
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
MINI REVIEW article
Front. Virol., 15 March 2024
Sec. Fundamental Virology
Volume 4 - 2024 | https://doi.org/10.3389/fviro.2024.1383261
This article is part of the Research TopicThe Viral Egress: Strategies for Therapeutic InterferenceView all articles
 Kyo Izumida1
Kyo Izumida1 Eiji Morita1,2*
Eiji Morita1,2*Hepatitis C virus (HCV) is a well-known virus that causes liver diseases such as liver cirrhosis and hepatocellular carcinoma. For several decades, numerous studies have been conducted to unravel the life cycle and molecular mechanisms of this virus with the aim of developing strategies to combat diseases caused by its infection. In this review, we summarize HCV assembly to budding, focusing on one of the structural proteins, the core, a viral capsid that binds both the viral genome and host membrane, along with the core-interacting host partners. The HCV core matures in the endoplasmic reticulum (ER), localizes at the lipid droplet (LD), and shuttles between the LD and ER to form viral particles. This process is controlled by many host factors known to binds core proteins, such as diacylglycerol acyltransferase-1 (DGAT-1), Rab18, μ subunit of the clathrin adaptor protein complex 2 (AP2M1), nuclear pore complex protein 98 (Nup98), Cortactin, group IVA phospholipase A2 (PLA2G4A) etc. Virion budding is thought to involve contributions from endosomal sorting complexes required for transport (ESCRT), similar to other envelope viruses. We delved into potential perspectives to enhance our understanding of the HCV mechanism by drawing insights from existing studies.
Hepatitis C virus (HCV) was first identified in 1989, and recent estimates suggest that approximately 58 million individuals currently experience chronic HCV infection (1). Currently, the primary sources of infection are identified through blood transfusions or needle sharing among drug users. The infection leads to liver diseases, including liver cirrhosis and hepatocellular carcinoma. Historically, treatments have faced challenges in terms of efficacy and intricate dosage regimens (2). However, with breakthroughs in the development of direct-acting antiviral agents (DAAs), treatment methods have recently become more effective and less complicated. Sustained virological response rates increase to 90–100% with appropriate treatment (3).
HCV is a positive-sense single-strand RNA virus with an envelope that belongs to the Flaviviridae family. Upon infection, virions exist as lipoviral particles (LVPs) composed of lipids and apolipoproteins, such as ApoB, ApoC, and ApoE (4). LVP attaches to glycosaminoglycan and low-density lipoprotein receptor (LDLR) and scavenger receptor B-I (SR-BI) followed by its interaction with CD81 on the cell surface (5). LVP then translocates to tight junctions and interacts with CLDN-1 and Occludin, initiating viral entry into cells through pH-dependent endocytosis (6, 7). During endocytosis, the viral envelope fuses with endosomal membranes, releasing viral genetic material into the cellular cytoplasm. Its genome comprises approximately 9600 nucleotides, encompassing the 5’-UTR with an internal ribosome entry site (IRES) and the 3’-UTR. IRES, along with cellular translation initiation factors (eIFs) and components of the cellular translational machinery, such as miR-122, play a role in the regulation of translation (8). Translation occurs in the rough endoplasmic reticulum (ER), which produces polyproteins. This polyprotein is processed by both viral and host signaling proteases, leading to the generation of ten distinct proteins. Viral proteins can be classified into two categories: structural proteins (Core, E1, and E2) and nonstructural proteins (p7, NS2, NS3, NS4A, NS4B, NS5A, and NS5B). The collaboration between HCV NS3 and NS5B proteins and host cell factors influences intracellular membranes forms, leading to the construction of a specific membrane structure known as the “membranous web (MW),” and/or “double membrane vesicles (DMV)” serving as the site for RNA replication (9). These structures create an environment separate from the cytosol, and it has been proposed that their function is to inhibit cellular innate immune responses (10). The replication of viral RNA within these structures is facilitated by HCV proteins, including the viral helicase, viral protease, and viral RNA-dependent RNA polymerase, in conjunction with various host cell factors (11–13). The replicated HCV genome is translocated by NS5A to the HCV core, which is located on lipid droplets (LD) (14). The HCV core shuttles between the LD and ER, and mature viral particles encompass E1 and E2. Viral particles budding at the ER membrane exit through the cellular secretion pathway.
