- 1National Risk Assessment Laboratory for Antimicrobial Resistance of Animal Original Bacteria, South China Agricultural University, Guangzhou, China
- 2Laboratory of Veterinary Pharmacology, College of Veterinary Medicine, South China Agricultural University, Guangzhou, China
- 3Guangdong Provincial Key Laboratory of Veterinary Pharmaceutics Development and Safety Evaluation, South China Agricultural University, Guangzhou, China
Salmonella enterica serotype Typhimurium is a major global food-borne pathogen and causes life-threatening infections. Although the resistance mechanisms to fluoroquinolones in S. Typhimurium had been well-defined, tolerance to fluoroquinolones and the associated mechanism for this are obscure. In the current work, we investigated an oqxAB-positive plasmid pHXY0908 and analyzed its role in S. Typhimurium tolerance to ciprofloxacin using time-kill, transcriptome sequencing and real-time PCR. S. Typhimurium ATCC14028 could survive under lethal concentrations of ciprofloxacin after acquiring plasmid pHXY0908. Transcriptome sequence analysis showed the chromosomal genes were systematically regulated after acquiring this plasmid suggesting an interaction between chromosome and plasmid. Additionally, the chromosomal efflux pump genes acrB, acrA, tolC, and yceE were up-regulated after acquiring plasmid pHXY0908 suggesting that these efflux pumps may contribute to the survival of ATCC14028 exposed to the lethal concentrations of ciprofloxacin. In conclusion, this is the first known report demonstrating that an IncHI2 type plasmid harboring oqxAB could assist S. Typhimurium survival under lethal concentrations of ciprofloxacin.
Introduction
Salmonella enterica Typhimurium is a major global food-borne pathogen, causing a wide spectrum of human and animal diseases including acute gastroenteritis, bacteremia, and extra intestinally localized infections involving many organs (Coburn et al., 2007). Poultry, pigs, cattle, and reptiles are S. Typhimurium reservoirs and humans generally become infected by eating undercooked or contaminated food (Gomez et al., 1997).
Although intestinal infections caused by non-typhoid Salmonella are usually self-limiting, effective antimicrobial therapy is essential if invasive infection occurs (Hohmann, 2011). The extensive use of antimicrobials in humans and animals has led to an increase in multi-drug resistance among numerous bacterial strains. In particular, multidrug resistant (MDR) Salmonella isolates such as S. Typhimurium monophasic variant (S. 4,[5],12:i:-), S. Typhimurium DT104 and S. Rissen are major global public health problems (Threlfall, 2000; Hopkins et al., 2010; Mather et al., 2013; Gomes-Neves et al., 2014). Due to increasing resistance to the conventional antimicrobial agents such as ampicillin, chloramphenicol, and trimethoprim/sulfonamides, fluoroquinolones such as ciprofloxacin for the treatment of severe invasive salmonellosis has become more common (Hohmann, 2011). The use of fluoroquinolones has also led to a rapid increase in reduced susceptibility of S. Typhimurium to these therapeutics. MDR S. Typhimurium with reduced ciprofloxacin susceptibility has become common in China (Li et al., 2013a; Wong and Chen, 2013).
Resistance to fluoroquinolones is mainly due to point mutations in the quinolone resistance-determining region (QRDR) of the gyrase (gyrA and gyrB) and topoisomerase IV (parC and parE) genes. The efflux pump AcrAB-TolC can decrease susceptibility to drugs from seven antibiotic classes such as fluoroquinolones, tetracyclines, rifamycins, oxazolidinones, macrolides, and so on (Schuster et al., 2017). The efflux function of AcrAB-TolC is not only used for antibiotics but also small molecules, such as metabolins, dyes, detergents, organic solutes and even bile salts (Nikaido, 1996; Pos, 2009; Oswald et al., 2016). In addition, plasmid-mediated quinolone resistance (PMQR), including derivatives of quinolone resistance proteins (Qnr), aminoglycoside acetyltransferase Aac(6′)-Ib-cr, and quinolone efflux pumps QepA and OqxAB, have also been described in fluoroquinolone-resistant S. Typhimurium isolates (Strahilevitz et al., 2009; Poirel et al., 2012). Additionally, tolerance to ciprofloxacin has been observed that was primarily linked to mutations in gyrA and parC (Dahiya et al., 2014). Even though tolerance of Salmonella is most often attributed to the action of efflux pumps the evidence linking PMQR genes and fluoroquinolone tolerance is scant (Webber et al., 2006; Thorrold et al., 2007).
Plasmids play an important role in the dissemination of antimicrobial resistance genes (Carattoli, 2009) and different incompatibility group plasmids have been examined for their roles in MDR S. Typhimurium (Fernandez et al., 2007; Zaidi et al., 2011). In our previous study, we identified that spread of oqxAB was predominately due to transferable MDR IncHI2 pHXY0908-like plasmids in S. Typhimurium (Li et al., 2013a). Interestingly, we found that possession of plasmid pHXY0908 was correlated with the treatment failure to avian salmonellosis using enrofloxacin at a routine dosage. This was in spite of the fact that this plasmid could directly confer only low-level fluoroquinolone resistance (Chen et al., 2016). In the current study, we examined potential mechanisms involved in S. Typhimurium survival to the ciprofloxacin selective pressure after acquisition of plasmid pHXY0908.
Materials and Methods
Bacterial Strains
S. Typhimurium ATCC14028 and S. Typhimurium ATCC14028-bearing plasmid pHXY0908 (ATCC14028-pHXY0908) were used as the test strains in the present study. IncHI2 plasmid pHXY0908 harboring oqxAB confers a multi-drug resistance phenotype. The ATCC14028-bearing plasmid pHXY0908 were obtained by electroporation of the transferable pHXY0908 into S. Typhimurium ATCC14028 as previously described (Chen et al., 2016, 2017).
Minimum Inhibitory Concentration (MIC) and Mutant Prevention Concentration (MPC) Determinations
The MIC of ciprofloxacin was determined for S. Typhimurium ATCC14028 and S. Typhimurium ATCC14028-bearing plasmid pHXY0908 by the standard broth microdilution methods according to the recommendations of the Clinical and Laboratory Standards Institute (CLSI) (M100-S25). The breakpoint criteria used to determine ciprofloxacin phenotype in Salmonella spp. was based on the CLSI breakpoint criteria [ ≤ 0.06 μg/mL (susceptible), 0.12–0.5 μg/mL (intermediate), and ≥1 μg/mL (resistant)]. E. coli ATCC25922 was used as a quality control strain.
The MPC values were determined as previously described (Allou et al., 2009). In summary, each strain was grown at 37°C in antibiotic-free Mueller Hinton (MH) broth for ~6 h, until an OD600 of ~1.0 was reached (corresponding to ~109 CFU/mL). Cultures (20 mL) were centrifuged at 4,000 × g for 15 min. The supernatant was discarded and the pellet containing ~1010 CFU/mL was suspended in 2 mL of sterile MH broth. MH agar plates containing ciprofloxacin at levels ranging from 0.002 to 32 μg/mL (diluted in log2 series) against each strain were inoculated with 100 μL of cell suspension and incubated at 37°C for 96 h. The MPC was recorded as the lowest ciprofloxacin concentration at which no colonies grew on an agar plate after 96 h.
