- 1Department of Veterinary Pathobiology, College of Veterinary Medicine, Oklahoma State University, Stillwater, OK, United States
- 2Oklahoma Center for Respiratory and Infectious Diseases, Oklahoma State University, Stillwater, OK, United States
- 3Department of Statistics, College of Arts and Science, Oklahoma State University, Stillwater, OK, United States
- 4Physiological Sciences, College of Veterinary Medicine, Oklahoma State University, Stillwater, OK, United States
- 5College of Veterinary Medicine, University of Missouri, Columbia, MO, United States
- 6Oklahoma Animal Disease Diagnostic Laboratory, College of Veterinary Medicine, Oklahoma State University, Stillwater, OK, United States
SARS-CoV-2 patients have been reported to have high rates of secondary Klebsiella pneumoniae infections. K. pneumoniae is a commensal that is typically found in the respiratory and gastrointestinal tracts. However, it can cause severe disease when a person’s immune system is compromised. Despite a high number of K. pneumoniae cases reported in SARS-CoV-2 patients, a co-infection animal model evaluating the pathogenesis is not available. In our cohort of COVID-19-positive human patients, 38% exhibited the presence of K. pneumoniae. Therefore we developed a mouse model to study the disease pathogenesis of SARS-CoV-2 and K. pneumoniae co-infection. BALB/cJ mice were inoculated with mouse-adapted SARS-CoV-2 followed by a challenge with K. pneumoniae. Mice were monitored for body weight change, clinical signs, and survival during infection. The bacterial load, viral titers, immune cell accumulation and phenotype, and histopathology were evaluated in the lungs. The co-infected mice showed severe clinical disease and a higher mortality rate within 48 h of K. pneumoniae administration. The co-infected mice had significantly elevated bacterial load in the lungs, however, viral loads were similar between co-infected and single-infected mice. Histopathology of co-infected mice showed severe bronchointerstitial pneumonia with copious intralesional bacteria. Flow cytometry analysis showed significantly higher numbers of neutrophils and macrophages in the lungs. Collectively, our results demonstrated that co-infection of SARS-CoV-2 with K. pneumoniae causes severe disease with increased mortality in mice.
Introduction
The COVID-19 pandemic caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has affected more than 700 million people with approximately 7 million deaths (1). Most individuals infected with SARS-CoV-2 were asymptomatic or had mild fever, headache, anosmia, fatigue and mild respiratory symptoms (2–4); however, a significant number of patients exhibited severe disease and required hospitalization. The hospitalized patients showed pneumonitis, hypoxia, acute respiratory distress syndrome (ARDS), and multiple organ failure (5, 6).
Viral respiratory tract infections such as influenza and SARS-CoV-2 are commonly accompanied by co- or secondary infections with other viruses, bacteria, and/or fungal pathogens (7, 8). Bacterial coinfections were responsible for high mortality during the 1918 influenza pandemic (9). Patients with severe respiratory signs are at a higher risk of secondary infections due to compromised respiratory epithelial barrier or dysregulated immune system (10, 11). Virus infections induce upregulation of pro-inflammatory cytokines, which can lead to epithelial cell damage or death (12), resulting in the breakage of the physical epithelial barrier and increase bacterial attachment and colonization (13). During the COVID-19 pandemic, high numbers of SARS-CoV-2 patients had detectable bacterial co-infection, and the most common bacterial agents identified were Staphylococcus aureus, Streptococcus pneumoniae, and Klebsiella pneumoniae (Kp) (14, 15). Even before the COVID-19 pandemic, secondary infections and coinfections were recognized as significant contributors to acute respiratory distress syndrome (ARDS) (16) and leading up to 80.5% fatality in these patients (17). In summary, the role of secondary and coinfections in worsening viral respiratory infections emphasizes the necessity of pathogenesis studies to effectively address these complications.
Kp causes hospital-acquired infections and is the third most common gram negative infection (18). Clinical studies have also identified Kp infection in COVID-19 patients with secondary bacterial pneumonia and mortality (14, 19, 20). Kp was detected in 37 to 55% of COVID-19 patients, and the highest incidence was noted during early days of SARS-CoV-2 infection (14, 21). These studies reveal that coinfections significantly increase severity and mortality, often leading to exacerbated respiratory symptoms, longer ICU stays, and a greater need for mechanical ventilation. Despite the high rate of co-infection, the specific impact of Kp on COVID-19 pathogenesis and clinical outcomes has yet to be thoroughly evaluated, highlighting the need for further research, including the use of mouse models. Murine models for SARS-CoV-2 have been established that exhibit similar symptoms and immune responses to human patients and are valuable tools for studying pathogenesis (22, 23). These models have been used to study SARS-CoV-2 co-infections, demonstrating the importance of secondary bacterial infections that can lead to high lethality during SARS-CoV-2 infection (24, 25).
In this study, we employed a mouse model to investigate the impact of SARS-CoV-2 and Kp co-infection. Our findings revealed a significantly higher mortality rate among co-infected mice compared to those infected with SARS-CoV-2 or Kp alone. Moreover, we observed a marked escalation in lung pathology, bacterial load, and infiltration of immune cells in co-infected mice. This mouse model can serve as a crucial tool for unraveling the intricate pathogenesis underlying both SARS-CoV-2 infection and secondary bacterial infections.
Results
Klebsiella pneumoniae is the most common coinfecting pathogen in COVID-19 patients
We conducted a PCR assay to identify the presence of multiple pathogens in samples from SARS-CoV-2 positive patients. We screened 50 samples for various viral, bacterial, and fungal agents using a PCR assay capable of detecting 32 of the most common respiratory pathogens. We identified seven different pathogens in 54% (27 of a total 50) of the samples tested. The remaining 23 samples were negative for the pathogens included in the assay. The seven pathogens were: Human Respiratory Syncytial Virus (HRSV) A and B, Staphylococcus aureus (Sa), Klebsiella pneumoniae, Enterovirus, Pneumocystis jirovecii, Salmonella sp., and Moraxella catarrhalis (Figure 1A). Out of the 27 samples, 26 samples contained at least one bacterial agent, 2 samples contained two viruses (HRSV and Enterovirus), and 1 sample contained a fungal agent (P. jirovecii) (Figure 1B). Of the SARS-CoV-2 positive samples (27 samples), sixteen samples contained a single infectious agent, 10 samples contained two pathogens, and 1 sample contained three pathogens (Figure 1C). The most common bacterial agents observed were Kp (38%) and Sa (28%) (Figure 1A). Additionally, we found that Kp was detected in 18% of samples as a potential standalone infection. Due to its implications in SARS-CoV-2 patients as a secondary bacterial infection (19, 20, 26), we focused on Kp for further studies.
