- 1Shandong Sino-Japanese Center for Collaborative Research of Carbon Nanomaterials, College of Chemistry and Chemical Engineering, College of Life Sciences, Qingdao University, Qingdao, China
- 2The Affiliated Hospital of Qingdao University Medical College, Qingdao, China
- 3Shandong Provincial Key Laboratory of Biochemical Engineering, College of Marine Science and Biological Engineering, Qingdao University of Science and Technology, Qingdao, China
Staphylococcus aureus (S. aureus) contamination in food safety has become a worldwide health problem. In this work, we utilized RNA one-step detection of denaturation bubble-mediated Strand Exchange Amplification (SEA) method to realize the detection of viable foodborne pathogen S. aureus. A pair of S. aureus specific primers were designed for the SEA reaction by targeting hypervariable V2 region of 16S rDNA and the amplification reaction was finished about 1 h. The results of amplification reaction could be observed by the naked eyes with a significant color change from light yellow to red to realize the colorimetric detection of S. aureus. Therefore, there only required an isothermal water bath, which was very popular for areas with limited resources. In real sample testing, although the SEA detection was so time-saving compared with the traditional plating method, the SEA method showed great consistency with the traditional plating method. In view of the above-described advantages, we provided a simple, rapid and equipment-free detection method, which had a great potential on ponit-of-care testing (POCT) application. Our method reported here will also provide a POCT detection platform for other food-borne pathogens in food, even pathogenic bacteria from other fields.
Introduction
Staphylococcus aureus (S. aureus), a facultative anaerobic Gram-positive coccus, can greatly threaten our health due to a combination of toxin-mediated virulence, invasiveness, and antibiotic resistance (Oliveira et al., 2011). For example, S. aureus is the main cause of nosocomial infections and community-acquired diseases, including deep-seated, endocarditis, abscesses and bacteria, which lead to toxic and septic shock syndromes (Abdalhai et al., 2014). S. aureus widely exists in the air, water, dust, human and animal excretions, which makes the food much easier to become contaminated (Yu et al., 2016). It has been reported that S. aureus contamination is a worldwide health problem. In the United States, nearly half a million hospitalizations and 50,000 deaths occur resulting from S. aureus each year (Schlecht et al., 2015). Likewise, 11 outbreaks of Staphylococcal food poisoning were reported between 2006 and 2009 in Shenzhen, China, which ranked the second most frequent cause of bacterial food poisoning (Yan et al., 2012). Therefore, an efficient S. aureus detection method is necessary for food safety and human health.
To date, S. aureus has been listed as a legal testing item in food safety of most countries worldwide (Xiong et al., 2016). The conventional S. aureus detection method is performed by the culture-based technique, which is time-consuming (4–7 days) (Roda et al., 2012). Immunological procedures, including immunoprecipitation, enzyme-labeled immunosorbent assay, and immunoblotting are also used for detecting S. aureus based on specific binding of antigen and antibody (Min et al., 2011). However, the sensitivity of these methods are low and their operations are usually very difficult and tedious, which make them hard to popularize (Kimura et al., 2013). Recently, with the development of molecular biology technology, polymerase chain reaction (PCR) (Niraj et al., 2016) has been widely used for the S. aureus nucleic acid detection. These methods derived from PCR require sophisticated instruments and lifting temperature, and they are not suitable for the point-of-care (POCT) testing. Considering the wide distribution and the harmful effects of S. aureus, therefore, a rapid and simple method for the detection of S. aureus was seriously required. Recently, a strand exchange amplification (SEA), mediated by denaturation bubbles, was established by Shi et al. (2016). Similarly to the traditional PCR technique, SEA employed a Bst DNA polymerase and a pair of primers to carry out an exponential DNA amplification under an isothermal condition. In addition, Bst DNA polymerase has intrinsic reverse transcriptase activity (Shi et al., 2015), which makes the SEA method able to detect RNA directly without additional reverse transcription. Therefore, in this work, we firstly utilized RNA one-step detection of SEA method to realize the detection of viable foodborne pathogen S. aureus. In particular, the sensitivity of SEA method was greatly improved in this work due to the high abundance of RNA in live bacteria. The whole SEA detection procedure for real samples took only 1–2 h. In addition, the colorimetric detection of S. aureus was also developed to realize the detection result read out by the naked eyes. We firstly utilized RNA one-step detection of SEA method to realize the detection of viable foodborne pathogen S. aureus. The sensitivity of SEA method especially, was greatly improved in this work due to the high abundance of RNA in live bacteria. The whole SEA detection procedure for real samples took only 1–2 h. In addition, the colorimetric detection of S. aureus was also developed to realize the detection result read out by the naked eyes. Therefore, there was only the need for a metal bath, which was very popular for areas with limited resources. In brief, we firstly used the SEA method for rapid and simple detection of foodborne pathogen S. aureus, even viable S. aureus, to further ensure food safety.