The crystal and NMR structures of various HCV proteins, including E1 and E2 ectodomain (with antibody 8FSJ), p7 (2M6X), NS2 (membrane segment 2JY0), NS3 (3O8B-D, 3O8R, and others), NS4A (complex 3KF2, 2A4Q, 4I31, and others), partial NS5A (4CL1, 3FQQ, 1ZH1), and NS5B (3FQK, 4KAI, and others), have been reported. However, the structure of the HCV core has not yet been elucidated, although a partial structure has been reported (e.g., 1CWX). The HCV core protein shares homology with the nucleocapsid proteins of other flaviviruses. It is highly conserved across different HCV strains (15) and is expected to play a vital role in the diagnosis of HCV infection by detecting specific anti-core protein antibodies and the core itself (16). The HCV core is composed of two alpha-helical modules in its C-terminal region (domain 2: D2) that are believed to be involved in membrane binding. In addition, it possesses a basic amino acid cluster in its N-terminal region (domain 1: D1), which is presumed to play a role in binding to viral genomic RNA (15) (Figure 1). These characteristics suggest its crucial role in surrounding and protecting genomic RNA as a viral capsid protein, as well as in genome packaging and virion assembly. In addition, it is involved in various cellular responses such as modulating host cell transcription, influencing apoptosis (inhibition or stimulation), and suppressing host immunity (17). Extensive investigations on the HCV core have yielded numerous published papers. In this review, we provide a comprehensive overview of the functions of HCV core and its host partners in viral assembly and particle formation.
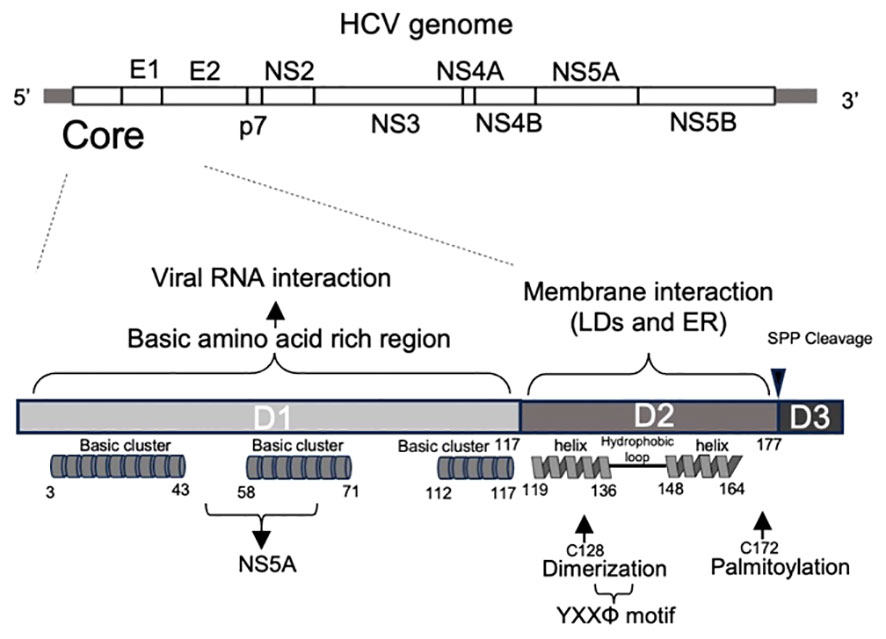
Figure 1 Primary structure of HCV core protein. Primary structure of the HCV genome (top) and core proteins (bottom). HCV core encoding region is located at 5’ end of HCV genome. The HCV core comprises of three domains. Domain 1 (D1) contains three basic amino acid-rich clusters, indicating RNA binding. Domain 2 (D2) is a hydrophobic region containing two alpha helices. It contains cysteine 128, which contributes to dimerization, and cysteine 172, which is palmitoylated. YXXΦ motif is also located in this domain. Domain 3 (D3): Domain 3 includes a signal sequence to the ER membrane and is cleaved by a signal peptide peptidase during the core maturation process.
Following cleavage of the transmembrane (TM) region (domain 3: D3) by the host signal peptide peptidase (SPP), the HCV core protein changes to its soluble mature form and is released into the cytoplasm. However, in its role as a viral capsid protein, the mature core protein has the potential to interact with the host cell membrane. The D2 domain is responsible for the association between ER membranes and LDs (18). Comprising two amphipathic α-helices, Helix I (119–136 aa) and Helix II (148–164 aa), separated by a hydrophobic loop (HL) (Figure 1), the D2 domain features a helix-turn-helix motif essential for the effective localization of the protein to LDs, possibly mediating an in-plane membrane interaction (18). Mutant core proteins that are incapable of associating with LDs undergo rapid degradation by the proteasome, suggesting that the D2 domain plays an indispensable role in folding and stability of the mature core (18). In addition to the D2 domain, the HCV core protein undergoes palmitoylation at the conserved cysteine residue 172. This modification is also important for facilitating the interaction of the core with the membrane and subsequent formation of viral particles (19).
