- 1State Key Laboratory of Crop Stress Biology for Arid Areas, College of Life Sciences, Northwest A&F University, Yangling, China
- 2Shaanxi Key Laboratory of Agricultural and Environmental Microbiology, College of Life Sciences, Northwest A&F University, Yangling, China
The type VI secretion system (T6SS) is a versatile secretion system widely distributed in Gram-negative bacteria that delivers multiple effector proteins into either prokaryotic or eukaryotic cells, or into the extracellular milieu. T6SS participates in various physiological processes including bacterial competition, host infection, and stress response. Three pathogenic Yersinia species, namely Yersinia pestis, Yersinia pseudotuberculosis, and Yersinia enterocolitica, possess different copies of T6SSs with distinct biological functions. This review summarizes the pathogenic, antibacterial, and stress-resistant roles of T6SS in Yersinia and the ion-transporting ability in Y. pseudotuberculosis. In addition, the T6SS-related effectors and regulators identified in Yersinia are discussed.
Introduction
The genus Yersinia comprises three species of bacterial pathogens, namely Y. pestis, Y. enterocolitica, and Y. pseudotuberculosis, that are causative agents of human diseases. Y. pestis is the etiological agent of plague, often transmitted by flea bites or aerosols. It infects regional lymph nodes or lungs and causes the highly lethal disease in humans. Y. pseudotuberculosis and Y. enterocolitica are enteric pathogens, which usually grow in the environment and can be transmitted to mammalian hosts through ingestion of contaminated food or water. They typically cause a broad range of gastrointestinal diseases, from enteritis to mesenteric lymphadenitis (Bibikova, 1977; Brubaker, 1991; Bottone, 1997; Putzker et al., 2001; Pujol and Bliska, 2005). These three bacterial species have been used in experimental models of infection to study their pathogenicity for the mammalian host.
Despite differences in pathogenesis, these virulent Yersinia species have several common virulence factors, including the 70-kb virulence plasmid (pCD1 in Y. pestis and pYV in enteropathogenic Yersinia) and the yersiniabactin (Ybt) system (Brubaker, 1991; Heesemann et al., 1993; Cornelis et al., 1998). The 70-kb plasmid contains dozens of genes encoding structural components of a type III secretion system (T3SS), and also encodes several T3SS effector proteins called Yersinia outer proteins (Yops) and their dedicated chaperones to subvert the innate immune system of the hosts (Bliska et al., 2013; Schwiesow et al., 2015). Since enzymes/toxins/effectors delivered by secretion systems play a crucial role in the interaction between pathogens and their hosts or competitors, the various types of secretion systems attract interest in the research on pathogenic bacteria.
Several virulence associated secretion (vas) genes in T6SS gene clusters were identified in bacterial pathogens before, but till the year of 2006 the Type VI secretion system was proposed and defined as a contact-dependent secretion mechanism (Mougous et al., 2006; Pukatzki et al., 2006). T6SS was found distributed in about 25% of all sequenced Gram-negative bacteria with high conservation (Boyer et al., 2009; Salomon and Orth, 2015). The T6SS is a proteinaceous machinery that directly injects effector proteins into target cells in a one-step process with a bacteriophage-like cell-puncturing device (Zoued et al., 2014), although the membrane puncturing process has not been directly observed to date. The versatile T6SS weapons allow bacteria to compete with other bacteria or attack simple or higher eukaryotes. The main function of T6SS was implicated in virulence, commensalism or symbiosis, and interbacterial competition (Jani and Cotter, 2010). Apart from the conserved components and similar structures, various functions of T6SSs have been exploited in the past decade, and the newly discovered effectors and their dedicated chaperones secreted by T6SS are also being updated. In the three pathogenic Yersinia species, different series of T6SS seem to possess different functions. This review focuses on the detailed description of T6SSs function in Yersinia.
Comparison of T6SS Gene Clusters in Yersinia Species
Component, Structure, and Energetics of T6SSs
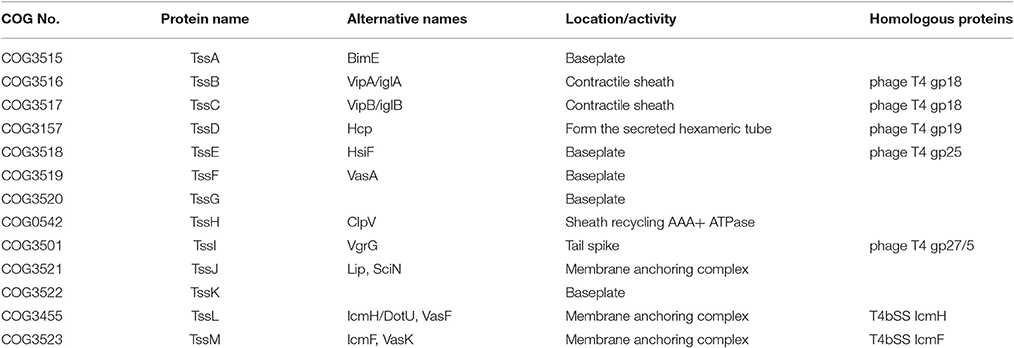
The core components of T6SS contain 13 subunits, which comprise the typical T6SS structure similar to the T4 bacteriophage, with tail, spike, sheath, hub or baseplate proteins (Boyer et al., 2009; Cascales and Cambillau, 2012). The typical T6SS structure is composed of three subunits: the membrane complex, the baseplate complex, and the injection apparatus. The membrane complex is composed of proteins TssJLM, which anchor the baseplate complex (TssAEFGK) to the membrane and provide structural support. The injection apparatus contains needle sheath (TssBC), tail tube (TssD/Hcp) and spike complex (TssI/VgrG and PAAR motifs) (Silverman et al., 2012; Brunet et al., 2015; Cianfanelli et al., 2016). Generally, various effectors and chaperones could bind to this injection apparatus when they were needed to be secreted out through the T6SS apparatus, and the secretion of Hcp or VgrG is often regarded as the hallmark of a functional T6SS in many bacterial species (Mougous et al., 2006; Pukatzki et al., 2006; Wang et al., 2011). The PAAR (Proline-Alanine-Alanine-aRginine) repeat containing proteins were regarded as an additional component of the T6SS machinery. PAAR-repeat could form a sharp conical extension on the tip of the VgrG spike, which is further involved in the effector recruitment and attaching effector domains to the spike (Shneider et al., 2013; Bondage et al., 2016). The 13 core components of typical T6SS are listed in Table 1.
The secretion of T6SS is a dynamic cycle of assembly/extension, contraction/puncture, and disassembly of the sheath, accompanying the energy supply and effector transportation (Basler et al., 2012). The initial stage of the T6SS assembly is to form the membrane complex, followed by the recruitment of baseplate proteins for anchor and extension of the outer sheath and the inner tube (Brunet et al., 2015). This indicated that the T6SS shares a common assembly pathway of tail tubes with bacteriophage (Brunet et al., 2014). During the attack process, the sheath-like structure propels an inner tube through contraction, and the membrane-puncturing spike is pierced into the target cells. Then effectors and chaperones are delivered into target cells with this expelled structure in a one-step manner (Cianfanelli et al., 2016).