Time-Kill Experiments
Time-kill curve kinetics assays were conducted using MH broth containing ciprofloxacin levels equaling 1 × MIC, 2 × MIC, 4 × MIC, and 8 × MIC of the strains tested. Antibiotic-free broth was evaluated in parallel as a control. Cultures were incubated at 37°C with shaking. Viable counts were determined by serial dilution after 0, 3, 6, 9, and 24 h of incubation and by plating 100 μL of the control, test cultures, or with dilutions at the indicated times onto MH agar plates. Colony counts were determined after 24 h of incubation.
Plasmid Sequencing
DNA of plasmid pHXY0908 was sequenced using the Single Molecule Real Time (SMRT) DNA Sequencing approach. After filtering S. Typhimurium ATCC14028 chromosomal DNA data, the remaining reads were assembled by HGAP2.2.0 method (Chin et al., 2013). Open reading frames (ORF) were predicted using ORF Finder (http://www.ncbi.nlm.nih.gov/gorf/gorf.html) and annotation was performed using RAST tools (Aziz et al., 2008). The sequence comparison and map generation was performed using BLAST (http://blast.ncbi.nlm.nih.gov) and Easyfig version 2.1 (Sullivan et al., 2011). The annotated sequence of pHXY0908 has been submitted to the GenBank nucleotide sequence database under the accession number KM877269.
Transcriptome Sequencing and Sequence Analysis
ATCC14028 and ATCC14028-pHXY0908 were cultured in LB broth with and without 1/2 × MIC levels of ciprofloxacin. Total RNA was extracted as previously described (Li et al., 2013b). A pooled sample from 3 independent experiments was used for RNA-seq. Ribo-Zero rRNA Removal Kit (Gram-Negative Bacteria) (Epicentre, Madison, WI, USA) was used to remove rRNA from total bacterial RNA. The library was constructed using an Illumina TruSeq RNA sample Prep Kit v2 as previously described (Wang et al., 2013). In brief, mRNA was fragmented into lengths of 200~300 bp and first and second strand cDNA was synthesized. The short cDNA fragments were purified and end repaired and tailed with single A (adenine) addition. Adapters were ligated to the A-tailed cDNA fragments and ligated. These cDNA fragments were enriched by 12 PCR cycles. Purified libraries were quantified using a by Qubit 2.0 Fluorometer (Invitrogen, Carlsbad, CA, USA) and validated using an Agilent 2100 Bioanalyzer (Agilent, Beijing, China). Libraries were sequence by the Illumina Hiseq-2000 for 90 cycles. The reads that passed the Illumina quality filter were kept for sequence analysis.
High quality reads were mapped to S. Typhimurium strain 14028 genome (downloaded from https://www.ncbi.nlm.nih.gov/nuccore/267991652/) and plasmid pHXY09080 sequence by using SOAP aligner/SOAP2 (Li et al., 2009) with 5 max alignment error. The mRNA abundance was normalized using rpkM (reads per kilobase per million reads) (Mortazavi et al., 2008). Gene differential expression analysis was performed as previously described (Audic and Claverie, 1997). The genes with <0.001 FDR and >2-fold change or <0.5-fold were detected as differentially expressed genes (DEG). The RNA-seq data had been submitted to SRA database (Accession number: PRJNA544622, https://www.ncbi.nlm.nih.gov/Traces/study/?acc=PRJNA544622).
Gene Ontology Enrichment Analysis, Kyoto Encyclopedia of Genes and Genomes (KEGG) Pathway Enrichment Analysis, and Functional Protein Association Networks Analysis
The DEGs were performed Gene Ontology enrichment analysis by MATLAB bioinformatics toolbox (MathWorks, Natick, MA, USA). The gene ontology annotations were downloaded from Gene Ontology Consortium (http://www.geneontology.org/page/download-go-annotations). KEGG pathway enrichment analysis was performed by ClueGO and CluePedia which are Cytoscape apps (Bindea et al., 2009, 2013). The functional protein association networks were constructed by STRING with high confidence and hided disconnected nodes in the network (Szklarczyk et al., 2017).
Validation the DEGs by qRT-PCR
Total bacterial RNA was obtained and reverse transcribed into cDNA as described (Li et al., 2013b). The qRT-PCR was performed with SYBR Premix Ex Taq (Takara, Dalian, China) in an iQ5 thermal cycler (Biorad, Hercules, CA) according to the manufacturer's instructions. The cycling conditions were as follows: 94°C for 5 min, followed by 35 cycles at 94°C for 1 min, at 55°C for 1 min and 72°C for 1 min with a final step of 72°C for 5 min. Melting curves were read from 60 to 95°C in steps of 1°C. Normalized expression levels of the target gene transcripts were calculated relative to 16S rRNA using the –ΔΔCT method. The primers used for gene amplification are listed in Table S1.
Statistics
All the in vitro experiments described above were repeated at least three times. Geometric means were used to express the results for MICs and MPCs and the means ± standard deviations (SD) were calculated for CFU counts.
Results
MIC, MPC, and Time-Kill Curve Assays
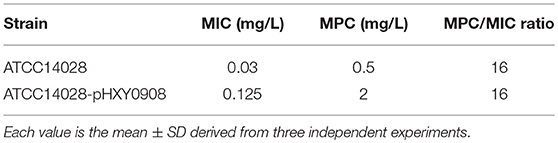
Ciprofloxacin MIC and MPC values were increased by 4-fold when S. Typhimurium ATCC14028 acquired plasmid pHXY0908. However, the MPC/MIC ratios between these strains did not differ (Table 1).
However, time-kill curve assays indicated a survival advantages at 1×, 2×, and 4× ciprofloxacin MIC levels for the plasmid bearing strain compared to the parental strain. In addition, the antibacterial activity of ciprofloxacin significantly decreased over the 3 h antibiotic exposure period against ATCC14028 strain harboring plasmid pHXY0908. Interestingly, the bacterial concentration of ATCC14028-pHXY0908 increased after 3 h and after 6 h under the 1 × MIC. The CFU of ATCC14028-pHXY0908 was almost identical with ATCC14028-pHXY0908 cultured without ciprofloxacin for 24 h (Figure 1).