The ER plays a crucial role as a foundational organelle for both HCV genome replication (4) and translation of viral proteins within an infected cell. Following viral entry, viral genome replication occurs on membrane structures derived from the ER (15). LDs, which are derived from ER membranes but are separated from ER membranes, are recognized as a viral assembly platform (15, 20). The presence of triglycerides (TGs) within LDs facilitates the folding of the core protein (21). Previous studies have suggested that the HCV core shuttles between LDs and the ER, facilitating the transfer of newly synthesized viral genomes to the site of viral particle assembly on LDs (Figure 2). Moreover, it aids in the transfer of assembled materials to the ER membrane for association with E1 and E2 proteins, contributing to particle envelopment (15, 22). Notably, the predominant localization of the core on LDs was evident when the core was expressed alone. In contrast, during infection with recombinant viruses, the core predominantly localizes to the ER membrane rather than to LDs (23, 24). This suggests that other viral proteins play a role in core shuttling between the ER and LDs. Boson et al. reported that among the viral proteins, p7 and NS2 act as HCV strain-specific factors that influence the recruitment of core proteins from LDs to ER assembly sites. To date, there has been no evidence of a direct interaction between the core protein and NS2 or p7. Other viral proteins, such as NS5A, which directly interacts with core proteins, may play a concerted role in these processes (25, 26).
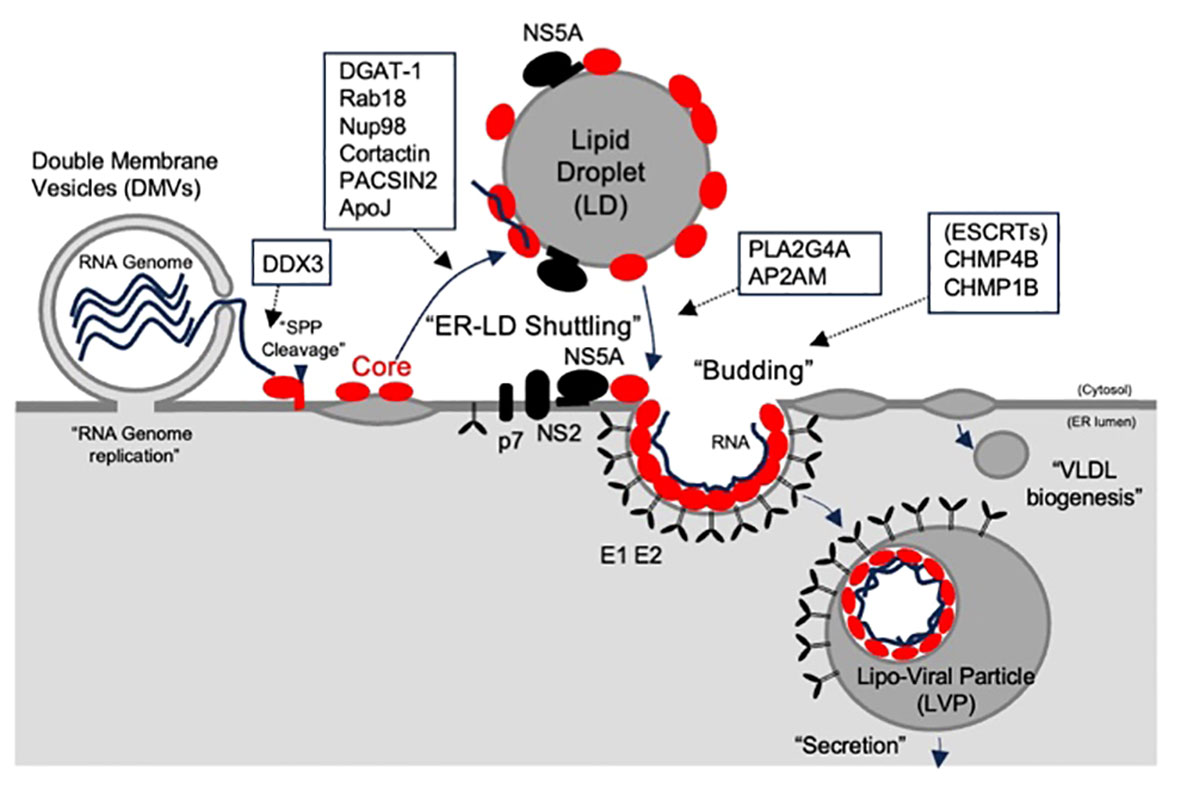
Figure 2 The role of core and its host partners on assembly and release of HCV particle. The HCV core, which is located at the N-terminus of the polypeptide, is cleaved by a signal peptidase and relocates to lipid droplets (LDs), where the virus is assembled. The HCV core shuttles to the endoplasmic reticulum (ER) to form viral particles with E1, and E2, enveloped proteins. Budding was linked to VLDV to form lipoviral particles (LVPs). The predicted roles of the core-binding host partners are indicated by the open boxes.
