- 1Department of Biology, Eastern Michigan University, Ypsilanti, MI, United States
- 2Division of Research, Innovation, and Economic Development, Tarleton State University, Stephenville, TX, United States
- 3Department of Soil and Crop Sciences, Texas A&M University, College Station, TX, United States
- 4Department of Microbiology, Immunology and Genetics, University of North Texas Health Science Center, Fort Worth, TX, United States
- 5Department of Biology and Biotechnology, University of Houston—Clear Lake, Houston, TX, United States
Hurricane Harvey caused widespread flooding along the Texas Gulf Coast in August 2017; some areas of Houston received >150 cm of rainfall within a few days. Due to concerns over fecal contamination of floodwaters, surface water samples were collected at six locations in the southeastern Houston area immediately before and after the hurricane and then every 1 to 2 weeks thereafter over a 2-month period. Total E. coli was enumerated using the IDEXX Quanti-Tray/2000 system. DNA extracted from water samples was analyzed via quantitative real-time PCR (qPCR) for general and source-specific total Bacteroidales and human Bacteroidales markers, and digital PCR (dPCR) for antibiotic resistance genes (ARG) and a plasmid (pBI143) associated with human waste. SourceTracker2 was used to determine human source contributions based on metagenomic analysis of PCR-amplified 16S rRNA gene fragments. Samples collected immediately after the hurricane had elevated levels of E. coli, ranging from 488 to 1,733 CFU 100 ml−1. After 1 week, E. coli levels decreased to <100 MPN 100 ml−1. Total Bacteroidales numbers were elevated immediately following the hurricane and remained high for 12 days. Human-source contributions, as assessed by PCR methods and metagenomic analysis, peaked within 12 days after the hurricane consistently across all sampling sites. Multiple regression analysis of environmental parameters, copies of ARG and pBI143, and metagenomic data confirmed that human waste caused the dramatic, short-term, high levels of fecal contamination of floodwaters generated by Hurricane Harvey. Fecal indicators approached normal background levels approximately 3 weeks after the rainfall ended.
1 Introduction
Originating as a tropical storm in the Atlantic Ocean, Hurricane Harvey made landfall as a Category 4 hurricane on August 25, 2017 in Rockport, Texas extending widespread destruction and damage across coastal Texas and Louisiana (Emanuel, 2017; National Weather Service, 2017; TCEQ, 2017). In the weeks that followed, Harvey brought in a record amount of rainfall of over 127 cm and was the second costliest hurricane in U.S. history (Emanuel, 2017; National Weather Service, 2017; National Hurricane Center, 2017; TCEQ, 2017). In the Houston metropolitan area, catastrophic flooding caused by Harvey accounted for at least 80 fatalities and 150 billion dollars in damages (Emanuel, 2017; National Hurricane Center, 2017; Scutti, 2017; TCEQ, 2017). The Houston metropolitan area and surroundings experienced overflows and spills from wastewater treatment plants and sanitary sewers into the environmental floodwater (Scutti, 2017; TCEQ, 2017; USEPA, 2017).
Sewage spills can heavily contaminate floodwaters, as indicated by high concentrations of fecal indicator bacteria (FIB; Schwab et al., 2007; Sinigalliano et al., 2007; Veldhuis et al., 2010). For over 40 years, FIBs have been measured routinely as indicators of potential pathogens that are serious risk concerns to humans and animals, and to assess environmental health (Cabelli et al., 1979; Cabelli et al., 1982; Griffith et al., 2009; National Nonpoint Source Monitoring Program, 2013; Pruss, 1998; Wade et al., 2003; USEPA, 2013). In addition to routine quantification of FIBs in contaminated water sources, identification and allocation of the leading specific sources of contamination is crucial for precise risk assessments and for identifying measures for restoration of such impaired water sources (Harwood et al., 2014; Mcquaig et al., 2012). Microbial source tracking (MST) is a contemporary tool that often targets common FIBs, such as Escherichia coli or Bacteroidales, to determine (1) the degree to which a body of water has been contaminated with fecal matter and (2) the specific source(s) of such contamination (Bernhard and Field, 2000; Field and Samadpour, 2007; Hagedorn et al., 1999; Harwood et al., 2014; Scott et al., 2005; Stoeckel and Harwood, 2007). Amongst the commonly used MST targets, Bacteroidales, Gram-negative, strictly anaerobic bacteria found in the human gut and in both human and animal feces, are widely used to track sources of fecal contamination in recreational waters (Ahmed et al., 2008; Ahmed et al., 2016; Bernhard and Field, 2000; Griffith et al., 2009; Kreader, 1995; Mcquaig et al., 2012; Wexler, 2007).
Research over the last few decades has shown floodwaters generated by extreme weather events can have high levels of FIB. Following Hurricanes Dennis, Floyd and Irene, Escherichia coli and total coliforms were elevated in agricultural soils impacted by the resulting floodwaters (Casteel et al., 2006). In 2005, immediately following Hurricanes Katrina and Rita, E. coli levels were five orders of magnitude higher than the USEPA maximum for freshwater beaches (Dobbs, 2007; Sinigalliano et al., 2007). Research studies thus far representing the catastrophic flooding caused by Hurricane Harvey have identified drastic changes in physical, chemical, and biological water quality standards, including increased abundance of FIBs, various sources of markers for microbial contamination, and overall changes in microbial community structures and functions across different sites throughout Harvey-affected areas (LaMontagne et al., 2022; Landsman et al., 2019; Kapoor et al., 2018; Moghadam et al., 2022; Pérez-Valdespino et al., 2021; Yang et al., 2021; Yu et al., 2018).
The key objective of this study was to examine the extent, duration, and sources of fecal contamination from Hurricane Harvey floodwaters in the Houston area. As detailed in a previous study (LaMontagne et al., 2022), flood water samples were collected around the Houston area immediately after Hurricane Harvey and again every 1 to 2 weeks for approximately 2 months and analyzed for FIBs and microbial community composition. We expand on results reported in that study by analyzing the samples for other fecal markers and indicators of human-specific fecal contamination. These analyses include digital PCR (dPCR) with primers and probes for antibiotic resistance genes (ARG), which are commonly associated with wastewater, and a recently described plasmid (pBI143) that abounds in waterways contaminated with human waste (Fogarty et al., 2024). High levels of contamination with human waste were found in all samples collected immediately after the Hurricane. Levels returned to pre-Hurricane levels within 2 weeks.