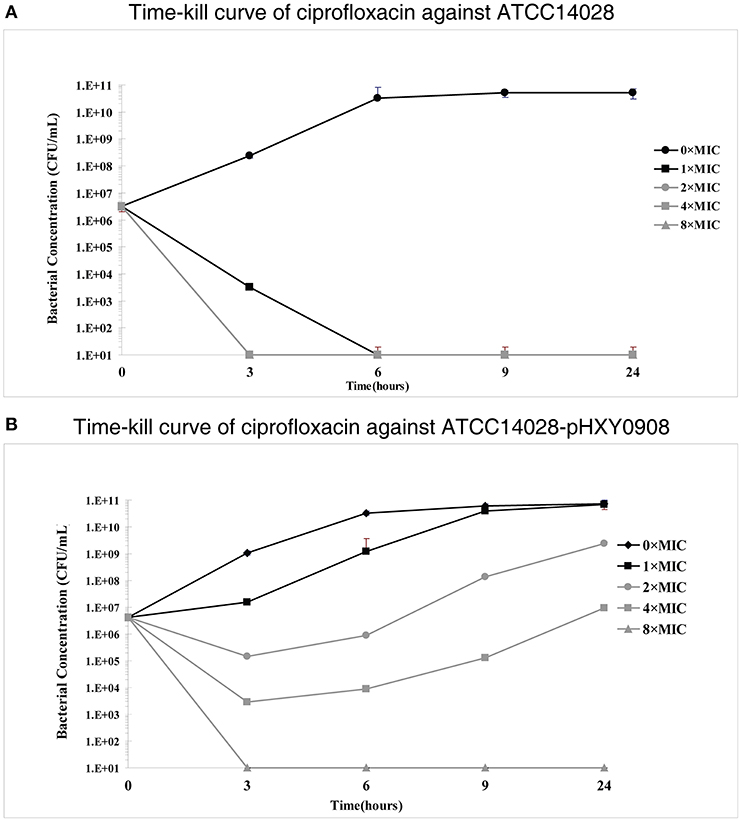
Figure 1. In vitro time-kill studies of ciprofloxacin (A) Salmonella Typhimurium ATCC14028 and (B) and ATCC14028-pHXY0908 1 × MIC is equivalent to 0.03125 mg/L for ATCC14028 and 0.125 mg/L for ATCC14028-pHXY0908. The lines and markers of 2 × MIC and 8 × MIC coincided with 4 × MIC in (A).
General Features of Plasmid pHXY0908
Plasmid pHXY0908 is a circular molecule of 249,144 bp with an average G + C content of 46.22% and harboring 265 predicted ORFs (Figure 2A). The backbone region of pHXY0908 is closely related to other IncHI2 plasmids including pHK0653 (KT334335) from Salmonella enterica, plasmid R478 (BX664015) from Serratia marcescens and pAPEC-O1-R (DQ517526) from an extraintestinal pathogenic Escherichia coli (Figure 2B). These backbone regions contained the replication gene repHI2, the plasmid maintenance and partitioning modules parA-parB and hipAB and two tra transfer regions. In addition to the typical backbone, the 52,876 bp MDR region of pHXY0908 contained 9 IS26 elements flanking four main segments (Figure S1). The first segment containing an oqxAB cassette flanked by IS26 elements was first identified in the IncX1 type plasmid pOLA52 from E. coli. The second segment between IS26-6 and IS26-7 contained the antibiotic resistance genes sul3, aadA1, cmlA, and aadA2 and was identical to plasmid pND11_107 (HQ114281). The third segment containing insertion sequences IS26-7 and IS26-8 and the structure tnp21-floR-sul2-IS4-aacC4-IS26, identical to that of pK1HV (HF545434) from Klebsiella pneumoniae. The fourth segment was similar to plasmids pSTA155 (NG041621) and pAPEC-O1-R (DQ517526) and included aac(6′)-Ib-cr adjacent to an unusual class 1 integron containing blaOXA−1, catB3, arr3, qacEΔ1, and sul1. Importantly, oqxAB, and aac(6′)-Ib-cr could confer low-level resistance to fluoroquinolones.
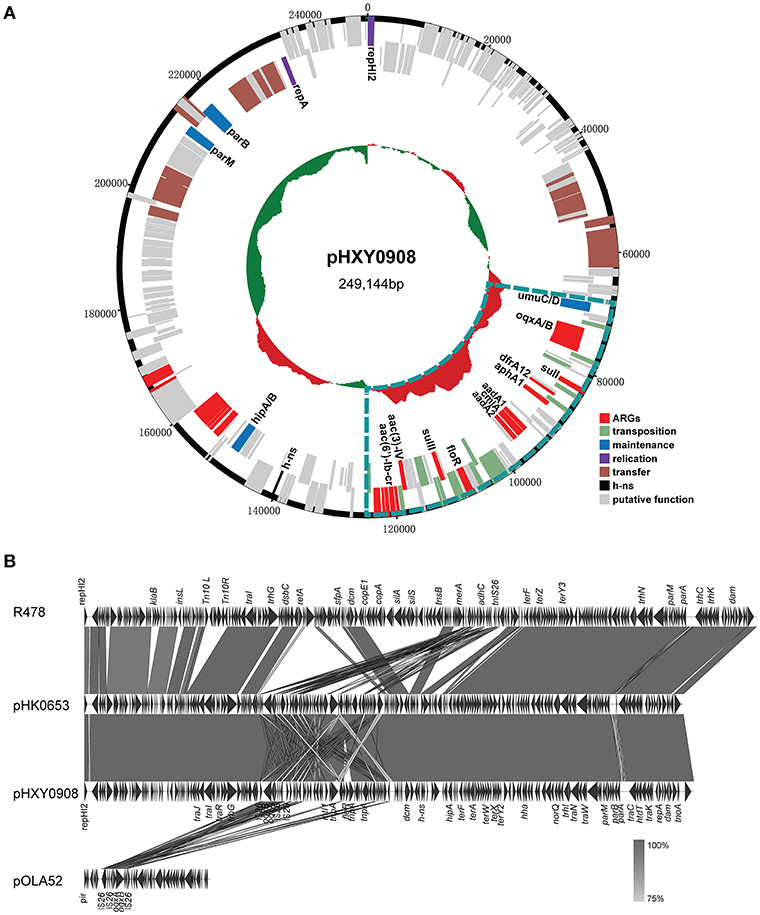
Figure 2. Genetic map and linear comparisons for pHXY0908. (A) Genetic map of pHXY0908. The central region of the map is G+C plot of the nucleotide sequence. Boxes represent ORFs predicted with RAST tools and are color coded after their predicted functions. The blue lines represent the multidrug-resistant regions. (B) Linear comparison of pHXY0908, R478, pHK0653, and oqxAB-positive plasmid pOLA52. The arrows represent ORF position and transcriptional direction. Regions of homology are shaded in gray and functional regions are indicated above and below the linear maps.
Transcriptional Regulation of ATCC14028 After Acquiring pHXY0908
The transcriptional profiles of ATCC14028 with and without pHXY0908 were obtained using transcriptome sequencing. Both strains were cultured in the presence and absence of 1/2 × MIC concentrations of ciprofloxacin. We quantified the levels of 4,687 and 4,671 genes in ATCC14028 and ATCC14028-pHXY0908, respectively. The 283 genes were only expressed in ATCC14028 and the 267 genes were only expressed in ATCC14028-pHXY0908 (Figure 3A). We found 411 chromosomal DEGs between ATCC14028-pHXY0908 and ATCC14028 (Figure 3C). And 223 plasmid genes were expressed in ATCC14028-pHXY0908. The plasmid situated efflux pump oqxA and oqxB genes were highly expressed in ATCC14028-pHXY0908. Interestingly, the chromosomal efflux pump gene acrB was significantly up-regulated (2.5-fold, P <10−32) in the plasmid-bearing strain. The acrA gene which product composed efflux pump with AcrB protein was also slightly up-regulated (1.6 fold, P <10−32). The tolC gene was also significantly up-regulated as same as acrB (2.16 fold, P = 1.5 ×10−11).
