tfh, T follicular helper; TGF-β, Transforming Growth Factor- β; Treg, regulatory T cell; tTreg, Thymic-derived regulatory T cells; UTR, untranslated region.
1. Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell (2004) 116:281–97. doi: 10.1016/S0092-8674(04)00045-5
2. Griffiths-Jones S, Saini HK, van Dongen S, Enright AJ. miRBase: tools for microRNA genomics. Nucleic Acids Res. (2008) 36:D154–8. doi: 10.1093/nar/gkm952
3. Londin E, Loher P, Telonis AG, Quann K, Clark P, Jing Y, et al. Analysis of 13 cell types reveals evidence for the expression of numerous novel primate- and tissue-specific microRNAs. Proc Natl Acad Sci USA. (2015) 112:E1106–15. doi: 10.1073/pnas.1420955112
4. Friedman RC, Farh KKH, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. (2009) 19:92–105. doi: 10.1101/gr.082701.108
5. Bronevetsky Y, Villarino AV, Eisley CJ, Barbeau R, Barczak AJ, Heinz GA, et al. T cell activation induces proteasomal degradation of Argonaute and rapid remodeling of the microRNA repertoire. J Exp Med. (2013) 210:417–32. doi: 10.1084/jem.20111717
6. Gutiérrez-Vázquez C, Rodríguez-Galán A, Fernández-Alfara M, Mittelbrunn M, Sánchez-Cabo F, Martínez-Herrera DJ, et al. miRNA profiling during antigen-dependent T cell activation: a role for miR-132-3p. Sci Rep. (2017) 7:3508. doi: 10.1038/s41598-017-03689-7
7. Jindra PT, Bagley J, Godwin JG, Iacomini J. Costimulation-dependent expression of MicroRNA-214 increases the ability of T cells to proliferate by targeting Pten. J Immunol. (2010) 185:990–7. doi: 10.4049/jimmunol.1000793
8. Wu H, Neilson JR, Kumar P, Manocha M, Shankar P, Sharp PA, et al. miRNA Profiling of Naïve, Effector and Memory CD8 T Cells. PLoS ONE (2007) 2:e1020. doi: 10.1371/journal.pone.0001020
9. Grigoryev YA, Kurian SM, Hart T, Nakorchevsky AA, Chen C, Campbell D, et al. MicroRNA regulation of molecular networks mapped by global MicroRNA, mRNA, and Protein Expression in Activated T Lymphocytes. J Immunol. (2011) 187:2233–43. doi: 10.4049/jimmunol.1101233
10. Sousa IG, do Almo MM, Simi KCR, Bezerra MAG, Andrade RV, Maranhão AQ, et al. MicroRNA expression profiles in human CD3+ T cells following stimulation with anti-human CD3 antibodies. BMC Res Notes (2017) 10:124. doi: 10.1186/s13104-017-2442-y
11. Teteloshvili N, Smigielska-Czepiel K, Kroesen B-J, Brouwer E, Kluiver J, Boots A, et al. T-cell Activation Induces Dynamic Changes in miRNA Expression Patterns in CD4 and CD8 T-cell Subsets. MicroRNA (2015) 4:117–22. doi: 10.2174/2211536604666150819194636
12. Bernstein E, Caudy AA, Hammond SM, Hannon GJ. Role for bidentate ribnuclease in the initiation site of RNA interference. Nature (2001) 409:363–6. doi: 10.1038/35053110
13. Hutvagner G. A cellular function for the RNA-interference enzyme dicer in the maturation of the let-7 small temporal RNA. Science (80-) (2001) 293:834–8. doi: 10.1126/science.1062961
14. Grishok A, Pasquinelli AE, Conte D, Li N, Parrish S, Ha I, et al. Genes and mechanisms related to RNA interference regulate expression of the small temporal RNAs that control C. elegans developmental timing. Cell. (2001) 106:23–34. doi: 10.1016/S0092-8674(01)00431-7
15. Bernstein E, Kim SY, Carmell MA, Murchison EP, Alcorn H, Li MZ, et al. Dicer is essential for mouse development. Nat Genet. (2003) 35:215–7. doi: 10.1038/ng1253