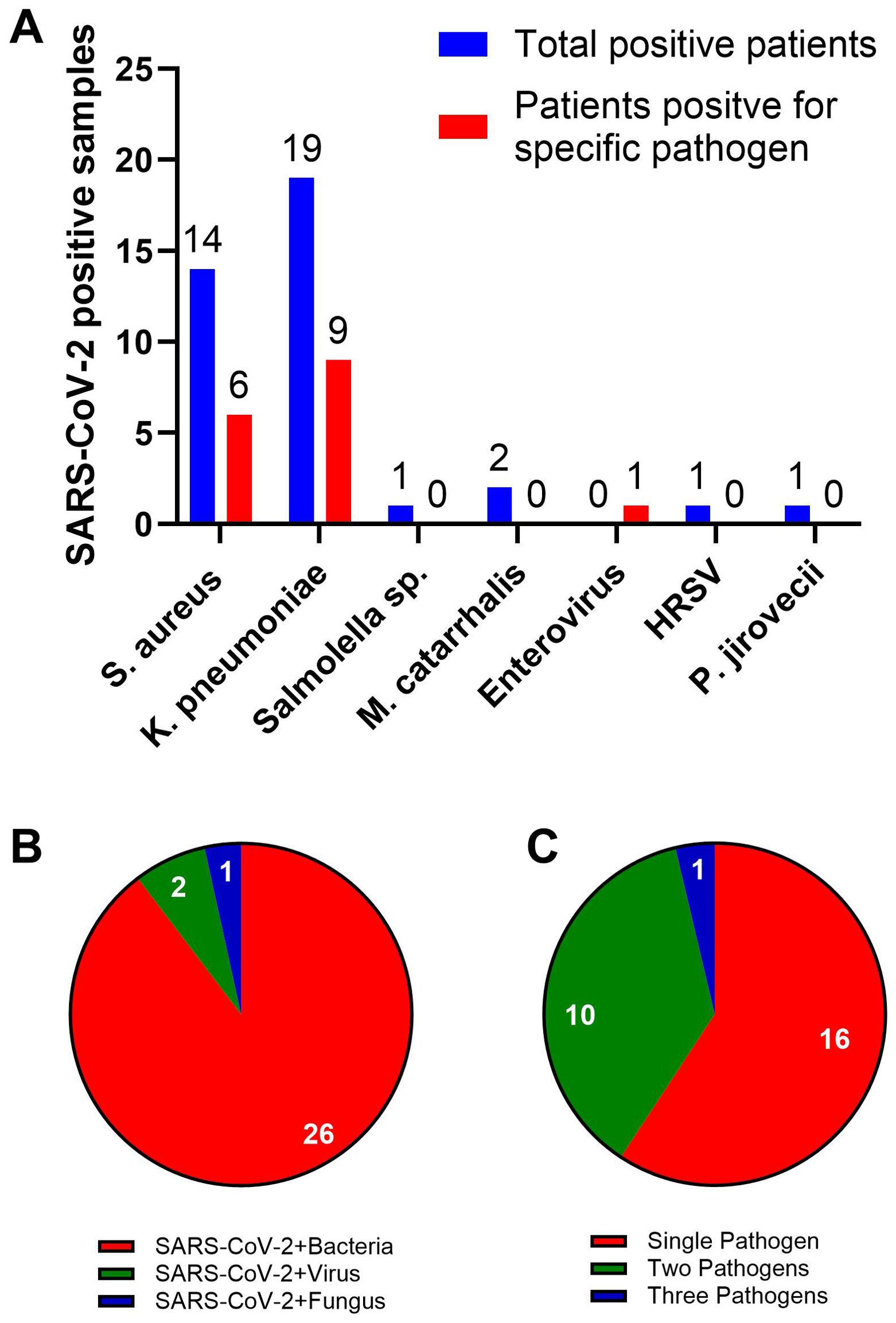
Figure 1. Most common co-infections in SARS-CoV-2 patients. (A) FTD-33 Real-time PCR assay demonstrating pathogens that are detected in SARS-CoV-2 positive nasopharyngeal swabs. The blue bars represent the total number of patient samples positive for SARS-CoV-2, the pathogen listed on the X-axis and other pathogens included in the assay. The red bars represent the number of patient samples positive for only the listed pathogen and SARS-CoV-2. Pie charts demonstrating distribution of co-infecting pathogen type (B) and multiples of infection (C) along with SARS-CoV-2.
Co-infected mice exhibit high rates of morbidity and mortality
To further understand the impact of Kp on SARS-CoV-2 pathogenesis, we conducted in vivo experiments with a mouse model. We infected BALB/cJ mice with SARS-CoV-2 on day 0 and subsequently infected them with Kp on day 4, assessing disease progression, clinical signs, and mortality rates in co-infected versus single-infected groups. We observed a similar pattern of weight loss until day 6 post-infection between the S-CoV-2 and S-CoV-2 + Kp groups (Figure 2A), with both groups of mice losing up to 20% of their initial body weight. However, the S-CoV-2 + Kp group, 87.5% of mice continued to lose body weight and succumbed to infection by day 7 (Figure 2C). In comparison, the S-CoV-2-only and Kp-alone group had only 12.5% and 16.67% of mice died, respectively, due to infection. The difference in outcome can be observed in Figure 2B, which shows a continued increase in clinical score for co-infected mice compared to S-CoV-2-only infection. These findings demonstrate that SARS-CoV-2 co-infection with Kp significantly increases clinical signs, disease severity, and mortality.
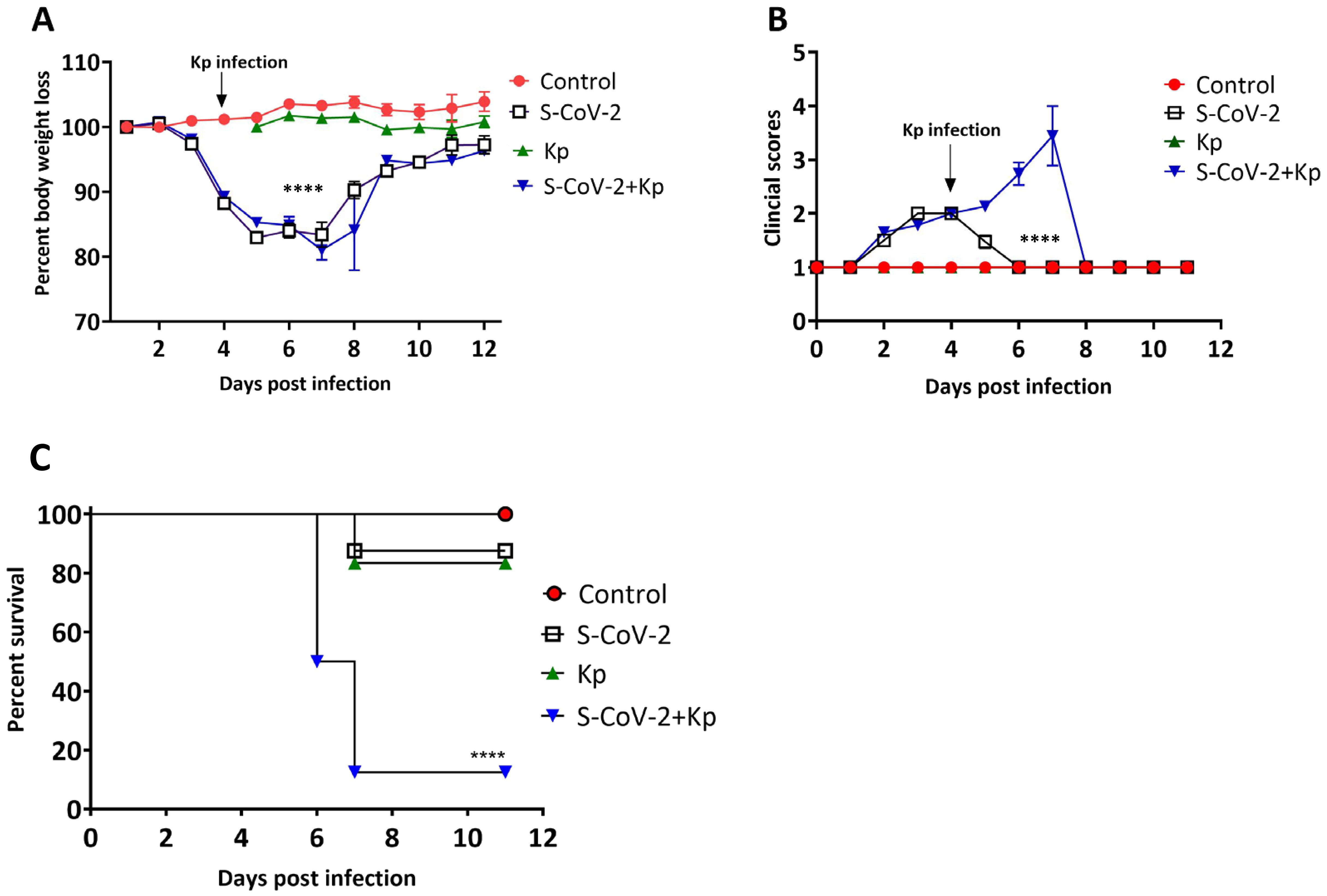
Figure 2. Co-infection with SARS-CoV-2 and Kp increases disease severity. Percent body weight loss calculated based on day 0 body weight is presented on y-axis. The data is represented as mean ± SEM. (B) Clinical scores for each group are represented as mean ± SEM (C) Survival of the animals was monitored for 12 days post infection. Data represent n = 18 to 31 mice per group with two-three independent experiments. Statistical significance was determined in (A) using the Tukey-Kramer mean separation technique (Control v.s. S-CoV-2 + Kp, Control v.s. S-CoV-2, S-CoV-2 v.s. S-CoV-2 + Kp; (B) Kruskal-Wallis test (Control v.s. S-CoV-2, Control v.s. S-CoV-2 + Kp, S-CoV-2 v.s. Kp, S-CoV-2 v.s. S-CoV-2 + Kp, Kp v.s. S-CoV-2 + Kp and (C) Kaplan-Meier survival curve with a Mantel-Cox test. ****P < 0.0001.
