Short interfering RNAs (siRNAs) are processed from long double-stranded RNA (dsRNA), and a guide strand is selected and incorporated into the RNA-induced silencing complex (RISC). Within RISC, a member of the Argonaute protein family directly binds the guide strand and the siRNA guides RISC to fully complementary sites on-target RNAs, which are then sequence-specifically cleaved by the Argonaute protein—a process commonly referred to as RNA interference (RNAi). In animals, endogenous microRNAs (miRNAs) function similarly but do not lead to direct cleavage of the target RNA but to translational inhibition followed by exonucleolytic decay. This is due to only partial complementarity between the miRNA and the target RNA. SiRNAs, however, can function as miRNAs, and partial complementarity can lead to miRNA-like off-target effects in RNAi applications. Since siRNAs are widely used not only for screening but also for therapeutics as well as crop protection purposes, such miRNA-like off-target effects need to be minimized. Strategies such as RNA modifications or pooling of siRNAs have been developed and are used to reduce off-target effects.
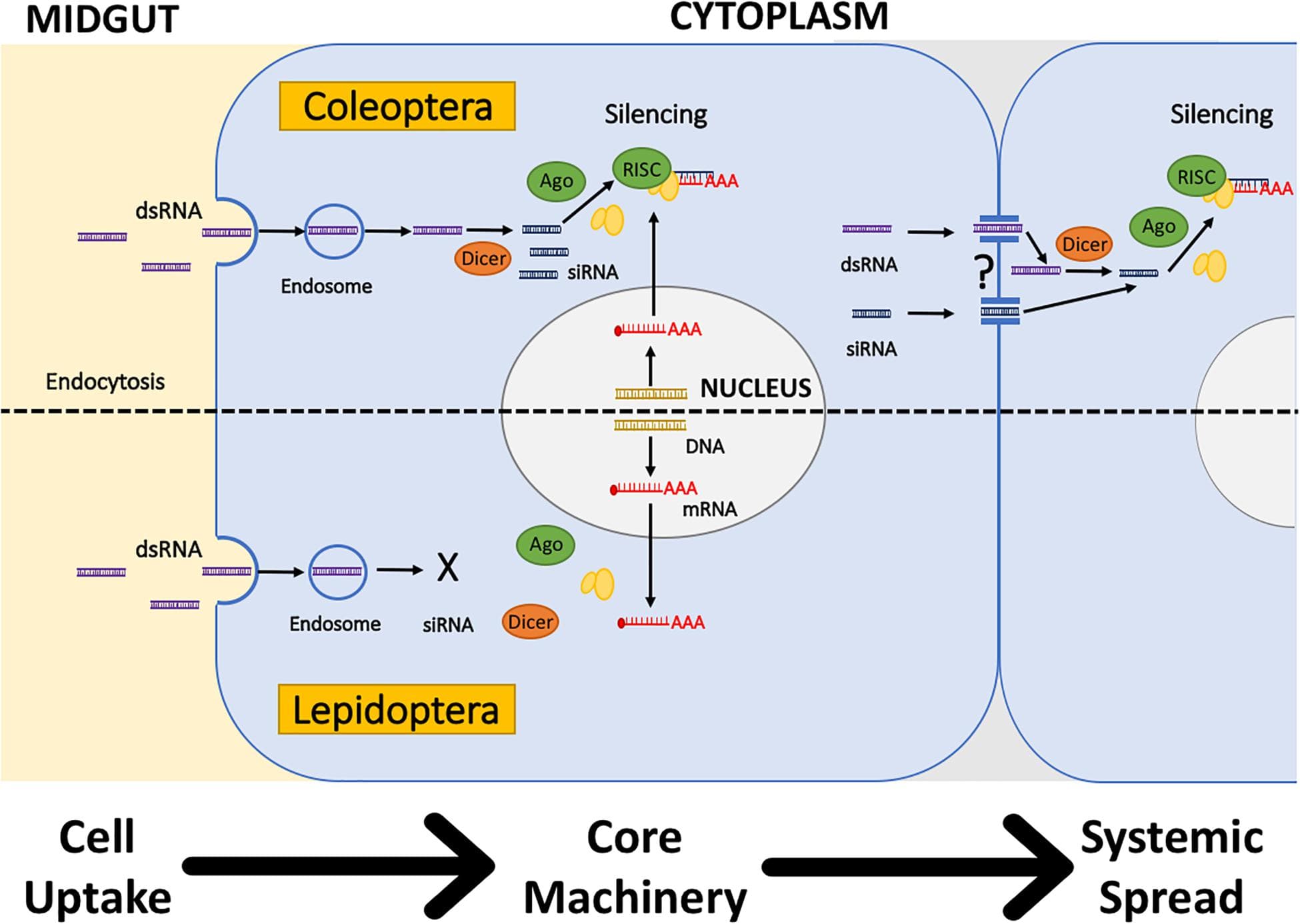
Canonical RNAi, one of the so-called RNA-silencing mechanisms, is defined as sequence-specific RNA degradation induced by long double-stranded RNA (dsRNA). RNAi occurs in four basic steps: (i) processing of long dsRNA by RNase III Dicer into small interfering RNA (siRNA) duplexes, (ii) loading of one of the siRNA strands on an Argonaute protein possessing endonucleolytic activity, (iii) target recognition through siRNA basepairing, and (iv) cleavage of the target by the Argonaute’s endonucleolytic activity. This basic pathway diversified and blended with other RNA silencing pathways employing small RNAs. In some organisms, RNAi is extended by an amplification loop employing an RNA-dependent RNA polymerase, which generates secondary siRNAs from targets of primary siRNAs. Given the high specificity of RNAi and its presence in invertebrates, it offers an opportunity for highly selective pest control. The aim of this text is to provide an introductory overview of key mechanistic aspects of RNA interference for understanding its potential and constraints for its use in pest control.