16. Marcais A, Blevins R, Graumann J, Feytout A, Dharmalingam G, Carroll T, et al. microRNA-mediated regulation of mTOR complex components facilitates discrimination between activation and anergy in CD4 T cells. J Exp Med. (2014) 211:2281–95. doi: 10.1084/jem.20132059
17. Muljo SA, Ansel KM, Kanellopoulou C, Livingston DM, Rao A, Rajewsky K. Aberrant T cell differentiation in the absence of Dicer. J Exp Med. (2005) 202:261–9. doi: 10.1084/jem.20050678
18. Chong MMW, Rasmussen JP, Rudensky AY, Littman DR. The RNAseIII enzyme drosha is critical in T cells for preventing lethal inflammatory disease. J Exp Med. (2008) 205:2005–17. doi: 10.1084/jem.20071219090508c
19. Steiner DF, Thomas MF, Hu JK, Yang Z, Babiarz JE, Allen CDC, et al. MicroRNA-29 Regulates T-Box transcription factors and interferon-γ production in helper T cells. Immunity (2011) 35:169–81. doi: 10.1016/j.immuni.2011.07.009
20. Kroesen B-J, Teteloshvili N, Smigielska-Czepiel K, Brouwer E, Boots AMH, van den Berg A, et al. Immuno-miRs: critical regulators of T-cell development, function and ageing. Immunology (2015) 144:1–10. doi: 10.1111/imm.12367
21. Jeker LT, Bluestone JA. MicroRNA regulation of T-cell differentiation and function. Immunol Rev. (2013) 253:65–81. doi: 10.1111/imr.12061
22. Liu J, Wu C-P, Lu B-F, Jiang J-T. Mechanism of T cell regulation by microRNAs. Cancer Biol Med. (2013) 10:131–7. doi: 10.7497/j.issn.2095-3941.2013.03.002
23. Baumjohann D, Ansel KM. MicroRNA-mediated regulation of T helper cell differentiation and plasticity. Nat Rev Immunol. (2013) 13:666–78. doi: 10.1038/nri3494
24. Podshivalova K, Salomon DR. MicroRNA regulation of T-lymphocyte immunity: modulation of molecular networks responsible for T-cell activation, differentiation, and development. Crit Rev Immunol. (2013) 33:435–76. doi: 10.1615/CritRevImmunol.2013006858
25. Cobb BS, Hertweck A, Smith J, O'Connor E, Graf D, Cook T, et al. A role for Dicer in immune regulation. J Exp Med. (2006) 203:2519–27. doi: 10.1084/jem.20061692
26. Liston A, Lu L-F, O'Carroll D, Tarakhovsky A, Rudensky AY. Dicer-dependent microRNA pathway safeguards regulatory T cell function. J Exp Med. (2008) 205:1993–2004. doi: 10.1084/jem.20081062
27. Zhou X, Jeker LT, Fife BT, Zhu S, Anderson MS, McManus MT, et al. Selective miRNA disruption in T reg cells leads to uncontrolled autoimmunity. J Exp Med. (2008) 205:1983–91. doi: 10.1084/jem.20080707
28. Zhang N, Bevan MJ. Dicer controls CD8+ T-cell activation, migration, and survival. Proc Natl Acad Sci USA. (2010) 107:21629–34. doi: 10.1073/pnas.1016299107
29. Boldin MP, Taganov KD, Rao DS, Yang L, Zhao JL, Kalwani M, et al. miR-146a is a significant brake on autoimmunity, myeloproliferation, and cancer in mice. J Exp Med. (2011) 208:1189–201. doi: 10.1084/jem.20101823
30. Yang L, Boldin MP, Yu Y, Liu CS, Ea C-K, Ramakrishnan P, et al. miR-146a controls the resolution of T cell responses in mice. J Exp Med. (2012) 209:1655–70. doi: 10.1084/jem.20112218
31. Rossi RL, Rossetti G, Wenandy L, Curti S, Ripamonti A, Bonnal RJP, et al. Distinct microRNA signatures in human lymphocyte subsets and enforcement of the naive state in CD4+ T cells by the microRNA miR-125b. Nat Immunol. (2011) 12:796–803. doi: 10.1038/ni.2057
32. Sun Y, Oravecz-Wilson K, Mathewson N, Wang Y, McEachin R, Liu C, et al. Mature T cell responses are controlled by microRNA-142. J Clin Invest. (2015) 125:2825–40. doi: 10.1172/JCI78753
