
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
MINI REVIEW article
Front. Virol., 03 June 2024
Sec. Fundamental Virology
Volume 4 - 2024 | https://doi.org/10.3389/fviro.2024.1386580
This article is part of the Research TopicInteraction between virus and host cell: How do viruses cause cellular dysregulation?View all 5 articles
Chikungunya virus (CHIKV) is a vector-born alphavirus responsible for chikungunya fever with clinical manifestation of polyarthritis transmitted by Aedes aegypti and Aedes albopictus. Establishing viral pathogenesis needs host machinery modulation, and the microRNAs (miRNA) modulate host cellular machinery to establish the infection or inhibit viral replication. miRNAs are the small noncoding RNA that control the gene expression. They are essential in cell differentiation, growth, development, and apoptosis. It also affects disease progression, cancer, and viral infection. CHIKV infection causes differential expression of miRNA, and miRNA has target genes involved in different cellular functions. These target genes may be crucial in CHIKV replication and cell growth. Suppression or overexpression of these miRNAs may have been linked with CHIKV pathogenesis by regulating immune and signaling pathways. Identification of biomarkers in disease progression through the study of circulating miRNAs during CHIKV infection is an emerging field. Therefore, understanding miRNAs’ differential expression and function during CHIKV infection is essential. The detailed studies on the miRNA-mediated regulatory network will provide new ways to develop miRNA-based therapies.
Chikungunya virus (CHIKV) is an arthropod-borne virus belonging to the family Togaviridae and the genus Alphavirus (1, 2). The CHIKV genome comprises a single-stranded, positive-sense RNA molecule with a length of ~11.8 kb, and the genome encodes for two Open Reading Frames (ORFs). The precursor polyproteins of the non-structural proteins (nsP1 to nsP4) are encoded in the first ORF at the 5′ end (1). The second ORF is located at the 3′ end, encoding the structural proteins: capsid (C), envelope proteins (E1, E2, E3), and 6K (3). The envelope glycoproteins E1 and E2 have a role in membrane fusion and viral entry (4, 5).
CHIKV is a disease primarily characterized by the onset of fever, arthralgia, nausea, vomiting, rash, and myalgia. Although the symptoms subside within a week, certain patients may experience prolonged joint pain (6). Patients with comorbidities, specific genetic disorders, and elderly individuals are more susceptible to developing acute disease conditions with a higher mortality rate (7).
The primary vectors of CHIKV transmission are Ae. aegypti. And Ae. Albopictus (8, 9). According to the phylogenetic analysis, based on geographic origin, three different genotypes emerged from the CHIKV African lineage, i.e., East/Central/South African (ECSA), West African (WA), and Asian. The East/Central/South African (ECSA) further diverged into the sublineage Indian Ocean lineage (IOL) (10, 11).
In the model organism Caenorhabditis elegans, Lee and colleagues first discovered the miRNA lin-4 in 1993 (12). miRNAs are small, noncoding RNAs essential in regulating gene expression by binding to the specific RNA post-transcriptionally (13). The miRNA targeting depends on the base pairing between seed regions, which is 2–7 nucleotides at the 5′ end of mature miRNA with 3′ untranslated regions (UTRs) of target mRNAs (14, 15). RNA polymerase is involved in the transcription of miRNA genes to produce the primary miRNA (pri-miRNA) (16). Pri-miRNA has 80-nt hairpin structures with a large terminal loop and 32 bp imperfect stem, recognized by Dgcr8, RNA binding protein, and binds to GGAC and other pri-miRNA motifs (17). Drosha cleaves the primary miRNA, producing a precursor miRNA (pre-miRNA) of ∼a 70-nt hairpin molecule with a 3’ overhang (18). The exportin-5 transports pre-miRNAs produced in the nucleus into the cytoplasm by a Ran-GTP-dependent mechanism (19). In cytoplasm Dicer, a second RNase II enzyme with TRBP removes the terminal loop from the pre-miRNA to produce the mature double-stranded ~22 bp miRNA intermediates with two nucleotide overhangs at each 3′ end (20). One strand (Guide strand) from the duplex RNA is introduced into the RNA-induced silencing complex (RISC) complex, having four different Argonaute proteins (AgoI- VI), of which Ago II protein has endonuclease activity and the ability to break bound target mRNA. Further, depending on miRNA and mRNA base pairing, it will either lead to translation inhibition or mRNA degradation. Meanwhile, another strand (passenger strand) is released and further degraded (21). (Supplementary Figure).
miRNAs have a role in various cellular processes such as cell differentiation, cell proliferation, signal transduction pathway, lipid metabolism, and apoptosis (22–24). miRNAs also play a crucial role in viral infection and pathogenesis (25). However, viruses can modify host miRNAs to escape the immune response. This review mainly focuses on the role of miRNAs in CHIKV life cycle.
Mosquito’s innate immune system can recognize various microorganisms and mount a strong immune response against them (26). They recognize foreign particles and then initiate phagocytosis, produce antimicrobial peptides, produce melanin to trap pathogens, form nodules, and promote wound healing (27). RNA interference is a defense strategy used by mosquitoes against viruses (28). miRNAs are involved in the posttranscriptional level gene regulation and might also be involved in the regulation of insect immune response (29). Previous studies have suggested that miRNAs may play an important role in regulating and establishing infection of arboviruses and Wolbachia (an endosymbiotic bacteria) in mosquitoes (30). However, there is limited knowledge regarding the specific targets of these miRNAs.
