- 1Department of Life Sciences, Faculty of Natural Sciences, Imperial College London, London, United Kingdom
- 2Department of Clinical, Pharmaceutical & Biological Science, School of Life and Medical Sciences, University of Hertfordshire, Hatfield, United Kingdom
- 3Crop Diseases Research Institute (CDRI), National Agricultural Research Centre, Islamabad, Pakistan
The number of documented viruses that infect fungi has increased during the past few decades. Mycoviruses that infect plant pathogenic fungi are the main focus of mycoviral research since some of them have the capacity to cause hypovirulence to their host and hence function as potential biocontrol agents. This article provides a comprehensive overview of mycoviruses infecting plant pathogenic fungus Rosellinia necatrix causing white root rot, including the prevalence of their occurrence, their taxonomic classification, their genomic organization and structure, impacts on their fungal host in terms of phenotype in general and virulence in particular, and their ecological interactions including transmission. The white root rot fungus is found to harbor diverse mycoviruses with double-stranded and positive-sense single-stranded RNA genomes from different families, including Spinareoviridae, Megabirnaviridae, Partitiviridae, Quadriviridae, Pseudototiviridae, Endornaviridae, Fusariviridae, Yadokariviridae, Hypoviridae, Fusagraviridae and Megatotiviridae. Some of these mycoviruses studied in R. necatrix or a heterologous host Cryphonectria parasitica revealed interesting virus-host interplays and appear to be promising agents for biological control applications against white root rot.
1 Introduction
Fungal viruses or mycoviruses are found in all major groups of fungi. Plant pathogenic fungi provide platforms for identifying such mycoviruses (1–4) and it is evident that mycovirus diversity is much greater than previously thought (5–9). Most mycoviruses lack an extracellular phase in their replication cycle and have exclusively intercellular routes of transmission, such as hyphal anastomosis or sexual and asexual sporulation (10–12).
Some mycoviruses are cryptic, i.e., have no obvious effects on their host, while others are associated with hypovirulence or hypervirulence. Hypovirulence, or debilitated fungal virulence, due to mycovirus infections has been reported for several phytopathogenic fungi including chestnut blight fungus Cryphonectria parasitica (13) and white root rot fungus Rosellinia necatrix (6). This phenomenon has attracted consideration due to the potential of hypovirulent strains of phytopathogenic fungi and their associated mycoviruses in biological control of fungal diseases in agriculture and forestry. Moreover, understanding the molecular mechanisms underpinning the effect of mycovirus infection is useful in developing effective control methods for fungal diseases (2). Hypovirus infection in C. parasitica serves as a model for studying virus/host and virus/virus interactions, and virus-infected R. necatrix also presents a tractable system. Several techniques have been recently developed that facilitate the investigation of virus evolution, replication, transmission and symptomatology in the mycovirus/R. necatrix system (6). In particular, the phenomenon of RNA silencing, which is a fundamental antiviral response in eukaryotic organisms including fungi, has been well studied in R. necatrix. The viral dsRNA is detected and processed by Dicer, an enzyme with endoribonuclease activity, producing fragments 19–22 nt in length and known as viral small (vs) RNAs. The vsRNAs associate with argonaute proteins, activate the RNA-induced silencing complex (RISC) and guide it towards the sequence-specific degradation of target RNA. Encapsidated dsRNA mycoviruses are thought to replicate within their particles, raising the question of whether they are triggers and targets of RNA silencing (14).
R. necatrix is a phytopathogenic soil-inhabiting ascomycete that belongs to class Sordariomycetes; order Xylariales and family Xylariaceae. It is common in temperate, subtropical, and tropical regions of all five continents, and has a worldwide distribution (15). The main ecological factor required by all Rosellinia spp. to flourish is moisture (16), while soil rich in organic matter and acidity contributes towards growth (17, 18). R. necatrix is the causative agent of white root rot disease in a wide range of plant species, including ornamental plants and fruits (19, 20). Infected plants normally show two types of symptoms, on the root system below ground and on the aerial part due to damaged roots (21).
This review focuses on the mycoviruses of R. necatrix, including the prevalence of their occurrence, their taxonomic classification, their genomic organization and structure, impacts on their fungal host in terms of phenotype in general and virulence in particular, and their ecological interactions including transmission. Table 1 summarizes information on known mycoviruses infecting R. necatrix isolates.
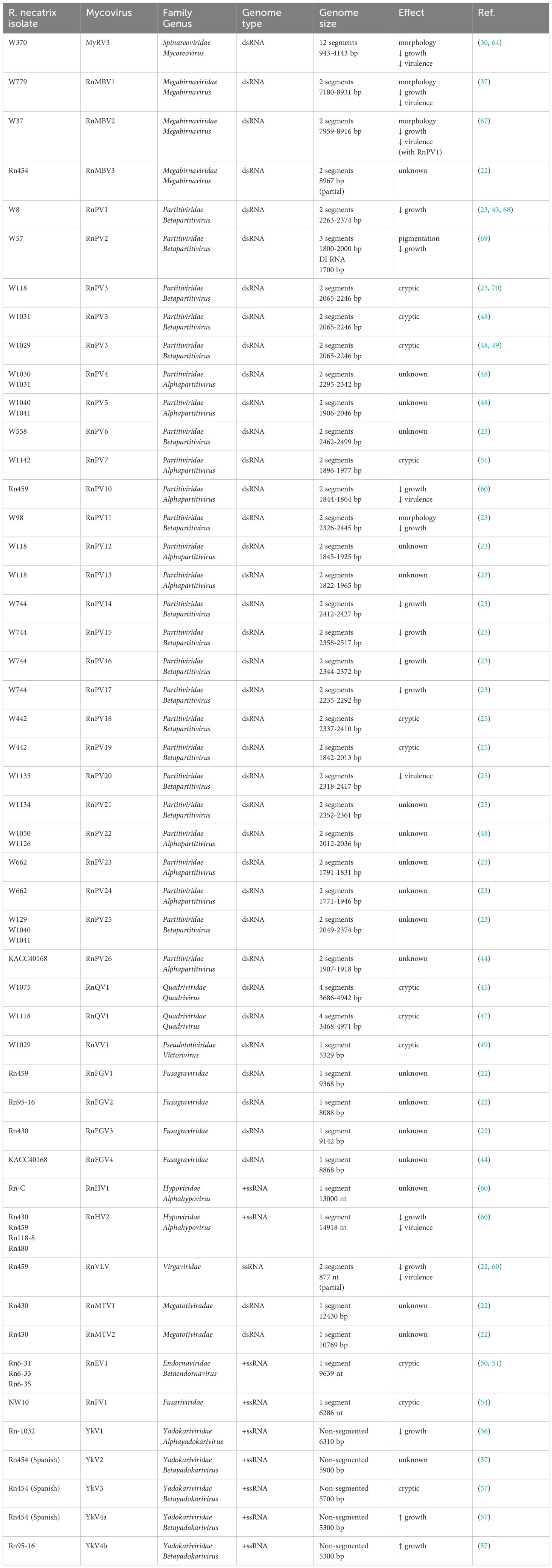
Table 1. Mycoviruses infecting Rosellinia necatrix isolates, their taxonomy, genome type and size, and effects on host.