33. Mildner A, Chapnik E, Varol D, Aychek T, Lampl N, Rivkin N, et al. MicroRNA-142 controls thymocyte proliferation. Eur J Immunol. (2017) 47:1142–52. doi: 10.1002/eji.201746987
34. Rodriguez A, Vigorito E, Clare S, Warren MV, Couttet P, Soond DR, et al. Requirement of bic/microRNA-155 for normal immune function. Science (80-). (2007) 316:608–11. doi: 10.1126/science.1139253
35. Jiang S, Li C, Olive V, Lykken E, Feng F, Sevilla J, et al. Molecular dissection of the miR-17-92 cluster's critical dual roles in promoting Th1 responses and preventing inducible Treg differentiation. Blood (2011) 118:5487–97. doi: 10.1182/blood-2011-05-355644
36. Hildeman DA, Zhu Y, Mitchell TC, Bouillet P, Strasser A, Kappler J, et al. Activated T cell death in vivo mediated by proapoptotic Bcl-2 family member Bim. Immunity (2002) 16:759–67. doi: 10.1016/S1074-7613(02)00322-9
37. Reynolds C, Roderick JE, LaBelle JL, Bird G, Mathieu R, Bodaar K, et al. Repression of BIM mediates survival signaling by MYC and AKT in high-risk T-cell acute lymphoblastic leukemia. Leukemia (2014) 28:1819–27. doi: 10.1038/leu.2014.78
38. Xiao C, Srinivasan L, Calado DP, Patterson HC, Zhang B, Wang J, et al. Lymphoproliferative disease and autoimmunity in mice with increased miR-17-92 expression in lymphocytes. Nat Immunol. (2008) 9:405–14. doi: 10.1038/ni1575
39. Haftmann C, Stittrich AB, Zimmermann J, Fang Z, Hradilkova K, Bardua M, et al. MiR-148a is upregulated by Twist1 and T-bet and promotes Th1-cell survival by regulating the proapoptotic gene Bim. Eur J Immunol. (2015) 45:1192–205. doi: 10.1002/eji.201444633
40. Rouquette-Jazdanian AK, Kortum RL, Li W, Merrill RK, Nguyen PH, Samelson LE, et al. miR-155 controls lymphoproliferation in LAT mutant mice by restraining T-cell apoptosis via SHIP-1/mTOR and PAK1/FOXO3/BIM pathways. PLoS ONE (2015) 10:1–27. doi: 10.1371/journal.pone.0131823
41. Sang W, Sun C, Zhang C, Zhang D, Wang Y, Xu L, et al. MicroRNA-150 negatively regulates the function of CD4+T cells through AKT3/Bim signaling pathway. Cell Immunol. (2016) 306–307:35–40. doi: 10.1016/j.cellimm.2016.05.007
42. Guerrero AD, Welschhans RL, Chen M, Wang J. Cleavage of Anti-Apoptotic Bcl-2 Family Members after TCR Stimulation Contributes to the Decision between T Cell Activation and Apoptosis. J Immunol. (2013) 190:168–73. doi: 10.4049/jimmunol.1201610
43. Rouse M, Rao R, Nagarkatti M, Nagarkatti PS. 3,3'-Diindolylmethane ameliorates experimental autoimmune encephalomyelitis by promoting cell cycle arrest and apoptosis in activated T cells through MicroRNA signaling pathways. J Pharmacol Exp Ther. (2014) 350:341–52. doi: 10.1124/jpet.114.214742
44. Lorenzi JCC, Brum DG, Zanette DL, de Paula Alves Souza A, Barbuzano FG, dos Santos AC, et al. miR-15a and 16-1 Are Downregulated in CD4+ T Cells of Multiple Sclerosis Relapsing Patients. Int J Neurosci. (2012) 122:466–71. doi: 10.3109/00207454.2012.678444
45. Cimmino A, Calin GA, Fabbri M, Iorio MV, Ferracin M, Shimizu M, et al. miR-15 and miR-16 induce apoptosis by targeting BCL2. Proc Natl Acad Sci USA. (2005) 102:13944–9. doi: 10.1073/pnas.0506654102
46. Chiang K, Sung T-L, Rice AP. Regulation of Cyclin T1 and HIV-1 Replication by MicroRNAs in Resting CD4+ T Lymphocytes. J Virol. (2012) 86:3244–52. doi: 10.1128/JVI.05065-11