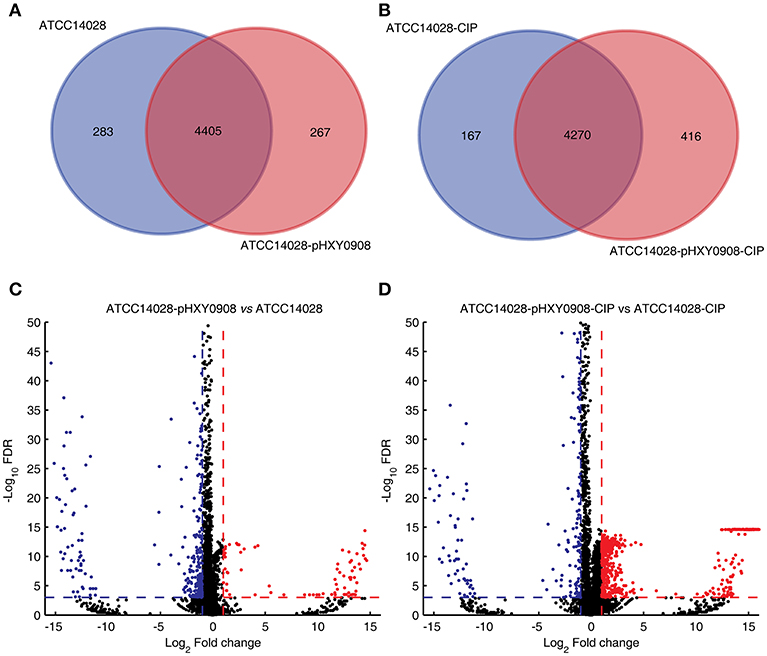
Figure 3. Venn diagram of expressed genes and volcano plot. (A) Venn diagram of expressed genes in ATCC14028 and ATCC14028-pHXY0908. (B) Venn diagram of expressed genes in ATCC14028-CIP and ATCC14028-pHXY0908-CIP. (C) Volcano plot of ATCC14028-pHXY0908 vs. ATCC14028. (D) Volcano plot of ATCC14028-pHXY0908-CIP vs. ATCC14028-CIP. The blue plot in volcano plot represented down-regulated genes (Log2 Fold change ≤ −1 and FDR <0.001). The red plot in volcano plot represented up-regulated genes (Log2 Fold change ≥ 1 and FDR <0.001).
The gene expression profiles of these strains also differed with the strains cultured in the presence of ciprofloxacin. We quantified the expression of 4,473 and 4,686 genes in ATCC14028-CIP and ATCC14028-pHXY0908-CIP, respectively. The 167 genes were only expressed in ATCC14028-CIP and the 416 genes were only expressed in ATCC14028-pHXY0908-CIP (Figure 3B). There were 1029 chromosomal DEGs between ATCC14028-pHXY0908-CIP and ATCC14028-CIP. And 224 plasmid genes were expressed in ATCC14028-pHXY0908-CIP. The acrB and tolC genes were up-regulated in ATCC14028-pHXY0908-CIP, but we found no difference for acrA. Furthermore, we found that many chromosomal efflux pump genes were up-regulated significantly in ATCC14028-pHXY0908-CIP compared with ATCC14028-CIP, for example, ydgF, ydgE, ybjY, yceE, sugE, and yohM (Figure 3D).
The GO and KEGG Enrichment Analysis of Chromosomal DEGs of ATCC14028 After Acquiring pHXY0908
The transcriptional profile of ATCC14028 was changed after acquiring pHXY0908. GO enrichment analysis indicated that chromosomal DEGs of ATCC14028-pHXY0908 vs. ATCC14028 were enriched for some biological processes. These included sulfur incorporation into metallo-sulfur clusters, sulfate assimilation, 6-sulfoquinovose (1-) metabolism, sulfur compounds in catabolic process, type III protein secretion (Figure 4A). Interestingly, six biological processes in the top ten enriched biological processes were related to sulfide. pHXY0908 contained the thiol:disulfide interchange protein DsbC that is responsible for the formation of disulfide bonds and this process is related to sulfur metabolism (Mangold et al., 2011). This suggests that ATCC14028 may regulate genes expression to support the plasmid function. In addition, sulfur-mediated biological process could provide a new way for ciprofloxacin biodegradation (Jia et al., 2018).
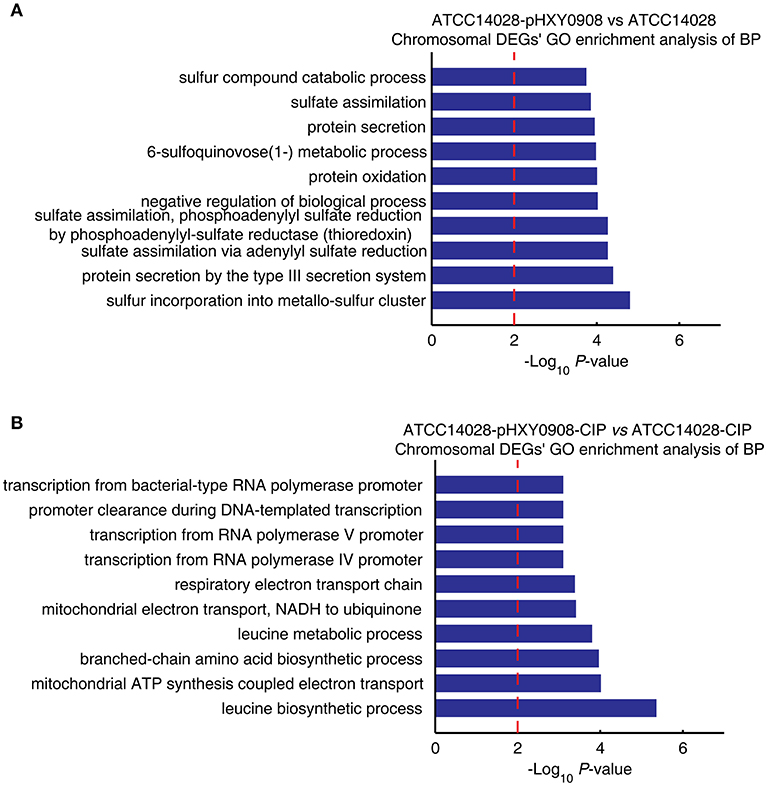
Figure 4. The chromosomal DEGs GO enrichment analysis of biological processes. (A) ATCC14028-pHXY0908 vs. ATCC14028 and (B) ATCC14028-pHXY0908-CIP vs. ATCC14028-CIP (B). The top 10 biological processes are shown. The dotted red line indicates the P-value threshold of 0.01.
The chromosomal DEGs of ATCC14028-pHXY0908-CIP vs. ATCC14028-CIP were enriched in four transcriptional biological processes and two electron transport chain processes (Figure 4B). After acquiring pHXY0908, ATCC14028 could activate transcription of resistance genes including efflux pump genes under ciprofloxacin selective pressure. This indicates that pHXY0908 can influence the gene expression of ATCC14028 to resist ciprofloxacin selective pressure.
The chromosomal DEGs of ATCC14028-pHXY0908 vs. ATCC14028 were enriched in four KEGG pathways, including phosphotransferase system (PTS), fructose and mannose metabolism, amino sugar and nucleotide sugar metabolism and citrate cycle (TCA cycle) (Figure 5A). Additionally, the KEGG pathways enrichment analysis for DEGs of ATCC14028-pHXY0908-CIP vs. ATCC14028-CIP is C5-branched dibasic acid metabolism and phosphotransferase system (PTS) (Figure 5B). The phosphotransferase system (PTS) serves as a complex protein kinase system that regulates a wide variety of transport, metabolic and mutagenic processes as well as the expression of numerous genes (Saier, 2015). That indicated the pHXY0908 influenced the gene expression, the substance metabolism and energy metabolism in the ATCC14028.