1.1 Prevalence of mycoviruses in R. necatrix
Several mycoviruses have been reported and characterized in R. necatrix since 2004, through detection of double-stranded (ds) RNA that may be the genome of dsRNA viruses or a replicative form of single-stranded (ss) RNA viruses. The overall incidence of mycoviruses in R. necatrix was determined in a study performed in 2015. A large-scale screening of over 1000 Japanese isolates revealed an overall dsRNA incidence of approximately 20% (3). However, the incidence of dsRNA in R. necatrix was found to be only 14% in a 2018 study, where out of 79 Mediterranean isolates (62 from Spain, 16 from Israel, and 1 from Italy), 11 tested positive for dsRNA (22). Therefore, mycovirus prevalence in R. necatrix differs depending on the sampling location, being approximately 20% in Japan and approximately 14% in the Mediterranean region. Nevertheless, it should be noted that these infection rates are based on investigating potentially the same or duplicate isolates. In an older 2004 study (23), R. necatrix isolates were assigned into mycelial compatibility groups (MCGs) and the detection frequency of dsRNA in each group was determined to prevent duplication. Out of the total of 186 MCGs investigated, 45 were tested positive for dsRNA illustrating a 24.2% infection rate. The frequencies of dsRNA prevalence in MCGs from cultivated and uncultivated lands were significantly different, respectively 24.2% and 11.8%. Other similar studies reported an incidence of mycoviruses in R. necatrix between 14 to 21% (23–25). Most of these viruses are latent but some cause visible phenotypic alterations.
1.2 Spinareoviridae in R. necatrix
Viruses from the family Spinareoviridae are not only found in fungi but are reported to infect mammals, birds, reptiles, fish, arthropods including crustaceans, molluscs and plants. Spinareoviruses form non-enveloped icosahedral particles 60–85 nm in diameter, composed of 1–3 concentric protein layers. Their segmented linear dsRNA genome is between 23 and 29 kbp in size, with each of their 9–12 segments ranging from 0.5 to 4.8 kbp in length (26, 27). The presence of reoviruses in a fungal host, namely C. parasitica, was reported for the first time in 1994 (28). The reoviruses were named mycoreovirus 1 (MyRV1) and 2 (MyRV2), but their genome sequences were not fully determined until 2004 (29). Later, another reovirus designated as Rosellinia necatrix mycoreovirus 3 (RnMRV3) was detected in the R. necatrix hypovirulent strain W370, obtained from a root of Japanese pear in Yamato, Hiroshima, and placed in the genus Mycoreovirus in the family Spinareoviridae (24). The 12 genomic dsRNA segments (S1–S12) of RnMYRV3 range from 943 bp to 4,143 bp in length and have a 3-3-6 electrophoretic profile on 5% (w/v) polyacrylamide gel electrophoresis (PAGE). Figure 1A shows the genome organization of RnMyRV3. All RnMyRV3 segments were genetically unique, each having one long open reading frame (ORF), flanked by 5’ and 3’ untranslated regions (UTRs). The largest segment encodes an RNA-dependent RNA polymerase (RdRP) and the second largest a putative capsid protein (CP). Two more segments encode a guanylyl transferase and an NTP-binding protein, while the rest encode proteins of unknown function. Interestingly, one of the RnMyRV3 segments, namely segment 8 that is absent in C. parasitica MyRV1, was occasionally lost during subculturing of infected fungal strains (30). Segment-specific panhandle structures were found in all segments, which may act as a guiding site for the RdRP. RnMyRV3 particles are 80 nm in diameter, non-enveloped and double-layered, with an inner and an outer capsid layer (31–33). The internal capsid of RnMyRV3 is 50 nm in diameter and has surface projections (turrets or spikes).
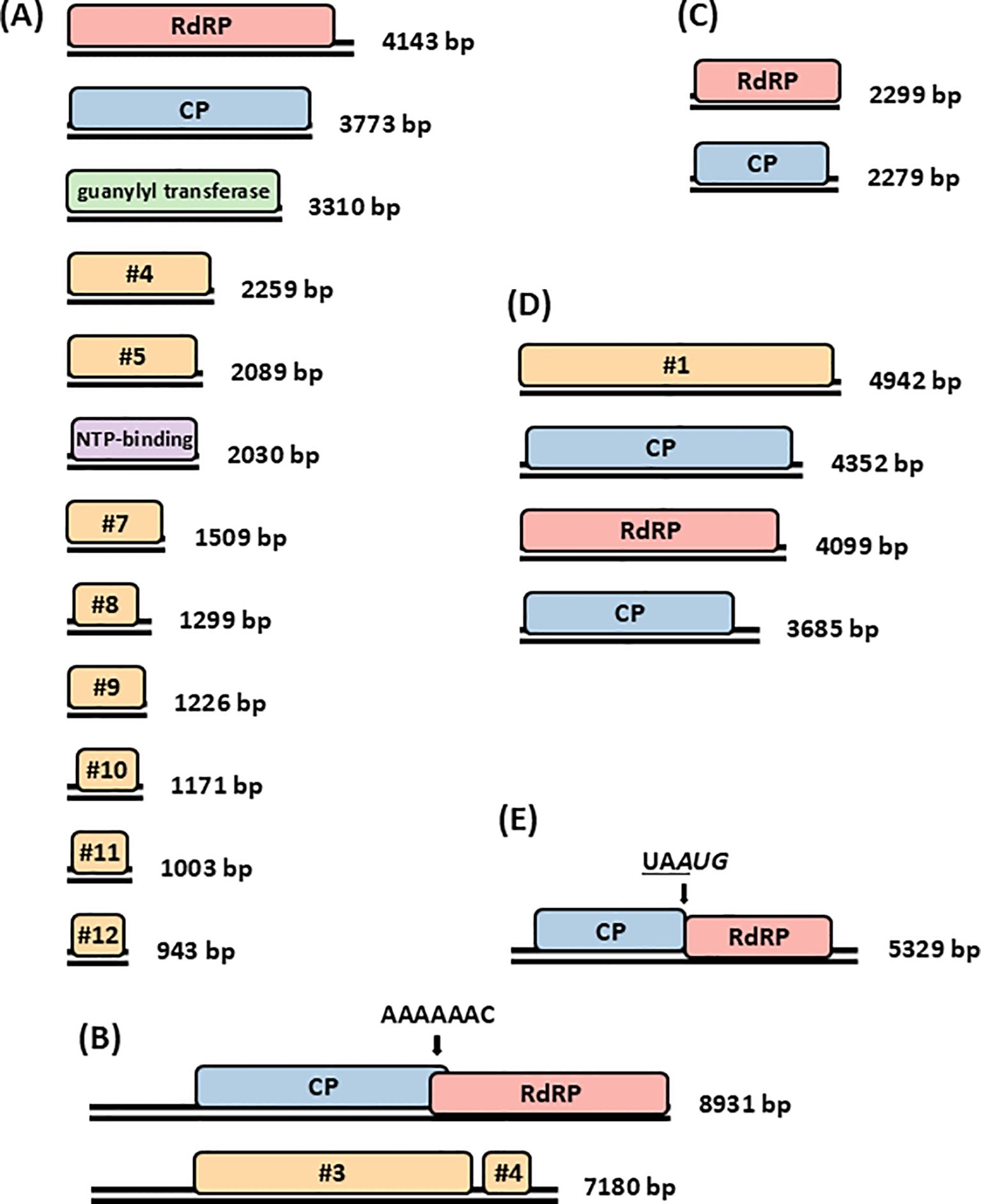
Figure 1. Mycoviruses with dsRNA genomes infecting R. necatrix. Genome organization of (A) dodecasegmented Rosellinia necatrix mycoreovirus 3 (RnMRV3); (B) bisegmented Rosellinia necatrix megabirnavirus 1 (RnMBV1); (C) bisegmented Rosellinia necatrix partitivirus 1 (RnPV1); (D) tetrasegmented Rosellinia necatrix quadrivirus 1 (RnQV1); and (E) monosegmented Rosellinia necatrix victorivirus 1 (RnVV1) W1029. Black double lines represent dsRNAs and coloured boxes represent ORFs. Sequences facilitating ribosomal frameshift in RnMBV1 and RnVV1 are indicated by arrows. RdRP, RNA-dependent RNA polymerase, CP, capsid protein.