SARS-CoV-2 infection promotes Kp growth in the lungs
Given the high morbidity and mortality in the co-infection group, we investigated the cause of this phenotype. We determined bacterial load in the lungs of single- (Kp) and co-infected mice (S-CoV-2 + Kp) and viral load in the lungs of S-CoV-2-only as well as co-infected mice (S-CoV-2 + Kp) on 6th dpi. The day 6 post S-CoV-2 was selected because of the significant percentage of death in co-infected mice (Figure 2C) on that day. We also wanted to examine whether these groups of mice had Kp in the bloodstream indicative of septicemia. The number of mice with Kp in their heart blood was higher in S-CoV-2 + Kp group (Figure 3C; Kp =1/8 vs S-CoV-2 + Kp =7/8). A comparison of bacterial load in the lungs of Kp and S-CoV-2 + Kp showed a significant difference (p = 0.0343) with Kp mice having mostly no bacterial growth in the lung (Figure 3A). Virus titer in the lungs of S-CoV-2 and S-CoV-2+ Kp groups, did not show significant difference (Figure 3B). Thus, the high mortality rate in the S-CoV-2 + Kp groups are associated with increased Kp replication rather than increased SARS-CoV-2 replication.
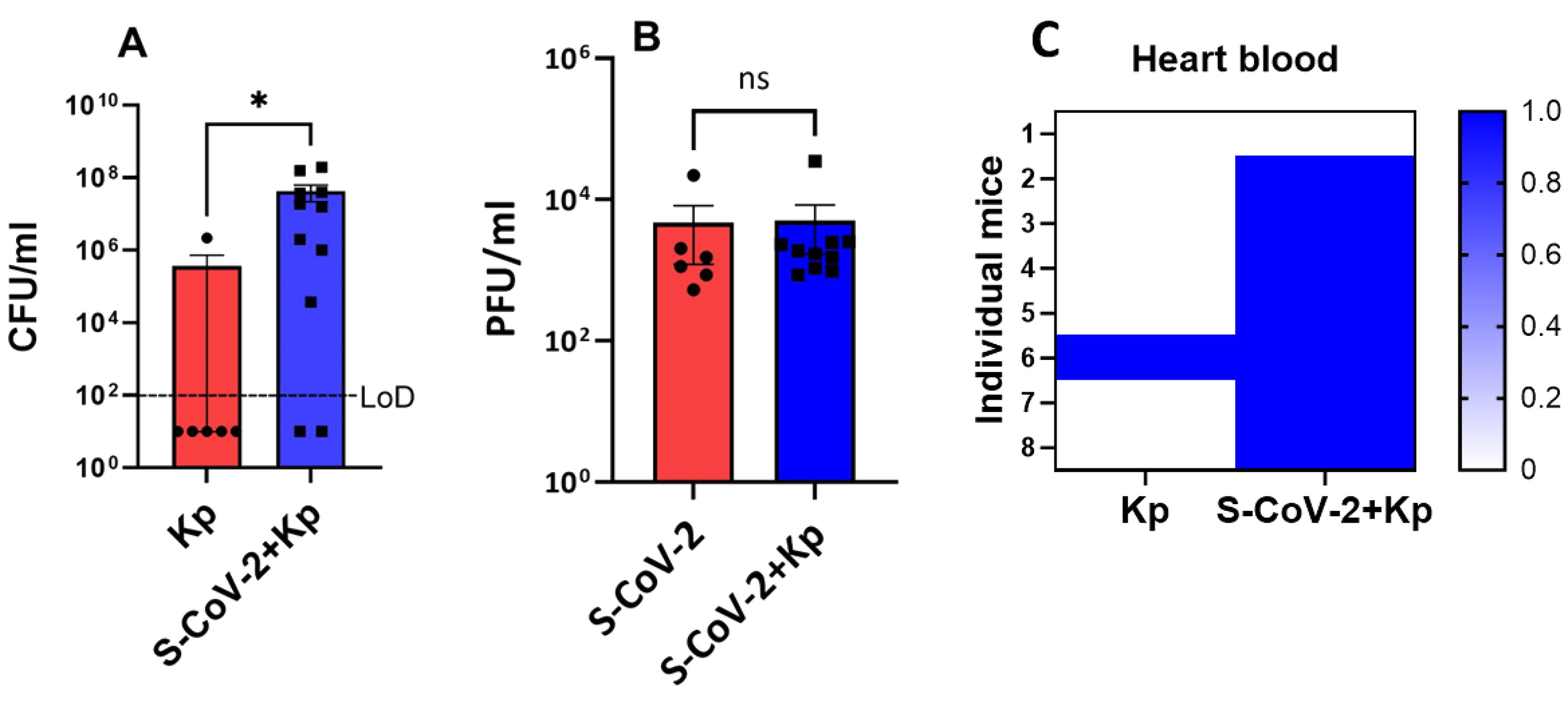
Figure 3. SARS-CoV-2 co-infected mice exhibit increased Kp propagation in the lungs. SARS-CoV-2 infected mice were coinfected with Kp or Kp-alone on day 4 and lungs were collected on day 6 to examine the viral and bacterial loads. Bacterial count (A) and viral titers (B) in the lungs of respected group are presented. Data is presented as mean ± SEM of 2 independent experiments (n = 6-11). For graph (A), the mice did not contain bacteria in the lungs were assigned a random value below the level of detection (LoD). (C) Presence of Kp in the heart blood; 1 = present and 0 = absent (n = 8). Y-axis represent individual mice in each group. Statistical significance was obtained with Welch’s test with *P < 0.05. ns, non significant.
Co-infection causes excessive infiltration of neutrophils and macrophages into the lungs
Since COVID-19 patients with ARDS contain a higher percentage of neutrophils (27) as well as their phagocytic role in bacterial infection, we wanted to quantify the neutrophil response in the lungs. Neutrophil recruitment in the lungs was significantly higher in the S-CoV-2 + Kp co-infection group compared to the other groups (Figures 4A–C). Furthermore, we wanted to quantify inflammatory macrophages/monocyte changes in the lungs due to infection because of their pathogenic role in mice infected with a lethal dose of SARS-CoV and MERS-CoV (28, 29). The percentage of inflammatory macrophages/monocytes (IMMs) was not significantly different between all the groups (Figures 4D, E), however, there were significantly higher numbers of IMMs in the S-CoV-2 + Kp group compared to all other groups (Figure 4F). These increases in inflammatory cells indicate a response to the infection, however, it appears that co-infected mice are recruiting more cells due to the secondary infection, which could be causing immunopathology to epithelial cells.