2 Materials and methods
2.1 Sampling sites and sample collection
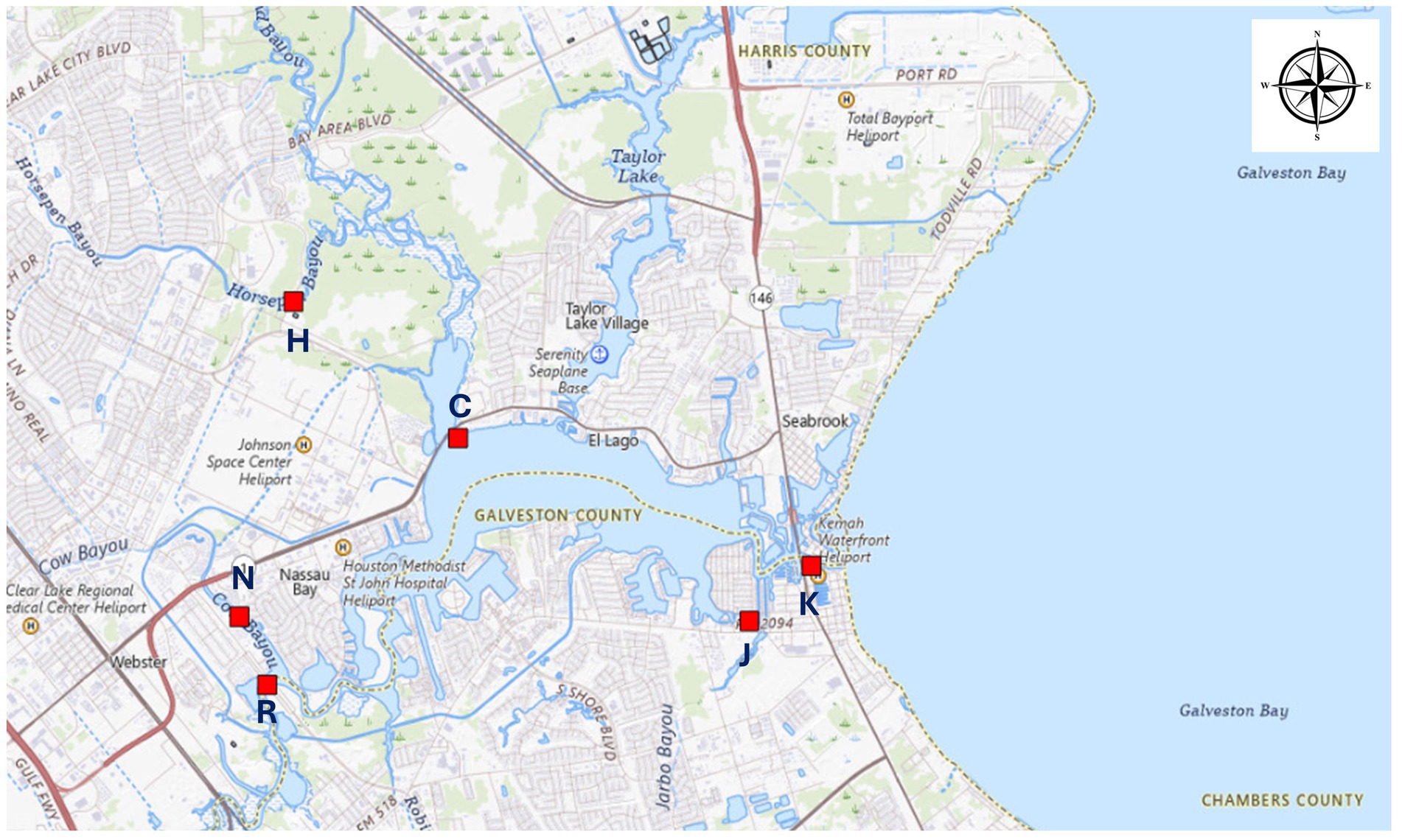
Six sampling sites were chosen around the Houston Clear Lake area (Figure 1) primarily based upon accessibility after initial rainfall and the subsequent receding of floodwaters. Sampling began as soon as sites were accessible and continued every 1–2 weeks for 2 months. Water samples were collected via a grab-sampling technique and stored inside sterile, plastic bottles (IDEXX, Westbrook, ME) and transported, on ice, to the lab for further analysis within 6 h of collection as described previously (LaMontagne et al., 2022).
2.2 Quantitative PCR analysis of human and total Bacteroidales
Relative quantities of total Bacteroidales and human Bacteroidales were measured via quantitative polymerase chain reaction (qPCR). Approximately 50–100 ml of water sample was filtered onto 0.22 μM Supor membrane filters of 47 mm diameter and stored at-80°C. DNA was extracted from the filters using the DNeasy PowerWater Kit (Qiagen, Hilden, Germany) and quantified using a nanodrop spectrophotometer (Thermo Scientific, Waltham, MA). The qPCR reaction conditions and thermal cycler programs used for both Total Bacteroidales and Human Bacteroidales assays are listed in Supplementary material (Bernhard and Field, 2000; Shanks et al., 2009; Shanks et al., 2016). For the total Bacteroidales assay, a gBlock standard based on the 16S rRNA gene of Bacteroides fragilis was developed (sequence in Supplementary Table S4) and used as standard for the qPCR assay (Integrated DNA Technologies, Newark, NJ). This standard was diluted to create a standard curve between concentrations ranging from 1.32 × 108 to 1.32 × 101 copies ml−1. For the Human Bacteroidales assay, a gBlock (Shanks et al., 2016; sequence in Supplementary material) diluted between 3.2 × 108–3.2 × 101 copies ml−1 was used to create a standard curve. Eppendorf LoBind® Tubes were used for all assays to ensure maximal recovery of nucleic acid molecules (Eppendorf, Framingham, MA). Gene copies were calculated and reported as copies per 100 ml of water filtered.
2.3 Digital PCR (dPCR) analysis of antibiotic resistance genes and pBI143
Absolute quantification of the ARGs intI1, aint1, and sul1 and the plasmid pBI143 was performed using an Applied Biosystems QuantStudio™ Absolute Q™ Digital PCR System per the manufacturer’s recommendations. Purified DNA samples, which were archived previously (LaMontagne et al., 2022, #4256), were analyzed for all four target genes in a single 4-plex reaction. Each 10 μl-PCR mixture consisted of 2.0 μl of 5X Absolute Q™ DNA Digital PCR Master Mix, 1 μM of each primer, 80 nM of each probe, 1 μl of DNA template, and a final concentration of 0.2 mg/ml of BSA. After centrifugation at 10,000 X g for 1 min, 9 μl of sample reactions were loaded into individual wells of an Absolute Q™ MAP16 Digital PCR plate and covered by 15 μl of isolation buffer. All multiplex reactions were run with the following thermocycler conditions: 10 min of initial denaturation at 98°C, followed by 45 cycles of 95°C for 30 s and 60°C for 1 min. Data analysis was performed on-instrument using provided Absolute Q™ Digital PCR Software (version 6.3.0-RC11). Primers and probes, described previously (Heuer and Smalla, 2007; Barraud et al., 2010; Quintela-Baluja et al., 2021; Fogarty et al., 2024) are shown in Supplementary Table S1. All labeled PCR probes (6,000 pmoles each) were purchased from Thermo Fisher Scientific. All primers and synthetic gBLOCK positive control sequences were purchased from IDT. Gene copies were calculated and reported as copies per 100 ml of water filtered.