Materials and Methods
Reagents and Bacterial Strains
SEA detection kits were obtained from Qingdao Navid Biotechnology Co. Ltd. (China). Twenty bp DNA marker, 6 × DNA loading buffer and sodium dodecyl sulfate (SDS) were purchased from Sangon Biotech (Shanghai, China). Acrylamide and methylene diacrylamide were purchased from Sigma-Aldrich (St Louis, MO, USA). Recombinant DNase I was purchased from Takara Bio (Beijing, China). RNA-Be-Gone was purchased from Sangon Biotech (Shanghai, China). The bacterial strains including S. aureus, Listeria monocytogenes (L. monocytogenes), Salmonella typhimurium (S. typhimurium), Shigella castellani (S. castellani), Vibrio parahemolyticus (V. parahemolyticus), and Escherichia coli (E. coli) were stored in our laboratory.
Primers Design and Synthesis
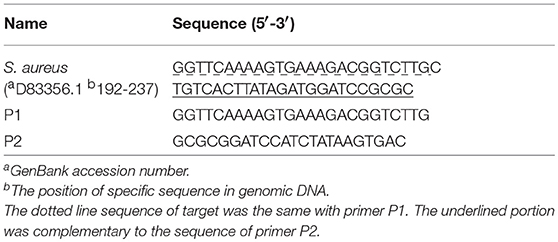
A pair of specific primers (Table 1) were designed in the hypervariable region of S. aureus 16S rDNA with the NUPACK software (http://www.nupack.org/) and DINAMelt Web Server (http://unafold.rna.albany.edu/?q=DINAMelt). The primers were synthesized and purified by high performance liquid chromatography (HPLC) in Sangon Biotech (Shanghai, China).
Extraction of Genomic DNA and RNA From S. aureus
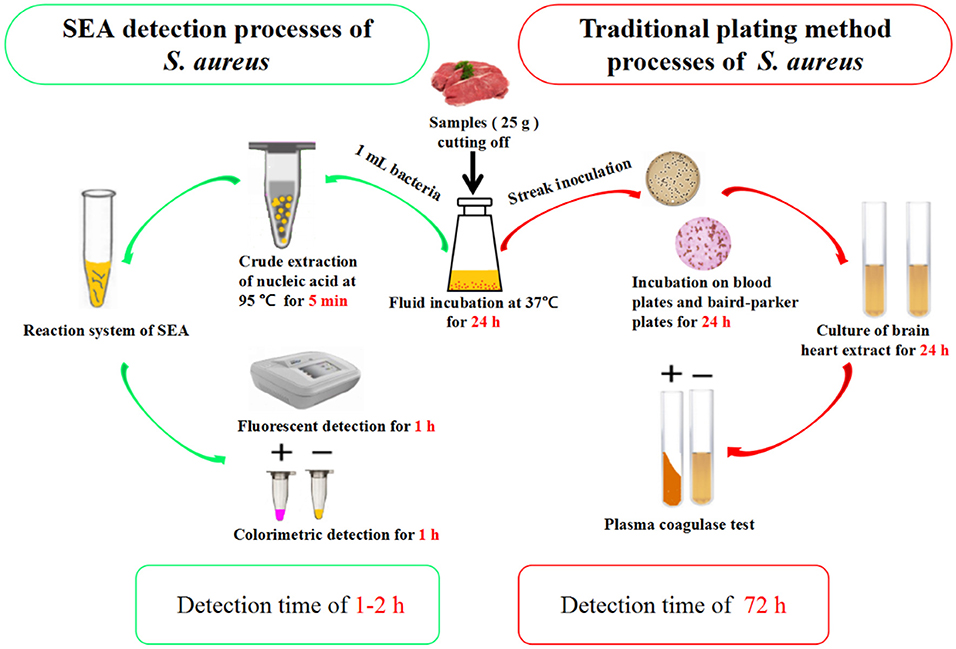
The genomic DNA of S. aureus was extracted with TIANamp bacteria DNA kits (Tiangen Biotech, Beijing, China) according to the literature (Ulrich and Hughes, 2010). The target for colorimetric assay, specificity and anti-interference experiments. After enrichment culture for 24 h at 37°C, 1 mL bacterial liquid was heated at 95°C for 5 min, then centrifugated. One micro liter supernatant was used as target for colorimetric assay, specificity and anti-interference experiments.