Different Series of T6SS in Three Yersinia Species
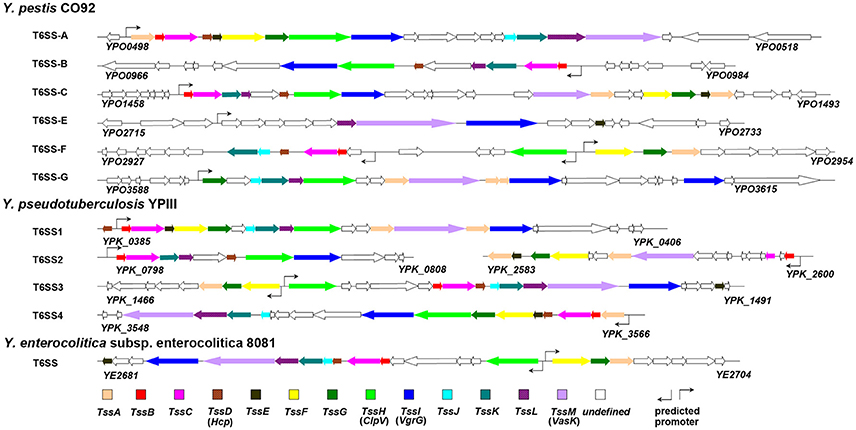
T6SS displays a single copy in majority of the bacterial species, yet multiple distinct copies are found in several bacterial species (Boyer et al., 2009). In the Yersinia species, six and four T6SS clusters were identified in Y. pestis (Andersson et al., 2017) and Y. pseudotuberculosis (Zhang et al., 2011b), respectively, while only one copy was found in Y. enterocolitica (Jaakkola et al., 2015). The T6SS gene clusters in Yersinia are shown in Figure 1.
It is noteworthy that the multiple distinct T6SS copies are not functionally redundant; for example, T6SS-1 and T6SS-5 in B. thailandensis mediate the bacterial antagonism and macrophage infection, respectively (Schwarz et al., 2010). In Y. pestis, the T6SS region YPO0498-YPO0516 (the T6SS Cluster A in Figure 1) is preferentially expressed at 26 vs. 37°C (Cathelyn et al., 2006; Robinson et al., 2009), which suggests this gene cluster may function in natural conditions rather than in its mammalian host. Deletion of the T6SS Cluster A locus reduced the uptake by J774.1 murine macrophages (Robinson et al., 2009). However, it had no effect on virulence in bubonic or pneumonic murine plague models compared to the parental Y. pestis CO92 strain (Robinson et al., 2009; Andersson et al., 2017). An Hcp-like protein encoded by ypo0502 was found to play important roles in autoagglutination (AA), indicating that the T6SS Cluster A is involved in intraspecies interaction of bacteria (Podladchikova et al., 2011). A similar thermoregulated gene cluster in Y. pseudotuberculosis was identified as T6SS4 (ypk_3548- ypk_3566), which is precisely regulated by temperature, growth phase, and AHL-dependent quorum sensing systems, and plays a crucial role in resistance to environmental stresses (Zhang et al., 2011b). The T6SS Cluster A in Y. pestis and T6SS4 in Y. pseudotuberculosis could both be induced by room temperature conditions (about 25°C). The phylogenetic analysis based on TssL of Yersinia T6SSs showed a close genetic distance between T6SS Cluster A in Y. pestis and T6SS4 in Y. pseudotuberculosis (Figure 2). Furthermore, the two T6SS clusters have similar genomic organization, suggesting they may both play crucial roles in environment adaptability. From Figure 1 it showed the T6SS-E and F clusters in Y. pestis only contain 4 and 9 T6SS genes, respectively. Considering these components seemed complementary, and the two T6SS clusters locate in vicinity to each other, it speculated they may represent an integral set of T6SS. It is worth noting that Y. pestis has 6 clusters but only one with a complete set of T6SS genes, while Y. pseudotuberculosis has 4 clusters with full set of genes. Note that some clusters encode an incomplete set of T6SS genes and could be non-functional. The function of these incomplete T6SSs should be experimental verified in the future.
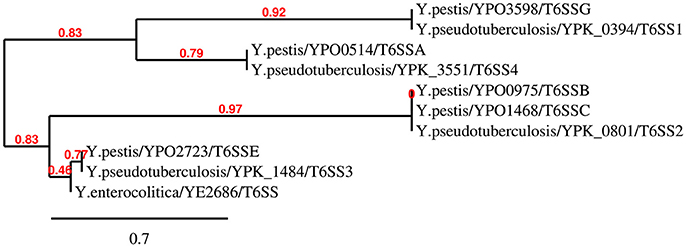
Figure 2. Neighbour-Joining tree of TssL proteins of Yersinia T6SSs. The phylogenetic tree was generated using BioNJ with 100 bootstrap replicates based on the alignment of TssL sequences (Gascuel, 1997). As the TssL is missed in T6SS gene cluster F in Y. pestis, the T6SS gene cluster F was not present in this N-J dendrogram.
The Functions of T6SS in Yersinia
Virulence
At the beginning, genes of T6SS cluster in Vibrio cholerae and Pseudomonas aeruginosa were identified as vas (virulence-associated secretion) genes for their roles in pathogenesis (Mougous et al., 2006; Pukatzki et al., 2006). Afterwards, the virulence-related functions of T6SS in the host were verified in several organisms including Edwardsiella tarda (Zheng and Leung, 2007), Aeromonas hydrophila (Suarez et al., 2008), Salmonella enterica (Blondel et al., 2010), Campylobacter jejuni (Lertpiriyapong et al., 2012), and Burkholderia pseudomallei (Schwarz et al., 2014).
In 2008, five T6SS gene clusters in Y. pestis KIM were identified through a phylogenetic analysis (Bingle et al., 2008), while six T6SS clusters in Y. pestis CO92 were predicted based on in silico analysis (Boyer et al., 2009). Actually, six T6SS clusters are distributed in genome of Y. pestis KIM and CO92 according to the SecReT6 database, which is a web-based resource for type VI secretion systems found in bacteria (Li et al., 2015). However, the virulence of T6SS in Y. pestis to hosts was confirmed through laboratory experiments till 2015. Using an in vivo signature-tagged mutagenesis (STM) screening approach, Ponnusamy et al. identified three T6SS genes with virulence potential in Y. pestis CO92. In-frame deletion of vasK (YPO3603, a component of the T6SS cluster G) in Y. pestis CO92 resulted in significant attenuation of the bacterium in murine models of infection, and this attenuation could be fully complemented (Ponnusamy et al., 2015). In addition, the five deletion mutants of T6SS clusters (B, C, E-G) showed varying levels of attenuation when evaluated in a mouse model of pneumonic plague, but not all T6SS clusters of Y. pestis are equally functional. Deletion mutants for Cluster C and Cluster F exhibited limited, 14%, or no attenuation, respectively, while deletion mutants for Cluster B, Cluster E, and Cluster G, exhibited significant levels of attenuation in infection of a bubonic plague model. A further augmented attenuation was detected in mutants lacking multiple T6SSs. The Δypo2720-2733Δhcp3 (Cluster E and hcp homolog 3) double deletion mutant showed 60% of attenuation in comparison to WT CO92. This result further supports the involvement of T6SSs in Y. pestis virulence (Andersson et al., 2017). Moreover, most of the attenuated T6SS deletion mutant strains showed decreased host cell cytotoxicity and intracellular survival in comparison to WT CO92 (Andersson et al., 2017).
Y. pseudotuberculosis YPIII mutants lacking T6SS-4 or yezP (ypk_3549, an effector gene in T6SS-4 cluster) are defective in virulence in mice (Wang et al., 2015). The enteropathogenic Y. enterocolitica causes infections similar to Y. pseudotuberculosis, but has several different characteristics related to epidemiology and ecology, and only one type of T6SS was found in the Y. enterocolitica genome. It is speculated that the lack of multiple T6SS might be beneficial for escaping the host immune system, because a T6SS with toxic effectors might encumber the invasion and survival of Y. enterocolitica cells in mammalian hosts (Jaakkola et al., 2015). This function can be regarded as anti-virulence, which has been discussed previously (Jani and Cotter, 2010).