The virus-infected R. necatrix hypovirulent strain W370 was cured by hyphal tipping and the resulting virus-free strain recovered its virulence, suggesting that RnMyRV3 attenuates fungal virulence and may be used to control white root rot (30). In another study, RnMyRV3 was also found to reduce mycelial growth rate and induce hypovirulence to the host fungus. When transmitted to R. necatrix strain RT37-1, RnMyRV3 infection resulted in hypovirulence on apple seedlings, with mortality ranging between 0 to 17% for seedlings inoculated with virus-infected strains and 50 to 100% for seedlings inoculated with virus-free strains.
RnMyRV3 led to upregulation of RNA silencing related genes and the abundance of vsRNAs derived from RnMyRV3 was found to be 13.0% (14). The counter-defense strategy of RnMyRV3 against RNA silencing was also investigated in a different study. As indicated by the RnMyRV3 suppressed RNA silencing of green fluorescent protein (GFP), reduced accumulation of GFP-small (s) RNAs and increased accumulation of GFP-dsRNA, observations suggesting that it interferes with dsRNA fragmentation and has RNA silencing suppressor activity (34).
1.3 Megabirnaviridae in R. necatrix
In 2004, a bisegmented dsRNA mycovirus was isolated from R. necatrix strain W779, originally collected from a bait twig buried in a Japanese pear orchard in Ibaraki prefecture (23). The mycovirus was designated as Rosellinia necatrix megabirnavirus 1 (RnMBV1) and a distinct virus family, Megabirnaviridae, was proposed for to accommodate it based on its biological and molecular attributes. Megabirnaviruses form non-enveloped, icosahedral particles, 50-55 nm in diameter. Their bisegmented linear dsRNA genome is approximately 16.1 kbp in size, with two segments ranging from 7.2 to 8.9 kbp in length (35). The word ‘mega’ (‘large’ in Greek, mégas) indicates the relative large genome size, while the word ‘bi’ (‘two’ in Latin) refers to bisegmented nature of the genome, similar to the families Birnaviridae and Picobirnaviridae (6). However, it is noteworthy that these three families, Megabirnaviridae, Birnaviridae and Picobirnaviridae, show different genomic organization with no considerable sequence similarity (6, 36). RnMBV1 has two genomic dsRNA segments 8,931 bp and 7,180 bp in length, as shown in Figure 1B. Both segments are bicistronic, possessing two ORFs each. ORF1 and ORF2 of the largest segment encode for CP and RdRP, respectively. Both segments exhibit substantial levels of sequence similarity in the lengthy 5′ untranslated region of roughly 1.6 kbp, aside from the rigorously conserved 5′ (24 nt) and 3′ (8 nt) terminal sequences. RnMBV1 particles are rigid, spherical and approximately 50 nm in diameter (37).
RnMBV1 is transmitted horizontally via hyphal anastomosis and is homogenously distributed within R. necatrix colonies. RnMBV1 infection results in severe reduction of both R. necatrix growth and virulence against plant hosts, and therefore has strong potential as a biocontrol agent of white root rot (37, 38). The factor(s) responsible for R. necatrix attenuation are unknown but RnMBV1 mutants lacking dsRNA2 result in milder impact on growth and virulence demonstrating the important role of RNA2. An RnMBV1 strain with genome rearrangement was isolated and named RnMBV1-RS1. In addition to dsRNA1 with two open reading frames (ORFs) encoding the CP and the RdRP, RnMBV1-RS1 had a new segment dsRNAS1, which emerged as a result of an ORF1 deletion and an ORF2 partial tandem duplication, retaining a much shorter 5’ untranslated region (UTR). R. necatrix transfected with RnMBV-RS1 virions maintained its virulence on host plants, in contrast to RnMBV1-infected R. necatrix. This suggests that dsRNAS1 is transcribed and packaged, and that dsRNA2, while dispensable for virus replication, is required for virulence reduction in R. necatrix (39).
Transcriptome profiling was performed to investigate gene expression alterations in the Japanese R. necatrix strain W97, which exhibited hypovirulence following transfection with RnMBV1 from strain W779 (37). In total, 545 and 615 genes were found to be up- and down-regulated, respectively, in virus-infected as compared to virus-free R. necatrix. Differential gene expression analysis suggested that primary and secondary metabolism is deregulated in virus-infected R. necatrix. Genes encoding transcriptional regulators, plant cell wall-degrading enzymes, and factors involved in toxin production, such as cytochalasin E, were also differentially expressed (40).
Similar to RnMyRV1, RnMBV1 infection led to upregulation of RNA silencing genes. The abundance of vsRNAs derived from RnMBV1 was found to be 24.9%, higher as compared to RnMyRV1 (section 1.2) and other mycoviruses infecting R. necatrix, including a partitivirus (section 1.4), a quadrivirus (section 1.5), and a victorivirus (section 1.6). Notably, RnMBV1 and RnMyRV1 were the only two mycoviruses examined that upregulated RNA silencing genes in R. necatrix, leading to vsRNA abundance over 10% (14).
1.4 Partitiviridae in R. necatrix
Members of the family Partitiviridae infect fungi and plants. Partitiviruses form non-enveloped, icosahedral particles, 25-43 nm in diameter. Their bisegmented, linear dsRNA genome with two segments ranging from 3.0 to 4.8 kb in length (41, 42). In addition to the families Megabirnaviridae and Reoviridae, members of the family Partitiviridae are also widespread in R. necatrix (6, 25, 43). Many R. necatrix strains tested were co-infected by multiple partitiviruses, but only a few of them have been molecularly and biologically characterized so far.
Rosellinia necatrix partitivirus 1 (RnPV1) was discovered infecting isolate W8, collected from grapevine in Okayama, Japan. Genome organization of RnPV1 is shown in Figure 1C, with the largest segment encoding RdRP and the smallest encoding CP. The UTRs of the two segments were similar with approximately 70% nt identity at the 5’ termini, but different at the 3’ termini, which contained adenosine-uracil rich elements (AREs) (44). Electron microscopy of purified RnPV1 particles indicated that they were approximately 25 nm in diameter. RnPV1 infection did not lead to upregulation of RNA silencing genes and the abundance of vsRNAs derived from RnPV1 was found to be 6.8% (41).
In a separate study, RnPV7 was discovered infecting isolate 7-11/W1142 following a controlled environment experiment (section 1.7) and was assigned to genus Alphapartitivirus. RnPV7 infection had no effect on host growth or virulence.
According to one report (25), a total of 20 partitiviruses from 16 R. necatrix strains, belonging to 5 previously reported species and 15 new species, for which the names Rosellinia necatrix partitivirus 11 to 25 were proposed and which were assigned to genera Alphapartitivirus and Betapartitivirus. For 13 of these novel partitiviruses, transfection experiments were performed to introduce purified virions into reference strains of the natural host R. necatrix and a heterologous host C. parasitica. The effects of partitivirus infection were assessed in R. necatrix W97, together with C. parasitica wild-type EP155 and its Δdcl2 KO mutant, an RNA silencing deficient strain. Initially, the quintuply infected (RnPV1, RnPV14-RnPV17) W97 transfectant displayed considerably reduced growth; its mycelium was fluffy and white in color, while the mycelium of the virus-free strain was off-white and more mat-like in appearance. Subsequent hyphal fusion with virus-free W97 led to the manifestation of a milder growth defect. Even after repeated coculturing, EP155 stably maintained single infection with RnPV11 from W98 or RnPV20 from W1134, and double infection with RnPV14 and RnPV16 from W744. All infected EP155 strains showed reduction pigmentation, growth rate, and aerial hyphae. Δdcl2, when singly infected by RnPV11 or RnPV20 and doubly infected by RnPV14 and RnPV16, also exhibited reduced growth with irregular margins. Conversely, EP155 infected by RnPV18 and RnPV19 was either asymptomatic or showed mild reduction in growth rate (25).