Interactions between the core proteins and NS5A have been reported to play a role in virion assembly (25). This interaction is facilitated by the N-terminal basic residues R50, K51, R59, and R62 in the D1 domain of the core, which engage with N-terminal domain I of NS5A (25). However, another study suggested that the highly phosphorylated serine cluster located in the C-terminal domain III of NS5A also plays a role in its interactions with the core protein (25). Various host factors regulate these interactions, and diacylglycerol acyltransferase-1 (DGAT1) is one such factor (27, 28). DGAT1 forms a complex with NS5A and its core region, thereby promoting interactions between these viral proteins. The presence of a catalytically inactive mutant of DGAT1 (H426A) disrupted the localization of NS5A (but not the core) to LDs in a dominant-negative manner and blocked the production of infectious viral particles (28). These results suggest that the LD association of the core and NS5A is not a random process but instead necessitates the formation of LDs mediated by active DGAT1. Besides DGAT-1, PACSIN2 (29), cortactin (30), osteopontin (OPN) (31), Rab18 (32), group IVA phospholipase A2 (PLA2G4A), (33) and Apolipoprotein J (Apo J) (34) have been identified as factors that promote the interaction between the core and NS5A in LDs. Depletion of any of the above proteins has been demonstrated to impair the release of infectious virus particles, coinciding with a reduction in the localization of the core and/or NS5A on the LDs and/or ER. Some of these factors have specific roles. Rab18 is associated with LDs and is thought to facilitate the physical interactions between LDs and ER membranes. Rab18 may play a role in enhancing the association between NS5A, other viral components, and LDs (32). Cortactin is an actin-binding protein crucial for cell migration and invasion. NS5A and its core region may induce cell migration by triggering cortactin phosphorylation, suggesting that this altered cellular behavior could contribute to the spread of viruses (30). PLA2G4A plays a pivotal role in the cleavage of lipids, particularly arachidonic acid, and is involved in the metabolism of prostaglandins and leukotrienes. Inhibition of PLA2G4A results in the generation of aberrant HCV particles, and their infectivity can be restored by the addition of arachidonic acid. This suggests that this specific lipid is crucial for the production of highly infectious HCV progeny, possibly by establishing a membrane environment conducive to the efficient incorporation of essential host and viral factors into the lipid envelope of the viral particles (33).
Several studies have demonstrated the pivotal roles of core proteins as potent nucleic acid chaperones. Analogous to other RNA chaperone proteins, the core protein directly guides the proper folding of viral genomic RNA, preventing misfolding, and resolving misfolded RNA structures. Specifically, the core has been observed to enhance the annealing of complementary sequences, facilitate strand exchanges, and promote the dimerization of the 3’-UTR sequence of the HCV genome (35–37). These effects are mediated by peptides comprising either all three core basic domains or a combination of the two (38). Some of these processes may involve a collaboration with cellular factors. DDX3, a DEAD-box RNA helicase 3, has been reported to interacts with core proteins (39–41). The N-terminal region of HCV core interacts with the C-terminal region of DDX3 (41). Depletion of DDX3 leads to a reduction in RNA genome replication and the release of the core into the culture supernatants (42). However, conflicting results exist as other reports have suggested that the core-DDX3 interaction does not contribute to genome replication (43). Instead, DDX3 is reported to interact with the HCV genome 3’UTR, leading to IKKα activation and subsequent core-associated lipid droplet generation (44). The precise role of DDX3 and HCV core interactions in the HCV life cycle remains unclear, necessitating further investigation.
Many enveloped viruses bud from the host cell membrane, utilizing the cellular endosomal sorting complexes required for transport (ESCRT) machinery. It was initially identified in a study on human immunodeficiency virus I (HIV-I). The structural proteins of HIV-1 Gag, including the viral capsid protein, directly bind to TSG101, a subunit of the ESCRT-I complex, and ALIX through its late domains. The ESCRT machinery comprises a modular pathway consisting of more than 30 factors and is known to function in sealing membranes in the cytoplasmic phase. Viral late domains comprising short peptide motifs play a crucial role in the recruitment of upstream ESCRT factors, including ESCRT-I and ALIX. Subsequently, these domains facilitate the recruitment of downstream ESCRTs such as ESCRT-III proteins (also known as CHMP family proteins), which are key players in membrane sealing. These late domains have been identified in other enveloped viruses, such as rhabdoviruses, filoviruses, and hepadna viruses, indicating that the ESCRT pathway is a common budding machinery for various enveloped viruses.
HCV is also an envelope virus, and reports have indicated the involvement of the ESCRT pathway in virion release (45–48). Inhibiting the ESCRT pathway through the expression of a dominant-negative form or siRNA transfection targeting the VPS4 family proteins ITCH or HRS markedly reduces the production of infectious viral particles. However, there is a divergence in perspectives regarding the role of ESCRT machinery in HCV production. Ariumi et al. and Corless et al. reported that ESCRT inhibition did not affect the production of intracellular viral particles. This suggests that the ESCRT pathway may be involved in the secretory pathway of the virion but not in the process of viral particle formation (45, 46). Conversely, Barouch-Bentov et al. and Deng et al. demonstrated that inhibition of the ESCRT pathway impairs the production of intracellular virions (47, 48). Additionally, Barouch-Bentov et al. conducted a proteinase K protection assay, providing evidence that the envelopment of core proteins was insufficient for ESCRT inhibition, indicating a role for ESCRT in viral particle formation (47).