2.4 SourceTracker analysis of 16S rRNA gene sequences
As an additional source tracking approach, we further analyzed 16S rRNA sequence data that was previously published for these same water samples. The samples were initially sequenced using a MiSeq Illumina platform as described by LaMontagne et al. (2022) and results deposited in the NCBI SRA under accession number/Bioproject ID: PRJNA795782. We used SourceTracker2 to reanalyze these data with a focus on human source contributions. Raw sequencing reads were collected and analyzed with the QIIME 2 software package version 2022.8 (https://qiime2.org; Bolyen et al., 2019) and plugins associated with this version. After importing raw reads to the Qiime2, paired-end sequencing reads were demultiplexed using the demux plugin.1 Quality control, filtering chimeric sequences and feature table construction were done using the q2-dada2 plugin (Callahan et al., 2016) with trimming parameters based on the demux output visualization files. Taxonomy classification was done against the SILVA 138 database (Quast et al., 2012; Yilmaz et al., 2014) using the “feature-classifier” plugin (Bokulich et al., 2018) with the “classify-sklearn.” Taxonomy-based filtering was conducted to remove mitochondria, chloroplast, Archaea and Eukaryota using “taxa-filter-table and taxa-filter-seqs” command from both taxonomic and rep-seqs tables. A taxonomic bar-plot was generated using the filtered table and assigned taxonomy file and visualize through the QIIME 2 view website. The resulting filtered taxonomy file was used to extract sample specific taxonomic classification. SourceTracker2 software with QIIME Gibbs Sampler plugin was used to determine source-sink proportions and taxonomic classification based on ASV abundance (Knights et al., 2011).
2.5 Multivariate regression analysis
Multivariate regression analysis was conducted on previously published (LaMontagne et al., 2022) water quality data [dissolved inorganic nutrients (DIN), salinity, temperature, oxygen and pH] and the metagenomic and dPCR data produced as described above. Values for missing a set of DIN measurements, which were not collected at station H (Figure 1) immediately before Hurricane Harvey made landfall, were imputed from DIN values pulled from a public archive water quality measurements for that water body2 and processed with scripts presented in Supplementary file S2. The R (R Core Team, 2021) package mice (v 3.16.0) was used to impute the missing values with scripts presented in Supplementary file S3. The input Supplementary file S4 in this analysis contained 29 data points from the public archive and 6 data points from LaMontagne et al. (2022).
Metagenomic data processed previously (LaMontagne et al., 2022) with functions available in DADA2 v1.32.0 (Callahan et al., 2016), and custom scripts available here https://github.com/MgLaMontagne/dada2-optimize, was subsetted to the samples collected on August 25th, August 30th and September 8th 2017. This sequence data, and the corresponding metadata (water quality and dPCR), was analyzed with scripts presented in Supplementary file S5. Analysis of variance (ANOVA) was conducted with base functions in R and further analyzed with functions available in agricolae v1.3-7 (de Mendiburu and Yaseen, 2020) and scripts also presented in Supplementary file S5. Metadata is available as Supplementary file S6. Multivariate analysis was conducted on vectors produced by non-metric multi-dimensional scaling (NMDS) with functions available in the R packages phyloseq v1.48.0 (McMurdie and Holmes, 2013) and vegan v2.6–6.1 (Oksanen et al., 2024) with scripts available in Supplementary file S7.
3 Results
3.1 Quantification of total Bacteroidales and human Bacteroidales
Total Bacteroidales copy numbers peaked in the immediate aftermath of Hurricane Harvey. Levels of this marker, across all sites, were highest on August 31 and September 8 (Figure 2A). At the first sampling after the rainfall ended, total Bacteroidales copies ranged between 5.7 × 104–2.4 × 105 copies 100 ml−1 across the six sites. The following week (September 8), total Bacteroidales copy numbers ranged between 4.2 × 103 and 1.5 × 106 copies 100 ml−1 across the six sites (Figure 2A). The average human Bacteriodales values ranged between 1.6 × 103 and 4.0 × 103 copies 100 ml−1 and peaked on September 8 (Figure 2B). Following the initial peak, total Bacteroidales remained somewhat high following the hurricane with some additional elevated values observed in October. In contrast, human Bacteroidales values leveled off following the peak observed during the September 8 sampling. As previously reported in LaMontagne et al. (2022), E. coli numbers in these water samples peaked on August 31, averaging 1,087 MPN ml−1 and then dropped to baseline within a week (Supplementary Figure S8).
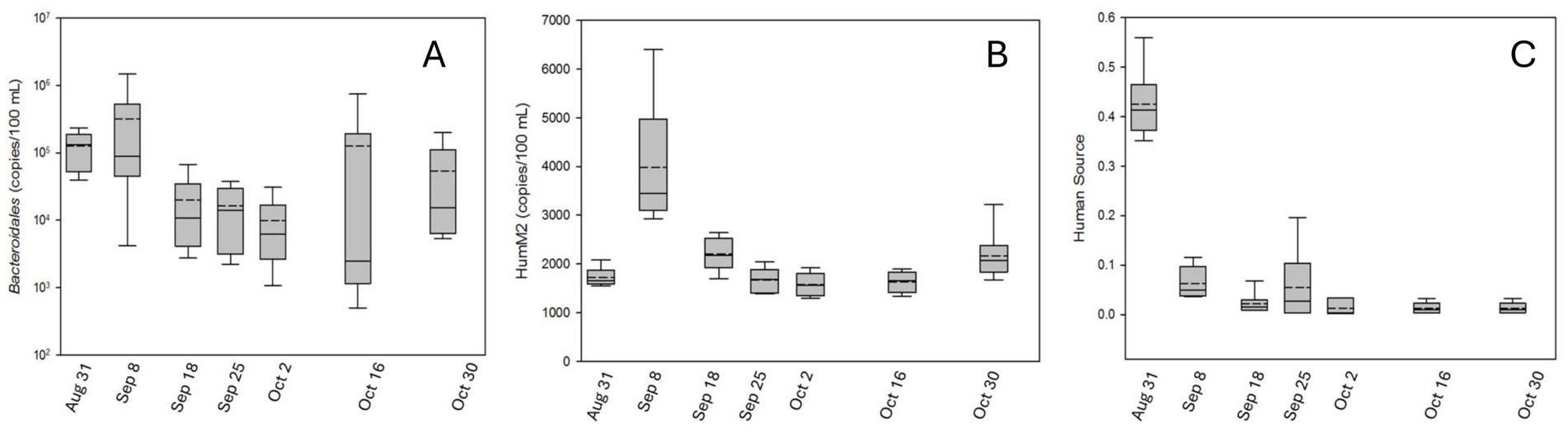
Figure 2. Box plot showing fecal indicator and source levels following Hurricane Harvey flooding at six locations around Houston, TX, USA. (A) Total Bacteroidales displayed in copies 100 ml−1 of water. (B) Human Bacteroidales displayed in copies 100 ml−1 of water. (C) SourceTracker results expressed as fraction of bacterial community from human sources.