47. Yu T, Zuo Q-F, Gong L, Wang L-N, Zou Q-M, Xiao B. MicroRNA-491 regulates the proliferation and apoptosis of CD8+ T cells. Sci Rep. (2016) 6:30923. doi: 10.1038/srep30923
48. Wells AC, Daniels KA, Angelou CC, Fagerberg E, Burnside AS, Markstein M, et al. Modulation of let-7 miRNAs controls the differentiation of effector CD8 T cells. Elife. (2017) 6:e26398. doi: 10.7554/eLife.26398
49. Chapman LM, Ture SK, Field DJ, Morrell CN. miR-451 limits CD4+ T cell proliferative responses to infection in mice. Immunol Res. (2017) 65:828–40. doi: 10.1007/s12026-017-8919-x
50. Zeng Z, Wang K, Li Y, Xia N, Nie S, Lv B, et al. Down-regulation of microRNA-451a facilitates the activation and proliferation of CD4+ T cells by targeting Myc in patients with dilated cardiomyopathy. J Biol Chem. (2017) 292:6004–13. doi: 10.1074/jbc.M116.765107
51. Liu Y, Zhang D, Liu X. mTOR Signaling in T Cell Immunity and Autoimmunity. Int Rev Immunol. (2015) 34:50–66. doi: 10.3109/08830185.2014.933957
52. Singh Y, Garden OA, Lang F, Cobb BS. MicroRNA-15b/16 enhances the induction of regulatory T cells by regulating the expression of rictor and mTOR. J Immunol. (2015) 195:5667–77. doi: 10.4049/jimmunol.1401875
53. Warth SC, Hoefig KP, Hiekel A, Schallenberg S, Jovanovic K, Klein L, et al. Induced miR-99a expression represses Mtor cooperatively with miR-150 to promote regulatory T-cell differentiation. Embo J. (2015) 34:1195–213. doi: 10.15252/embj.201489589
54. Chen L, Flies DB. Molecular mechanisms of T cell co-stimulation and co-inhibition. Nat Rev Immunol. (2013) 13:227–42. doi: 10.1038/nri3405
55. Pratama A, Srivastava M, Williams NJ, Papa I, Lee SK, Dinh XT, et al. MicroRNA-146a regulates ICOS-ICOSL signalling to limit accumulation of T follicular helper cells and germinal centres. Nat Commun. (2015) 6:1–14. doi: 10.1038/ncomms7436
56. Di Yu, Tan AHM, Hu X, Athanasopoulos V, Simpson N, Silva DG, et al. Roquin represses autoimmunity by limiting inducible T-cell co-stimulator messenger RNA. Nature (2007) 450:299–303. doi: 10.1038/nature06253
57. Li Q-J, Chau J, Ebert PJR, Sylvester G, Min H, Liu G, et al. miR-181a Is an Intrinsic Modulator of T Cell Sensitivity and Selection. Cell (2007) 129:147–61. doi: 10.1016/j.cell.2007.03.008
58. Sang W, Wang Y, Zhang C, Zhang D, Sun C, Niu M, et al. MiR-150 impairs inflammatory cytokine production by targeting ARRB-2 after blocking CD28/B7 costimulatory pathway. Immunol Lett. (2016) 172:1–10. doi: 10.1016/j.imlet.2015.11.001
59. Taganov KD, Boldin MP, Chang K-J, Baltimore D. NF- B-dependent induction of microRNA miR-146, an inhibitor targeted to signaling proteins of innate immune responses. Proc Natl Acad Sci. USA (2006) 103:12481–6. doi: 10.1073/pnas.0605298103
60. Lu Y, Hippen KL, Lemire AL, Gu J, Wang W, Ni X, et al. miR-146b antagomir-treated human Tregs acquire increased GVHD inhibitory potency. Blood. (2016) 128:1424–35. doi: 10.1182/blood-2016-05-714535
61. Xue Q, Guo ZY, Li W, Wen WH, Meng YL, Jia LT, et al. Human activated CD4+ T lymphocytes increase IL-2 expression by downregulating microRNA-181c. Mol Immunol. (2011) 48:592–9. doi: 10.1016/j.molimm.2010.10.021
62. Weitzel RP, Lesniewski ML, Haviernik P, Kadereit S, Leahy P, Greco NJ, et al. microRNA 184 regulates expression of NFAT1 in umbilical cord blood CD4+ T cells. Blood. (2009) 113:6648–57. doi: 10.1182/blood-2008-09-181156
