
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Genet., 20 December 2019
Sec. Computational Genomics
Volume 10 - 2019 | https://doi.org/10.3389/fgene.2019.01276
 Songtao Jiu1
Songtao Jiu1 Yan Xu1
Yan Xu1 Jiyuan Wang1
Jiyuan Wang1 Lei Wang1
Lei Wang1 Shiping Wang1
Shiping Wang1 Chao Ma1
Chao Ma1 Le Guan2
Le Guan2 Muhammad Abdullah1
Muhammad Abdullah1 Maoxiang Zhao1
Maoxiang Zhao1 Wenping Xu1
Wenping Xu1 Wenli Ma3
Wenli Ma3 Caixi Zhang1*
Caixi Zhang1*The TEOSINTE BRANCHED 1/CYCLOIDEA/PROLIFERATING CELL FACTORS (TCP) protein, belonging to a plant-specific transcription factors (TFs) family, participates in the control of plant growth and development by regulating cell proliferation. Until now, a comprehensive study of concerning the TCP gene family and their roles in grapevine (Vitis vinifera L.) has not been completed. Using bioinformatics approaches, 17 VvTCP genes were identified and further classified into two classes, designated class I (PCF subclass) and class II (CIN and CYC/TB1 subclass), which was further supported by exon-intron organizations and conserved motif analysis. Promoter analysis demonstrated that VvTCPs have numerous cis-acting elements related to plant growth and development, phytohormone, and abiotic/biotic stress responses. The singleton duplication of grapevine TCP genes contributed to this gene family expansion. The syntenic analyses among Vitis vinifera, Arabidopsis, and Oryza sativa showed that these genes located in corresponding syntenic blocks arose before the divergence of V. vinifera, Arabidopsis, and O. sativa. The expression levels of 17 VvTCPs were determined in different tissues and fruit developmental stages, and abscisic acid (ABA) treatment. Seventeen VvTCPs exhibited distinct tissue-specific expression patterns, potentially illustrating the functional divergence of VvTCPs in all tested tissues. Eleven VvTCPs were down-regulated in five berry developmental stages, while three VvTCPs were up-regulated. Additionally, many members were strongly modulated by ABA treatment, suggesting these VvTCPs have important and diverse regulatory roles in ABA treatment. Our results provide valuable information on the evolution and functions of the VvTCPs, pave the way for further functional verification of these VvTCPs in grapevine.
The TEOSINTE BRANCHED 1/CYCLOIDEA/PROLIFERATING CELL FACTORS (TCP) gene is a plant-specific and widespread transcription factor (TF) family, which plays versatile functions in multiple biological processes, such as lateral branch (Takeda et al., 2003; Vadde et al., 2018), embryo growth (Tatematsu et al., 2008; Balsemao-Pires et al., 2013), leaf morphogenesis (Vadde et al., 2018; Mao et al., 2014), flower development (Xu et al., 2013; Juntheikki-Palovaara et al., 2014), circadian rhythm (Giraud et al., 2010), mitochondrial biogenesis (Hammani et al., 2011), seed sprout (Tatematsu et al., 2008; Rueda-Romero et al., 2012), and hormonal transduction pathways (Guo et al., 2010; Viola et al., 2011). According to the previous reports, TCP gene family is characterized as holding a basic helix-loop-helix (bHLH) motif of 59 amino acid (AA), involving in protein-protein interaction, DNA binding, and protein nuclear localization (Martin-Trillo and Cubas, 2010). TCP gene family can be divided into two subfamilies, class I (PCF subclass) and class II (CYC/TB1 and CIN subclass), on the basis of differences in AA sequences, particularly in the basic region of the TCP domains (Navaud et al., 2007; Martin-Trillo and Cubas, 2010). Compared with class II, the most significant difference is a four-amino-acid deletion of the TCP domain in class I. According to the bioinformatic analysis, some TCP members of class II also have an arginine-rich R domain outside the conservative TCP domain with unclear function, which is deemed to contribute to protein-protein interaction (Lupas et al., 1991; Cubas et al., 1999). The screening assays of DNA-binding site demonstrated that the two TCP subclasses can specifically recognize and bind to GC-rich DNA sequences with slightly different. To be specific, GGNCCCAC is the DNA binding sequence for class I, whereas class II prefers to bind to the motif G(T/C)GGNCCC (Kosugi and Ohashi, 1997; Schommer et al., 2008; Viola et al., 2011).
In general, the two class (class I and class II) genes are proven to play an antagonistical role in the specific biological process. TCP genes belonging to class I, such as AtTCP20 and OsPCF1/OsPCF2, play a vital role in cell propagation and growth (Li et al., 2005; Martin-Trillo and Cubas, 2010; Danisman et al., 2012). In practice, single mutants of most PCF-type TCP genes do not display any noticeable phenotypic difference compared with wild type (WT) plants, which might be functional redundancies. For instance, growing evidence indicates that AtTCP9 and AtTCP20 function redundantly in the regulation of leaf senescence by jasmonate signaling pathway (Danisman et al., 2012). Moreover, AtTCP14 and AtTCP15 showed overlapping functions in repressing endoreduplication through regulating the cell-cycle gene expression to further affect the growth and development of organ and cell (Peng et al., 2015). However, double mutants of tcp14–tcp15 showed altered sepal and blade shape, shortened internode length, as well as reduced the capability of seed germination in comparison with the WT plants (Kieffer et al., 2011; Resentini et al., 2015). Additionally, the expression levels of CYCA1:1, SHOOT-MERISTEMLESS, and BP in quintuple mutants of tcp8-tcp15-tcp21-tcp22-tcp23 were up-regulated, which resulted in larger leaves (Aguilar Martinez and Sinha, 2013).
Class II can be further divided into two subclades: CYC/TB1 and CIN (Martin-Trillo and Cubas, 2010). Contrast with class I, phenotypic analysis about mutants show that the TCP genes belonging to class II usually inhibit cell growth and proliferation. In the cin-type mutants of Arabidopsis and tomato, leaf blade cells are able to continuously divide for longer period in comparison with the WT plants, and thus the mutants showed bigger leaves with altered shape and/or crinkled surface (Palatnik et al., 2003; Crawford et al., 2004; Ori et al., 2007; Efroni et al., 2008; Masuda et al., 2008). Additionally, many TCP genes of TB1-type function as axillary bud-specific regulators, such as AtBRC1 and AtBRC2 (Aguilar-Martinez et al., 2007), OsFC1/OsTB1 (Takeda et al., 2003; Minakuchi et al., 2010), ZmTB1 (Doebley et al., 1997), and PsBRC1 (Braun et al., 2012). The loss-of-function of the above genes resulted in superabundant shoot branching, which are revealing of a negative function of these TCP genes on bud activity (Takeda et al., 2003; Minakuchi et al., 2010; Braun et al., 2012). Additionally, miR319a-mediated TCPs interacted with ASYMMETRIC LEAVES2 and ensured normal development of leaves via suppressing the expressions of KNAT2 and BP (Li et al., 2012).
Recently, TCP gene family has been reported in various plants, for instance, 24 TCP genes in Arabidopsis, 30 in Lycopersicon esculentum, 28 in Oryza sativa, 33 in Populus euphratica, 36 in Populus trichocarpa, 27 in Citrullus lanatus, 19 in Prunus mume (Martin-Trillo and Cubas, 2010; Parapunova et al., 2014; Ma et al., 2016; Shi et al., 2016; Zhou et al., 2016). The grape is an ideal model for understanding the berry development in perennial fruit species. The availability of grapevine genome sequencing (Jaillon et al., 2007; Velasco et al., 2007) make it possible and feasible to examine and identify gene families in Vitis vinifera. Although TCP family genes have important regulatory roles in plant growth and development, but still lack of detail analysis in grapevine. Hence, we performed a comprehensive bioinformatics analysis and expression analysis of the TCP gene family in grapevine. The present study involved in the identification of putative VvTCPs via genome-wide searches, the investigation of their phylogenetic relationships, chromosomal distribution, exon-intron organizations, conserved motif, promoter, as well as syntenic analysis. In addition, the expression patterns of VvTCPs have been investigated by RNA sequencing data and quantitative reverse-transcription PCR (qRT-PCR) assay. The findings of the current investigations will assist in better comprehending of the classification and potential functions of VvTCPs and pave the way for further functional verification of these VvTCP genes in grapevine.
The grape (the hybrid of V. vinifera and Vitis labrusca cv. “Jumeigui”) plants, planted at farm of Nanjing Agricultural University (Nanjing, China) under standard field conditions, were used as experimental materials. Three biological replicates each consisting of three clusters were sampled at each sampling date. The developmental stages were defined in term of the criterion containing the color, size, sugar content, and softening degree of the berries (Boss et al., 1996). The berry samples were harvested at five development stages, including small green berry (SGB), big green berry (BGB), veraison berry (VB), post-VB (PVB), and ripening berry (RB). Additionally, initial flowering (IF), full flowering (FF), young stems (YS), roots (Ro), buds (Bu), young leaves (YL), medium leaves (ML), old leaves (OL), and tendrils (Te) were also collected. Five-year-old uniformly grapevine plants were chosen for the ABA treatments. ABA treatments were performed by spraying the grape berry before veraison with a solution containing 50 and 150 ppm abscisic acid (ABA), while the control berries were only treated with distilled water. The berry was collected at veraison, post-veraison, and ripening. For making one sample, nine berries were collected from three different grapevines with respect to each sampling point and treatment. All of the samples were collected in triplicate, and then immediately frozen under liquid nitrogen, and kept in refrigerator at −80°C until further analysis.
The extraction of total RNA was carried out from grapevine samples by following cetyltrimethylammonium bromide method (Jiu et al., 2016; Jiu et al., 2018). The concentration of total RNA from each sample was evaluated using NanoDrop (Thermo Fisher Scientific Inc., USA) and OD260/280 ratios were close to 2.0 and OD260/230 ratios were >2.0 for all the samples. RNA integrity was also assessed using standard denaturing agarose gel electrophoresis. The samples of total RNA were digested with DNase I (Takara, Japan) against genomic DNA contamination. Then, manufacturer's protocol was followed for construction of cDNA libraries with 1.0 g of RNA samples (PrimeScriptTM First Strand cDNA synthesis kit, Takara, Japan).
The sequence of grapevine genome was download from National Center for Biotechnology Information website. A local BLASTn searches against the grapevine genome database using 24 known AtTCPs acquired by The Arabidopsis Information Resource website (http://www.arabidopsis.org), setting the threshold of the E-value as 1e−10 to confirm the detection of all VvTCP genes in the grape genome database (http://genomes.cribi.unipd.it/grape/index.php). Additionally, we also used the seed file of TCP domain (PF03634) from Pfam online software (http://pfam.xfam.org/) to acquire the Hidden Markov Model (HMM) sequences, then we carried out HMM searches using HMMER3 software against the grapevine protein sequences. Subsequently, each candidate VvTCP was used to further confirm by the Pfam database. To eliminate repetitive genes, all potential VvTCPs were aligned using the DNAMAN6.0 software and manually collated. All the no-overlapping VvTCPs were used for further analysis. The sequences of TCP family members in C. lanatus, O. sativa, Brassica rapa for constructing phylogenetic tree were obtained from PlantTFDB TF database (http://planttfdb.cbi.pku.edu.cn/, v3.0). The number of TCP family genes were obtained in Arabidopsis thaliana (Wei et al., 2016), O. sativa (Yao et al., 2007), L. esculentum (Parapunova et al., 2014), P. mume (Zhou et al., 2016), C. lanatus (Shi et al., 2016), turnip (Du et al., 2017), Chinese cabbage (Liu et al., 2018), and Gossypium raimondii (Ma et al., 2014).
Multiple sequence alignments were performed on the AA sequences of TCP proteins in V. vinifera, A. thaliana, C. lanatus, O. sativa, and B. rapa genomes using the CLUSTALW program with default parameters. Subsequently, MEGA7.0 program was used to construct a phylogenetic tree based on the alignments using the neighbor-joining (NJ) method and bootstrap tests replicated 1,000 time, which was performed using the p-distance model (Ogrodzki and Forsythe 2015). Maximum likelihood (ML) method was also used for the construction of phylogenetic tree to confirm the result from the NJ method. Additionally, the prediction of subcellular localization of all identified VvTCP genes was completed by WoLF PSORT Prediction PSORT II (http://wolfpsort.org/) and TargetP (http://www.cbs.dtu.dk/services/TargetP) software (Emanuelsson et al., 2000).
The AA sequences of 17 VvTCPs were analyzed by the MEME program (http://meme-suite.org/) using the following parameters (optimum width, 15–60; number of repetitions, any; and maximum number of motifs, 15) to identify the conserved motifs (Bailey and Elkan, 1994). The conserved motifs of VvTCP proteins were confirmed by InterPro software (http://www.ebi.ac.uk/interpro/). The gene structure display server 2.0 (GSDS, http://gsds.cbi.pku.edu.cn) (Hu et al., 2014) was employed to display exon-intron organization for the 17 VvTCPs by the comparison of the cDNAs with their genomic DNA sequences.
The nucleotide sequences of VvTCP family genes were obtained from the grapevine genome database (http://genomes.cribi.unipd.it/grape/index.php) in this study. The upstream 2,000-bp region from the start codon for all VvTCPs was regarded as the promoter sequence (Jiu et al., 2015; Jiu et al., 2018). The putative cis-acting elements of promoters were identified by PlantCARE online software (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/, Lescot et al., 2002). The putative cis-acting elements involved in plant growth and development, plant hormone responses, as well as biotic and abiotic stress responses are summarized.
A total of 17 VvTCPs were mapped to grapevine chromosomes by analyzing their chromosomal localization refer to the information available at the grapevine genome database (http://genomes.cribi.unipd.it/grape/index.php). The duplication events in the grapevine genome were acquired using MCscanX software. For syntenic analysis, synteny blocks within the grapevine genome and between grapevine and A. thaliana genomes, as well as between grapevine and O. sativa genomes were obtained from the Plant Genome Duplication Database (PGDD) website (http://chibba.agtec.uga.edu/duplication/index/downloads) and visualized by Circos software (http://circos.ca/) (Krzywinski et al., 2009).
The expression values normalized in numerous tissues, were obtained from the RNA sequencing data (Fasoli et al., 2012). The primers used for amplifying 17 VvTCP genes, designed by using Primer-BLAST online software (https://www.ncbi.nlm.nih.gov/tools/primer-blast/index.cgi?LINK_LOC=BlastHome), are listed in Supplemental Table S1. Polymerase chain reaction (PCR) were used for screening all primer pairs. The expression levels were detected by qRT-PCR assay using a Bio-Rad System (Bio-Rad, CA, USA). Each PCR mixture (20 µl) consist of 2×TB Green™ Premix Ex Taq™ II (10 µl), dilute cDNA (1 µl), each primer (0.4 µl), and RNase-free water (8.2 µl). The condition of qRT-PCR was as follows: pre-incubate (95°C for 30 s) and then 40 cycles (95°C for 5 s, 60°C for 30 s). To detect the relative fold differences for each gene in each experiment, the Ct value of the genes was normalized to the Ct value for the reference genes, and the relative expression levels were calculated using the formula 2−ΔΔCT. KyActin1 was used as the internal reference controls. The lowest expression values of the samples were manually set to 1.