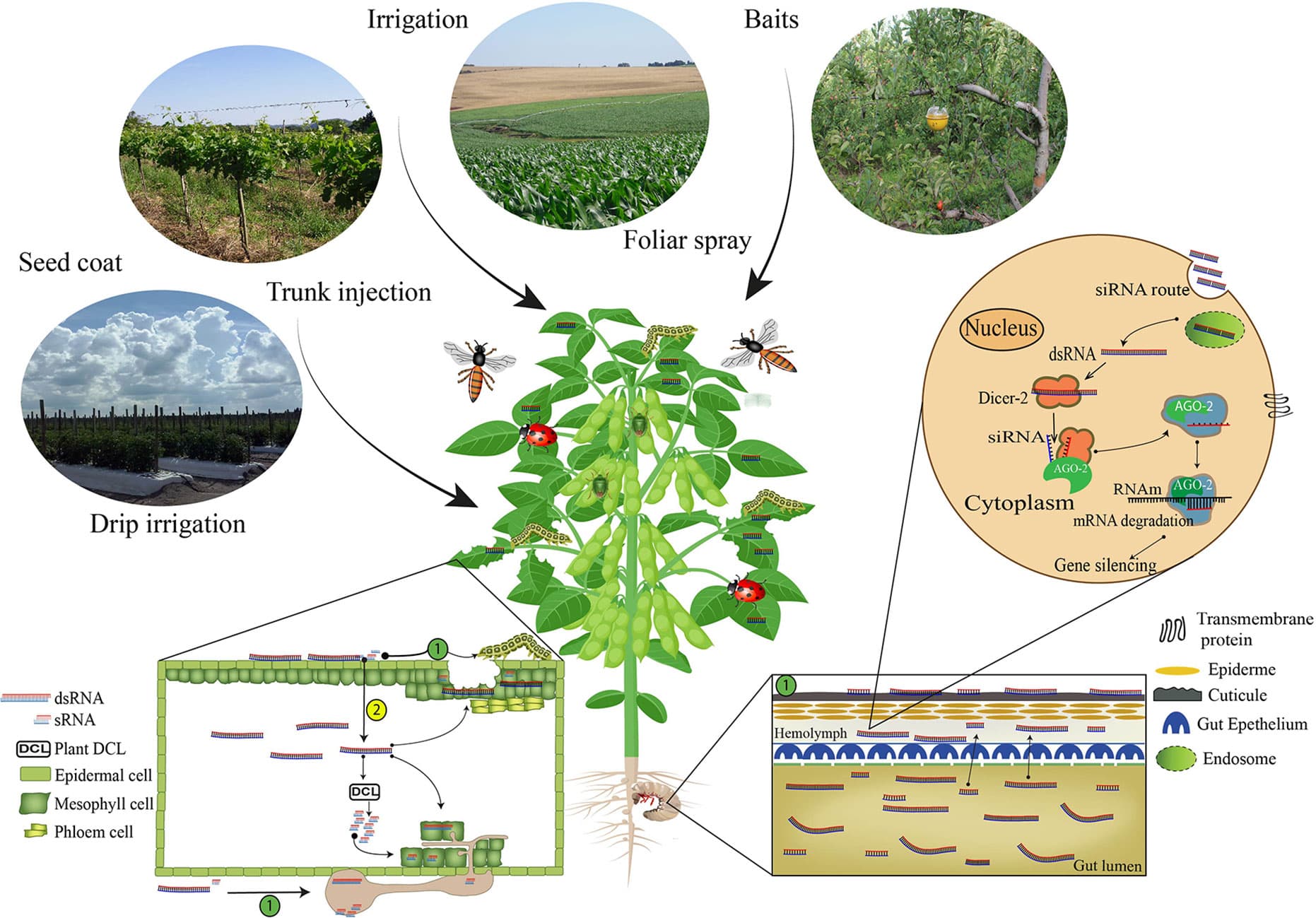
Exploiting the RNA interference (RNAi) gene mechanism to silence essential genes in pest insects, leading to toxic effects, has surfaced as a promising new control strategy in the past decade. While the first commercial RNAi-based products are currently coming to market, the application against a wide range of insect species is still hindered by a number of challenges. In this review, we discuss the current status of these RNAi-based products and the different delivery strategies by which insects can be targeted by the RNAi-triggering double-stranded RNA (dsRNA) molecules. Furthermore, this review also addresses a number of physiological and cellular barriers, which can lead to decreased RNAi efficacy in insects. Finally, novel non-transgenic delivery technologies, such as polymer or liposomic nanoparticles, peptide-based delivery vehicles and viral-like particles, are also discussed, as these could overcome these barriers and lead to effective RNAi-based pest control.