In a recent study of isolates from South Korea, R. necatrix KACC 40168 was reported to be infected with two viruses, one of which was a new member of the genus Alphapartitivirus in the family Partitiviridae and hereafter designated as RnPV26 (44).
1.5 Quadriviridae in R. necatrix
A tetrasegmented dsRNA virus was reported in R. necatrix strain W1075 from a Japanese pear orchard in the Saga prefecture in 2012 (45). Similar to RnMBV1, a new family Quadriviridae was established to accommodate Rosellinia necatrix quadrivirus 1 (RnQV1). Quadriviruses form non-enveloped, icosahedral particles, 45 nm in diameter. Their tetrasegmented linear dsRNA genome is between 16.8 and 17.1 kbp in size, with four segments ranging from 3.5 to 5.0 kbp in length (46). An agarose gel electrophoretic profile of the four genomic dsRNAs of RnQV1 is very much similar to that of a member of the family Chrysoviridae, Helminthosporium victoriae virus 145S (HvV145S), although their size ranges are different: 3.9–4.9 kbp for RnQV1 and 2.8–3.6 kbp for HvV145S (6). All 4 dsRNA segments, i.e., dsRNAs 1 to 4, possess a single large ORF flanked by 5’ and 3’ UTRs. RnQV1 dsRNAs 2 and 4 encode for CPs while dsRNA3 encodes for RdRP as shown in Figure 1D. The infected mycelia were found to contain rigid spherical particles, approximately 45 nm in diameter (45). RnQV1 infection did not lead to upregulation of RNA silencing genes and the abundance of vsRNAs derived from RnQV1 was found to be 1.2% (41).
Later in the same year, the biological and molecular characterization of a second quadrivirus strain termed Rosellinia necatrix quadrivirus 1 strain W1118 (RnQV1-W1118) was reported. Commonalities with the first quadrivirus (RnQV1-W1075) included its spherical particle morphology, quadripartite genome structure, 72–82% sequence similarity between homologous proteins, terminal sequence heterogeneity, and ability to cause a latent infection. Conversely, distinguishing features included different conserved terminal sequences and the degree of susceptibility to proteolytic degradation of the two major capsid proteins, which is thought to occur during virion purification. In particular, the numbers of the strictly conserved sequences are different between the two viruses: 9 vs 7 nt and 12 vs 14 nt for the 5’- and 3’-terminal sequences of RnQV1-W1118 and RnQV1-W1075, respectively. In RnQV1-W1118, the 5’-terminal sequences are more conserved than the 3’-terminal sequences, while the opposite is observed for Rn W1075. Several highly conserved sequence stretches are detected at both ends in addition to the strictly shared sequences (47).
1.6 Pseudototiviridae in R. necatrix
Members of the family Pseudototiviridae infect fungi and protozoa. Pseudototiviruses form non-enveloped, icosahedral particles, 40 nm in diameter, and have monosegmented, linear dsRNA genomes, ranging from 4.6 to 7.0 kbp in length (2023 Release, MSL #39). R. necatrix strains W1028 to W1030 from the experimental orchard of apple trees (Nagano Fruit Tree Experiment Station) at Suzaka, Nagano prefecture, Japan, were confirmed to be coinfected by members of the genus Victorivirus, family Pseudototiviridae, together with partitiviruses (48, 49). Figure 1E shows the genome organization of the monosegmented Rosellinia necatrix victorivirus 1 (RnVV1) with two large overlapping ORFs, encoding CP and RdRP. Similarities and differences to other reported victoriviruses include respectively the presence of a UAAUG pentamer sequence element, facilitating translation termination/reinitiation at the junction between two ORFs and sequence divergence with 41% RdRP amino acid sequence identity to its closest relative, Botryotinia fuckeliana totivirus 1. The RnVV1 particles were spherical, approximately 40 nm in diameter.
Molecular and biological characterization of RnVV1-W1029 has been reported (49). The effects of RnVV1 on the phenotype of the natural host R. necatrix were negligible. The colony morphology of four strains with the same host background was compared: R. necatrix W563 (virus-free field isolate believed to be isogenic to W1029), W1029 (virus-infected), W1029-T20 and W1029-T31 (partially cured via hyphal tipping; RnPV3-free). No effects on appearance or growth rate were observed regardless of whether the host fungus was uninfected (in case of W563) or infected with RnVV1 alone (T20 and T31) or in combination with RnPV3 (W1029). In another experiment, RnVV1 particles were purified from W1029 and used to transfect strain W97, which belongs to a different vegetative compatibility group. Three transfectants (Tf1, Tf2 and Tf3) were compared to parental virus-free W97 strain, concluding that RnVV1 infected R. necatrix asymptomatically under lab conditions.
Further experiments were performed to expand the RnVV1 (and RnPV3) host range and study its phenotype in C. parasitica. Viruses purified from W1029 were used to transfect C. parasitica Δdcl-2 mutant. Three types of transfectants were obtained: A26 doubly infected with RnVV1 and RnPV3; A12, singly infected with RnVV1; and A6, singly infected with RnPV3. In C. parasitica, phenotypic changes were observed in the Δdcl-2 strain singly infected with RnVV1 which showed altered phenotype and reduced growth. Interestingly, comparison of the RNA silencing-competent (wild-type EP155) and -defective (Δdcl-2) strains of C. parasitica infected with RnVV1 showed that RNA silencing acted against the virus to repress its replication, which was restored by coinfection with Cryphonectria parasitica hypovirus 1 (CHV1) or transgenic expression of an RNA silencing suppressor, CHV1 p29. This was the first study reporting host range expansion and RNA silencing of a member of the family Pseudototiviridae (49). Interestingly, RnVV1 infection did not lead to upregulation of RNA silencing genes and the abundance of vsRNAs derived from RnQV1 was found to be 0.3% in R. necatrix (41).
1.7 Endornaviridae in R. necatrix
Members of the family Endornaviridae infect plants, fungi and oomycetes. Endornaviruses are capsidless and have monosegmented, linear, positive-sense (+) ssRNA genomes, ranging from 9.7 to17.6 kb in length. This family has two genera, Alphaendornavirus and Betaendornavirus, which accommodate viruses on the basis of genome size, respectively greater than 11.9 kb and less than 10.7 kb, and fungal host (50). Three R. necatrix isolates, designated as 6-31, 6-33 and 6-35, were reported to be infected with an endornavirus for the first time. Rosellinia necatrix endornavirus 1 (RnEV1) was placed in genus Betaendornavirus because of its relatively short genome of 9,369 nt. A large ORF encoded a protein with a methyltransferase domain, a cysteine rich region, an RNA helicase domain and an RdRP domain, as shown in Figure 2A. RnEV1 infection had no effect on host growth or virulence (50, 51).

Figure 2. Mycoviruses with +ssRNA genomes infecting R. necatrix. Genome organization of (A) monosegmented Rosellinia necatrix endornavirus 1 (RnEV1); (B) monosegmented Rosellinia necatrix fusarivirus 1 (RnFV1), excluding its poly(A) tail; (C) monosegmented Yado-kari virus 1 (YkV1). Black single lines represent ssRNAs and coloured boxes represent ORFs. MET, methyltransferase, CR, cysteine rich region, HEL, RNA helicase, and RdRP, RNA-dependent RNA polymerase.
