
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Water , 16 May 2022
Sec. Water and Human Health
Volume 4 - 2022 | https://doi.org/10.3389/frwa.2022.883282
This article is part of the Research Topic Resiliency and Sustainability of Urban Water Systems View all 4 articles
With their large, diverse microbial communities chronically exposed to sub-inhibitory antibiotic concentrations, wastewater treatment works (WWTW) have been deemed hotspots for the emergence and dissemination of antimicrobial resistance, with growing concern about the transmission of antibiotic resistance genes (ARGs) and antibiotic resistant bacteria (ARB) into receiving surface waters. This study explored (1) the prevalence of ARG and ARB in local WWTW, (2) the effect of sub-inhibitory antimicrobial exposure on ARG copy numbers in pure cultures from WWTW, and (3) two WWTW with different treatment configurations. For each WWTW, qPCR determined the prevalence of mcr3, sul1, sul2, and blaKPC during the treatment process, and culture methods were used to enumerate and identify ARB. Bacterial colonies isolated from effluent samples were identified by 16S rDNA sequencing and their respective minimum inhibitory concentrations (MIC) were determined. These were compared to the MICs of whole community samples from the influent, return activated sludge, and effluent of each WWTW. Resistance genes were quantified in 11 isolated cultures before and after exposure to sub-MIC concentrations of target antibiotics. The numbers of ARG and ARB in both WWTW effluents were notably reduced compared to the influent. Sul1 and sul2 gene copies increased in cultures enriched in sub-MIC concentrations of sulfamethoxazole, while blaKPC decreased after exposure to amoxicillin. It was concluded, within the parameters of this study, that WWTW assist in reducing ARG and ARB, but that sub-inhibitory exposure to antimicrobials has a varied effect on ARG copy number in pure cultures.
Antibiotics have cured many life-threatening bacterial infections and can be used preventatively against the onset of nosocomial infections after surgeries, as well as prophylactically in animal husbandry (Salkind and Rao, 2011; Ling et al., 2015). Their use has been a fundamental part of public health since the mid-1900s (Ling et al., 2015). However, antimicrobial resistance (AMR) emerged as a global health-care crisis, resulting in numerous investigations into factors that promote AMR emergence in the environment. With population growth, urbanization, and the additional crisis of an increased demand for water, with diminished freshwater resources (Visser, 2018), wastewater reclamation for agricultural and industrial processes has become a necessity (Palomo-Briones et al., 2016; Fito and Van Hulle, 2020). Wastewater treatment works (WWTW) are designed to remove a large variety of inorganic and organic matter. Rapid urbanization and densification necessitate upgrades to increase capacity and maintain an adequate standard of water treatment, which includes the elimination of microbes that pose a risk to public health. These environments contain extremely diverse microbial communities due to high, varying nutrient loads and the implementation of microbes in bioremediation (Singh et al., 2006; Osunmakinde et al., 2019), in addition to the entry of microbes from external environments. The AMR has been frequently detected in WWTW and they have consequently been deemed hotspots for AMR dissemination (Hultman et al., 2018; Pazda et al., 2019).
Spreading of antibiotic resistant bacteria (ARB) into environments downstream of WWTW is an ecological concern, as well as a risk for entering the food chain through livestock drinking contaminated water, potentially containing AMR zoonotic pathogens (Sano et al., 2016; Caicedo et al., 2019). Crops being fertilized or irrigated with reclaimed WWTW waste sludge and effluent water create another pathway for AMR into the food chain. In addition, antibiotic resistance genes (ARGs) have been more prevalent in soil treated with poultry and cattle manure compared to untreated soils and transmission pathways of these genes into the food chain from the rhizosphere have been suggested (Hu et al., 2016; Zhang et al., 2019). Multi-drug resistant (MDR) and extensively drug resistant (XDR) diseases are becoming increasingly prevalent (Pérez-Rodríguez and Mercanoglu Taban, 2019). With MDR bacteria also being isolated from WWTW (Adefisoye and Okoh, 2016), the movement of AMR into the food chain has the potential to place significant pressure on the race to develop novel antimicrobial compounds.
Except for studies applying metagenomics, most investigations on AMR in WWTW focus primarily on single species that have been isolated from WWTW, or indicator organisms such as Escherichia coli (Ekwanzala et al., 2018; Fouz et al., 2020). However, less common bacterial species, and multi-species interactions, are likely to influence the outcome of AMR in these diverse environments (Osunmakinde et al., 2019). With different population dynamics, locations and proximity to hospitals, and agriculture or industrial settings, conditions for the effective reduction of ARG and ARB may differ between WWTW. Therefore, the treatment strategies that work for one location may not work for another (Jasim, 2020). Thus, two different WWTW discharges in Cape Town, South Africa (SA) were selected to gain insight into the dissemination of AMR in local contexts, to provide future solutions for preventing AMR transmission through WWTW. This study aimed to determine the persistence of bacterial resistance to four antibiotics [amoxicillin (AMX), gentamicin (GM), sulfamethoxazole (SMX), and colistin (CST)] and associated resistance genes (blaKPC, blaOXA−48, sul1, sul2, and mcr3) within the WWTW, as well as the effect of sub-inhibitory antibiotic concentrations on resistance gene prevalence in resistant isolates from WWTW. Antibiotics were selected for this study based on (1) their frequency of prescription and use in the Western Cape (National Institute for Communicable Diseases, n.d.), (2) their status as “critical” or “highly important” antimicrobials (World Health Organisation, 2011), and (3) the resistance of clinically relevant pathogens to the relevant compounds (National Institute for Communicable Diseases, n.d.). Due to the rise in resistance to β-lactam and last resort antibiotics such as carbapenems and CST, blaKPC, blaOXA−48, and mcr3 resistance genes were selected (Sekyere, 2016). Many HIV positive patients are treated with low doses of co-trimoxazole (a SMX and trimethoprim combination) to prevent opportunistic infections (Kaplan et al., 2009). With 13.5% of the SA population living with HIV (Stats, 2019), sul1 and sul2 are important genes to study in this context.
Two WWTW located in Cape Town, SA were selected for this study (Supplementary Figure 1). Both WWTW made use of conventional activated sludge treatment; however, after screening and grit removal, WWTW1 made use of primary sedimentation and a mixed flow bioreactor setup, followed by secondary sedimentation and chlorination, and finally discharged into maturation ponds. In contrast, WWTW2 made use of plug-flow treatment followed by the discharge of clarified water into a maturation pond prior to chlorination. The two WWTW serve approximately 248,000 people and 1,102,000 people respectively as calculated by Archer et al. (2021).
Wastewater grab samples were collected in 100 ml sterile Schott bottles from the influent (post grit screens), return activated sludge (RAS), and treated effluent (post-chlorination in WWTW1; post-chlorination and post-maturation pond in WWTW2). All samples were stored on ice during transport to the laboratory and stored at 4°C until processed, within 2 h of arriving at the laboratory.
Plasmid-mediated antibiotic resistance genes conferring resistance to AMX (blaOXA−48, blaKPC), SMX (sul1 and sul2), and CST (mcr3) were quantified with qPCR in influent, RAS, and effluent WWTW samples.
Total DNA was extracted from the respective samples using the Quick-DNA Fecal/Soil Microbe Kit (ZymoResearch, n.d.) according to manufacturer's specifications. Positive target gene controls were synthesized according to the sequences obtained from NCBI accession numbers (Supplementary Table 1) that were flanked by the selected primers shown in Table 1. Synthesized genes were inserted into a pBluescript II SK (+) cloning vector, resulting in a 1,498 bp plasmid containing all target genes. All genes, primers, and probes were synthesized by Inqaba Biotech. Gene copies were normalized with 16S rDNA gene copies. The generation of standard curves and calculation of gene copy numbers are described in Supplementary Materials (Supplementary Text S1, Supplementary Table 2).