3.2 SourceTracker analysis
Human source contributions, as assessed by metagenomic analysis, were greatest immediately after Hurricane Harvey (August 31) and then decreased precipitously and remained relatively low for all subsequent sampling dates (Figure 2C). To further confirm the presence of enteric-like bacteria, we extracted the sample-specific ASVs and determined taxonomy based on the Silva database (Quast et al., 2012; Yilmaz et al., 2014). Based on the bacterial abundance counts >5, bacteria similar to that found in the sewage source samples (Gammaproteobacteria and Bacteroidia) dominated all water samples collected immediately after flooding (August 31; LaMontagne et al., 2022).
3.3 Quantification of pBI143 and ARG
Copy numbers of pBI143, intI1 and aint1 were significantly higher in water samples collected immediately after Hurricane Harvey than before or after the storm (Supplementary Table S3). The highest copy number for pBI143 (9.4 × 104 copies/100 ml, Figure 3) was observed in samples collected immediately after Hurricane Harvey at station H (Figure 3). Lowest copies (6 copies/100 ml) for this plasmid were observed before Hurricane Harvey at station C, which is in Clear Lake (Figure 3). The highest copy number for intI1 (1.2 × 105 copies/100 ml, Figure 3) was observed in samples collected immediately after Hurricane Harvey at station N. Lowest copies (7,008 copies/100 ml) for this ARG were observed before Hurricane Harvey at station K, which is also in Clear Lake (Figure 1). The highest copy number for aint1 (9,131/100 ml) was observed in samples collected immediately after Hurricane Harvey at station H (Supplementary Figure S1). Lowest copies (620 copies/100 ml) for this ARG were observed before Hurricane Harvey at station K. Copy numbers of sul1 did not differ significantly between sampling dates (Supplementary Figure S1).
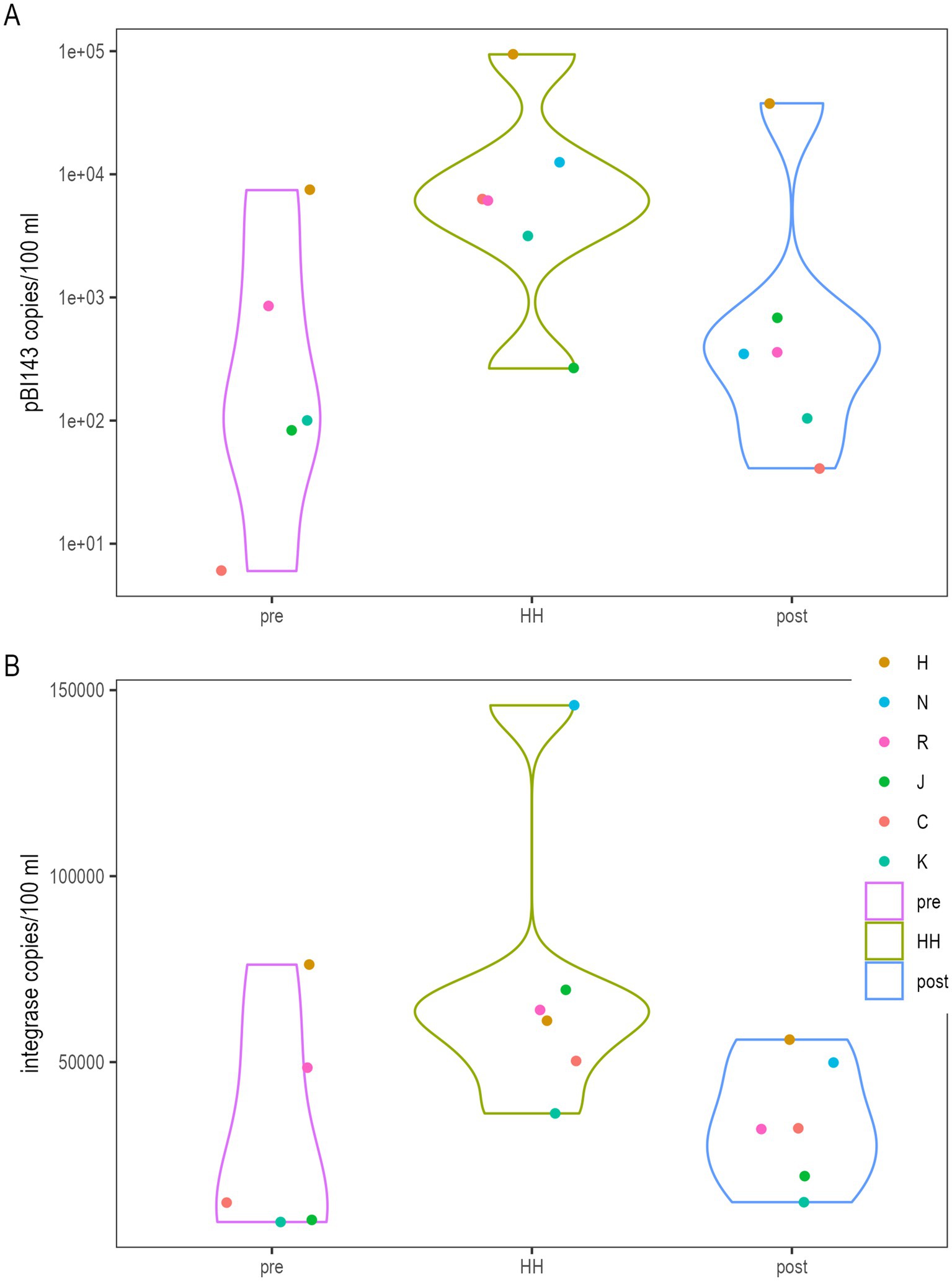
Figure 3. Quantification of pBI143 (A) and integrase (intI1) (B) by dPCR. Sample types (“pre,” “HH” and “post”) correspond to samples collected before Hurricane Harvey (August 25) made landfall in the Houston area, immediately after (August 30th) and on September 8th, respectively. Letters in legend correspond to stations in Figure 1. Quantification by dPCR is described in Methods. Figure was generated with scripts in Supplementary file S5.