In a study by Shrinet et al., 2014, it was observed that following the infection of CHIKV, there were significant changes in signaling and metabolic pathways, including protein synthesis in the endoplasmic reticulum and the immunity pathways. KEGG and KOBAS analysis also indicated that altered miRNA expression may regulate the cellular pathways. The authors further evaluated the role of vector miRNA in CHIKV pathogenesis. Different miRNAs, namely aae-miR-1000, aae-miR-190–5p, aae-miR-2b, and aae-miR-2c were downregulated in CHIKV-infected Ae. albopictus Singh’s cell line. Among these miRNAs, aae-miR-1000, aae-miR-2b, and aae-miR-2c have been suggested to be involved in ribosome biogenesis. Similarly, aae-miR-927–5p, aae-miR-305–3p, aae-miR-283, and aae-miR-100 were upregulated in Ae. albopictus Singh’s cell line upon CHIKV infection. The miRNA target prediction analysis revealed that aae-miR-305–3p, aae-miR-100, and aae-miR-283 target protein processing pathways and cell-mediated cytotoxicity. The aae-miR-305–3p and aae-miR-927 play a role in the vesicular transport of the SNARE (Soluble N-ethylmaleimide-sensitive factor activating protein receptor) protein interaction. (Supplementary Table 1) Also, the aae-miR-305–3p plays a role in virus ECM (extracellular matrix) receptor-mediated interaction and endocytosis pathways (31). Similar to CHIKV, differential expression of aae-miR-927 was observed in Dengue viruses (DENV) -infected C6/36 cells (32). Azlan et al., 2022, showed that aae-miR-927 acts as a proviral factor in DENV1-infected C6/36. Further, it was found that the aae-miR-927 targets filamin (FLN), which is part of the Toll signaling pathway (33). (Supplementary Table 1) Certain miRNAs, such as aae-miR-927, may be involved in the pathogenesis of multiple arboviruses.
In contrast, Dubey et al., 2017 reported that in CHIKV-infected Aag-2 (Ae. aegypti cell line), the expression of aae-miR-2b was upregulated. aae-miR-2b has two target genes: URMs (ubiquitin-like modifier) and ubiquitine. (Supplementary Table 1) The expression of these URM was significantly controlled by aae-miR-2b in Aag 2 (Ae. aegypti cell line), affecting CHIKV replication (34). Studies have demonstrated URM’s role in the thiolation of some transfer RNA (tRNA), and some viruses have developed diverse strategies to optimize tRNA utilization by using host tRNA for translation of viral proteins, packaging of virions, and priming reverse transcription reaction of their genome or some viruses encode their own tRNA (35). CHIKV infected Ae. aegypti showed enhanced levels of aae-miR-2b, leading to the downregulation of the URM, which controls the CHIKV replication in the vector (34) (Figure 1).
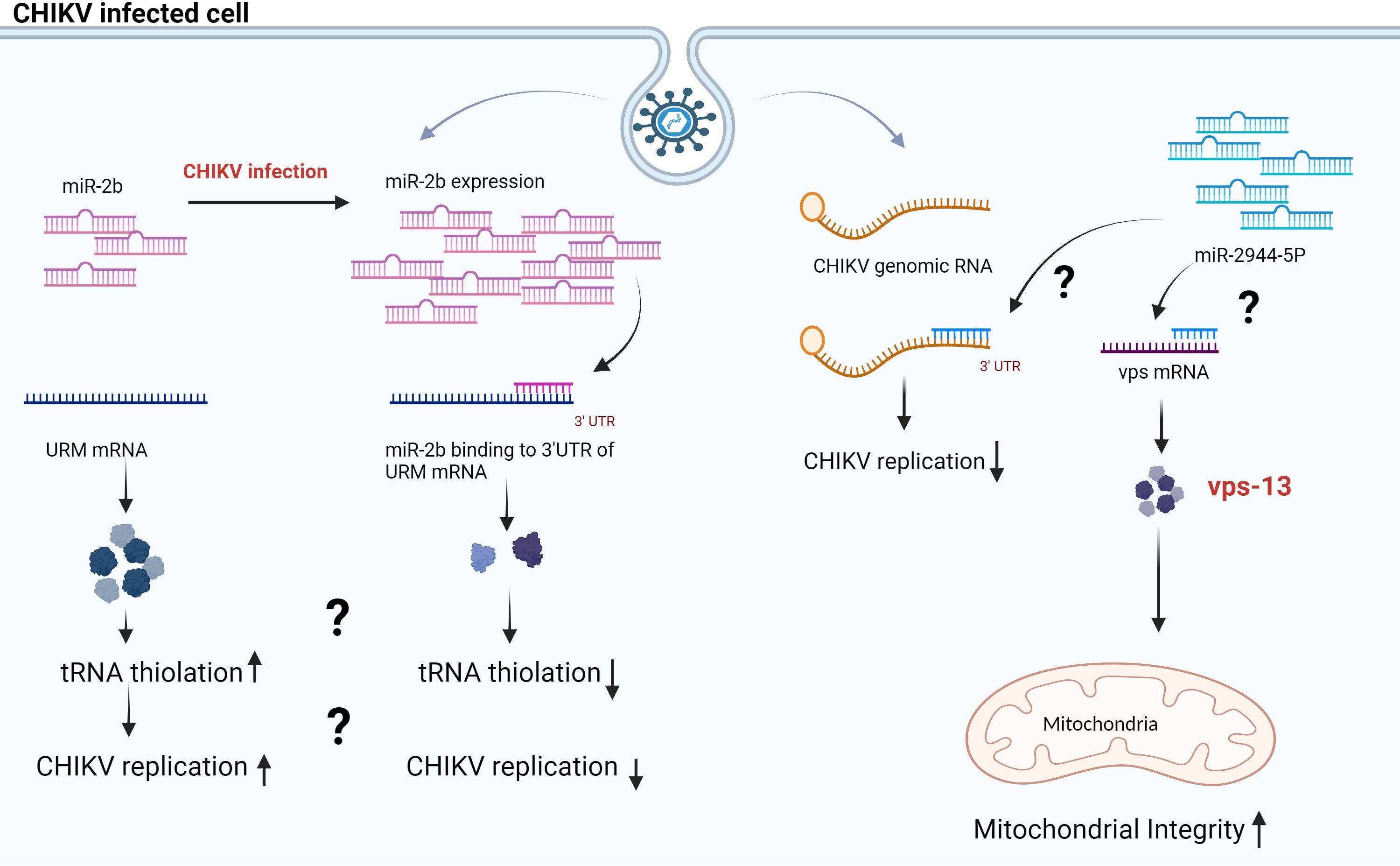
Figure 1 Proposed model for miR-2b and miR-2944–5b during CHIKV infection in Aag 2 cells. The scheme illustrates that miR-2b and miR-2944–5b control the CHIKV replication in the Aag-2 cell line by binding at the 3’UTR of CHIKV RNA. The miR-2b and miR-2944b-5p have target URM and vps mRNA, respectively. URM has a role in tRNA thiolation, and vps maintains the membrane potential in mitochondria (34, 36).