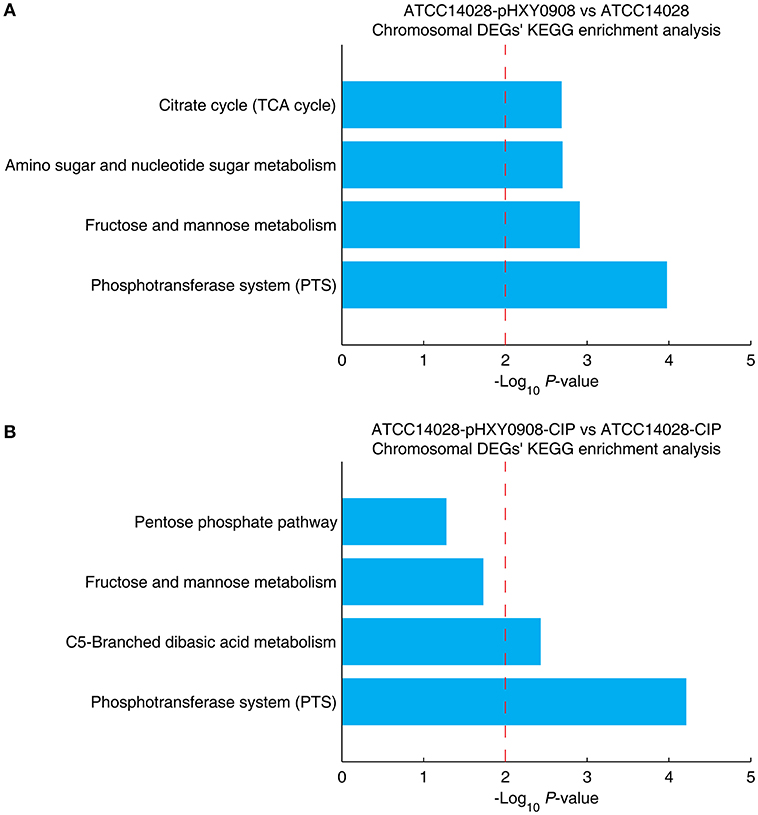
Figure 5. The chromosomal DEGs KEGG pathway enrichment analysis. (A) ATCC14028-pHXY0908 vs. ATCC14028 and (B) ATCC14028-pHXY0908-CIP vs. ATCC14028-CIP (B). The dotted red line indicates the P-value threshold of 0.01.
The Functional Protein Association Networks Analysis for DEGs of ATCC14028 After Acquiring pHXY0908
The functional protein association networks were constructed by chromosomal DEGs of ATCC14028 after acquiring pHXY0908. A majority of DEGs were constructed into two main networks. This suggests pHXY0908 may systematically regulate ATCC14028 functional protein association networks rather than at an independent gene expression level. The largest sub-network that included the genes spal, invA, sicA, and sipB is related to invasion and type III secretion systems. Another sub-network that included cysA, cysH, and cysl was related to sulfur metabolism. These two main functional networks were consistent with the GO enrichment analysis and pHXY0908 function (Figure 6). Additionally, the networks of ATCC14028-pHXY0908-CIP vs. ATCC14028-CIP DEGs also contained the sub-networks related to invasion and type III secretion systems, sulfur metabolism, energy metabolism, and other substance metabolism (Figure S2).
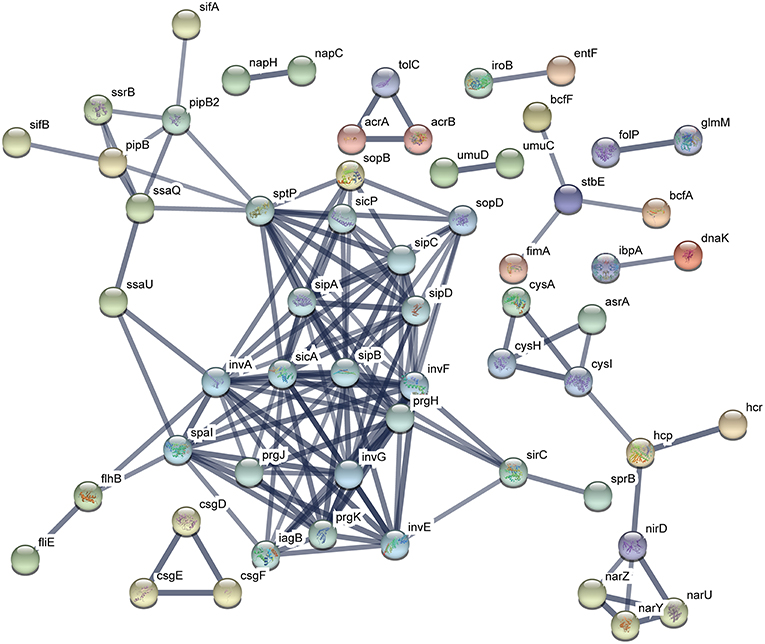
Figure 6. Functional protein association networks of chromosomal DEGs after acquiring pHXY0908. Each node is a protein. Each edge represents protein-protein associations. The thicker edges indicate higher levels of confidence.
RNA-seq Data Validated by qRT-PCR
Five randomly selected DEGs that including dam, csgD, csgE, STM14-1453, and acrB, and five genes which we interested, including acrA, tolC, yceE, sicA, and cysA, were validated by using qRT-PCR. The qRT-PCR results had a good correlation with different expression analysis (Figure S3, R = 0.9793, P = 7.7606 ×10−7 in ATCC14028-pHXY0908 vs. ATCC14028; R = 0.9761, P = 1.3851 ×10−6 in ATCC14028-pHXY0908-CIP vs. ATCC14028-CIP). This indicated that the RNA-seq and the associated data analysis represented actual gene expression changes.
Discussion
This study demonstrated that an MDR IncHI2 type plasmid encoding OqxAB could contribute to the survival under ciprofloxacin selective pressure of S. Typhimurium strains that had acquired plasmid pHXY0908. Although the MIC and MPC increased after acquiring pHXY0908, the same MPC/MIC ratios for both ATCC14028 and ATCC14028-pHXY0908 indicated that pHXY0908 did not increase ability of ciprofloxacin for selecting resistant mutation in ATCC14028. The time-kill curve assays demonstrated that ciprofloxacin concentrations >MIC values for S. Typhimurium ATCC14028 were lethal while the plasmid-bearing strain could survive up to 4 × MIC. Thus, we further explored the implied survival mechanism of S. Typhimurium under drug pressure after acquiring an oqxAB-positive IncHI2 type plasmid. The same MPC/MIC suggested that the genes expression regulation may be the potential mechanism instead of genes mutation.
Firstly, we determined the complete nucleotide sequence of plasmid pHXY0908.