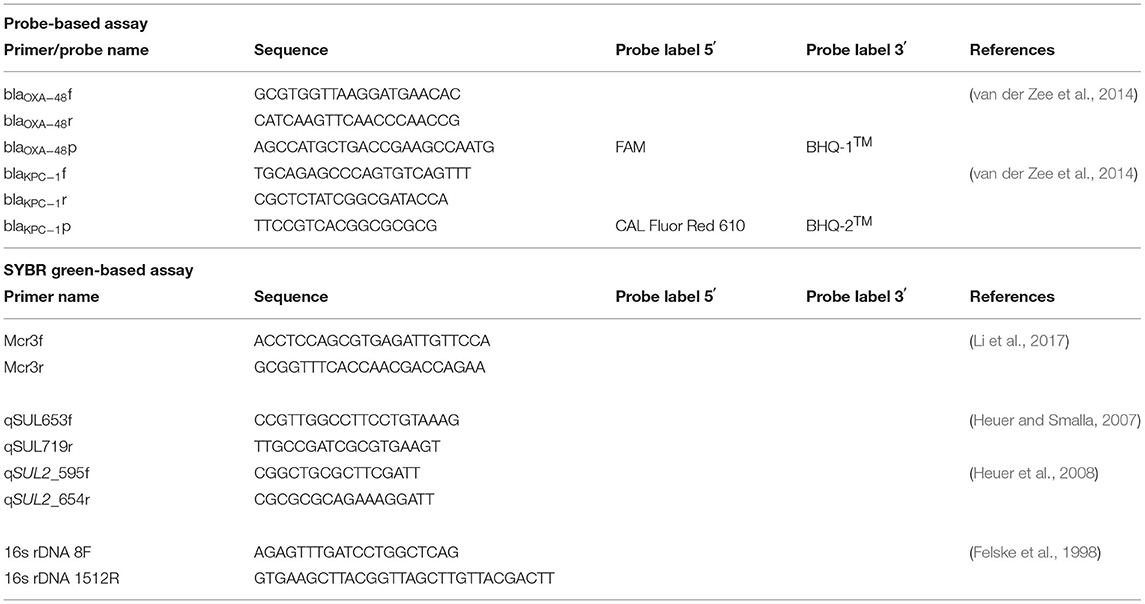
Table 1. Sequences and references of primers and probes used in both probe-based and SYBR Green-based qPCR assays.
The sul1, sul2, mcr3, and 16s rDNA genes were quantified from WWTW samples using a SYBR green assay. Samples were run in duplicate and the reaction composition included the following final concentrations: 1X Luna® Universal qPCR Master Mix (New England Biolabs Inc.), 0.25 μM of each primer, 1 μl template DNA (concentrations in Supplementary Table 3), and sterile milli-Q water to make the final reaction volume up to 20 μl. Cycling conditions for the SMX and CST resistance genes were according to the master mix manufacturer's instructions with 50 cycles of the 2-step amplification used. Cycling conditions for 16S rDNA involved an initial denaturation 94°C for 30 s, followed by 25 cycles of 94°C for 20 s, 58°C for 40 s, and 68°C for 90 s. A final extension at 68°C for 5 min was performed (protocol adapted from manufacturers recommendations for OneTaq® 2X Master Mix with Standard Buffer, New England Biolabs, Inc.).
The β-lactam (AMX) resistance genes were quantified using a probe-based assay containing 1X Luna® Universal Probe qPCR Master Mix (New England Biolabs Inc.), 0.4 μM of each primer, 0.2 μM probe, 1 μl template DNA, and sterile milli-Q water to make the final reaction volume up to 20 μl. Three technical repeats and two biological repeats were performed. The cycling conditions were performed according to manufacturer's specifications.
Amplification was conducted using a LightCycler® 96 (Roche) system with excitation/emission wavelengths of 470/514 nm selected for the SYBR green reactions and FAM-labeled probe, and 577/620 nm selected for the CAL Fluor Red 610-labeled probe. Melting peaks for the SYBR green reactions are shown in Supplementary Figures 3–5. Negative controls containing sterile milliQ water instead of template DNA were included in all assays for each resistance gene.
Antibiotic resistant bacteria were enumerated, and representative colonies were isolated and identified from two WWTW using two different methods to reduce potential culture bias and obtain a more diverse range of isolates.
To classify a particular organism as antibiotic resistant, its minimum inhibitory concentrations (MIC) to a particular antibiotic needs to be higher than the resistance breakpoint concentration set by the Clinical and Laboratory Standards Institute (Clinical and Laboratory Standards Institute, 2015) and European Committee on Antimicrobial Susceptibility Testing (European Committee on Antimicrobial Susceptibility Testing, 2017). If the MIC is lower than the breakpoint, the organism would be deemed susceptible to that antibiotic. As a result, the CLSI and EUCAST resistance breakpoints were used to select starting concentrations of the selected antibiotics to incorporate into culture media for the selection of antibiotic resistant bacteria. As these documents obtain their values from pure cultures (i.e., laboratory conditions/strains) by microbroth dilution susceptibility assays, there was a need to determine MICs for environmental whole communities to select for highly resistant organisms. A microbroth dilution susceptibility assay was performed in 96-well microtiter plates (Sigma Aldrich). As CST is only active against Gram-negative bacteria, MacConkey (MAC) broth (Merck) (selective for Gram-negative bacteria) was used, while Mueller Hinton Broth (MHB) (Merck) was used for the other broad-spectrum antibiotics. Antibiotic concentrations were sequentially doubled in 5 increments as described by Andrews (2001), for colistin sulfate (CST; 1–16 μg/ml), GM (4–64 μg/ml), AMX (32–512 μg/ml), and SMX (64–1,024 μg/ml). The RAS samples obtained from the two WWTW were diluted in phosphate buffered saline (PBS) to obtain a 0.5 McFarland standard turbidity equivalent and used as the inoculum for the susceptibility testing. After incubation at 37°C for 18–24 h with shaking, optical density was measured at 600 nm, and OD600 ≤ 0.02 indicated the MIC. The antibiotic concentrations that were 2-fold lower than the determined RAS MICs (shown in Supplementary Figure 6) were selected as the sub-MIC concentrations for the enumeration and selection of antibiotic resistant bacteria in WWTW.
Five milliliters of the homogenized water samples from the WWTW influent, RAS, and effluent, respectively, were treated with 0.001% cycloheximide (w/v) to inhibit the growth of fungi and were subsequently serially diluted (1:10) with PBS to dilute solid matter in the raw samples. A droplet (20 μl) of each dilution was inoculated in duplicate into 96-well microtiter plates (Sigma Aldrich) containing MAC broth supplemented with 8 μg/ml CST, and MHB supplemented in individual wells with 32 μg/ml GM, 512 μg/ml AMX, and 512 μg/ml SMX, respectively. Blank controls containing only MAC and MHB media were also included. After 18–24 h of incubation at 37°C, the optical density was measured at 600 nm (Bio–Rad XMark microplate absorbance spectrophotometer). Twenty milliliters of the respective cultures obtained in the 10−1 dilution were sub-cultured overnight in fresh broth containing the same antibiotic conditions for enrichment, a total of three times, after which serial dilutions were enumerated on agar plates containing the same antibiotic concentrations and incubated for 18–24 h at 37°C. Four single colonies from each antibiotic plate were selected at random and streaked to purity. They were subsequently cultured in Tryptic Soy Broth (TSB) overnight and DNA was extracted using a method adapted from Crouse and Amorese (1987) (amendments to protocol described in Supplementary Text 2) and stored at −20°C, with long-term storage at −80°C (40% glycerol).
The abundance of antibiotic resistant bacteria in the WWTW effluent was lower than in the influent, in contrast to findings in previous studies which indicated that WWTW promoted the dissemination of AMR (Czekalski et al., 2012; Rizzo et al., 2013; Adefisoye and Okoh, 2016; Sharma and Sharma, 2021). Therefore, a second sampling event was conducted to test the validity of this unexpected result. Samples were collected and processed as previously described, however, only CST and GM were supplemented into MAC and MHB, respectively, to simplify the experiment. In addition to broth enrichment, the samples were spot-plated onto respective antibiotic plates before and after enrichment and incubated for 18–24 h at 37°C to obtain plate counts. AMX and SMX were omitted here to simplify the experiment. Colonies from all plates before and after enrichment were selected at random and DNA was extracted from each pure colony and stored as described previously.
The DNA extracted from each isolate was thawed and PCR targeting the 16S rDNA gene was performed using universal primers 8F and 1512R (sequences in Table 1). The reaction components were combined to obtain final concentrations of 1.25-units OneTaq Polymerase (New England Biolabs), 200-μM dNTPs, 0.2 μM of each primer mentioned here, 1 × OneTaq Buffer, 4-μl DNA template (concentrations in Supplementary Table 4), and sterile milliQ in a 50-μl volume. Cycling conditions were as described previously for the qPCR reactions. Amplicons were sequenced (3730XL DNA Analyzer; Applied Biosystems) and sequence chromatograms were trimmed and consensus sequences for each primer pair were constructed using BioEdit (v. 7.05); BLASTn was performed for each sequence with the database selected for 16S ribosomal RNA sequences (bacteria and archaea). The species of isolates were identified based on query cover and identity using a threshold of 99%.
Whole community and pure culture MICs were determined to identify the effect of antibiotic exposure in sub-MIC concentrations on the copy number of ARG present in isolates obtained from WWTW.
Whole-community and pure culture MICs were established. The 30 glycerol stocks previously isolated were revived in TSB overnight at 37°C. Susceptibility testing was performed in duplicate by microbroth dilution as described here; however, the doubling dilution antibiotic concentration ranges used were increased to accommodate highly resistant isolates: CST at 1–512 μg/ml, GM at 2–1,024 μg/ml, and both AMX and SMX at 32–16,384 μg/ml. After overnight incubation at 37°C, the culture from wells containing growth at the highest concentration before inhibition (sub-MIC) was sub-cultured overnight at 37°C in TSB. The DNA was extracted from these cultures as mentioned previously and stored at −20°C.
