
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
METHODS article
Front. Water, 23 May 2022
Sec. Environmental Water Quality
Volume 4 - 2022 | https://doi.org/10.3389/frwa.2022.870453
This article is part of the Research TopicCrowdsourced Understanding of Global River Organic Matter Composition through the Lens of Ecological TheoryView all 5 articles
 Mikayla A. Borton1
Mikayla A. Borton1 Sarah M. Collins2
Sarah M. Collins2 Emily B. Graham1
Emily B. Graham1 Vanessa A. Garayburu-Caruso1
Vanessa A. Garayburu-Caruso1 Amy E. Goldman1
Amy E. Goldman1 Michaela de Melo3
Michaela de Melo3 Lupita Renteria1
Lupita Renteria1 James C. Stegen1* and WHONDRS Crowdsourced Consortium
James C. Stegen1* and WHONDRS Crowdsourced ConsortiumThough community-based scientific approaches are becoming more common, many scientific efforts are conducted by small groups of researchers that together develop a concept, analyze data, and interpret results that ultimately translate into a publication. Here, we present a community effort that breaks these traditional boundaries of the publication process by engaging the scientific community from initial hypothesis generation to final publication. We leverage community-generated data from the Worldwide Hydrobiogeochemistry Observation Network for Dynamic River Systems (WHONDRS) consortium to study organic matter composition through the lens of ecological theory. This community endeavor will use a suite of paired physical and chemical datasets collected from 97 river corridors across the globe. With our first step aimed at ideation, we engaged a community of scientists from over 20 countries and 60 institutions, spanning disciplines and career stages by holding a virtual workshop (April 2021). In the workshop, participants generated content for questions, hypotheses, and proposed analyses based on the WHONDRS dataset. These ideation efforts resulted in several narratives investigating different questions led by different teams, which will be the basis for research articles in a Frontiers in Water collection. Currently, the community is collectively analyzing, interpreting, and synthesizing these data that will result in six crowdsourced articles using a single, existing WHONDRS dataset. The use of a shared dataset across articles not only lowers barriers for broad participation by not requiring generation of new data, but also provides unique opportunities for emergent learning by connecting outcomes across studies. Here we will explain methods used to enable this community endeavor aimed to promote a greater diversity of thinking on river corridor biogeochemistry through crowdsourced science.
River corridors act as an important nutrient and carbon link between terrestrial and aquatic ecosystems and play a key role in global biogeochemical cycling (McClain et al., 2003). Substantial amounts of nutrients and carbon are processed within the sediment and water column components of river corridors, a majority of which is dissolved and complexed with other elements as organic matter (OM) (Findlay and Sinsabaugh, 1999). A key challenge to understanding and predicting river corridor hydrobiogeochemistry is that governing processes are variable across environmental conditions and are sensitive to anthropogenic disturbance (Powers et al., 2016; Gómez-Gener et al., 2020; Juhls et al., 2020; Maavara et al., 2020). Nonetheless, this is a challenge that needs to be addressed because disturbances to river corridors have the potential to alter sustainable energy production, agricultural yields, climate change mitigation strategies, and environmental and human health (Palmer et al., 2009). This highlights the need for a robust understanding of river corridor hydrobiogeochemistry in general and OM chemistry in particular across diverse environmental settings.
The OM pool comprises a complex mixture of organic molecules that are products and drivers of carbon and nutrient cycling in river corridors, thereby influencing water quality and rates of greenhouse gas production and evasion (Tranvik et al., 2009; Yamashita et al., 2011; Tanentzap et al., 2019). For heterotrophic microbes and numerous macroinvertebrate species, OM provides the energy needed to fuel metabolism and the raw materials for biomass production. Various biotic and abiotic processes influence the quantity and composition of OM during transport along the river corridor, playing an essential role in whole ecosystem metabolic processes. However, due, in part, to high OM molecular complexity (thousands or even millions of different molecules), the identification of isomeric structures of each molecule is still far from any instrumental capabilities. There exists no analytical method that can detect all OM components beyond bulk concentrations which contain no information on molecular composition. Further, there is limited understanding of how detailed properties of OM (e.g., thermodynamic traits, diversity of molecular formulae) vary across river corridors and how hydrobiogeochemical processes shape the patterns and distribution of OM. These limitations highlight the need for studies employing high-resolution OM chemistry techniques across large spatial extents and diverse environmental settings.
In river corridors, OM comes from multiple allochthonous sources including terrestrial vegetation, surface runoffs, groundwater discharge, sewage, and also internal primary production (Tank et al., 2010). The relative contributions of these sources vary substantially within and across stream networks, driven by local source distribution and environmental conditions (Mosher et al., 2015; Wang et al., 2019; Casas-Ruiz et al., 2020). Further, the interplay between biotic and abiotic processes influence the quantity and chemistry of OM in river corridors via microbial degradation and biotransformation (Riedel et al., 2016; Kamjunke et al., 2020; Fudyma et al., 2021), riparian root exudation (Wagner et al., 2014), sorption/desorption dynamics with mineral surfaces (Riedel et al., 2012, 2013), as well as photooxidation-driven transformations (Gao and Zepp, 1998; Gonsior et al., 2009; Berg et al., 2019; Wilske et al., 2020). This broad suite of roles, sources, and influential processes and their inter-linked relationships leads to complex mixtures of river corridor OM with a wide range of reactivity that varies through space and time and can have a profound influence on river corridor hydrobiogeochemistry.
The complex and dynamic nature of river corridor OM makes it difficult to characterize, understand, and mechanistically link to biogeochemical functions. Fourier Transform Ion Cyclotron Resonance Mass Spectrometry (FTICR-MS) is one ultra-high resolution approach, among others such as Orbitrap (Hawkes et al., 2016; Simon et al., 2018), that is increasingly used to study OM chemistry in the environment (Stegen et al., 2018; Danczak et al., 2020, 2021). With the highest resolving power and mass accuracy, molecular and compositional information can be obtained from FTICR-MS based on accurate mass without chromatographic separation or fragmentation. Its primary strength is the ability to differentiate (by mass) hundreds of thousands of low concentration organic molecules in environmental samples. FTICR-MS is therefore a powerful tool for untargeted characterization of OM, but it also has limitations (Cooper et al.). Specifically, FTICR-MS is semi-quantitative at best, because not all components of the OM mixture are quantitatively represented in the mass spectrum and there are unknown ionization efficiencies that vary across OM components. The technique also has a specific analytical window, where small molecules (e.g., glucose or molecules beneath 100/150 Da) can go undetected (Cooper et al.). Moreover, some studies have demonstrated limitations in extraction efficiencies and exclusion of semi-polar components (Kong et al., 2021).
Despite its limitations, FTICR-MS has a great potential to characterize OM and answer diverse questions concerning OM dynamics across multiple spatial and temporal scales. The untargeted OM composition data are particularly well-suited to answer questions formulated through an ecological lens due to conceptual parallels between OM assemblages and ecological communities (Danczak et al., 2020). In community ecology, the fundamental data take the form of a species-by-site matrix capable of describing the abundance (or presence/absence) of each biological species in each site and their distribution in a community. This simple matrix is used to address an incredible range of questions in community ecology and is often coupled to diverse metadata (e.g., environmental conditions, spatial relationships, time, and spatiotemporal scales, microbial composition and activity). Similarly, FTICR-MS data can be set up as a species-by-site matrix, but in this case “species” refers to a unique molecular mass observed in the mass spectrometer (Osterholz et al., 2016). Consequently, FTICR-MS data can help understand mechanistic implications of OM chemistry for biogeochemical functions by employing theoretical analysis analogous to ecological concepts concerning biological species. For example, ecological communities are assembled through a combination of organismal birth, death, and dispersal events. In comparison, recent investigations demonstrated that OM is assembled through conceptually analogous processes (Danczak et al., 2020, 2021). That is, OM is a collection of molecules that are assembled through molecular production (analogous to birth), transformation (analogous to death), and transport (analogous to dispersal). Therefore, the “community structure” of OM composition can likely be used to study the underlying hydrobiogeochemical processes regulating the OM assemblage. These conceptual parallels and congruent data structures provide a strong motivation to study OM chemistry by asking questions comparable to those pursued in community ecology.
The parallels between ecology and OM chemistry have been recognized and pursued for decades (Vannote et al., 1980; Sleighter and Hatcher, 2008; Hur et al., 2010; Sleighter et al., 2010; Kellerman et al., 2014; Medeiros et al., 2015; Seidel et al., 2015; Mentges et al., 2017; Valle et al., 2018, 2020; Zark and Dittmar, 2018; Roth et al., 2019; Tanentzap et al., 2019; Merder et al.), though the focus has been primarily on OM in river surface water with less attention to other parts of the river corridor. For example, the river continuum concept represents an early attempt to frame water column OM chemistry through an ecological lens (Vannote et al., 1980), positing that diversity of OM decreases moving down a stream network. There has been some direct support for this hypothesis (Mosher et al., 2015; Kamjunke et al., 2019), but additional work is needed to understand variability in OM diversity gradients. OM chemistry has also been viewed through the lens of functional trait diversity, which is a powerful approach often used in community ecology (McGill et al., 2006; Carmona et al., 2016). For example (Kida et al., 2021), used methods adapted from trait-based ecology Mentges et al. (2017) to reveal downstream gradients in functional trait diversity tied to OM elemental stoichiometries. OM functional traits also underlie the recent extension of community assembly theory to OM chemistry, revealing that more biochemically active compounds have a smaller amount of variance explained by stochasticity relative to biochemically inactive molecules (Danczak et al., 2020). Furthermore, OM chemogeography has been studied as an analog to biogeography (Danczak et al., 2020, 2021; Garayburu-Caruso et al., 2020). Such studies have found both consistency and divergence in OM molecular properties across globally distributed river corridors and this was recently extended to riverbed sediments, in which Garayburu-Caruso et al. (2020) revealed continental-scale shifts in molecular stoichiometries.