In a study by Dubey et al., 2019, the authors studied the interactions of aae-miR-2944b-5p with CHIKV 3’ UTR. In addition, this miRNA targets cellular factor vacuolar protein sorting-13 (vps-13). (Supplementary Table 1) Additionally, the cellular factor vps-13 contributes to balancing mitochondrial membrane potential in Ae. aegypti during CHIKV infection (36). The authors demonstrated that replication of CHIKV decreased in Aag2 cells (Ae. Aegypti cell line) and Ae. aegypti mosquitoes when vps-13 was silenced. Conversely, CHIKV replication increased in Aag2 cells when aae-miR-2944b-5b was silenced with anti-miR-2944–5p (36). In summary, research indicates that both aae-miR-2944b-5b and vsp-13 play crucial roles in the CHIKV lifecycle in insect hosts. Earlier studies have demonstrated that viruses like herpes simplex virus 1 (HSV-1) deplete the host’s mitochondrial DNA, while others, such as the human immunodeficiency virus (HIV), manipulate the host’s mitochondrial proteins to function entirely inside the host cell (37). The aae-miR-2944b-5p is restricted to insects and absent in mammals. The study shows that, by binding to the 3’UTR of CHIKV, aae-miR-2944b-5p downregulates CHIKV replication in mosquitoes. Also, it could maintain mitochondrial membrane potential during CHIKV infection by targeting cellular factor vps-13 (36). (Figure 1) This study indicates that the mosquito cells may possess defense mechanisms that regulate viral proliferation, or the virus may exert control over the mosquito cells to ensure its survival within the cells. Thus, further studies are warranted to understand the detailed function and interactions of aae-miR-2b and aae-miR-2944b-5p. In summary, the findings suggest that manipulating the activity of specific miRNAs can be an effective method to restrict the transmission of arboviruses.
The saliva of mosquitoes consists of various proteins and enzymes that play roles in the modulation of blood coagulation, inflammation, platelet aggregation, and vascular constriction (38). It creates a favorable microenvironment for virus establishment and disease progression. Exosomes are the extracellular vesicles that are responsible for the transportation of miRNA in saliva and serum (39). A study by Fiorillo et al., 2022, demonstrated that certain miRNAs are enriched in the saliva and midgut of Ae. Aegypti (38). In another study by Maharaj et al., 2014, the authors identified 103 miRNAs in mosquito saliva using the next-generation sequencing platform. Several of these miRNAs were specifically expressed in the context of CHIKV infection. Authors used synthetic miRNA inhibitors to study the effect of inhibition of specific miRNA. They chose five distinct miRNAs (aae-miR-12, aae-miR-125, aae-miR-184, aae-miR-375, and aae-miR-2492) based on their relative abundance levels in CHIKV-infected saliva and prior studies. Aag-2 cells were treated with miRNA inhibitors, and the replication of CHIKV was assessed in these cells. Suppression of all five miRNAs exhibited declined CHIKV titer in Aag-2 cells. Notably, the suppression of aae-miR-184, aae-miR-375, and aae-miR-2492 resulted in a reduction in CHIKV titer in both Aag-2 (insect cells) and BHK-21 cells (Mammalian cells) (40). It is suggested that mosquito salivary miRNAs may potentially influence CHIKV pathogenesis across different species.
CHIKV mainly infects peripheral tissue but also affects the central nervous system in newborns and adults (41). The peripheral tissues are the first site of the virus encounter and the location of different immune cells (42). CHIKV initially replicates in the skin and fibroblast cells and then circulates via blood to the liver and joints (43). CHIKV infection leads to the activation of various signaling pathways in host cells (44–46). miRNAs are known to be involved in the regulation of viral infection, inflammation, and immune response (47). To date, the function of miRNA in CHIKV infection remains unclear.
Sharma et al., 2015, showed significant upregulation of hsa-miR-409–3p in CHIKV-infected fibroblast cells, and it was predicted to target topoisomerase IIβ (48) (Supplementary Table 2). Topoisomerase IIβ is a DNA gyrase that has an essential role in the transcription and replication of DNA (49, 50).
Saxena et al., 2013, revealed that the cluster of hsa-miR-15b/16, hsa-miR-17–92, hsa-miR-23a/24, and hsa-miR-106b/miR25 were found to be upregulated in CHIKV-infected Human Embryonic Kidney 293 (HEK293T) cells (51). The miRNA clusters hsa-miR-17–92, hsa-miR-23a, and hsa-miR-106b regulate the TGF-b (JUN, SMAD6) signaling pathway (52, 53). TGF-b, endocytosis, and adherens junction pathways were upregulated during CHIKV infection, while the cell cycle, proteasome, and lysosome pathways were significantly downregulated (51). The hsa-miR-15/16 cluster targets BCL2 and triggers apoptosis (54). (Supplementary Table 2) hsa-miR-15 and hsa-miR-16 act as regulators in Rheumatic arthritis (RA). The hsa-miR-15 was shown to be downregulated in arthritic synovial tissue, whereas hsa-miR-16 was upregulated in synovial fluid of RA patients (55, 56). Study showed that the apoptosis pathway and CHIKV-engulfed macrophages are the main contributors to prolonged arthritis (46). Studying the role of these miRNAs in CHIKV infection can aid in developing biomarkers to predict arthritis or treatments for CHIKV infection.
Agrawal et al., 2020 studied the differential regulation of miRNA in CHIKV-infected primary human synovial fibroblast cells and observed that upregulation of hsa‐miR‐1264, hsa‐miR‐4717‐3p, hsa‐miR‐ 4299, and hsa‐miR‐21. The miRNA target prediction analysis indicates that hsa‐miR‐4717‐3p targets the AKT3 (57). (Supplementary Table 2) Many viruses exploit PI3K/Akt/mTOR for survival in host cells (58). Reports indicate that CHIKV infection relies on the PI3K/Akt/mTOR pathway for its replication and survival in the Vero cells. (Origin: African green monkey kidney epithelial cells) (44). The PI3K/Akt/mTOR pathway plays a vital role in protein translation, metabolism, survival, and cell growth. AKT is a crucial signaling molecule for these pathways (59). It has a role in developing T cells and regulates transcription factors NF-κB and FOXO3, which are responsible for T cell activation (60, 61). During CHIKV infection, AKT activation leads to the mTOR’s phosphorylation, which activates S6K and 4EBP1, which is responsible for the translation of the viral and host mRNAs (44). The hsa-miR-21 also targets the AKT, PTEN, and PELI1 (62). PELI is an E3 ubiquitin ligase that regulates innate immunity by ubiquitylation of IRAK1 (63). Also, the TRIM and SOCS 7 are potential targets of hsa-miR-1264 and hsa-miR-4299, respectively (57). (Supplementary Table 2) TRIM is an E3 ubiquitin ligase that targets the transcription factor IRF3, which is mainly involved in IFN- β production (64). The SOCS family of proteins inhibits cytokine-induced signaling through the JAK/STAT pathway (65). SOCS 7 is a less-studied member of this family, and it inhibits the growth hormone and prolactin signaling pathway via STAT3 and STAT5 proteins (66). Hence, this miRNA may suppress the inflammatory response in CHIKV infection by targeting host signaling pathways.