A critical aspect of comprehending the mechanics of ESCRT-mediated viral particle formation is identifying the factors responsible for their recruitment. Although no discernible late domains have been identified in the HCV genome, there is information about the physiological interactions between HCV proteins and ESCRT factors (46, 47). Interaction of core proteins with CHMP4B and CHMP1A. Barouch-Bentov et al. highlighted the significance of the interaction between ubiquitinated NS2 and the ubiquitin-interacting motif (UIM) of HRS in the HCV budding process. The interaction between the core protein and ESCRT-III proteins may also play a role in viral particle formation (47, 49).
The HCV core protein is also known to interact with other cellular proteins, such as μ subunit of the clathrin adaptor protein complex 2 (AP2M1) (50) and nuclear pore complex protein 98 (Nup98) (51), to facilitate efficient virus release. Neveu et al. identified a conserved, YXXΦ motif (where Φ represents a hydrophobic residue) within the core protein. This motif, which corresponds to sorting signals found in host cargo proteins, binds to AP2M1. Mutations in this core motif and the inhibition of AP2M1 function significantly impede both intra- and extracellular infectivity without any discernible effect on HCV RNA replication. This implies that they play pivotal roles in viral assembly (50). AP2M1 is recognized for mediating the sorting of cargo proteins containing YXXΦ into clathrin-coated vesicles (CCVs). These vesicles either form at the cell surface and are destined for fusion with early endosomes or trafficking along lysosomal transport pathways (52). These results imply that core proteins exploit these intracellular membrane trafficking pathways to facilitate translocation between LDs and the ER for particle formation.
Boson et al. reported that the expression of the core protein induces the relocalization of Nup98, an essential component of the nuclear pore, from annulate lamellae to LDs (51). The recruitment of Nup98 by the core to viral assembly sites may play a crucial role in enhancing the local concentration of the RNA genome, thereby facilitating its incorporation into nascent virions (51). To date, a direct interaction between the core and Nup98 has not been conclusively demonstrated. Understanding the involvement of Nup98 in the assembly of HCV particles is crucial for elucidating this aspect of the viral life cycle.
In this review, we summarize the interactions between HCV core and host cell proteins, with a particular focus on their roles in the assembly and budding of HCV virions. Numerous host–cell interactions with the core have been reported, but limitations exist in definitively establishing direct binding. Furthermore, the contributions of certain interactions to the viral life cycle and their impact on cellular activity have not been fully elucidated. Recently, we developed a method for purifying core proteins under non-denaturing conditions (53), enabling more detailed analysis of the interactions between host cell proteins and HCV core. To date, most interactions have been demonstrated in cells or cell lysates, suggesting the presence of intermediate factors, such as nucleic acids or host membranes. Purified core proteins offer the opportunity to obtain more concrete evidence supporting the direct interactions between them. We also need to note that HCV assembly is derived from complex interactions between host cell factors and other HCV proteins as well. One example is the E2-SPCS1-NS2 interaction, which influences the assembly of infectious HCV particles (54). A comprehensive understanding of viral assembly is essential. Even with the recent development of HCV treatments, a thorough understanding of the HCV lifecycle, host cell-protein interactions, and mechanisms would be valuable, not only for HCV, but also for gaining insights into other viruses.
KI: Writing – original draft, Writing – review & editing. EM: Writing – original draft, Writing – review & editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This research was supported by the JST CREST (Grant Number JPMJCR17H4), Japan, and the Takeda Medical Research Foundation. This work was also supported by JSPS KAKENHI (Grant Numbers 23790503, 26460555, 16H01188, 17H06413, 20K21874, 22K18378, 22H02873, 22H00553) and AMED Grant (Grant Number 20333747, 19fk0108168h0001, 20he0622012h0001, 22fk0108527s0101). This work was also supported by The Research Foundation for Microbial Diseases of Osaka University (BIKEN).
