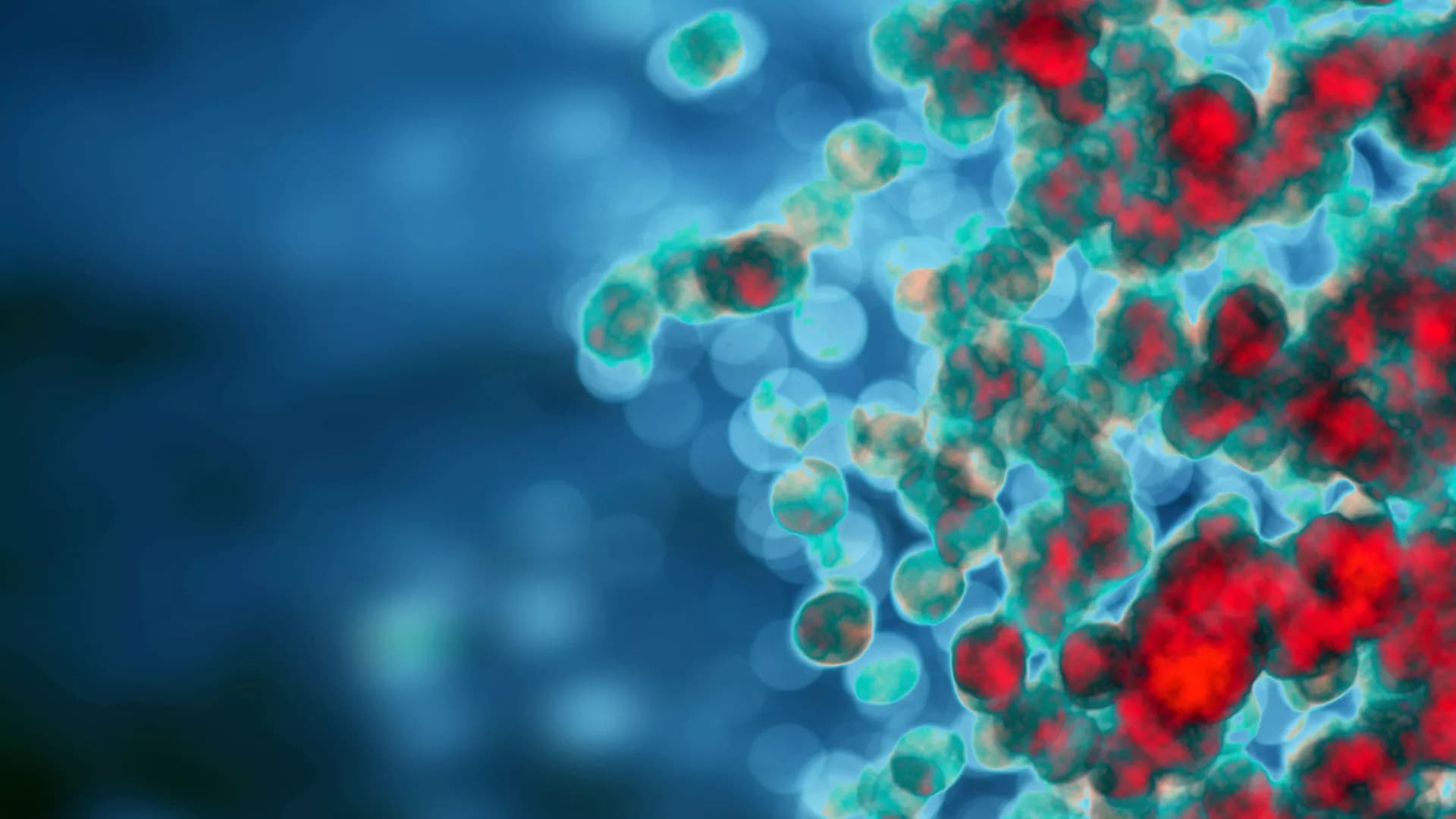
Phagocytosis is the cellular defense mechanism used to eliminate antigens derived from dysregulated or damaged cells, and microbial pathogens. Phagocytosis is therefore a pillar of innate immunity, whereby foreign particles are engulfed and degraded in lysolitic vesicles. In hexacorallians, phagocytic mechanisms are poorly understood, though putative anthozoan phagocytic cells (amoebocytes) have been identified histologically. We identify and characterize phagocytes from the coral Pocillopora damicornis and the sea anemone Nematostella vectensis. Using fluorescence-activated cell sorting and microscopy, we show that distinct populations of phagocytic cells engulf bacteria, fungal antigens, and beads. In addition to pathogenic antigens, we show that phagocytic cells engulf self, damaged cells. We show that target antigens localize to low pH phagolysosomes, and that degradation is occurring within them. Inhibiting actin filament rearrangement interferes with efficient particle phagocytosis but does not affect small molecule pinocytosis. We also demonstrate that cellular markers for lysolitic vesicles and reactive oxygen species (ROS) correlate with hexacorallian phagocytes. These results establish a foundation for improving our understanding of hexacorallian immune cell biology.