Copy numbers of the plasmid pBI143 and ARGs (intI1, sul1 and aint1) all differed significantly (Supplementary Table S3) between stations. These differences between locations did not show a significant interaction with the date (pre/Hurricane Harvey/post, Supplementary Table S3). The highest copy number for pBI143, sul1 and aint1 were observed for samples collected at station H, where the geometric means across all dates for these markers were 29,775, 70,594 and 4,881 copies/100 ml, respectively. Copy numbers for pBI143 and sul1 at stations C and K (within Clear Lake) were at least an order of magnitude lower. The highest copy number for intI1 was observed for samples collected at station N, where the geometric mean across all dates at that station was 85,348/100 ml. These counts were significantly higher than the geometric mean for intI1 for samples collected at stations C and K.
3.4 Multivariate analysis
NMDS analysis of metagenomic data showed a coherent cluster containing the six samples collected immediately after Hurricane Harvey (Figure 4). This cluster appeared driven by contamination with human waste, as assessed by copies of the plasmid pBI143 and the ARG intI1. That is these two parameters, which were measured by dPCR, corresponded to the structure of the microbial community, as assessed by sequencing of 16S amplicons. Salinity, temperature, total dissolved solids and oxygen levels also show significant relationships with bacterial community structure; however, dissolved inorganic nutrients and pH did not.
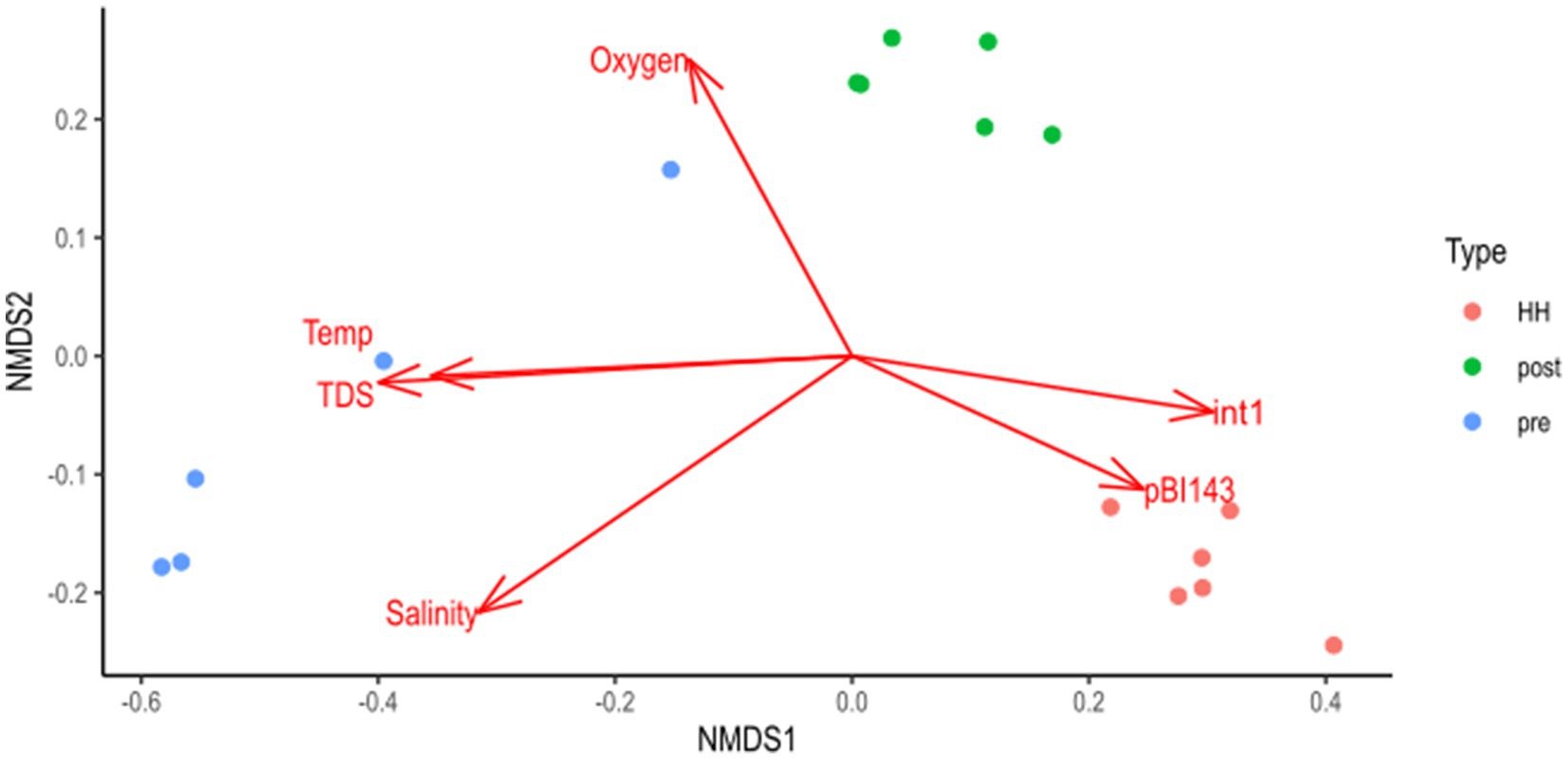
Figure 4. Non-metric multidimensional scaling (NMDS) analysis of targeted (16S rRNA amplicons) metagenomic data. NMDS results were fit to water quality parameters: temperature (temp), salinity, dissolved inorganic nutrients, pH, oxygen, total dissolved solids (TDS) and the number of copies of ARG and the plasmid pBI143. Vectors show fit of variables. Only vectors that were significant at p < 0.1 are shown. Figure was generated with scripts in Supplementary file S7.