For whole communities, to avoid initial culturing bias, the collected wastewater samples were used as inoculum. The conventional 0.5 McFarland standard turbidity could not be used to standardize inoculum density in wastewater samples due to the effluent samples having lower turbidity than required, and a large volume would have been required for filtration to concentrate the samples. Inoculum density was made uniform across all wastewater samples (influent, RAS, and effluent), by measuring the OD600 of the effluent (cleanest sample) and diluting the other samples with PBS to obtain a similar OD600 (0.018 ± 0.015). Microbroth dilutions were performed as for the single colonies. The DNA from the remaining influent, RAS, and effluent samples was extracted and stored at −20°C.
The sul1, sul2, and blaKPC genes were quantified in pure cultures isolated during the second sampling event prior to antibiotic exposure, and after sub-MIC antibiotic exposure during the MIC experiments. Quantification was performed with qPCR as described previously to determine gene copy numbers.
Data obtained from culturing was analyzed in GraphPad Prism (v. 6) using a multiple t-test to determine significance between the two WWTW in influent, RAS, and effluent samples. Student's t-tests for independent means were used to determine significant differences in resistance gene copy numbers between WWTW and in single colonies before and after exposure to sub-MIC concentrations of antibiotics. Significance was determined at a confidence interval of 95% (p < 0.05).
The percentage reduction of target resistance genes between influent and effluent ranged from 69 to 100% as shown in Figure 1. The percentage reductions were higher in WTWW2 effluent compared to WWTW1 for three out of the four target genes, with more than a 99% decrease of mcr3, sul1, and sul2. The mcr3 gene was completely removed from the effluent of WWTW2. The negative percentage reduction observed for blaKPC in WWTW2 indicates an 80.9% increase in gene copies from influent to effluent. All samples were negative for blaOXA−48 genes and are therefore excluded. Copy numbers for each resistance gene are shown in Supplementary Figure 7.
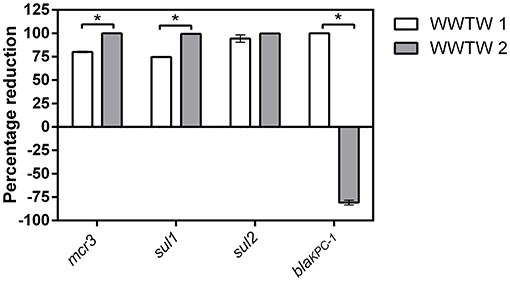
Figure 1. Percentage reduction of target antibiotic resistance genes from influent to effluent in two WWTW. Significant differences (p < 0.05) in percentage reduction between WWTW, identified using a t-test, are shown by the asterisk (*).
The abundance of highly resistant bacteria was similar for all antibiotics within each sample matrix, except for SMX, which had a visibly greater effect on the bacteria within the samples obtained from sampling event 1 (Figures 2A,B). The abundance of ARB in both WWTW effluents was lower compared to the influent and RAS, with the abundance of bacteria resistant to AMX, GM, and CST in WWTW2 effluent lower than in WWTW1 (Figures 2A,B). This is also the case for CST CFU/ml in sampling event 2 (Figures 3A,B). The number of GM-resistant bacteria did not change in sampling event 2 between matrices or WWTW (Figures 3A,B). Upon enrichment in antibiotics, the abundance of ARB to all antibiotics increased in both sampling events 1 (Figures 2C,D) and 2 (Figures 3C,D). Most samples enriched in POS MAC and CST resulted in higher abundance (1.5–2.2) in WWTW1 compared to WWTW2 (0.5–1.8) (Figures 2C,D, respectively). A higher OD600 in POS MAC broth compared to POS MHB was observed for most of the sample matrices (Figures 2A,D).
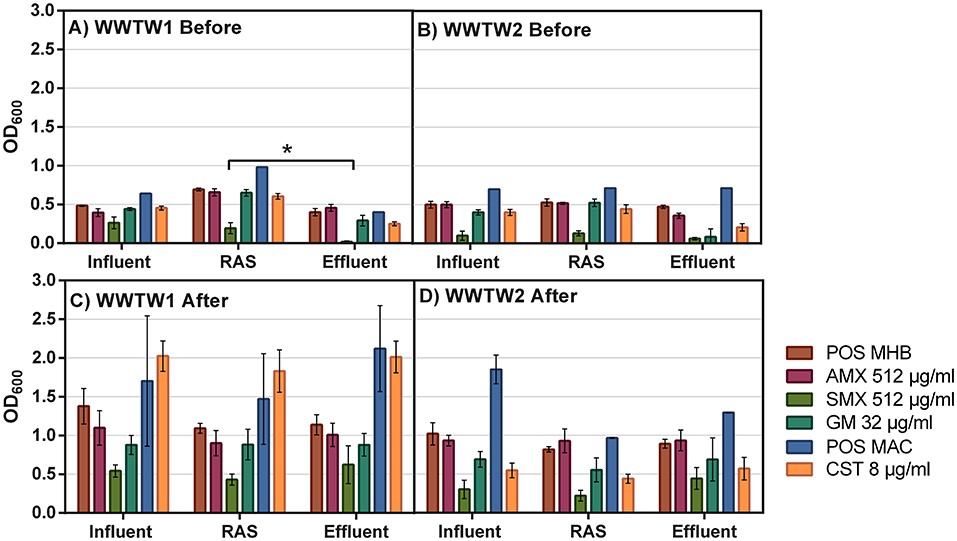
Figure 2. Abundance of bacteria (OD600) in the presence of AMX (512μg/ml), SMX (512μg/ml), GM (32μg/ml), and CST (8μg/ml) from influent, RAS, and effluent samples in two WWTW [WWTW1: (A,C); WWTW2: (B,D)] in sampling event 1 before (A,B) and after (C,D) being enriched in antibiotics. MacConkey media was used as a positive control (POS MAC) for CST while MHB was used as a positive control (POS MHB) for the other antibiotics. Significant differences (p < 0.0.5) between matrices are indicated by the black bar and asterisk (*).
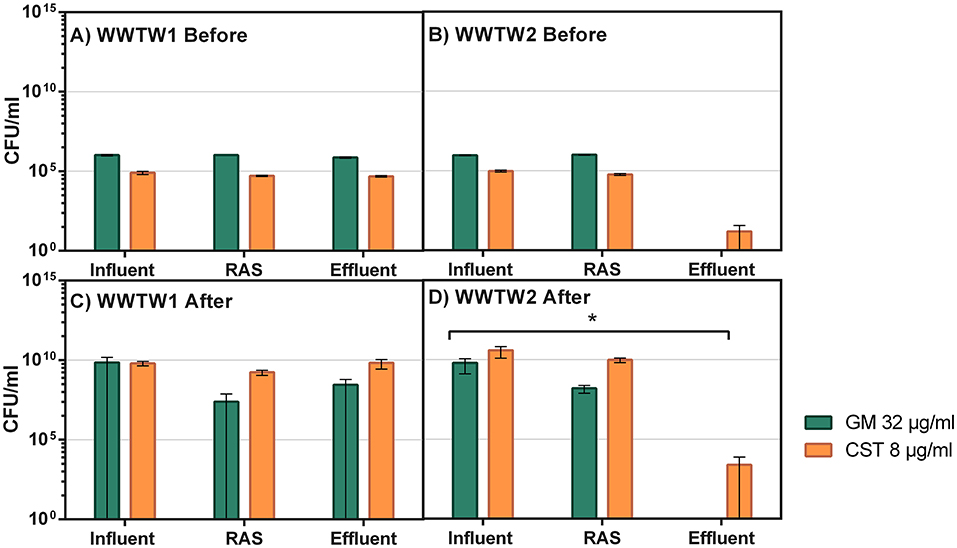
Figure 3. Viable bacterial cells in the presence of GM (32μg/ml) and CST (8μg/ml) from influent, RAS, and effluent samples in two WWTW [WWTW1: (A,C), WWTW2: (B,D)] in sampling event 2 before (A,B) and after (C,D) being enriched in antibiotics. Significant differences (p < 0.0.5) between matrices are indicated by the black bar and asterisk (*).
Of the randomly selected single colonies that were isolated from both sampling events (n = 30), Morganella morganii was the most dominant (20%), followed by Escherichia fergusonii/Shigella flexneri (13.3%), while Citrobacter freundii, Pseudomonas indoloxydans, Klebsiella pneumoniae, Klebsiella quasipneumoniae, and Enterobacter tabaci – each contributed 6.7% to the composition of the selected isolates. The remaining isolates each make up 3.3% of the sample (Figure 4). Figure 4 shows that there was a notable shift in community composition after enrichment in antibiotics.