Given the numerous conceptual analogs between OM assemblages and ecological communities, here we described an ecologically inspired investigation of OM dynamics. Many applications of ecological topics applied to OM can be explored such as (i) spatial scaling of OM functional diversity, (ii) chemical similarities/differences between “core” molecules found in nearly all river corridors and “satellite” molecules that are rarely observed, (iii) the degree to which OM chemistry is related to co-located surface water and sediments, (iv) variation in the structure of biochemical transformation networks, (v) variation in the sources of observed organic molecules, and (vi) variation in the balance between deterministic and stochastic assembly processes. Below we describe an effort that aims to investigate each of these topics in detail across global watersheds.
To investigate these ecological topics in the context of river OM chemistry, OM data derived from global river surface water and sediments are being examined through an ecological lens using a crowdsourced approach (Uhlmann et al., 2019; Graham and Smith, 2020; Graham et al., 2021). Here we outline methods for data collection and interpretation currently being used to generate a collection of at least six crowdsourced manuscripts investigating river corridor OM using the dataset outlined in Garayburu-Caruso et al. (2020). This dataset was generated as a part of the Worldwide Hydrobiogeochemistry Observation Network for Dynamic River Systems (WHONDRS) (Stegen and Goldman, 2018) by researchers across the globe. A team centered at Pacific Northwest National Laboratory (PNNL) leads WHONDRS, which is a consortium of researchers using an Integrated, Coordinated, Open, and Networked (ICON) science (Goldman et al., 2021) approach to elucidate general principles underlying variation in hydrobiogeochemical function across diverse river corridors. WHONDRS is funded by the U.S. Department of Energy's Environmental System Science program, and it ascribes to the perspective that science advances more rapidly through open data policies, thus data generated by the community belongs to the community. Crowdsourced WHONDRS samples have enabled this collection, which leverages global capabilities from data collection to data interpretation and publication. This is a recipe for major scientific advancement while simultaneously pioneering a new way of doing science designed to benefit all those that choose to engage.
This crowdsourced collection was designed to use ICON principles (Stegen et al., 2019; Goldman et al., 2021) based on intentionally building research efforts that are Integrated across disciplines and scales, Coordinated through use of consistent protocols/methods, Open throughout the research lifecycle, and Networked across stakeholders to achieve mutual benefit (Goldman et al., 2021). For example, writing teams were encouraged to Integrate the FTICR-MS OM chemistry data with other contextual data types such as physical properties of landscapes and biological variables such as vegetation cover. The collection is also Coordinated through the use of a common dataset that all contributions use to ask scientific questions. This allows for “conceptual interoperability” in the sense that outcomes of each analysis can be combined together to draw out knowledge that may be more than the sum of the individual analyses. The collection is globally Open throughout the research lifecycle, using an open dataset (see WHONDRS 2019 dataset overview below) based on crowdsourced samples and allowing anyone interested to join or engage in the process (https://github.com/WHONDRS-Crowdsourced-Manuscript-Effort). The initial ideation workshop was globally open, and all manuscript teams are still open for contributions to conceptual directions, data analysis/interpretation, and manuscript development. In addition, the collection is open for the addition of more teams focused on additional scientific questions. Lastly, the collection is Networked to be mutually beneficial to all that choose to engage. This mutual benefit is achieved through multiple mechanisms for everyone's perspective/voice to be heard and encouraged. For example, if two divergent opinions emerge in the interpretation of certain analyses, both interpretations will be captured and discussed in the associated manuscript. In addition, open sharing of analysis outcomes can be used to mutually inform conceptual interpretations across studies to collectively elevate the scientific impact of all the manuscripts. Furthermore, there is increasing recognition of the need for efforts to accommodate and overcome accessibility challenges (e.g., providing closed captioning, transcripts, and/or translations of workshop discussions). Below we provide additional details on the crowdsourced methods used to employ a global sampling campaign of river corridors and engage the scientific community to develop several manuscripts surrounding the theme of river OM understood through the lens of ecologically inspired concepts and analyses.
Crowdsourced scientific approaches are becoming more common and have been shown to accelerate the progress of science and improve the quality of scientific research (Gilbert et al., 2014; Breuer et al., 2015; Kopf et al., 2015; Leeuw and Boss, 2018; Uhlmann et al., 2019; Graham and Smith, 2020; Landy et al., 2020). These approaches deviate from traditional vertical approaches where science is driven by small groups of specified researchers that together develop a concept, analyze data, and interpret results that ultimately translate into a publication (Uhlmann et al., 2019; Landy et al., 2020). Instead, crowdsourced approaches aim to maximize the use of available resources, democratize who contributes to science, enhance efficiency, and increase transparency by distributing tasks horizontally across large groups of unspecified researchers (Uhlmann et al., 2019). This alternative approach has proven to enable the execution of more ambitious projects that would not be possible under the standard model (Thompson et al., 2017; Goldman et al., 2021; Graham et al., 2021). Beyond collaboration with individuals outside of a particular institution or funded projects, crowdsourcing efforts attempt to maximize the benefit of multiple perspectives by recruiting contributors from across the globe to participate in research on a much larger scale, including participants from multiple institutions, countries, and disciplines.
Crowdsource efforts involve different components of the research process, including those that generate research questions and conceptual models, increase sample collection, or collectively analyze a dataset. For example, crowdsourcing offers a way to increase environmental sampling across the globe using consistent protocols to meet sampling needs that are unattainable by individuals or small teams working in isolation. This method was employed to sample Earth's microbiomes with huge success, collecting and generating microbiome data from more than 20,000 samples (Gilbert et al., 2014; Thompson et al., 2017). Crowdsourcing research in river corridors has also sampled environmental variables including stream flow, soil moisture, and water quality (Breuer et al., 2015; Castilla et al., 2015; Leeuw and Boss, 2018; Sermet et al., 2020). Crowdsourcing at the manuscript stage is less common, but recent efforts have shown that this too leads to impactful outcomes (Graham and Krause, 2020; Graham and Smith, 2020; Goldman et al., 2021; Graham et al., 2021). Here we describe a method that builds on these previous efforts and uses crowdsourced approaches throughout the entire research lifecycle.
In July and August 2019 (Figure 1), the WHONDRS consortium initiated an ICON-based study that aimed to Integrate physical, chemical, and biological properties of global river corridors. After initial conceptualization of the study, several Open discussions were held with the research community to collect feedback on the study design and initial sampling protocol to achieve an approach Coordinated with established methods; identify strengths and weaknesses; recruit samplers; and embody a Networked approach to identify changes that could make the dataset more valuable to individuals, the research community, and other stakeholders. The initial discussion occurred at a town hall during an American Geophysical Union (AGU) conference in December 2018. Follow-on discussions were recorded and uploaded to the WHONDRS YouTube channel (https://www.youtube.com/watch?v=z1AV5MVIWNc) to provide a continued venue for communication. The study approach was modified using the crowdsourced input to add or alter sample types, metadata collection, and analyses. WHONDRS built standardized kits to collect surface water and shallow hyporheic zone sediments. Sampling kits were sent to global researchers (at no cost to them) interested in collecting samples for the study. Garayburu-Caruso et al. (2020) provides details of the kits; briefly, kits included basic sampling equipment such as 0.2 μm filters for water and microbial collection and stainless steel scoops for sediments, along with detailed instructions for sampling, preservation, and return shipping to PNNL in Richland, WA. Samplers across 100 global sites aimed to collect samples within a 6-week period in summer 2019. Ultimately 97 sites were sampled across eight countries following detailed written and video protocols to ensure consistency in collection methods (Garayburu-Caruso et al., 2020; Goldman et al., 2020) (Figure 2).
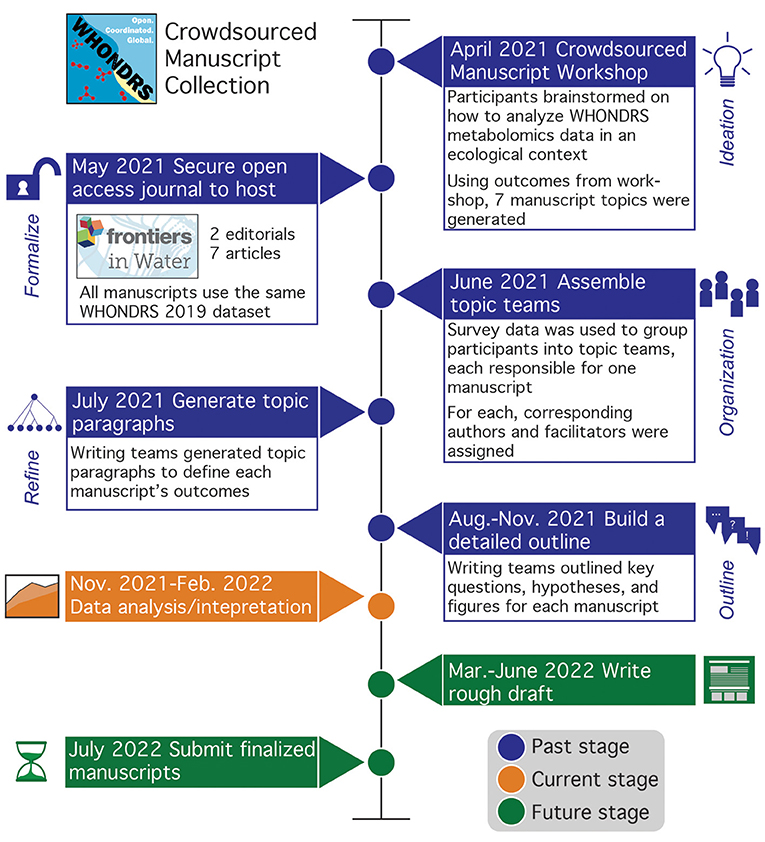
Figure 1. Timeline of crowdsourced manuscript collection, with boxes detailing key phases of the manuscript process. Boxes are colored by past (blue), current (orange), and future (green) to highlight phases and corresponding deliverables that have been completed. To date, the initial workshop, topic paragraphs, and manuscript outlines have been completed with materials posted in the shared GitHub (https://github.com/WHONDRS-Crowdsourced-Manuscript-Effort/Overview). Currently, manuscript teams are analyzing and interpreting data using manuscript outlines as a guide. Next steps include manuscript writing and submission. New participants can join by engaging on the Frontiers manuscript collection page (https://www.frontiersin.org/research-topics/23353/crowdsourced-understanding-of-global-river-organic-matter-composition-through-the-lens-of-ecological) or by emailing WHONDRS@pnnl.gov.