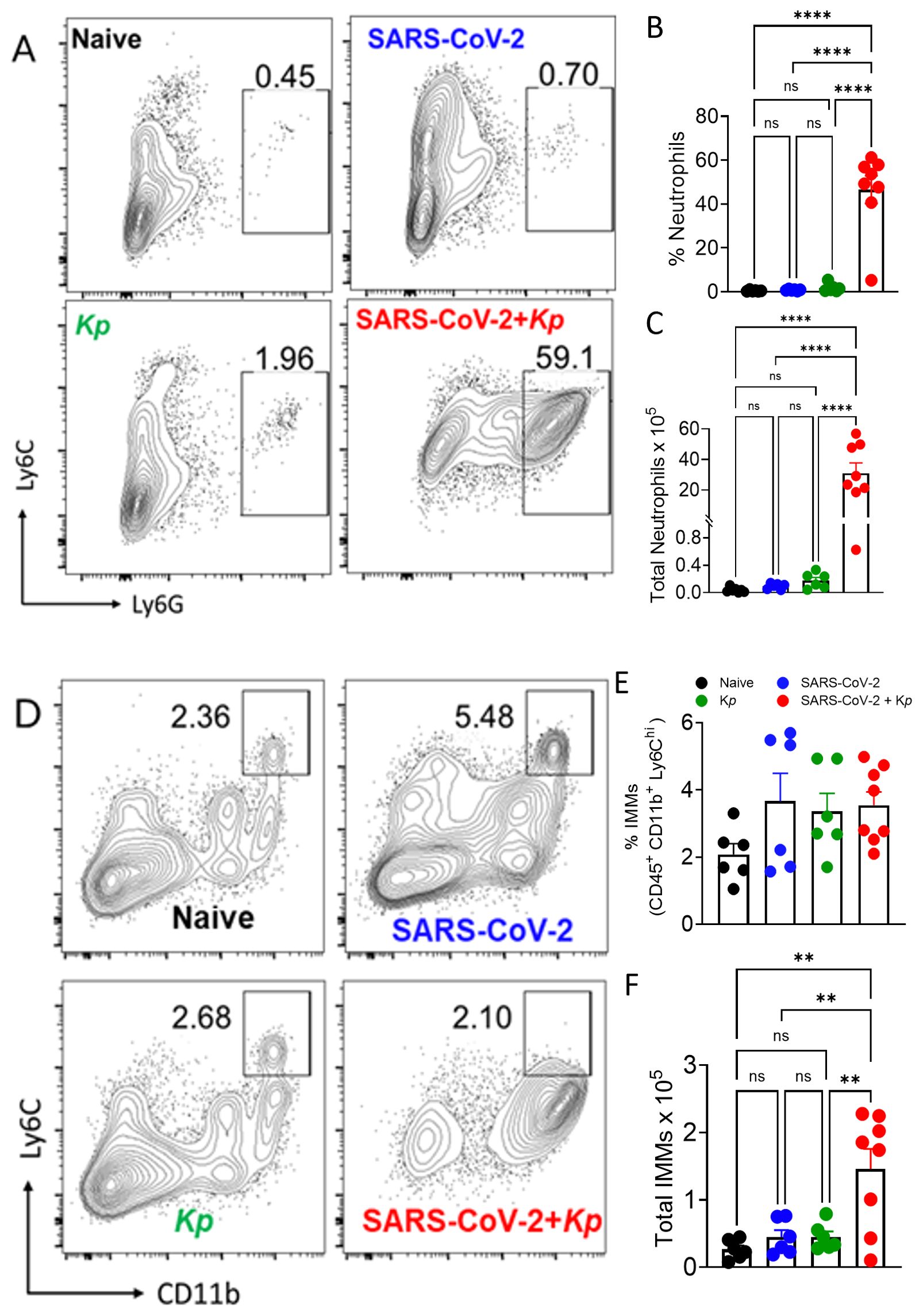
Figure 4. Neutrophil and inflammatory monocytes and macrophages (IMM) responses to coinfection. Representative FACS plots (A, D), quantification of percent (B) and total neutrophils (C), percent CD11b+Ly6Chi IMMs (E) and total CD11b+Ly6Chi IMMs (F) in the lungs on day 6. Data were pooled from 2 independent experiments with 3 to5 mice/group/experiment. **P ≤ 0.01, and ****P ≤ 0.001, by One-Way ANOVA. ns, non significant.
Co-infection of SARS-CoV-2 and Kp induces severe inflammation in the lungs
Following the gross diagnosis of bronchointerstitial pneumonia as well as the recruitment of large numbers of inflammatory cells to the lungs (Figure 4F), we performed histopathology on the lung at 6 dpi (2 days post-Kp infection). Bronchointerstitial pneumonia, thrombosis, fibrin, epithelial cell necrosis, perivascular cuffing, edema, and hemorrhage were the most common lung lesions (Figures 5A–D). The lesions were graded on a scale of 0 to 4, with 0 being nonexistent and 4 being severe. Control and Kp-infected mice lungs had open alveoli and few histopathological changes (Figures 5A, C). The lung infected with S-CoV-2 had multifocal, moderate interstitial pneumonia (black triangle, Figure 5B) and perivascular cuffing (black arrow, Figure 5B). The alveoli showed moderate edema and fibrin deposition. Conversely, the pathologic lesions in the co-infected mice were the most severe (Figure 5D). The alveoli and bronchioles were densely packed with neutrophils and macrophages and were admixed with edema and fibrin (bronchopneumonia, black arrowhead, Figure 5D). There was multifocal thrombosis in small caliber vessels. Innumerable bacteria were also observed in the lesions (white arrow). Overall, S-CoV-2 + Kp mice showed significantly higher lesion scores than control, Kp, and S-CoV-2 (Figure 5E).
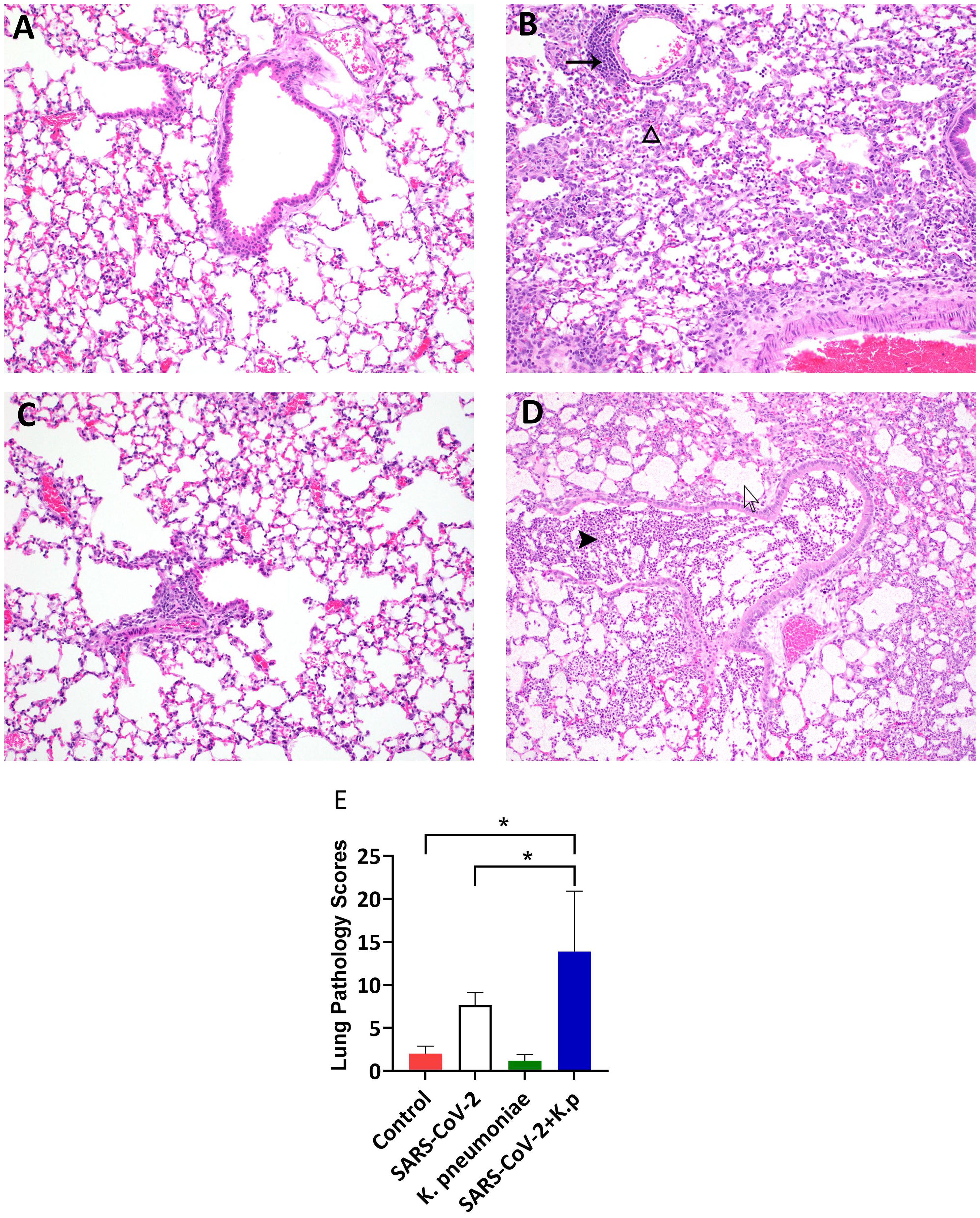
Figure 5. Lung pathology in co-infected mice: Photomicrographs of Lungs (A: Control, B: S-CoV-2, C: Kp, and D: S-CoV-2 +Kp at day 6 post-infection demonstrating lung pathology (Hematoxylin and Eosin stain). (B) black arrow indicates perivascular cuffing and blank arrowhead indicates interstitial pneumonia. (D) black arrowhead indicates neutrophil and blank arrow indicates intralesional bacteria. (E) Average of total lung pathology scores from two board-certified pathologists, n = 3-8 mice per group. *P value < 0.05 based on an ANOVA with a Tukey-Kramer mean separation, S-CoV-2 vs S-CoV-2 + Kp, Control vs S-CoV-2 + Kp.