We thank the laboratory members for useful discussions.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
KI belonged to the “Laboratory of Viral Infection, Research Institute for Microbial Diseases, Osaka University” at the time dedicated to related study but had no correlation with their current employer.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
1. Lopera L, Posada C, Montoya-Guzmán M, Muñoz D, Navas MC. Treatment of hepatitis C virus infection: from therapy with interferon to direct-acting antivirals. Iatreia. (2023) 189:1122–32. doi: 10.17533/udea.iatreia.183
2. Millman AJ, Nelson NP, Vellozzi C. Hepatitis C: review of the epidemiology, clinical care, and continued challenges in the direct-acting antiviral era. Curr Epidemiol Rep. (2017) 4:174–855. doi: 10.1007/s40471-017-0108-x
3. Zeuzem S. Treatment options in hepatitis C - the current state of the art. Deutsches Arzteblatt Int. (2017) 114:11–21. doi: 10.3238/arztebl.2017.0011
4. Morozov VA, Lagaye S. Hepatitis C virus: morphogenesis, infection and therapy. World J Hepatol. (2018) 10:186–212. doi: 10.4254/wjh.v10.i2.186
5. Wong MT, Chen SSL. Emerging roles of interferon-stimulated genes in the innate immune response to hepatitis C virus infection. In Cellular and Molecular Immunology. Chinese Soc Immunology. (2016) 13(1):11–35. doi: 10.1038/cmi.2014.127
6. Colpitts CC, Tsai PL, Zeisel MB. Hepatitis C virus entry: an intriguingly complex and highly regulated process. Int J Mol Sci. (2020) 21. doi: 10.3390/ijms21062091
7. Zeisel MB, Fofana I, Fafi-Kremer S, Baumert TF. Hepatitis C virus entry into hepatocytes: molecular mechanisms and targets for antiviral therapies. J Hepatol. (2011) 54(3):566–76. doi: 10.1016/j.jhep.2010.10.014
8. Kunden RD, Khan JQ, Ghezelbash S, Wilson JA. The role of the liver-specific microrna, mirna-122 in the hcv replication cycle. Int J Mol Sci. (2020) 21. doi: 10.3390/ijms21165677
9. Li H-C, Yang C-H, Lo S-Y. Cellular factors involved in the hepatitis C virus life cycle. World J Gastroenterol. (2021) 27:4555–81. doi: 10.3748/wjg.v27.i28.4555
10. Neufeldt CJ, Joyce MA, Van Buuren N, Levin A, Kirkegaard K, Gale M, et al. The hepatitis C virus-induced membranous web and associated nuclear transport machinery limit access of pattern recognition receptors to viral replication sites. PloS Pathog. (2016) 12. doi: 10.1371/journal.ppat.1005428
11. Bartenschlager R, Cosset FL, Lohmann V. Hepatitis C virus replication cycle. J Hepatol. (2010) 53:583–5. doi: 10.1016/j.jhep.2010.04.015
12. Tabata K, Neufeldt CJ, Bartenschlager R. Hepatitis C virus replication. Cold Spring Harbor Perspect Med. (2020) 10. doi: 10.1101/cshperspect.a037093
13. Li HC, Yang CH, Lo SY. Hepatitis c viral replication complex. Viruses. (2021) 13. doi: 10.3390/v13030520
14. Alazard-Dany N, Denolly Solène, Boson B, Cosset FrançoisLoïc. Overview of hcv life cycle with a special focus on current and possible future antiviral targets. Viruses. (2019) 11. doi: 10.3390/v11010030
15. Gawlik K, Gallay PA. HCV core protein and virus assembly: what we know without structures. Immunologic Res. (2014) 60:1–105. doi: 10.1007/s12026-014-8494-3
16. Wang Y, Jie W, Ling J, Yuanshuai H. HCV core antigen plays an important role in the fight against HCV as an alternative to HCV-RNA detection. J Clin Lab Anal. (2021) 35. doi: 10.1002/jcla.23755
17. Mani H, Yen J-H, Hsu H-J, Chang C-C, Lioua J-W. Hepatitis C virus core protein not just a nucleocapsid building block, but an immunity and inflammation modulator. Tzu Chi Med J (2021) 34:139–47. doi: 10.4103/tcmj.tcmj_97_21
18. Boulant S, Montserret R, Graham Hope R, Ratinier M, Targett-Adams P, Lavergne JP, et al. Structural determinants that target the hepatitis C virus core protein to lipid droplets. J Biol Chem. (2006) 281:22236–47. doi: 10.1074/jbc.M601031200
19. Majeau N, Frometin Rémi, Savard C, Duval M, . Tremblay MJ, Leclerc D. Palmitoylation of hepatitis C virus core protein is important for virion production. J Biol Chem. (2009) 284:33915–25. doi: 10.1074/jbc.M109.018549
20. Jones DM, McLauchlan J. Hepatitis C virus: assembly and release of virus particles. J Biol Chem. (2010) 285:22722–9. doi: 10.1074/jbc.R110.133017