4 Discussion
Results from this study show that the overall amounts of E. coli, human-Bacteroidales, the human-associated plasmid pBI143 and two ARGs (intI1 and aint1) were elevated following landfall of Hurricane Harvey and returned to lower levels within a few weeks; total Bacteroidales also saw a similar spike after the hurricane followed by a decrease although levels were generally more variable over the next 2 months of sampling than those observed for human-Bacteroidales or E. coli. These results indicate a spike in fecal contamination occurred within approximately the first 1–2 weeks after initial flooding, and this subsequently decreased for following sampling events. The initial rainfall, from Hurricane Harvey, would have been expected to flush fecal material (e.g., from pets, wildlife, etc.) from the surrounding area into the water thus increasing amounts of E. coli and Bacteroidales. However, multiple microbial source tracking (MST) methods suggest that the high levels of E. coli and total Bacteroidales, measured immediately following Hurricane Harvey, resulted from contamination of floodwaters with human waste. In particular, SourceTracker results and dPCR copy numbers for pBI143 and two ARG (intI1 and aint1) indicated that the greatest human fecal contributions occurred immediately following Hurricane Harvey. These changes corresponded to changes in microbial community structure (Figure 4) and the highest levels (9.4 × 104 copies/100 ml) of pBI143 in floodwaters measured herein are in the range recently reported for freshwater systems moderately to highly contaminated with human waste (Fogarty et al., 2024). In contrast, the HumM2 qPCR marker showed highest concentrations over a week later. The highest level of sul1 was observed at station H, which is about 10 meters downstream of a municipal wastewater treatment plant (Supplementary file S8) before and a week after Hurricane Harvey (Supplementary Figure S1). Dilution could explain this pattern. Copies of this ARG decrease with the dilution of wastewater, as assessed by distance from an outfall pipe (Haenelt et al., 2023). Once the floodwaters receded, the HumM2 marker and sul1 both spiked (September 8). Unfortunately, due to the flooding overwhelming surrounding gaging stations, we cannot determine total loading which might help corroborate whether this apparent delay in spiking was caused by initial dilution effects from the extreme rainfall.
The differences between the relative levels of total Bacteroidales and HumM2 at times could suggest that the majority of initial fecal contamination was from non-human sources, but this may be complicated by the differential environmental persistence of the qPCR markers (Haenelt et al., 2023) and the quantitative differences observed between qPCR and dPCR assessments of MST markers (Stachler et al., 2019). Overall, the input from domesticated livestock animals appears relatively unimportant for this study area. The watersheds sampled are in suburbs between Houston and Galveston. These areas are dominated by residential and urban populations and do not have clear livestock sources, like concentrated animal feeding operations (TCEQ, 2024), and only a few small pastures where a handful of cattle are raised. We cannot discount input from companion animals and wildlife but the homogeneity of the community structure of the floodwaters sampled immediately after the deluge (Figure 4), suggests the floodwaters were all contaminated with microbes from the same source. Human populations are the common denominator of all watersheds that are coupled to the waterways sampled herein. In any case, the levels of all fecal indicator bacteria returned to lower levels within approximately 3 weeks.
A handful of other studies (e.g., Kapoor et al., 2018; Yang et al., 2021) also reported a similar increase in FIB and/or human fecal markers after Hurricane Harvey that returned to normal after a few weeks; however, it should be noted that our first sampling occurred weeks before the first sampling events in those studies. Based on our results, it appears that the greatest impacts from flooding likely occurred within the first 2 weeks after the hurricane, so the initial impacts may have been even greater than reported in the other studies. The results of this study reinforce the need for sampling as soon as possible following extreme weather events such as hurricanes to capture maximum level of impacts on water quality.
5 Conclusion
• Fecal indicator and several MST marker levels were elevated immediately after the hurricane but decreased within 3 weeks.
• Multiple MST markers and microbial community structure changes suggest that human waste was a major contaminant of water samples immediately after hurricane landfall.
• Rapid decrease in MST parameters after flooding reinforces the need for rapid sampling after perturbations to capture temporally dynamic environmental effects.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found at: https://ncbi.nlm.nih.gov/bioproject/795782.
Author contributions
MM: Formal analysis, Investigation, Methodology, Writing – original draft, Writing – review & editing. MH: Formal analysis, Methodology, Writing – original draft, Writing – review & editing. JB: Writing – original draft, Writing – review & editing. YZ: Formal analysis, Methodology, Validation, Writing – review & editing. MA: Conceptualization, Funding acquisition, Supervision, Writing – review & editing, Resources. ML: Conceptualization, Formal analysis, Funding acquisition, Investigation, Methodology, Project administration, Supervision, Writing – review & editing, Resources, Validation. TG: Conceptualization, Formal analysis, Funding acquisition, Investigation, Methodology, Project administration, Supervision, Validation, Writing – original draft, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. Funding provided by NSF Rapid Grant Project #CBET-1759540-1759542, EPA grant #02D18322, and USDA National Institute of Food and Agriculture, Hatch project W5170: Beneficial Use of Residuals to Improve Soil Health and Protect Public and Ecosystem Health.
Acknowledgments
We thank Heidi Mjelde for help with laboratory sample processing in this study.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/frwa.2024.1466377/full#supplementary-material
Abbreviations
FIB, Fecal indicator bacteria; qPCR, Quantitative polymerase chain reaction; CFU, Colony forming unit.
Footnotes
References
Ahmed, W., Hughes, B., and Harwood, V. J. (2016). Current status of marker genes of Bacteroides and related taxa for identifying sewage pollution in environmental waters. Water 8:231. doi: 10.3390/w8060231
Ahmed, W., Stewart, J., Powell, D., and Gardner, T. (2008). Evaluation of Bacteroides markers for the detection of human fecal pollution. Lett. Appl. Microbiol. 46, 237–242. doi: 10.1111/j.1472-765X.2007.02287.x
Barraud, O., Baclet, M. C., Denis, F., and Ploy, M. C. (2010). Quantitative multiplex real-time PCR for detecting class 1, 2 and 3 integrons. J. Antimicrob. Chemother. 65, 1642–1645. doi: 10.1093/jac/dkq167
Bernhard, A. E., and Field, K. G. (2000). A PCR assay to discriminate human and ruminant feces on the basis of host differences in Bacteroides-Prevotella genes encoding 16S rRNA. Appl. Environ. Microbiol. 66, 4571–4574. doi: 10.1128/AEM.66.10.4571-4574.2000
Bokulich, N. A., Kaehler, B. D., Rideout, J. R., Dillon, M., Bolyen, E., Knight, R., et al. (2018). Optimizing taxonomic classification of marker-gene amplicon sequences with QIIME 2’s q2-feature-classifier plugin. Microbiome 6:90. doi: 10.1186/s40168-018-0470-z
Bolyen, E., Rideout, J. R., Dillon, M. R., Bokulich, N. A., Abnet, C. C., Al-Ghalith, G. A., et al. (2019). Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 37, 852–857. doi: 10.1038/s41587-019-0209-9
Cabelli, V. J., Dufour, A. P., Levin, M. A., McCabe, L. J., and Haberman, P. W. (1979). Relationship of microbial indicators to health effects at marine bathing beaches. Am. J. Public Health 69, 690–696. doi: 10.2105/AJPH.69.7.690
Cabelli, V. J., Dufour, A. P., McCabe, L. J., and Levin, M. A. (1982). Swimming-associated gastroenteritis and water quality. Amer. J. Epidemiol. 115, 606–616. doi: 10.1093/oxfordjournals.aje.a113342
Callahan, B. J., McMurdie, P. J., Rosen, M. J., Han, A. W., Johnson, A. J. A., and Holmes, S. P. (2016). DADA2: high-resolution sample inference from Illumina amplicon data. Nat. Methods 13, 581–583. doi: 10.1038/nmeth.3869
Casteel, M. J., Sobsey, M., and Mueller, J. P. (2006). Fecal contamination of agricultural soils before and after hurricane-associated flooding in North Carolina. J. Environ. Sci. Health Part A 41, 173–184. doi: 10.1080/10934520500351884
de Mendiburu, F., and Yaseen, M. (2020). Statistical procedures for agricultural research. 1.40 ed2020.