The full-length cDNA of the VvTCP9 and VvTCP15 were amplified using the primers (Supplemental Table S1) and cloned into the binary vector PHB containing two cauliflower mosaic virus (CaMV) 35S promoter, a translation enhancer and a GFP fluorescent protein tag, respectively, to generate two fusion constructs (p35S-VvTCP9-GFP and p35S-VvTCP15-GFP). After identified the sequence, the two fusion constructs and the control vector (PHB) were transformed into Agrobacterium tumefaciens GV3101 strains and subsequently agroinfiltrated into the leaves of 3 to 5-week-old Nicotiana benthamiana plants. Localization of fluorescent proteins was observed 3–7 days after infiltration, the period when GFP fluorescence was optimal, by using a confocal laser scanning microscope (Zeiss LSM 780, Germany) according to the manufacturer's instructions.
The experiment was arranged in a completely randomized design (CRD) with three replications in this study and collected data were statistically analyzed with multivariate analysis methods using SAS computer software (SAS Version 9.2, Institute). Analysis of variance (ANOVA) was used to determine the overall statistical significance of the data at level of P < 0.05 and data were represented as average ±STDEV (n = 3).
The availability of the grapevine genome sequences provides the sources in the genome-wide identification of grapevine genes (Jaillon et al., 2007). To identify TCP family genes in grapevine, AtTCP proteins sequences from Arabidopsis were used as the query for a local BLAST search against the grapevine Genome Database. A sum of 17 putative VvTCP proteins, which had the conserved TCP domain, were identified (Table 1). Here, we designated the VvTCP1 to VvTCP17 genes in grapevine according to the Arabidopsis TCP proteins with the highest sequence similarity and following the classification of gene terminology used in the Arabidopsis. The AA sequence lengths of the 17 identified VvTCP proteins ranged from 204 to 549 followed for an average of 370.65 AA. The molecular weights (Mw) of these potential VvTCP TFs ranged from 51.58 kDa (VvTCP5) to 136.89 kDa (VvTCP16), and the isoelectric points (pI) ranged from 4.89 to 5.10. Other characteristics of the VvTCPs, including the chromosome location, CIN or CYC/TB1 type, and the subcellular localization are showed in Table 1. The VvTCPs can be divided into the two TCP classes according to the differences within their TCP domains: 9 VvTCPs (VvTCPs 3, 5, 6, 7, 9, 14, 15, 16, and 17) belong to class I characterized as the existence of a four-AA-deletion in the basic domain compared with the members of class II; the 8 class II VvTCPs can be further clustered into the two subclasses (CYC/TB1 and CIN) (Figure 1).
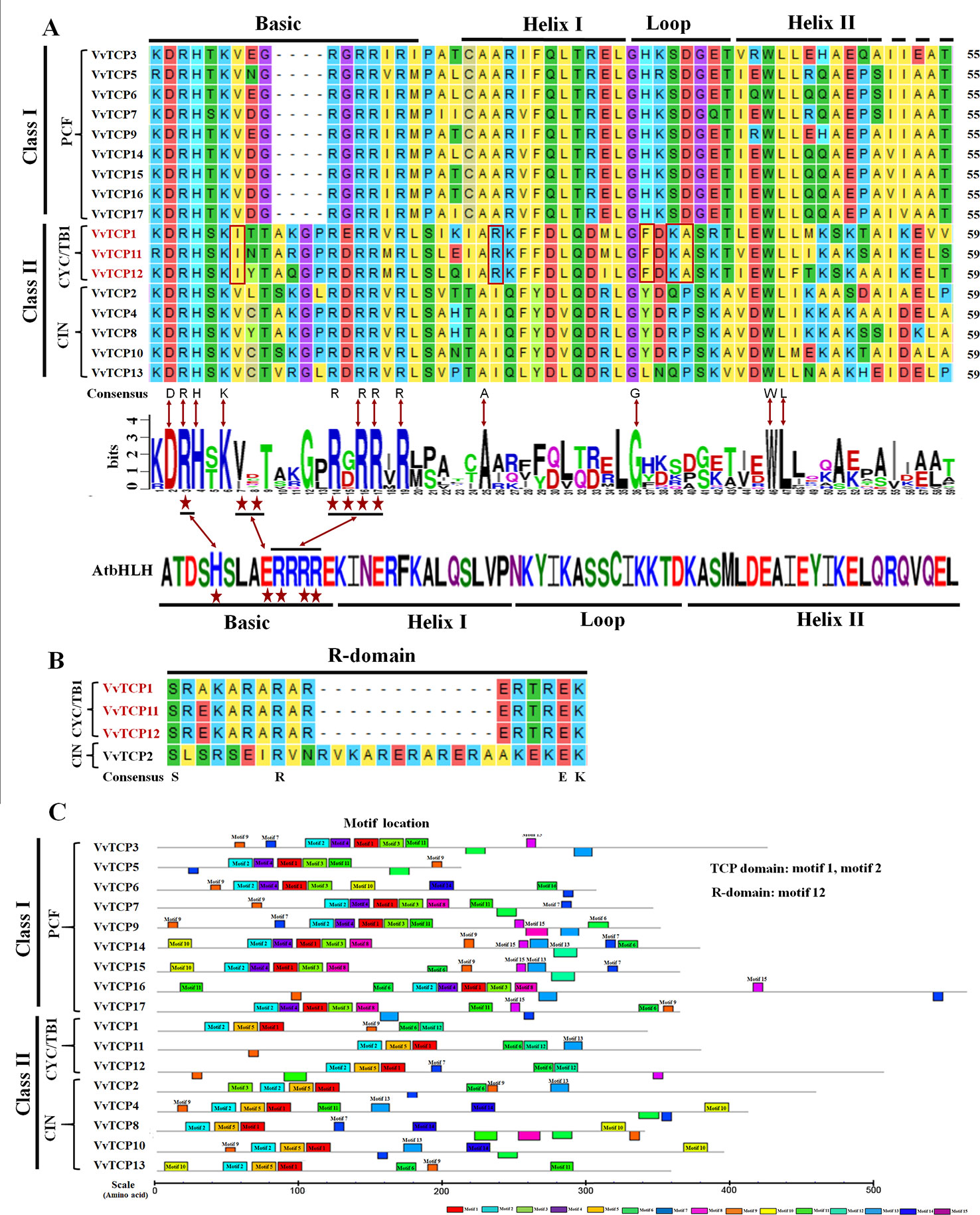
Figure 1 Conservative structural domains analysis of VvTCPs family in grapevine. (A) Multiple sequence alignment and protein sequence signs of the TCP domain. Multiple sequence alignment was performed with DNAMAN6.0 software. The sequence signs were constructed using WebLogo software. The stacks height indicated the sequence preservation at that position. (B) Multiple sequence alignment of the R domain. (C) Motif distribution of TCP genes in Vitis vinifera. Motifs were predicted using MEME online software. A colored block with motif number was consistent with each motif (color figure online). The lengths and positions of motifs in the AA sequences of 17 VvTCPs were exhibited by the lengths and positions of the blocks.
The TCP domain is vital for the catalysis activity of TCP proteins. In this study, the InterPro and MEME programs were employed to confirm the conserved motifs of VvTCP proteins in V. vinifera (Bailey and Elkan, 1994). A total of 15 potential motifs were identified and designated motifs 1–15 in grapevine (Supplemental Figure S1). Motifs 1, 2, 9, and 11 were identified on the functional domains of all 17 VvTCP proteins (Figure 1), indicating their importance for the VvTCP proteins in grapevine. TCP domains have been expounded to involved in the dimerization and DNA binding by a bHLH-structure with usually 59 AA residues (Cubas et al., 1999). In this study, TCP domains have been identified and consisted of 55 or 59 AA residues with a bHLH-structure in all VvTCPs (Figure 1). In the VvTCP members of class I, the deletion of four AA was found compared with that in the VvTCP members of class II at the bHLH structure. AA sequences of VvTCPs between class I and class II were definitely different in the loop, helix I, and helix II regions, however a conserved tandem of tryptophan (W) and leucine (L) was found in helix II. The above results illuminated that TCP members belonging to different classes are supposed to have complementary functions, while those in the same class seem to display redundancies.
The chromosomal location of each VvTCP in grapevine genome is shown in Figure 2. A total of 17 VvTCPs are unevenly distributed on 10 of 19 chromosomes. No genes were located on chromosomes (chrs) 3, 4, 5, 6, 7, 9, 11, 13 and 16. Whereas, one gene was located on chrs 2, 8, 15, 18, and 19, respectively. Furthermore, the chrs 1, 12, and 17 each had two genes. Meanwhile, three genes were located on chrs 10 and 14, respectively. The direction of transcription (arrows) and position of each VvTCP were determined on grape chromosomes available at Grape Genome Browser (Genoscope, 12× coverage). Moreover, further study displayed that the distributions of nine class I VvTCPs were irregular and all of them located on different chromosomes. In addition, the distribution of eight class II VvTCPs was also irregular and all class II VvTCPs located on different chromosomes except VvTCP1 and VvTCP13 on same chr14, as well as VvTCP2 and VvTCP10 on same chr10. To better comprehend the structure of VvTCP genes, the exon-intron organization were analyzed, which play an important role in the evolution of numerous gene families. The present study showed that most VvTCPs (12 out of 17) had only one exon organization, and four VvTCP members (VvTCPs 1, 3, 9, and 12) contained two exons separated by one intron (Figure 3). Moreover, VvTCP11 had four exons separated by three introns. Most of VvTCP genes possessed similar exon-intron organizations and distribution feature in the same VvTCP subclass, which were verified by the phylogenetic analysis (Figure 4).
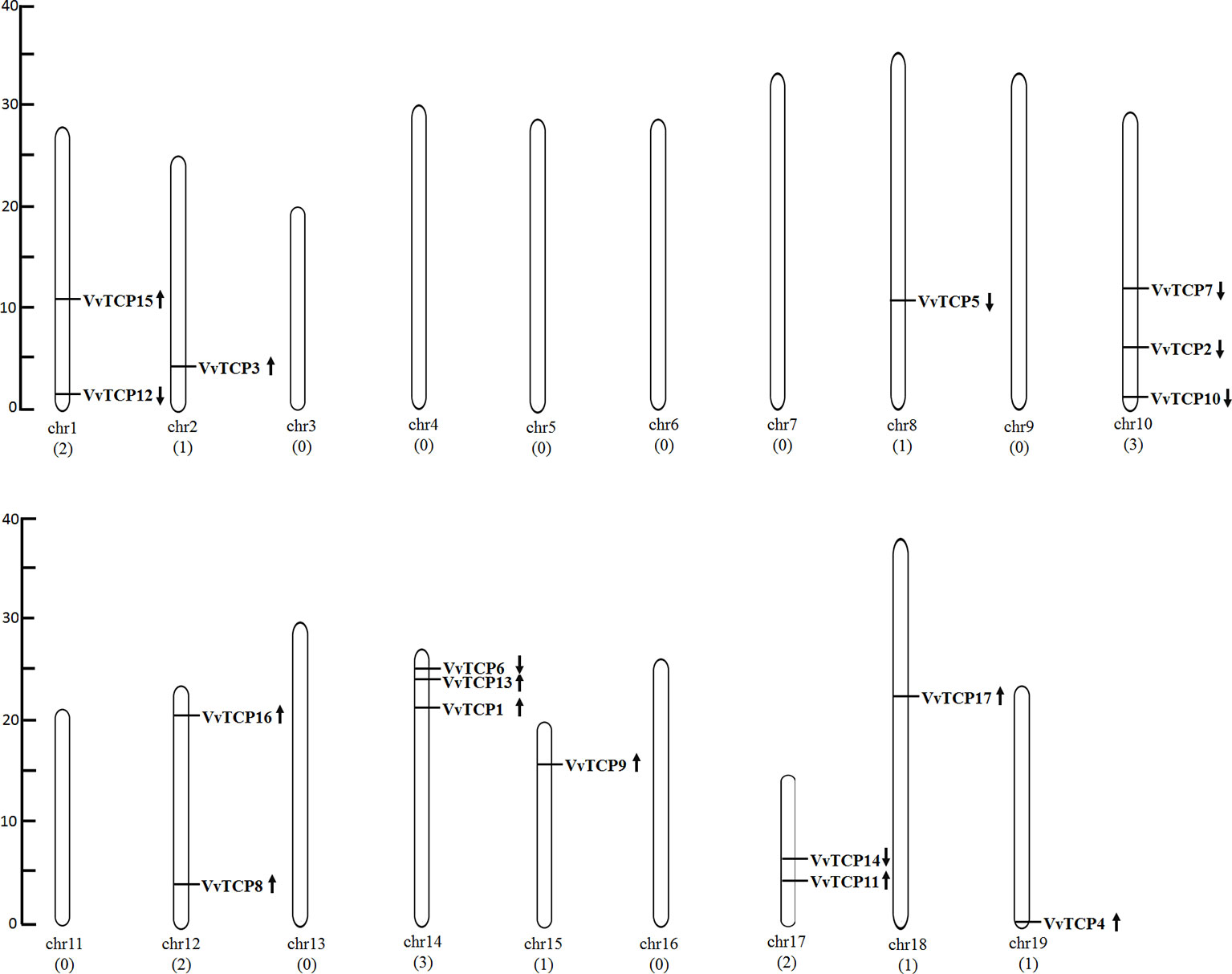
Figure 2 The chromosomal location of TCP genes from Vitis vinifera. The scale represents 40 Mb chromosomal distance. The VvTCP numbers are marked on the bottom of chromosomes. The arrows close proximity to VvTCPs indicated the direction of transcription. The numbers on the left side of the bars designated the approximate physical position of the first exon of corresponding VvTCPs on grapevine chromosomes.