Isothermal Amplification Reaction
The SEA reaction was performed according to the manufacturers' instructions under optimal conditions. The fluorescence signal of the SEA reaction was detected by the Gentier 48S Isothermal amplification fluorescence detection system at 1 min intervals. Gel images were recorded by the ChampGel 5,000 system (Saizhi Innovation Technology Co. Ltd, Beijing, China). The reaction temperature was optimized at 57°C, 58.2°C, 60.5°C, 62°C, 63.5°C, 65.3°C, 66.6°C, and 67°C, respectively.
Feasibility and Sensitivity of SEA Detection Method
The genomic DNA of S. aureus was used to demonstrate the feasibility of the SEA method. The sensitivity of the SEA detection method for S. aureus was evaluated with different dilutions of genomic DNA and DNA fragments. The culture fluids of S. aureus with L. monocytogenes, S. typhimurium, V. parahemolyticus, S. castellani, and E. coli were used to demonstrate the specificity and anti-jamming capacity of the SEA method.
Colorimetric Assay by SEA Method
Colorimetric kits were obtained from Qingdao Navid Biotechnology Co. Ltd. (China). Rapid nucleic acid extraction from bacterial fluids was used to demonstrate the feasibility of SEA colorimetric method.
Detection of S. aureus in Real Samples
The beef, pork, chicken, dried fish, and ham sausage real samples were collected from the small farmers markets (Qingdao, China). Samples were divided into 25 g each with a sterile knife. Then each sample was transferred to 225 mL 7.5% NaCl and then treated according to the literature with slight modifications (Abdalhai et al., 2014). After enrichment at 37°C for 24 h, 1 mL enrichment solution was taken from each sample for SEA and traditional plating detection, respectively.
Results and Discussion
The Design of SEA for the Detection of S. aureus
SEA was based on the single-stranded denaturation bubbles of dsDNA at the reaction temperature, including a pair of primers, Bst DNA polymerase, and a constant temperature. The specific primers (P1 and P2) were designed with 50 bp amplification fragment in the hypervariable V2 region of 16S rDNA (Table 1), which has a high copy number in bacteria. Briefly, one pair of specific primers bound with the targeted DNA fragments by invading the denaturing bubbles and induced DNA polymerase to extend the chain. In addition, according to the Tm values of primers, the reaction temperature was optimized using the SEA kit with S. aureus genomic DNA as the template. Finally, 62°C was chosen as the optimal reaction temperature (Figure S1) in this work.
The Feasibility of SEA Method to Detect S. aureus
Genomic DNA of S. aureus was used as the template to demonstrate the feasibility of SEA method to detect S. aureus. As shown in Figure 1, compared with the no target control (NTC) group, the fluorescence signal with the addition of genomic DNA significantly increased, which indicated that the SEA method could effectively detect genomic DNA of S. aureus. This result was consistent with the targeted 50 bp amplification products in the gel electrophoresis (Figure 1 inserted), which further confirmed the feasibility of SEA method to detect S. aureus.
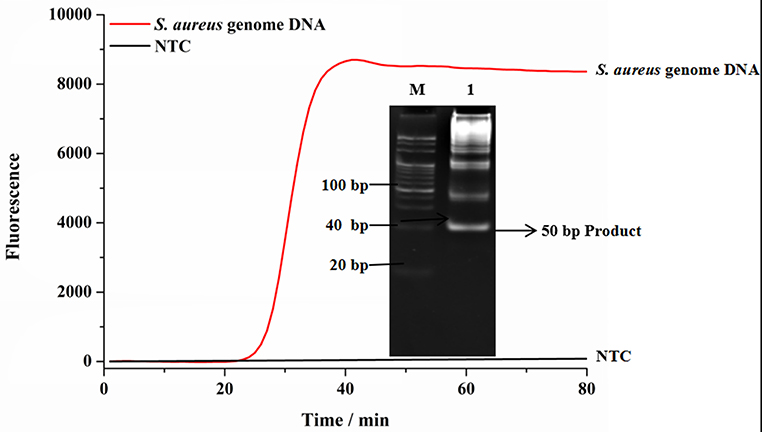
Figure 1. The feasibility of SEA to detect S. aureus. The red line represented that the targets were 1.0 × 10−11 M genomic DNA; The black line represented no targets control (NTC); Inset represented the amplified products in the gel electrophoresis was 50 bp.