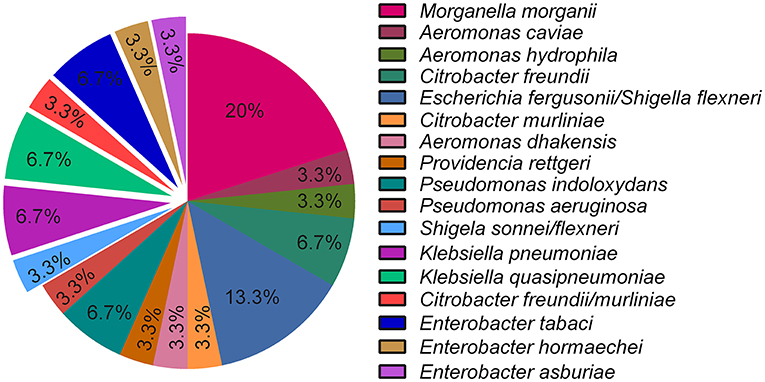
Figure 4. Percentage of isolated species obtained from effluent samples that were previously exposed to antibiotics (n = 19) and isolates obtained from various parts of the WWTW (n = 11) (exploded slices). Isolates that indicate multiple species, had similar percentage identities to the query sequence when using BLAST.
Species composition varied between WWTW as seen in Figure 5. Of the randomly surveyed species isolated before and after antibiotic exposure, M. morganii was a dominant species in both WWTW, with E. fergusonii/S. flexneri also dominant in WWTW1. Other species isolated from WWTW2 effluent were found in equal proportions.
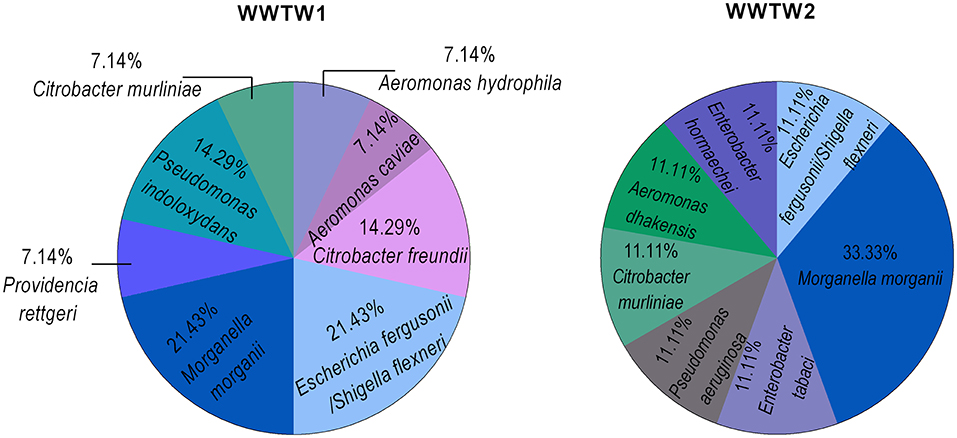
Figure 5. Species composition from different WWTW effluent samples previously exposed to antibiotics (WWTW1: n = 12; WWTW2: n = 7), and not previously exposed to antibiotics (WWTW1: n = 2; WWTW2: n = 2).
Figure 6 shows that the concentrations of the selected antibiotics (AMX, SMX, and CST) required to inhibit the growth of bacteria in WWTW samples were at least 2-fold lower in the effluent compared to influent and RAS of WWTW2. The MIC for CST and SMX was consistent throughout WWTW1 and between influent and RAS for WWTW2. The AMX MIC in the RAS of WWTW1 (16,384 μg/ml) was double that of the rest of the WWTW samples (8,192 μg/ml). The MIC for GM in WWTW2 effluent was 4-fold lower than that of the other WWTW2 matrices and all WWTW1 matrices.
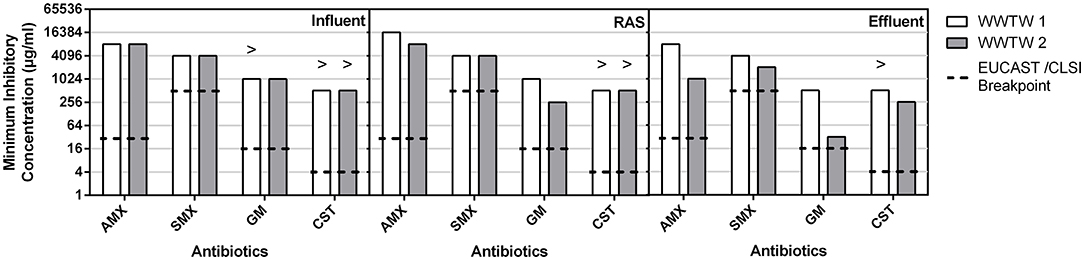
Figure 6. The MICs of whole communities in influent, RAS, and effluent samples from two WWTW for AMX, SMX, GM and CST. The greater than symbol (>) indicates growth still occurred in the highest concentrations used and as a result the MIC is greater than the value depicted by the bar.
In addition to the whole-community MICs depicted in Figure 6, Supplementary Table 4 shows the individual MICs of AMX, SMX, GM, and CST for the isolates randomly selected during the study. The MICs (Figure 7) for most isolates were higher than CLSI and EUCAST breakpoints for resistance (16,384 μg/ml, 4,096 μg/ml, and 2,048 μg/ml for AMX, SMX, and CST, respectively). The mode MIC for GM is on par with clinical breakpoints, although most of the other isolates have MICs above this value (16 μg/ml) (Figure 7). Comparing the modes of isolate MICs (Figure 7) to whole community MICs (Figure 6), MICs of AMX and CST were higher for isolates than for whole communities, those of GM were lower for isolates than whole communities, and those of SMX were the same for single colonies and whole communities.
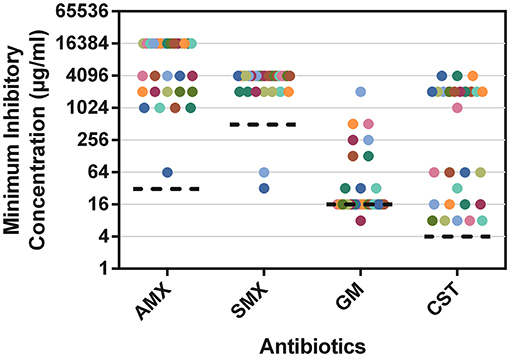
Figure 7. The MICs of randomly selected isolates from WWTW, where each data point represents a single isolate. Mode MICs appear where data points cluster. Individual MICs, species identification and antibiotic exposure can be observed in Supplementary Table 2.
The sul1, sul2, and blaKPC genes were observed in all organisms isolated on antibiotic-free media (Figure 8). Except for two isolates showing exponentially higher (1 × 1013) sul2 copies, most isolates cluster around the median. A paired Student's t-test determined that changes in median gene copies after antibiotic exposure for all ARGs were not significant (p > 0.05). However, median gene copy trends were similar between sul1 and sul2 and increased after exposure to sub-MIC concentrations of SMX (average of 1.3 × 108 copies before, increasing to 6.8 × 108 copies after), while sub-MIC exposure to AMX resulted in decreasing trends in the median copy numbers of blaKPC(3.9 × 108 to 1 × 108 gene copies/16S rDNA).
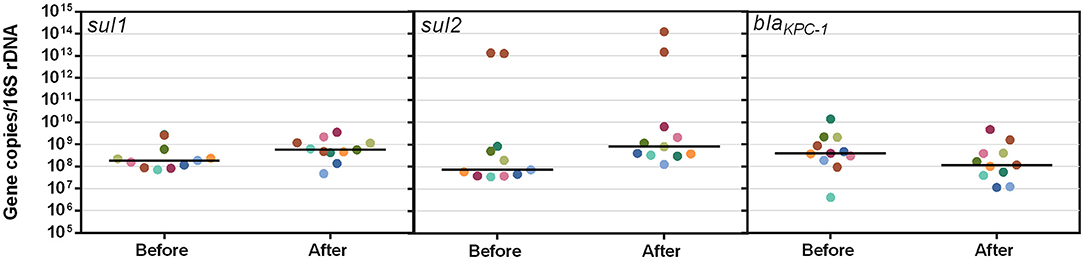
Figure 8. Copy number of antibiotic resistance genes/16S rDNA copies in colonies isolated on antibiotic free media before and after sub-MIC exposure to SMX (sul1 and sul2) and AMX (blaKPC-1). The line for each data set represents the median.