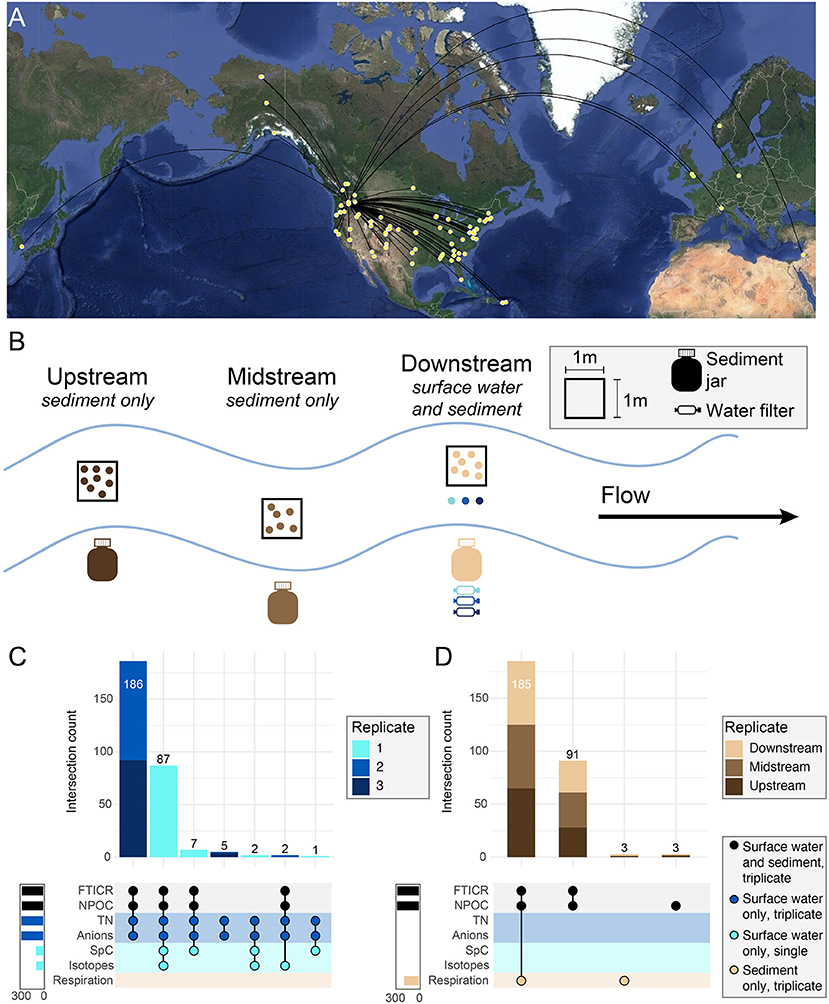
Figure 2. Experimental design overview for WHONDRS S19S including map and types of data available. (A) Points on map show sampling locations for the WHONDRS 2019 dataset. At each location, collaborators selected sampling sites within 100 m of a sensor station that measured river discharge, height, or pressure. (B) Stream diagram represents a single location. Within each sampling location, three depositional zones were identified for sediment collection and labeled as upstream, midstream, or downstream, and were situated within a single stream reach (~10–100 m). Surface water was sampled in triplicate prior to sediment sampling in the most downstream site. Surface water was collected only at the downstream site before collecting the sediments to make sure the water collected was not affected by sediment debris mobilized during water or sediment sampling at upstream locations. Sediments were collected from all three zones. FTICR-MS data were generated from both sediment and surface water and are the basis for this crowdsourced manuscript collection. (C,D) Intersection plots show data type inventory for surface water (C) and sediment (D) samples including the number of samples with overlapping data types (upper stacked bar charts) and total samples within each data type (solid bar charts).
The locations that were sampled were primarily based on where researchers were already working. One goal was to maximize the spatial and environmental extent sampled, but without systematically targeting specific locations. Existing research networks were leveraged where possible, such as the watershed testbed sites funded by the United States (US) Department of Energy and the National Ecological Observation Network funded by the US National Science Foundation. Additional sites were added by individual investigators, often using their existing field sites. Sites in the US were limited to those with existing permits or other access agreements held by the samplers. This requirement was needed to remain in compliance with federal law, but inherently decreased inclusiveness of the sampling campaign. In addition, sampling locations were limited to the Northern Hemisphere to maintain summer conditions that were near base flow across sites. Future efforts may consider alternative approaches that maximize inclusiveness (e.g., both hemispheres) and/or target extreme weather events (e.g., drought, flood) while being compliant with regulations.
Following collection, samples were shipped on a rolling basis as they were collected and processed immediately in the WHONDRS home laboratory at PNNL. A large laboratory team (10 individuals working simultaneously) was needed to efficiently process the kits. Samples were prepared for storage and later analysis or when possible, analyzed immediately. Certain sub-samples were shipped to collaborators who had volunteered to generate additional data types across the 97 sites. Data were published in two data packages on ESS-DIVE (Goldman et al., 2020), and the data packages have been updated with new data as it has become available. Certain data sets are still being processed (e.g., elemental composition of sediment minerals) and will be made public as they become available. The focal data type for this set of crowdsourced manuscripts is detailed characterization of dissolved (water) and water soluble (sediment) OM via FTICR-MS. Garayburu-Caruso et al. (2020), provides details of all protocols/methods, and the data are published on ESS-DIVE (see Data Availability Statement).
To initiate this crowdsourced effort, a virtual workshop was held in April 2021 to generate questions and hypotheses that could be evaluated using the WHONDRS FTICR-MS dataset. The science focus was on applying the ecological concept of core and satellite “species” to OM chemistry of global rivers, and the logistical focus was on how to best engage with participants in a crowdsourcing context. This event was advertised to people using multiple strategies that included email lists, twitter, posting on the ASLO blog (https://www.aslo.org/for-and-by-the-global-science-community/). The recruitment was successful, and we gathered more than 100 subscriptions to the project, including undergraduate, masters and PhD students, postdocs, researchers and professors from 67 institutions and 21 countries. During this workshop (April 30th, 2021), the leadership team provided information about the WHONDRS dataset to be used in the collection and how a crowdsource process works. Subsequently, workshop participants divided themselves into collaborative teams to brainstorm ideas for scientific questions, hypotheses, figures, and analyses; we also discussed how to optimize the use of the crowdsource approach.
The workshop was divided into two blocks: (1) generation of questions and hypotheses for applying the concept of core and satellite species to organic molecules and (2) brainstorm how to effectively use crowdsourcing to elevate a diverse set of voices. In the first block, participants were asked to develop questions, concepts, and hypotheses surrounding the core-satellite concept applied to OM chemistry by brainstorming individually, in small groups, and as a whole group. Subsequently, participants were asked to generate a specific conceptual figure that illustrated their question and hypothesis. Participants worked in shared Google jamboards to capture the workshop content generated. Similar to block 1, block 2 also used Google jamboards to capture content as well as asked participants to brainstorm individually, in small groups, and as a whole group. However, in this block, several topics surrounding the crowdsourced approach were posed including: (1) diversification of participation, (2) ensuring all voices are heard during the crowdsourcing process, (3) continuous engagement of participants, and (4) tracking progress. Participants were encouraged to pick a topic to focus on. All content generated in this workshop by way of Google jamboards has been made available on a shared github repository (https://github.com/WHONDRS-Crowdsourced-Manuscript-Effort).
Following the workshop, the leadership team collated and organized the information contained in the Google jamboards into themes. Initially, the crowdsourced effort was aimed at the core-satellite concept applied to OM chemistry. The goal was to generate a single crowdsourced manuscript about the chemistry of core and satellite organic molecules. However, multiple scientific narratives emerged during the workshop that went beyond the core-satellite concept (see results below). In turn, the leadership team elected to pursue a manuscript collection to allow space for multiple manuscripts generated by different crowdsourced teams pursuing distinct questions.
Participants of the workshop and others that engaged with the effort through asynchronous communication were invited to join the crowdsourced collection by filling out a survey. Each individual filled out a survey indicating their interest in different topics, their ideal timeline for completing benchmarks, and whether they were willing to serve as a corresponding author or facilitator. Corresponding authors were tasked with leading the manuscript development, and facilitators were responsible for coordinating meetings and task completion. Individuals who were interested in corresponding author and facilitator roles based on their areas of interest were contacted by the leadership team to confirm their participation. In the process of identifying corresponding authors, we attempted to encourage a diverse group of individuals from varying career stages, job types, and personal demographics.