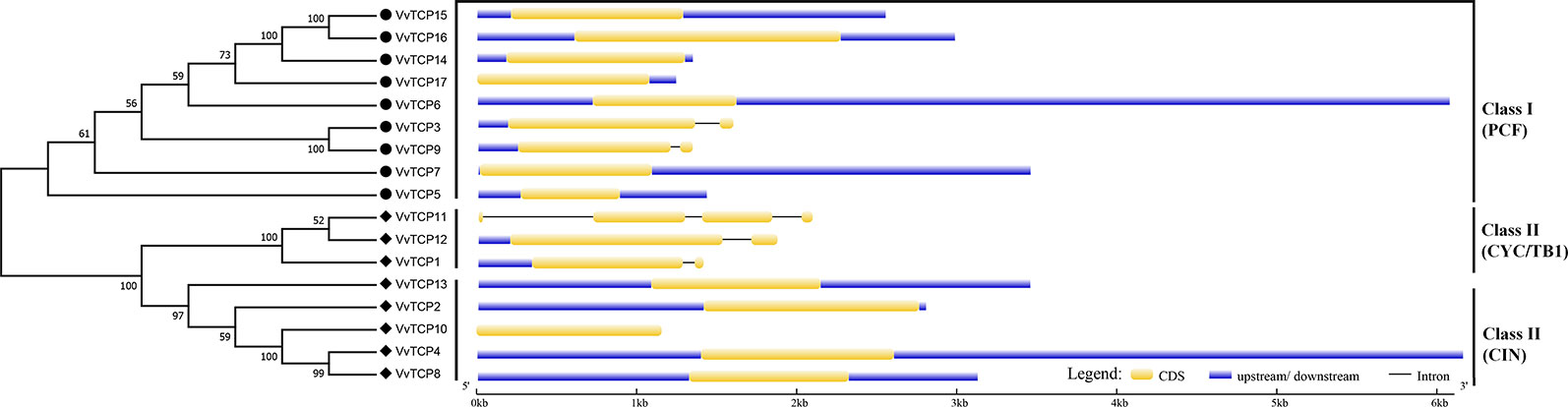
Figure 3 Exon-intron structure of 17 VvTCPs identified in Vitis vinifera. Shown is a graphic symbol of the gene models of all 17 VvTCPs identified in the present study. Exons are labeled using yellow boxes, introns are labeled using black lines, and untranslated regions are labeled using blue boxes.
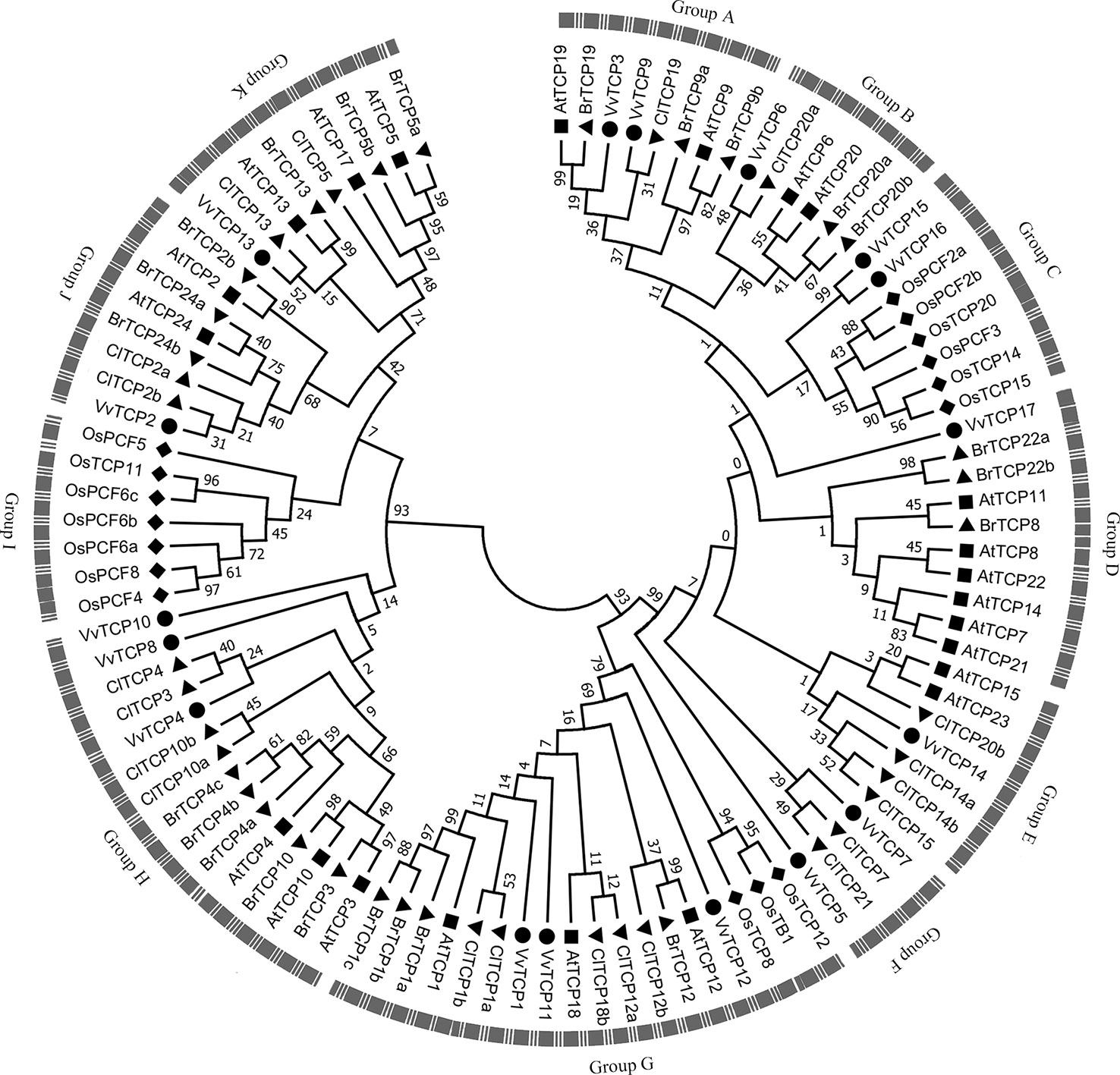
Figure 4 The phylogenetic analysis of TCP gene family among Vitis vinifera, Arabidopsis, Oryza sativa, Citrullus Lanatus, and Brassica rapa. The unrooted phylogenetic tree was constructed using the full-length protein sequences of TCPs from five species by the neighbor-joining (NJ) method with 1,000 bootstrap replicates.
To better understand the phylogenetic relationships of TCP genes in V. vinifera, Arabidopsis, O. sativa, C. lanatus, and B. rapa, a NJ phylogenetic tree was created based on multiple sequence alignment of 17 VvTCPs, 23 AtTCPs, 16 OsTCPs, 21 ClTCPs, and 23 BrTCPs using MEGA 7.0 software (Figure 4). The previous reports showed that bootstrap values for some nodes of the ML phylogenetic tree were low because of quite large number of TCP genes (Yao et al., 2007; Martin-Trillo and Cubas, 2010). Therefore, we also constructed ML phylogenetic tree and further sought extra evidences to validate the dependability of our phylogenetic tree. The phylogenetic trees constructed by NJ and ML methods mentioned above were almost identical with only tiny differences at some branches, indicating that the two methods were largely identify with one another. In addition, the analysis of conservative domains and motif and gene structure also further verified the rationality of our phylogenetic trees. In view of the great similarity among these tree topologies exactly as previous reports (Yao et al., 2007; Martin-Trillo and Cubas, 2010), the NJ phylogenetic tree was selected for deep analysis. According to the NJ phylogenetic tree, the TCP genes family was divided into 11 categories designated as group A to group K (Figure 4). Based on their sequence structures both outside and within the TCP domain, TCPs in group G belonged to the class II subclass CYC/TB1-type, groups H, I, J, and K belonged to the class II subclass CIN-type, whereas TCPs in the remaining groups A–F belonged to class I subclass PCF-type. Group G, the largest clade among all groups, had 18 TCP memberships, holding 18.0% of the overall TCP genes; while group H is the second largest clade, consisting of 15 TCP members. The smallest clade is group F, which contains only four memberships. In general, the TCP genes displayed a scattered distribution in the most of groups, suggesting that the expansion of TCP gene family have been performed before the lineage evolutionary divergence times. It is interesting to note that group I only contained seven TCP members of O. sativa. Meanwhile, the O. sativa TCP genes were absent in several groups, such as groups A, B, D, E, F, H, J, and K.
The evaluation of cis-elements in promoters is a key factor for comprehending transcriptional regulation and gene function. To recognize the putative cis-regulatory elements of the VvTCPs, the promoter sequences of the 17 VvTCPs were searched in the Vitis genomic database (http://genomes.cribi.unipd.it/grape/index.php). In the present study, a 2-kb was considered as a promoter region for 16 VvTCPs, whereas, a <2 kb promoter sequence for the VvTCP3 was also identified because of the existence of another gene situated at the <2 kb upstream (Supplemental Data Set 1). A total of 92 putative cis-elements involved in plant hormones [e.g., auxin, ABA, methyl jasmonate (MeJA), ethylene, gibberellin (GA), and salicylic acid (SA)] responses, biotic and abiotic stress response, and plant growth and development were identified using PlantCARE online database (Supplementary Table S2). As shown in Supplemental Table S2, the predicted cis-elements differed among the 17 VvTCPs, and two cis-elements (CAAT-box and TATA-box) were the most abundant, which had the largest number in all 17 VvTCPs. It is quite interesting to recognize that these unique cis-elements (AuxRR-core, MBSI, RY-element, circadian, etc.) were only present in one of 17 VvTCPs implying that these gene-specific cis-elements might play the special role in the regulating some biological process.
As shown in Figure 5, some phytohormone-related cis-regulatory elements including the ABA response element (ABRE), the MeJA response element (CGTCA motif), the SA-responsive element (TCA element), the ethylene-responsive element (ERE) and the auxin-responsive element (AuxRR-core and TGA-element) and the GA-responsive element (TATC-box, GARE motif, and P-box) were found in the promoters of 11, 8, 11, 10, 5, and 14 VvTCPs, respectively. A mass of phytohormone-responsive elements were recognized in the VvTCP promoters, suggesting they could play an important role in regulating the growth and development in plants (Figure 5). In several cis-elements related to plant growth and development, the circadian regulator element (circadian), the leaf development-related cis-regulatory element (HD-Zip1), the zein-metabolism regulation element (O2 site), the seed-specific regulation element (RY element), the meristem expression element (CAT-box), the meristem specific activation element (CCGTCC-box), and the endosperm-specific expression element (GCN4_motif) were identified in the promoters of 1, 3, 7, 1, 8, 3, and 4 VvTCPs, respectively. Additionally, some stress-related (e.g., low-temperatures, drought, and pathogens) cis-regulatory elements were also observed in the putative promoter sequences of the VvTCPs (Figure 5).
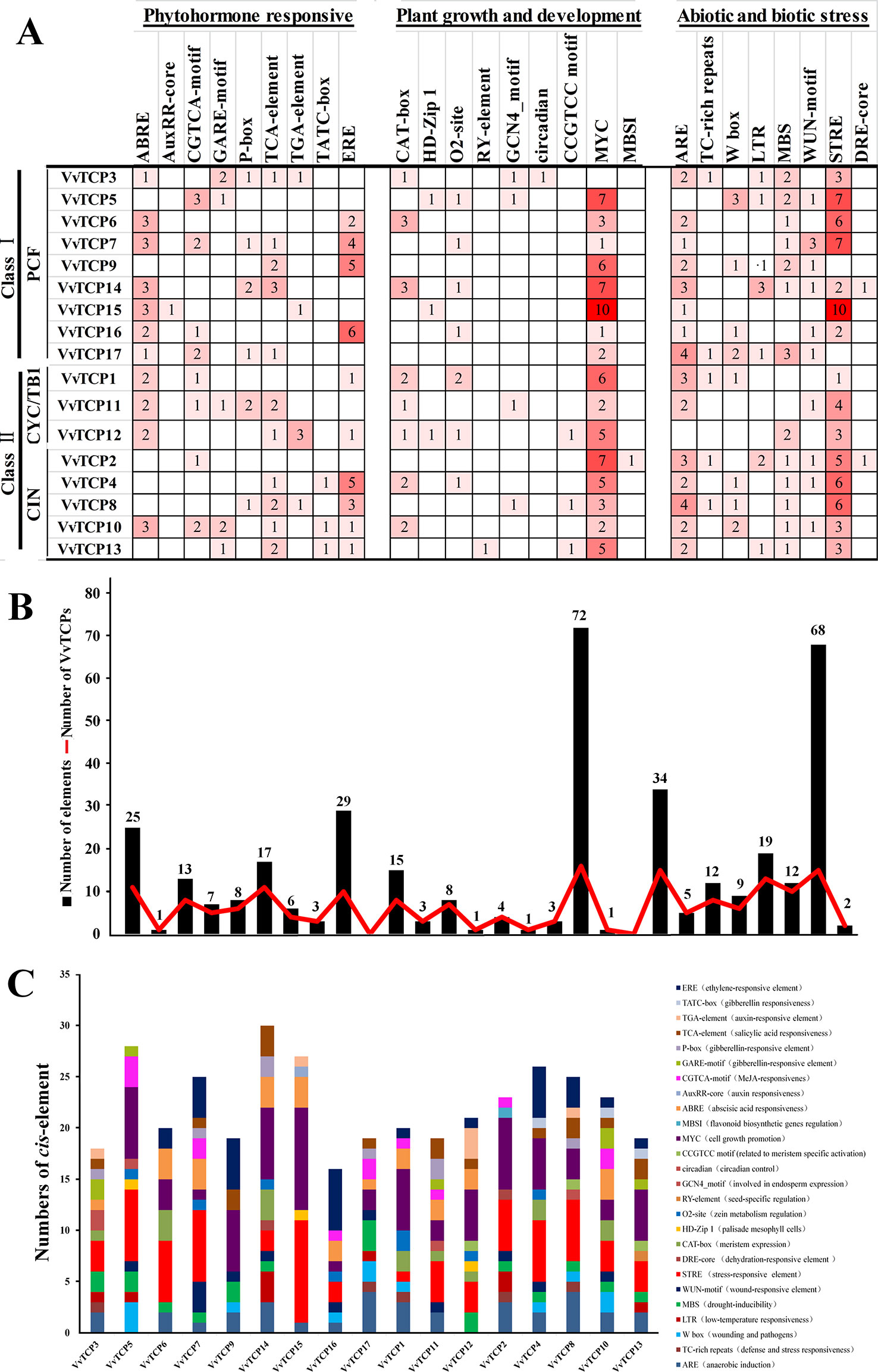
Figure 5 Cis-regulatory element analysis of the promoter regions of VvTCP genes in grapevine. (A) Number of every cis-regulatory element in the promoter region of VvTCPs. (B) Statistics for the overall number of VvTCPs, together with the corresponding cis-regulatory elements (red dot) and the overall number of cis-regulatory elements in VvTCP gene family grapevine (black box). Based on the functional annotation, the cis-regulatory elements were divided into three main categories: phytohormone responsive, plant growth and development, abiotic and biotic stresses-related cis-regulatory elements (detailed results shown in Supplementary Table S2). (C) Number of cis-acting elements in the promoter of VvTCPs genes that are related to stress responses and plant growth and development.