Selvamani et al., 2014 demonstrated the role of hsa-miR-146a in the CHIKV life cycle. The levels of hsa-miR-146a were increased in CHIKV-infected primary human synovial fibroblasts. Upregulation of hsa-miR-146a led to decreased expression of IRAK1, TRAF6, and IRAK2, ultimately inhibiting the function of nuclear transcription factor kB (NF-kB). The inhibition of nuclear transcription factor kB (NF-kB) may reduce proinflammatory cytokine production. (Figure 2) Hence, CHIKV may be using hsa-miR-146a to modulate the antiviral response (45). Similarly, hsa-miR-146a has been reported to disrupt the type I interferon and RIG signaling pathways in the case of vesicular stomatitis virus (VSV) (68). Primary hsa-miR-146a and mature hsa-miR-146a have been detected in RA synovial tissue (69). This study indicates that hsa-miR-146a may modulate the host antiviral response to establish CHIKV pathogenesis in primary human synovial fibroblasts.
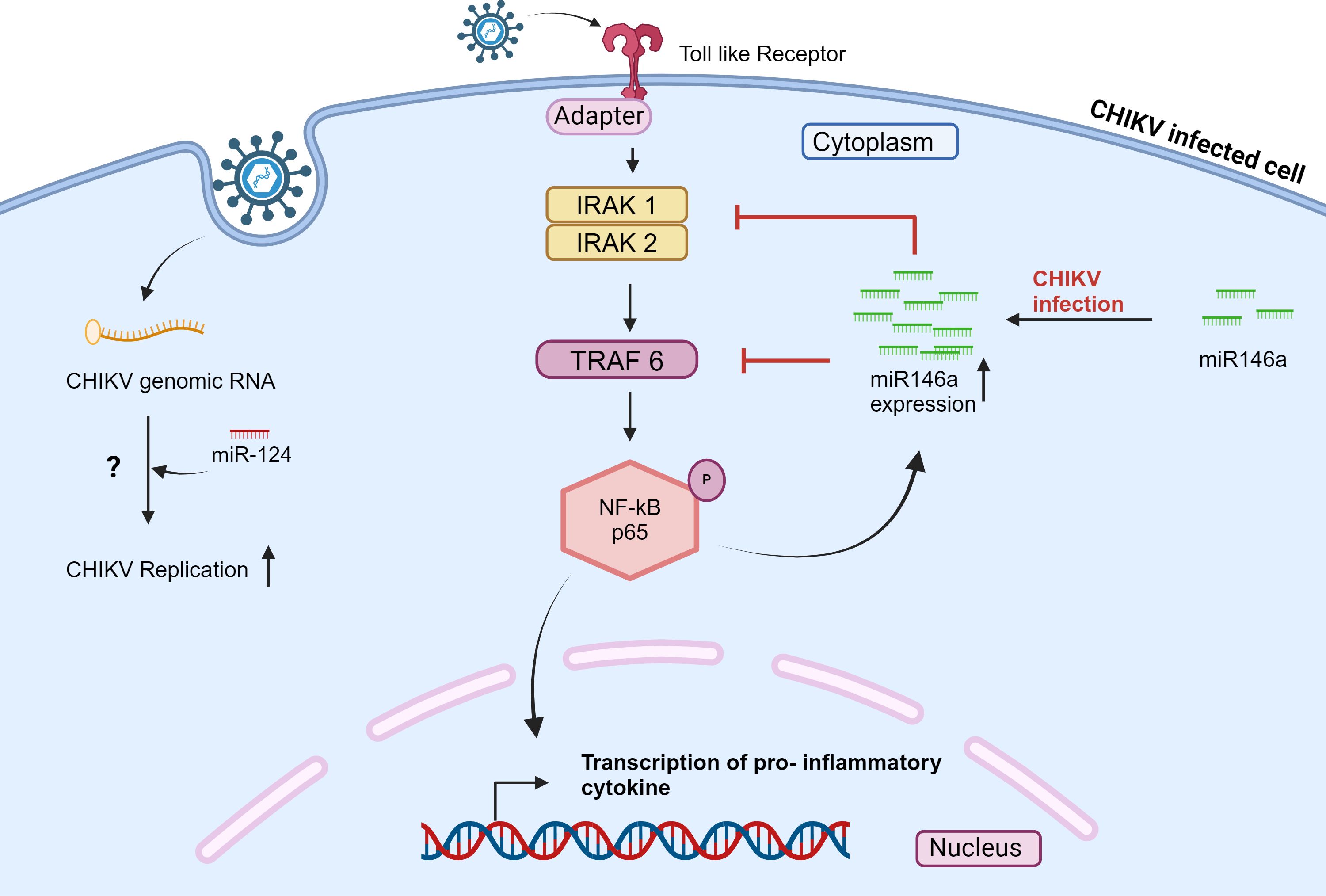
Figure 2 CHIKV infection modulates the expression of miRNAs in mammalian cells and affects the signaling pathway. Also, cellular miRNA interacts with the CHIKV genome. The scheme shows that during CHIKV infection, miR-124 directly interacts with the CHIKV genome and regulates the CHIKV expression in a host cell; the mechanism is still unclear. Also, NF-kB is activated in response to CHIKV infection; this activation elevates the level of miR-146a, which in turn inhibits the expression of IRAK1, IRAK2, and TRAF6, thereby downregulating NF-kB activity (45, 67).
Kansakar et al., 2022 have demonstrated that hsa-miR-142 targets TIM-1 in Human Endothelial cells (70). TIM-1 serves as the receptor for several viruses, including CHIKV (71). Authors suggested that hsa-miR-142 may play an important role in the life cycle of viruses such as CHIKV (70). Overall, it is suggested that hsa-miR-142 may be exploited further as a biomarker as well as a therapeutic candidate.