Discussion
Respiratory co-infections pose a significant risk of exacerbating disease processes, primarily due to the overwhelming burden placed on the immune system by multiple pathogens. Each pathogen elicits a unique immune response, and the dysregulation that occurs in co-infections can impair the body’s ability to mount an effective defense against both viral and bacterial invaders. This further complicates accurate diagnosis and treatment (30), as the clinical manifestations of co-infections can be intricate and challenging to interpret. Therefore, gaining insight into the influence of co-infections on disease progression becomes crucial in guiding appropriate diagnostic and therapeutic approaches. Given the prevalence and severity of secondary bacterial infection during COVID-19, there is a growing need to understand the pathogenesis and impact of co-infections. In this study, we aimed to investigate the pathogenesis of co-infection with SARS-CoV-2 and Kp using a mouse model. We chose Kp for coinfection studies based on its prevalence in multiple hospital reports (19, 20, 31) and our patient cohort (Figure 1) as well as its potential for drug resistance (18). We discovered that sublethal doses of Kp and SARS-CoV-2 can be lethal to mice when co-infected (SARS-CoV-2 followed by Kp). The mice exhibited severe clinical signs and weight loss along with increased mortality after 2-4 days of Kp infection. Histopathology demonstrated severe pneumonia along with a large influx of neutrophils, macrophages, and innumerable bacteria. Flow cytometric analysis also demonstrated a significant increase in the neutrophil and inflammatory monocytes/macrophage population.
In the case of COVID-19, coinfection with other respiratory viruses or bacteria can increase the risk of respiratory failure and death (17). Furthermore, co-infection with SARS-CoV-2 and other viruses may also result in changes in the way the body responds to treatment. For example, studies have suggested that influenza virus co-infection may interfere with the effectiveness of SARS-CoV-2 antiviral medications (32).
Gram-positive bacteria such as Streptococcus pneumoniae (Sp) are commonly found in COVID-19 patients (33). Similarly, gram-negative bacteria were detected in 64% of COVID-19 patients with pulmonary bacterial infection (26), which highlights their potential to cause secondary bacterial pneumonia. K. pneumoniae is a gram-negative bacterium capable of causing various diseases, such as pneumonia, urinary tract infections, and sepsis. It is often found in healthcare settings where it can be transmitted through contact with contaminated surfaces or medical equipment. Coinfection with both SARS-CoV-2 and Kp has been shown to cause severe illness (15, 19, 20, 31). The emergence of drug-resistant Carbapenemase-producing Kp is a growing concern, with a high prevalence (34%) in intensive care unit patients in Italian hospitals (19). Additionally, there have been reports of patients hospitalized for SARS-CoV-2 infection who subsequently developed hypervirulent Kp strain infection, resulting in fatal outcomes (20, 34). The global spread of antimicrobial resistance Kp (20, 35) and frequent reports of hospital-acquired infection (36, 37), underscores the need to study Kp pathogenesis in the context of COVID-19.
Mouse models of co-infection are key to understand the complex interactions between viruses, bacteria, and host innate and adaptive immunity. Several pathogens, such as influenza virus and respiratory syncytial virus (38), Mycobacteria (39), Human immunodeficiency virus (40), and Streptococcus pneumoniae (41), have been studied for co-infection with SARS-CoV-2. However, there are few animal models developed to understand the pathogenesis of SARS-CoV-2 and coinfections (24, 25, 42, 43). Recent co-infection studies demonstrated increased susceptibility and lethality due to Sp and SARS-CoV-2 co-infection (24, 25). In these studies, mice showed reduced survival rates due to increased bacterial loads in the lungs. Notably, mortality was observed regardless of whether the SARS-CoV-2 challenge occurred before or after Sp infection (24). The increased bacterial levels after co-infection were associated with reduced alveolar macrophages, making co-infection more severe than either infection alone (24). Neutrophil and macrophage/monocyte dysfunction have been reported in SARS-CoV-2 infections (44). These cells are at the forefront of an antibacterial immune response; thus, dysfunctional phenotype in these cells predisposes COVID-19 patients to secondary bacterial infections. Neutrophils and monocytes isolated from critically ill SARS-CoV-2 patients failed to clear Sp and Sa (45), demonstrating impairment of antibacterial function (46). These findings suggest that viral infection can impair antibacterial defense and enhance bacterial load, and this effect seems to be independent of whether the bacteria are gram-positive or gram-negative (24, 25). Hence, we anticipate that the dysfunctional phenotype of inflammatory cells may also contribute to the pathogenesis of SARS-CoV-2 and Kp coinfection, necessitating further investigations to elucidate the underlying mechanistic basis.
Respiratory viruses can trigger a cytokine storm, which can result in damage to the alveoli increasing leakage of fluid from the vessel (47) as well as systemic effects of inflammation and damage to other organs (48). In severe cases of SARS-CoV-2 pneumonia, 42% patients had ARDS, leading to hypoxia (49). Individuals with severe COVID-19 demonstrate interstitial pneumonia with alveolar damage and hyaline membrane formation (50). We observed rubbery firm lungs (gross pathology) with interstitial pneumonia, lung consolidation, infiltration of mononuclear cells, perivascular cuffing, and thickened alveolar septa in the lungs of SARS-CoV-2-infected mice. These lesions were consistent with other mouse models of SARS-CoV-2 infection (23, 51) indicating that our co-infection model largely mimicked the lung pathology associated with COVID-19. The absence of typical ARDS-like changes in the SARS-CoV-2-only group may be attributed to two factors: firstly, mice seldom exhibit hyaline membrane formation due to the relatively shorter duration of the disease; secondly, this represents a sublethal challenge, with the morphological features of the disease varying based on the severity and duration of the infection. Nevertheless, in the case of co-infection with Kp, the pathological alterations became notably severe with the lungs exhibiting a substantial presence of inflammatory cells, along with an abundance of fibrin, numerous bacteria, and necrosis.
In summary, we have developed a mouse model of co-infection with SARS-CoV-2 and Kp and our study provides insights into the disease pathogenesis. The findings highlight the significance of monocyte and neutrophil dysfunction in secondary bacterial infections, leading to increased lung pathology and mortality. Strategies aimed at restoring the antibacterial activity of these cells may hold promise in preventing clinical complications associated with secondary bacterial infections in COVID-19 patients. Further research is needed to uncover the underlying mechanisms of COVID-19 severity following co-infection and develop targeted therapeutic approaches to mitigate the impact of secondary bacterial infections in COVID-19 patients.
Experimental procedures
Respiratory pathogens PCR array
Deidentified and leftover SARS-CoV-2 positive nasopharyngeal swabs (n = 50) collected from human patients (IRB-20-357-STW) received at the Oklahoma Animal Disease Diagnostic Laboratory (OADDL) for COVID-19 testing between April 2020 – July 2020 were used in this study. Total RNA was extracted from these patient samples using the KingFisher Flex platform (Thermo Fisher Scientific, MA) and a commercially available kit (MagMax viral/pathogen nucleic acid isolation kit; Thermo Fisher Scientific) following the FDA Emergency Use Authorization protocol provided by the manufacturer. A commercially available multiplex real-time PCR-based kit (Fast Track Diagnostics, Sliema, Malta) was used for detecting other respiratory pathogens. The list of pathogens includes influenza A virus; influenza B virus; influenza C virus; influenza A (H1N1) virus (swine-lineage); human parainfluenza viruses 1, 2, 3, and 4; human coronaviruses NL63, 229E, OC43, and HKU1; human metapneumoviruses A/B; human rhinovirus; human respiratory syncytial viruses A/B (HRSV); human adenovirus; enterovirus; human parechovirus; human bocavirus; Pneumocystis jirovecii; Mycoplasma pneumoniae; Chlamydophila pneumoniae; Streptococcus pneumoniae; Haemophilus influenzae B; Staphylococcus aureus; Moraxella catarrhalis; Bordetella spp.; Klebsiella pneumoniae; Legionella pneumophila/longbeachae; Salmonella spp. The PCR results were interpreted as detected or non-detected.
Bacteria
K. pneumoniae subspecies pneumoniae (Schroter) Trevisan (Cat #8045 ATCC, Manassas, VA) was used for mice infection studies. Bacteria were grown in nutrient broth (BD, Franklin Lakes, NJ) to an exponential growth curve (OD600 = 0.6). The bacterial stock was diluted in phosphate-buffered saline (PBS) pH 7.4 (Gibco, Evansville, IN) and prepared to a final range of 100-200 colony-forming units (CFU) per mouse (in 50 µl) and transported on ice until inoculation. After mice infection, 50 μL of remaining stock was plated on nutrient agar (NA) (BD, Franklin Lakes, NJ) and grown at 37°C overnight to obtain an estimate of bacteria delivered to each mouse.