Interestingly, natural RnEV1 infection occurred when the virus-free R. necatrix strain W97 was incubated in a soil sample from an apple tree in Suzaka under glasshouse conditions, giving rise isolates 6-31, 6-33 and 6-35. A similar phenomenon was observed with the virus-free R. necatrix strain W370T1, which was found to be infected with RnPV7 (section 1.4) and gave rise to isolate 7-11/W1142, following incubation in a soil sample from a forest in Morioka under glasshouse conditions. This observation raises an important question: how did these mycoviruses get transmitted to R. necatrix? No R. necatrix genetically different from the original strains W97 and W370T1 were isolated from soil after incubation, suggesting that these mycoviruses could not have been transmitted from infected R. necatrix already present in the soil samples. Therefore, it was hypothesized that the spread of these mycoviruses may have been facilitated by another fungus or through interactions with soil organisms such as nematodes or micro-arthropods. No horizontal transmission of RnEV1 and RnPV7 between incompatible R. necatrix strains was observed on agar plates during in vitro experiments (50, 51). However, the experiment was performed in soil samples that may have contained plant exudates such as proline. Proline was recently shown to weaken incompatibility between fungi and improve mycovirus transmission (52). This may also help explain mycovirus infection of R. necatrix, potentially through cross-species transmission of mycoviruses.
1.8 Fusariviridae in R. necatrix
Members of the family Fusariviridae infect fungi and oomycetes. Fusariviruses are capsidless and have monosegmented, linear +ssRNA genomes, which range from 5.9 to 10.7 kb in length and may be monocistronic, bicistronic, tricistronic or quadricistronic (53). R. necatrix strain NW10 from an apple tree in Nagano, Japan, was found to be infected Rosellinia necatrix fusarivirus 1 (RnFV1), related to Fusarium graminearum virus 1 (FgV1) and hence placed in the newly established family Fusariviridae. The 6,286 nt genome of RnFV1 has two ORFs, as shown in Figure 2B. The largest encoding a protein with the RdRP and RNA helicase domains typical of fusariviruses. Both of these domains showed moderate level of sequence identity to each other. The smallest ORF encoded a conserved protein with unknown function, which contained a motif from a structural maintenance of chromosomes (SMC) protein. RnFV1 was successfully transferred between incompatible fungal strains using a zinc ion-based technique. RnFV1 caused a latent infection, and therefore has no potential to be used as a biocontrol agent (54).
1.9 Yadokariviridae in R. necatrix
The family Yadokariviridae, which includes the genera Alphayadokarivirus and Betayadokarivirus, accommodates capsidless, non-segmented, linear +ssRNA viruses, 3.3 to 6.3 kb in length that hijack capsids from phylogenetically distant dsRNA viruses, resulting in spherical and non-enveloped particles 33–50 nm in diameter. Yadokariviruses likely replicate within the hijacked heterocapsids using their own RdRP, imitating the replication process of dsRNA viruses. Through their interaction with the capsid-donating dsRNA viruses, yadokariviruses can influence their fungal hosts in both positive and negative ways (55).
Co-infection with the 6.3 kb long alphayadokarivirus Yado-kari virus 1 shown in Figure 2C and its unclassified capsid donor, Yado-nushi virus 1, led to a growth defect in strain Rn-1032 from Japan but also promoted the accumulation of the donor virus (48, 56). Conversely, the betayadokarivirus Yado-kari virus 4a from strain Rn-454, reduced the levels of its capsid donor dsRNA virus and alleviated the growth defect caused by the associated dsRNA virus. Another betayadokarivirus, Yado-kari virus 3 from strain Rn-454, has no impact on either its capsid donor or the host fungus (57).
These findings contribute to our understanding of viral ecology and virus-host interactions in fungal systems. The effects shown by yadokariviruses on their capsid donors and host fungi illustrate how mycoviruses can exhibit a wide range of behaviors, from promoting viral replication to mitigating harmful effects on the host fungus. This variability could be driven by genetic differences between viruses, their ability to take over the host machinery, or the ecological context in which the viruses exist. Furthermore, these results raise interesting questions about the potential for viral interactions to influence fungal disease dynamics, especially in phytopathogenic fungi like R. necatrix.
1.10 Unclassified viruses and mixed infections
A study was performed to screen R. necatrix strains for viral infections and it was reported that different strains of R. necatrix were infected with at least six different viral families. The viral sequences represent families such as Megabirna-, Partiti-, Fusari-, Yado-kariviridae, described in previous sections, but also Hypo-, Fusagra-, and Megatotiviridae. Hypoviruses are capsidless viruses with monosegmented, linear, +ssRNA genomes ranging from 7.3 to 18.3 kb in length and may be monocistronic or bicistronic (58). Conversely, fusagraviruses form non-enveloped, icosahedral capsids approximately 30 nm in diameter, have monosegmented, linear, dsRNA genomes ranging from 8 to 11 kb in length and are bicistronic (59). Finally, megatotiviruses form non-enveloped, spherical particles 40-50 nm in diameter and have monosegmented, linear, dsRNA genomes ranging from 10-12.5 kb in length (Taxon Details | ICTV) (22). The hypovirulent effect of mycovirus infection in the Mediterranean R. necatrix strain Rn459, causing root rot in avocado, was reported (22, 60). Rn459 was found to be infected with RnPV10 (section 1.4) and at least three other viruses: Rosellinia necatrix hypovirus 2 (RnHV2), Rosellinia necatrix fusagravirus 1 (RnFGV1), and Rosellinia necatrix virga-like virus (RnVLV). The latter resembles members of the family Virgaviridae, plant viruses with non-enveloped rod-shaped capsids 17-24 nm in diameter and up to 300 nm in length, and monosegmented, linear, +ssRNA genomes ranging from 6.3 to 13.0 kb in length (61).
Following spontaneous loss of RnFGV1 from the original strain, hyphal tipping was used to cure the cultures of virus infection in order to examine the effects on growth in vitro and virulence on avocado plants. The fungal strain obtained, Rn459_PV10F/VLVF, was confirmed to be cured of RnPV10 and RnVLV infection but still retained RnHV2. Rn459_PV10F/VLVF manifested a phenotype different from the original Rn459 strain, growing faster in vitro and being more virulent on avocado plants. These collective results suggest that RnPV10 and RnVLV, alone or in combination, contribute to confer hypovirulence to R. necatrix.
A mixed dsRNA mycovirus infection in a Korean R. necatrix isolate, including a fusagravirus and a partitivirus was reported (44). The two mycoviruses were detected by next-generation sequencing analysis of purified dsRNAs samples. The first dsRNA virus had a complete genome sequence of 8,868 bp in size and contained two large ORFs 1 and 2, overlapped by 22 bp containing a canonical (−1) slippery heptanucelotide sequence of UUUAAAC. Phylogenetic analysis showed that it clustered with RnFGV3 and other fusagraviruses and showed similarity with the RnFGV3 hypothetical protein and RdRP. The virus was named Rosellinia necatrix fusagravirus 4 due to its genomic organization, sequence similarity, and phylogenetic analysis, which indicate that it belongs to a novel species of the family Fusagraviridae.
2 Conclusion
This review provides a comprehensive summary of viruses infecting different R. necatrix strains. The investigation of mycoviruses that infect Rosellinia necatrix provides important information about the intricate relationships between fungal diseases and related viruses. Mycoviruses have demonstrated the ability to decrease virulence of R. necatrix, opening up new avenues for biocontrol methods to control this harmful plant pathogen. Understanding the molecular mechanics of viral infection and the possibility of horizontal transmission has advanced significantly, but there are still a number of unanswered concerns. Further research is needed to explore the diversity of mycoviruses in natural populations, their precise modes of action, and how they can be effectively used in agricultural settings. Given the increasing challenges posed by R. necatrix in various crops, understanding the role of mycoviruses in modulating fungal virulence and their potential for integrated pest management could pave the way for sustainable and environmentally friendly disease control methods.