López et al., 2020, studied the role of hsa-miR-124–3p in CHIKV-infected Huh7.5.1 cell line. The hsa-miR-124–3p binding site was identified on both CHIKV and SINV genomes. When hsa-miR-124–3p was overexpressed in Huh7.5.1 cells, a significantly increased CHIKV replication was observed. While inhibition of hsa-miR-124–3p led to a drop in CHIKV titer (67). (Figure 2) However, the hsa-miR-124–3p is a nervous system-specific miRNA (72). Thus, it would be interesting to examine whether hsa-miR-124–3p is associated with neuropathology during CHIKV infection.
Scheel et al. (2017) analyzed the interaction of miRNAs with fifteen infectious RNA viruses, including CHIKV, using the AGO-CLIP (Argonaute-crosslinking immunoprecipitation) method and miRNA target chimera analyses. Their study suggests many miRNA can potentially interact with CHIKV genome such as miR-21, miR-122, let -7 and miR-181 (73). Patil et al. (2020) further predicted the presence of a hsa-miR-214–3p binding site on the CHIKV genome (74). However, the role of these miRNAs in the CHIKV life cycle is still unknown. In summary, these miRNAs could be exploited for the development of broad-spectrum therapeutics. Therefore, it is essential to study the role of miRNAs in both host signaling pathways and virus infection.
Identifying novel biomarkers and developing miRNA-based therapies to treat viral infection is an emerging field of research. Thus, it is interesting to study the role of miRNA during CHIKV infection. Computational studies have shown that the CHIKV genome has miRNA binding sites for various miRNAs, suggesting the key role of miRNA in CHIKV replication and pathogenesis (34, 36, 67, 74). The miRNA–based therapeutics for CHIKV infection can be developed by targeting virus replication or modulating host factors to mitigate CHIKV symptoms (75). Nevertheless, our current knowledge of the function of miRNAs in the host cellular system and viral life cycle remains limited.
Bioinformatics tools and high-throughput screening methods are available to predict miRNA during pre-clinical studies. Researchers are currently working on designing more scientific and precise methods for predicting miRNA targets (76). Furthermore, various in vitro cell cultures and in vivo mouse models are present to study the toxicity, efficacy, and safety of miRNA therapeutics.
Several miRNAs have been suggested as biomarkers and are linked to certain disease stages or diseases. For example, to detect the H1N1 influenza virus, hsa-miR-1254 and hsa-miR-181c-5p were suggested as biomarkers. Still, further research is necessary to fully establish the use of miRNAs as a biomarker to diagnose viral infection and predict outcomes.
Miravirsen, an antimiR developed by Santaris Pharma, USA, was the first miRNA-targeting drug to enter clinical trials (77). Miravirsen targets hsa-miR-122, which is abundant in the liver cells and helps HCV replication (78, 79). Further, RG-101, which is also an inhibitor of hsa-miR-122, was used in a clinical trial but showed adverse effects, so the trial was discontinued (80). Therefore, understanding the functions of miRNAs in CHIKV infection and pathogenesis will provide a way to design and develop targeted therapies.
RN: Writing – review & editing, Writing – original draft, Methodology, Data curation, Conceptualization. VG: Writing – review & editing, Writing – original draft, Supervision, Methodology, Investigation, Data curation. YK: Writing – review & editing, Writing – original draft, Visualization, Validation, Supervision, Project administration, Methodology, Funding acquisition, Formal analysis, Data curation, Conceptualization.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. We acknowledge “Science and Engineering Research Board”(SERB), New Delhi, Government of India, for funding. (Project no: CRG/2020/001153 awarded to YK).
All Authors thank the Director of ARI and MACS-ARI, Pune, for all the support. Rohini A. Nangare acknowledges the University Grants Commission (UGC) for a senior research fellowship. All the figures were created with BioRender.com.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fviro.2024.1386580/full#supplementary-material
1. Strauss JH, Strauss EG. The alphaviruses: gene expression, replication, and evolution. Microbiol Rev. (1994) 58:491–562. doi: 10.1128/mr.58.3.491-562.1994
2. Fauquet CM, Mayo MA, Maniloff J, Desselberger U, Ball LA, Boston A, et al. Virus Taxonomy Classification and Nomenclature of Viruses Eighth Report of the International Committe on the Taxonomy of Viruses (2005). Available online at: http://www.elsevier.com.