Raw data are publicly available in two datasets on the U.S. DOE's data repository ESS-DIVE (https://data.ess-dive.lbl.gov/data): one for surface water, https://data.ess-dive.lbl.gov/view/doi: 10.15485/1603775 (Toyoda et al., 2020) and one for sediment, https://data.ess-dive.lbl.gov/view/doi: 10.15485/1729719 (Goldman et al., 2020). Processed data, including the presence and absence of formula per sample, can be found on the shared github repository (https://github.com/WHONDRS-Crowdsourced-Manuscript-Effort) in the Overview>Data folder. Both datasets include non-purgeable organic carbon (NPOC) and FTICR-MS. The surface water dataset includes anions, total nitrogen, specific conductance, and oxygen and hydrogen stable water isotopes. The sediment dataset includes site photographs and aerobic respiration calculated from laboratory incubations. Data that will be added to the datasets as it is generated and reviewed for quality include sediment bulk carbon-to-nitrogen, water and sediment microbial cell counts, sediment elemental composition, and hydrographs.
The workshop began with discussions around the concept of core-satellite species and developed to span diverse questions and hypotheses all centered around ecological concepts applied to OM. Questions and hypotheses generated by workshop participants spanned topics such as understanding the origin of molecules, biochemical transformations, ecological assembly, species-area scaling, and molecular signatures of underlying processes. The crowdsourced collection is based on manuscripts pursuing each of these themes using the same dataset, which are expanded upon below.
The summary paragraphs (Topics 1–6) and associated figures (see below), which are meant to be a visual representation of the topic to be explored in each manuscript, were developed by crowdsourced participants including corresponding authors, facilitators, and group participants based on outcomes from the initial April 2021 workshop. Following the timeline in Figure 1, each group will create a specific list of questions and outline the sections, analyses, and figures for their manuscript. The leadership team will ensure papers are complementary and do not address overlapping questions. After detailed outlines are finished and have been reviewed by the leadership team, groups will continue to conduct analyses and produce draft manuscripts described below. Groups are working in shared google drive folders and documenting analyses on a communal Github repository (https://github.com/WHONDRS-Crowdsourced-Manuscript-Effort). We emphasize that the collection is not limited to these contributions; additional crowdsourced manuscripts that include analyses of the WHONDRS FTICR-MS dataset are strongly encouraged.
In Summer 2019, the WHONDRS crowdsourced campaign collected surface water and sediment data from 97 rivers corridors. Here, we present the dissolved organic matter (DOM) composition of a wide range of riverine settings measured at ultra-high resolution (FTICR-MS) and discuss how riverine DOM assemblages—complex mixtures of substrates and metabolic products—are organized. To develop a transferable understanding of controls of DOM composition across sites and environments, we utilize the concept of core and satellite species, indicating coherences and similarities in substrates, dominant inputs or accumulated products. We explore various conventional and emergent definitions of core-satellite species and demonstrate their performance on riverine DOM classification. We discuss the patterns in terms of compound groups, lability, molecular size, and watershed characteristics for both surface water and sediments. We anticipate that this community-led effort will enhance our understanding of how organic matter is structured across global riverine systems and identify underlying factors that structure the riverine DOM composition. Figure 3 shows our approach for understanding core and satellite DOM features, where metabolites will be identified as core, satellite, or overlapping. These three pools will then be evaluated for molecular characteristics of interest.
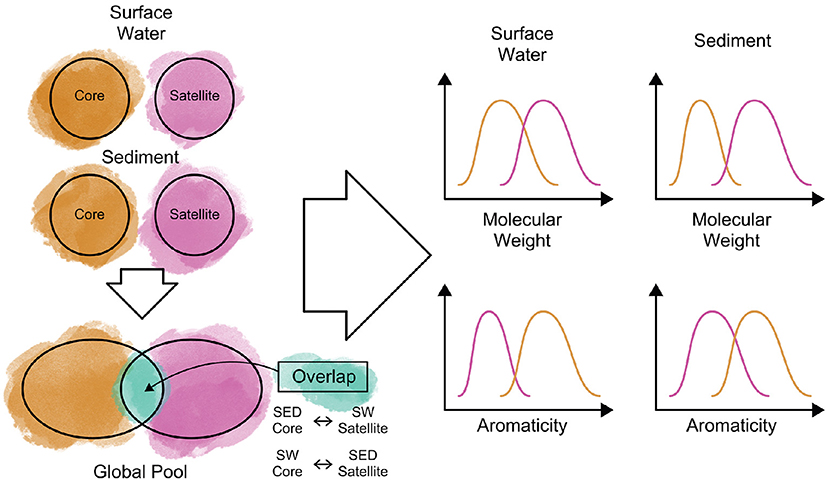
Figure 3. Schematic overview of our approach to identify core (molecular formulae that are broadly distributed across our dataset) and satellite (molecular formulae with a sparser spatial distribution) dissolved organic matter (DOM) in surface water and sediment samples. Metabolites that are identified as core in one system (e.g., sediment) but classified as satellite in the other (e.g., surface water) or vice versa, will be evaluated separately as “overlap” metabolites. Once metabolites are classified into the three pools, we will compare the molecular characteristics among these groupings to evaluate their similarities and differences and discuss potential underlying causes for the observed patterns.
Understanding the origin of organic molecules across ecosystems remains challenging and requires data integration across a wide range of quantitative and qualitative measurements. To identify the patterns of organic matter composition related to its source, requires the analysis of large data sets (>100 s of samples) across vast spatial scales. Here we combine measurements of dissolved organic matter (DOM) composition in surface waters from 97 river corridors worldwide with a variety of hydrological and spatial catchment characteristics to answer the questions: “What molecular composition signatures are associated with different autochthonous and allochthonous sources of organic matter?” and “Under what ecological conditions are these sources active and/or hydrologically connected?” With this approach, we intend to identify both co-occurring event-based processes (e.g., the activation of different DOM sources, compound specific transport and transformation, biological degradation, photochemical degradation) that structure the molecular composition of DOM across large spatial scales and how that information can be used to predict the fate of organic molecules in an environment. Figure 4 shows one mechanism we will use to understand the origin of organic molecules (allochthonous vs. autochthonous sources) through the relation of hydrogen to carbon and oxygen to carbon ratios (H:C and O:C).
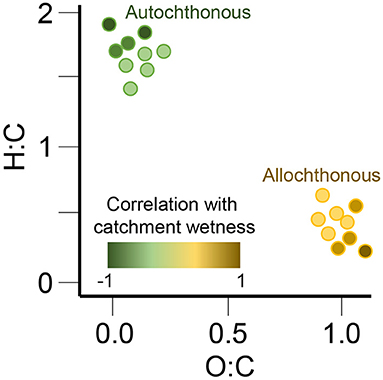
Figure 4. We will relate molecular traits, such as hydrogen to carbon and oxygen to carbon ratios (H:C and O:C), to classifications of the inferred origins of organic compounds, that is, allochthonous vs. autochthonous. These analyses will be informed by the physical, chemical, and biological characteristics of rivers and their catchments, such as the hydrological wetness index shown by the color ramp in this example analysis. Together, these approaches will test the hypothesis that autochthonous and allochthonous compounds have distinct molecular traits and their abundances across sites correlate with different environmental predictors.
Rivers are biogeochemical hotspots, but the environmental and biological factors that govern the transformation of organic matter (OM) flowing through them are still poorly constrained. The analysis of the data from the WHONDRS campaign will allow us to investigate broad continental-scale trends in OM composition and localized phenomena that influence biogeochemical transformations. Two different compartments, sediments and surface water, are analyzed to identify different responses in processes relevant to OM transformations, such as the methylation or oxidation of OM (Van Krevelen and Kendrick mass plots). By using dimensional reduction techniques, we have begun to identify how putative biogeochemical transformations vary across sediment and surface water. We have gained new insight on how spatial (latitude), physical (stream order, slope) and biogeochemical (respiration rates, dissolved organic carbon, anions) factors may influence the reactivity of the OM pool. Figure 5 shows one example how we will analyze watershed characteristics, such as stream order, relative to biogeochemical transformation data.
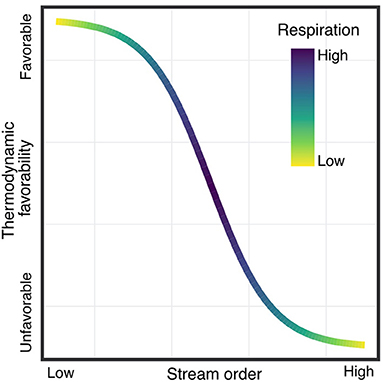
Figure 5. Relationships between stream order and the thermodynamic favorability of metabolic reactions involving Organic Matter (OM) pools. We aim to link watershed characteristics (e.g., stream order, geomorphology, and biological indicators) to biogeochemical reactivity of OM, using a combination of direct measurements (e.g., respiration rates, DOC, etc.) and OM pool compositional features derived from FT-IRCMS data.
Dissolved organic matter (DOM) assemblages in freshwaters are formed from complex mixtures of compounds that are highly variable across time and space due to the environmental heterogeneity of river networks and contribution of diverse allochthonous and autochthonous OM sources. We propose that ecologically significant DOM assemblages can be extracted from a continental-scale dataset of fluvial FTICR-MS organic matter molecular characterization, nutrient levels, stable water isotopes (2H and 18O), and other physicochemical parameters. To identify assemblages and trends, we will apply artificial neural networks and machine learning on DOM samples collected across the continental US. Once DOM assemblages are identified we will use variation across space to investigate metabolomic processes in metacommunity ecology and community metabolomes. We will address questions about the origins of DOM assemblages in streams and rivers. Specifically, we will examine the relative importance of biotic vs. abiotic processes, watershed characteristics, coupled nutrient cycles, and sediment metabolism on DOM assemblage formation. We argue that this data-driven approach will reveal common continental-scale DOM assemblages and the metabolomic processes that generate these groups of compounds. Figure 6 shows how we aim to identify assemblages of organic molecules that comprise DOM and relate them to environmental features using artificial neural networks and other machine learning approaches.