Bacteria-Bacteria Interaction
Bacteria have evolved various mechanisms to kill bacterial competitors and defense against predators in their environment. The T6SS has membrane-penetrating tails similar to bacteriophages, suggesting that T6SS may be involved in cell-cell interactions of bacteria (Pell et al., 2009). Bacteria could utilize T6SS to intimately interface with other bacteria, efficiently killing or inhibiting competitors with T6SS toxins and protecting itself with immune proteins, which has been reported in P. aeruginosa (HSI-1) (Hood et al., 2010), B. thailandensis (T6SS-1) (Schwarz et al., 2010), V. cholerae (MacIntyre et al., 2010; Ishikawa et al., 2012), and Serratia marcescens (Murdoch et al., 2011). Different from the traditional offensive and defensive model of T6SS mediated bacterial competition (Russell et al., 2012; Lien and Lai, 2017; Yang et al., 2018), swarming Proteus mirabilis discriminate non-identical population via T6SS-dependent delivery of toxic effectors, thus forming a visible demarcation line (Dienes line) between different Proteus isolates (Alteri et al., 2013). Although this phenomenon of self-recognition during swarming in P. mirabilis has been known for decades it was only ascribed to the T6SS recently.
Through DNA microarrays analysis of Y. pestis CO92, an IAHP (IcmF-Associated Homologous Protein) locus (YPO0498-0516) was found regulated by RovA, which was specifically required for bubonic plague (Cathelyn et al., 2006). However, another study indicated that mutation in this locus had no effect in laboratory models of infected oriental rat flea X. cheopis, or in virulence using bubonic or pneumonic murine plague models (Robinson et al., 2009). Similarly, in-frame deletion of YPO0498 did not affect virulence in vivo (Ponnusamy et al., 2015). Recently, this locus was classified as T6SS cluster A in Y. pestis CO92, and was confirmed not required for pathogenesis (Andersson et al., 2017). Nevertheless, deletion of the T6SS Cluster A locus resulted in increased uptake and intracellular growth of Y. pestis in the macrophage-like J774.A1 cells, indicating a role of T6SS in interaction with host cells (Robinson et al., 2009). Among this T6SS cluster, YPO0502, identified as a Hcp-like protein that forms pilus-like structure, determines Y. pestis autoagglutination (AA). This suggests that at least one (T6SS Cluster A) of the 6 T6SS clusters in Y. pestis is involved in bacterial interaction (Podladchikova et al., 2011). However, whether Yersinia T6SSs are involved in bacterial competition and Dienes line formation needs to be verified in the future.
Stress Response
Many of the earliest studies on T6SS focused on their roles in virulence and the interactions between pathogens and hosts. In 2009, it was reported that a T6SS in Vibrio anguillarum acts as a sensor for an unknown extracytoplasmic signal that modulates RpoS (Weber et al., 2009). RpoS is the main regulator of gene expression during stationary phase and stress conditions, and it positively regulates VanT linking quorum sensing (QS) to stress response and general physiology of V. anguillarum (Weber et al., 2008). Therefore, this study suggested a new function for T6SS in the ecology of bacteria, that T6SS played a crucial role in bacterial stress response and cell survival after exposure to various environmental challenges (Weber et al., 2009).
In Y. pseudotuberculosis, RpoS was found to be important in resistance to multiple stressors including oxidative stress, acid stress, osmotic stress, and 42°C heat shock, and was also crucial for motility, biofilm formation, and T6SS expression. The electrophoretic mobility shift assay (EMSA) showed that RpoS regulates T6SS4 expression by directly binding to the T6SS4 promoter (Guan et al., 2015). However, compared with the two reported YpsRI QS system (Atkinson et al., 2008) and the OmpR regulator (Zhang et al., 2013), RpoS plays a less crucial role in T6SS4 activation. T6SS4 is only partly responsible for the stress-resistance activity of RpoS, consistent with the relatively weak role of RpoS in activating T6SS4 expression in Y. pseudotuberculosis (Guan et al., 2015). Similarly, the PppA-PpkA pair of H1-T6SS in P. aeruginosa plays roles in bacterial resistance to oxidative and osmotic stress, and pppA-ppkA deletion affects the expression of the RpoS and QS regulons (Goldova et al., 2011).
In 2013, an OmpR-T6SS4 regulation pathway for stress resistance in Y. pseudotuberculosis was characterized in two studies, both of which confirmed the direct binding of OmpR on the T6SS-4 promoter region in vitro (Gueguen et al., 2013; Zhang et al., 2013). In an earlier study, OmpR was found to be essential for low pH adaptation by positively regulating the expression of urease in Y. pseudotuberculosis (Hu et al., 2009). Here, it was indicated that T6SS4 contributed to acid survival by maintaining a steady-state intracellular pH in Y. pseudotuberculosis. The acid resistance ability is dependent upon the ATPase activity of ClpV4 that participates in H+ extrusion, and this process is positively regulated by the global regulator OmpR (Zhang et al., 2013).
In addition, the T6SS-4 expression mediated by OmpR is also induced in high osmolarity conditions or in the presence of sodium deoxycholate. Besides T6SS4 gene cluster, OmpR activates additional genes involved in tolerance to high osmolarity, because the ompR transposon mutant cells were more severely affected than the tssF4 cells (Gueguen et al., 2013). Similarly, an earlier study showed that deletion of T6SS increased the susceptibility of C. jejuni to a bile salt, deoxycholic acid (DCA), and the reason for which was attributed to an increase in the intracellular influx of DCA mediated by T6SS (Lertpiriyapong et al., 2012).
Ion Transport
Recently, T6SS4 in Y. pseudotuberculosis was found to have a notable function of transporting zinc ions (Zn2+) from the environment into bacterial cells to mitigate the detrimental hydroxyl radicals induced by multiple stressors (Wang et al., 2015). Generally, bacteria utilize the classical ZnuABC transport system to acquire Zn2+ to maintain zinc balance. In Y. pseudotuberculosis, the ZnuABC transporter contributes to reduce the intracellular ROS levels and thus prevents oxidative damage to cells (Wang et al., 2016). While in Y. pestis, the yersiniabactin (Ybt, a zincophore for Zn2+ acquisition) synthetase HMWP2, together with ZnuABC, are critical for lethal infections in a mouse model of septicemic plague (Bobrov et al., 2014). Interestingly, in Y. pseudotuberculosis a T6SS4-mediated substrate YezP (YPK_3549), a novel Zn2+-binding protein, has the capacity to rescue the sensitivity to oxidative stress exhibited by T6SS4 mutants when added to extracellular milieu. Zn2+ acquisition achieved by YezP is co-regulated with T6SS4 by OxyR, which is a global oxidative stress regulator that senses diverse environmental cues (Wang et al., 2015).
Similar capability of ion transport by T6SS was found in B. thailandensis and P. aeruginosa. In B. thailandensis, a proteinaceous zincophore (TseZ) secreted through T6SS4 interacts with the outer membrane heme transporter HmuR to acquire zinc under oxidative stress (Si et al., 2017a). In addition, the T6SS4 in B. thailandensis facilitated the uptake of Mn2+ by secreting a Mn2+-binding effector TseM during oxidative stress. The Mn2+ load on TseM could be delivered to MnoT, a Mn2+-specific TonB-dependent outer membrane transporter, and then transported into cells to fulfill the increased cellular demand for Mn2+ under oxidative stress (Si et al., 2017b). While in P. aeruginosa, a H3-T6SS secreted effector TseF (PA2374) is involved in iron uptake by interacting with outer membrane vesicles (OMVs) and the Pseudomonas Quinolone Signal (PQS) system (Lin et al., 2017). These studies highlight the newly characterized function of ion transport through T6SSs. Whether and how Yersinia T6SSs transport other metal ions besides Zn2+ needs to be investigated in the future.
Other Potential Functions
More functions were identified in other bacterial species in addition to Yersinia. T6SS is also implicated in biofilm formation in several bacterial pathogens. For example, Hcp participates in biofilm formation in P. aeruginosa PAO1 (Southey-Pillig et al., 2005), and H1-T6SS is associated with biofilm-specific antibiotic resistance in P. aeruginosa PA14 (Zhang et al., 2011a). In Acidovorax citrulli, four mutants of T6SS components were reduced in biofilm formation (Tian et al., 2015). Recently, T6SS was shown to be associated with cell autophagy. VgrG2, a translocon of T6SS2 in Vibrio parahaemolyticus, induces autophagy in macrophages (Yu et al., 2015). A Type VI secretion PGAP1-like effector TplE, belonging to Tle4 phospholipase family in P. aeruginosa (Russell et al., 2013), induced autophagy through disruption of endoplasmic reticulum (ER) homeostasis (Jiang et al., 2016). In addition, the T6SS in P. mirabilis plays a role in self-recognition during swarming via delivery of toxic effectors (Alteri et al., 2013). Further research and analysis need to be performed to verify these potential T6SS functions in Yersinia species.