DNA sequencing further confirmed that this plasmid belonged to the IncHI2 group and showed high similarity to the backbone region of R478, one of the prototypes of IncHI2 plasmid groups (Garcia-Fernandez and Carattoli, 2010). In addition to the conserved backbone, pHXY0908 harbored a multidrug resistance region composed of oqxAB, aac(6′)-Ib-cr as well as nine other ARGs and included a wide range of mobile genetic elements and notably, IS26. Except for oqxAB and aac(6′)-Ib-cr, none of the other known genes are involved in fluoroquinolone resistance although many hypothetical proteins were found on pHXY0908. In addition, pHXY0908 showed a surprisingly high degree of homology (100% coverage, 99% identity) with the IncHI2 plasmid pHK0653 from an S. Typhimurium strain of human origin and pHK0653-like plasmids that were identified as the key vectors responsible for oqxAB transmission among Salmonella species (Wong et al., 2016).
Plasmid pHXY0908 harbored diverse ARGs including oqxAB and aac(6′)-Ib-cr that could confer only low-level fluoroquinolone resistance. However, the gene features of pHXY0908 cannot explain the reason for ATCC14028 survival under the lethal concentrations of ciprofloxacin up to 4 × MIC. To study the influence of pHXY0908, we measured the transcriptional profiles of ATCC14028 with and without pHXY0908 cultured in the presence and absence of 1/2 MIC concentrations of ciprofloxacin. ATCC14028 could not grow at >1 × MIC, so we selected a 1/2 MIC level of ciprofloxacin to study the transcriptional regulation of ATCC14028 by pHXY0908. The 1/2 MIC concentrations allowed bacterial growth but also induced stress responses that are sub-MIC and often used in the study of antibiotic resistance (Patkari and Mehra, 2013; Heo et al., 2014; Zhong et al., 2015; Aedo and Tomasz, 2016). The transcriptome data show that 283 chromosomal genes were not expressed after ATCC14028 acquiring pHXY0908. Similarly, there are 167 chromosomal genes were not expressed in ATCC14028-pHXY0908-CIP compared with ATCC14028-CIP. The bacteria will synthesize proteins encoded by plasmid leading the cost of resources. The bacteria may turn off some chromosomal genes expression to fit this situation. These genes were included in DEGs to analyze the function. Due to the ciprofloxacin inhibits the activity of gyrase and topoisomerase IV, we analyzed the expression of gyrA, gyrB, parC, and parE. These four genes were not differently expressed in ATCC14028-pHXY0908 compared with ATCC14028. The gyrA was down-regulated in ATCC14028-pHXY0908-CIP compared with ATCC14028-CIP. However, other three genes were not differently expressed in ATCC14028-pHXY0908-CIP compared with ATCC14028-CIP. The gyrA down-regulated in ATCC14028-pHXY0908-CIP compared with ATCC14028-CIP may due to the resistant proteins encoded by pHXY0908 could decreased the ciprofloxacin pressure in the cell. The GO enrichment analysis of DEGs indicated that pHXY0908 influenced chromosomal gene expression to support plasmid function and resist ciprofloxacin selective pressure. This indicated an interaction between the chromosome and plasmid. And the KEGG pathway enrichment analysis of DEGs indicated that pHXY0908 influence ATCC14028 genes expression and metabolism. Furthermore, the pHXY0908 systematically regulated ATCC14028 functional protein association networks. Intriguingly, the chromosomal efflux pump genes were up-regulated after acquiring the plasmid and these included acrB, acrA, and tolC. Except for AcrAB-TolC efflux pump, the ydgF, ydgE, ybjY, yceE, sugE, and yohM were especially up-regulated when cultured under the 1/2 MIC levels of ciprofloxacin. The multidrug efflux pump AcrAB-tolC system decreases susceptibility to fluoroquinolones (Piddock, 1999) and yceE encoding the multidrug transporter subunit MdtG contributes to fluoroquinolone-resistance (Fàbrega et al., 2010). The ydgF and ydgE encoded MdtI and MdtJ, respectively. These two proteins compose multidrug efflux pump MdtIJ which excretes polyamine (Higashi et al., 2008; Leuzzi et al., 2015). The ybjY codes macrolide transporter subunit MacA. The sugE was reported the contribution for tributyltin (TBT) and antimicrobial resistance (He et al., 2011; Cruz et al., 2013). And the yohM encodes a membrane-bound polypeptide conferring increased nickel and cobalt resistance (Rodrigue et al., 2005). The up-regulation of chromosomal efflux pump genes may be caused by the accumulation of intermediate metabolites for plasmid DNA replication and proteins synthesis. In addition, another potential mechanism is some plasmid proteins regulated chromosomal genes. It is worthy of further investigations. The chromosomal efflux pump up-regulated by pHXY0908 may be the one of the reasons for survival under the lethal concentrations of ciprofloxacin. Excepted the efflux function, the synthesis of chromosomal efflux pump proteins, the activity of efflux pumps and the encoding of plasmid genes may cost much energy, so that the growth of ATCC14028 may be influenced. The slow grown speed lead to the tolerance to fluoroquinolone (Evans et al., 1991; Brauner et al., 2016). The change of ATP level and grown rate of the ATCC14028 after acquiring pHXY0908 may be measured in the further researches. However, the specific molecular mechanism of the tolerance phenotype is not clear. In the further investigations, we will use functional genome technologies combined our plasmid gene expression profiles to detected the plasmid gene caused the tolerance to ciprofloxacin.
Taken together, we found that chromosomal genes were systematically regulated after ATCC14028 acquired the IncHI2 type MDR plasmid pHXY0908. ATCC14028 could survive under the ciprofloxacin lethal concentrations may be attributed to an up-regulation of chromosomal efflux pump genes after acquiring pHXY0908.
Data Availability
The raw data supporting the conclusions of this manuscript will be made available by the authors, without undue reservation, to any qualified researcher.
Author Contributions
XLL carried out the study design, data analysis, and manuscript writing. XW carried out the sequence experiments and was involved in the preparation of the manuscript. XL and JX carried out the RT-qPCR experiments. LF, JS, XPL, and YL contributed to the study design. All authors approved it for publication.