For the gene family expansion and evolution of new functions, gene duplication and divergence are essential steps in the plant genome. Vitis spp. has suffered from whole-genome duplications (WGD) during its generative history (Jaillon et al., 2007). Moreover, several gene duplication events including tandem duplication, WGD/segmental, and rearrangements at the chromosomal and gene level, drive the evolution of gene families encoding proteins (Maher et al., 2006). To evaluate the effect of duplications on the VvTCPs family, we firstly analyzed the origins of duplicate genes for the VvTCPs family in the grapevine genome utilizing the MCScanX software. Each member of the VvTCPs was allocated to one of five duplication events (dispersed, singleton, tandem, proximal, and WGD/segmental). The results showed that 76.47% (13) of the VvTCPs in grapevine were duplicated from singleton event, compared with only 17.65% (3) from dispersed, and 5.88% (1) from tandem event (Table 1). However, no WGD/segmental and proximal duplication events were found in the grapevine VvTCP genes. The results demonstrated that singleton duplication played a vital role in the expansion of the VvTCPs family in grapevine.
To further explore the origin and probable evolutionary mechanisms of the VvTCPs family, we also investigated the syntenic blocks between Vitis and other plant species, and detected that syntenic relationship was observed between grapevine and A. thaliana, meanwhile between grapevine and rice (O. sativa) based on the results obtained from PGDD online database. The syntenic analysis demonstrated that eight syntenic gene pairs within grapevine genome included: VvTCP1-VvTCP12/VvTCP11, VvTCP3-VvTCP9, VvTCP4-VvTCP8/VvTCP10, VvTCP8-VvTCP10, VvTCP11-VvTCP12, VvTCP14-VvTCP15. The syntenic analysis showed that nine syntenic gene pairs between grapevine and A. thaliana included: VvTCP3-AtTCP9 (AT2G45680)/AtTCP19 (AT5G51910), VvTCP7-AtTCP21 (AT5G08330)/AtTCP7 (AT5G23280), VvTCP9-AtTCP9 (AT2G45680), VvTCP14-AtTCP15 (AT1G69690)/AtTCP14 (AT3G47620), VvTCP15-AtTCP15 (AT1G69690), VvTCP16-AtTCP8 (AT1G58100). Moreover, the syntenic analysis showed that five syntenic gene pairs between grapevine and O. sativa included: VvTCP3-OsPCF2-like (LOC_Os09g34950), VvTCP4-OsTCP22 (LOC_Os01g55100), VvTCP14-OsTCP14 (LOC_Os02g51280), VvTCP15-OsTCP15 (LOC_Os06g12230)/OsTCP14 (LOC_Os02g51280) (Figure 6). The syntenic analyses among grapevine, A. thaliana, and rice showed that these genes located in corresponding syntenic blocks arose before the divergence of grapevine, A. thaliana, and rice.
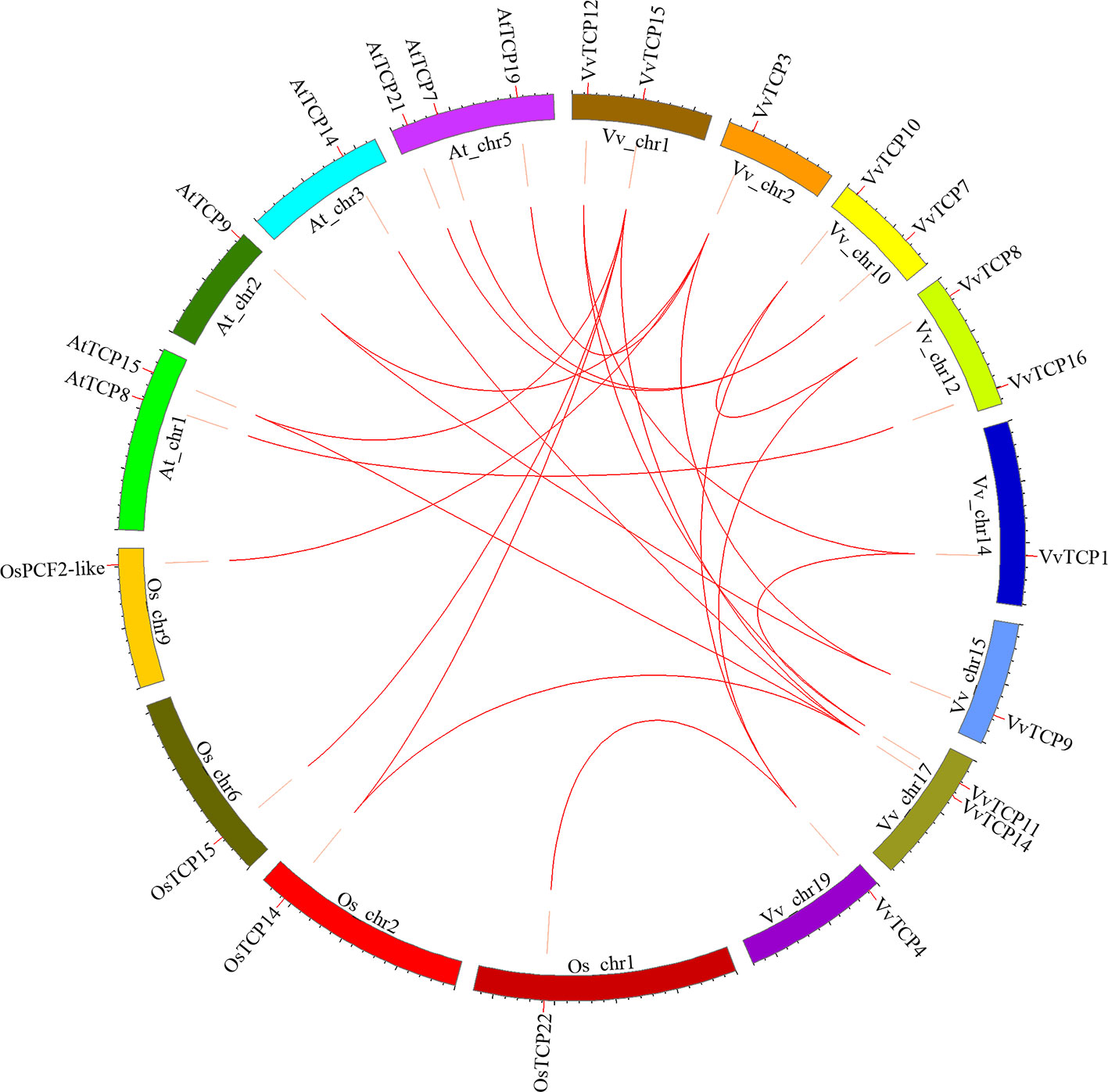
Figure 6 Syntenic block among TCPs from Vitis vinifera, Arabidopsis thaliana, and Oryza sativa. Chromosomes of V. vinifera, A. thaliana, and O. sativa are shown in different colors and in circular form. The approximate positions of the VvTCPs, OsTCPs, and AtTCPs are labeled with a short red line on the circle. Colored curves represented the syntenic relationships between grapevine, O. sativa, and A. thaliana TCP genes.
In V. vinifera, three vvi-miR159 family members (vvi-miR159a, b, and c) and four miR319 family members (vvi-miR319b, c, f, and g) stored in miRBase (http://www.mirbase.org/summary.shtml?org=vvi). A result of two putative miR159-target genes (VvTCPs 2 and 13) was attained from the analysis of psRNATarget software with the full-length nucleotide sequences of VvTCPs and the mature sequence of vvi-miR159a, and b. However, four putative miR159-targeted TCP genes (VvTCPs 2, 4, 8, and 13) were predicted based on the mature vvi-miR159c sequence in the psRNATarget online application. And all miR159-targeted TCP genes belonged to CIN subclass. Meanwhile, five putative miR319-targeted TCP genes (VvTCPs 2, 4, 8, 10, and 13) were predicted with the full-length nucleotide sequences of VvTCPs and the mature sequence of vvi-miR319b, c, g and f. And all target genes for four vvi-miR319, which belonged to CIN subclass. Likewise, five TCP genes (AtTCPs 2, 3, 4, 10, and 24) were target by miR319 in Arabidopsis, and all of them were also CIN subclass. The phenomenon showed that the miR319-target sites were reserved during the diversification and evolution of the plants. Previous reports have shown that the mature sequence of miR159 and miR319 were very similar, however, they were encoded by different precursors (Schommer et al., 2012). Sometimes, they were assigned in the same family. In V. vinifera, the vvi-miR159 sequence was discordant with miR319 aside from six same base (Figures 7A, B). Thus, we considered vvi-miR159 and vvi-miR319 is not belong same miRNA family in V. vinifera. The alignments of the reverse vvi-miR159/319 sequence and VvTCP genes showed that these VvTCPs are complementary to the vvi-miR159/319 sequence (Figures 7C, D). Though, the mismatch of vvi-miR159/319 and their targets raised to 3–8 bases, the regulation of TCP TFs through vvi-miR159/319 was considered to be possible in grapevine.
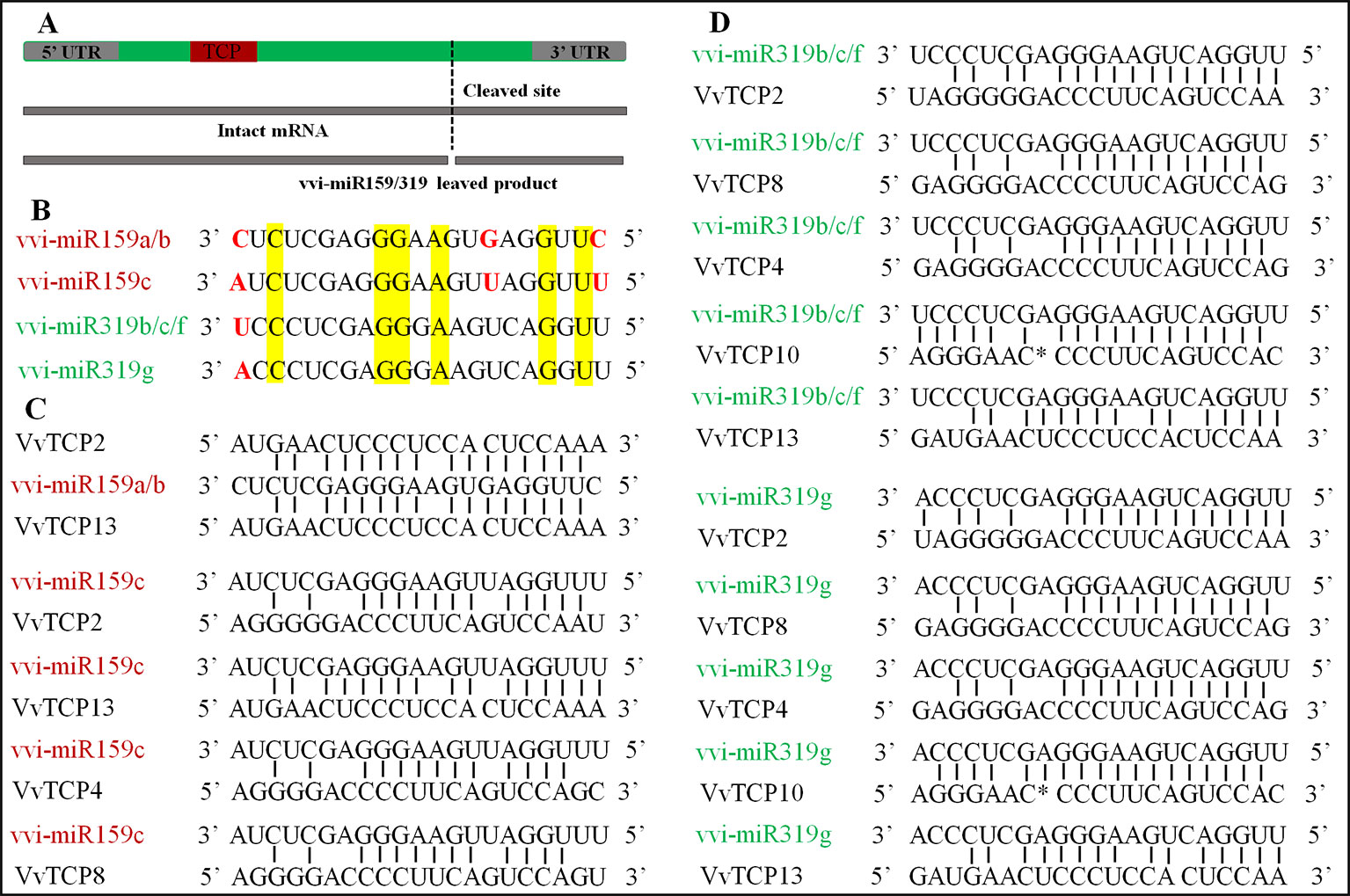
Figure 7 The miR319/159 binding sites of TCP gene in Vitis vinifera. (A) The TCP gene structure displayed the coding region (green), the TCP domain (red). (B) The sequence alignment of vvi-miR319 and vvi-miR159 in grapevine. (C) Alignment of vvi-miR159 complementary sequences with V. vinifera TCP genes. (D) Alignment of vvi-miR319 complementary sequences with V. vinifera TCP genes.
To explore the tissue-specific expression patterns of TCP genes in V. vinifera, we analyzed the transcripts of VvTCPs utilizing qRT-PCR analysis of different tissues, including fruits, leaves, roots, buds, stems, tendrils and flowers from the ‘Jumeigui' grapevine. As shown in Figure 8, 17 VvTCPs exhibited tissue-specific expression patterns, potentially showing the functional divergence of VvTCPs in all tested tissues. For example, VvTCP11 and VvTCP12, the members of CYC/TB1 subclass, are both highly expressed in flower and bud, whereas poorly expressed in YS, Ro, RB, leaves, and Te. As shown in Figure 3, VvTCP11 and VvTCP12 possessed the closest relationship in the phylogenetic tree of VvTCPs, further implying their functional similarity on regulating the growth and development of various tissues. The expression level of VvTCP1 belonging to CYC/TB1 subclass, was higher in the YS, Ro, ML than other selected tissues. For the CIN subclass, the transcript accumulation levels of VvTCP8 and VvTCP13, were higher in ML and OL, however low expression levels were observed in Ro, displaying that they might play a vital role in the growth and development of ML and OL. VvTCP4 and VvTCP10 presented very high expression levels in Bu and FF, however low expression levels in Ro, YS, RB, YL, and Te. In particular, VvTCP2 were preferentially expressed at high levels in FF, which indicated that it might make a difference in flower growth and development. Additionally, PCF subclass displayed more extensive tissue-specific expression patterns; such as, VvTCP6, VvTCP14, and VvTCP16 were hugely expressed in Te; VvTCP7 and VvTCP9 displayed high relative transcript levels in Te, ML and OL; and VvTCP3 and VvTCP15 are highly expressed in YS, Te, and ML, VvTCP17 exhibited higher expression levels in flowers. Remarkably, qRT-PCR analysis showed that VvTCP5 preferentially expressed at high levels in flowers, which indicated that they might play a vital role in flower growth and development. The results urge us to explore the expression level of VvTCPs during different berry developmental stages.