Effectors Secreted by T6SS in Yersinia
With a bacteriophage tail-like structure, T6SS apparatus can deliver various effectors into bacterial competitors for interbacterial competition (Cianfanelli et al., 2016), into eukaryotic hosts for pathogenesis (Durand et al., 2014) or into extracellular milieu for nutritional demands (Wang et al., 2015; Lin et al., 2017; Si et al., 2017a). The antibacterial T6SS effector usually has a chaperone immunity protein that provides protection to the organism itself. These effectors could be divided into three categories as targeting the membrane, cell wall, or nucleic acid (Russell et al., 2014). The eukaryotic host-targeted T6SS effectors are diverse in biological and biochemical functions. For example, VgrG-1 in V. cholerae can cause actin crosslinking for intestinal inflammation (Ma and Mekalanos, 2010), VgrG2b in P. aeruginosa interacts with microtubules for successful invasion of epithelial cells (Sana et al., 2015), and VgrG-5 in B. thailandensis could induce the host cell membrane fusions by interacting directly with a neighboring host cell receptor or by engaging in homotypic interactions within or between cells (Schwarz et al., 2014). A recent publication summarizes the T6SS effectors identified in recent years (Lien and Lai, 2017).
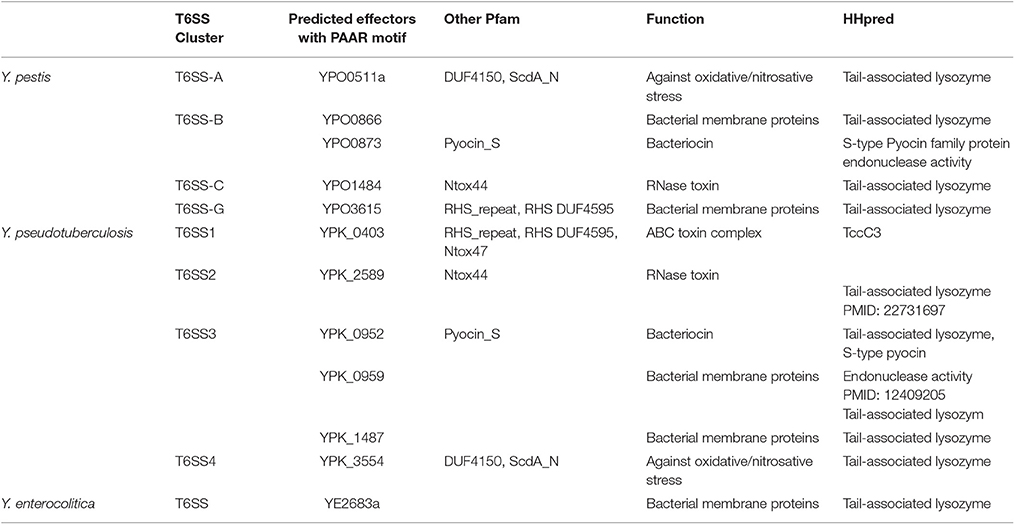
Many T6SS secretion effectors have been identified in bacterial organisms. However, apart from the hallmark of T6SS effectors Hcp or VgrG, only two effectors were functionally characterized in Yersinia species according to published papers. They are YezP (YPK_3549) in Y. pseudotuberculosis, which is a novel Zn2+-binding protein substrate (Wang et al., 2015), and YPO0502 in Y. pestis CO92, which is a low-molecular-weight component with siderophore activity and regarded as the autoagglutination factor (Podladchikova et al., 2011). In addition, three T6SS associated proteins, y3673/Hcp, y3674 and y3675, were detected in the outer-membrane proteome in Y. pestis KIM6+ using 2DGE gel electrophoresis (Pieper et al., 2009). However, the y3674/y3675 genes possess VipB/VipA motif according to the KEGG database, which suggests these two proteins may comprise the contractile sheath of T6SS. With high-throughput signature-tagged mutagenesis (STM) approach, three T6SS components or effector-encoding genes (vasK/ypo3603, ypo0498, and ypo1484) were identified in Y. pestis CO92 (Ponnusamy et al., 2015). VasK was confirmed to be important during Y. pestis infection in mouse models of plague (Andersson et al., 2017). With a conserved “MxiM” motif, YPO0498 may be a lipoprotein, while YPO1484 is likely to be an RNase toxin with a sharp conical extension complex of “PAAR” (Proline-Alanine-Alanine-aRginine) (Shneider et al., 2013). However, more experiments need to be performed to explain the connection between these proteins and T6SS apparatus. Recently, Andersson et al generated three PAAR motif repeat-containing T6SS effector encoding gene deletion mutants (Δypo0873, Δypo1484, and Δypo3615). Only Δypo1484 exhibited a limited level of attenuation, 20% survivability, in comparison to WT CO92 in a mouse model of pneumonic plague. This attenuation was similar to the level of attenuation, 14%, reported in the T6SS Cluster C (ypo1458-1484) deletion mutant, which contains the ypo1484 gene (Andersson et al., 2017). As multiple copies of T6SS have been confirmed in Yersinia species, it seems that more potential undiscovered effectors remain to be identified. Here, we presented the hypothetical effectors with “PAAR” motif in Yersinia T6SS clusters (as shown in Table 2).
T6SS Regulation in Yersinia
T6SS gene clusters are found both in pathogenic and non-pathogenic bacteria, which are distributed in various environments including marine, soil, rhizosphere, higher plants and mammalian hosts, implying their diverse functions (Boyer et al., 2009). As the assembly, contraction, and disassembly cycle of the T6SS is likely to be energetically costly to the bacterial cell, the T6SS organelle is tightly regulated when organisms alternate in various environmental conditions (Silverman et al., 2012). It has been reported that T6SS in bacterial species responds to various environmental factors including salinity (Salomon et al., 2013), iron-concentration (Chakraborty et al., 2011), temperature (Pieper et al., 2009), QS-dependent cell density (Zheng et al., 2010; Kitaoka et al., 2011), host immune system, and other stressors (Brooks et al., 2013).
Temperature is a common environmental factor for most pathogens, and changes widely when organisms alternate in various environmental conditions. The expression of T6SS cluster A (YPO0498-YPO0516 in Y. pestis CO92 or y3658-y3677 in Y. pestis KIM) is induced by lowering the temperature from 37 to 26°C (Pieper et al., 2009; Robinson et al., 2009). In Y. pseudotuberculosis, all the four T6SS clusters are differentially thermoregulated. In addition to the induction of T6SS4 at 26°C, the T6SS1 expression is obviously induced at 37°C, while T6SS2, T6SS3, and T6SS4 was completely repressed at 37°C, which suggested that the T6SS1 played a more important role in bacterial virulence during mammalian host infection than other 3 T6SS loci. In addition, T6SS is precisely regulated by growth phase and acylated homoserine lactones (AHLs) dependent quorum sensing (QS) systems (Zhang et al., 2011b). Furthermore, T6SS4 in Y. pseudotuberculosis has been shown to respond to oxidative stress (Wang et al., 2015).