Dobbs, F. C. (2007). Après le déluge: microbial landscape of New Orleans after the hurricanes. Proc. Natl. Acad. Sci. 104, 9103–9104. doi: 10.1073/pnas.0703191104
Emanuel, K. (2017). Assessing the present and future probability of hurricane Harvey’s rainfall. Proc. Natl. Acad. Sci. 114, 12681–12684. doi: 10.1073/pnas.1716222114
Field, K. G., and Samadpour, M. (2007). Fecal source tracking, the indicator paradigm, and managing water quality - identifying sources of fecal pollution. Water Res. 41, 3517–3538. doi: 10.1016/j.watres.2007.06.056
Fogarty, E. C., Schechter, M. S., Lolans, K., Sheahan, M. L., Veseli, I., Moore, R. M., et al. (2024). A cryptic plasmid is among the most numerous genetic elements in the human gut. Cell 187, 1206–1222.e16. doi: 10.1016/j.cell.2024.01.039
Griffith, J. F., Yiping, C., McGee, C. D., and Weisberg, S. B. (2009). Evaluation of rapid methods and novel indicators for assessing microbiological beach water quality - cross-validation of detection methods for pathogens and fecal indicators. Water Res. 43, 4900–4907. doi: 10.1016/j.watres.2009.09.017
Haenelt, S., Wang, G., Kasmanas, J. C., Musat, F., Richnow, H. H., da Rocha, U. N., et al. (2023). The fate of sulfonamide resistance genes and anthropogenic pollution marker intI1 after discharge of wastewater into a pristine river stream. Front. Microbiol. 14:1058350. doi: 10.3389/fmicb.2023.1058350
Hagedorn, C., Robinson, S. L., Filtz, J. R., Grubbs, S. R., Angier, T. A., and Reneau, R. B. (1999). Determining sources of fecal pollution in a rural Virginia watershed with antibiotic resistance patterns in fecal streptococci. Appl. Environ. Microbiol. 65, 5522–5531. doi: 10.1128/AEM.65.12.5522-5531.1999
Harwood, V. J., Staley, C., Badgley, B. D., Borges, K., and Korajkic, A. (2014). Microbial source tracking markers for detection of fecal contamination in environmental waters: relationships between pathogens and human health outcomes. FEMS Microbiol. Rev. 38, 1–40. doi: 10.1111/1574-6976.12031
Heuer, H., and Smalla, K. (2007). Manure and sulfadiazine synergistically increased bacterial antibiotic resistance in soil over at least two months. Environ. Microbiol. 9, 657–666. doi: 10.1111/j.1462-2920.2006.01185.x
Kapoor, V., Gupta, I., Pasha, T. A. B. M., and Phan, D. (2018). Real-time quantitative PCR measurements of fecal Indicator Bacteria and human-associated source tracking markers in a Texas River following hurricane Harvey. Environ. Sci. Tech. Lett. 5, 322–328. doi: 10.1021/acs.estlett.8b00237
Knights, D., Kuczynski, J., Charlson, E. S., Zaneveld, J., Mozer, M. C., Collman, R. G., et al. (2011). Bayesian community-wide culture-independent microbial source tracking. Nat. Methods 8:761–763. doi: 10.1038/nmeth.1650
Kreader, C. A. (1995). Design and evaluation of Bacteroides DNA probes for the specific detection of human fecal pollution. Appl. Environ. Microbiol. 61, 1171–1179. doi: 10.1128/aem.61.4.1171-1179.1995
LaMontagne, M. G., Zhang, Y., Guillen, G. J., Gentry, T. J., and Allen, M. S. (2022). Hurricane Harvey impacts on water quality and microbial communities in Houston, TX waterbodies. Front. Microbiol. 13:875234. doi: 10.3389/fmicb.2022.875234
Landsman, M. R., Rowles, L. S., Brodfuehrer, S. H., Maestre, J. P., Kinney, K. A., Kirisits, M. J., et al. (2019). Impacts of hurricane Harvey on drinking water quality in two Texas cities. Environ. Res. Lett. 14:124046. doi: 10.1088/1748-9326/ab56fb
McMurdie, P. J., and Holmes, S. (2013). Phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS One 8:e61217. doi: 10.1371/journal.pone.0061217
Mcquaig, S., Griffith, J., and Harwood, V. J. (2012). Association of fecal indicator bacteria with human viruses and microbial source tracking markers at coastal beaches impacted by nonpoint source pollution. Appl. Environ. Microbiol. 78, 6423–6432. doi: 10.1128/AEM.00024-12
Moghadam, S. V., Vadde, K. K., Phan, D. C., Jafarzadeh, A., and Kapoor, V. (2022). Assessing the impact of flooding on bacterial community structure and occurrence of potentially pathogenic bacteria in Texas rivers after hurricane Harvey. J. Hazard. Mater. Lett. 3:100058.
National Hurricane Center (2017). Costliest U.S. tropical cyclones. Available at: https://www.nhc.noaa.gov/news/UpdatedCostliest.pdf [Accessed April 10, 2024]
National Nonpoint Source Monitoring Program (2013). Monitoring for microbial pathogens and indicators. Available at: https://www.epa.gov/sites/default/files/2016-05/documents/tech_notes_9_dec2013_pathogens.pdf [Accessed April 10, 2024]
National Weather Service (2017). Major hurricane Harvey - August 25-29, 2017. Available at: https://www.weather.gov/crp/hurricane_harvey [Accessed April 10, 2024].
Oksanen, J., Simpson, G., Blanchet, F., Kindt, R., Legendre, P., Minchin, P., et al. (2024). Vegan: community ecology package R package version 2.6-6.1.