Different studies indicate that RnMyRV3 and RnMBV1are two mycoviruses in R. necatrix with biocontrol potential. RnMyRV3 is less stable and unevenly distributed, but causes hypovirulence. Conversely, RnMBV1 is a better biocontrol candidate since it is more stable and efficiently spreads throughout MCGs, therefore it is a better candidate for biocontrol applications (62). RnMBV1 may be used as a specialized biocontrol agent against particular fungal strains in various MCGs, as evidenced by trials employing RnMBV1-infected hypovirulent strains that demonstrated decreased growth in apple roots (37, 63). Unexpected behaviors were also noted by various field researches: following R. necatrix inoculation of apple orchards, the fungi reisolated after two to three years were genetically identical, but they harbored five new viruses that were not present originally, indicating the possibility of spontaneous viral infections (48). Subsequent tests on R. necatrix cultivated in soil verified comparable spontaneous infections by new viruses (51). These findings suggest that mycoviruses may have the ability to naturally infect fungi, potentially across various species in soil ecosystem. Supporting this notion, experiments using protoplasts showed that RnPV1 and RnMyRV3 could infect and replicate in different types of fungi, including C. parasitica and Diaporthe species. These findings suggest that the host range of mycoviruses could be broader, and they may have the potential to infect different fungi in nature (62, 64).
Evidence for horizontal virus transmission within and between fungal species of the family Xylariaceae from avocado was observed (65). Despite vegetative incompatibility among Entoleuca isolates and different MCGs in R. necatrix, both fungal species shared the same set of mycoviruses, suggesting potential horizontal transmission. This is further supported by polymorphisms observed in Entoleuca hypovirus 1 (EnHV1), which was not linked to host origin. Previous research has demonstrated both natural and artificial mycovirus transfection between fungal species, and experimental transmission of a virus between R. necatrix isolates was achieved using zinc ions (64, 66). Interestingly, Entoleuca partitivirus (EnPV) 1 and 2 were detected in a Fusarium sp. isolate from the same avocado orchards, indicating the possible involvement of mycophagous species or other vectors in the transmission of mycoviruses. These findings suggests that virocontrol of R. necatrix in avocado could be a practical approach if a virus capable of inducing hypovirulence is identified. Additionally, the use of high-throughput sequencing technologies will be crucial in determining the complete viral infection status in fungal hosts and exploring virocontrol strategies for fungal diseases, while advancing our understanding of virus evolution and their interactions with fungal and plant hosts. More field research is required to look for factors influencing mycovirus transmission and virulence (65).
Author contributions
SH: Conceptualization, Investigation, Visualization, Writing – original draft, Writing – review & editing. RC: Supervision, Writing – review & editing. AJ: Supervision, Writing – review & editing. IKL: Conceptualization, Visualization, Supervision, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. SH is very grateful to the British Council Women in STEM programme, offering her a fellowship at Imperial College London.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Pearson MN, Beever RE, Boine B, Arthur K. Mycoviruses of filamentous fungi and their relevance to plant pathology. Mol Plant Pathol. (2009) 10:115–28. doi: 10.1111/j.1364-3703.2008.00503.x
2. Xie J, Jiang D. New insights into mycoviruses and exploration for the biological control of crop fungal diseases. Annu Rev Phytopathol. (2014) 52:45–68. doi: 10.1146/annurev-phyto-102313-050222
3. Ghabrial SA, Castón JR, Jiang D, Nibert ML, Suzuki N. 50-plus years of fungal viruses. Virology. (2015) 479:356–68. doi: 10.1016/j.virol.2015.02.034
4. Hillman BI, Annisa A, Suzuki N. Viruses of plant-interacting fungi. Adv Virus Res. (2018) 100:99–116. doi: 10.1016/bs.aivir.2017.10.003
5. Yu X, Li B, Fu Y, Jiang D, Ghabrial SA, Li G, et al. A geminivirus-related DNA mycovirus that confers hypovirulence to a plant pathogenic fungus. Proc Natl Acad Sci. (2010) 107:8387–92. doi: 10.1073/pnas.0913535107
6. Kondo H, Kanematsu S, Suzuki N. Viruses of the white root rot fungus, Rosellinia necatrix. Adv Virus Res. (2013) 86:177–214. doi: 10.1016/B978-0-12-394315-6.00007-6
7. Liu L, Xie J, Cheng J, Fu Y, Li G, Yi X, et al. Fungal negative-stranded RNA virus that is related to bornaviruses and nyaviruses. Proc Natl Acad Sci. (2014) 111:12205–10. doi: 10.1073/pnas.1401786111
8. Kanhayuwa L, Kotta-Loizou I, Özkan S, Gunning AP, Coutts RH. A novel mycovirus from Aspergillus fumigatus contains four unique dsRNAs as its genome and is infectious as dsRNA. Proc Natl Acad Sci. (2015) 112:9100–5. doi: 10.1073/pnas.1419225112
9. Zhang Y, Gao J, Li Y. Diversity of mycoviruses in edible fungi. Virus Genes. (2022) 58:377–91. doi: 10.1007/s11262-022-01908-6
10. Xie J, Wei D, Jiang D, Fu Y, Li G, Ghabrial S, et al. Characterization of debilitation-associated mycovirus infecting the plant-pathogenic fungus Sclerotinia sclerotiorum. J Gen Virol. (2006) 87:241–9. doi: 10.1099/vir.0.81522-0
11. Chu Y-M, Lim W-S, Yea S-J, Cho J-D, Lee Y-W, Kim K-H. Complexity of dsRNA mycovirus isolated from Fusarium graminearum. Virus Genes. (2004) 28:135–43. doi: 10.1023/B:VIRU.0000012270.67302.35
12. Bridge P, Couteaudier Y, Clarkson J. “Molecular variability of fungal pathogens”. In: Buck KW, Editor. Molecular variability of viruses of fungi. (1998). p. xiv+-319.
13. Dawe AL, Nuss DL. Hypoviruses and chestnut blight: exploiting viruses to understand and modulate fungal pathogenesis. Annu Rev Genet. (2001) 35:1–29. doi: 10.1146/annurev.genet.35.102401.085929
14. Yaegashi H, Shimizu T, Ito T, Kanematsu S. Differential inductions of RNA silencing among encapsidated double-stranded RNA mycoviruses in the white root rot fungus Rosellinia necatrix. J Virol. (2016) 90:5677–92. doi: 10.1128/JVI.02951-15
15. Kulshrestha S, Seth CA, Sharma M, Sharma A, Mahajan R, Chauhan A. Biology and control of Rosellinia necatrix causing white root rot disease: a review. J Pure Appl Microbiol. (2014) 8:1803–14.