Multiple authors state that WWTW are hotspots for the transmission of ARG and ARB (Berendonk et al., 2015; Guo et al., 2017; Dafale et al., 2020). This study shows that while these environments do indeed have waters that are contaminated with ARG and ARB, all tested genes, except blaKPC in WWTW2, were less prominent in the effluent than in the influent (Figure 1). The increase in K. pneumoniae carbapenemase (KPC) is a concern as it confers resistance to carbapenems, which, along with the last resort antibiotics, is of greater concern than AMX (Feil, 2016). However, all other ARG were removed by the WWTW processes. As total DNA was extracted from the WWTW samples, the ARGs detected in this part of the study are not necessarily only from bacterial origin but also potentially from other sources including fungi, algae, protozoa, and viruses as suggested by the DNA extraction kit manufacturers (ZymoResearch, n.d.). Similarly, Figures 2, 3 mostly show no net increase in ARB between the influent and effluent of the WWTW and while not statistically significant, there were notable decreases in ARB abundance. Similar results have been found in other studies (Duong et al., 2008; Lupan et al., 2017). This does not suggest that the ARG and ARB released into the environment from the WWTW effluent do not play a role in the transmission of AMR downstream but demonstrates that the WWTW included in this study led to reduced numbers of ARG and ARB that entered the environment. In this case, without wastewater treatment, ARG and ARB would be more prominent in environmental waters.
The antibiotic concentrations used for ARB selection in this study were selected based on resistance breakpoints (μg/ml range) and, as a result, were much higher than concentrations observed in WWTW. Antibiotic levels in WWTW vary depending on the location, type, and size of the facility, as well as operational efficiency. Antibiotic concentrations in local WWTW range from 1,344.8 ng/L−3,612 ng/L for SMX in WWTW effluent (Suzuki et al., 2015; Archer et al., 2017); 1,400 ng/L for AMX in influent (Watkinson et al., 2009); and 0.019–7.6 ng/L for GM (Löffler and Ternes, 2003; Tahrani et al., 2016), while, to our knowledge, no quantification of CST in WWTW has been published. The concentration of a particular antibiotic that does not select for antibiotic resistance in bacteria is termed the predicted no-effect concentration (PNEC). Some PNECs have been identified by Bengtsson-Palme and Larsson (2016) with respective values for AMX, SMX, GM, and CST of 250, 16,000, 1,000, and 2,000 ng/L. Therefore, it is reasonable to question whether concentrations of antibiotics in WWTW would need to be higher than these PNEC values to drive bacteria to develop resistance mechanisms and select for resistant communities in WWTW – a point that requires further analyses. Figures 2C,D shows that the positive controls (without antibiotics) also had increased abundance of bacteria after enrichment in antibiotic-free media, similar to trends observed for abundance of ARB enriched in antibiotic media. This could be expanded to a WWTW setting where the highly labile nutrient source present is the driving factor for bacterial proliferation that selects for organisms with higher growth rates and opportunists, which multiply rapidly and produce many progenies that are less adapted for their particular environment opposed to extensive proliferation of organisms with ARG when exposed to sub-inhibitory concentrations of antibiotics (Vadstein et al., 2018). A suppressive pressure on whole-community growth rate (OD600) for AMX, SMX, and GM antibiotic enrichments in comparison to the POS MHB control was observed (Figure 2). In contrast, the OD600 for POS MAC in Figure 2C indicated that exposure to CST resulted in a higher growth rate in influent and RAS microbial communities from WWTW1 compared to the control. This may indicate a highly adaptive community, with lower competitive pressure in these matrices. However, wavelength interference of the acidity indicator in MacConkey media may also play a role in the observed CST results. The complex interactions in wastewater microbial metabolism could influence wavelength transmission in all media types. Thus, culturing was selected as a quantification metric for subsequent analyses.
The percentage of ARGs removed from influent to effluent was significantly higher in WWTW2 for mcr3, sul1, and sul2 (Figure 1). It was surprising that no blaOXA−48 genes were identified despite successful amplification of the lab-synthesized blaOXA−48 control gene, as it has been previously identified in various countries, in multiple plasmid types, and in SA environments (Bonomo, 2017; Ebomah and Okoh, 2020; Gibbon et al., 2021). Differences in capacity, sources of influent, and type of effluent treatment strategies between the two WWTW may contribute to the variation in ARB and ARG abundance and the dominant species observed between WWTW. The first WWTW uses a mixed flow approach for tertiary treatment, has a 200 Ml/day capacity, and was sampled upstream of the maturation pond. Due to the layout of this WWTW, a sample point downstream of the maturation pond was inaccessible. In contrast, WWTW2 uses a plug flow system for tertiary treatment, has a 38 Ml/day capacity, and was sampled downstream of the maturation pond. In a mixed flow system, contaminants such as antibiotics, ARG, and ARB, are immediately distributed throughout the system. This reduces the concentrations of these contaminants and promotes persistence and proliferation. In contrast, in a plug flow there is less mixing, and contaminants in treated water are thus “pushed out” of WWTW within its hydraulic retention time as new contaminants enter (Rumbaugh, 2014). Due to the fact that samples were collected after the maturation pond in WWTW2, aggregates of suspended solids may have settled in the maturation ponds, and may not be present in the collected sample, further reducing the ARG and ARB seen in the effluent of WWTW2 (Figures 1, 3). This study did not sample other parts of wastewater treatment such as solid waste prior to grit screening or waste sludge. Thus, ARG and ARB may still enter the environment including rivers, the ocean, and sediments through different avenues despite seeming reduced in liquid wastewater effluent, as suggested by Sabri et al. (2020). Through this dissemination, other organisms such as bacteriophages and other vectors could acquire these genes, increasing the spread of resistance (Czekalski et al., 2012; Fouz et al., 2020; Wang et al., 2021; Zare et al., 2021). However, the amount of ARB in sludge has also been found to reduce significantly after anaerobic digestion due to the heat generated during the process that kills bacteria (Zhao and Liu, 2019).
The most prevalent species identified from a randomly isolated footprint of colonies, M. morganii (Figure 4), is intrinsically (innately) resistant to AMX and CST (Liu et al., 2016), both of which were used as a selective pressure in this study and could explain why this organism was the most prominent in the samples. Several other identified genera (Aeromonas, Escherichia, Citrobacter, and Providencia, etc.) in Figure 4 are commensal bacteria in humans and/or animals and are frequently found in WWTW, regardless of the treatment strategy (Lee et al., 2009; Janda and Abbott, 2010; Abbott, 2011; Liu et al., 2016, 2017; Bassetti et al., 2018). This is evident in Figure 5 where these commensal genera are present in both WWTW1 and WWTW2. These organisms can be present in different environments and are not exclusive to the human gut, however, WWTW are receivers of human (and other) waste, and the fact that antibiotic resistant commensal organisms are dominant in these environments suggests that humans and animals might be a source of entry of ARB into WWTW and subsequently play a major role in the emergence of AMR in the environment. This is further supported by the increased abundance of ARB in WWTW samples after antibiotic exposure (Figures 2, 3C,D). Higher concentrations of antibiotics used in treating a patient or animal in a clinical or agricultural setting (or using them as supplements in animal feed) applies a higher selection pressure within the microbiome which has been shown to increase antibiotic resistance at a faster rate than lower antibiotic concentrations (Oz et al., 2014; Dafale et al., 2020). As a result, selection of intrinsically resistant pathogens and resistant microbial populations are more likely to occur in these microbial communities which may be excreted and enter WWTW in addition to surface waters. These organisms that persist in the WWTW effluent may have detrimental implications downstream of the WWTW. For example, the sprinkler systems used for irrigation have been found to play a role in the spread of aerosolized pathogens (Adegoke et al., 2018) with several bacterial and viral outbreaks being reported due to crops being contaminated after irrigation with treated wastewater (Figueras Salvat and Ashbolt, 2019). These incidences reinforce the potential of antibiotic resistant organisms to disseminate into the environment and through the food chain if they are present in WWTW discharge.
The mode MICs for pure cultures for CST, AMX, SMX, and GM (2,048, 16,384, 4,096, and 16 μg/ml, respectively) (Figure 7) are 256, 2,048, 8 times, and equal to the respective EUCAST MIC resistant breakpoint values, indicating that the microbes in these WWTW, as a collective, are resistant to all the antibiotics tested in this study (European Committee on Antimicrobial Susceptibility Testing, 2017; Clinical Laboratory Standards Institute., 2018). High MICs for bacteria isolated from wastewater have also been reported in numerous studies (Ovejero et al., 2017; Obayiuwana et al., 2018). Mode MICs for individual isolates (Figure 7) were found to be higher than the MICs for the whole community wastewater samples for AMX and CST (Figure 6). MICs for SMX were similar between whole community and pure cultures, and MICs for GM were higher in whole communities. Increased prevalence of β-lactamase-producing bacteria has led to AMX having poor activity against Enterobacteriaceae and Pseudomonas spp. when used without a β-lactamase inhibitor (European Committee on Antimicrobial Susceptibility Testing., 2010; González-Bello et al., 2020). However, referring to the previous example of resistance emergence occurring in the gut microbiome of animals and humans, individual members or a sub-group of these microbial communities that enter the environment through excretion may develop higher MICs after being exposed to higher concentrations in the gut, compared to organisms in the environment exposed to trace antibiotic concentrations that are lower than expected PNEC values (Rolain, 2013; Bengtsson-Palme and Larsson, 2016; Booth et al., 2020) that could be diluted further by rainfall and stormwater before entering WWTW. The lack of a significant change in resistant gene copy number in various isolates from WWTW, after sub-inhibitory antibiotic exposure for 18–24 h, emphasizes this as well; however, it needs to be acknowledged that these results are indicative of the ARG copy numbers in bacterial isolates, and the presence of some ARGs in bacteriophages as a result of HGT are not represented here (Figure 8). It has been shown previously that exposure of E. coli isolates to sub-MIC concentrations of various antibiotics result in increased MICs within 21 days of exposure through serial passage and that organisms exposed to higher concentrations (0.5 × MIC) had a higher occurrence of cross-resistance compared to isolates exposed to lower concentrations (1/8 × MIC). As a result, whole communities that dominate regions of WWTW for biological treatment may have varying proportions of species intrinsically resistant to certain antibiotics. The abundance of these species may be lower and as a result the MICs may appear to be lower than single isolates that have the necessary resistance mechanisms. This may explain the trends seen for the whole community and pure culture MICs in Figures 6, 7. Thus, curbing AMR in the environment cannot be addressed by confronting WWTW alone and needs to be stewarded carefully from the point of usage. The wide range of antibiotic reactivities, resistance genes, and mechanisms that would respond differently to various treatments lead to a range of AMR issues. Potential outbreaks in humans or animals with organisms identified in this study would be untreatable with the target antibiotics due to the MIC values seen in Figure 7.