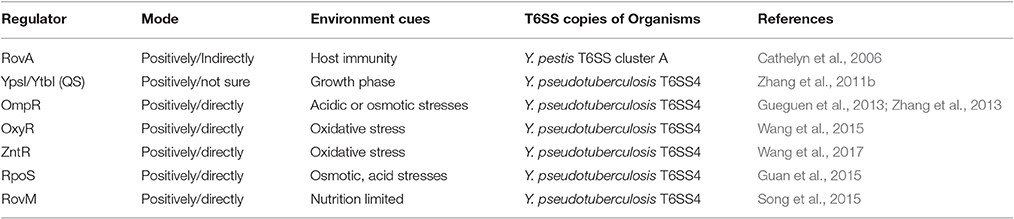
T6SS could be activated by different transcriptional regulators to combat multiple stresses and host immunity, and several transcriptional regulators have been identified in Yersinia species. In Y. pseudotuberculosis, the OmpR-regulated T6SS4 plays important roles under acidic and osmotic stress conditions (Gueguen et al., 2013; Zhang et al., 2013). It was proposed that T6SS4 was activated by RpoS and partly mediated the roles of RpoS in osmotic and acid resistance (Guan et al., 2015). Furthermore, T6SS4 could be regulated by OxyR or ZntR to cope with oxidative stress by removing ROS produced by host immunity or harmful environments (Wang et al., 2015, 2017). We have summarized the identified regulators of T6SS in Yersinia in Table 3.
In addition to the research on Yersinia, other transcriptional regulators of T6SSs have been identified in more bacterial species. In P. aeruginosa, a T6SS is controlled by the global virulence regulator proteins RetS and LadS (Mougous et al., 2006). In Salmonella enterica, T6SS genes were controlled by the SsrA/SsrB two-component regulatory system (Parsons and Heffron, 2005). Furthermore, T6SS is regulated by AggR in enteroaggregative Escherichia coli (Dudley et al., 2006), by VirA/G in Burkholderia mallei (Schell et al., 2007), and similarly by quorum sensing mechanism in the plant pathogen Pectobacterium atrosepticum (Liu et al., 2008). By contrast, few post-transcriptional regulators have been identified so far. Lon, a posttranslational regulator, is known to affect a variety of physiological traits in many bacteria. It suggested that T6SS was regulated by LonA in V. cholerae, as well as biofilm formation, swimming motility, intracellular levels of cyclic diguanylate (Rogers et al., 2016). In P. aeruginosa, a screen to identify T6SS regulatory elements found that the posttranscriptional regulator RsmA imposes a concerted repression on all three T6SS clusters. RsmA and a transcriptional regulator AmrZ orchestrate the assembly of all three T6SSs in P. aeruginosa (Allsopp et al., 2017).
Conclusions
Over the past decade, pathogenic bacteria P. aeruginosa and V. cholerae have been widely used to study T6SS, and significant advancements have been made on the understanding of their structure, activation signals, and regulatory pathways (Chen et al., 2015; Joshi et al., 2017). As important pathogenic bacteria, three Yersinia species also attracted increasing attention because of their multiple T6SS copies. As a summary, we drew a schematic diagram based on the current achievement of research on T6SSs in Yersinia (Figure 3). However, many questions need to be addressed. We believe that the future work in Yersinia T6SS research will involve at least six points: 1, The T6SS activation conditions; 2, The T6SS regulators and the regulation network; 3, Identification of T6SS secreted effectors and their new functions; 4, Based on the knowledge of T6SS attack and protection ability, the ways to prevent the T6SS+ pathogens; 5, The different functions and connection of different T6SS clusters; 6, The exploitation of new methods or techniques for further T6SS studies. The progress of Yersinia T6SS research will provide new knowledge and challenges, as well as further our understanding of the T6SS complexities in other important human pathogens. This research will also influence clinical medicine, microbial community ecology, and physiology.
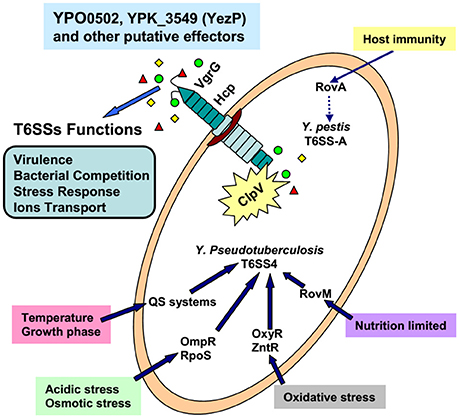
Figure 3. The confirmed environmental factors, transcriptional regulators, and functions of T6SSs in Yersinia.
Author Contributions
XY, JP, and YW collected and assessed the references; YW and XS contributed in the proposal and guideline of the review; XY, JP, and XS wrote the paper.
Funding
The authors thank the National Natural Science Foundation of China (Nos. 31725003, 31670053, and 31370150) for the financial support.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
Allsopp, L. P., Wood, T. E., Howard, S. A., Maggiorelli, F., Nolan, L. M., Wettstadt, S., et al. (2017). RsmA and AmrZ orchestrate the assembly of all three type VI secretion systems in Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. U.S.A. 114, 7707–7712. doi: 10.1073/pnas.1700286114
Alteri, C. J., Himpsl, S. D., Pickens, S. R., Lindner, J. R., Zora, J. S., Miller, J. E., et al. (2013). Multicellular bacteria deploy the type VI secretion system to preemptively strike neighboring cells. PLoS Pathog. 9:e1003608. doi: 10.1371/journal.ppat.1003608
Andersson, J. A., Sha, J., Erova, T. E., Fitts, E. C., Ponnusamy, D., Kozlova, E. V., et al. (2017). Identification of new virulence factors and vaccine candidates for Yersinia pestis. Front. Cell. Infect. Microbiol. 7:448. doi: 10.3389/fcimb.2017.00448
Atkinson, S., Chang, C. Y., Patrick, H. L., Buckley, C. M., Wang, Y., Sockett, R. E., et al. (2008). Functional interplay between the Yersinia pseudotuberculosis YpsRI and YtbRI quorum sensing systems modulates swimming motility by controlling expression of flhDC and fliA. Mol. Microbiol. 69, 137–151. doi: 10.1111/j.1365-2958.2008.06268.x
Basler, M., Pilhofer, M., Henderson, G. P., Jensen, G. J., and Mekalanos, J. J. (2012). Type VI secretion requires a dynamic contractile phage tail-like structure. Nature 483, 182–186. doi: 10.1038/nature10846
Bibikova, V. A. (1977). Contemporary views on the interrelationships between fleas and the pathogens of human and animal diseases. Ann. Rev. Entomol. 22, 23–32. doi: 10.1146/annurev.en.22.010177.000323
Bingle, L. E., Bailey, C. M., and Pallen, M. J. (2008). Type VI secretion: a beginner's guide. Curr. Opin. Microbiol. 11, 3–8. doi: 10.1016/j.mib.2008.01.006
Bliska, J. B., Wang, X., Viboud, G. I., and Brodsky, I. E. (2013). Modulation of innate immune 387 responses by Yersinia type III secretion system translocators and effectors. Cell. Microbiol. 15, 1622–1631. doi: 10.1111/cmi.12164
Blondel, C. J., Yang, H. J., Castro, B., Chiang, S., Toro, C. S., Zaldivar, M., et al. (2010). Contribution of the type VI secretion system encoded in SPI-19 to chicken colonization by Salmonella enterica serotypes Gallinarum and Enteritidis. PLoS ONE 5:e11724. doi: 10.1371/journal.pone.0011724
Bobrov, A. G., Kirillina, O., Fetherston, J. D., Miller, M. C., Burlison, J. A., and Perry, R. D. (2014). The Yersinia pestis siderophore, yersiniabactin, and the ZnuABC system both contribute to zinc acquisition and the development of lethal septicaemic plague in mice. Mol. Microbiol. 93, 759–775. doi: 10.1111/mmi.12693
Bondage, D. D., Lin, J. S., Ma, L. S., Kuo, C. H., and Lai, E. M. (2016). VgrG C terminus confers the type VI effector transport specificity and is required for binding with PAAR and adaptor-effector complex. Proc. Natl. Acad. Sci. U.S.A. 113, E3931–E3940. doi: 10.1073/pnas.1600428113
Bottone, E. J. (1997). Yersinia enterocolitica: the charisma continues. Clin. Microbiol. Rev. 10, 257–276.