17. Garcı́a RAM, Ten Hoopen GM, Kass DC, Garita VAS, Krauss U. Evaluation of mycoparasites as biocontrol agents of Rosellinia root rot in cocoa. Biol Control. (2003) 27:210–27. doi: 10.1016/S1049-9644(03)00014-8
18. Ofong A, Pearce R. Toxin production by Rosellinia desmazieresii (Berk. & Br.) Sacc. Eur J For Pathol. (1991) 21:57–63. doi: 10.1111/j.1439-0329.1991.tb00302.x
19. ten Hoopen GM, Krauss U. Biology and control of Rosellinia bunodes, Rosellinia necatrix and Rosellinia pepo: a review. Crop Protect. (2006) 25:89–107. doi: 10.1016/j.cropro.2005.03.009
20. Pliego C, López-Herrera C, Ramos C, Cazorla FM. Developing tools to unravel the biological secrets of Rosellinia necatrix, an emergent threat to woody crops. Mol Plant Pathol. (2012) 13:226–39. doi: 10.1111/j.1364-3703.2011.00753.x
21. Delatour C, Guillaumin J. Importance des pourridiés dans les regions tempérées. Eur J For Pathol. (1985) 15:258–63. doi: 10.1111/j.1439-0329.1985.tb01097.x
22. Arjona-Lopez JM, Telengech P, Jamal A, Hisano S, Kondo H, Yelin MD, et al. Novel, diverse RNA viruses from Mediterranean isolates of the phytopathogenic fungus, Rosellinia necatrix: insights into evolutionary biology of fungal viruses. Environ Microbiol. (2018) 20:1464–83. doi: 10.1111/1462-2920.14065
23. Ikeda K-I, Nakamura H, Arakawa M, Matsumoto N. Diversity and vertical transmission of double-stranded RNA elements in root rot pathogens of trees, Helicobasidium mompa and Rosellinia necatrix. Mycol Res. (2004) 108:626–34. doi: 10.1017/S0953756204000061
24. Arakawa M, Nakamura H, Uetake Y, Matsumoto N. Presence and distribution of double-stranded RNA elements in the white root rot fungus Rosellinia necatrix. Mycoscience. (2002) 43:21–6. doi: 10.1007/s102670200004
25. Telengech P, Hisano S, Mugambi C, Hyodo K, Arjona-López JM, López-Herrera CJ, et al. Diverse partitiviruses from the phytopathogenic fungus, Rosellinia necatrix. Front Microbiol. (2020) 11:1064. doi: 10.3389/fmicb.2020.01064
26. Matthijnssens J, Attoui H, Bányai K, Brussaard CP, Danthi P, Del Vas M, et al. ICTV virus taxonomy profile: Sedoreoviridae 2022. J Gen Virol. (2022) 103:001782. doi: 10.1099/jgv.0.001782
27. Matthijnssens J, Attoui H, Bányai K, Brussaard CP, Danthi P, Del Vas M, et al. ICTV virus taxonomy profile: Spinareoviridae 2022. J Gen Virol. (2022) 103:001781. doi: 10.1099/jgv.0.001781
28. Enebak S, Hillman B, MacDonald W. A hypovirulent isolate of Cryphonectria parasitica with multiple, genetically unique dsRNA segments. Mol Plant Microbe Interact. (1994) 7:590–5. doi: 10.1094/MPMI-7-0590
29. Hillman BI, Suzuki N. Viruses in the chestnut blight fungus. Adv Virus Res. (2004) 63:423–72. doi: 10.1016/S0065-3527(04)63007-7
30. Kanematsu S, Arakawa M, Oikawa Y, Onoue M, Osaki H, Nakamura H, et al. A reovirus causes hypovirulence of Rosellinia necatrix. Phytopathology. (2004) 94:561–8. doi: 10.1094/PHYTO.2004.94.6.561
31. Osaki H, Wei CZ, Arakawa M, Iwanami T, Nomura K, Matsumoto N, et al. Nucleotide sequences of double-stranded RNA segments from a hypovirulent strain of the white root rot fungus Rosellinia necatrix: possibility of the first member of the Reoviridae from fungus. Virus Genes. (2002) 25:101–7. doi: 10.1023/A:1020182427439
32. Wei C, Osaki H, Iwanami T, Matsumoto N, Ohtsu Y. Complete nucleotide sequences of genome segments 1 and 3 of Rosellinia anti-rot virus in the family Reoviridae. Arch Virol. (2004) 149:773–7. doi: 10.1007/s00705-003-0259-6
33. Wei CZ, Osaki H, Iwanami T, Matsumoto N, Ohtsu Y. Molecular characterization of dsRNA segments 2 and 5 and electron microscopy of a novel reovirus from a hypovirulent isolate, W370, of the plant pathogen Rosellinia necatrix. J Gen Virol. (2003) 84:2431–7. doi: 10.1099/vir.0.19098-0
34. Yaegashi H, Yoshikawa N, Ito T, Kanematsu S. A mycoreovirus suppresses RNA silencing in the white root rot fungus, Rosellinia necatrix. Virology. (2013) 444:409–16. doi: 10.1016/j.virol.2013.07.010
35. Sato Y, Miyazaki N, Kanematsu S, Xie J, Ghabrial SA, Hillman BI, et al. ICTV virus taxonomy profile: Megabirnaviridae. J Gen Virol. (2019) 100:1269–70. doi: 10.1099/jgv.0.001297
36. Delmas B, Attoui H, Ghosh S, Malik YS, Mundt E, Vakharia VN, et al. ICTV virus taxonomy profile: Picobirnaviridae. J Gen Virol. (2019) 100:133–4. doi: 10.1099/jgv.0.001186
37. Chiba S, Salaipeth L, Lin Y-H, Sasaki A, Kanematsu S, Suzuki N. A novel bipartite double-stranded RNA mycovirus from the white root rot fungus Rosellinia necatrix: molecular and biological characterization, taxonomic considerations, and potential for biological control. J Virol. (2009) 83:12801–12. doi: 10.1128/JVI.01830-09
38. Mata CP, Rodríguez JM, Suzuki N, Castón JR. Structure and assembly of double-stranded RNA mycoviruses. Adv Virus Res: Elsevier. (2020) 108:213–47. doi: 10.1016/bs.aivir.2020.08.001
39. Kanematsu S, Shimizu T, Salaipeth L, Yaegashi H, Sasaki A, Ito T, et al. Genome rearrangement of a mycovirus Rosellinia necatrix megabirnavirus 1 affecting its ability to attenuate virulence of the host fungus. Virology. (2014) 450:308–15. doi: 10.1016/j.virol.2013.12.002
40. Shimizu T, Kanematsu S, Yaegashi H. Draft genome sequence and transcriptional analysis of Rosellinia necatrix infected with a virulent mycovirus. Phytopathology. (2018) 108:1206–11. doi: 10.1094/PHYTO-11-17-0365-R
41. Nibert ML, Ghabrial SA, Maiss E, Lesker T, Vainio EJ, Jiang D, et al. Taxonomic reorganization of family Partitiviridae and other recent progress in partitivirus research. Virus Res. (2014) 188:128–41. doi: 10.1016/j.virusres.2014.04.007
42. Vainio EJ, Chiba S, Ghabrial SA, Maiss E, Roossinck M, Sabanadzovic S, et al. ICTV virus taxonomy profile: Partitiviridae. J Gen Virol. (2018) 99:17–8. doi: 10.1099/jgv.0.000985
43. Sasaki A, Miyanishi M, Ozaki K, Onoue M, Yoshida K. Molecular characterization of a partitivirus from the plant pathogenic ascomycete Rosellinia necatrix. Arch Virol. (2005) 150:1069–83. doi: 10.1007/s00705-005-0494-0
44. Chun J, Kim D-H. Co-infection of a novel fusagravirus and a partitivirus in a Korean isolate of Rosellinia necatrix KACC40168. Virus Genes. (2021) 57:121–6. doi: 10.1007/s11262-020-01809-6
45. Lin Y-H, Chiba S, Tani A, Kondo H, Sasaki A, Kanematsu S, et al. A novel quadripartite dsRNA virus isolated from a phytopathogenic filamentous fungus, Rosellinia necatrix. Virology. (2012) 426:42–50. doi: 10.1016/j.virol.2012.01.013