While the problem areas within the wastewater treatment process in terms of AMR should be identified, and alternative technologies that promote more effective removal of all harmful contaminants should be implemented, it is important to recall that WWTW are the receiving bodies rather than the original source of antibiotics and ARB that carry ARG. In addition to the entry of ARB, other industrial chemicals may contribute to AMR. Chlorine treatment has been found to increase the permeability of bacterial cells, allowing increased horizontal gene transfer (HGT) of ARGs between donor and recipient cells (Guo et al., 2015; Wu et al., 2022). The entry of chlorine and other disinfectants used for cleaning hospitals, abattoirs, swimming pools, etc., into WWTW may increase HGT in WWTW with a dense microbial load, such as activated sludge, or promote gene transfer at the original source of contamination. In addition, extracellular ARGs have been found to adsorb to microplastics which are ubiquitous environmental contaminants and would play a role in AMR dissemination in the environment outside of a WWTW (Ross et al., 2021; Cheng et al., 2022).
It is evident that antimicrobial resistant (AMR) microorganisms are abundant in WWTW regardless of the stage of the treatment process. While ARB and ARG were present in chlorinated effluent, the numbers were reduced compared to the influent, suggesting that WWTW assist in reducing dissemination of AMR in the environment as opposed to contributing to it. It appears plausible that the sub-MIC exposure in WWTW had little effect on the emergence of ARB and ARG. As a result, a more proactive solution in tackling AMR would be to mitigate the entry of these ARB and ARG into the WWTW altogether and to encourage the development of antibiotics that are metabolized more readily to prevent excretion of the parent compound and active metabolites into WWTW. In addition, providing access to basic hygiene and sanitation amenities in developing countries, promoting food-animal health, and implementing potential incentives or consequences to encourage adherence to disposal legislation and to reduce the use of antibiotics, where possible, would help combat the issue. Education on AMR and daily routines that play a role in the functioning of the WWTW could aid in the removal efficiency of antimicrobials and other contaminants and promote a proactive mindset in addressing the issue of AMR in the environment.
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author/s.
KT conceptualized and conducted the study, analyzed the results, and wrote the original draft of the manuscript that formed part of a PhD thesis. WS had significant input into the conceptualization, methodology, and analysis of the qPCR work, as well as reviewing and editing the manuscript. GW assisted in analyzing the results and edited the manuscript. MB and EF reviewed and contributed to the manuscript. All authors contributed to the article and approved the submitted version.
KT was supported in part by the National Research Foundation of South Africa (Grant numbers: 118159 and 130527) and the Water Research Commission (Project No. K5/2733). WS was supported by Biogeochemistry Research Infrastructure Platform (BIOGRIP), funded by the Department of Science and Innovation of South Africa.
The study reflects only the authors' views and is independent of the funders. This work was part of a PhD thesis by KT (Tucker, 2021).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The authors wish to thank Edward Archer, Mercia Volschenk, and the City of Cape Town for assisting with and permitting sampling of WWTW, the Central Analytical Facility at Stellenbosch University for DNA sequencing support, and the financial assistance from the NRF, WRC, and BIOGRIP.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/frwa.2022.883282/full#supplementary-material
Abbott, S. L (2011). “Klebsiella, enterobacter, citrobacter, serratia, plesiomonas, and other enterobacteriaceae,” in Manual of Clinical Microbiology, (New York, NY: American Society of Microbiology), 639–657.
Adefisoye, M. A., and Okoh, A. I. (2016). Identification and antimicrobial resistance prevalence of pathogenic Escherichia coli strains from treated wastewater effluents in Eastern Cape, South Africa. Microbiol. open 5, 143–151. doi: 10.1002/mbo3.319
Adegoke, A. A., Amoah, I. D., Stenström, T. A., Verbyla, M. E., and Mihelcic, J. R. (2018). Epidemiological evidence and health risks associated with agricultural reuse of partially treated and untreated wastewater: A review. Frontiers in Public Health (Vol. 6, Issue DEC, p. 337). Frontiers Media S.A. doi: 10.3389/fpubh.2018.00337
Andrews, J (2001). Determination of minimum inhibitory concentrations. J. Antimicrob. Chemother. 48, 5–16. doi: 10.1093/jac/48.suppl_1.5
Archer, E., Petrie, B., Kasprzyk-Hordern, B., and Wolfaardt, G. M. (2017). The fate of pharmaceuticals and personal care products (PPCPs), endocrine disrupting contaminants (EDCs), metabolites and illicit drugs in a WWTW and environmental waters. Chemosphere 174, 437–446. doi: 10.1016/j.chemosphere.2017.01.101
Archer, E., Volschenk, M., Brocker, L., and Wolfaardt, G. M. (2021). A two-year study of emerging micro-pollutants and drugs of abuse in two Western Cape wastewater treatment works (South Africa). Chemosphere 285, 131460. doi: 10.1016/J.CHEMOSPHERE.2021.131460
Bassetti, M., Vena, A., Croxatto, A., Righi, E., and Guery, B. (2018). How to manage Pseudomonas aeruginosa infections. Drugs Context 7, 212527. doi: 10.7573/dic.212527
Bengtsson-Palme, J., and Larsson, D. G. J. (2016). Concentrations of antibiotics predicted to select for resistant bacteria: proposed limits for environmental regulation. Environ. Int. 86, 140–149. doi: 10.1016/j.envint.2015.10.015
Berendonk, T. U., Manaia, C. M., Merlin, C., Fatta-Kassinos, D., Cytryn, E., Walsh, F., et al. (2015). Tackling antibiotic resistance: the environmental framework. Nat. Rev. Microbiol. 13, 310. doi: 10.1038/nrmicro3439
Bonomo, R. A (2017). “β-Lactamases: a focus on current challenges,” in Cold Spring Harbor Perspectives in Medicine (London: Cold Spring Harbor Laboratory Press), p. 284.
Booth, A., Aga, D. S., and Wester, A. L. (2020). Retrospective analysis of the global antibiotic residues that exceed the predicted no effect concentration for antimicrobial resistance in various environmental matrices. Environ. Int. 141, 105796. doi: 10.1016/J.ENVINT.2020.105796
Caicedo, C., Rosenwinkel, K. H., Exner, M., Verstraete, W., Suchenwirth, R., Hartemann, P., et al. (2019). “Legionella occurrence in municipal and industrial wastewater treatment plants and risks of reclaimed wastewater reuse: review,” in Water Research (London: Elsevier Ltd), 21–34.
Cheng, Y., Lu, J., Fu, S., Wang, S., Senehi, N., and Yuan, Q. (2022). Enhanced propagation of intracellular and extracellular antibiotic resistance genes in municipal wastewater by microplastics. Environ. Pollut. 292, 8284. doi: 10.1016/J.ENVPOL.2021.118284
Clinical and Laboratory Standards Institute. (2015). Performance Standards for Antimicrobial Susceptibility Testing. 25th supplement.
Clinical and Laboratory Standards Institute. (2018). M100 Performance Standards for Antimicrobial Susceptibility Testing.
Crouse, J., and Amorese, D. (1987). Ethanol precipitation: ammonium acetate as an alternative to sodium acetate. Focus 9, 3–5.