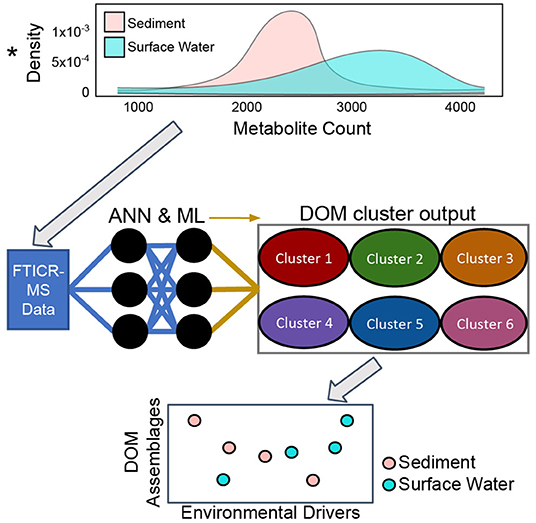
Figure 6. Conceptual overview of Dissolved Organic Matter (DOM) cluster identification from water and sediment Fourier Transform Ion Cyclotron Resonance Mass Spectrometry (FTICR-MS) data with artificial neural networks (ANN) and other machine learning (ML) tools. We aim to identify the environmental drivers, reactivity and chemical characteristics of these DOM assemblages in surface water and sediment samples.
Complex and heterogeneous biogeochemical processes across landscapes introduce a fundamental problem to macro ecological theory—how to scale measured properties to maximize predictive abilities of state-of-the-art process models? Scaling frameworks can include both allometric (rarity scales with intensity) and environmental (rarity scales with the spatial area) scaling properties. To determine universal macroecological patterns associated with organic matter type and distribution, quantitative and qualitative scaling laws for prediction across biomes and latitudes are required. We examine relationships of commonness (as measured by richness and evenness) and rarity (dominance and rarity) using a continental-scale compilation of dissolved organic matter molecular formulae data generated by the WHONDRS Project. Specifically, we look at how commonness/rarity scales with hydrological scaling properties of catchments. We hypothesize that these rates are similar to environmental scaling properties found within each watershed and across latitudes, and that universal patterns exist across allometric and environmental scaling parameters. Taking advantage of the natural hierarchical scales of aggregation found in watershed river corridors across latitudes, we hypothesize that the rarity of molecules will be reduced as watersheds expand, and that solar radiation is a driving factor in rarity/abundance/diversity of compounds from persistent or seasonal degradation mechanisms. We also plan to examine the effects within compound classes (lignin, tannin, etc.) to assess how scaling laws may predict the relative abundance of formulae across chemical groups, mapping roughly onto ecological function across spatial scales. We ask the fundamental question: Are scaling laws universal such that molecular scaling laws show similar behavior and co-variability to spatial scaling laws? Figure 7 depicts our hypothesis for the scaling of OM metrics with watershed characteristics.
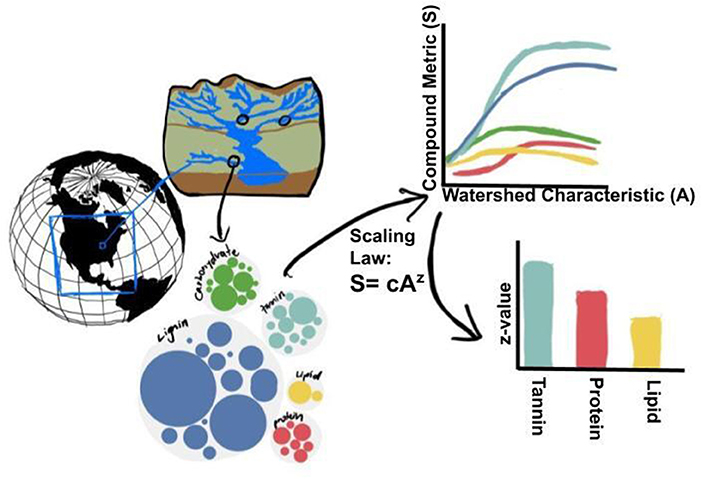
Figure 7. We hypothesize that when complex mixtures of compounds from sediment and stream water are decomposed into compounded classes, the direction and shape at which compound metrics (S, such as: rarity, dominance, compound evenness, and compound richness) scale with watershed characteristics (A, such as: area or stream branching complexity) will be modified. Further, exponents of scaling equations (i.e., z in the generalized scaling equation: S = cAz) should vary among compound classes revealing generalizable patterns that reflect common degradation patterns across global to regional scales including across latitudes, continents, and watersheds.
Organic matter (OM) assemblages in surface water and sediment are determined by complex biogeochemical processes, including those governed by microbes, and thus highly variable in spatiotemporal spaces. This continental-scale, ultrahigh-resolution river metabolomic dataset provides an opportunity to investigate global trends of riverine OM characteristics (e.g., source, mobility, bioavailability, reactivity, parent vs. product compounds) through both data-driven and physics-based modeling. In particular, models applying thermodynamic theory will allow better synthesis and identification of key OM signatures (e.g., thermodynamic favorability, reaction energy efficiency) that control their biogeochemical transformation (e.g., oxidative respiration rate), and can help improve the representation of biogeochemical mechanisms in more integrated models such as reactive transport models. Further, when coupled with existing meta-data of climate, hydrology, geology, and ecology, OM thermodynamics will allow better predictions of biogeochemical and ecosystem dynamics under changing climate. Finally, through data-driven approaches like Sparse Identification of Non-linear Dynamics (SINDy), we will identify key compound sets that control OM thermodynamics and aerobic respiration rates. The results could be analyzed along with microbiome data to better understand microbe-metabolite interactions provided that microbiome data will be collected from the same locations in future investigations. Figure 8 highlights our model-driven approach to identify controls on biogeochemical transformations in river corridors.
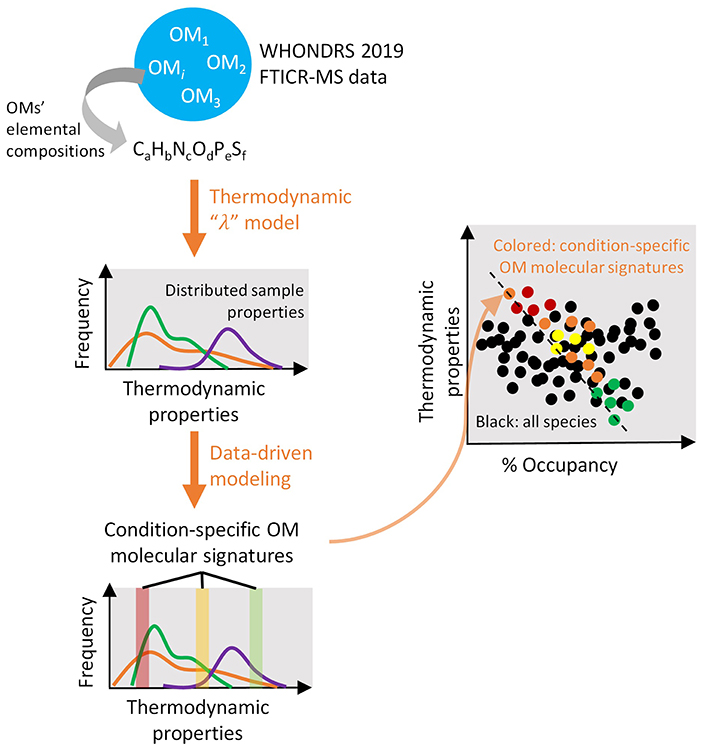
Figure 8. Our model-driven approach employs a thermodynamics-based substrate-explicit “λ” model on ultra-high-resolution Fourier transform ion cyclotron resonance mass spectrometry (FTICR-MS) data of organic molecules to synthesize distributions of thermodynamic properties among samples that represent varying environmental conditions (first orange arrow). Molecular signatures of organic molecules are extracted from thermodynamic property distributions using an advanced data-driven sparse identification of non-linear dynamics (SINDy) framework (second orange arrow). Comparisons of the identified molecular signatures across spatial (surface vs. sediment, along stream orders), climatic (intermittent vs. perennial streams), and ecological (vegetation vs. bare) domains allow a better mechanistic understanding and prediction of organic matter biogeochemistry under a changing environment (third orange arrow). In the last sub-figure, colored dots represent condition-specific clusters of organic matter molecular signatures.
The second block of the workshop held in April focused on effective crowdsourcing strategies, with the intention of gaining feedback on how to ensure all voices are heard, keep participants engaged, and track progress. Feedback gained from the second block of the workshop guided how this manuscript collection has been managed, with key points outlined below.
Participants suggested that dividing people into smaller writing teams would allow inclusion of all perspectives and contributions. Following the workshop, which was originally meant to guide a single manuscript, we realized that the idea of smaller teams along with the number of topics that could be explored in this dataset would allow for several manuscripts which would focus on different aspects. As such, we opted to allow the group of crowdsource participants to self-select into topic teams that would contribute a manuscript to the collection. At present, these topic teams are small enough (~15 participants) that all voices can be heard and have influence over the scientific trajectory of each team's work.