3. Simizu B, Yamamoto K, Hashimoto K, Ogata T. Structural proteins of chikungunya virus. J Virol. (1984) 51:254–8. doi: 10.1128/jvi.51.1.254-258.1984
4. Bréhin AC, Rubrecht L, Navarro-Sanchez ME, Maréchal V, Frenkiel MP, Lapalud P, et al. Production and characterization of mouse monoclonal antibodies reactive to Chikungunya envelope E2 glycoprotein. Virology. (2008) 371:185–95. doi: 10.1016/j.virol.2007.09.028
5. Kielian M, Rey FA. Virus membrane-fusion proteins: More than one way to make a hairpin. Nat Rev Microbiol. (2006) 4:67–76. doi: 10.1038/nrmicro1326
6. Mason PJ, Haddow AJ. An epidemic of virus disease in southern province. tanganyika territory. Trans R Soc Trop Med Hyg. (1957) 51:238–40. doi: 10.1016/0035-9203(57)90022-6
7. Badawi A, Ryoo SG, Vasileva D, Yaghoubi S. Prevalence of chronic comorbidities in chikungunya: A systematic review and meta-analysis. Int J Infect Dis. (2018) 67:107–13. doi: 10.1016/j.ijid.2017.12.018
8. Ross RW. The newala epidemic: III. The virus: Isolation, pathogenic properties and relationship to the epidemic. J Hyg. (1956) 54:177–91. doi: 10.1017/S0022172400044442
9. Schuffenecker I, Iteman I, Michault A, Murri S, Frangeul L, Vaney MC, et al. Genome microevolution of chikungunya viruses causing the Indian ocean outbreak. PloS Med. (2006) 3:e263. doi: 10.1371/journal.pmed.0030263
10. Khan AH, Morita K, Del Carmen M, Hasebe F, Mathenge EGM, Igarashi A. Printed in Great Britain Complete nucleotide sequence of chikungunya virus and evidence for an internal polyadenylation site. J Gen Virol. (2002) 83:3075–84. doi: 10.1099/0022-1317-83-12-3075
11. Volk SM, Chen R, Tsetsarkin KA, Adams AP, Garcia TI, Sall AA, et al. Genome-scale phylogenetic analyses of chikungunya virus reveal independent emergences of recent epidemics and various evolutionary rates. J Virol. (2010) 84:6497–504. doi: 10.1128/JVI.01603-09
12. Lee RC, Feinbaum RL, Ambrost V. The C. elegans Heterochronic Gene lin-4 Encodes Small RNAs with Antisense Complementarity to &II-14. Cell. (1993) 75:843–54. doi: 10.1016/0092-8674(93)90529-Y
13. Lau NC, Lim LP, Weinstein EG, Bartel DP. An abundant class of tiny RNAs with probable regulatory roles in caenorhabditis. Science. (2001) 294:858–62. doi: 10.1126/science.1065062
14. Wightman B, Ha L, Ruvkun G. Posttranscriptional Regulation of the Heterochronic Gene lin-14 by W-4 Mediates Temporal Pattern Formation in C. elegans. Cell. (1993) 75:855–62. doi: 10.1016/0092-8674(93)90530-4
15. Wightman B, Biirglin TR, Gatto J, Arasu P, Ruvkun G. Negative regulatory sequences in the lin-14 3’-untranslated region are necessary to generate a temporal switch during Caenorhabditis elegans development. Genes Dev. (1991) 5:1813–24. doi: 10.1101/gad.5.10.1813
16. Lee Y, Kim M, Han J, Yeom KH, Lee S, Baek SH, et al. MicroRNA genes are transcribed by RNA polymerase II. EMBO J. (2004) 23:4051–60. doi: 10.1038/sj.emboj.7600385
17. Landthaler M, Yalcin A, Tuschl T. The Human DiGeorge Syndrome Critical Region Gene 8 and Its D. melanogaster Homolog Are Required for miRNA Biogenesis other genes known to be involved in nuclear and cyto-plasmic miRNA processing. DsRNA of about 500 base. Curr Biol. (2004) 14:2162–7. doi: 10.1016/j.cub.2004.11.001
18. Lee Y, Ahn C, Han J, Choi H, Kim J, Yim J, et al. The nuclear RNase III Drosha initiates microRNA processing (2003). Available online at: www.nature.com/nature.
19. Yi R, Qin Y, Macara IG, Cullen BR. Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes Dev. (2003) 17:3011–6. doi: 10.1101/gad.1158803
20. Chendrimada TP, Gregory RI, Kumaraswamy E, Norman J, Cooch N, Nishikura K, et al. TRBP recruits the Dicer complex to Ago2 for microRNA processing and gene silencing. Nature. (2005) 436:740–4. doi: 10.1038/nature03868
21. Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. (1998) 391:806–11. doi: 10.1038/35888
22. Song L, Tuan RS. MicroRNAs and cell differentiation in mammalian development. Birth Defects Res C Embryo Today. (2006) 78:140–9. doi: 10.1002/bdrc.20070
23. Adlakha YK, Saini N. MicroRNA: a connecting road between apoptosis and cholesterol metabolism. Tumor Biol. (2016) 37:8529–54. doi: 10.1007/s13277-016-4988-z
24. Hagen JW, Lai EC. microRNA control of cell-cell signaling during development and disease. Cell Cycle (Georgetown, Tex.) (2008) 7:2327–32. doi: 10.4161/cc.6447
25. Bruscella P, Bottini S, Baudesson C, Pawlotsky JM, Feray C, Trabucchi M. Viruses and miRNAs: More friends than foes. Front Microbiol. (2017) 8. doi: 10.3389/fmicb.2017.00824
26. Sim S, Jupatanakul N, Dimopoulos G. Mosquito immunity against arboviruses. Viruses. (2014) 6:4479–504. doi: 10.3390/v6114479
27. Lemaitre B, Hoffmann J. The host defense of Drosophila melanogaster. Annu Rev Immunol. (2007) 25:697–743. doi: 10.1146/annurev.immunol.25.022106.141615
28. Blair CD, Olson KE. The role of RNA interference (RNAi) in arbovirus-vector interactions. Viruses. (2015) 7:820–43. doi: 10.3390/v7020820
29. Choi IK, Hyun S. Conserved microRNA miR-8 in fat body regulates innate immune homeostasis in Drosophila. Dev Comp Immunol. (2012) 37:50–4. doi: 10.1016/j.dci.2011.12.008
30. Bishop C, Hussain M, Hugo LE, Asgari S. Analysis of Aedes aEgypti microRNAs in response to Wolbachia wAlbB infection and their potential role in mosquito longevity. Sci Rep. (2022) 12:15245. doi: 10.1038/s41598-022-19574-x
31. Shrinet J, Jain S, Jain J, Bhatnagar RK, Sunil S. Next Generation Sequencing Reveals Regulation of Distinct Aedes microRNAs during Chikungunya Virus Development. PloS Negl Trop Dis. (2014) 8:32. doi: 10.1371/journal.pntd.0002616