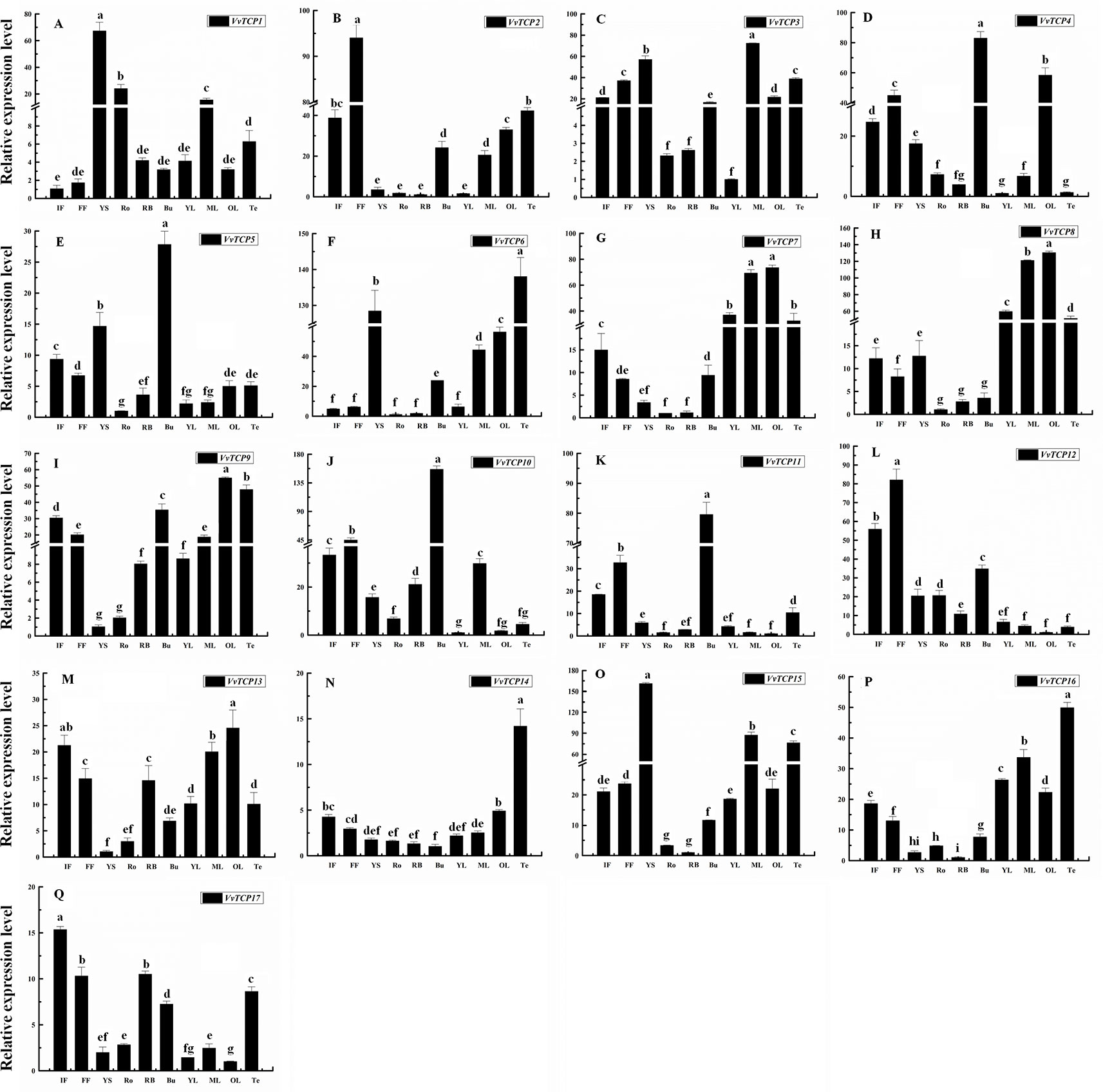
Figure 8 Relative expression levels of VvTCPs in different organs of ‘Jumeigui' grapevine. Values were normalized against the expression data of KyActin1 and are given as means ± standard error among three biological replicates. Different letters indicate significant differences between genes (p < 0.05). The expression levels were calculated based on the 2−ΔΔCt method.
To illuminate their functions in fruit development, qRT-PCR were employed to explore the expression levels of 17 VvTCP genes at different fruit developmental stages. As indicated in Figures 9 and 10, the expression level of eight VvTCPs (VvTCPs 2, 6, 7, 9, 10, 14, 15, and 16) were down-regulated toward fruit maturity, which suggested that they might function mainly in the early period of berry development. As shown in Supplementary Table S2, VvTCPs 3, 8, and 11 harbored GCN4_motif cis-elements involving in endosperm expression, which is consistent with high expression level of three VvTCPs in early period of fruit development. On the contrary, the transcripts of three genes (VvTCPs 4, 12, and 17) were up-regulated during the ripening process, which suggested that they might play a vital important role in fruit maturation. Same expression levels were observed for VvTCP5, which indicated that VvTCP5 participate in the whole process of berry development. VvTCP1 showed a jump expression patterns during different fruit developmental stages. VvTCP1 showed high expression levels at BGB and PVB in comparison with the other stages. The expression patterns of VvTCP13 during SGB period was significantly high as compared to the other stages. The relative expression level of VvTCP13 showed a declined trend from SGB to BGB and then exhibited a continuous increasing trend as fruit proceeds toward maturity, indicating that VvTCP13 played different roles in different stages of fruit development.
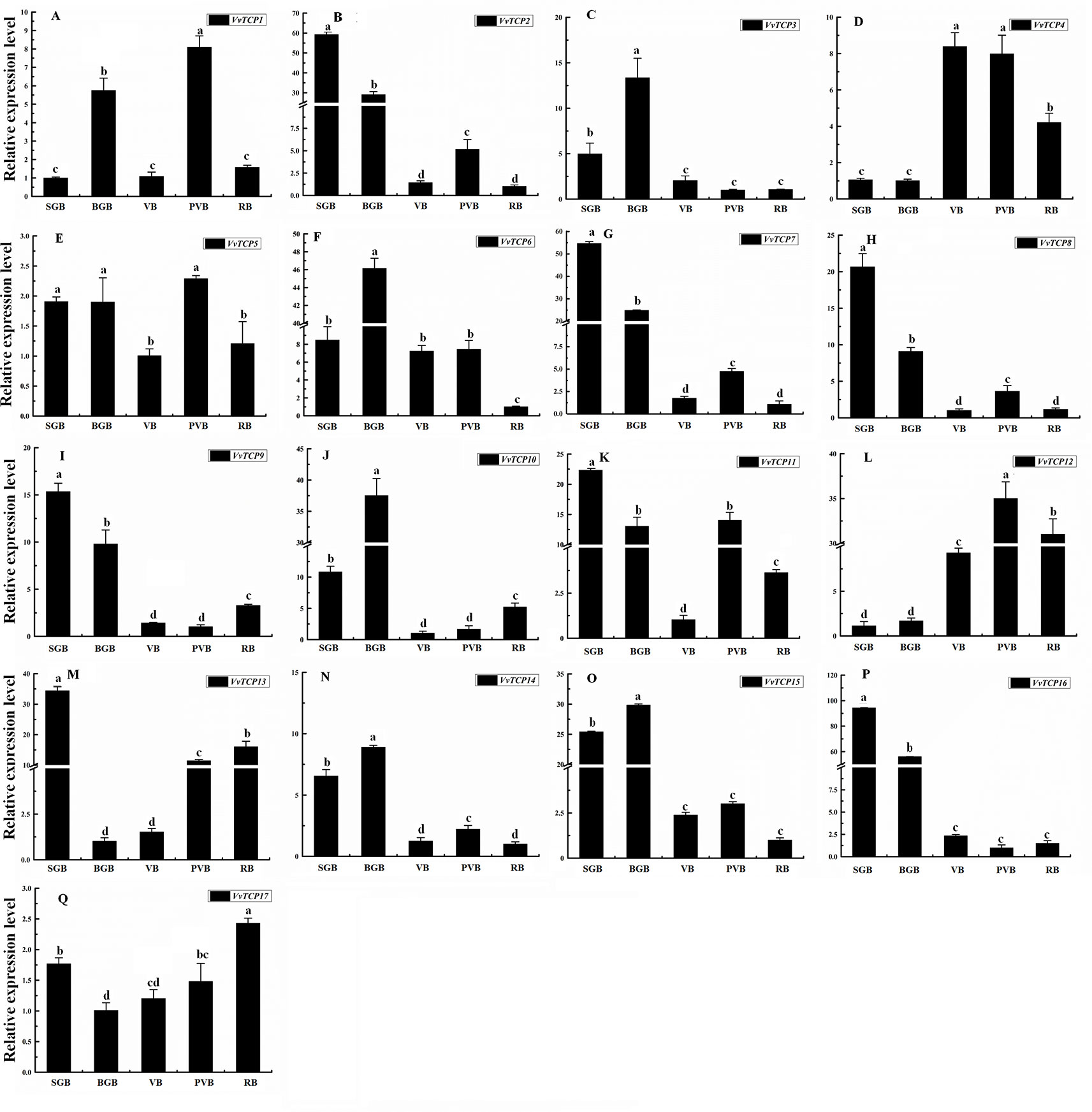
Figure 9 Relative expression levels of VvTCPs in different fruit development of ‘Jumeigui' grapevine. Values were normalized against the expression data of KyActin1 and are given as means ± standard error among three biological replicates. Different letters indicate significant differences between genes (p < 0.05). The expression levels were calculated based on the 2−ΔΔCt method.
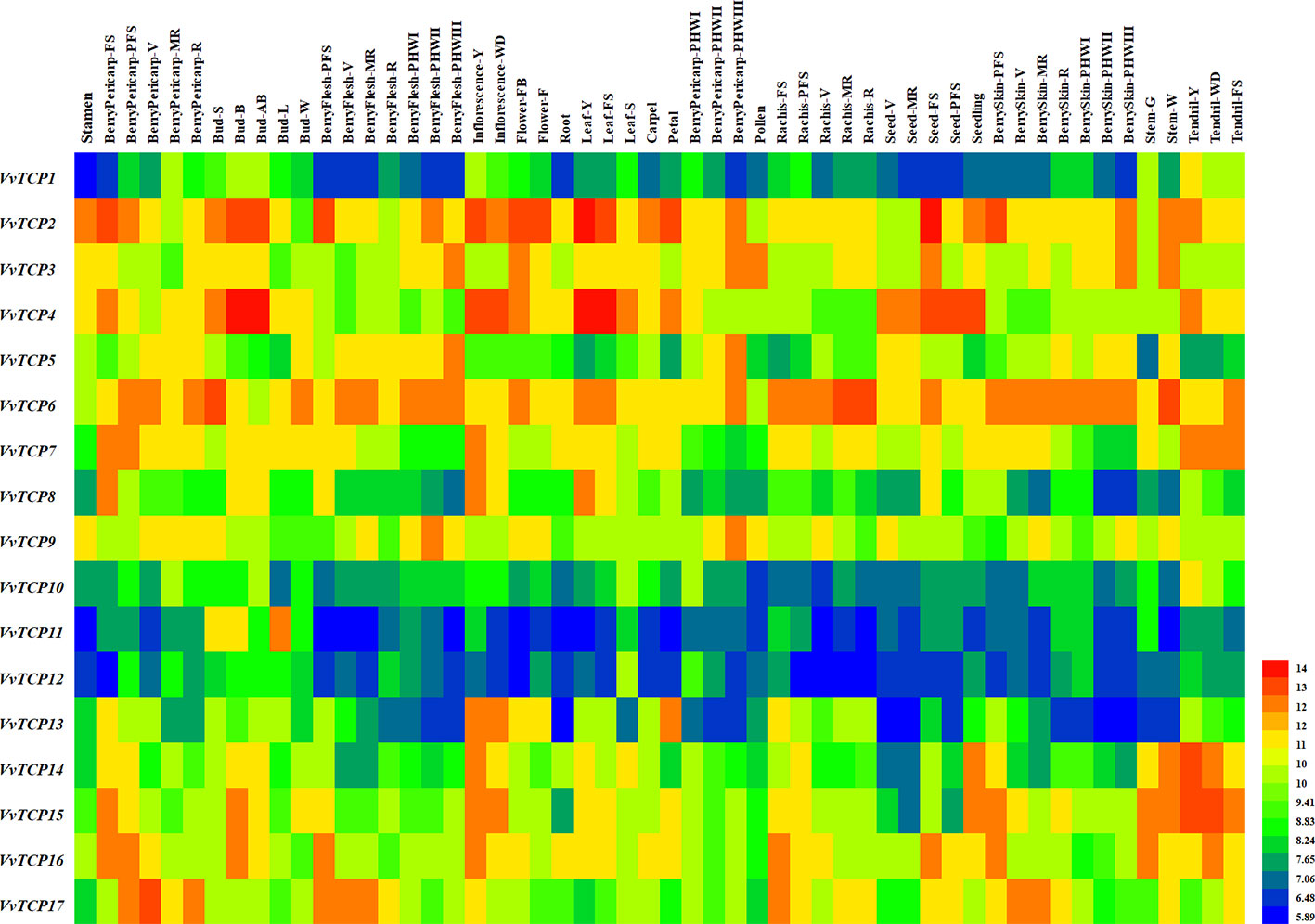
Figure 10 Expression profiles of VvTCP genes in the V. vinifera cv. ‘Corvina' atlas (GEO accession: GSE36128). Data were normalized based on the mean expression value of each gene in all tissues analyzed. The mean expression values were again normalized using logarithm with the base of 2 using the Heml software. Blue and red boxes show low and high expression levels, respectively, for every gene.