63. Li W, Kong L, Li J-T, Guo Z-Y, Xue Q, Yang T, et al. MiR-568 inhibits the activation and function of CD4+ T cells and Treg cells by targeting NFAT5. Int Immunol. (2014) 26:269–81. doi: 10.1093/intimm/dxt065
64. Xin Y, Cai H, Lu T, Zhang Y, Yang Y, Cui Y. miR-20b Inhibits T cell proliferation and activation via NFAT Signaling Pathway in Thymoma-Associated Myasthenia Gravis. Biomed Res Int. (2016) 2016:9595718. doi: 10.1155/2016/9595718
65. Helms WS, Jeffrey JL, Holmes DA, Townsend MB, Clipstone NA, Su L. Modulation of NFAT-dependent gene expression by the RhoA signaling pathway in T cells. J Leukoc Biol. (2007) 82:361–9. doi: 10.1189/jlb.0206120
66. Fan W, Liang D, Tang Y, Qu B, Cui H, Luo X, et al. Identification of microRNA-31 as a novel regulator contributing to impaired interleukin-2 production in T cells from patients with systemic lupus erythematosus. Arthritis Rheum. (2012) 64:3715–25. doi: 10.1002/art.34596
67. Xue F, Li H, Zhang J, Lu J, Xia Y, Xia Q. MiR-31 regulates interleukin 2 and kinase suppressor of ras 2 during T cell activation. Genes Immun. (2013) 14:127–31. doi: 10.1038/gene.2012.58
68. Thiele S, Wittmann J, Jäck H-M, Pahl A. miR-9 enhances IL-2 production in activated human CD4+ T cells by repressing Blimp-1. Eur J Immunol. (2012) 42:2100–8. doi: 10.1002/eji.201142203
69. Curtale G, Citarella F, Carissimi C, Goldoni M, Carucci N, Fulci V, et al. An emerging player in the adaptive immune response: microRNA-146a is a modulator of IL-2 expression and activation-induced cell death in T lymphocytes. Blood (2010) 115:265–73. doi: 10.1182/blood-2009-06-225987
70. Fayyad-Kazan H, Hamade E, Rouas R, Najar M, Fayyad-Kazan M, El Zein N, et al. Downregulation of microRNA-24 and−181 parallels the upregulation of IFN-γ secreted by activated human CD4 lymphocytes. Hum Immunol. (2014) 75:677–85. doi: 10.1016/j.humimm.2014.01.007
71. Sang W, Zhang C, Zhang D, Wang Y, Sun C, Niu M, et al. MicroRNA-181a, a potential diagnosis marker, alleviates acute graft versus host disease by regulating IFN-γ production. Am J Hematol. (2015) 90:998–1007. doi: 10.1002/ajh.24136
72. Chandran PA, Keller A, Weinmann L, Adel Seida A, Braun M, Andreev K, et al. The TGF-β-inducible miR-23a cluster attenuates IFN- levels and antigen-specific cytotoxicity in human CD8+ T cells. J Leukoc Biol. (2014) 96:633–45. doi: 10.1189/jlb.3A0114-025R
73. Ma F, Xu S, Liu X, Zhang Q, Xu X, Liu M, et al. The microRNA miR-29 controls innate and adaptive immune responses to intracellular bacterial infection by targeting interferon-γ. Nat Immunol. (2011) 12:861–9. doi: 10.1038/ni.2073
74. Lu TX, Hartner J, Lim E-J, Fabry V, Mingler MK, Cole ET, et al. MicroRNA-21 limits in vivo immune response-mediated activation of the IL-12/IFN- Pathway, Th1 Polarization, and the severity of delayed-type hypersensitivity. J Immunol. (2011) 187:3362–73. doi: 10.4049/jimmunol.1101235
75. Banerjee A, Schambach F, DeJong CS, Hammond SM, Reiner SL. Micro-RNA-155 inhibits IFN-γ signaling in CD4+ T cells. Eur J Immunol. (2009) 40:225–31. doi: 10.1002/eji.200939381
76. McAdam AJ, Chang TT, Lumelsky AE, Greenfield EA, Boussiotis VA, Duke-Cohan JS, et al. Mouse Inducible Costimulatory Molecule (ICOS) expression is enhanced by CD28 costimulation and regulates differentiation of CD4+ T cells. J Immunol. (2000) 165:5035–40. doi: 10.4049/jimmunol.165.9.5035