Virus
Mouse-adapted (MA) SARS-CoV-2 strain obtained from Dr. Stanley Perlman (University of Iowa) was used in this study. The virus was propagated in Vero E6 TMPRSS2 T2A ACE2 cells (Cat #NR-54970 BEI, Manassas, VA) which were maintained in Dulbecco’s Modified Eagle’s Medium supplemented with 10% fetal bovine serum (GIBCO, Evansville, IN) and 10 µg per mL puromycin (InviovGen, San Diego, CA) at 37°C in a humidified 5% CO2 incubator. The infected cells were clarified by centrifuging at 2000 rpm for 5 minutes. Serial dilutions were prepared from supernatants and 100 µl was added to the Vero E6 cell (ATCC, CRL-1586) monolayers for 1 hour with gentle agitation every 10 minutes. After an hour of incubation, the media was removed and replaced with 1 ml of overlay composed 2.4% Avicel (Du Pont), 2XDMEM (Millipore Corp., Burlington, MA), and DMEM with a 10% FBS (Gibco, Evansville, IN). The ratio of each component was 1:1:1. Cells were incubated at 37°C in a humidified 5% CO2 incubator for 3 days. After 3 days of incubation, Avicel overlay was removed, and cells were fixed with 10% formalin for 5 minutes and stained with 0.05% crystal violet to visualize plaques. All tests were performed in duplicate. The viral load in mouse lungs was estimated using a similar plaque assay. The left lung lobe was homogenized in 500 μL of Opti-MEM using a bead mill and centrifuged at 2000 rpm for 5 minutes. Ten-fold serial dilutions were used for plaque assay. To prevent the growth of Kp in the Vero E6 cells during plaque assay, 40 μg/mL of Gentamicin (GIBCO, Evansville, IN) was used during virus incubation and in overlay media.
Mice studies
Eight to 9-weeks-old female BALB/cJ mice were purchased from Jackson Laboratories (Jackson Research Laboratories, Bar Harbor, ME). Mice were housed in biosafety cages in an animal BSL-3 facility at Oklahoma State University. All the experiments were approved by the Oklahoma State University Institutional Animal Care and Use Committee (Protocol #20-68). Mice were divided into the following groups: control (PBS), S-CoV-2 (S-CoV-2), K. pneumoniae (Kp), and S-CoV-2+ K. pneumoniae (S-CoV-2 + Kp). On day 0 under isoflurane anesthesia, the S-CoV-2 and S-CoV-2 + Kp groups were intranasally inoculated with 250–1000 PFU of MA-SARSCoV-2, whereas the control and Kp groups were administered 50 μL PBS. Days post infection (dpi) were based on SARS-CoV-2 inoculation day. On 4 dpi, the Kp and S-CoV-2 + Kp groups were inoculated intranasally with 100–135 CFU of Kp under isoflurane anesthesia. Mice were weighed and scored daily for clinical signs for 12 dpi. Clinical score parameters were: 1 = normal skin and active in the cage before handling; 2 = ruffled fur and alert; 3 = hunched posture, skin tent and decreased resistance to handling; 4 = piloerection with severe skin tent, only moves when touched; 5 = failure to right itself, nonresponsive. Mice were humanely euthanized if they lost more than 25% of their starting weight or if they had a clinical score of 4 or more. Survival was defined as an animal not losing 25% of the starting body weight, sick, or dead. Animals assigned a day for sacrifice were included in the count if they fit the criteria. Necropsies were performed on sick mice and mice that were assigned for sample collection. The lung lobes were collected for histopathology, flow cytometry, and bacterial, and viral load analysis.
Determination of bacterial load in lungs and heart blood
Approximately 10 μL of heart blood was collected aseptically and plated directly on NA plates during necropsy. The plates were incubated at 37°C and 5% CO2 overnight. The left lung was homogenized using a bead mill (Fisher, Hampton, NH) in 500 μL of Opti-MEM (Gibco, Evansville, IN). Ten microliters (in triplicate) of 10-fold serially diluted lung homogenates were plated on NA plates. The plates were incubated overnight at 37°C and 5% CO2 and CFUs were counted.
Flow cytometry
The phenotypic profile of lung-infiltrating immune cells was analyzed in the left lung lobe. For this, lungs were treated with collagenase-D and DNAse1, and isolated cells were surface-immunolabeled for alveolar macrophages (AM) (CD45+ CD11b- CD11c+ SiglecF+), neutrophil (CD45+ CD11b+ Ly6Ghi), inflammatory monocyte (CD45+ CD11b+ Ly6chi), dendritic cell (CD45+ CD11c+ MHCII+), natural killer cell (CD45+ CD3− NKP46+), and T-cell markers, and analyzed by flow cytometry. For cell surface staining, lung cells were labeled with the following fluorochrome-conjugated monoclonal antibodies: PECy7 α-CD45 (clone: 30-F11); FITC α-Ly6G (clone: 1A8, BD Biosciences); PE/PerCp-Cy5.5 α-Ly6C (clone: AL-21 [BD Biosciences or clone: HK1.4); V450 α-CD11b (clone: M1/70); APC α-F4/80 (clone: BM8) (unless otherwise stated, all from eBioscience). The labeling of the cell surface and intracellular markers was performed as previously (52). All fluorochrome-conjugated antibodies were used at a final concentration of 1:200 (antibody: the FACS buffer), except for FITC-labeled antibodies used at 1:100 concentration.
Histopathology
The lung lobes were perfused with 200 µl of 10% formalin and stored for 72 hours to inactivate the virus. Lungs were then trimmed and processed for hematoxylin and eosin (H&E) staining. Histopathological lesions were scored by two American College of Veterinary Pathology Board-certified pathologists in a blinded fashion. One H&E section per mouse was used for analysis. The lesions categories scored included bronchiolitis, thrombosis, fibrin, necrosis, interstitial pneumonia, edema, and hemorrhages. The lesions were scored on a scale of 0-4; 0 = no lesion, 1 = minimum, 2 = mild, 3 = moderate, and 4 = severe. The sum of all lesion category scores for each mouse was used for data plotting and analysis.
Data analysis
The data for the body weight was collected over time, therefore, we employed a repeated measures design. Before comparing the significance of mean body weight percentages and clinical score changes for each treatment group using ANOVA (Analysis of variance), we carefully examined the variance-covariance structure of the dataset. An autoregressive variance-covariance structure was recommended as it yielded smaller AIC (Akaike Information Criterion), AICC (Corrected AIC for smaller samples), BIC (Bayesian Information Criterion), and other indicator values. The Tukey-Kramer mean separation technique was used to determine the significant differences in body weight and Kruskal-Wallis test for clinical score changes. Analyses were performed using SAS 9.4 (Cary, NC) and GraphPad Prism 9 (San Diego, CA). Respective statistical tests are mentioned in the corresponding figure legends.
Data availability statement
The original contributions presented in the study are included in the article/supplementary material. Further inquiries can be directed to the corresponding authors.
Ethics statement
The studies involving human samples were approved by Oklahoma State University Institutional Review Board and was exempted. The human samples used in this study were acquired from COVID-19 diagnostic testing conducted at the Oklahoma Animal Diseases Diagnostic Laboratory. The samples were de-identified. Written informed consent for participation was not required from the participants or the participants’ legal guardians/next of kin in accordance with the national legislation and institutional requirements. The animal study was approved by Oklahoma State University Institutional Animal Care and Use Committee. The study was conducted in accordance with the local legislation and institutional requirements (Institutional Biosafety Committee).
Author contributions
CV: Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Software, Validation, Writing – original draft, Writing – review & editing. GP: Investigation, Methodology, Writing – review & editing. SN: Investigation, Methodology, Writing – review & editing. RG: Investigation, Methodology, Writing – review & editing. DC: Investigation, Methodology, Writing – review & editing. TS: Investigation, Writing – review & editing. AR: Investigation, Writing – review & editing, Conceptualization, Methodology. RC: Conceptualization, Investigation, Methodology, Writing – review & editing, Formal analysis. SM: Conceptualization, Formal analysis, Investigation, Methodology, Writing – review & editing, Data curation, Funding acquisition, Project administration, Resources, Software, Supervision, Validation, Visualization, Writing – original draft. NS: Data curation, Formal analysis, Methodology, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. Research reported in this publication was supported by the National Institute Of General Medical Sciences of the National Institutes of Health under Award Number P20GM103648. In addition, start-up funds from the College of Veterinary Medicine at Oklahoma State University were used for this study. The content is solely the authors’ responsibility and does not necessarily represent the official views of the National Institutes of Health.