Boyer, F., Fichant, G., Berthod, J., Vandenbrouck, Y., and Attree, I. (2009). Dissecting the bacterial type VI secretion system by a genome wide in silico analysis: what can be learned from available microbial genomic resources? BMC Genomics 10:104. doi: 10.1186/1471-2164-10-104
Brooks, T. M., Unterweger, D., Bachmann, V., Kostiuk, B., and Pukatzki, S. (2013). Lytic activity of the Vibrio cholerae type VI secretion toxin VgrG-3 is inhibited by the antitoxin TsaB. J. Biol. Chem. 288, 7618–7625. doi: 10.1074/jbc.M112.436725
Brubaker, R. R. (1991). Factors promoting acute and chronic diseases caused by yersiniae. Clin. Microbiol. Rev. 4, 309–324. doi: 10.1128/CMR.4.3.309
Brunet, Y. R., Henin, J., Celia, H., and Cascales, E. (2014). Type VI secretion and bacteriophage tail tubes share a common assembly pathway. EMBO Rep. 15, 315–321. doi: 10.1002/embr.201337936
Brunet, Y. R., Zoued, A., Boyer, F., Douzi, B., and Cascales, E. (2015). The type VI secretion TssEFGK-VgrG phage-like baseplate is recruited to the TssJLM membrane complex via multiple contacts and serves as assembly platform for tail tube/sheath polymerization. PLoS Genet. 11:e1005545. doi: 10.1371/journal.pgen.1005545
Cascales, E., and Cambillau, C. (2012). Structural biology of type VI secretion systems. Philos. Trans. R. Soc. Lond. B Biol. Sci. 367, 1102–1111. doi: 10.1098/rstb.2011.0209
Cathelyn, J. S., Crosby, S. D., Lathem, W. W., Goldman, W. E., and Miller, V. L. (2006). RovA, a global regulator of Yersinia pestis, specifically required for bubonic plague. Proc. Natl. Acad. Sci. U.S.A. 103, 13514–13519. doi: 10.1073/pnas.0603456103
Chakraborty, S., Sivaraman, J., Leung, K. Y., and Mok, Y. K. (2011). Two-component PhoB-PhoR regulatory system and ferric uptake regulator sense phosphate and iron to control virulence genes in type III and VI secretion systems of Edwardsiella tarda. J. Biol. Chem. 286, 39417–39430. doi: 10.1074/jbc.M111.295188
Chen, L., Zou, Y., She, P., and Wu, Y. (2015). Composition, function, and regulation of T6SS in Pseudomonas aeruginosa. Microbiol. Res. 172, 19–25. doi: 10.1016/j.micres.2015.01.004
Cianfanelli, F. R., Monlezun, L., and Coulthurst, S. J. (2016). Aim, load, fire: the type VI Secretion System, a bacterial nanoweapon. Trends Microbiol. 24, 51–62. doi: 10.1016/j.tim.2015.10.005
Cornelis, G. R., Boland, A., Boyd, A. P., Geuijen, C., Iriarte, M., Neyt, C., et al. (1998). The virulence plasmid of Yersinia, an antihost genome. Microbiol. Mol. Biol. Rev. 62, 1315–1352.
Dudley, E. G., Thomson, N. R., Parkhill, J., Morin, N. P., and Nataro, J. P. (2006). Proteomic and microarray characterization of the AggR regulon identifies a pheU pathogenicity island in enteroaggregative Escherichia coli. Mol. Microbiol. 61, 1267–1282. doi: 10.1111/j.1365-2958.2006.05281.x
Durand, E., Cambillau, C., Cascales, E., and Journet, L. (2014). VgrG, Tae, Tle, and beyond: the versatile arsenal of type VI secretion effectors. Trends Microbiol. 22, 498–507. doi: 10.1016/j.tim.2014.06.004
Gascuel, O. (1997). BIONJ: an improved version of the NJ algorithm based on a simple model of sequence data. Mol. Biol. Evol. 14, 685–695. doi: 10.1093/oxfordjournals.molbev.a025808
Goldova, J., Ulrych, A., Hercik, K., and Branny, P. (2011). A eukaryotic-type signalling system of Pseudomonas aeruginosa contributes to oxidative stress resistance, intracellular survival and virulence. BMC Genomics 12:437. doi: 10.1186/1471-2164-12-437
Guan, J., Xiao, X., Xu, S., Gao, F., Wang, J., Wang, T., et al. (2015). Roles of RpoS in Yersinia pseudotuberculosis stress survival, motility, biofilm formation and type VI secretion system expression. J. Microbiol. 53, 633–642. doi: 10.1007/s12275-015-0099-6
Gueguen, E., Durand, E., Zhang, X. Y., d'Amalric, Q., Journet, L., and Cascales, E. (2013). Expression of a Yersinia pseudotuberculosis type VI secretion system is responsive to envelope stresses through the ompr transcriptional activator. PLoS ONE 8:e66615. doi: 10.1371/journal.pone.0066615
Heesemann, J., Hantke, K., Vocke, T., Saken, E., Rakin, A., Stojiljkovic, I., et al. (1993). Virulence of Yersinia-Enterocolitica is closely associated with siderophore production, expression of an iron-repressible outer-membrane polypeptide of 65000 Da and pesticin sensitivity. Mol. Microbiol. 8, 397–408. doi: 10.1111/j.1365-2958.1993.tb01583.x
Hood, R. D., Singh, P., Hsu, F., Guvener, T., Carl, M. A., Trinidad, R. R., et al. (2010). A type VI secretion system of Pseudomonas aeruginosa targets a toxin to bacteria. Cell Host Microbe 7, 25–37. doi: 10.1016/j.chom.2009.12.007
Hu, Y., Lu, P., Wang, Y., Ding, L., Atkinson, S., and Chen, S. (2009). OmpR positively regulates urease expression to enhance acid survival of Yersinia pseudotuberculosis. Microbiology 155, 2522–2531. doi: 10.1099/mic.0.028381-0
Ishikawa, T., Sabharwal, D., Broms, J., Milton, D. L., Sjostedt, A., Uhlin, B. E., et al. (2012). Pathoadaptive conditional regulation of the type VI secretion system in Vibrio cholerae O1 strains. Infect. Immun. 80, 575–584. doi: 10.1128/IAI.05510-11
Jaakkola, K., Somervuo, P., and Korkeala, H. (2015). Comparative genomic hybridization analysis of Yersinia enterocolitica and Yersinia pseudotuberculosis identifies genetic traits to elucidate their different ecologies. Biomed. Res. Int. 2015:760494. doi: 10.1155/2015/760494
Jani, A. J., and Cotter, P. A. (2010). Type VI secretion: not just for pathogenesis anymore. Cell Host Microbe 8, 2–6. doi: 10.1016/j.chom.2010.06.012
Jiang, F., Wang, X., Wang, B., Chen, L., Zhao, Z., Waterfield, N. R., et al. (2016). The Pseudomonas aeruginosa type VI secretion PGAP1-like effector induces host autophagy by activating endoplasmic reticulum stress. Cell Rep. 16, 1502–1509. doi: 10.1016/j.celrep.2016.07.012
Joshi, A., Kostiuk, B., Rogers, A., Teschler, J., Pukatzki, S., and Yildiz, F. H. (2017). Rules of engagement: the type VI secretion system in Vibrio cholerae. Trends Microbiol. 25, 267–279. doi: 10.1016/j.tim.2016.12.003
Kitaoka, M., Miyata, S. T., Brooks, T. M., Unterweger, D., and Pukatzki, S. (2011). VasH is a transcriptional regulator of the type VI secretion system functional in endemic and pandemic Vibrio cholerae. J. Bacteriol. 193, 6471–6482. doi: 10.1128/JB.05414-11