Funding
This work was supported by the International Cooperation and Exchange of the National Natural Science Foundation of China (Grant No. 31520103918), the National Key Research and Development Program of China (2016YFD0501300), and the Foundation for Innovation and Strengthening School Project of Guangdong, China (Grant No. 2016KCXTD010).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2019.00242/full#supplementary-material
References
Aedo, S., and Tomasz, A. (2016). Role of the stringent stress response in the antibiotic resistance phenotype of methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 60, 2311–2317. doi: 10.1128/AAC.02697-15
Allou, N., Cambau, E., Massias, L., Chau, F., and Fantin, B. (2009). Impact of low-level resistance to fluoroquinolones due to qnrA1 and qnrS1 genes or a gyrA mutation on ciprofloxacin bactericidal activity in a murine model of Escherichia coli urinary tract infection. Antimicrob. Agents Chemother. 53, 4292–4297. doi: 10.1128/AAC.01664-08
Audic, S., and Claverie, J. M. (1997). The significance of digital gene expression profiles. Genome Res. 7:986. doi: 10.1101/gr.7.10.986
Aziz, R. K., Bartels, D., Best, A. A., Dejongh, M., Disz, T., Edwards, R. A., et al. (2008). The RAST server: rapid annotations using subsystems technology. BMC Genomics 9, 75–75. doi: 10.1186/1471-2164-9-75
Bindea, G., Galon, J., and Mlecnik, B. (2013). CluePedia Cytoscape plugin: pathway insights using integrated experimental and in silico data. Bioinformatics 29, 661–663. doi: 10.1093/bioinformatics/btt019
Bindea, G., Mlecnik, B., Hackl, H., Charoentong, P., Tosolini, M., Kirilovsky, A., et al. (2009). ClueGO: a Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics 25, 1091–1093. doi: 10.1093/bioinformatics/btp101
Brauner, A., Fridman, O., Gefen, O., and Balaban, N. Q. (2016). Distinguishing between resistance, tolerance and persistence to antibiotic treatment. Nat. Rev. Microbiol. 14, 320–330. doi: 10.1038/nrmicro.2016.34
Carattoli, A. (2009). Resistance plasmid families in Enterobacteriaceae. Antimicrob. Agents Chemother. 53, 2227–2238. doi: 10.1128/AAC.01707-08
Chen, Y., Hu, D., Zhang, Q., Liao, X. P., Liu, Y. H., and Sun, J. (2017). Efflux pump overexpression contributes to tigecycline heteroresistance in Salmonella enterica serovar Typhimurium. Front. Cell Infect. Microbiol. 7:37. doi: 10.3389/fcimb.2017.00037
Chen, Y., Sun, J., Liao, X. P., Shao, Y., Li, L., Fang, L. X., et al. (2016). Impact of enrofloxacin and florfenicol therapy on the spread of OqxAB gene and intestinal microbiota in chickens. Vet. Microbiol. 192, 1–9. doi: 10.1016/j.vetmic.2016.05.014
Chin, C. S., Alexander, D. H., Marks, P., Klammer, A. A., Drake, J., Heiner, C., et al. (2013). Nonhybrid, finished microbial genome assemblies from long-read SMRT sequencing data. Nat. Methods 10:563. doi: 10.1038/nmeth.2474
Coburn, B., Grassl, G. A., and Finlay, B. B. (2007). Salmonella, the host and disease: a brief review. Immunol. Cell Biol. 85, 112–118. doi: 10.1038/sj.icb.7100007
Cruz, A., Micaelo, N., Felix, V., Song, J. Y., Kitamura, S., Suzuki, S., et al. (2013). sugE: a gene involved in tributyltin (TBT) resistance of Aeromonas molluscorum Av27. J. Gen. Appl. Microbiol. 59, 39–47. doi: 10.2323/jgam.59.47
Dahiya, S., Kapil, A., Lodha, R., Kumar, R., Das, B. K., Sood, S., et al. (2014). Induction of resistant mutants of Salmonella enterica serotype Typhi under ciprofloxacin selective pressure. Indian J. Med. Res. 139, 746–753.
Evans, D. J., Allison, D. G., Brown, M. R., and Gilbert, P. (1991). Susceptibility of Pseudomonas aeruginosa and Escherichia coli biofilms towards ciprofloxacin: effect of specific growth rate. J. Antimicrob. Chemother. 27, 177–184. doi: 10.1093/jac/27.2.177
Fàbrega, A., Martin, R. G., Rosner, J. L., Tavio, M. M., and Vila, J. (2010). Constitutive SoxS expression in a fluoroquinolone-resistant strain with a truncated SoxR protein and identification of a new member of the marA-soxS-rob regulon, mdtG. Antimicrob. Agents Chemother. 54, 1218–1225. doi: 10.1128/AAC.00944-09
Fernandez, A., Cloeckaert, A.A, Bertini, A., Praud, K., Doublet, B., Weill, F., et al. (2007). Comparative analysis of IncHI2 plasmids carrying blaCTX-M-2 or blaCTX-M-9 from Escherichia coli and Salmonella enterica strains isolated from poultry and humans. Antimicrob. Agents Chemother. 51, 4177–4180. doi: 10.1128/AAC.00603-07
Garcia-Fernandez, A., and Carattoli, A. (2010). Plasmid double locus sequence typing for IncHI2 plasmids, a subtyping scheme for the characterization of IncHI2 plasmids carrying extended-spectrum beta-lactamase and quinolone resistance genes. J. Antimicrob. Chemother. 65, 1155–1161. doi: 10.1093/jac/dkq101
Gomes-Neves, E., Antunes, P., Manageiro, V., Gärtner, F., Caniça, M., Da, C. J., et al. (2014). Clinically relevant multidrug resistant Salmonella enterica in swine and meat handlers at the abattoir. Vet. Microbiol. 168, 229–233. doi: 10.1016/j.vetmic.2013.10.017
Gomez, T. M., Motarjemi, Y., Miyagawa, S., Käferstein, F. K., and Stöhr, K. (1997). Foodborne salmonellosis. World Health Stat. Q. 50:81.
He, G. X., Zhang, C., Crow, R. R., Thorpe, C., Chen, H., Kumar, S., et al. (2011). SugE, a new member of the SMR family of transporters, contributes to antimicrobial resistance in Enterobacter cloacae. Antimicrob. Agents Chemother. 55, 3954–3957. doi: 10.1128/AAC.00094-11
Heo, A., Jang, H. J., Sung, J. S., and Park, W. (2014). Global transcriptome and physiological responses of Acinetobacter oleivorans DR1 exposed to distinct classes of antibiotics. PLoS ONE 9:e110215. doi: 10.1371/journal.pone.0110215
Higashi, K., Ishigure, H., Demizu, R., Uemura, T., Nishino, K., Yamaguchi, A., et al. (2008). Identification of a spermidine excretion protein complex (MdtJI) in Escherichia coli. J. Bacteriol. 190, 872–878. doi: 10.1128/JB.01505-07
Hohmann, E. L. (2011). Nontyphoidal salmonellosis. Clin. Infect. Dis. 32, 128–136. doi: 10.1016/B978-0-7020-3935-5.00017-3
Hopkins, K. L., Kirchner, M., Guerra, B., Granier, S. A., Lucarelli, C., Porrero, M. C., et al. (2010). Multiresistant Salmonella enterica serovar 4,[5],12:i:- in Europe: a new pandemic strain? Euro Surveill. 15:19580.