Acknowledgments
The following reagent was obtained through BEI Resources, NIAID, NIH: Cercopithecus aethiops Kidney Epithelial Cells Expressing Transmembrane Protease, Serine 2 and Human Angiotensin-Converting Enzyme 2 (Vero E6-TMPRSS2-T2A-ACE2), cat no. NR-54970. We also thank Dr. Dr. Stanley Perlman (University of Iowa) for providing Mouse-adapted SARS-CoV-2 strain.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. WHO COVID-19 dashboard. (2024). Available online at: https://data.who.int/dashboards/covid19/cases (accessed September 1, 2024).
2. Couture A, Iuliano AD, Chang HH, Patel NN, Gilmer M, Steele M, et al. Estimating COVID-19 hospitalizations in the United States with surveillance data using a bayesian hierarchical model: modeling study. JMIR Public Health Surveill. (2022) 8:e34296. doi: 10.2196/34296
3. da Rosa Mesquita R, Francelino Silva Junior LC, Santos Santana FM, Farias de Oliveira T, Campos Alcântara R, Monteiro Arnozo G, et al. Clinical manifestations of COVID-19 in the general population: systematic review. Wien Klin Wochenschr. (2021) 133:377–82. doi: 10.1007/s00508-020-01760-4
4. Walker A, Pottinger G, Scott A, Hopkins C. Anosmia and loss of smell in the era of covid-19. Br Med J. (2020) 370:m2808. doi: 10.1136/bmj.m2808
5. Hasan SS, Capstick T, Ahmed R, Kow CS, Mazhar F, Merchant HA, et al. Mortality in COVID-19 patients with acute respiratory distress syndrome and corticosteroids use: a systematic review and meta-analysis. Expert Rev Respir Med. (2020) 14:1149–63. doi: 10.1080/17476348.2020.1804365
6. Mokhtari T, Hassani F, Ghaffari N, Ebrahimi B, Yarahmadi A, Hassanzadeh G. COVID-19 and multiorgan failure: A narrative review on potential mechanisms. J Mol Histol. (2020) 51:613–28. doi: 10.1007/s10735-020-09915-3
7. Morris DE, Cleary DW, Clarke SC. Secondary bacterial infections associated with influenza pandemics. Front Microbiol. (2017) 8:1041–1. doi: 10.3389/fmicb.2017.01041
8. Chong WH, Saha BK, Ramani A, Chopra A. State-of-the-art review of secondary pulmonary infections in patients with COVID-19 pneumonia. Infection. (2021) 49:591–605. doi: 10.1007/s15010-021-01602-z
9. Morens DM, Taubenberger JK, Fauci AS. Predominant role of bacterial pneumonia as a cause of death in pandemic influenza: implications for pandemic influenza preparedness. J Infect Dis. (2008) 198:962–70. doi: 10.1086/591708
10. Phetsouphanh C, Darley DR, Wilson DB, Howe A, Munier CML, Patel SK, et al. Immunological dysfunction persists for 8 months following initial mild-to-moderate SARS-CoV-2 infection. Nat Immunol. (2022) 23:210–6. doi: 10.1038/s41590-021-01113-x
11. Didierlaurent A, Goulding J, Patel S, Snelgrove R, Low L, Bebien M, et al. Sustained desensitization to bacterial Toll-like receptor ligands after resolution of respiratory influenza infection. J Exp Med. (2008) 205:323–9. doi: 10.1084/jem.20070891
12. Kash JC, Tumpey TM, Proll SC, Carter V, Perwitasari O, Thomas MJ, et al. Genomic analysis of increased host immune and cell death responses induced by 1918 influenza virus. Nature. (2006) 443:578–81. doi: 10.1038/nature05181
13. Denney L, Ho LP. The role of respiratory epithelium in host defence against influenza virus infection. BioMed J. (2018) 41:218–33. doi: 10.1016/j.bj.2018.08.004
14. Zhu X, Ge Y, Wu T, Zhao K, Chen Y, Wu B, et al. Co-infection with respiratory pathogens among COVID-2019 cases. Virus Res. (2020) 285:198005. doi: 10.1016/j.virusres.2020.198005
15. Sreenath K, Batra P, Vinayaraj EV, Bhatia R, SaiKiran K, Singh V, et al. Coinfections with other respiratory pathogens among patients with COVID-19. Microbiol Spectr. (2021) 9:e0016321. doi: 10.1128/Spectrum.00163-21
16. Luyt CE, Bouadma L, Morris AC, Dhanani JA, Kollef M, Lipman J, et al. Pulmonary infections complicating ARDS. Intensive Care Med. (2020) 46:2168–83. doi: 10.1007/s00134-020-06292-z
17. Kao K-C, Chiu LC, Hung CY, Chang CH, Yang CT, Huang CC, et al. Coinfection and mortality in pneumonia-related acute respiratory distress syndrome patients with bronchoalveolar lavage: A prospective observational study. Shock. (2017) 47:615–20. doi: 10.1097/SHK.0000000000000802
18. Effah CY, Sun T, Liu S, Wu Y. Klebsiella pneumoniae: an increasing threat to public health. Ann Clin Microbiol Antimicrob. (2020) 19:1. doi: 10.1186/s12941-019-0343-8
19. Arcari G, Raponi G, Sacco F, Bibbolino G, Di Lella FM, Alessandri F, et al. Klebsiella pneumoniae infections in COVID-19 patients: a 2-month retrospective analysis in an Italian hospital. Int J antimicrobial Agents. (2021) 57:106245–5. doi: 10.1016/j.ijantimicag.2020.106245
20. Hosoda T, Harada S, Okamoto K, Ishino S, Kaneko M, Suzuki M, et al. COVID-19 and fatal sepsis caused by hypervirulent klebsiella pneumoniae, Japan, 2020. Emerg Infect Dis. (2020) 27:556–9. doi: 10.3201/eid2702.204662
21. Said KB, Alsolami A, Moussa S, Alfouzan F, Bashir AI, Rashidi M, et al. COVID-19 clinical profiles and fatality rates in hospitalized patients reveal case aggravation and selective co-infection by limited gram-negative bacteria. Int J Environ Res Public Health. (2022) 19:5270. doi: 10.3390/ijerph19095270
22. Dinnon KH 3rd, Leist SR, Schäfer A, Edwards CE, Martinez DR, Montgomery SA, et al. A mouse-adapted model of SARS-CoV-2 to test COVID-19 countermeasures. Nature. (2020) 586:560–6. doi: 10.1038/s41586-020-2708-8
23. Wong LR, Zheng J, Wilhelmsen K, Li K, Ortiz ME, Schnicker NJ, et al. Eicosanoid signalling blockade protects middle-aged mice from severe COVID-19. Nature. (2022) 605:146–51. doi: 10.1038/s41586-022-04630-3
24. Barman TK, Singh AK, Bonin JL, Nafiz TN, Salmon SL, Metzger DW. Lethal synergy between SARS-CoV-2 and Streptococcus pneumoniae in hACE2 mice and protective efficacy of vaccination. JCI Insight. (2022) 7(11):e159422. doi: 10.1172/jci.insight.159422
25. Smith AP, Williams EP, Plunkett TR, Selvaraj M, Lane LC, Zalduondo L, et al. Time-dependent increase in susceptibility and severity of secondary bacterial infections during SARS-coV-2. Front Immunol. (2022) 13:894534. doi: 10.3389/fimmu.2022.894534
26. Yahya RO. Problems associated with co-infection by multidrug-resistant klebsiella pneumoniae in COVID-19 patients: A review. Multidisciplinary Digital Publishing Institute (2022) 10(12):2412. doi: 10.3390/healthcare10122412
27. Holliday ZM, Alnijoumi MM, Reed MA, Earhart AP, Schrum AG, Allen LH, et al. Neutrophils and secondary infections in COVID-19 induced acute respiratory distress syndrome. New Microbes New Infect. (2021) 44:100944. doi: 10.1016/j.nmni.2021.100944