32. Avila-Bonilla RG, Yocupicio-Monroy M, Marchat LA, De Nova-Ocampo MA, del Ángel RM, Salas-Benito JS. Analysis of the miRNA profile in C6/36 cells persistently infected with dengue virus type 2. Virus Res. (2017) 232:139–51. doi: 10.1016/j.virusres.2017.03.005
33. Azlan A, Yunus MA, Abdul Halim M, Azzam G. Revised Annotation and Characterization of Novel Aedes albopictus miRNAs and Their Potential Functions in Dengue Virus Infection. Biol (Basel). (2022) 11(10):1536. doi: 10.3390/biology11101536
34. Dubey SK, Shrinet J, Jain J, Ali S, Sunil S. Aedes aEgypti microRNA miR-2b regulates ubiquitin-related modifier to control chikungunya virus replication. Sci Rep. (2017) 7:17666. doi: 10.1038/s41598-017-18043-0
35. Isaacson MK, Ploegh HL. Ubiquitination, ubiquitin-like modifiers, and deubiquitination in viral infection. Cell Host Microbe. (2009) 5:559–70. doi: 10.1016/j.chom.2009.05.012
36. Dubey SK, Shrinet J, Sunil S. Aedes aEgypti microRNA, miR-2944b-5p interacts with 3’UTR of chikungunya virus and cellular target vps-13 to regulate viral replication. PloS Negl Trop Dis. (2019) 13:e0007429. doi: 10.1371/journal.pntd.0007429
37. Anand SK, Tikoo SK. Viruses as modulators of mitochondrial functions. Adv Virol. (2013) 2013:738794. doi: 10.1155/2013/738794
38. Fiorillo C, Yen PS, Colantoni A, Mariconti M, Azevedo N, Lombardo F, et al. MicroRNAs and other small RNAs in Aedes aEgypti saliva and salivary glands following chikungunya virus infection. Sci Rep. (2022) 12:9536. doi: 10.1038/s41598-022-13780-3
39. Schneider BS, Higgs S. The enhancement of arbovirus transmission and disease by mosquito saliva is associated with modulation of the host immune response. Trans R Soc Trop Med Hyg. (2008) 102:400–8. doi: 10.1016/j.trstmh.2008.01.024
40. Maharaj PD, Widen SG, Huang J, Wood TG, Thangamani S. Discovery of mosquito saliva microRNAs during CHIKV infection. PloS Negl Trop Dis. (2015) 9:e0003386. doi: 10.1371/journal.pntd.0003386
41. Arpino C, Curatolo P, Rezza G. Chikungunya and the nervous system: What we do and do not know. Rev Med Virol. (2009) 19:121–9. doi: 10.1002/rmv.606
42. Lane RS, Lund AW. Non-hematopoietic control of peripheral tissue t cell responses: Implications for solid tumors. Front Immunol. (2018) 9. doi: 10.3389/fimmu.2018.02662
43. Sourisseau M, Schilte C, Casartelli N, Trouillet C, Guivel-Benhassine F, Rudnicka D, et al. Characterization of reemerging chikungunya virus. PloS Pathog. (2007) 3:e89. doi: 10.1371/journal.ppat.0030089
44. Das I, Basantray I, Mamidi P, Nayak TK, Pratheek BM, Chattopadhyay S, et al. Heat shock protein 90 positively regulates Chikungunya virus replication by stabilizing viral non-structural protein nsP2 during infection. PloS One. (2014) 9:e100531. doi: 10.1371/journal.pone.0100531
45. Selvamani SP, Mishra R, Singh SK. Chikungunya virus exploits miR-146a to regulate NF-κB pathway in human synovial fibroblasts. PloS One. (2014) 9:e103624. doi: 10.1371/journal.pone.0103624
46. Krejbich-Trotot P, Denizot M, Hoarau J, Jaffar-Bandjee M, Das T, Gasque P. Chikungunya virus mobilizes the apoptotic machinery to invade host cell defenses. FASEB J. (2011) 25:314–25. doi: 10.1096/fj.10-164178
47. Barbu MG, Condrat CE, Thompson DC, Bugnar OL, Cretoiu D, Toader OD, et al. MicroRNA involvement in signaling pathways during viral infection. Front Cell Dev Biol. (2020) 8. doi: 10.3389/fcell.2020.00143
48. Sharma A, Balakathiresan NS, Maheshwari RK. Chikungunya virus infection alters expression of microRNAs involved in cellular proliferation, immune response and apoptosis. Intervirology. (2015) 58:332–41. doi: 10.1159/000441309
49. Gellert M, Mizuuchi K, O’dea MH, Nasht HA. DNA gyrase: An enzyme that introduces superhelical turns into DNA (Escherichia coli/ATP-dependent reaction/superhelix density). Proc Natl Acad Sci USA. (1976) 73:3872–6. doi: 10.1073/pnas.73.11.3872
50. Liu LF, Liu CC, Alberts BM. Type II DNA topoisomerases: Enzymes that can unknot a topologically knotted DNA molecule via a reversible double-strand break. Cell. (1980) 19:697–707. doi: 10.1016/S0092-8674(80)80046-8
51. Saxena T, Tandon B, Sharma S, Chameettachal S, Ray P, Ray AR, et al. Combined miRNA and mRNA signature identifies key molecular players and pathways involved in Chikungunya virus infection in human cells. PloS One. (2013) 8:e79886. doi: 10.1371/journal.pone.0079886
52. Chhabra R, Adlakha YK, Hariharan M, Scaría V, Saini N. Upregulation of miR-23a∼27a∼24–2 cluster induces caspase-dependent and -independent apoptosis in human embryonic kidney cells. PloS One. (2009) 4:e5848. doi: 10.1371/journal.pone.0005848
53. Petrocca F, Vecchione A, Croce CM. Emerging role of miR-106b-25/miR-17–92 clusters in the control of transforming growth factor β signaling. Cancer Res. (2008) 68:8191–4. doi: 10.1158/0008-5472.CAN-08-1768
54. Cimmino A, Calin GA, Fabbri M, Iorio MV, Ferracin M, Shimizu M, et al. miR-15 and miR-16 induce apoptosis by targeting BCL2. Proc Natl Acad Sci USA. (2005) 102:13944–9. doi: 10.1073/pnas.0506654102
55. Li G, Qiu Z. Deletion of miR-15 Protects Against Rheumatoid Arthritis via Deregulating its Target Gene BCL2L2 and Repressing NF-κB Pathway. Ann Clin Lab Sci. (2019) 49:581–9. doi: 0091-7370/19/0500-581
56. Filková M, Aradi B, Šenolt L, Ospelt C, Vettori S, Mann H, et al. Association of circulating miR-223 and miR-16 with disease activity in patients with early rheumatoid arthritis. Ann Rheum Dis. (2014) 73:1898–904. doi: 10.1136/annrheumdis-2012-202815