Jia, Y., Khanal, S. K., Shu, H., Zhang, H., Chen, G. H., and Lu, H. (2018). Ciprofloxacin degradation in anaerobic sulfate-reducing bacteria (SRB) sludge system: mechanism and pathways. Water Res. 136, 64–74. doi: 10.1016/j.watres.2018.02.057
Leuzzi, A., Di Martino, M. L., Campilongo, R., Falconi, M., Barbagallo, M., Marcocci, L., et al. (2015). Multifactor regulation of the MdtJI polyamine transporter in shigella. PLoS ONE 10:e0136744. doi: 10.1371/journal.pone.0136744
Li, L., Liao, X., Yang, Y., Sun, J., Liu, B., Yang, S., et al. (2013a). Spread of oqxAB in Salmonella enterica serotype Typhimurium predominantly by IncHI2 plasmids. J. Antimicrob. Chemother. 68, 2263–2268. doi: 10.1093/jac/dkt209
Li, L., Yang, Y. R., Liao, X. P., Lei, C. Y., Sun, J., Li, L. L., et al. (2013b). Development of ceftriaxone resistance affects the virulence properties of Salmonella enterica serotype Typhimurium strains. Foodborne Pathog. Dis. 10, 28–34. doi: 10.1089/fpd.2012.1216
Li, R., Yu, C., Li, Y., Lam, T. W., Yiu, S. M., Kristiansen, K., et al. (2009). SOAP2: an improved ultrafast tool for short read alignment. Bioinformatics 25, 1966–1967. doi: 10.1093/bioinformatics/btp336
Mangold, S., Valdes, J., Holmes, D. S., and Dopson, M. (2011). Sulfur metabolism in the extreme acidophile acidithiobacillus caldus. Front. Microbiol. 2:17. doi: 10.3389/fmicb.2011.00017
Mather, A. E., Reid, S. W., Maskell, D. J., Parkhill, J., Fookes, M. C., Harris, S. R., et al. (2013). Distinguishable epidemics of multidrug-resistant Salmonella typhimurium DT104 in different hosts. Science 341, 1514–1517. doi: 10.1126/science.1240578
Mortazavi, A., Williams, B. A., Mccue, K., Schaeffer, L., and Wold, B. (2008). Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods 5:621. doi: 10.1038/nmeth.1226
Nikaido, H. (1996). Multidrug efflux pumps of gram-negative bacteria. J. Bacteriol. 178, 5853–5859. doi: 10.1128/jb.178.20.5853-5859.1996
Oswald, C., Tam, H. K., and Pos, K. M. (2016). Transport of lipophilic carboxylates is mediated by transmembrane helix 2 in multidrug transporter AcrB. Nat. Commun. 7:13819. doi: 10.1038/ncomms13819
Patkari, M., and Mehra, S. (2013). Transcriptomic study of ciprofloxacin resistance in Streptomyces coelicolor A3(2). Mol. Biosyst. 9, 3101–3116. doi: 10.1039/c3mb70341j
Piddock, L. J. (1999). Mechanisms of fluoroquinolone resistance: an update 1994–1998. Drugs 58 (Suppl. 2), 11–18. doi: 10.2165/00003495-199958002-00003
Poirel, L., Cattoir, V., and Nordmann, P. (2012). Plasmid-mediated quinolone resistance; interactions between human, animal, and environmental ecologies. Front. Microbiol. 3:24. doi: 10.3389/fmicb.2012.00024
Pos, K. M. (2009). Drug transport mechanism of the AcrB efflux pump. Biochim. Biophys. Acta 1794, 782–793. doi: 10.1016/j.bbapap.2008.12.015
Rodrigue, A., Effantin, G., and Mandrand-Berthelot, M. A. (2005). Identification of rcnA (yohM), a nickel and cobalt resistance gene in Escherichia coli. J. Bacteriol. 187, 2912–2916. doi: 10.1128/JB.187.8.2912-2916.2005
Saier, M. H. Jr. (2015). The bacterial phosphotransferase system: new frontiers 50 years after its discovery. J. Mol. Microbiol. Biotechnol. 25, 73–78. doi: 10.1159/000381215
Schuster, S., Vavra, M., Schweigger, T. M., Rossen, J. W. A., Matsumura, Y., and Kern, W. V. (2017). Contribution of AcrAB-TolC to multidrug resistance in an Escherichia coli sequence type 131 isolate. Int. J. Antimicrob. Agents 50, 477–481. doi: 10.1016/j.ijantimicag.2017.03.023
Strahilevitz, J., Jacoby, G. A., Hooper, D. C., and Robicsek, A. (2009). Plasmid-mediated quinolone resistance: a multifaceted threat. Clin. Microbiol. Rev. 22:664. doi: 10.1128/CMR.00016-09
Sullivan, M. J., Petty, N. K., and Beatson, S. A. (2011). Easyfig: a genome comparison visualizer. Bioinformatics 27, 1009–1010. doi: 10.1093/bioinformatics/btr039
Szklarczyk, D., Morris, J. H., Cook, H., Kuhn, M., Wyder, S., Simonovic, M., et al. (2017). The STRING database in 2017: quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Res. 45, D362–D368. doi: 10.1093/nar/gkw937
Thorrold, C. A., Letsoalo, M. E., Duse, A. G., and Marais, E. (2007). Efflux pump activity in fluoroquinolone and tetracycline resistant Salmonella and E. coli implicated in reduced susceptibility to household antimicrobial cleaning agents. Int. J. Food Microbiol. 113, 315–320. doi: 10.1016/j.ijfoodmicro.2006.08.008
Threlfall, E. J. (2000). Epidemic Salmonella typhimurium DT 104–a truly international multiresistant clone. J. Antimicrob. Chemother. 46:7. doi: 10.1093/jac/46.1.7
Wang, T., Cui, Y., Jin, J., Guo, J., Wang, G., Yin, X., et al. (2013). Translating mRNAs strongly correlate to proteins in a multivariate manner and their translation ratios are phenotype specific. Nucleic Acids Res. 41, 4743–4754. doi: 10.1093/nar/gkt178
Webber, M., Buckley, A. M., Randall, L. P., Woodward, M. J., and Piddock, L. J. (2006). Overexpression of marA, soxS and acrB in veterinary isolates of Salmonella enterica rarely correlates with cyclohexane tolerance. J. Antimicrob. Chemother. 57, 673–679. doi: 10.1093/jac/dkl025
Wong, M. H., Chan, E. W., Xie, L., Li, R., and Chen, S. (2016). IncHI2 plasmids are the key vectors responsible for oqxAB transmission among Salmonella species. Antimicrob. Agents Chemother. 60, 6911–6915. doi: 10.1128/AAC.01555-16
Wong, M. H. Y., and Chen, S. (2013). First detection of oqxAB in Salmonella spp. isolated from food. Antimicrob. Agents Chemother. 57:658. doi: 10.1128/AAC.01144-12
Zaidi, M. B., Freddy, C., Cevallos, M. A., Marcos, F. M., Edmundo, C., Magdalena, W., et al. (2011). Salmonella typhimurium ST213 is associated with two types of IncA/C plasmids carrying multiple resistance determinants. BMC Microbiol. 11:9. doi: 10.1186/1471-2180-11-9
Keywords: oqxAB, IncHI2, plasmids, Salmonella, tolerance
Citation: Lian X, Wang X, Liu X, Xia J, Fang L, Sun J, Liao X and Liu Y (2019) oqxAB-Positive IncHI2 Plasmid pHXY0908 Increase Salmonella enterica Serotype Typhimurium Strains Tolerance to Ciprofloxacin. Front. Cell. Infect. Microbiol. 9:242. doi: 10.3389/fcimb.2019.00242
Received: 26 March 2019; Accepted: 18 June 2019;
Published: 03 July 2019.
Edited by:
Tracy Raivio, University of Alberta, CanadaReviewed by:
Travis Bourret, Creighton University, United StatesGuoqiang Zhu, Yangzhou University, China
Copyright © 2019 Lian, Wang, Liu, Xia, Fang, Sun, Liao and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jian Sun, amlhbnN1bkBzY2F1LmVkdS5jbg==; Xiaoping Liao, eHBsaWFvQHNjYXUuZWR1LmNu; Yahong Liu, bHloQHNjYXUuZWR1LmNu
†These authors have contributed equally to this work
 Xinlei Lian
Xinlei Lian Xiran Wang
Xiran Wang Xiao Liu
Xiao Liu Jing Xia
Jing Xia Liangxing Fang1,2,3
Liangxing Fang1,2,3 Jian Sun
Jian Sun Xiaoping Liao
Xiaoping Liao Yahong Liu
Yahong Liu