28. Channappanavar R, Fehr AR, Vijay R, Mack M, Zhao J, Meyerholz DK, et al. Dysregulated type I interferon and inflammatory monocyte-macrophage responses cause lethal pneumonia in SARS-coV-infected mice. Cell Host Microbe. (2016) 19:181–93. doi: 10.1016/j.chom.2016.01.007
29. Channappanavar R, Fehr AR, Zheng J, Wohlford-Lenane C, Abrahante JE, Mack M, et al. IFN-I response timing relative to virus replication determines MERS coronavirus infection outcomes. J Clin Invest. (2019) 130:3625–39. doi: 10.1172/JCI126363
30. McArdle AJ, Turkova A, Cunnington AJ. When do co-infections matter? Curr Opin Infect Dis. (2018) 31:209–15. doi: 10.1097/QCO.0000000000000447
31. Lai C-C, Chen SY, Ko WC, Hsueh PR. Increased antimicrobial resistance during the COVID-19 pandemic. Int J antimicrobial Agents. (2021) 57:106324–4. doi: 10.1016/j.ijantimicag.2021.106324
32. Cheemarla NR, Mihaylova VT, Watkins TA, Foxman EF. Counterintuitive effect of antiviral therapy on influenza A-SARS-CoV-2 coinfection due to viral interference. bioRxiv. (2023). doi: 10.1101/2023.02.07.527372
33. Parker AM, Jackson N, Awasthi S, Kim H, Alwan T, Wyllie AL, et al. Association of upper respiratory streptococcus pneumoniae colonization with severe acute respiratory syndrome coronavirus 2 infection (SARS-coV-2) among adults. Clin Infect Dis. (2022) 1(1):166. doi: 10.1093/cid/ciac907
34. Falcone M, Tiseo G, Arcari G, Leonildi A, Giordano C, Tempini S, et al. Spread of hypervirulent multidrug-resistant ST147 Klebsiella pneumoniae in patients with severe COVID-19: an observational study from Italy, 2020-21. J Antimicrob Chemother. (2022) 77:1140–5. doi: 10.1093/jac/dkab495
35. Suay-García B, Pérez-Gracia MT. Present and future of carbapenem-resistant enterobacteriaceae (CRE) infections. Antibiot (Basel). (2019) 8:122. doi: 10.3390/antibiotics8030122
36. Hlophe ST, McKerrow NH. Hospital-acquired Klebsiella pneumoniae infections in a paediatric intensive care unit: research. SAJCH. (2014) 8(4):125–8. doi: 10.7196/sajch.747
37. Le T, Wang L, Zeng C, Fu L, Liu Z, Hu J. Clinical and microbiological characteristics of nosocomial, healthcare-associated, and community-acquired Klebsiella pneumoniae infections in Guangzhou, China. Antimicrob Resist Infect Control. (2021) 10:41. doi: 10.1186/s13756-021-00910-1
38. Swets MC, Russell CD, Harrison EM, Docherty AB, Lone N, Girvan M, et al. SARS-CoV-2 co-infection with influenza viruses, respiratory syncytial virus, or adenoviruses. Lancet. (2022) 399:1463–4. doi: 10.1016/S0140-6736(22)00383-X
39. Shah T, Shah Z, Yasmeen N, Baloch Z, Xia X. Pathogenesis of SARS-coV-2 and mycobacterium tuberculosis coinfection. Front Immunol. (2022) 13:909011. doi: 10.3389/fimmu.2022.909011
40. Nagarakanti SR, Okoh AK, Grinberg S, Bishburg E. Clinical outcomes of patients with COVID-19 and HIV coinfection. J Med Virol. (2021) 93:1687–93. doi: 10.1002/jmv.26533
41. Pal C, Przydzial P, Chika-Nwosuh O, Shah S, Patel P, Madan N. Streptococcus pneumoniae coinfection in COVID-19: A series of three cases. Case Rep Pulmonol. (2020) 2020:8849068. doi: 10.1155/2020/8849068
42. Hildebrand RE, Chandrasekar SS, Riel M, Touray BJB, Aschenbroich SA, Talaat AM. Superinfection with SARS-CoV-2 Has Deleterious Effects on Mycobacterium bovis BCG Immunity and Promotes Dissemination of Mycobacterium tuberculosis. Microbiol Spectr. (2022) 10(5):e03075–22. doi: 10.1128/spectrum.03075-22
43. Rosas Mejia O, Gloag ES, Li J, Ruane-Foster M, Claeys TA, Farkas D, et al. Mice infected with Mycobacterium tuberculosis are resistant to acute disease caused by secondary infection with SARS-CoV-2. PloS Pathog. (2022) 18:e1010093. doi: 10.1371/journal.ppat.1010093
44. Mairpady Shambat S, Gómez-Mejia A, Schweizer TA, Huemer M, Chang CC, Acevedo C, et al. Hyperinflammatory environment drives dysfunctional myeloid cell effector response to bacterial challenge in COVID-19. PloS Pathog. (2022) 18:e1010176. doi: 10.1371/journal.ppat.1010176
45. Mairpady Shambat S, Gómez-Mejia A, Schweizer TA, Huemer M, Chang C-C, Acevedo C, et al. Hyperinflammatory environment drives dysfunctional myeloid cell effector response to bacterial challenge in COVID-19. PLoS Pathog. (2022) 18(1):e1010176. doi: 10.1371/journal.ppat.1010176
46. Mirzaei R, Goodarzi P, Asadi M, Soltani A, Aljanabi HAA, Jeda AS, et al. Bacterial co-infections with SARS-coV-2. IUBMB Life. (2020) 72(10):2097–111. doi: 10.1002/iub.2356
47. Fajgenbaum DC, June CH. Cytokine storm. N Engl J Med. (2020) 383(23):2255–73. doi: 10.1056/NEJMra2026131
48. Kopańska M, Barnaś E, Błajda J, Kuduk B, Łagowska A, Banaś-Ząbczyk A. Effects of SARS-coV-2 inflammation on selected organ systems of the human body. Int J Mol Sci. (2022) 23:4178. doi: 10.3390/ijms23084178
49. Gibson PG, Qin L, Puah SH. COVID-19 acute respiratory distress syndrome (ARDS): clinical features and differences from typical pre-COVID-19 ARDS. Med J Aust. (2020) 213:54–56.e1. doi: 10.5694/mja2.50674
50. Hammoud H, Bendari A, Bendari T, Bougmiza I. Histopathological findings in COVID-19 cases: A systematic review. Cureus. (2022) 14:e25573. doi: 10.7759/cureus.25573
51. Leist SR, Dinnon KH 3rd, Schäfer A, Tse LV, Okuda K, Hou YJ, et al. A mouse-adapted SARS-coV-2 induces acute lung injury and mortality in standard laboratory mice. Cell. (2020) 183:1070–1085.e12. doi: 10.1016/j.cell.2020.09.050
Keywords: SARS-CoV-2, Klebsiella pneumoniae, co-infection, COVID-19, secondary bacterial infection of viral respiratory disease
Citation: Villalva C, Patil G, Narayanan SS, Ghimire R, Chanda D, Samarakoon N, Snider T, Ramachandran A, Channappanavar R and More S (2024) Klebsiella pneumoniae co-infection leads to fatal pneumonia in SARS-CoV-2-infected mice. Front. Virol. 4:1426728. doi: 10.3389/fviro.2024.1426728
Received: 02 May 2024; Accepted: 16 September 2024;
Published: 18 October 2024.
Edited by:
Antonio Caruz, University of Jaén, SpainReviewed by:
Wibke Bayer, Essen University Hospital, GermanyVeronica Ueckermann, University of Pretoria, South Africa
Copyright © 2024 Villalva, Patil, Narayanan, Ghimire, Chanda, Samarakoon, Snider, Ramachandran, Channappanavar and More. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sunil More, c3VuaWwubW9yZUBva3N0YXRlLmVkdQ==; Rudragouda Channappanavar, cmNoYW5uYUBva3N0YXRlLmVkdQ==
 Crystal Villalva1,2
Crystal Villalva1,2 Girish Patil
Girish Patil Sai Sankara Narayanan
Sai Sankara Narayanan Roshan Ghimire
Roshan Ghimire Debarati Chanda
Debarati Chanda Akhilesh Ramachandran
Akhilesh Ramachandran Rudragouda Channappanavar
Rudragouda Channappanavar Sunil More
Sunil More