Czekalski, N., Berthold, T., Caucci, S., Egli, A., and Bürgmann, H. (2012). Increased levels of multiresistant bacteria and resistance genes after wastewater treatment and their dissemination into Lake Geneva, Switzerland. Front. Microbiol. 3, 1–18. doi: 10.3389/fmicb.2012.00106
Dafale, N. A., Srivastava, S., and Purohit, H. J. (2020). Zoonosis: an emerging link to antibiotic resistance under “one health approach.” Indian J. Microbiol. 60, 139–152. doi: 10.1007/s12088-020-00860-z
Duong, H. A., Pham, N. H., Nguyen, H. T., Hoang, T. T., Pham, H. V., Pham, V. C., et al. (2008). Occurrence, fate and antibiotic resistance of fluoroquinolone antibacterials in hospital wastewaters in Hanoi, Vietnam. Chemosphere 72, 968–973. doi: 10.1016/j.chemosphere.2008.03.009
Ebomah, K. E., and Okoh, A. I. (2020). Enterobacter cloacae harbouring bla NDM-1, bla KPC-1, and bla OXA-48-like carbapenem-resistant genes isolated from different environmental sources in South Africa. Int. J. Environ. Stud. 20, 1–14. doi: 10.1080/00207233.2020.1778274
Ekwanzala, M. D., Dewar, J. B., Kamika, I., and Momba, M. N. B. (2018). Systematic review in South Africa reveals antibiotic resistance genes shared between clinical and environmental settings. Infect. Drug Resist. 11, 1907–1920. doi: 10.2147/IDR.S170715
European Committee on Antimicrobial Susceptibility Testing. (2010). Amoxicillin: Rationale for the Clinical Breakpoints. Available online at: http://www.eucast.org (accessed October 7, 2019).
European Committee on Antimicrobial Susceptibility Testing. (2017). Breakpoint Tables for Interpretation of MICs and Zone Diameters. Version 7. Available online at: http://www.eucast.org (accessed November 25, 2019).
Feil, E. J (2016). Enterobacteriaceae: joining the dots with pan-European epidemiology. Lancet Infect. Dis. 17, 118–119. doi: 10.1016/s1473-3099(16)30333-4
Felske, A., Akkermans, A. D., and De Vos, W. M. (1998). Quantification of 16S rDNAs in complex bacterial communities by multiple competitive reverse transcription-PCR in temperature gradient gel electrophoresis fingerprints. Appl. Environ. Microbiol. 64, 4581–4587.
Figueras Salvat, M. J., and Ashbolt, N. (2019). “Aeromonas,” in Global Water Pathogen Project, eds A. Pruden, N. Ashbolt, and J. Miller (London: Michigan State University).
Fito, J., and Van Hulle, S. W. H. (2020). “Wastewater reclamation and reuse potentials in agriculture: towards environmental sustainability,” in Environment, Development and Sustainability (New York, NY: Springer), pp. 1–24.
Fouz, N., Pangesti, K. N. A., Yasir, M., Al-Malki, A. L., Azhar, E. I., Hill-Cawthorne, G. A., et al. (2020). “The contribution of wastewater to the transmission of antimicrobial resistance in the environment: implications of mass gathering settings,” in Tropical Medicine and Infectious Disease.
Gibbon, M. J., Couto, N., David, S., Barden, R., Standerwick, R., Jagadeesan, K., et al. (2021). A high prevalence of blaOXA−48 in Klebsiella (Raoultella) ornithinolytica and related species in hospital wastewater in South West England. Microb. Genom. 7, 509. doi: 10.1099/mgen.0.000509
González-Bello, C., Rodríguez, D., Pernas, M., Rodríguez, Á., and Colchón, E. (2020). β-Lactamase inhibitors to restore the efficacy of antibiotics against superbugs. J. Med. Chemistr. 63, 1859–1881. doi: 10.1021/ACS.JMEDCHEM.9B01279/SUPPL_FILE/JM9B01279_SI_001.PDF
Guo, J., Li, J., Chen, H., Bond, P. L., and Yuan, Z. (2017). Metagenomic analysis reveals wastewater treatment plants as hotspots of antibiotic resistance genes and mobile genetic elements. Water Res. 123, 468–478. doi: 10.1016/j.watres.2017.07.002
Guo, M. T., Yuan, Q., and Bin, Yang, J. (2015). Distinguishing effects of ultraviolet exposure and chlorination on the horizontal transfer of antibiotic resistance genes in municipal wastewater. Environ. Sci. Technol. 49, 5771–5778. doi: 10.1021/acs.est.5b00644
Heuer, H., Focks, A., Lamshöft, M., Smalla, K., Matthies, M., Spiteller, M., et al. (2008). Fate of sulfadiazine administered to pigs and its quantitative effect on the dynamics of bacterial resistance genes in manure and manured soil. Soil Biol. Biochemistr. 40, 1892–1900. doi: 10.1016/j.soilbio.2008.03.014
Heuer, H., and Smalla, K. (2007). Manure and sulfadiazine synergistically increased bacterial antibiotic resistance in soil over at least two months. Environ. Microbiol. 9, 657–666. doi: 10.1111/j.1462-2920.2006.01185.x
Hu, H.-W., Han, X.-M., Shi, X.-Z., Wang, J.-T., Han, L.-L., Chen, D., et al. (2016). Temporal changes of antibiotic-resistance genes and bacterial communities in two contrasting soils treated with cattle manure. FEMS Microbiol. Ecol. 92, 16. doi: 10.1093/femsec/fiv169
Hultman, J., Tamminen, M., Pärnänen, K., Cairns, J., Karkman, A., and Virta, M. (2018). Host range of antibiotic resistance genes in wastewater treatment plant influent and effluent. FEMS Microbiol. Ecol. 94, fiy038–fiy038. doi: 10.1093/femsec/fiy038
Janda, J. M., and Abbott, S. L. (2010). The genus aeromonas: taxonomy, pathogenicity, and infection. Clinic. Microbiol. Rev. 23, 35–73. doi: 10.1128/cmr.00039-09
Jasim, N. A (2020). The design for wastewater treatment plant (WWTP) with GPS X modelling. Cogent Eng. 7, 1. doi: 10.1080/23311916.2020.1723782
Kaplan, J. E., Benson, C., Holmes, K. K., Brooks, J. T., Pau, A., and Masur, H. (2009). Guidelines for prevention and treatment of opportunistic infections in HIV-infected adults and adolescents: recommendations from CDC, the National Institutes of Health, and the HIV Medicine Association of the Infectious Diseases Society of America. MMWR Recomm Rep. 58, 1–207.
Lee, C.-Y., Lee, H.-F., Huang, F.-L., and Chen, P.-Y. (2009). Haemorrhagic bullae associated with a chicken scratch. Annals Tropic. Paediatr. 29, 309–311. doi: 10.1179/027249309X12547917869168
Li, J., Shi, X., Yin, W., Wang, Y., Shen, Z., Ding, S., et al. (2017). A Multiplex SYBR green real-time pcr assay for the detection of three colistin resistance genes from cultured bacteria, feces, and environment samples. Front. Microbiol. 8, 2078. doi: 10.3389/fmicb.2017.02078
Ling, L. L., Schneider, T., Peoples, A. J., Spoering, A. L., Engels, I., Conlon, B. P., et al. (2015). A new antibiotic kills pathogens without detectable resistance. Nature 517, 455–459. doi: 10.1038/nature14098
Liu, H., Zhu, J., Hu, Q., and Rao, X. (2016). Morganella morganii, a non-negligent opportunistic pathogen. Int. J. Infect. Dis. 50, 10–17. doi: 10.1016/j.ijid.2016.07.006
Liu, L., Lan, R., Liu, L., Wang, Y., Zhang, Y., Wang, Y., et al. (2017). Antimicrobial resistance and cytotoxicity of Citrobacter spp. in Maanshan Anhui Province, China. Front. Microbiol. 8, 1357. doi: 10.3389/fmicb.2017.01357
Löffler, D., and Ternes, T. A. (2003). Analytical method for the determination of the aminoglycoside gentamicin in hospital wastewater via liquid chromatography–electrospray-tandem mass spectrometry. J. Chromatograph. A, 1000, 583–588. doi: 10.1016/S0021-9673(03)00059-1
Lupan, I., Carpa, R., Oltean, A., Kelemen, B. S., and Popescu, O. (2017). Release of antibiotic resistant bacteria by a waste treatment plant from Romania. Microbes Environ. 32, 219–225. doi: 10.1264/jsme2.ME17016
National Institute for Communicable Diseases. (n.d.). AMR Surveillance - National (incl Pvt). Available online at: https://mstrweb.nicd.ac.za/MicroStrategy/asp/Main.aspx (accessed March 31 2022)
Obayiuwana, A., Ogunjobi, A., Yang, M., and Ibekwe, M. (2018). Characterization of bacterial communities and their antibiotic resistance profiles in wastewaters obtained from pharmaceutical facilities in Lagos and Ogun states, Nigeria. Int. J. Environ. Res. Public Health 15, 1365. doi: 10.3390/ijerph15071365