Participants asked for clear deadlines and semi-regular meetings to keep participants engaged. For deadlines, we had participants fill out surveys for the timeline they felt comfortable with for each task (e.g., generating an outline, finalizing analyses, first draft of manuscript). We then set deadlines outlined in Figure 1 based on the average of these time frames. Participants are regularly reminded of deadlines by the leadership team. Further the facilitator assigned to each topic team was tasked with keeping their team on schedule. In regard to semi-regular meetings, topic teams have been meeting regularly on their own schedule. Not only does having smaller groups of participants working toward a common goal enable all voices to be heard, but also enables flexibility with meeting schedules given the smaller number of schedule conflicts and time zones. In addition to individual team meetings, the leadership team is organizing additional workshops for all teams to participate in. The goal is to exchange ideas, summarize outcomes, identify any emergent science opportunities, and gather feedback on practical modifications needed to maximize the mutual benefit of this crowdsourced effort.
Based on feedback from participants, we have opted to keep the manuscripts publicly available. As such, topic teams are operating in google docs and open GitHub repositories. We also use these mechanisms to track progress through each of the deadlines, with key deliverables from each deadline posted in GitHub.
ICON-based science is designed to complement approaches that are based on relatively small teams that pursue their work without direct input or help from the broader science and stakeholder communities. The intentional use of these principles can help increase the utility of generated products (e.g., data, protocols) to those beyond the immediate research team (e.g., regional stakeholders). ICON principles can also be leveraged to address challenges requiring knowledge and data that span a diverse suite of systems while increasing access to scientific resources (e.g., instrumentation, collegial networks, data) for any researcher interested in engaging.
This crowdsourced collection is just one example of how to use ICON principles to bring researchers together to openly pursue mutually beneficial outcomes. There are other ways to use ICON principles in such collections and in primary research projects. Intentionally designing research efforts to use ICON principles can be challenging as it requires a significant amount of coordination, planning, and community engagement to ensure outcomes are beneficial across a diverse suite of stakeholders (e.g., local communities impacted by produced data) without being too focused on any one stakeholder.
The ICON Science Cooperative (https://icon-science.pnnl.gov) was launched to help the science community use and improve ICON science principles. The cooperative's mission is to “to enable researchers from all science domains to implement ICON science in a way that is mutually beneficial across a broad range of stakeholders, thereby accelerating the pursuit of transferable results and enhancing scientific equity.” The cooperative is set up to engage with researchers at any career stage and at any point during the research lifecycle, including in the development of proposals and/or in the modification of existing projects to increase the use of ICON principles. The cooperative can also host visiting scientists to enable mutual learning about how to best implement ICON science across diverse scientific settings. The cooperative aims to use these mechanisms of engagement to achieve its vision: A future world in which researchers across all of science study, improve, and use ICON principles. Future crowdsourced efforts (e.g., sampling, manuscript collections, etc.) can use the ICON Science Cooperative to facilitate their efforts.
Participation in this crowdsourced science effort remains open and we welcome scientists of all levels and disciplines who are excited about ecological principles, OM, and/or river corridor science to join. Participants can propose an additional manuscript that complements the science already being pursued in the collection by submitting an abstract to the collection. Participants can also join an existing team by selecting their topic of interest and begin engaging in the manuscript process by emailing V0hPTkRSU0Bwbm5sLmdvdg==. New team members can be added up until a given manuscript team has finalized their data analyses and moved into the interpretation stage. This limit is imposed to ensure substantial contributions from named authors on each manuscript. Overall, the effort aims to advance our understanding of OM chemistry in river corridors through crowdsourced methods that incorporate diverse perspectives and voices.
The efforts summarized above leverage the power of crowdsourcing to provide resources, opportunities, and knowledge to the scientific community. The conceptual directions of the contributed manuscripts are rich and are broader than they would be if only one pre-existing team analyzed the data and interpreted outcomes. We encourage all interested researchers to join one of the manuscript efforts summarized above and/or initiate an additional, complementary manuscript. Additional manuscripts will also analyze the WHONDRS dataset and follow a crowdsourced approach with an open invitation to the global science community to join. This effort will advance understanding of river corridor OM while helping to develop new ways of doing science based on the use of ICON principles. That is, we consider this collection a foundation to build from, not the final goal, and expect continued innovation in our understanding of OM and around what ICON principles are and how they are used across all of science.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: Raw FTICR-MS data and corresponding metadata for this study and collection can be found in ESS-DIVE under doi: 10.15485/1603775 and doi: 10.15485/1729719. Processed FTICR-MS data, crowdsourced workshop materials, authorship agreements, and manuscript outlines can be accessed via https://github.com/WHONDRS-Crowdsourced-Manuscript-Effort.
• Abdallah, Rehab Z., The American University in Cairo, Egypt
• Ahamed, Firnaaz, University of Nebraska-Lincoln, USA
• Atique, Usman, Chungnam National University, South Korea
• Anak Richard, Florina S., Swinburne University of Technology Sarawak Campus, Malaysia
• Barnard, Malcolm, University of North Carolina at Chapel Hill, USA
• Bice, Kadir, University of Georgia, USA
• Bier, Raven, University of Georgia, USA
• Bladon, Kevin D., Oregon State University, USA
• Bottos, Eric, Thompson Rivers University, Canada
• Brooks, Scott, Oak Ridge National Laboratory, USA
• Burgin, Amy, University of Kansas, USA
• Buser-Young, Jessica, Oregon State University, USA
• Chen, Shuo, Idaho State University; University of Georgia, USA
• Cyle, K. Taylor, North Carolina State University, USA
• D'Andrilli, Juliana, Louisiana Universities Marine Consortium, USA
• Freeman, Erika C., University of Cambridge, UK
• Garayburu-Caruso, Vanessa A., Pacific Northwest National Laboratory, USA
• Garcia, Patricia, INIBIOMA-CONICET, Argentina
• González-Pinzón, Ricardo, University of New Mexico, USA
• Grosbois, Guillaume, Forest research institute université du Québec en Abitibi-Témiscamingue, Canada
• Hosen, Jacob, Purdue University, USA
• Kaplan, Daniel I., Savannah River National Laboratory, USA
• Kominoski, John, Florida International University, USA
• Krause, Stefan, University of Birmingham, UK
• Lee Chang, Miko M., Swinburne University of Technology Sarawak Campus, Malaysia
• Linkhorst, Annika, Federal Institute of Hydrology, Germany
• Lønborg, Christian, Aarhus University, Roskilde, Denmark
• Lu, Yuehan, University of Alabama, USA
• Malard-Adam, Julien, G-EAU/Institut de recherche pour le développement (IRD), Montpellier, France,
• Mateus-Barros, Erick, Federal University of São Carlos (DHb - UFSCar), BRA
• McClure, Emily Ann, Dartmouth College, USA
• Meile, Christof, University of Georgia, USA
• Mendoza-Lera, Clara, University of Koblenz-Landau, Germany
• Mudunuru, Maruti, Pacific Northwest National Laboratory, USA
• Müller, Moritz, Swinburne University of Technology Sarawak Campus, Malaysia
• Newcomer, Michelle, Lawrence Berkeley National Laboratory, USA
• Pena, Jasquelin, University of California Davis, USA
• Plont, Stephen, Virginia Polytechnic Institute and State University, USA
• Raji, Saheed, Federal University of Petroleum Resources Effurun., Nigeria
• Regier, Peter, Pacific Northwest National Laboratory, USA
• Repasch, Marisa, Lawrence Livermore National Laboratory, USA
• Richard, Florina, Swinburne University of Technology Sarawak Campus, Malaysia
• Schrenk, Matthew, Michigan State University, USA
• Shi, Cheng, Oregon State University, USA
• Song, Hyun-Seob, University of Nebraska-Lincoln, USA
• Stadler, Masumi, Université du Québec à Montréal, Canada
• Summers, Betsy, University of New Mexico, USA
• Tanentzap, Andrew J, University of Cambridge, UK
• Veach, Allison, University of Texas at San Antonio, USA
• Ward, Nicholas, Pacific Northwest National Laboratory, USA
• Ward, Christopher, Bowling Green State University, USA
• Wisnoski, Nathan I., University of Wyoming, USA
• Wollheim, Wilfred, University of New Hampshire, USA
• Wymore, Adam, University of New Hampshire, USA
• You, Yaqi, SUNY College of Environmental Science and Forestry, USA
• Zheng, Jianqiu, Pacific Northwest National Laboratory, USA
MB, SC, EG, AG, MM, and JS: conceptualization and methodology. VG-C, AG, LR, and JS: investigation. MB, VG-C, AG, LR, and JS: data curation. MB, SC, EG, MM, and JS: project administration. All authors: writing and data visualization. All authors contributed to the article and approved the submitted version.
This research was supported by the U.S. Department of Energy, Office of Science, Office of Biological and Environmental Research, and Environmental System Science (ESS) Program. This contribution originates from the River Corridor Scientific Focus Area (SFA) project at Pacific Northwest National Laboratory (PNNL). PNNL is operated by Battelle Memorial Institute for the U.S. Department of Energy under Contract No. DE-AC05-76RL01830.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Data used in this manuscript and for future manuscripts in this collection are from the WHONDRS' 2019 crowdsourced global sampling campaign and we thank those that participated in the design and implementation of that effort.