Lertpiriyapong, K., Gamazon, E. R., Feng, Y., Park, D. S., Pang, J., Botka, G., et al. (2012). Campylobacter jejuni type VI secretion system: roles in adaptation to deoxycholic acid, host cell adherence, invasion, and in vivo colonization. PLoS ONE 7:e42842. doi: 10.1371/journal.pone.0042842
Li, J., Yao, Y., Xu, H. H., Hao, L., Deng, Z., Rajakumar, K., et al. (2015). SecReT6: a web-based resource for type VI secretion systems found in bacteria. Environ. Microbiol. 17, 2196–2202. doi: 10.1111/1462-2920.12794
Lien, Y. W., and Lai, E. M. (2017). Type VI secretion effectors: methodologies and biology. Front. Cell. Infect. Microbiol. 7:254. doi: 10.3389/fcimb.2017.00254
Lin, J., Zhang, W., Cheng, J., Yang, X., Zhu, K., Wang, Y., et al. (2017). A Pseudomonas T6SS effector recruits PQS-containing outer membrane vesicles for iron acquisition. Nat. Commun. 8:14888. doi: 10.1038/ncomms14888
Liu, H., Coulthurst, S. J., Pritchard, L., Hedley, P. E., Ravensdale, M., Humphris, S., et al. (2008). Quorum sensing coordinates brute force and stealth modes of infection in the plant pathogen Pectobacterium atrosepticum. PLoS Pathog. 4:e1000093. doi: 10.1371/journal.ppat.1000093
Ma, A. T., and Mekalanos, J. J. (2010). In vivo actin cross-linking induced by Vibrio cholerae type VI secretion system is associated with intestinal inflammation. Proc. Natl. Acad. Sci. U.S.A. 107, 4365–4370. doi: 10.1073/pnas.0915156107
MacIntyre, D. L., Miyata, S. T., Kitaoka, M., and Pukatzki, S. (2010). The Vibrio cholerae type VI secretion system displays antimicrobial properties. Proc. Natl. Acad. Sci. U.S.A. 107, 19520–19524. doi: 10.1073/pnas.1012931107
Mougous, J. D., Cuff, M. E., Raunser, S., Shen, A., Zhou, M., Gifford, C. A., et al. (2006). A virulence locus of Pseudomonas aeruginosa encodes a protein secretion apparatus. Science 312, 1526–1530. doi: 10.1126/science.1128393
Murdoch, S. L., Trunk, K., English, G., Fritsch, M. J., Pourkarimi, E., and Coulthurst, S. J. (2011). The opportunistic pathogen Serratia marcescens utilizes type VI secretion to target bacterial competitors. J. Bacteriol. 193, 6057–6069. doi: 10.1128/JB.05671-11
Parsons, D. A., and Heffron, F. (2005). sciS, an icmF homolog in Salmonella enterica serovar Typhimurium, limits intracellular replication and decreases virulence. Infect. Immun. 73, 4338–4345. doi: 10.1128/IAI.73.7.4338-4345.2005
Pell, L. G., Kanelis, V., Donaldson, L. W., Howell, P. L., and Davidson, A. R. (2009). The phage lambda major tail protein structure reveals a common evolution for long-tailed phages and the type VI bacterial secretion system. Proc. Natl. Acad. Sci. U.S.A. 106, 4160–4165. doi: 10.1073/pnas.0900044106
Pieper, R., Huang, S. T., Robinson, J. M., Clark, D. J., Alami, H., Parmar, P. P., et al. (2009). Temperature and growth phase influence the outer-membrane proteome and the expression of a type VI secretion system in Yersinia pestis. Microbiology 155, 498–512. doi: 10.1099/mic.0.022160-0
Podladchikova, O., Antonenka, U., Heesemann, J., and Rakin, A. (2011). Yersinia pestis autoagglutination factor is a component of the type six secretion system. Int. J. Med. Microbiol. 301, 562–569. doi: 10.1016/j.ijmm.2011.03.004
Ponnusamy, D., Fitts, E. C., Sha, J., Erova, T. E., Kozlova, E. V., Kirtley, M. L., et al. (2015). High-throughput, signature-tagged mutagenic approach to identify novel virulence factors of Yersinia pestis CO92 in a mouse model of infection. Infect. Immun. 83, 2065–2081. doi: 10.1128/IAI.02913-14
Pujol, C., and Bliska, J. B. (2005). Turning Yersinia pathogenesis outside in: subversion of macrophage function by intracellular yersiniae. Clin. Immunol. 114, 216–226. doi: 10.1016/j.clim.2004.07.013
Pukatzki, S., Ma, A. T., Sturtevant, D., Krastins, B., Sarracino, D., Nelson, W. C., et al. (2006). Identification of a conserved bacterial protein secretion system in Vibrio cholerae using the Dictyostelium host model system. Proc. Natl. Acad. Sci. U.S.A. 103, 1528–1533. doi: 10.1073/pnas.0510322103
Putzker, M., Sauer, H., and Sobe, D. (2001). Plague and other human infections caused by Yersinia species. Clin. Lab. 47, 453–466.
Robinson, J. B., Telepnev, M. V., Zudina, I. V., Bouyer, D., Montenieri, J. A., Bearden, S. W., et al. (2009). Evaluation of a Yersinia pestis mutant impaired in a thermoregulated type VI-like secretion system in flea, macrophage and murine models. Microb. Pathog. 47, 243–251. doi: 10.1016/j.micpath.2009.08.005
Rogers, A., Townsley, L., Gallego-Hernandez, A. L., Beyhan, S., Kwuan, L., and Yildiz, F. H. (2016). The LonA protease regulates biofilm formation, motility, virulence, and the type VI secretion system in Vibrio cholerae. J. Bacteriol. 198, 973–985. doi: 10.1128/JB.00741-15
Russell, A. B., LeRoux, M., Hathazi, K., Agnello, D. M., Ishikawa, T., Wiggins, P. A., et al. (2013). Diverse type VI secretion phospholipases are functionally plastic antibacterial effectors. Nature 496, 508–512. doi: 10.1038/nature12074
Russell, A. B., Peterson, S. B., and Mougous, J. D. (2014). Type VI secretion system effectors: poisons with a purpose. Nat. Rev. Microbiol. 12, 137–148. doi: 10.1038/nrmicro3185
Russell, A. B., Singh, P., Brittnacher, M., Bui, N. K., Hood, R. D., Carl, M. A., et al. (2012). A widespread bacterial type VI secretion effector superfamily identified using a heuristic approach. Cell Host Microbe 11, 538–549. doi: 10.1016/j.chom.2012.04.007
Salomon, D., Gonzalez, H., Updegraff, B. L., and Orth, K. (2013). Vibrio parahaemolyticus type VI secretion system 1 is activated in marine conditions to target bacteria, and is differentially regulated from system 2. PLoS ONE 8:e61086. doi: 10.1371/journal.pone.0061086
Salomon, D., and Orth, K. (2015). Type VI secretion system. Curr. Biol. 25, R265–R266. doi: 10.1016/j.cub.2015.02.031
Sana, T. G., Baumann, C., Merdes, A., Soscia, C., Rattei, T., Hachani, A., et al. (2015). Internalization of Pseudomonas aeruginosa strain PAO1 into epithelial cells is promoted by interaction of a T6SS effector with the microtubule network. MBio 6:e00712. doi: 10.1128/mBio.00712-15
Schell, M. A., Ulrich, R. L., Ribot, W. J., Brueggemann, E. E., Hines, H. B., Chen, D., et al. (2007). Type VI secretion is a major virulence determinant in Burkholderia mallei. Mol. Microbiol. 64, 1466–1485. doi: 10.1111/j.1365-2958.2007.05734.x
Schwarz, S., Singh, P., Robertson, J. D., LeRoux, M., Skerrett, S. J., Goodlett, D. R., et al. (2014). VgrG-5 is a Burkholderia type VI secretion system-exported protein required for multinucleated giant cell formation and virulence. Infect. Immun. 82, 1445–1452. doi: 10.1128/IAI.01368-13