21. Ajjaji D, M’barek KB, Boson B, Omrane M, Gassama-Diagne A, Blaud M, et al. Hepatitis C virus core protein uses triacylglycerols to fold onto the endoplasmic reticulum membrane. Traffic. (2022) 23:63–80. doi: 10.1111/tra.12825
22. Galli A, Ramirez S, Bukh J. Lipid Droplets Accumulation during Hepatitis C Virus Infection in Cell-Culture Varies among Genotype 1-3 Strains and Does Not Correlate with Virus Replication. Viruses (2021) 13. doi: 10.3390/v13030389
23. Boson B, Granio Ophélia, Bartenschlager R, CossetFranç FrançoisLoïc. A concerted action of hepatitis C virus P7 and nonstructural protein 2 regulates core localization at the endoplasmic reticulum and virus assembly. PloS Pathog. (2011) 7. doi: 10.1371/journal.ppat.1002144
24. Vieyres G, Pietschmann T. HCV Pit Stop at the Lipid Droplet: Refuel Lipids and Put on a Lipoprotein Coat before Exit. Cells. (2019) 8. doi: 10.3390/cells8030233
25. Masaki T, Suzuki R, Murakami K, Aizaki H, Ishii K, Murayama A, et al. Interaction of hepatitis C virus nonstructural protein 5A with core protein is critical for the production of infectious virus particles. J Virol. (2008) 82. doi: 10.1128/jvi.00826-08
26. Popescu CI, Callens N, Trinel D, Roingeard P, Moradpour D, Descamps Véronique, et al. NS2 protein of hepatitis C virus interacts with structural and non-structural proteins towards virus assembly. PloS Pathog. (2011) 7. doi: 10.1371/journal.ppat.1001278
27. Herker E, Harris C, Hernandez Céline, Carpentier A, Kaehlcke K, . Rosenberg AR, et al. Efficient hepatitis C virus particle formation requires diacylglycerol acyltransferase-1. Nat Med. (2010) 16:1295–8. doi: 10.1038/nm.2238
28. Camus G, Herker E, . Modi AA, . Haas JT, . Ramage HR, . Farese RV, et al. Diacylglycerol acyltransferase-1 localizes hepatitis C virus NS5A protein to lipid droplets and enhances NS5A interaction with the viral capsid core. J Biol Chem. (2013) 288:9915–23. doi: 10.1074/jbc.M112.434910
29. Nguyen LP, Tran S, Suetsugu S, Lim Y-S, . Hwang SB. PACSIN2 interacts with nonstructural protein 5A and regulates hepatitis C virus assembly. J Virol. (2020) 94. doi: 10.1128/jvi.01531-19
30. Nguyen LP, Nguyen TTT, Nguyen HC, Pham HT, Han KM, Choi DH, et al. Cortactin interacts with hepatitis C virus core and NS5A proteins: implications for virion assembly. J Virol. (2020) 94. doi: 10.1128/jvi.01306-20
31. Iqbal J, Sarkar-Dutta M, McRae S, Ramachandran A, Kumar B, Waris G. Osteopontin regulates hepatitis C virus (HCV) replication and assembly by interacting with HCV proteins and lipid droplets and by binding to receptors AVβ3 and CD44. J Virol. (2018) 92. doi: 10.1128/jvi.02116-17
32. Salloum S, Wang H, Ferguson C, . Parton RG, . Tai AW. Rab18 binds to hepatitis C virus NS5A and promotes interaction between sites of viral replication and lipid droplets. PloS Pathog. (2013) 9. doi: 10.1371/journal.ppat.1003513
33. Menzel N, Fischl W, Hueging K, Bankwitz D, Frentzen A, Haid S, et al. MAP-kinase regulated cytosolic phospholipase A2 activity is essential for production of infectious hepatitis C virus particles. PloS Pathog. (2012) 8:215. doi: 10.1371/journal.ppat.1002829
34. Lin CC, Tsai P, Sun HYu, Hsu MC, Lee JC, Wu IC, et al. Apolipoprotein J, a glucose-upregulated molecular chaperone, stabilizes core and NS5A to promote infectious hepatitis C virus virion production. J Hepatol. (2014) 61:984–93. doi: 10.1016/j.jhep.2014.06.026
35. Cristofari Gaël, Ivanyi-Nagy R, Gabus C, Boulant S, Lavergne JP, Penin François, et al. The hepatitis C virus core protein is a potent nucleic acid chaperone that directs dimerization of the viral (+) strand RNA in vitro. Nucleic Acids Res. (2004) 32:2623–31. doi: 10.1093/nar/gkh579
36. Ivanyi-Nagy R, Kanevsky I, Gabus C, Lavergne JP, Ficheux D, Penin François, et al. Analysis of hepatitis C virus RNA dimerization and core-RNA interactions. Nucleic Acids Res. (2006) 34:2618–33. doi: 10.1093/nar/gkl240
37. Sharma KK, Rocquigny HDe, Darlix JL, Lavergne JP, Pénin F, Lessinger JM, et al. Analysis of the RNA chaperoning activity of the hepatitis C virus core protein on the conserved 3′X region of the viral genome. Nucleic Acids Res. (2012) 40:2540–53. doi: 10.1093/nar/gkr1140