77. Milner JD. TCR signaling abnormalities in human Th2-associated atopic disease. Front Immunol. (2018) 9:2–6. doi: 10.3389/fimmu.2018.00719
78. Guerau-De-Arellano M, Smith KM, Godlewski J, Liu Y, Winger R, Lawler SE, et al. Micro-RNA dysregulation in multiple sclerosis favours pro-inflammatory T-cell-mediated autoimmunity. Brain (2011) 134:3575–86. doi: 10.1093/brain/awr262
79. Cho S, Wu C-J, Yasuda T, Cruz LO, Khan AA, Lin L-L, et al. miR-23~27~24 clusters control effector T cell differentiation and function. J Exp Med. (2016) 213:235–49. doi: 10.1084/jem.20150990
80. Simpson LJ, Patel S, Bhakta NR, Choy DF, Brightbill HD, Ren X, et al. A microRNA upregulated in asthma airway T cells promotes TH2 cytokine production. Nat Immunol. (2014) 15:1162–70. doi: 10.1038/ni.3026
81. Chen Z, Tato CM, Muul L, Laurence A, O'Shea JJ. Distinct regulation of interleukin-17 in human T helper lymphocytes. Arthritis Rheum. (2007) 56:2936–46. doi: 10.1002/art.22866
82. Gomez-Rodriguez J, Sahu N, Handon R, Davidson TS, Anderson SM, Kirby MR, et al. Differential Expression of Interleukin-17A and−17F is coupled to T Cell Receptor Signaling via Inducible T Cell Kinase. Immunity (2009) 31:587–97. doi: 10.1016/j.immuni.2009.07.009
83. Purvis HA, Stoop JN, Mann J, Woods S, Kozijn AE, Hambleton S, et al. Low-strength T-cell activation promotes Th17 responses. Blood (2016) 116:4829–38. doi: 10.1182/blood-2010-03-272153
84. Zhu E, Wang X, Zheng B, Wang Q, Hao J, Chen S, et al. miR-20b Suppresses Th17 differentiation and the pathogenesis of experimental autoimmune encephalomyelitis by targeting ROR t and STAT3. J Immunol. (2014) 192:5599–609. doi: 10.4049/jimmunol.1303488
85. Wang H, Flach H, Onizawa M, Wei L, Mcmanus MT, Weiss A. Negative regulation of Hif1a expression and TH 17 differentiation by the hypoxia-regulated microRNA miR-210. Nat Immunol. (2014) 15:393–401. doi: 10.1038/ni.2846
86. Mycko MP, Cichalewska M, Machlanska A, Cwiklinska H, Mariasiewicz M, Selmaj KW. MicroRNA-301a regulation of a T-helper 17 immune response controls autoimmune demyelination. Proc Natl Acad Sci USA. (2012) 109:E1248–57. doi: 10.1073/pnas.1114325109
87. Nakahama T, Hanieh H, Nguyen NT, Chinen I, Ripley B, Millrine D, et al. Aryl hydrocarbon receptor-mediated induction of the microRNA-132/212 cluster promotes interleukin-17-producing T-helper cell differentiation. Proc Natl Acad Sci USA. (2013) 110:11964–9. doi: 10.1073/pnas.1311087110
88. Escobar TM, Kanellopoulou C, Kugler DG, Kilaru G, Nguyen CK, Nagarajan V, et al. miR-155 Activates Cytokine Gene Expression in Th17 Cells by Regulating the DNA-Binding Protein Jarid2 to Relieve Polycomb-Mediated Repression. Immunity (2014) 40:865–79. doi: 10.1016/j.immuni.2014.03.014
89. Hu R, Huffaker TB, Kagele DA, Runtsch MC, Bake E, Chaudhuri AA, et al. MicroRNA-155 confers encephalogenic potential to Th17 cells by promoting effector gene expression. J Immunol. (2013) 190:5972–80. doi: 10.4049/jimmunol.1300351
90. Du C, Liu C, Kang J, Zhao G, Ye Z, Huang S, et al. MicroRNA miR-326 regulates TH-17 differentiation and is associated with the pathogenesis of multiple sclerosis. Nat Immunol. (2009) 10:1252–9. doi: 10.1038/ni.1798
91. Li Z, Wu F, Brant SR, Kwon JH. IL-23 receptor regulation by Let-7f in human CD4+ memory T cells. J Immunol. (2011) 186:6182–90. doi: 10.4049/jimmunol.1000917