Pérez-Valdespino, A., Pircher, R., Pérez-Domínguez, C. Y., and Mendoza-Sanchez, I. (2021). Impact of flooding on urban soils: changes in antibiotic resistance and bacterial community after hurricane Harvey. Sci. Tot. Environ. 766:142643. doi: 10.1016/j.scitotenv.2020.142643
Pruss, A. (1998). Review of epidemiological studies on health effects from exposure to recreational water. Int. J. Epidemiol. 27, 1–9. doi: 10.1093/ije/27.1.1
Quast, C., Pruesse, E., Yilmaz, P., Gerken, J., Schweer, T., Yarza, P., et al. (2012). The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 41, D590–D596. doi: 10.1093/nar/gks1219
Quintela-Baluja, M., Frigon, D., Abouelnaga, M., Jobling, K., Romalde, J. L., Gomez-Lopez, M., et al. (2021). Dynamics of integron structures across a wastewater network – implications to resistance gene transfer. Water Res. 206:117720. doi: 10.1016/j.watres.2021.117720
R Core Team (2021). R: A language and environment for statistical computing. R foundation for statistical computing, Vienna, Austria. Available at: https://www.R-project.org/ [Accessed September 9, 2022].
Schwab, K.J., Gibson, K.E., Williams, D.E., and Kulbicki, K.M., Paul L.C, Mihalic, J.N., Breysse, P.N., Curriero, F.C., and Geyh, A.S. (2007). Microbial and chemical assessment of regions within New Orleans, LA impacted by hurricane Katrina. Environ. Sci. Technol. 41, 2401–2406, doi: 10.1021/es062916x
Scott, T. M., Jenkins, T. M., Lukasik, J., and Rose, J. B. (2005). Potential use of a host associated molecular marker in Enterococcus faecium as an index of human fecal pollution. Environ. Sci. Technol. 39, 283–287. doi: 10.1021/es035267n
Scutti, S . (2017). Sewage, fecal bacteria in hurricane Harvey floodwaters. Available at: https://www.cnn.com/2017/09/01/health/houston-flood-water-contamination/index.html [Accessed April 10, 2024].
Shanks, O. C., Kelty, C. A., Oshiro, R., Haugland, R. A., Madi, T., Brooks, L., et al. (2016). Data acceptance criteria for standardized human-associated fecal source identification quantitative real-time PCR methods. Appl. Environ. Microbiol. 82, 2773–2782. doi: 10.1128/AEM.03661-15
Shanks, O. C., Kelty, C. A., Sivaganesan, M., Varma, M., and Haugland, R. A. (2009). Quantitative PCR for genetic markers of human fecal pollution. Appl. Environ. Microbiol. 75, 5507–5513. doi: 10.1128/AEM.00305-09
Sinigalliano, C. D., Gidley, M. L., Shibata, T., Whitman, D., Dixon, T. H., Laws, E., et al. (2007). Impacts of hurricanes Katrina and Rita on the microbial landscape of the New Orleans area. Proc. Natl. Acad. Sci. 104, 9029–9034. doi: 10.1073/pnas.0610552104
Stachler, E., Crank, K., and Bibby, K. (2019). Co-occurrence of crassphage with antibiotic resistance genes in an impacted urban watershed. Environ. Sci. Tech. Lett. 6, 216–221. doi: 10.1021/acs.estlett.9b00130
Stoeckel, D. M., and Harwood, V. J. (2007). Performance, design, and analysis in microbial source tracking studies. Appl. Environ. Microbiol. 73, 2405–2415. doi: 10.1128/AEM.02473-06
TCEQ (2017). Hurricane Harvey after action review. Available at: https://www.tceq.texas.gov/assets/public/response/hurricanes/hurricane-harvey-after-action-review-report.pdf. [Accessed April 10, 2024]
TCEQ (2024). Water quality general permits search - advanced search. Available at: https://www2.tceq.texas.gov/wq_dpa/index.cfm [Accessed September 9, 2024].
USEPA (2013). Recreational water quality criteria and methods - reports and assessments. Available at: https://www.epa.gov/wqc/recreational-water-quality-criteria-and-methods [Accessed April 10, 2024].
USEPA (2017). Status of water Systems in Areas Affected by Harvey. Available at: https://www.epa.gov/archive/epa/newsreleases/status-water-systems-areas-affected-harvey.html [Accessed April 10, 2024].
Veldhuis, J., Ten, A. E., Clemens, F. H. L. R., Sterk, G., and Berends, B. R. (2010). Microbial risks associated with exposure to pathogens in contaminated urban flood water. Water Res. 44, 2910–2918. doi: 10.1016/j.watres.2010.02.009
Wade, T. J., Pai, N., Eisenberg, J. N. S., and Colford, J. M. (2003). Do U.S. Environmental Protection Agency water quality guidelines for recreational waters prevent gastrointestinal illness? A systematic review and meta-analysis. Environ. Health Perspect. 111, 1102–1109. doi: 10.1289/ehp.6241
Wexler, H. M. (2007). Bacteroides: the good, the bad, and the nitty-gritty. Clin. Microbiol. Rev. 20, 593–621. doi: 10.1128/CMR.00008-07
Yang, S. H., Chen, C. H., and Chu, K. H. (2021). Fecal indicators, pathogens, antibiotic resistance genes, and ecotoxicity in Galveston Bay after hurricane Harvey. J. Hazard. Mater. 411:124953. doi: 10.1016/j.jhazmat.2020.124953
Yilmaz, P., Parfrey, L. W., Yarza, P., Gerken, J., Pruesse, E., Quast, C., et al. (2014). The SILVA and ‘all-species living tree project (LTP)’ taxonomic frameworks. Nucleic Acids Res. 42, D643–D648. doi: 10.1093/nar/gkt1209
Keywords: E. coli, fecal indicator bacteria, coliform, hurricane, Harvey, NMDS, pBI143, microbial source tracking
Citation: Mukherjee M, Hossain MS, Boswell J, Zhang Y, Allen MS, LaMontagne MG and Gentry TJ (2024) Large, but short-term, increase in fecal indicator bacteria following extreme flooding from Hurricane Harvey in Houston, TX. Front. Water. 6:1466377. doi: 10.3389/frwa.2024.1466377
Edited by:
Antonio Bucci, University of Molise, ItalyReviewed by:
Fulvio Celico, University of Parma, ItalyErmanno Federici, University of Perugia, Italy
Copyright © 2024 Mukherjee, Hossain, Boswell, Zhang, Allen, LaMontagne and Gentry. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Maitreyee Mukherjee, bWFpdHJleWVlMjVAZ21haWwuY29t; Michael G. LaMontagne, bGFtb250YWduZUB1aGNsLmVkdQ==; Terry J. Gentry, dGpnZW50cnlAdGFtdS5lZHU=
 Maitreyee Mukherjee
Maitreyee Mukherjee Md Shakhawat Hossain
Md Shakhawat Hossain John Boswell3
John Boswell3 Michael G. LaMontagne
Michael G. LaMontagne Terry J. Gentry
Terry J. Gentry