38. Sharma KK, Didier P, Darlix JL, de Rocquigny H, Bensikaddour H, Lavergne JP, et al. Kinetic analysis of the nucleic acid chaperone activity of the hepatitis C virus core protein. Nucleic Acids Res. (2010) 38:3632–42. doi: 10.1093/nar/gkq094
39. Mamiya N, Worman HJ. Hepatitis C virus core protein binds to a DEAD box RNA helicase. J Biol Chem. (1999) 274:15751–6. doi: 10.1074/jbc.274.22.15751
40. Owsianka AM, Patel AH. Hepatitis C virus core protein interacts with a human DEAD box protein DDX3. Virology. (1999) 257:330–40. doi: 10.1006/viro.1999.9659
41. You L-R, Chen C-M, Yeh T-S, Tsai T-Y, Mai R-T, Lin C-H, et al. Hepatitis C virus core protein interacts with cellular putative RNA helicase. J Virol. (1999) 73. doi: 10.1128/jvi.73.4.2841-2853.1999
42. Ariumi Y, Kuroki M, Abe K-i, Dansako H, Ikeda M, Wakita T, et al. DDX3 DEAD-box RNA helicase is required for hepatitis C virus RNA replication. J Virol. (2007) 81. doi: 10.1128/jvi.01517-07
43. Angus AG, Dalrymple D, Boulant S, McGivern DR, Clayton RF, Scott MJ, et al. Requirement of cellular DDX3 for hepatitis C virus replication is unrelated to its interaction with the viral core protein. J General Virol. (2010) 91(1):122–32. doi: 10.1099/vir.0.015909-0
44. Li Q, Pène Véronique, Krishnamurthy S, Cha H, Liang TJ. Hepatitis C virus infection activates an innate pathway involving IKK-α in lipogenesis and viral assembly. Nat Med. (2013) 19:722–295. doi: 10.1038/nm.3190
45. Corless L, Crump CM, Griffin SDC, Harris M. Vps4 and the ESCRT-III complex are required for the release of infectious hepatitis C virus particles. J Gen Virol. (2010) 91:362–72. doi: 10.1099/vir.0.017285-0
46. Ariumi Y, Kuroki M, Maki M, Ikeda M, Dansako H, Wakita T, et al. The ESCRT system is required for hepatitis C virus production. PloS One. (2011) 6. doi: 10.1371/journal.pone.0014517
47. Barouch-Bentov R, Neveu G, Xiao F, Beer M, Bekerman E, Schor S, et al. Hepatitis C virus proteins interact with the endosomal sorting complex required for transport (ESCRT) machinery via ubiquitination to facilitate viral envelopment. MBio. (2016) 7:e01456–16. doi: 10.1128/mBio.01456-16
48. Deng L, Liang Y, Ariffianto A, Matsui C, Abe T, Muramatsu M, et al. Hepatitis C virus-induced ROS/JNK signaling pathway activates the E3 ubiquitin ligase itch to promote the release of HCV particles via polyubiquitylation of VPS4A. J Virol. (2022) 96. doi: 10.1128/jvi.01811-21
49. Kumar S, Barouch-Bentov R, Xiao F, Schor S, Pu S, Biquand E, et al. MARCH8~Ubiquitinates the hepatitis C virus nonstructural 2 protein and mediates viral envelopment. Cell Rep. (2019) 26:1800–1814.e5. doi: 10.1016/j.celrep.2019.01.075
50. Neveu G, Barouch-Bentov R, Ziv-Av A, Gerber D, Jacob Y, Einav S. Identification and targeting of an interaction between a tyrosine motif within hepatitis C virus core protein and AP2M1 essential for viral assembly. PloS Pathog. (2012) 8. doi: 10.1371/journal.ppat.1002845
51. Boson B, Mialon Chloé, Schichl K, Denolly Solène, Cosset FrançoisLoïc. Nup98 is subverted from annulate lamellae by hepatitis C virus core protein to foster viral assembly. MBio. (2022) 13. doi: 10.1128/mbio.02923-21
52. Owen DJ, Collins BM, Evans PR. Adaptors for clathrin coats: structure and function. Annu Rev Cell Dev Biol. (2004) 20:153–91. doi: 10.1146/annurev.cellbio.20.010403.104543
53. Izumida K, Hara Y, Furukawa Y, Ishida K, Tabata K, Morita E. Purification of hepatitis C virus core protein in non-denaturing condition. J Virological Methods. (2024) 323:114852. doi: 10.1016/j.jviromet.2023.114852
Keywords: hepatitis C virus (HCV), core, host partners, assembly, budding
Citation: Izumida K and Morita E (2024) The roles of HCV core protein and its binding host factor in virus assembly and release. Front. Virol. 4:1383261. doi: 10.3389/fviro.2024.1383261
Received: 07 February 2024; Accepted: 23 February 2024;
Published: 15 March 2024.
Edited by:
Akio Adachi, Tokushima University, JapanReviewed by:
Kohji Moriishi, University of Yamanashi, JapanCopyright © 2024 Izumida and Morita. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Eiji Morita, bW9yaXRhZUBoaXJvc2FraS11LmFjLmpw
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.