Sensitivity of SEA to Detect DNA Fragments and Genomic DNA of S. aureus
To evaluate the sensitivity of SEA detection method for S. aureus (Zhang et al., 2018), different concentrations of DNA fragments and genomic DNA of S. aureus were detected. As shown in Figure 2A, the fluorescence signals gradually increased with the increasing concentrations of targeted DNA fragments ranging from 1.0 × 10−11 M to 1.0 × 10−14 M. In addition, as shown in Figure 2B, the threshold time (Tt) value increased linearly with the increasing negative logarithm (lg) value of concentrations of S. aureus DNA fragments, ranging from 1.0 × 10−11 M to 1.0 × 10−14 M. The regression equation was Tt = 13.299 (-lgCDNA)−134.35 (CDNA was the concentration of S. aureus DNA fragments, R2 = 0.9796). Moreover, SEA was also carried out using 10-fold serial dilutions of genomic DNA extracted from S. aureus. The results showed that SEA could be used to detect the concentration of S. aureus genomic DNA as low as 400 pg/μL (Figure 2C). As shown in Figure 2D, the Tt value increased linearly with the increasing lg of S. aureus genomic DNA concentrations in the range from 40 ng/μL to 400 pg/μL, which yielded a correlation equation of Tt = 13.32 (-lgCDNA)+49.68 (CDNA was the concentration of S. aureus genomic DNA, R2 = 0.9993). In conclusion, the SEA method showed good linearity and sensitivity both in detecting genome DNA and DNA fragments.
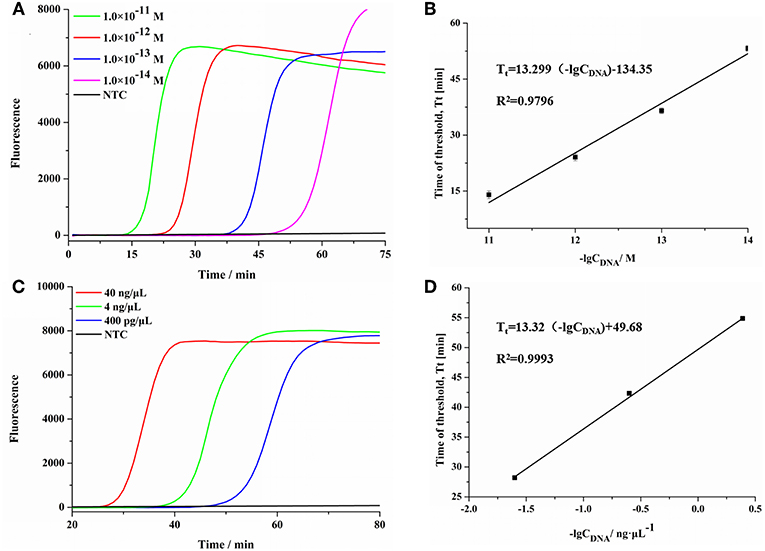
Figure 2. The sensitivity of SEA method for S. aureus. (A) The real-time fluorescence curves for different concentrations of S. aureus DNA fragment. (B) Relationship between the Tt values and the negative logarithmic values of the concentration of S. aureus DNA targets (DNA fragments concentration). Error bars showed mean standard deviations of three determinations. (C) The real-time fluorescence curves for different concentrations of S. aureus genomic DNA. (D) Relationship between the Tt values and the negative logarithmic values of the concentration of S. aureus DNA targets (DNA concentration). Error bars showed mean standard deviations of three determinations.