Abscisic acid (ABA) have engrained roles in fruit developmental processes as well as stress signaling networks (Bari and Jones, 2009). As shown in Supplementary Table S2, ABA-responsive cis-element, ABRE, were observed in several VvTCPs. In order to comprehend how VvTCPs express in response to ABA treatment, qRT-PCR was employed to analyze VvTCP transcripts respond to ABA, which has three concentration levels (0, 50, 150 ppm). The expression levels of four VvTCPs (VvTCPs 1, 10, 14, and 16) were all enhanced with 50 ppm, 150 ppm ABA compared with the 0 ppm ABA at VB, PVB, and RB period, which is consistent with presence of ABA-responsive cis-element (ABRE) in the promoter of these VvTCPs, further implying ABA may play an important role in berry ripening (Figure 11). On the contrary, the expression levels of five VvTCPs (VvTCPs 2, 6, 7, 15, and 17) were all suppressed with 50 ppm, 150 ppm ABA compared with the 0 ppm ABA at VB, PVB, and RB period. VvTCP11 transcript was enhanced with 150 ppm ABA compared with the 0 ppm ABA at VB and PVB period, whereas it was suppressed with 150 ppm ABA compared with the 0 ppm ABA at RB period. The expression levels of two VvTCPs (VvTCPs 5 and 13) were all enhanced with 150 ppm ABA compared with the 0 ppm ABA at veraison, however they were suppressed with 50 and 150 ppm ABA compared with the 0 ppm ABA at PVB and RB period. The expression levels of two VvTCPs (VvTCPs 9 and 12) were strongly enhanced with 50 ppm ABA at PVB period compared with the 0 and 150 ppm ABA at veraison and RB period.
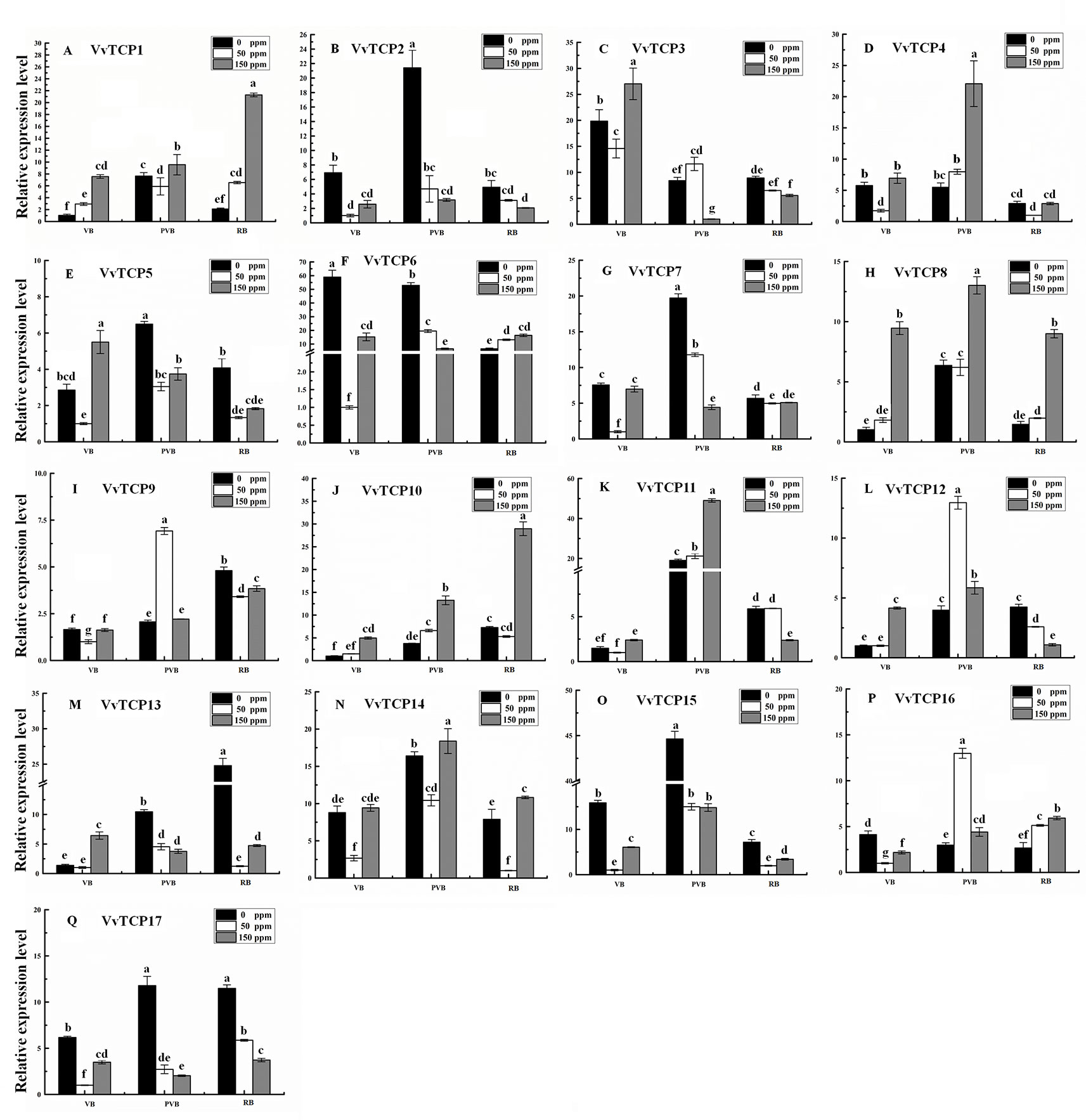
Figure 11 Relative expression levels of VvTCPs under abscisic acid treatment. Values were normalized against the expression data of KyActin1 and are given as means ± standard error among three biological replicates. The expression levels were calculated based on the 2−ΔΔCt method.
To investigate the function of VvTCPs, their subcellular localizations were measured by the fluorescent protein-tagging method. Firstly, the full-length open reading frames (ORFs) lacking the stop codon of two VvTCPs were merged to the N-terminal of green fluorescence protein (GFP) protein driven by CaMV 35S promoter, generating fusion proteins p35S-VvTCP9-GFP and p35S-VvTCP15-GFP were agroinfiltrated into leaves of 3 to 5-week-old N. benthamiana plants. Fluorescence microscopy exhibited that the PHB-GFP was homogenously disseminated throughout the whole cell, the fusion proteins of p35S-VvTCP9-GFP, and p35S-VvTCP15-GFP were observed in the plasma membrane and nucleus (Figure 12). The results demonstrated that VvTCP9 and VvTCP15 were both nuclear- and cytomembrane-localized protein. Different from other nuclear-localized TFs, VvTCP9, and VvTCP15 also localized in the cytomembrane, which indicated that two VvTCPs (VvTCP9 and VvTCP15), as well as other single nuclear-localized TCP TFs may possessed diversity function.
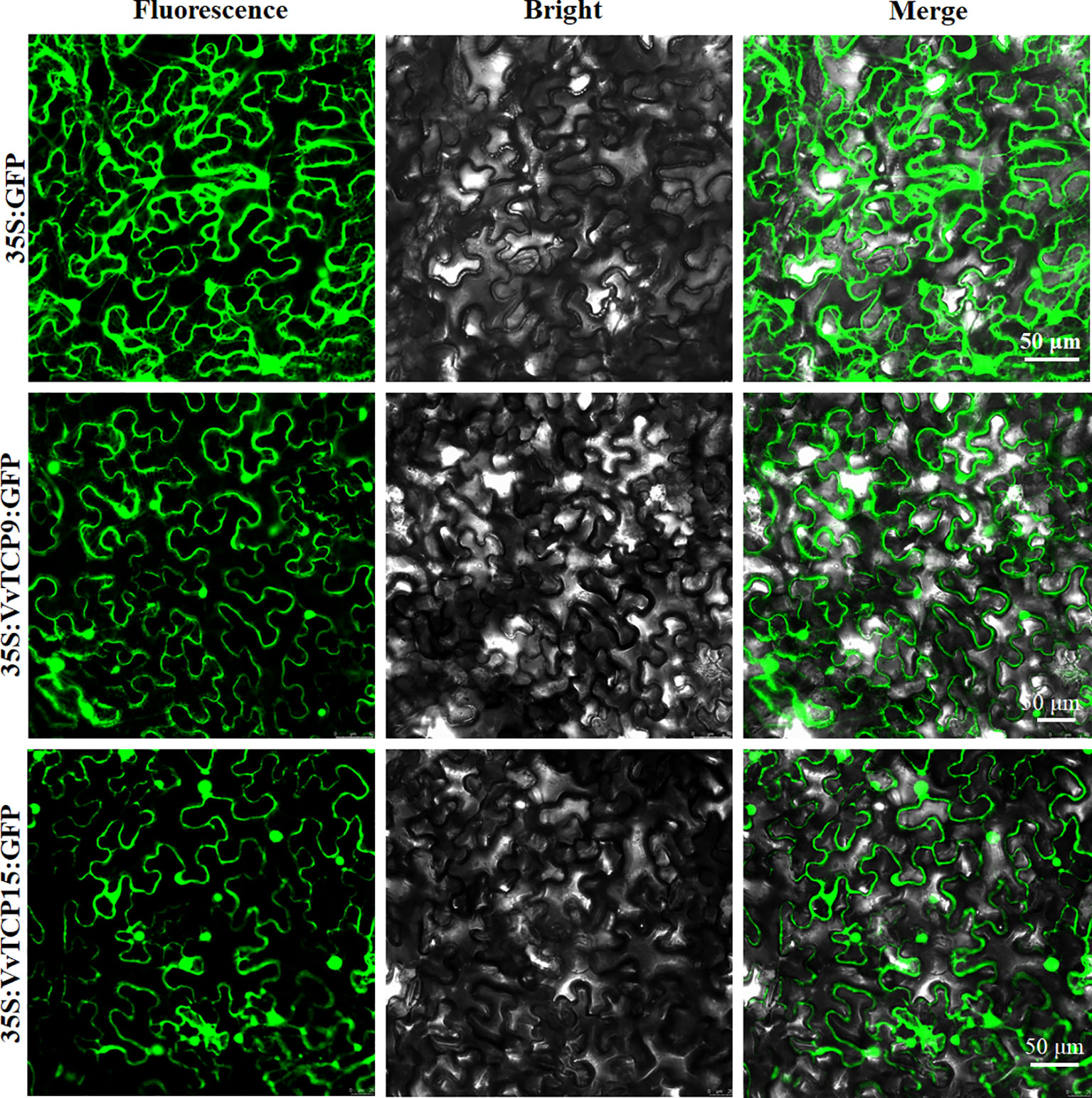
Figure 12 The leaves of 3 to 5-week-old Nicotiana benthamiana plants were transiently transformed with control, 35S-VvTCP9-GFP, and 35S-VvTCP15-GFP. Images under blight field (middle), fluorescence (left), and the merged images are shown on the right. Bar: 50 μm.
TCP proteins are a type of plant-specific TFs, which participate in numerous biological activities throughout the growth and development in plants (Manassero et al., 2013). To date, many TCP genes have been reported in some studies with genome-wide characterization, such as, G. raimondii (Ma et al., 2014), Malus domestica (Xu et al., 2014), L. esculentum (Parapunova et al., 2014), Arabidopsis (Li, 2015), C. lanatus (Shi et al., 2016), and B. rapa (Du et al., 2017). Nevertheless, no comprehensive analyses of TCP TFs in V. vinifera have been done. In the present study, we performed a comprehensive analysis of the VvTCPs family in grapevine by exploring their evolutionary relations, exon-intron organization, conserved motifs, cis-regulatory elements, gene duplication and syntenic analysis, and expression patterns in various tissues and developmental phases and under ABA treatment situations, and sub-cellular localizations.
In the present study, a sum of 17 VvTCP genes were identified from the grapevine genome, and the TCPs number of V. vinifera was less than those of Arabidopsis, O. sativa, L. esculentum, P. mume, C. lanatus, turnip, Chinese cabbage, and G. raimondii, which had 24, 22, 30, 19, 27, 39, 39, and 38 TCP members, respectively (Table 2). The quantity of TCP genes in turnip and G. raimondii are almost 2.29 and 2.24 times more than that in V. vinifera, which are inconsistent regarding the coding genes in turnip genome (41,174 genes, http://brassicadb.org/brad/speciesinfo.php) and G. raimondii genome (40,976 genes) are about 1.56 and 1.55 times more than that in grapevine (26,346 genes) (Wang et al., 2012). It is observed that many BrTCP genes contain two or more counterparts in turnip and G. raimondii, suggesting that the enlargement of TCP gene family in turnip and G. raimondii might be due to genome duplication events. Therefore, we consider that the difference of genome duplication events may be the main reason for this inconsistency. The phylogenetic tree among V. vinifera, Arabidopsis, O. sativa, C. lanatus, and B. rapa supported the previous described division of TCP genes family into two subfamilies: class I (PCF) and class II (CYC/TB1 and CIN) (Navaud et al., 2007; Martin-Trillo and Cubas, 2010). TCP gene family in V. vinifera also consisted of these three types: PCF with nine members, CIN with five members, and CYC/TB1 with three members (Table 2). Nearly all the VvTCPs had the conserved motif 1 and motif 2 in V. vinifera. Besides conservative motif 1 and motif 2, the VvTCPs of PCF type usually has motif 4 in front of motif 1 in proteins. Motif 5 was only identified in the VvTCPs of class II. Moreover, motif 12 was only found in VvTCP1, VvTCP11, and VvTCP12 of the CYC/TB1 type, which contained an R domain (Figure 1B). Furthermore, motifs 1–2 were specific to the TCP domain, which was formed by a bHLH-motif of 54–59 AA remains. These showed that VvTCPs could play vital roles in plant growth and development, even though they belong to identical TCP subclass. Moreover, most of VvTCP proteins are belong to the same subclass that not only shared similar conserved domain but also similar gene structure.
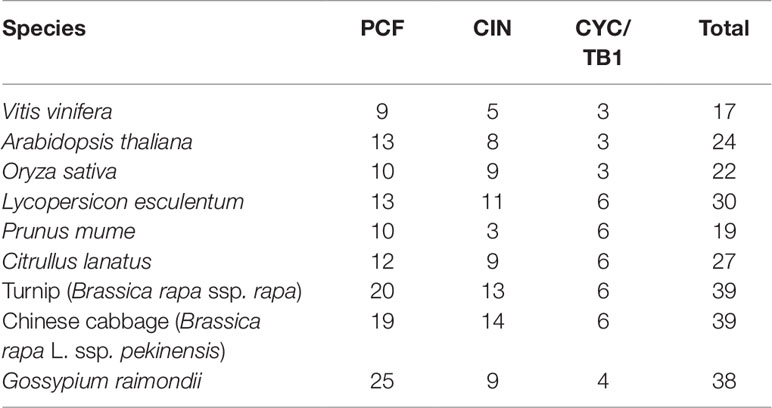
Table 2 The number of TCP family genes in Vitis vinifera, Arabidopsis thaliana, Oryza sativa, Lycopersicon esculentum, Prunus mume, Citrullus lanatus, turnip, Chinese cabbage, and Gossypium raimondii.