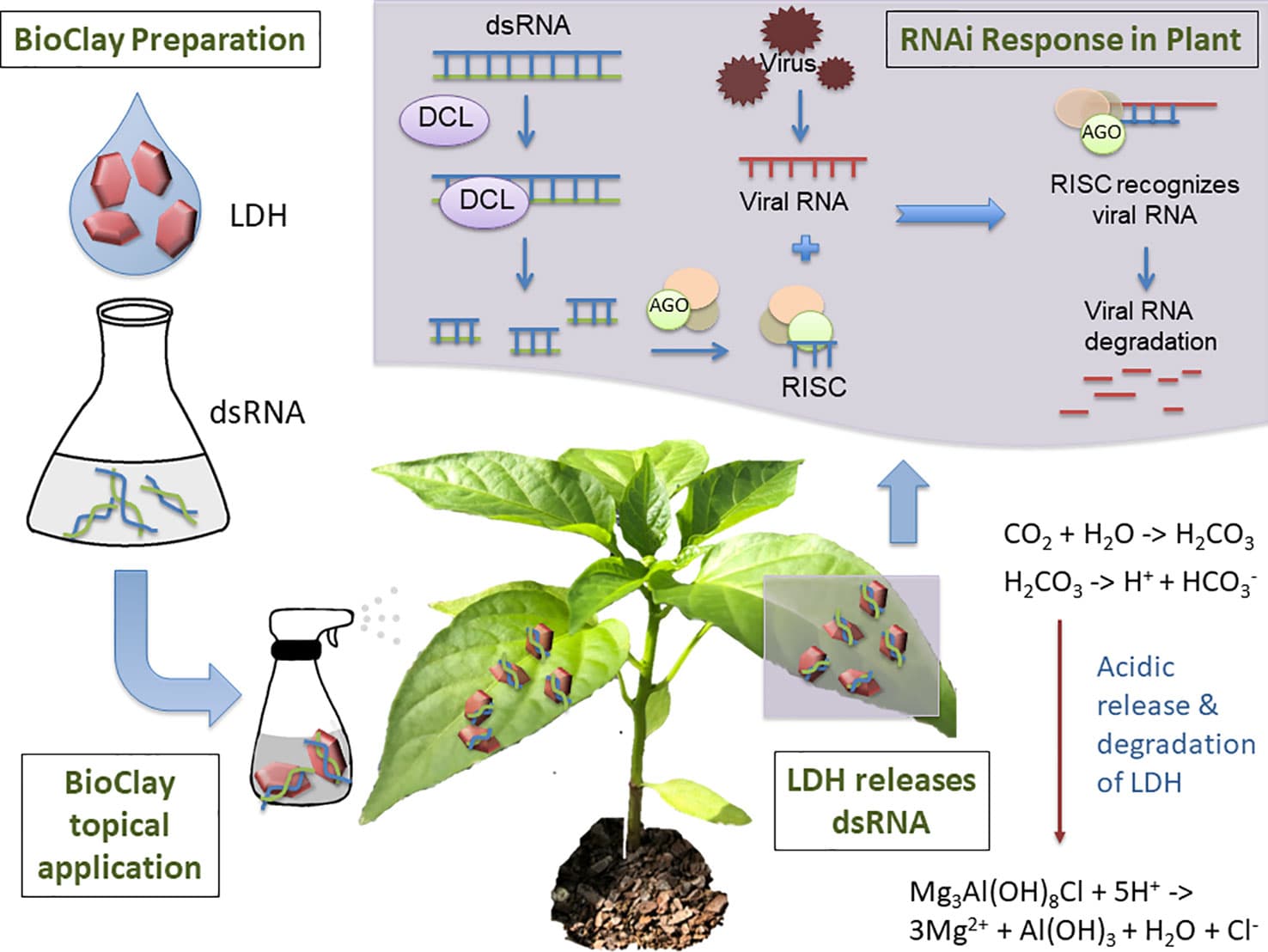
Sustainable agriculture relies on practices and technologies that combine effectiveness with a minimal environmental footprint. RNA interference (RNAi), a eukaryotic process in which transcript expression is reduced in a sequence-specific manner, can be co-opted for the control of plant pests and pathogens in a topical application system. Double-stranded RNA (dsRNA), the key trigger molecule of RNAi, has been shown to provide protection without the need for integration of dsRNA-expressing constructs as transgenes. Consequently, development of RNA-based biopesticides is gaining momentum as a narrow-spectrum alternative to chemical-based control measures, with pests and pathogens targeted with accuracy and specificity. Limitations for a commercially viable product to overcome include stable delivery of the topically applied dsRNA and extension of the duration of protection. In addition to the research focus on delivery of dsRNA, development of regulatory frameworks, risk identification, and establishing avoidance and mitigation strategies is key to widespread deployment of topical RNAi