Berg, S. M., Whiting, Q. T., Herrli, J. A., Winkels, R., Wammer, K. H., and Remucal, C. K. (2019). The role of dissolved organic matter composition in determining photochemical reactivity at the molecular level. Environ. Sci. Technol. doi: 10.1021/acs.est.9b03007
Breuer, L., Hiery, N., Kraft, P., Bach, M., Aubert, A. H., and Frede, H.-G. (2015). HydroCrowd: a citizen science snapshot to assess the spatial control of nitrogen solutes in surface waters. Sci. Rep. 5, 16503. doi: 10.1038/srep16503
Carmona, C. P., Bello, F., de Mason, N. W. H., and Lepš, J. (2016). Traits without borders: integrating functional diversity across scales. Trends Ecol. Evol. 31, 382–394. doi: 10.1016/j.tree.2016.02.003
Casas-Ruiz, J. P., Spencer, R. G. M., Guillemette, F., von Schiller, D., Obrador, B., Podgorski, D. C., et al. (2020). Delineating the continuum of dissolved organic matter in temperate river networks. Glob. Biogeochem. Cycles 34, e2019GB006495. doi: 10.1029/2019GB006495
Castilla, E. P., Cunha, D. G. F., Lee, F. W. F., Loiselle, S., Ho, K. C., and Hall, C. (2015). Quantification of phytoplankton bloom dynamics by citizen scientists in urban and peri-urban environments. Environ. Monit. Assess. 187, 690. doi: 10.1007/s10661-015-4912-9
Cooper, W. T., Chanton, J. C., D'Andrilli, J., Hodgkins, S. B., Podgorski, D. C., and Stenson, A. C. A history of molecular level analysis of natural organic matter by FTICR mass spectrometry the paradigm shift in organic geochemistry. Mass Spectrom. Rev.
Danczak, R. E., Chu, R. K., Fansler, S. J., Goldman, A. E., Graham, E. B., Tfaily, M. M., et al. (2020). Using metacommunity ecology to understand environmental metabolomes. Nat. Commun. 11, 6369. doi: 10.1038/s41467-020-19989-y
Danczak, R. E., Goldman, A. E., Chu, R. K., Toyoda, J. G., Garayburu-Caruso, V. A., Tolić, N., et al. (2021). Ecological theory applied to environmental metabolomes reveals compositional divergence despite conserved molecular properties. Sci. Total Environ. 788, 147409. doi: 10.1016/j.scitotenv.2021.147409
Findlay, S., and Sinsabaugh, R. L. (1999). Unravelling the sources and bioavailability of dissolved organic matter in lotic aquatic ecosystems. Mar. Freshw. Res. 50, 781–790. doi: 10.1071/MF99069
Fudyma, J. D., Chu, R. K., Grachet, N., Stegen, J. C., and Tfaily, M. M. (2021). Coupled biotic-abiotic processes control biogeochemical cycling of dissolved organic matter in the columbia river hyporheic zone. Front. Water 2. doi: 10.3389/frwa.2020.574692
Gao, H., and Zepp, R. G. (1998). Factors influencing photoreactions of dissolved organic matter in a coastal river of the southeastern United States. Environ. Sci. Technol. 32, 2940–2946. doi: 10.1021/es9803660
Garayburu-Caruso, V. A., Danczak, R. E., Stegen, J. C., Renteria, L., Mccall, M., Goldman, A. E., et al. (2020). Using community science to reveal the global chemogeography of river metabolomes. Metabolites 10, 518. doi: 10.3390/metabo10120518
Gilbert, J. A., Jansson, J. K., and Knight, R. (2014). The Earth Microbiome project: successes and aspirations. BMC Biol. 12, 69. doi: 10.1186/s12915-014-0069-1
Goldman, A. E., Chu, R. K., Danczak, R. E., Daly, R. A., Fansler, S., Garayburu-Caruso, V. A., et al (2020). WHONDRS Summer 2019 Sampling Campaign: Global River Corridor Sediment FTICR-MS, NPOC, and Aerobic Respiration. Available online at: https://data.ess-dive.lbl.gov/view/doi: 10.15485/1729719 (accessed September 11, 2021).
Goldman, A. E., Emani, S. R., Pérez-Angel, L. C., Rodríguez-Ramos, J. A., and Stegen, J. C. (2021). Integrated, coordinated, open, and networked (ICON) science to advance the geosciences: introduction and synthesis of a special collection of commentary articles. Earth Space Sci. Open Arch. doi: 10.1002/essoar.10508554.1
Gómez-Gener, L., Lupon, A., Laudon, H., and Sponseller, R. A. (2020). Drought alters the biogeochemistry of boreal stream networks. Nat. Commun. 11, 1795. doi: 10.1038/s41467-020-15496-2
Gonsior, M., Peake, B. M., Cooper, W. T., Podgorski, D., D'Andrilli, J., and Cooper, W. J. (2009). Photochemically induced changes in dissolved organic matter identified by ultrahigh resolution fourier transform ion cyclotron resonance mass spectrometry. Environ. Sci. Technol. 43, 698–703. doi: 10.1021/es8022804
Graham, E. B., Averill, C., Bond-Lamberty, B., Knelman, J. E., Krause, S., Peralta, A. L., et al. (2021). Toward a generalizable framework of disturbance ecology through crowdsourced science. Front. Ecol. Evol. 9, 76. doi: 10.3389/fevo.2021.588940
Graham, E. B., and Krause, S. (2020). Social media sows consensus in disturbance ecology. Nature 577, 170–170. doi: 10.1038/d41586-020-00006-7
Graham, E. B., and Smith, A. P. (2020). Crowdsourcing global perspectives in ecology using social media.
Hawkes, J. A., Dittmar, T., Patriarca, C., Tranvik, L., and Bergquist, J. (2016). Evaluation of the orbitrap mass spectrometer for the molecular fingerprinting analysis of natural dissolved organic matter. Anal. Chem. 88, 7698–7704. doi: 10.1021/acs.analchem.6b01624
Hur, M., Yeo, I., Park, E., Kim, Y. H., Yoo, J., Kim, E., et al. (2010). Combination of statistical methods and fourier transform ion cyclotron resonance mass spectrometry for more comprehensive, molecular-level interpretations of petroleum samples. Anal. Chem. 82, 211–218. doi: 10.1021/ac901748c
Juhls, B., Stedmon, C. A., Morgenstern, A., Meyer, H., Hölemann, J., Heim, B., et al. (2020). Identifying drivers of seasonality in lena river biogeochemistry and dissolved organic matter fluxes. Front. Environ. Sci. 8, 53. doi: 10.3389/fenvs.2020.00053
Kamjunke, N., Hertkorn, N., Harir, M., Schmitt-Kopplin, P., Griebler, C., Brauns, M., et al. (2019). Molecular change of dissolved organic matter and patterns of bacterial activity in a stream along a land-use gradient. Water Res. 164, 114919. doi: 10.1016/j.watres.2019.114919
Kamjunke, N., Lechtenfeld, O. J., and Herzsprung, P. (2020). Quality of dissolved organic matter driven by autotrophic and heterotrophic microbial processes in a large river. Water 12, 1577. doi: 10.3390/w12061577
Kellerman, A. M., Dittmar, T., Kothawala, D. N., and Tranvik, L. J. (2014). Chemodiversity of dissolved organic matter in lakes driven by climate and hydrology. Nat. Commun. 5, 3804. doi: 10.1038/ncomms4804
Kida, M., Fujitake, N., Kojima, T., Tanabe, Y., Hayashi, K., Kudoh, S., et al. (2021). Dissolved organic matter processing in Pristine Antarctic streams. Environ. Sci. Technol. 55, 10175–10185. doi: 10.1021/acs.est.1c03163
Kong, X., Jendrossek, T., Ludwichowski, K.-U., Marx, U., and Koch, B. P. (2021). Solid-phase extraction of aquatic organic matter: loading-dependent chemical fractionation and self-assembly. Environ. Sci. Technol. 55, 15495–15504. doi: 10.1021/acs.est.1c04535
Kopf, A., Bicak, M., Kottmann, R., Schnetzer, J., Kostadinov, I., Lehmann, K., et al. (2015). The ocean sampling day consortium. GigaScience 4. doi: 10.1186/s13742-015-0066-5
Landy, J. F., Jia, M. (Liam), Ding, I. L., Viganola, D., Tierney, W., Dreber, A., et al. (2020). Crowdsourcing hypothesis tests: Making transparent how design choices shape research results. Psychol. Bull. 146, 451–479. doi: 10.1037/bul0000220
Leeuw, T., and Boss, E. (2018). The HydroColor app: above water measurements of remote sensing reflectance and turbidity using a smartphone camera. Sensors 18, 256. doi: 10.3390/s18010256
Maavara, T., Chen, Q., Van Meter, K., Brown, L. E., Zhang, J., Ni, J., et al. (2020). River dam impacts on biogeochemical cycling. Nat. Rev. Earth Environ. 1, 103–116. doi: 10.1038/s43017-019-0019-0
McClain, M. E., Boyer, E. W., Dent, C. L., Gergel, S. E., Grimm, N. B., Groffman, P. M., et al. (2003). Biogeochemical hot spots and hot moments at the interface of terrestrial and aquatic ecosystems. Ecosystems 6, 301–312. doi: 10.1007/s10021-003-0161-9
McGill, B. J., Enquist, B. J., Weiher, E., and Westoby, M. (2006). Rebuilding community ecology from functional traits. Trends Ecol. Evol. 21, 178–185. doi: 10.1016/j.tree.2006.02.002
Medeiros, P. M., Seidel, M., Ward, N. D., Carpenter, E. J., Gomes, H. R., Niggemann, J., et al. (2015). Fate of the Amazon River dissolved organic matter in the tropical Atlantic Ocean. Glob. Biogeochem. Cycles 29, 677–690. doi: 10.1002/2015GB005115
Mentges, A., Feenders, C., Seibt, M., Blasius, B., and Dittmar, T. (2017). Functional molecular diversity of marine dissolved organic matter is reduced during degradation. Front. Mar. Sci. 4. doi: 10.3389/fmars.2017.00194
Merder, J., Röder, H., Dittmar, T., Feudel, U., Freund, J. A., Gerdts, G, et al. Dissolved organic compounds with synchronous dynamics share chemical properties origin. Limnol. Oceanogr.