SEA Detection for Viable S. aureus
RNA is readily degradable in the environment of pathogens in vitro, a biomarker of live bacteria, has been considered as a more suitable target for live bacteria detection. Therefore, using the SEA method to detect RNA and DNA in one step can not only take advantage of the abundance of RNA in live bacteria, but also make up for the low sensitivity of DNA detection. In this assay, total S. aureus nucleic acid (DNA and RNA), DNA and RNA were used as targets, respectively, to elucidate the nucleic acid detection efficiency of the SEA method. As shown in Figure 3, the SEA method could realize the detection of viable S. aureus by one-step detection of RNA by the fluorescence signal. We also found that the occurrence of the fluorescence signal was significantly delayed with DNA or RNA elimination, which indicated that detecting RNA and DNA simultaneously with the SEA method showed higher sensitivity than that of DNA or RNA independently. This result further verified that SEA method can not only detect DNA, but also RNA by one-step.
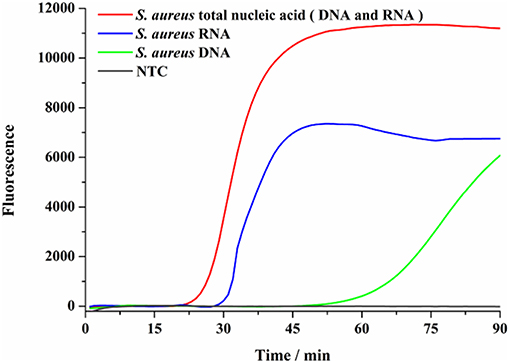
Figure 3. SEA detection on nucleic acids. Total nucleic acid was conducted to SEA detection of S. aureus (red line). The total nucleic acid was treated with DNA scavenger of recombinant DNase I before SEA detection of S. aureus (blue line). The total nucleic acid was treated with RNA scavenger of RNA-Be-Gone before SEA detection of S. aureus (green line).
In addition, the targeted RNA fragments are relatively short in the SEA assay, so that both RNA and most of its incompletely-degraded RNA fragments could be used as amplification templates. This would undoubtedly improve the stability and sensitivity of SEA method for RNA detection.
Detection of S. aureus by the Colorimetric Assay
Considering the wide existence in nature and severe effects of S. aureus, we have to make surveillance on it in various foods to ensure our health. At present, most rapid detection methods of S. aureus depend on complex and large-scale precision instruments, so that they are not suitable for developing regions. Therefore, it is necessary to develop a simple and rapid detection method for S. aureus. We developed a colorimetric method combined with SEA to detect S. aureus. As shown in Figure 4, the reaction mixture color could change from light yellow to red for positive samples and stayed light yellow for negative samples. Most importantly, the whole colorimetric detection procedure took no more than 50 min and was read out by the naked eyes, which was extremely simple and convenient. As shown in Figure 4 inset, the positive group changed its color from light yellow to red, while the NTC group remained the original yellow, which was consistent with the fluorescence results (Figure 4). Consequently, colorimetric detection results of the SEA method could be readily observed by the naked eyes instead of using the large and costly fluorescence detection equipment (Wang et al., 2014). In a word, the colorimetric SEA results could be performed without any complicated detection instruments or well-trained staff, which was especially applicable for the field detection of S. aureus in resource-limited environments.
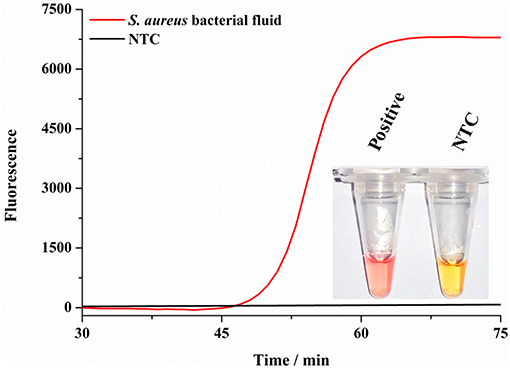
Figure 4. Detection of S. aureus by colorimetry. Fluorescence curves of the SEA reaction for S. aureus culture fluids and NTC; Inset represented the corresponding colorimetric result of SEA method to detect culture fluids of S. aureus and NTC.