Schwarz, S., West, T. E., Boyer, F., Chiang, W. C., Carl, M. A., Hood, R. D., et al. (2010). Burkholderia type VI secretion systems have distinct roles in eukaryotic and bacterial cell interactions. PLoS Pathog. 6:e1001068. doi: 10.1371/journal.ppat.1001068
Schwiesow, L., Lam, H., Dersch, P., and Auerbuch, V. (2015). Yersinia type III secretion system master regulator LcrF. J. Bacteriol. 198, 604–614. doi: 10.1128/JB.00686-15
Shneider, M. M., Buth, S. A., Ho, B. T., Basler, M., Mekalanos, J. J., and Leiman, P. G. (2013). PAAR-repeat proteins sharpen and diversify the type VI secretion system spike. Nature 500, 350–353. doi: 10.1038/nature12453
Si, M., Wang, Y., Zhang, B., Zhao, C., Kang, Y., Bai, H., et al. (2017a). The type VI secretion system engages a redox-regulated dual-functional heme transporter for zinc acquisition. Cell Rep. 20, 949–959. doi: 10.1016/j.celrep.2017.06.081
Si, M., Zhao, C., Burkinshaw, B., Zhang, B., Wei, D., Wang, Y., et al. (2017b). Manganese scavenging and oxidative stress response mediated by type VI secretion system in Burkholderia thailandensis. Proc. Natl. Acad. Sci. U.S.A. 114, E2233–E2242. doi: 10.1073/pnas.1614902114
Silverman, J. M., Brunet, Y. R., Cascales, E., and Mougous, J. D. (2012). Structure and regulation of the type VI secretion system. Ann. Rev. Microbiol. 66, 453–472. doi: 10.1146/annurev-micro-121809-151619
Song, Y., Xiao, X., Li, C., Wang, T., Zhao, R., Zhang, W., et al. (2015). The dual transcriptional regulator RovM regulates the expression of AR3- and T6SS4-dependent acid survival systems in response to nutritional status in Yersinia pseudotuberculosis. Environ. Microbiol. 17, 4631–4645. doi: 10.1111/1462-2920.12996
Southey-Pillig, C. J., Davies, D. G., and Sauer, K. (2005). Characterization of temporal protein production in Pseudomonas aeruginosa biofilms. J. Bacteriol. 187, 8114–8126. doi: 10.1128/JB.187.23.8114-8126.2005
Suarez, G., Sierra, J. C., Sha, J., Wang, S., Erova, T. E., Fadl, A. A., et al. (2008). Molecular characterization of a functional type VI secretion system from a clinical isolate of Aeromonas hydrophila. Microb. Pathog. 44, 344–361. doi: 10.1016/j.micpath.2007.10.005
Tian, Y., Zhao, Y., Wu, X., Liu, F., Hu, B., and Walcott, R. R. (2015). The type VI protein secretion system contributes to biofilm formation and seed-to-seedling transmission of Acidovorax citrulli on melon. Mol. Plant Pathol. 16, 38–47. doi: 10.1111/mpp.12159
Wang, M., Luo, Z., Du, H., Xu, S., Ni, B., Zhang, H., et al. (2011). Molecular characterization of a functional type VI secretion system in Salmonella enterica serovar Typhi. Curr. Microbiol. 63, 22–31. doi: 10.1007/s00284-011-9935-z
Wang, T., Chen, K., Gao, F., Kang, Y., Chaudhry, M. T., Wang, Z., et al. (2017). ZntR positively regulates T6SS4 expression in Yersinia pseudotuberculosis. J. Microbiol. 55, 448–456. doi: 10.1007/s12275-017-6540-2
Wang, T., Si, M., Song, Y., Zhu, W., Gao, F., Wang, Y., et al. (2015). Type VI secretion system transports Zn2+ to combat multiple stresses and host immunity. PLoS Pathog. 11:e1005020. doi: 10.1371/journal.ppat.1005020
Wang, T. T., Yang, X. B., Gao, F., Zhao, C., Kang, Y. W., Wang, Y., et al. (2016). Zinc acquisition via ZnuABC in Yersinia pseudotuberculosis facilitates resistance to oxidative stress. Ann. Microbiol. 66, 1189–1197. doi: 10.1007/s13213-016-1205-7
Weber, B., Croxatto, A., Chen, C., and Milton, D. L. (2008). RpoS induces expression of the Vibrio anguillarum quorum-sensing regulator VanT. Microbiology 154, 767–780. doi: 10.1099/mic.0.2007/014167-0
Weber, B., Hasic, M., Chen, C., Wai, S. N., and Milton, D. L. (2009). Type VI secretion modulates quorum sensing and stress response in Vibrio anguillarum. Environ. Microbiol. 11, 3018–3028. doi: 10.1111/j.1462-2920.2009.02005.x
Yang, X., Long, M., and Shen, X. (2018). Effector(-)immunity pairs provide the T6SS nanomachine its offensive and defensive capabilities. Molecules 23:E1009. doi: 10.3390/molecules23051009
Yu, Y., Fang, L., Zhang, Y., Sheng, H., and Fang, W. (2015). VgrG2 of type VI secretion system 2 of Vibrio parahaemolyticus induces autophagy in macrophages. Front. Microbiol. 6:168. doi: 10.3389/fmicb.2015.00168
Zhang, L., Hinz, A. J., Nadeau, J. P., and Mah, T. F. (2011a). Pseudomonas aeruginosa tssC1 links type VI secretion and biofilm-specific antibiotic resistance. J. Bacteriol. 193, 5510–5513. doi: 10.1128/JB.00268-11
Zhang, W., Wang, Y., Song, Y., Wang, T., Xu, S., Peng, Z., et al. (2013). A type VI secretion system regulated by OmpR in Yersinia pseudotuberculosis functions to maintain intracellular pH homeostasis. Environ. Microbiol. 15, 557–569. doi: 10.1111/1462-2920.12005
Zhang, W., Xu, S., Li, J., Shen, X., Wang, Y., and Yuan, Z. (2011b). Modulation of a thermoregulated type VI secretion system by AHL-dependent quorum sensing in Yersinia pseudotuberculosis. Arch. Microbiol. 193, 351–363. doi: 10.1007/s00203-011-0680-2
Zheng, J., and Leung, K. Y. (2007). Dissection of a type VI secretion system in Edwardsiella tarda. Mol. Microbiol. 66, 1192–1206. doi: 10.1111/j.1365-2958.2007.05993.x
Zheng, J., Shin, O. S., Cameron, D. E., and Mekalanos, J. J. (2010). Quorum sensing and a global regulator TsrA control expression of type VI secretion and virulence in Vibrio cholerae. Proc. Natl. Acad. Sci. U.S.A. 107, 21128–21133. doi: 10.1073/pnas.1014998107
Keywords: pathogenic Yersinia, type VI secretion system (T6SS), function, effectors, regulation
Citation: Yang X, Pan J, Wang Y and Shen X (2018) Type VI Secretion Systems Present New Insights on Pathogenic Yersinia. Front. Cell. Infect. Microbiol. 8:260. doi: 10.3389/fcimb.2018.00260
Received: 02 May 2018; Accepted: 13 July 2018;
Published: 31 July 2018.
Edited by:
Matthew S. Francis, Umeå University, SwedenReviewed by:
Cristina Elisa Alvarez-Martinez, Universidade Estadual de Campinas, BrazilErwan Gueguen, Université de Lyon, France
Christopher Alteri, University of Michigan, United States
Copyright © 2018 Yang, Pan, Wang and Shen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xihui Shen, eGlodWlzaGVuQG53c3VhZi5lZHUuY24=
†These authors have contributed equally to this work.
 Xiaobing Yang
Xiaobing Yang Junfeng Pan
Junfeng Pan Yao Wang
Yao Wang Xihui Shen
Xihui Shen