As the key unit of transcriptional regulation, cis-elements participated in the regulation of molecular networks in many biological processes (Ibraheem et al., 2010). In terms of cis-regulatory element sequences, the promoters of VvTCPs have vastly repetitive regions and some common motifs. The minimum 25 cis-regulatory elements (ACE, GT1-motif, MRE, G-Box, etc.) in this study are necessary for light-driven transcriptional regulation in the promoter of VvTCPs, which are in accord with the important role of VvTCPs in the light-responsive process. Moreover, the existence of the circadian element shows the circadian regulation is very important for the VvTCP genes expression. Some VvTCPs (e.g., VvTCP1, VvTCP8, VvTCP10, VvTCP14, and VvTCP16) existed in ABA-responsive element (ABRE), which were in accord with the increased expression level of these VvTCPs under the ABA treatment. The results of cis-elements analyses showed the presence of unique cis-regulatory elements in the promoter of only one TCP gene, hence deducing the specificity of these genes expression. The unique cis-regulatory elements were recognized due to the length of their sequences (6–15 bases) but not usually easy to generate some nucleotide variabilities. Interestingly, out of the 23 unique cis-elements, four were identified only in the VvTCP1 promoter, followed by three in VvTCP2, two in VvTCP3, VvTCP11, VvTCP14, and VvTCP16, and only one in VvTCP5, VvTCP9, VvTCP10, VvTCP13, and VvTCP16. As we all known, gene duplication is the initial driving force for new functions in the development of genomic and genetic systems, and is one of the main evolutionary mechanisms further leading to species formation and differentiation (Lynch and Conery, 2000). Previous reports indicated that the enlargement of gene families is largely caused by gene duplication events on several types, including WGD, tandem duplication, segmental duplication, and other transposition events (Moore and Purugganan, 2003). Our results demonstrate that singleton duplication is a predominant type for VvTCP genes and the key factor to the enlargement of VvTCPs family in grapevine.
Previous reports indicated that more than half of the target genes of miRNA are TFs, such as MYB, TCP, AP2, etc. (Aukerman and Sakai, 2003; Martin-Trillo and Cubas, 2010; Xu et al., 2015). Concerning V. vinifera TCP genes, four of them (VvTCPs 2, 4, 8, and 13) were predicted to be targeted by vvi-miR159. Additionally, five VvTCP genes (VvTCPs 2, 4, 8, 10, and 13) were predicted to be targeted by vvi-miR319. In V. vinifera, the sequence of vvi-miR319 was obviously different with respect to vvi-miR159, but they are predicted to enable to regulate the same VvTCP genes. However, miR159 and miR319 possessed similar mature sequences, whereas they targeted different genes in Arabidopsis. Though miR159 did not target the TCP genes, it had same role with miR319 in regulating flower development in plants (Hong and Jackson, 2015). Current studies showed that five VvTCP genes (VvTCPs 2, 4, 8, 10, and 13) were targeted by vvi-miR319. The mRNA fragments of TCP gene are produced by a miR319a-mediated cleavage and this have been verified by in vivo experiment (Palatnik et al., 2003). In plants, the miR319 family involved in regulating floral development with the miR319-mediated-TCP model. The flaws in stamen and petal development of Arabidopsis were found in the mutants of miR319 loss-of-function (Nag et al., 2009). Similar to miR159, over-expression of miR319 leads to male sterile and stamen imperfections (Palatnik et al., 2007). In this study, four target genes (VvTCPs 2, 4, 10, and 13) were all expressed strongly in flower, especially at FF time. Thus, VvTCPs could possess an interactional and coefficient relations in the floral grow and development process of V. vinifera.
Many evidences have already suggested that TCP genes participate in regulating cell propagation and growth, and playing multiple roles in the process of growth and development in plants (Martin-Trillo and Cubas, 2010). The genes of CYC/TB1 subclass are mostly participated in the growth and development of axillary meristems that are responsible for the growth of either lateral shoots or flowers. AtTCP1, belonging to CYC/TB1 subclass, is participated in the longitudinal elongation of some tissues, such as inflorescent stems, rosette leaves, and petioles. AtTCP1 is expressed strongly in the distal region of growing rosette leaves, the lower part of the inflorescence stem, and the midrib region of the petiole and blade (Koyama et al., 2010). In addition, the mutant plants of gain-of-tcp1-1D have lengthened petioles and leaves, while the plants of TCP1-SRDX show reduced statures and petioles, as well as epinastic and rounded leaves (Guo et al., 2010). VvTCP1, which is closely related to AtTCP1, was highly expressed in YS, ML, and root (Figure 8). AtTCP18 (BRC1, BRANCHED1), a homolog of TB1 in Arabidopsis, was highly expressed in some tissues that mainly included axillary buds, such as stem, silique, inflorescence, and leaf base (Aguilar-Martinez et al., 2007). AtTCP18 acts downstream of strigolactone and auxin to regulate the outgrowth of axillary bud, and the mutant plants with decreased activity of either gene display an increasing amount of the rosette branches. Conversely, the inhibition effect in lateral branches were observed in plants due to the up-regulated expression of AtTCP18. VvTCP11, which is closely related to AtTCP18, exhibits a significantly higher relative expression level in the bud (Figure 8). To some extent, this result is in accord with the expression level of AtTCP18 and indicates that VvTCP11 likely play a significant role in axillary bud development, similar to AtTCP18, in grapevine. Additionally, the TCPs of CYC/TB1-type might be less ancient than CIN subclass, and CIN-type TCPs play a vital role in the leaf growth and development. For instance, eight CIN-type genes (AtTCPs 2, 3, 4, 5, 10, 13, 17, and 24) displayed relatively high expression levels in leaves and were capable for the regulation of leaf growth and development in Arabidopsis (Palatnik et al., 2003; Ori et al., 2007). Likewise, five CIN-subclass genes in V. vinifera except VvTCP10 were highly expressed in ML or OL. The above results showed that the regulation of leaf growth and development by TCP genes that are homologous to those in grapevine might be similar in Arabidopsis.
In VvTCPs of PCF-type, all genes displayed widespread expression level, implying these genes might play important roles in numerous different development phases. In Arabidopsis, AtTCP14 involved in regulating embryonic growth development through seed sprouting, which was closely linked to ABA responses (Tatematsu et al., 2008). In the present study, VvTCP14, and VvTCP17 were phylogenetically close to AtTCP14. Among them, VvTCP14 exhibited higher transcript expression levels after 150 ppm ABA treatment, however, VvTCP17 was down-regulated expression after ABA treatment at different berry development stages, implying they might play distinct roles in berry development in response to ABA. In grapevine, the endosperm plays a vital role in fruit setting as well as early stage of berry development. Promoter analysis showed that four (VvTCPs 3, 5, 8, and 11) of 17 VvTCP genes harbored GCN4_motif cis-elements participated in endosperm expression (Figure 5A and Supplementary Table S2). Hence, it concluded that these VvTCP genes are responsible for fruit development and ripening in grapevine. In tomato, some SlTCPs, for instance, SlTCP18, SlTCP15, and SlTCP12, are preferentially expressed in fruits. Furthermore, these genes are controlled by SlAP2a (APETALA2a), SlCNR (COLORLESS NON-RIPENING), and SlRIN (RIPENING INHIBITOR) proteins, which are important TFs with vital roles in ripening, implying an important role in fruit development or ripening of tomato (Parapunova et al., 2014). In grapevine, VvTCP6 and VvTCP7, which are homologs of SlTCP18 and SlTCP15, respectively, also exhibited high expression levels before the version of berry, demonstrating that VvTCP6 and VvTCP7 might play important roles in early berry development of grapevine (Figures 9 and 10).
Publicly available datasets were analyzed in this study. This data can be found here: http://genomes.cribi.unipd.it/grape/index.php; http://www.arabidopsis.org; http://planttfdb.cbi.pku.edu.cn/, v3.0.
SJ designed and performed most of the experiments and drafted the manuscript. YX and JW provided technical assistance. SW, CZ, MA, and LG revised the manuscript. LW and CM participated in the sequence alignment. MZ, WX, and WM performed the statistical analysis. CZ agrees to serve as the author responsible for contact and ensures communication. All authors reviewed and approved the final submission.
The study was supported by grants from National Postdoctoral Program for Innovative Talents (Grant No. BX20180199), China Postdoctoral Science Foundation (Grant No. 2018M642028), the Special Funds of Modern Industrial Technology System for Agriculture (Grant No. CARS-29-zp-7), National Key Research and Development Plan project (Grant No. 2018YFD0201305).
WM was employed by the company Agriculture, Forestry and Animal Husbandry Technology Promotion Service Center of Ningxia Agricultural Group.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The reviewer YXP and handling Editor declared their shared affiliation.
We thank the reviewers and editor for their careful reading and helpful comments on this manuscript.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2019.01276/full#supplementary-material
Aguilar Martinez, J. A., Sinha, N. R. (2013). Analysis of the role of Arabidopsis class I TCP genes AtTCP7, AtTCP8, AtTCP22 and AtTCP23 in leaf development. Front. Plant Sci. 4, 406. doi: 10.3389/fpls.2013.00406
Aguilar-Martinez, J. A., Poza-Carrion, C., Cubas, P. (2007). Arabidopsis branched1 acts as an integrator of branching signals within axillary buds. Plant Cell Online 19 (2), 458–472. doi: 10.1105/tpc.106.048934
Aukerman, M. J., Sakai, H. (2003). Regulation of flowering time and floral organ identity by a microRNA and its APETALA2-like target genes. Plant Cell 15 (11), 2730–2741. doi: 10.1105/tpc.016238
Bailey, T. L., Elkan, C. (1994). Fitting a mixture model by expectation maximization to discover motifs in biopolymers (Menlo Park.: The AAAI Press), 28–36.
Balsemao-Pires, E., Andrade, L. R., Sachetto-Martins, G. (2013). Functional study of TCP23 in Arabidopsis thaliana during plant development. Plant Physiol. 67, 120–125. doi: 10.1016/j.plaphy.2013.03.009
Bari, R., Jones, J. D. (2009). Role of plant hormones in plant defence responses. Plant Mol. Biol. 69, 473–488. doi: 10.1007/s11103-008-9435-0
Boss, P. K., Davies, C., Robinson, S. P. (1996). Analysis of the expression of anthocyanin pathway genes in developing Vitis vinifera L. cv shiraz grape berries and the implications for pathway regulation. Plant Physiol. 111, 1059–1066. doi: 10.1104/pp.111.41059
Braun, N., de Saint Germain, A., Pillot, J., Boutet-Mercey, S., Dalmais, M., Antoniadi, I., et al. (2012). The pea TCP transcription factor PsBRC1 acts downstream of strigolactones to control shoot branching. Plant Physiol. 158, 225–238. doi: 10.1104/pp.111.182725
Crawford, B., Nath, U., Carpenter, R., Coen, E. (2004). CINCINNATA controls both cell differentiation and growth in petal lobes and leaves of Antirrhinum. Plant Physiol. 135, 244–253. doi: 10.1104/pp.103.036368
Cubas, P., Lauter, N., Doebley, J., Coen, E. (1999). The TCP domain: a motif found in proteins regulating plant growth and development. Plant J. 18, 215–222. doi: 10.1046/j.1365-313X.1999.00444.x
Danisman, S., van der Wal, F., Dhondt, S., Waites, R., de Folter, S., Bimbo, A., et al. (2012). Arabidopsis class I and class II TCP transcription factors regulate jasmonic acid metabolism and leaf development antagonistically. Plant Physiol. 159, 1511–1523. doi: 10.1104/pp.112.200303
Doebley, J., Stec, A., Hubbard, L. (1997). The evolution of apical dominance in maize. Nature 386, 485–488. doi: 10.1038/386485a0
Du, J., Hu, S., Yu, Q., Wang, C., Yang, Y., Sun, H., et al. (2017). Genome-wide identification and characterization of BrrTCP transcription factors in Brassica rapa ssp. rapa. Front. Plant Sci. 8, 1588. doi: 10.3389/fpls.2017.01588
Efroni, I., Blum, E., Goldshmidt, A., Eshed, Y. (2008). A protracted and dynamic maturation schedule underlies Arabidopsis leaf development. Plant Cell 20, 2293–2306. doi: 10.1105/tpc.107.057521
Emanuelsson, O., Nielsen, H., Brunak, S., von Heijne, G. (2000). Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J. Mol. Biol. 300, 1005–1016. doi: 10.1006/jmbi.20003903
Fasoli, M., Santo, S. D., Zenoni, S., Tornielli, G. B., Farina, L., Zamboni, A., et al. (2012). The grapevine expression atlas reveals a deep transcriptome shift driving the entire plant into a maturation program. Plant Cell 24, 3489–3505. doi: 10.1105/tpc.112.100230
Fung, R. W., Gonzalo, M., Fekete, C., Kovacs, L. G., He, Y., Marsh, E., et al. (2008). Powdery mildew induces defense-oriented reprogramming of the transcriptome in a susceptible but not in a resistant grapevine. Plant Physiol. 146 (1), 236–249. doi: 10.1104/pp.107.108712
Giraud, E., Ng, S., Carrie, C., Duncan, O., Low, J., Lee, C. P., et al. (2010). TCP transcription factors link the regulation of genes encoding mitochondrial proteins with the circadian clock in Arabidopsis thaliana. Plant Cell 22, 3921–3934. doi: 10.1105/tpc.110.074518