Detection of S. aureus in Real Samples
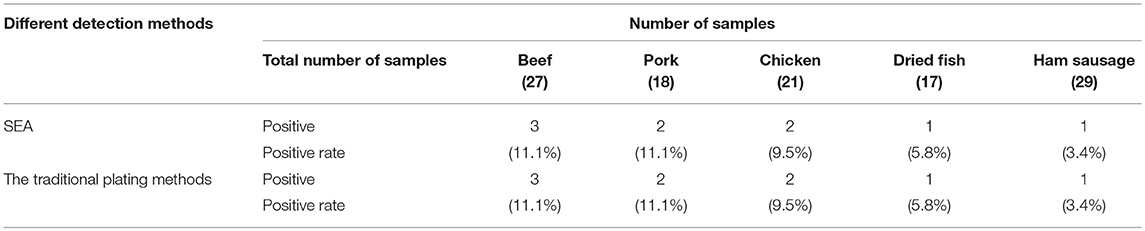
Five other bacterial fluids, including L. monocytogenes, S. typhimurium, S. castellani, V. parahemolyticus, and E. coli were used to verify the specificity and anti-interference of the SEA method (Yang et al., 2016). As shown in Figure S2, the fluorescence signal only occurred in detecting S. aureus and no obvious effects were observed with the existence of other bacterial mixture, showing the good specificity and anti-interference of the SEA method. Thereafter, we carried out further artificially S. aureus-contaminated pork tests. The results showed that the detection limit of the SEA method on S. aureus was 100 cfu/g (Figure S3), which met the requirements that S. aureus must not be detected out in foods. Furthermore, different meat real samples collected from the small farmers market were detected by the traditional plating method and SEA method for S. aureus simultaneously. As shown in Figure 5, the SEA method has a great advantage over the traditional plating method in detection time. The SEA method took only 1–2 h to detect real samples simply and rapidly, however, the traditional plating method need to spend as long as 72 h. In addition, positive rates of the SEA detection (Table 2) for S. aureus among those samples were consistent with that of the traditional plating method.
Conclusions
A rapid and simple SEA method for the detection of S. aureus was developed in this work. SEA method to detect RNA of S. aureus by one step took advantage of the high abundance of RNA in live bacteria, which made SEA method not only able to detect viable S. aureus, but also to greatly improve the low sensitivity of DNA detection. Furthermore, the colorimetric detection could be conducted at a constant temperature and then read out results by the naked eyes. The whole SEA detection procedure for real samples took only 1–2 h, which was so time-saving compared with the traditional plating method taking as long as 72 h. Thus, the SEA method provided a simple, rapid and equipment-free detection platform. It was also expected to be incorporated into microfluidic chips to realize the sample-to-answer diagnostic in a single device for further POCT purpose.
Author Contributions
CS conceived and designed the experiments. CL performed the experiments. ML and MW analyzed the data. CM and ZW wrote the paper. CS revised and approved the final version.
Funding
This study was financially supported by grants from the National Natural Science Foundation of China (21475071, 31670868, 21675094), the Taishan Scholar Program of Shandong (ts201511027), and the Shandong Province Natural Science Fund Major Basic Research Project (ZR2017ZC0123).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fchem.2019.00124/full#supplementary-material
Abbreviations
S. aureus, Staphylococcus aureus; SEA, strand exchange amplification; POCT, point-of-care testing; PCR, polymerase chain reaction; ELISA, enzyme linked immunosorbent assay; NASBA, nucleic acid sequence-based amplification; LAMP, loop-mediated isothermal amplification; SDS, sodium dodecyl sulfate; HPLC, high performance liquid chromatography; NTC, no target control; PAGE, polyacrylamide gel electrophoresis.