92. Fayyad-Kazan H, Rouas R, Fayyad-Kazan M, Badran R, El Zein N, Lewalle P, et al. MicroRNA profile of circulating CD4-positive regulatory T cells in human adults and impact of differentially expressed microRNAs on expression of two genes essential to their function. J Biol Chem. (2012) 287:9910–22. doi: 10.1074/jbc.M111.337154
93. Zhang Y, Sun E, Li X, Zhang M, Tang Z, He L, et al. miR-155 contributes to Df1-induced asthma by increasing the proliferative response of Th cells via CTLA-4 downregulation. Cell Immunol. (2017) 314:1–9. doi: 10.1016/j.cellimm.2017.01.005
94. Sonkoly E, Janson P, Majuri M-L, Savinko T, Fyhrquist N, Eidsmo L, et al. MiR-155 is overexpressed in patients with atopic dermatitis and modulates T-cell proliferative responses by targeting cytotoxic T lymphocyte–associated antigen 4. J Allergy Clin Immunol. (2010) 126:581–589.e20. doi: 10.1016/j.jaci.2010.05.045
95. Wei J, Nduom EK, Kong LY, Hashimoto Y, Xu S, Gabrusiewicz K, et al. MiR-138 exerts anti-glioma efficacy by targeting immune checkpoints. Neuro Oncol. (2016) 18:639–48. doi: 10.1093/neuonc/nov292
96. Cibrián D, Sánchez-Madrid F. CD69: from activation marker to metabolic gatekeeper. Eur J Immunol. (2017) 47:946–53. doi: 10.1002/eji.201646837
97. Kumar V, Torben W, Kenway CS, Schiro FR, Mohan M. Longitudinal examination of the intestinal lamina propria cellular compartment of simian immunodeficiency virus-infected rhesus macaques provides broader and deeper insights into the link between aberrant MicroRNA expression and persistent immune activati. J Virol. (2016) 90:5003–19. doi: 10.1128/JVI.00189-16
98. Jean S, Kiger AA. Classes of phosphoinositide 3-kinases at a glance. J Cell Sci. (2014) 127:923–8. doi: 10.1242/jcs.093773
99. Li G, Yu M, Lee WW, Tsang M, Krishnan E, Weyand CM, et al. Decline in miR-181a expression with age impairs T cell receptor sensitivity by increasing DUSP6 activity. Nat Med. (2012) 18:1518–24. doi: 10.1038/nm.2963
100. He W, Wang C, Mu R, Liang P, Huang Z, Zhang J, et al. MiR-21 is required for anti-tumor immune response in mice: an implication for its bi-directional roles. Oncogene (2017) 36:4212–23. doi: 10.1038/onc.2017.62
101. Ding S, Liang Y, Zhao M, Liang G, Long H, Zhao S, et al. Decreased microRNA-142-3p/5p expression causes CD4+ T cell activation and B cell hyperstimulation in systemic lupus erythematosus. Arthritis Rheum. (2012) 64:2953–63. doi: 10.1002/art.34505
102. Guan H, Fan D, Mrelashvili D, Hao H, Singh NP, Singh UP, et al. MicroRNA let-7e is associated with the pathogenesis of experimental autoimmune encephalomyelitis. Eur J Immunol. (2013) 43:104–14. doi: 10.1002/eji.201242702
103. Jiang L, Cheng Z, Qiu S, Que Z, Bao W, Jiang C, et al. Altered let-7 expression in Myasthenia gravis and let-7c mediated regulation of IL-10 by directly targeting IL-10 in Jurkat cells. Int Immunopharmacol. (2012) 14:217–23. doi: 10.1016/j.intimp.2012.07.003
104. Swaminathan S, Suzuki K, Seddiki N, Kaplan W, Cowley MJ, Hood CL, et al. Differential regulation of the Let-7 family of MicroRNAs in CD4+ T cells alters IL-10 expression. J Immunol. (2012) 188:6238–46. doi: 10.4049/jimmunol.1101196
105. Sharma A, Kumar M, Aich J, Hariharan M, Brahmachari SK, Agrawal A, et al. Posttranscriptional regulation of interleukin-10 expression by hsa-miR-106a. Proc Natl Acad Sci USA. (2009) 106:5761–6. doi: 10.1073/pnas.0808743106