46. Chiba S, Castón JR, Ghabrial SA, Suzuki N, Consortium IR. ICTV virus taxonomy profile: Quadriviridae. J Gen Virol. (2018) 99:1480–1. doi: 10.1099/jgv.0.001152
47. Lin Y-H, Hisano S, Yaegashi H, Kanematsu S, Suzuki N. A second quadrivirus strain from the phytopathogenic filamentous fungus Rosellinia necatrix. Arch Virol. (2013) 158:1093–8. doi: 10.1007/s00705-012-1580-8
48. Yaegashi H, Nakamura H, Sawahata T, Sasaki A, Iwanami Y, Ito T, et al. Appearance of mycovirus-like double-stranded RNAs in the white root rot fungus, Rosellinia necatrix, in an apple orchard. FEMS Microbiol Ecol. (2013) 83:49–62. doi: 10.1111/j.1574-6941.2012.01454.x
49. Chiba S, Lin Y-H, Kondo H, Kanematsu S, Suzuki N. A novel victorivirus from a phytopathogenic fungus, Rosellinia necatrix, is infectious as particles and targeted by RNA silencing. J Virol. (2013) 87:6727–38. doi: 10.1128/JVI.00557-13
50. Valverde RA, Khalifa ME, Okada R, Fukuhara T, Sabanadzovic S, Consortium IR. ICTV virus taxonomy profile: Endornaviridae. J Gen Virol. (2019) 100:1204–5. doi: 10.1099/jgv.0.001277
51. Yaegashi H, Kanematsu S. Natural infection of the soil-borne fungus Rosellinia necatrix with novel mycoviruses under greenhouse conditions. Virus Res. (2016) 219:83–91. doi: 10.1016/j.virusres.2015.11.004
52. Hai D, Li J, Jiang D, Cheng J, Fu Y, Xiao X, et al. Plants interfere with non-self recognition of a phytopathogenic fungus via proline accumulation to facilitate mycovirus transmission. Nat Commun. (2024) 15:4748. doi: 10.1038/s41467-024-49110-6
53. Chiba S, Suzuki N, Velasco L, Ayllón MA, Lee-Marzano S-Y, Sun L, et al. ICTV virus taxonomy profile: Fusariviridae 2024. J Gen Virol. (2024) 105:001973. doi: 10.1099/jgv.0.001973
54. Zhang R, Liu S, Chiba S, Kondo H, Kanematsu S, Suzuki N. A novel single-stranded RNA virus isolated from a phytopathogenic filamentous fungus, Rosellinia necatrix, with similarity to hypo-like viruses. Front Microbiol. (2014) 5:360. doi: 10.3389/fmicb.2014.00360
55. Sato Y, Das S, Velasco L, Turina M, Osaki H, Kotta-Loizou I, et al. ICTV virus taxonomy profile: Yadokariviridae 2023. J Gen Virol. (2023) 104:001826. doi: 10.1099/jgv.0.001826
56. Zhang R, Hisano S, Tani A, Kondo H, Kanematsu S, Suzuki N. A capsidless ssRNA virus hosted by an unrelated dsRNA virus. Nat Microbiol. (2016) 1:1–6. doi: 10.1038/nmicrobiol.2015.1
57. Sato Y, Hisano S, López-Herrera CJ, Kondo H, Suzuki N. Three-layered complex interactions among capsidless (+) ssRNA yadokariviruses, dsRNA viruses, and a fungus. Mbio. (2022) 13:e01685–22. doi: 10.1128/mbio.01685-22
58. Chiba S, Velasco L, Ayllón MA, Suzuki N, Lee-Marzano S-Y, Sun L, et al. ICTV virus taxonomy profile: Hypoviridae 2023. J Gen Virol. (2023) 104:001848. doi: 10.1099/jgv.0.001848
59. Das S, Hisano S, Eusebio-Cope A, Kondo H, Suzuki N. A transfectable fusagravirus from a Japanese strain of Cryphonectria carpinicola with spherical particles. Viruses. (2022) 14:1722. doi: 10.3390/v14081722
60. Arjona-López JM, Telengech P, Suzuki N, López-Herrera CJ. Coinfection of Rosellinia necatrix by a partitivirus and a virga-like virus is associated with hypovirulence. Eur J Plant Pathol. (2020) 158:111–9. doi: 10.1007/s10658-020-02058-x
61. Adams MJ, Adkins S, Bragard C, Gilmer D, Li D, MacFarlane SA, et al. ICTV virus taxonomy profile: Virgaviridae. J Gen Virol. (2017) 98:1999–2000. doi: 10.1099/jgv.0.000884
62. Kanematsu S. Studies on the interactions of Rosellinia necatrix and mycoviruses for sustainable management of white root rot of fruit trees. J Gen Plant Pathol. (2023) 89:357–60. doi: 10.1007/s10327-023-01148-y
63. Yaegashi H, Sawahata T, Ito T, Kanematsu S. A novel colony-print immunoassay reveals differential patterns of distribution and horizontal transmission of four unrelated mycoviruses in Rosellinia necatrix. Virology. (2011) 409:280–9. doi: 10.1016/j.virol.2010.10.014
64. Kanematsu S, Sasaki A, Onoue M, Oikawa Y, Ito T. Extending the fungal host range of a partitivirus and a mycoreovirus from Rosellinia necatrix by inoculation of protoplasts with virus particles. Phytopathology. (2010) 100:922–30. doi: 10.1094/PHYTO-100-9-0922
65. Velasco L, Arjona-Girona I, Cretazzo E, Lopez-Herrera C. Viromes in Xylariaceae fungi infecting avocado in Spain. Virology. (2019) 532:11–21. doi: 10.1016/j.virol.2019.03.021
66. Ikeda K, Inoue K, Kida C, Uwamori T, Sasaki A, Kanematsu S, et al. Potentiation of mycovirus transmission by zinc compounds via attenuation of heterogenic incompatibility in Rosellinia necatrix. Appl Environ Microbiol. (2013) 79:3684–91. doi: 10.1128/AEM.00426-13
67. Sasaki A, Nakamura H, Suzuki N, Kanematsu S. Characterization of a new megabirnavirus that confers hypovirulence with the aid of a co-infecting partitivirus to the host fungus, Rosellinia necatrix. Virus Res. (2016) 219:73–82. doi: 10.1016/j.virusres.2015.12.009
68. Sasaki A, Kanematsu S, Onoue M, Oyama Y, Yoshida K. Infection of Rosellinia necatrix with purified viral particles of a member of Partitiviridae (RnPV1-W8). Arch Virol. (2006) 151:697–707. doi: 10.1007/s00705-005-0662-2
69. Chiba S, Lin Y-H, Kondo H, Kanematsu S, Suzuki N. Effects of defective interfering RNA on symptom induction by, and replication of, a novel partitivirus from a phytopathogenic fungus, Rosellinia necatrix. J Virol. (2013) 87:2330–41. doi: 10.1128/JVI.02835-12
70. Kipkemboi TP. Molecular and biological properties of diverse fungal and plant partitiviruse (2021). Available online at: https://ousar.lib.okayamau.ac.jp/files/public/6/62962/20211203104040370553/K0006511_summary.pdf (Accessed October 2024).
Keywords: Rosellinia necatrix, mycoviruses, Spinareoviridae, Megabirnaviridae, Partitiviridae, Quadriviridae, Pseudototiviridae, Endornaviridae
Citation: Hassan S, Coutts RHA, Jamal A and Kotta-Loizou I (2025) A comprehensive review of mycoviruses infecting the plant pathogenic fungus Rosellinia necatrix. Front. Virol. 5:1513097. doi: 10.3389/fviro.2025.1513097
Received: 17 October 2024; Accepted: 21 January 2025;
Published: 05 February 2025.
Edited by:
Nobuhiro Suzuki, Okayama University, JapanCopyright © 2025 Hassan, Coutts, Jamal and Kotta-Loizou. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ioly Kotta-Loizou, aS5rb3R0YS1sb2l6b3UyQGhlcnRzLmFjLnVr; aS5rb3R0YS1sb2l6b3UxM0BpbXBlcmlhbC5hYy51aw==
 Sidra Hassan
Sidra Hassan Robert H. A. Coutts
Robert H. A. Coutts Atif Jamal
Atif Jamal Ioly Kotta-Loizou
Ioly Kotta-Loizou