References
Abdalhai, M. H., Fernandes, A. M., Bashari, M., Ji, J., He, Q., and Sun, X. (2014). Rapid and sensitive detection of foodborne pathogenic bacteria (Staphylococcus aureus) using an electrochemical DNA genomic biosensor and its application in fresh beef. J. Agric. Food Chem. 62, 12659–12667. doi: 10.1021/jf503914f
Kimura, K., Yanagisawa, H., Wachino, J., Shibayama, K., and Arakawa, Y. (2013). Rapid and reliable loop-mediated isothermal amplification method for detecting Streptococcus agalactiae. Jpn. J. Infect. Dis. 66, 546–548. doi: 10.7883/yoken.66.546
Min, R., Niu, C. G., Zeng, G. M., Qin, P. Z., Wang, X. Y., Huang, D. W., et al. (2011). Rapid detection of Staphylococcus aureus via a sensitive DNA hybridization assay based on a long-lifetime luminescent europium marker. Microchimica Acta 175, 105–112. doi: 10.1007/s00604-011-0654-8
Niraj, G., Prashant, T., Bhise, P. R., and Sonali, G. (2016). Colorimetric method for rapid detection of Oxacillin resistance in Staphylococcus aureus and its comparison with PCR for mecA gene. Sci. Rep. 6:23013. doi: 10.1038/srep23013
Oliveira, L. P. D., Soares, E. B. L. S., Silva, V. C., and Cirqueira, M.G. (2011). Study of Staphylococcus aureus in raw and pasteurized milk consumed in the Reconcavo area of the State of Bahia, Brazil. J. Food Process. Technol. 2. doi: 10.4172/2157-7110.1000128
Roda, A., Roda, B., Bonvicini, F., Colliva, C., and Reschiglian, P. (2012). Recent developments in rapid multiplexed bioanalytical methods for foodborne pathogenic bacteria detection. Microchimica Acta 178, 7–28. doi: 10.1007/s00604-012-0824-3
Schlecht, L. M., Peters, B. M., Krom, B. P., Freiberg, J. A., Hänsch, G. M., Filler, S. G., et al. (2015). Systemic Staphylococcus aureus infection mediated by Candida albicans hyphal invasion of mucosal tissue. Microbiology 161, 168–181. doi: 10.1099/mic.0.083485-0
Shi, C., Shang, F., Zhou, M., Zhang, P., Wang, Y., and Ma, C. (2016). Triggered isothermal PCR by denaturation bubble-mediated strand exchange amplification. Chem. Commun. 52, 11551–11554. doi: 10.1039/C6CC05906F
Shi, C., Shen, X., Niu, S., and Ma, C. (2015). Innate reverse transcriptase activity of DNA polymerase for isothermal RNA direct detection. J. Am. Chem. Soc. 137, 13804–13806. doi: 10.1021/jacs.5b08144
Ulrich, R. L., and Hughes, T. A. (2010). A rapid procedure for isolating chromosomal DNA from Lactobacillus species and other Gram-positive bacteria. Lett. Appl. Microbiol. 32, 52–56. doi: 10.1111/j.1472-765X.2001.00866.x
Wang, R., Wu, J., Zhang, F., Wang, L., and Ji, F. (2014). On-point detection of GM rice in 20 minutes with pullulan as CPA acceleration additive. Anal. Methods 6, 9198–9201. doi: 10.1039/C4AY02427C
Xiong, J., Wang, W., Zhou, Y., Kong, W., Wang, Z., and Fu, Z. (2016). Ultra-sensitive chemiluminescent detection of Staphylococcus aureus based on competitive binding of Staphylococcus protein A-modified magnetic beads to immunoglobulin G. Microchimica Acta 183, 1507–1512. doi: 10.1007/s00604-016-1769-8
Yan, X., Wang, B., Tao, X., Hu, Q., Cui, Z., Zhang, J., et al. (2012). Characterization of Staphylococcus aureus strains associated with food poisoning in Shenzhen, China. Appl. Environ. Microbiol. 78, 6637–6642. doi: 10.1128/AEM.01165-12
Yang, X., Zhang, J., Yu, S., Wu, Q., Guo, W., Huang, J., et al. (2016). Prevalence of Staphylococcus aureus and methicillin-resistant Staphylococcus aureus in retail ready-to-eat foods in China. Front. Microbiol. 7:816. doi: 10.3389/fmicb.2016.00816
Yu, J., Zhang, Y., Zhang, Y., Li, H., Yang, H., and Wei, H. (2016). Sensitive and rapid detection of staphylococcus aureus in milk via cell binding domain of lysin. Biosens. Bioelectron. 77, 366–371. doi: 10.1016/j.bios.2015.09.058
Keywords: strand exchange amplification, isothermal amplification, Staphylococcus aureus, rapid detection, point-of-care testing
Citation: Liu C, Shi C, Li M, Wang M, Ma C and Wang Z (2019) Rapid and Simple Detection of Viable Foodborne Pathogen Staphylococcus aureus. Front. Chem. 7:124. doi: 10.3389/fchem.2019.00124
Received: 26 December 2018; Accepted: 18 February 2019;
Published: 12 March 2019.
Edited by:
Shusheng Zhang, Linyi University, ChinaReviewed by:
Qingjiang Wang, East China Normal University, ChinaDechen Jiang, Nanjing University, China
Copyright © 2019 Liu, Shi, Li, Wang, Ma and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zonghua Wang, d2FuZ3pvbmdodWFAcWR1LmVkdS5jbg==
†These authors have contributed equally to this work
 Caiyan Liu
Caiyan Liu Chao Shi
Chao Shi Mengzhe Li1
Mengzhe Li1