Guo, Z., Fujioka, S., Blancaflor, E. B., Miao, S., Gou, X., Li, J. (2010). TCP1 modulates brassinosteroid biosynthesis by regulating the expression of the key biosynthetic gene DWARF4 in Arabidopsis thaliana. Plant Cell 22, 1161–1173. doi: 10.1105/tpc.109.069203
Hammani, K., Gobert, A., Hleibieh, K., Choulier, L., Small, I., Giegé, P. (2011). An Arabidopsis dual-localized pentatricopeptide repeat protein interacts with nuclear proteins involved in gene expression regulation. Plant Cell 23 (2), 730–740. doi: 10.1105/tpc.110.081638
Hong, Y., Jackson, S. (2015). Floral induction and flower formation–the role and potential applications of miRNAs. Plant Biotechnol. J. 13, 282–292. doi: 10.1111/pbi.12340
Hu, B., Jin, J., Guo, A. Y., Zhang, H., Luo, J., Gao, G. (2014). GSDS 2.0: an upgraded gene feature visualization server. Bioinformatics 31 (8), 1296–1297. doi: 10.1093/bioinformatics/btu817
Ibraheem, O., Botha, C. E., Bradley, G. (2010). In silico analysis of cis-acting regulatory elements in 5' regulatory regions of sucrose transporter gene families in rice (Oryza sativa Japonica) and Arabidopsis thaliana. Comput. Biol. Chem. 34 (5), 268–283. doi: 10.1016/j.compbiolchem.2010.09.003
Jaillon, O., Aury, J. M., Noel, B., Policriti, A., Clepet, C., Casagrande, A., et al. (2007). The grapevine genome sequence suggests ancestral hexaploidization in major angiosperm phyla. Nature 449, 463–467. doi: 10.1038/nature06148
Jiu, S., Zhu, X., Wang, J., Zhang, C., Mu, Q., Wang, C., et al. (2015). Genome-wide mapping and analysis of grapevine microRNAs and their potential target genes. Plant Genome 8 (2), 127–132. doi: 10.3835/plantgenome2014.120091
Jiu, S., Wang, C., Zheng, T., Liu, Z., Leng, X., Pervaiz, T., et al. (2016). Characterization of VvPAL-like promoter from grapevine using transgenic tobacco plants. Funct. Integr. Genomics 16 (6), 595–617. doi: 10.1007/s10142-016-0516-x
Jiu, S., Haider, M. S., Kurjogi, M. M., Zhang, K., Zhu, X., Fang, J. (2018). Genome-wide characterization and expression analysis of sugar transporter family genes in woodland strawberry. Plant Genome 11 (3). doi: 10.3835/plantgenome2017.110103
Juntheikki-Palovaara, I., Tahtiharju, S., Lan, T., Broholm, S. K., Rijpkema, A. S., Ruonala, R., et al. (2014). Functional diversification of duplicated CYC2 clade genes in regulation of inflorescence development in Gerbera hybrida (Asteraceae). Plant J. Cell Mol. Biol. 79, 783–796. doi: 10.1111/tpj.12583
Kieffer, M., Master, V., Waites, R., Davies, B. (2011). TCP14 and TCP15 affect internode length and leaf shape in Arabidopsis. Plant J. 68, 147–158. doi: 10.1111/j.1365-313X.2011.04674.x
Kosugi, S., Ohashi, Y. (1997). PCF1 and PCF2 specifically bind to cis-elements in the rice proliferating cell nuclear antigen gene. Plant Cell 9, 1607–1619. doi: 10.1105/tpc.9.91607
Koyama, T., Mitsuda, N., Seki, M., Shinozaki, K., Ohme-Takagi, M. (2010). TCP transcription factors regulate the activities of ASYMMETRIC LEAVES1 and miR164, as well as the auxin response, during differentiation of leaves in Arabidopsis. Plant Cell 22 (11), 3574–3588. doi: 10.1105/tpc.110.075598
Krzywinski, M., Schein, J., Birol, I., Connors, J., Gascoyne, R., Horsman, D., et al. (2009). Circos: an information aesthetic for comparative genomics. Genome Res. 19 (9), 1639–1645. doi: 10.1101/gr.092759.109
Lescot, M., Déhais, P., Thijs, G., Marchal, K., Moreau, Y., de Peer, Y., et al. (2002). PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 30 (1), 325–327. doi: 10.1093/nar/30.1.325
Li, C., Potuschak, T., Colón-Carmona, A., Gutiérrez, R. A., Doerner, P. (2005). Arabidopsis TCP20 links regulation of growth and cell division control pathways. Proc. Natl. Acad. Sci. 102 (36), 12978–12983. doi: 10.1073/pnas.0504039102
Li, Z., Li, B., Shen, W. H., Huang, H., Dong, A. (2012). TCP transcription factors interact with AS2 in the repression of class-I KNOX genes in Arabidopsis thaliana. Plant J. 71, 99–107. doi: 10.1111/j.1365-313X.2012.04973.x
Li, S. (2015). The Arabidopsis thaliana TCP transcription factors: A broadening horizon beyond development. Plant Signaling Behav. 10, e1044192. doi: 10.1080/15592324.2015.1044192
Liu, Y., Guan, X., Liu, S., Yang, M., Ren, J., Guo, M., et al. (2018). Genome-wide identification and analysis of TCP transcription factors involved in the formation of leafy head in Chinese cabbage. Int. J. Mol. Sci. 19 (3), 847. doi: 10.3390/ijms19030847
Lupas, A., Vandyke, M., Stock, J. (1991). Predicting coiled coils from protein sequences. Science 252, 1162–1164. doi: 10.1126/science.252.50091162
Lynch, M., Conery, J. S. (2000). The evolutionary fate and consequences of duplicate genes. Science 290 (5494), 1151–1155. doi: 10.1126/science.290.54941151
Ma, J., Wang, Q., Sun, R., Xie, F., Jones, D. C., Zhang, B. (2014). Genome-wide identification and expression analysis of TCP transcription factors in Gossypium raimondii. Sci. Rep. 4, 6645. doi: 10.1038/srep06645
Ma, X., Ma, J., Fan, D., Li, C., Jiang, Y., Luo, K. (2016). Genome-wide identification of TCP family transcription factors from Populus Euphratica and their involvement in leaf shape regulation. Sci. Rep. 6, 32795. doi: 10.1038/srep32795
Maher, C., Stein, L., Ware, D. (2006). Evolution of Arabidopsis microRNA families through duplication events. Genome Res. 16, 510–519. doi: 10.1101/gr.4680506
Manassero, N. G., Viola, I. L., Welchen, E., Gonzalez, D. H. (2013). TCP transcription factors: architectures of plant form. Biomolecular Concepts 4, 111–127. doi: 10.1515/bmc-2012-0051
Mao, Y., Wu, F., Yu, X., Bai, J., Zhong, W., He, Y. (2014). MicroRNA319a-targeted Brassica rapa ssp. pekinensis TCP genes modulate head shape in Chinese cabbage by differential cell division arrest in leaf regions. Plant Physiol. 164, 710–720. doi: 10.1104/pp.113.228007
Martin-Trillo, M., Cubas, P. (2010). TCP genes: a family snapshot ten years later. Trends Plant Sci. 15, 31–39. doi: 10.1016/j.tplants.2009.11.003
Masuda, H., Cabral, L., De Veylder, L., Tanurdzic, M., de Almeida, E. J., Geelen, D., et al. (2008). ABAP1 is a novel plant Armadillo BTB protein involved in DNA replication and transcription. EMBO J. 27, 2746–2756. doi: 10.1038/emboj.2008.191
Minakuchi, K., Kameoka, H., Yasuno, N., Umehara, M., Luo, L., Kobayashi, K., et al. (2010). FINE CULM1 (FC1) works downstream of strigolactones to inhibit the outgrowth of axillary buds in rice. Plant Cell Physiol. 51, 1127–1135. doi: 10.1093/pcp/pcq083
Moore, R. C., Purugganan, M. D. (2003). The early stages of duplicate gene evolution. Proc. Natl. Acad. Sci. 100 (26), 15682–15687. doi: 10.1073/pnas.2535513100
Nag, A., King, S., Jack, T. (2009). MiR319a targeting of TCP4 is critical for petal growth and development in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 106, 22534–22539. doi: 10.1073/pnas.0908718106
Navaud, O., Dabos, P., Carnus, E., Tremousaygue, D., Herve, C. (2007). TCP transcription factors predate the emergence of land plants. J. Mol. Evol. 65, 23–33. doi: 10.1007/s00239-006-0174-z
Ogrodzki, P., Forsythe, S. (2015). Capsular profiling of the Cronobacter genus and the association of specific Cronobacter sakazakii and C. malonaticus capsule types with neonatal meningitis and necrotizing enterocolitis. BMC Genomics 16 (1), 1–15. doi: 10.1186/s12864-015-1960-z
Ori, N., Cohen, A., Etzioni, A., Brand, A., Yanai, O., Shleizer, S., et al. (2007). Regulation of LANCEOLATE by miR319 is required for compound-leaf development in tomato. Nat. Genet. 39, 787–791. doi: 10.1038/ng2036
Palatnik, J. F., Allen, E., Wu, X., Schommer, C., Schwab, R., Carrington, J. C., et al. (2003). Control of leaf morphogenesis by microRNAs. Nature 425, 257–263. doi: 10.1038/nature01958
Palatnik, J. F., Wollmann, H., Schommer, C., Schwab, R., Boisbouvier, J., Rodriguez, R., et al. (2007). Sequence and expression differences underlie functional specialization of Arabidopsis microRNAs miR159 and miR319. Dev. Cell 13, 115–125. doi: 10.1016/j.devcel.2007.04.012
Parapunova, V., Busscher, M., Busscher-Lange, J., Lammers, M., Karlova, R., Bovy, A. G., et al. (2014). Identification, cloning and characterization of the tomato TCP transcription factor family. BMC Plant Biol. 14, 157. doi: 10.1186/1471-2229-14-157
Peng, Y., Chen, L., Lu, Y., Wu, Y., Dumenil, J., Zhu, Z., et al. (2015). The ubiquitin receptors DA1, DAR1, and DAR2 redundantly regulate endoreduplication by modulating the stability of TCP14/15 in Arabidopsis. Plant Cell 27, 649–662. doi: 10.1105/tpc.114.132274
Resentini, F., Felipo-Benavent, A., Colombo, L., Blazquez, M. A., Alabadi, D., Masiero, S. (2015). TCP14 and TCP15 mediate the promotion of seed germination by gibberellins in Arabidopsis thaliana. Mol. Plant 8, 482–485. doi: 10.1016/j.molp.2014.11.018
Rueda-Romero, P., Barrero-Sicilia, C., Gomez-Cadenas, A., Carbonero, P., Onate-Sanchez, L. (2012). Arabidopsis thaliana DOF6 negatively affects germination in non-after-ripened seeds and interacts with TCP14. J. Exp. Bot. 63, 1937–1949. doi: 10.1093/jxb/err388
Schommer, C., Palatnik, J. F., Aggarwal, P., Chételat, A., Cubas, P., Farmer, E. E., et al. (2008). Control of jasmonate biosynthesis and senescence by miR319 targets. PloS Biol. 6 (9), e230. doi: 10.1371/journal.pbio.0060230
Schommer, C., Bresso, E. G., Spinelli, S. V., Palatnik, J. F. (2012). “Role of microRNA miR319 in plant development,” in MicroRNAs in plant development and stress responses. (Stillwater, USA: Springer Publisher, Oklahoma State University) 29–47. doi: 10.1007/978-3-642-27384-1_2
Shi, P., Guy, K. M., Wu, W., Fang, B., Yang, J., Zhang, M., et al. (2016). Genome-wide identification and expression analysis of the ClTCP transcription factors in Citrullus lanatus. BMC Plant Biol. 16, 85. doi: 10.1186/s12870-016-0765-9
Takeda, T., Suwa, Y., Suzuki, M., Kitano, H., Ueguchi-Tanaka, M., Ashikari, M., et al. (2003). The OsTB1 gene negatively regulates lateral branching in rice. Plant J. Cell Mol. Biol. 33, 513–520. doi: 10.1046/j.1365-313X.2003.01648.x
Tatematsu, K., Nakabayashi, K., Kamiya, Y., Nambara, E. (2008). Transcription factor AtTCP14 regulates embryonic growth potential during seed germination in Arabidopsis thaliana. Plant J. Cell Mol. Biol. 53, 42–52. doi: 10.1111/j.1365-313X.2007.03308.x
Vadde, B. V. L., Challa, K. R., Nath, U. (2018). The TCP 4 transcription factor regulates trichome cell differentiation by directly activating GLABROUS INFLORESCENCE STEMS in Arabidopsis thaliana. Plant J. 93 (2), 259–269. doi: 10.1111/tpj.13772
Velasco, R., Zharkikh, A., Troggio, M., Cartwright, D. A., Cestaro, A., Pruss, D., et al. (2007). A high quality draft consensus sequence of the genome of a heterozygous grapevine variety. PloS One 2 (12), e1326. doi: 10.1371/journal.pone.0001326
Viola, I. L., Uberti Manassero, N. G., Ripoll, R., Gonzalez, D. H. (2011). The Arabidopsis class I TCP transcription factor AtTCP11 is a developmental regulator with distinct DNA-binding properties due to the presence of a threonine residue at position 15 of the TCP domain. Biochem. J. 435, 143–155. doi: 10.1042/BJ20101019
Wang, K., Wang, Z., Li, F., Ye, W., Wang, J., Song, G., et al. (2012). The draft genome of a diploid cotton Gossypium raimondii. Nat. Genet. 44 (10), 1098. doi: 10.1038/ng2371
Wei, W., Hu, Y., Cui, M., Han, Y., Gao, K., Feng, J. (2016). Identification and transcript analysis of the tcp transcription factors in the diploid woodland strawberry Fragaria vesca. Front. Plant Sci. 07,1937. doi: 10.3389/fpls.2016.01937
Xu, S., Luo, Y., Cai, Z., Cao, X., Hu, X., Yang, J., et al. (2013). Functional diversity of CYCLOIDEA-like TCP genes in the control of zygomorphic flower development in Lotus japonicus. J. Integr. Plant Biol. 55, 221–231. doi: 10.1111/j.1744-7909.2012.01169.x
Xu, R., Sun, P., Jia, F., Lu, L., Li, Y., Zhang, S., et al. (2014). Genome-wide analysis of TCP transcription factor gene family in Malus domestica. J. Genet. 93, 733–746. doi: 10.1007/s12041-014-0446-0
Xu, Z., Sun, L., Zhou, Y., Yang, W., Cheng, T., Wang, J., et al. (2015). Identification and expression analysis of the SQUAMOSA promoter-binding protein (SBP)-box gene family in Prunus Mume. Mol. Genet. Genomics 290, 1701–1715. doi: 10.1007/s00438-015-1029-3
Yao, X., Ma, H., Wang, J., Zhang, D. (2007). Genome-wide comparative analysis and expression pattern of TCP gene families in Arabidopsis thaliana and Oryza sativa. J. Integr. Plant Biol. 49, 885–897. doi: 10.1111/j.1744-7909.2007.00509.x
Keywords: genome-wide analysis, TCP gene family, phylogeny analysis, expression pattern, grapevine
Citation: Jiu S, Xu Y, Wang J, Wang L, Wang S, Ma C, Guan L, Abdullah M, Zhao M, Xu W, Ma W and Zhang C (2019) Genome-Wide Identification, Characterization, and Transcript Analysis of the TCP Transcription Factors in Vitis vinifera. Front. Genet. 10:1276. doi: 10.3389/fgene.2019.01276
Received: 30 August 2019; Accepted: 19 November 2019;
Published: 20 December 2019.
Edited by:
Xiyin Wang, North China University of Science and Technology, ChinaReviewed by:
Milind B. Ratnaparkhe, ICAR Indian Institute of Soybean Research, IndiaCopyright © 2019 Jiu, Xu, Wang, Wang, Wang, Ma, Guan, Abdullah, Zhao, Xu, Ma and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Caixi Zhang, YWNhaXpoQHNqdHUuZWR1LmNu
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.