Osunmakinde, C. O., Selvarajan, R., Mamba, B. B., and Msagati, T. A. M. (2019). Profiling bacterial diversity and potential pathogens in wastewater treatment plants using high-throughput sequencing analysis. Microorganisms 7, 506. doi: 10.3390/microorganisms7110506
Ovejero, C. M., Delgado-Blas, J. F., Calero-Caceres, W., Muniesa, M., and Gonzalez-Zorn, B. (2017). Spread of mcr-1-carrying Enterobacteriaceae in sewage water from Spain. J. Antimicrob. Chemotherap. 72, 1050–1053. doi: 10.1093/jac/dkw533
Oz, T., Guvenek, A., Yildiz, S., Karaboga, E., Tamer, Y. T., Mumcuyan, N., et al. (2014). Strength of selection pressure is an important parameter contributing to the complexity of antibiotic resistance evolution. Mol. Biol. Evol. 31, 2387–2401. doi: 10.1093/molbev/msu191
Palomo-Briones, R., Ovando-Franco, M., Razo-Flores, E., Celis, L. B., Rangel-Méndez, J. R., Vences-Álvarez, E., et al. (2016). An overview of reclaimed wastewater reuse in gold heap leaching. Mineral Process. Extract. Metallurg. Rev. 37, 274–285. doi: 10.1080/08827508.2016.1190356
Pazda, M., Kumirska, J., Stepnowski, P., and Mulkiewicz, E. (2019). Antibiotic resistance genes identified in wastewater treatment plant systems—a review. Sci. Total Environ. 697, 134023. doi: 10.1016/j.scitotenv.2019.134023
Pérez-Rodríguez, F., and Mercanoglu Taban, B. (2019). A state-of-art review on multi-drug resistant pathogens in foods of animal origin: risk factors and mitigation strategies. Front. Microbiol. 10, 2091. doi: 10.3389/fmicb.2019.02091
Rizzo, L., Manaia, C., Merlin, C., Schwartz, T., Dagot, C., Ploy, M. C., et al. (2013). Urban wastewater treatment plants as hotspots for antibiotic resistant bacteria and genes spread into the environment: a review. Sci. Total Environ. 447, 345–360. doi: 10.1016/j.scitotenv.2013.01.032
Rolain, J. M (2013). “Food and human gut as reservoirs of transferable antibiotic resistance encoding genes,” Front. Microbiol. 4, 173. doi: 10.3389/fmicb.2013.00173
Ross, P. S., Chastain, S., Vassilenko, E., Etemadifar, A., Zimmermann, S., Quesnel, S. A., et al. (2021). Pervasive distribution of polyester fibres in the Arctic Ocean is driven by Atlantic inputs. Nat. Commun. 12, 1–9. doi: 10.1038/s41467-020-20347-1
Rumbaugh, E (2014). Plug Flow, Complete Mix Which is Better? Biological Waste Expert. Available online at: https://www.biologicalwasteexpert.com/blog/plug-flow-complete-mix-which-is-better
Sabri, N. A., van Holst, S., Schmitt, H., van der Zaan, B. M., Gerritsen, H. W., Rijnaarts, H. H. M., et al. (2020). Fate of antibiotics and antibiotic resistance genes during conventional and additional treatment technologies in wastewater treatment plants. Sci. Total Environ. 741, 199. doi: 10.1016/j.scitotenv.2020.140199
Salkind, A. R., and Rao, K. C. (2011). Antibiotic prophylaxis to prevent surgical site infections. Am. Fam. Physic. 83, 585.
Sano, D., Amarasiri, M., Hata, A., Watanabe, T., and Katayama, H. (2016). Risk management of viral infectious diseases in wastewater reclamation and reuse: review. Environ. Int. 91, 220–229. doi: 10.1016/j.envint.2016.03.001
Sekyere, J. O (2016). Current state of resistance to antibiotics of last-resort in South Africa: a review from a public health perspective. Front. Public Health 4, 209. doi: 10.3389/FPUBH.2016.00209
Sharma, N., and Sharma, S. K. (2021). Wastewater treatment plants as emerging source of antibiotic resistance. Green Chemistr. Water Remediat. Res. Applicat. 6, 239–269. doi: 10.1016/B978-0-12-817742-6.00008-6
Singh, R., Paul, D., and Jain, R. K. (2006). Biofilms: implications in bioremediation. Trends Microbiol. 14, 389–397. doi: 10.1016/j.tim.2006.07.001
Stats, S. A (2019). STATISTICAL RELEASE P0302: Mid-Year Population Estimates. Available online at: https://www.statssa.gov.za/publications/P0302/P03022019.pdf
Suzuki, S., Ogo, M., Koike, T., Takada, H., and Newman, B. (2015). Sulfonamide and tetracycline resistance genes in total- and culturable-bacterial assemblages in South African aquatic environments. Front. Microbiol. 6, 1–8. doi: 10.3389/fmicb.2015.00796
Tahrani, L., Van Loco, J., Ben Mansour, H., and Reyns, T. (2016). Occurrence of antibiotics in pharmaceutical industrial wastewater, wastewater treatment plant and sea waters in Tunisia. J. Water Health 14, 208. doi: 10.2166/wh.2015.224
Tucker, K. S (2021). Antibiotic resistance in surface waters and biofilm- response to environmental contaminants [dissertation/doctoral thesis] [Stellenbosch University]. Available online at: https://scholar.sun.ac.za:443/handle/10019.1/123739
Vadstein, O., Attramadal, K. J. K., Bakke, I., and Olsen, Y. (2018). K-selection as microbial community management strategy: a method for improved viability of larvae in aquaculture. Front. Microbiol. 9, 2730. doi: 10.3389/fmicb.2018.02730
van der Zee, A., Roorda, L., Bosman, G., Fluit, A. C., Hermans, M., Smits, P. H. M., et al. (2014). Multi-centre evaluation of real-time multiplex PCR for detection of carbapenemase genes OXA-48, VIM, IMP, NDM and KPC. BMC Infectious Dis. 14, 27. doi: 10.1186/1471-2334-14-27
Visser, W. P (2018). A perfect storm: the ramifications of Cape Town's drought crisis. J. Transdiscipl. Res. Southern Africa 14, 10. doi: 10.4102/td.v14i1.567
Wang, R., Ji, M., Guo, Y., Zhai, H., and Liu, Y. (2021). National high technology research and development program, tianjin city science and technology support program project, Tianjin research program of application foundation and advanced technology view project. Sci. Total Environ. 796, 148919. doi: 10.1016/j.scitotenv.2021.148919
Watkinson, A. J., Murby, E. J., Kolpin, D. W., and Costanzo, S. D. (2009). The occurrence of antibiotics in an urban watershed: from wastewater to drinking water. Sci. Total Environ. 407, 2711–2723. doi: 10.1016/j.scitotenv.2008.11.059
World Health Organisation. (2011). Critically Important Antimicrobials for Human Medicine. WHO. Available online at: internal-pdf://249.38.26.136/WHO Critically Important Antimicrobials.pdf
Wu, Y. H., Wang, Y. H., Xue, S., Chen, Z., Luo, L. W., Bai, Y., et al. (2022). Increased risks of antibiotic resistant genes (ARGs) induced by chlorine disinfection in the reverse osmosis system for potable reuse of reclaimed water. Sci. Total Environ. 815, 152860. doi: 10.1016/J.SCITOTENV.2021.152860
Zare, S., Derakhshandeh, A., Mohammadi, A., and Noshadi, M. (2021). Abundance of antibiotic resistance genes in bacteria and bacteriophages isolated from wastewater in Shiraz. Mol. Biol. Res. Commun. 10, 73. doi: 10.22099/MBRC.2021.39468.1584
Zhang, Y. J., Hu, H. W., Chen, Q. L., Singh, B. K., Yan, H., Chen, D., et al. (2019). Transfer of antibiotic resistance from manure-amended soils to vegetable microbiomes. Environ. Int. 130, 104912. doi: 10.1016/J.ENVINT.2019.104912
Zhao, Q., and Liu, Y. (2019). Is anaerobic digestion a reliable barrier for deactivation of pathogens in biosludge? Sci. Total Environ. 668, 893–902. doi: 10.1016/j.scitotenv.2019.03.063
Keywords: antibiotic resistance genes, colistin, sulfamethoxazole, wastewater effluent, sub-inhibitory antibiotic concentrations, β-lactamases
Citation: Tucker K, Stone W, Botes M, Feil EJ and Wolfaardt GM (2022) Wastewater Treatment Works: A Last Line of Defense for Preventing Antibiotic Resistance Entry Into the Environment. Front. Water 4:883282. doi: 10.3389/frwa.2022.883282
Received: 24 February 2022; Accepted: 08 April 2022;
Published: 16 May 2022.
Edited by:
Sartaj Ahmad Bhat, Gifu University, JapanReviewed by:
Takashi Azuma, Osaka Medical College, JapanCopyright © 2022 Tucker, Stone, Botes, Feil and Wolfaardt. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Keira Tucker, a2VpcmF0dWNrQGdtYWlsLmNvbQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.