106. Qin A, Wen Z, Zhou Y, Li Y, Li Y, Luo J, et al. MicroRNA-126 regulates the induction and function of CD4+ Foxp3+ regulatory T cells through PI3K/AKT pathway. J Cell Mol Med. (2013) 17:252–64. doi: 10.1111/jcmm.12003
107. Stagakis E, Bertsias G, Verginis P, Nakou M, Hatziapostolou M, Kritikos H, et al. Identification of novel microRNA signatures linked to human lupus disease activity and pathogenesis: MiR-21 regulates aberrant T cell responses through regulation of PDCD4 expression. Ann Rheum Dis. (2011) 70:1496–506. doi: 10.1136/ard.2010.139857
108. Chen CH, Seguin-Devaux C, Burke NA, Oriss TB, Watkins SC, Clipstone N, et al. Transforming growth factor beta blocks Tec kinase phosphorylation, Ca2+ influx, and NFATc translocation causing inhibition of T cell differentiation. J Exp Med. (2003) 197:1689–99. doi: 10.1084/jem.20021170
109. Li MO, Sanjabi S, Flavell RA. Transforming growth factor-β controls development, homeostasis, and tolerance of T cells by regulatory T cell-dependent and -independent mechanisms. Immunity (2006) 25:455–71. doi: 10.1016/j.immuni.2006.07.011
110. Tu E, Chia CPZ, Chen W, Zhang D, Park SA, Jin W, et al. T cell receptor-regulated TGF-β Type I receptor expression determines T cell quiescence and activation. Immunity (2018) 48:745–759.e6. doi: 10.1016/j.immuni.2018.03.025
111. Li MO, Flavell RA. TGF-β: a master of all T cell trades. Cell (2008) 134:392–404. doi: 10.1016/j.cell.2008.07.025
112. Becker W, Nagarkatti M, Nagarkatti PS. miR-466a targeting of TGF-β2 contributes to FoxP3+regulatory T cell differentiation in a murine model of allogeneic transplantation. Front Immunol. (2018) 9:688. doi: 10.3389/fimmu.2018.00688
113. Gauthy E, Cuende J, Stockis J, Huygens C, Lethé B, Collet JF, et al. GARP is regulated by miRNAs and controls latent TGF-β1 production by human regulatory T cells. PLoS ONE (2013) 8:e76186. doi: 10.1371/journal.pone.0076186
114. Zhou Q, Haupt S, Prots I, Thummler K, Kremmer E, Lipsky PE, et al. miR-142-3p Is involved in CD25+ CD4 T cell proliferation by targeting the expression of glycoprotein A repetitions predominant. J Immunol. (2013) 190:6579–88. doi: 10.4049/jimmunol.1202993
115. Meira M, Sievers C, Hoffmann F, Rasenack M, Kuhle J, Derfuss T, et al. Unraveling natalizumab effects on deregulated miR-17 expression in CD4+ T cells of patients with relapsing-remitting multiple sclerosis. J Immunol Res. (2014) 2014:897249. doi: 10.1155/2014/897249
Keywords: T cell activation, microRNAs (miRNAs), immunoregulatory molecules, miRNA signature, CD4, CD8, T lymphocyte
Citation: Rodríguez-Galán A, Fernández-Messina L and Sánchez-Madrid F (2018) Control of Immunoregulatory Molecules by miRNAs in T Cell Activation. Front. Immunol. 9:2148. doi: 10.3389/fimmu.2018.02148
Received: 18 July 2018; Accepted: 30 August 2018;
Published: 25 September 2018.
Edited by:
Alexandre M. Carmo, i3S, Instituto de Investigação e Inovação em Saúde, PortugalReviewed by:
Isabel Castro, i3S, Instituto de Investigação e Inovação em Saúde, PortugalCopyright © 2018 Rodríguez-Galán, Fernández-Messina and Sánchez-Madrid. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Francisco Sánchez-Madrid, ZnNtYWRyaWRAc2FsdWQubWFkcmlkLm9yZw==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.