Pattern recognition receptors (PRRs) are evolutionarily ancient and crucial components of innate immunity, recognizing danger-associated molecular patterns (DAMPs) and activating host defenses. Basal non-bilaterian animals such as cnidarians must rely solely on innate immunity to defend themselves from pathogens. By investigating cnidarian PRR repertoires we can gain insight into the evolution of innate immunity in these basal animals. Here we utilize the increasing amount of available genomic resources within Cnidaria to survey the PRR repertoires and downstream immune pathway completeness within 15 cnidarian species spanning two major cnidarian clades, Anthozoa and Medusozoa. Overall, we find that anthozoans possess prototypical PRRs, while medusozoans appear to lack these immune proteins. Additionally, anthozoans consistently had higher numbers of PRRs across all four classes relative to medusozoans, a trend largely driven by expansions in NOD-like receptors and C-type lectins. Symbiotic, sessile, and colonial cnidarians also have expanded PRR repertoires relative to their non-symbiotic, mobile, and solitary counterparts. Interestingly, cnidarians seem to lack key components of mammalian innate immune pathways, though similar to PRR numbers, anthozoans possess more complete immune pathways than medusozoans. Together, our data indicate that anthozoans have greater immune specificity than medusozoans, which we hypothesize to be due to life history traits common within Anthozoa. Overall, this investigation reveals important insights into the evolution of innate immune proteins within these basal animals.
The animal immune system mediates host-microbe interactions from the host perspective. Pattern recognition receptors (PRRs) and the downstream signaling cascades they induce are a central part of animal innate immunity. These molecular immune mechanisms are still not fully understood, particularly in terms of baseline immunity vs induced specific responses regulated upon microbial signals. Early-divergent phyla like sponges (Porifera) can help to identify the evolutionarily conserved mechanisms of immune signaling. We characterized both the expressed immune gene repertoire and the induced response to lipopolysaccharides (LPS) in Halichondria panicea, a promising model for sponge symbioses. We exposed sponges under controlled experimental conditions to bacterial LPS and performed RNA-seq on samples taken 1h and 6h after exposure. H. panicea possesses a diverse array of putative PRRs. While part of those PRRs was constitutively expressed in all analyzed sponges, the majority was expressed individual-specific and regardless of LPS treatment or timepoint. The induced immune response by LPS involved differential regulation of genes related to signaling and recognition, more specifically GTPases and post-translational regulation mechanisms like ubiquitination and phosphorylation. We have discovered individuality in both the immune receptor repertoire and the response to LPS, which may translate into holobiont fitness and susceptibility to stress. The three different layers of immune gene control observed in this study, - namely constitutive expression, individual-specific expression, and induced genes -, draw a complex picture of the innate immune gene regulation in H. panicea. Most likely this reflects synergistic interactions among the different components of immunity in their role to control and respond to a stable microbiome, seawater bacteria, and potential pathogens.
Frontiers in Immunology
Innate immunity in fish: responses and adaptations to diverse aquatic environments