57. Agrawal M, Pandey N, Rastogi M, Dogra S, Singh SK. Chikungunya virus modulates the miRNA expression patterns in human synovial fibroblasts. J Med Virol. (2020) 92:139–48. doi: 10.1002/jmv.25588
58. Diehl N, Schaal H. Make yourself at home: Viral hijacking of the PI3K/Akt signaling pathway. Viruses. (2013) 5:3192–212. doi: 10.3390/v5123192
59. Bartok B, Boyle DL, Liu Y, Ren P, Ball ST, Bugbee WD, et al. PI3 kinase δ is a key regulator of synoviocyte function in rheumatoid arthritis. Am J Pathol. (2012) 180:1906–16. doi: 10.1016/j.ajpath.2012.01.030
60. Abdullah L, Hills LB, Winter EB, Huang YH. Diverse roles of akt in T cells. Immunometabolism. (2021) 3:e210007. doi: 10.20900/immunometab20210007
61. Kane LP, Shapiro VS, Stokoe D, Weiss A. Induction of NF-κ κB by the akt/PKB kinase. Curr. Biol. (1999) 9:601–4. doi: 10.1016/S0960-9822(99)80265-6
62. Yang CH, Yue J, Fan M, Pfeffer LM. IFN induces miR-21 through a signal transducer and activator of transcription 3-dependent pathway as a suppressive negative feedback on IFN-induced apoptosis. Cancer Res. (2010) 70:8108–16. doi: 10.1158/0008-5472.CAN-10-2579
63. Moynagh PN. The roles of Pellino E3 ubiquitin ligases in immunity. Nat Rev Immunol. (2014) 14:122–31. doi: 10.1038/nri3599
64. Wang P, Zhao W, Zhao K, Zhang L, Gao C. TRIM26 negatively regulates interferon-β Production and antiviral response through polyubiquitination and degradation of nuclear IRF3. PloS Pathog. (2015) 11:1–22. doi: 10.1371/journal.ppat.1004726
65. Liau NPD, Laktyushin A, Lucet IS, Murphy JM, Yao S, Whitlock E, et al. The molecular basis of JAK/STAT inhibition by SOCS1. Nat Commun. (2018) 9:1558. doi: 10.1038/s41467-018-04013-1
66. Martens N, Uzan G, Wery M, Hooghe R, Hooghe-Peters EL, Gertler A. Suppressor of cytokine signaling 7 inhibits prolactin, growth hormone, and leptin signaling by interacting with STAT5 or STAT3 and attenuating their nuclear translocation. J Biol Chem. (2005) 280:13817–23. doi: 10.1074/jbc.M411596200
67. López P, Girardi E, Mounce BC, Weiss A, Chane-Woon-Ming B, Messmer M, et al. High-throughput fluorescence-based screen identifies the neuronal microRNA miR-124 as a positive regulator of alphavirus infection. J Virol. (2020) 94(9):e02145–19. doi: 10.1128/JVI
68. Hou J, Wang P, Lin L, Liu X, Ma F, An H, et al. MicroRNA-146a feedback inhibits RIG-I-dependent type I IFN production in macrophages by targeting TRAF6, IRAK1, and IRAK2. J Immunol. (2009) 183:2150–8. doi: 10.4049/jimmunol.0900707
69. Nakasa T, Miyaki S, Okubo A, Hashimoto M, Nishida K, Ochi M, et al. Expression of MicroRNA-146 in rheumatoid arthritis synovial tissue. Arthritis Rheumatol. (2008) 58:1284–92. doi: 10.1002/art.23429
70. Kansakar U, Gambardella J, Varzideh F, Avvisato R, Jankauskas SS, Mone P, et al. miR-142 targets TIM-1 in human endothelial cells: potential implications for stroke, COVID-19, zika, ebola, dengue, and other viral infections. Int J Mol Sci. (2022) 23:10242. doi: 10.3390/ijms231810242
71. Kirui J, Abidine Y, Lenman A, Islam K, Gwon YD, Lasswitz L, et al. The phosphatidylserine receptor tim-1 enhances authentic chikungunya virus cell entry. Cells. (2021) 10(7):1828. doi: 10.3390/cells10071828
72. Ponomarev ED, Veremeyko T, Barteneva N, Krichevsky AM, Weiner HL. MicroRNA-124 promotes microglia quiescence and suppresses EAE by deactivating macrophages via the C/EBP-α-PU.1 pathway. Nat Med. (2011) 17:64–70. doi: 10.1038/nm.2266
73. Scheel TKH, Luna JM, Liniger M, Nishiuchi E, Rozen-Gagnon K, Shlomai A, et al. A broad RNA virus survey reveals both miRNA dependence and functional sequestration. Cell Host Microbe. (2016) 19:409–23. doi: 10.1016/j.chom.2016.02.007
74. Patil RN, Karpe YA. Uncovering the roles of miR-214 in hepatitis E virus replication. J Mol Biol. (2020) 432:5322–42. doi: 10.1016/j.jmb.2020.07.015
75. Drury RE, O’Connor D, Pollard AJ. The clinical application of MicroRNAs in infectious disease. Front Immunol. (2017) 8. doi: 10.3389/fimmu.2017.01182
76. Huang W. MicroRNAs: biomarkers, diagnostics, and therapeutics. Methods Mol Biol. (2017) 1617:57–67. doi: 10.1007/978–1-4939–7046-9_4
77. Chahal J, Gebert LFR, Camargo C, Macrae IJ, Sagan SM. miR-122-based therapies select for three distinct resistance mechanisms based on alterations in RNA structure. Proc Natl Acad Sci USA. (2021) 118:e2103671118. doi: 10.1073/pnas.2103671118
78. Chang J, Nicolas E, Marks D, Sander C, Lerro A, Buendia MA, et al. miR-122, a mammalian liver-specific microRNA, is processed from hcr mRNA and may downregulate the high affinity cationic amino acid transporter CAT-1. RNA Biol. (2004) 1:106–13. doi: 10.4161/rna.1.2.1066
79. Jopling CL, Yi M, Lancaster AM, Lemon SM, Sarnow P. Modulation of hepatitis C virus RNA abundance by a liver-specific microrna. Sci (1979). (2005) 309:1577–81. doi: 10.1126/science.1113329
Keywords: Chikungunya virus, miRNA, RNA virus, Arbovirus, RNA virus replication
Citation: Nangare RA, Gajbhiye V and Karpe YA (2024) Role of miRNAs in the Chikungunya virus replication and pathogenesis. Front. Virol. 4:1386580. doi: 10.3389/fviro.2024.1386580
Received: 15 February 2024; Accepted: 15 May 2024;
Published: 03 June 2024.
Edited by:
Dasja Pajkrt, Academic Medical Center, NetherlandsReviewed by:
Israel Guerero-Arguero, Texas Biomedical Research Institute, United StatesCopyright © 2024 Nangare, Gajbhiye and Karpe. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yogesh A. Karpe, eWFrYXJwZUBhcmlwdW5lLm9yZw==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.