Mosher, J. J., Kaplan, L. A., Podgorski, D. C., McKenna, A. M., and Marshall, A. G. (2015). Longitudinal shifts in dissolved organic matter chemogeography and chemodiversity within headwater streams: a river continuum reprise. Biogeochemistry 124, 371–385. doi: 10.1007/s10533-015-0103-6
Osterholz, H., Singer, G., Wemheuer, B., Daniel, R., Simon, M., Niggemann, J., et al. (2016). Deciphering associations between dissolved organic molecules and bacterial communities in a pelagic marine system. ISME J. 10, 1717–1730. doi: 10.1038/ismej.2015.231
Palmer, M. A., Lettenmaier, D. P., Poff, N. L., Postel, S. L., Richter, B., and Warner, R. (2009). Climate change and river ecosystems: protection and adaptation options. Environ. Manage. 44, 1053–1068. doi: 10.1007/s00267-009-9329-1
Powers, S. M., Bruulsema, T. W., Burt, T. P., Chan, N. I., Elser, J. J., Haygarth, P. M., et al. (2016). Long-term accumulation and transport of anthropogenic phosphorus in three river basins. Nat. Geosci. 9, 353–356. doi: 10.1038/ngeo2693
Riedel, T., Biester, H., and Dittmar, T. (2012). Molecular fractionation of dissolved organic matter with metal salts. Environ. Sci. Technol. 46, 4419–4426. doi: 10.1021/es203901u
Riedel, T., Zak, D., Biester, H., and Dittmar, T. (2013). Iron traps terrestrially derived dissolved organic matter at redox interfaces. Proc. Natl. Acad. Sci. U. S. A. 110, 10101–10105. doi: 10.1073/pnas.1221487110
Riedel, T., Zark, M., Vähätalo, A. V., Niggemann, J., Spencer, R. G. M., Hernes, P. J., et al. (2016). Molecular signatures of biogeochemical transformations in dissolved organic matter from ten world rivers. Front. Earth Sci. 4, 85. doi: 10.3389/feart.2016.00085
Roth, V.-N., Lange, M., Simon, C., Hertkorn, N., Bucher, S., Goodall, T., et al. (2019). Persistence of dissolved organic matter explained by molecular changes during its passage through soil. Nat. Geosci. 12, 755–761. doi: 10.1038/s41561-019-0417-4
Seidel, M., Yager, P. L., Ward, N. D., Carpenter, E. J., Gomes, H. R., Krusche, A. V., et al. (2015). Molecular-level changes of dissolved organic matter along the Amazon River-to-ocean continuum. Mar. Chem. 177, 218–231. doi: 10.1016/j.marchem.2015.06.019
Sermet, Y., Villanueva, P., Sit, M. A., and Demir, I. (2020). Crowdsourced approaches for stage measurements at ungauged locations using smartphones. Hydrol. Sci. J. 65, 813–822. doi: 10.1080/02626667.2019.1659508
Simon, C., Roth, V.-N., Dittmar, T., and Gleixner, G. (2018). Molecular signals of heterogeneous terrestrial environments identified in dissolved organic matter: a comparative analysis of orbitrap and ion cyclotron resonance mass spectrometers. Front. Earth Sci. 6, 138. doi: 10.3389/feart.2018.00138
Sleighter, R. L., and Hatcher, P. G. (2008). Molecular characterization of dissolved organic matter (DOM) along a river to ocean transect of the lower Chesapeake Bay by ultrahigh resolution electrospray ionization Fourier transform ion cyclotron resonance mass spectrometry. Mar. Chem. 110, 140–152. doi: 10.1016/j.marchem.2008.04.008
Sleighter, R. L., Liu, Z., Xue, J., and Hatcher, P. G. (2010). Multivariate statistical approaches for the characterization of dissolved organic matter analyzed by ultrahigh resolution mass spectrometry. Environ. Sci. Technol. 44, 7576–7582. doi: 10.1021/es1002204
Stegen, J., Brodie, E., Wrighton, K., Bayer, P., Lesmes, D., Emani, S., et al. (2019). Open Watershed Science by Design: Leveraging Distributed Research Networks to Understand Watershed Systems: Workshop Report. USDOE Office of Science (SC) (United States).
Stegen, J. C., and Goldman, A. E. (2018). WHONDRS: a community resource for studying dynamic river corridors. mSystems 3. doi: 10.1128/mSystems.00151-18
Stegen, J. C., Johnson, T., Fredrickson, J. K., Wilkins, M. J., Konopka, A. E., Nelson, W. C., et al. (2018). Influences of organic carbon speciation on hyporheic corridor biogeochemistry and microbial ecology. Nat. Commun. 9, 585. doi: 10.1038/s41467-018-02922-9
Tanentzap, A. J., Fitch, A., Orland, C., Emilson, E. J. S., Yakimovich, K. M., Osterholz, H., et al. (2019). Chemical and microbial diversity covary in fresh water to influence ecosystem functioning. Proc. Natl. Acad. Sci. U. S. A. 116, 24689–24695. doi: 10.1073/pnas.1904896116
Tank, J. L., Rosi-Marshall, E. J., Griffiths, N. A., Entrekin, S. A., and Stephen, M. L. (2010). A review of allochthonous organic matter dynamics and metabolism in streams. J. N. Am. Benthol. Soc. 29, 118–146. doi: 10.1899/08-170.1
Thompson, L. R., Sanders, J. G., McDonald, D., Amir, A., Ladau, J., Locey, K. J., et al. (2017). A communal catalogue reveals Earth's multiscale microbial diversity. Nature 551, 457–463. doi: 10.1038/nature24621
Toyoda, J., Goldman, A. E., Chu, R. K., Danczak, R. E., Daly, R. A., Garayburu-Caruso, V. A., et al. (2020). WHONDRS Consortium T (2020): WHONDRS Summer 2019 Sampling Campaign: Global River Corridor Surface Water FTICR-MS, NPOC, and Stable Isotopes.
Tranvik, L. J., Downing, J. A., Cotner, J. B., Loiselle, S. A., Striegl, R. G., Ballatore, T. J., et al. (2009). Lakes and reservoirs as regulators of carbon cycling and climate. Limnol. Oceanogr. 54, 2298–2314. doi: 10.4319/lo.2009.54.6_part_2.2298
Uhlmann, E. L., Ebersole, C. R., Chartier, C. R., Errington, T. M., Kidwell, M. C., Lai, C. K., et al. (2019). Scientific Utopia III: crowdsourcing science. Perspect. Psychol. Sci. 14, 711–733. doi: 10.1177/1745691619850561
Valle, J., Gonsior, M., Harir, M., Enrich-Prast, A., Schmitt-Kopplin, P., Bastviken, D., et al. (2018). Extensive processing of sediment pore water dissolved organic matter during anoxic incubation as observed by high-field mass spectrometry (FTICR-MS). Water Res. 129, 252–263. doi: 10.1016/j.watres.2017.11.015
Valle, J., Harir, M., Gonsior, M., Enrich-Prast, A., Schmitt-Kopplin, P., Bastviken, D., et al. (2020). Molecular differences between water column and sediment pore water SPE-DOM in ten Swedish boreal lakes. Water Res. 170, 115320. doi: 10.1016/j.watres.2019.115320
Vannote, R. L., Minshall, G. W., Cummins, K. W., Sedell, J. R., and Cushing, C. E. (1980). The river continuum concept. Can. J. Fish. Aquat. Sci. 37, 130–137. doi: 10.1139/f80-017
Wagner, K., Bengtsson, M. M., Besemer, K., Sieczko, A., Burns, N. R., Herberg, E. R., et al. (2014). Functional and structural responses of hyporheic biofilms to varying sources of dissolved organic matter. Appl. Environ. Microbiol. 80, 6004–6012. doi: 10.1128/AEM.01128-14
Wang, X., Wu, Y., Bao, H., Gan, S., and Zhang, J. (2019). Sources, transport, and transformation of dissolved organic matter in a large river system: illustrated by the Changjiang River, China. J. Geophys. Res. Biogeosci. 124, 3881–3901. doi: 10.1029/2018JG004986
Wilske, C., Herzsprung, P., Lechtenfeld, O. J., Kamjunke, N., and von Tümpling, W. (2020). Photochemically induced changes of dissolved organic matter in a humic-rich and forested stream. Water 12, 331. doi: 10.3390/w12020331
Yamashita, Y., Kloeppel, B. D., Knoepp, J., Zausen, G. L., and Jaffé, R. (2011). Effects of watershed history on dissolved organic matter characteristics in headwater streams. Ecosystems 14, 1110–1122. doi: 10.1007/s10021-011-9469-z
Keywords: hydrobiogeochemistry, ecological theory, sediments, FTICR-MS, WHONDRS, ICON science, surface water
Citation: Borton MA, Collins SM, Graham EB, Garayburu-Caruso VA, Goldman AE, de Melo M, Renteria L, Stegen JC and WHONDRS Crowdsourced Consortium (2022) It Takes a Village: Using a Crowdsourced Approach to Investigate Organic Matter Composition in Global Rivers Through the Lens of Ecological Theory. Front. Water 4:870453. doi: 10.3389/frwa.2022.870453
Received: 06 February 2022; Accepted: 18 April 2022;
Published: 23 May 2022.
Edited by:
Karsten Rinke, Helmholtz Association of German Research Centres (HZ), GermanyReviewed by:
Peter Herzsprung, Helmholtz Association of German Research Centres (HZ), GermanyCopyright © 2022 Borton, Collins, Graham, Garayburu-Caruso, Goldman, de Melo, Renteria, Stegen and WHONDRS Crowdsourced Consortium. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: James C. Stegen, amFtZXMuc3RlZ2VuQHBubmwuZ292
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.