technologies. Once in place, these measures will provide the crop protection industry with the certainty necessary to expend resources on the development of innovative dsRNA-based products. Readily evident risks to human health appear minimal, with multiple barriers to uptake and a long history of consumption of dsRNA from plant material. Unintended impacts to the environment are expected to be most apparent in species closely related to the target. Holistic design practices, which incorporate bioinformatics-based dsRNA selection along with experimental testing, represent important techniques for elimination of adverse impacts.



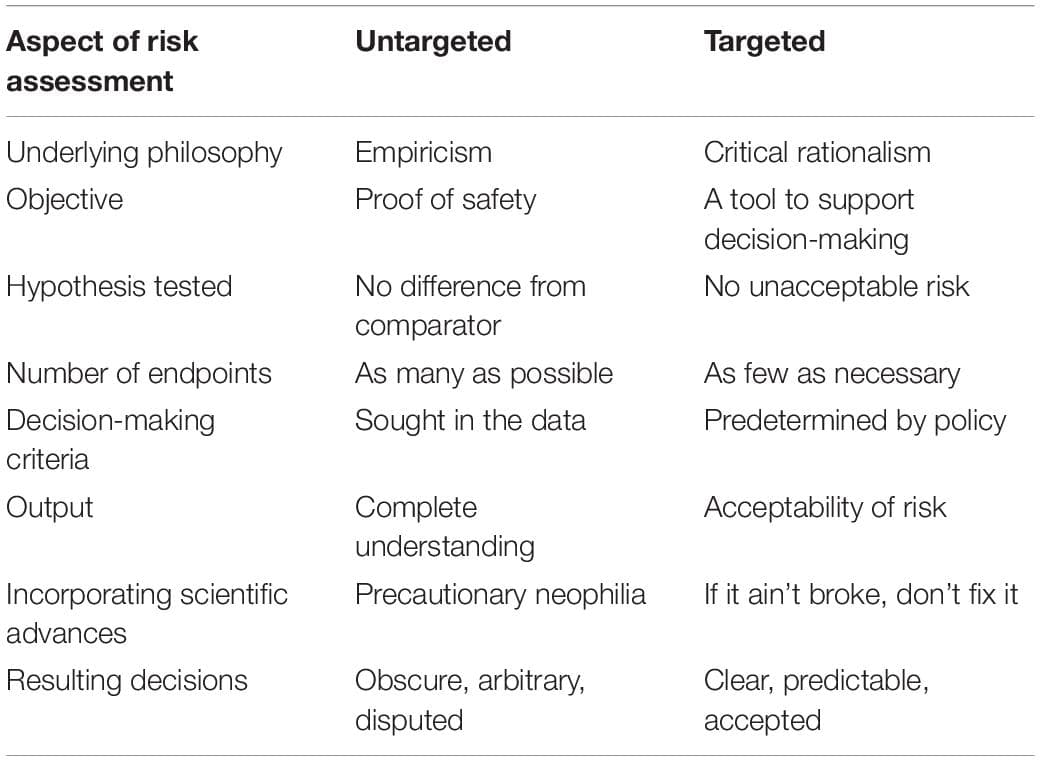
![Risk assessment approach for genetically modified plants [reprinted with permission from EFSA’s infographic (Available at https://www.efsa.europa.eu/en/discover/infographics/risk-assessment-genetically-modified-plants; ISBN 978-92-9199-913-2 | doi: 10.2805/240762 | TM-02-17-009-EN-N)].](https://www.frontiersin.org/_rtmag/_next/image?url=https%3A%2F%2Fwww.frontiersin.org%2Ffiles%2FArticles%2F522254%2Ffpls-11-00445-HTML%2Fimage_m%2Ffpls-11-00445-g001.jpg&w=3840&q=75)