
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
BRIEF RESEARCH REPORT article
Front. Virol. , 15 April 2024
Sec. Viral Disease Investigation
Volume 4 - 2024 | https://doi.org/10.3389/fviro.2024.1381001
This article is part of the Research Topic Enterovirus Surveillance in Europe and Beyond View all 6 articles
Enterovirus A71 (EV-A71) is among the most neuropathogenic non-polio enterovirus types and, in rare instances, can lead to severe or even fatal outcomes, particularly in children under 5 years of age. This case study presents clinical and microbiological findings from the initial documented severe pediatric EV-A71 case in Finland, identified in May 2019. The near-complete genome sequence confirms that the EV-A71 strain belongs to the newly identified recombinant C1-like EV-A71 genetic lineage, which emerged in 2015 and has since been circulating in Europe, causing severe cases among children in various European countries. Enhanced environmental surveillance revealed widespread circulation of EV-A71 in Finland in 2019. However, the overall number of EV clinical cases remained lower than in previous years.
Enteroviruses (EV), belonging to the Picornaviridae family, are highly contagious and can lead to a spectrum of clinical manifestations. While many individuals may experience asymptomatic or mild infections, certain populations, such as neonates, young children, and individuals with compromised immune systems, are at an increased risk of developing severe complications. The clinical presentation of severe EV infections is diverse, ranging from severe respiratory distress and neurological complications to acute flaccid paralysis (1).
Enteroviruses have a single-stranded, positive-sense RNA genome more than 7000 nucleotides long. Enterovirus genus consists of more than three hundred different types in 15 species, Enterovirus A to L and Rhinovirus A to C. In humans, diseases are caused by Enterovirus species A - D and Rhinoviruses. The most common enteroviruses associated with severe infections include poliovirus, enterovirus D68 and enterovirus A71 (EV-A71) (2). Since 1990s, EV-A71, from Enterovirus species A, has caused large hand-foot-and-mouth-disease (HFMD) epidemics in Asia-Pacific region (3). While most HFMD cases recover in 1-2 weeks, severe CNS complications, such as meningitis, brainstem encephalitis, encephalomyelitis and/or acute flaccid paralysis, have been seen especially in patients under 5 years of age. Due to the emergence of large EV-A71 epidemics, with substantial proportion of severe or fatal cases, EV-A71 is considered as the most neuropathogenic non-polio enterovirus globally (4, 5). EV-A71 is classified into seven genotypes (from A up to G) (6, https://www.rivm.nl/mpf/typingtool/enterovirus/). The most prevalent genotypes B and C are further divided into multiple sub-genotypes (1).
In Europe, the emergence of severe EV-A71 infections took place in 2015 and coincided with the appearance of a new C1-like genetic lineage. This novel multirecombinant virus was initially identified in Germany (7–10), with subsequent reports of severe cases in France (11, 12). In 2016, an outbreak involving approximately 300 patients occurred in Spain (13–15). Since then, the C1-like genotype has been consistently detected in many European countries, including Denmark (16, 17) and Poland (18).
We present here clinical and microbiological findings of a fatal EV-A71 case, which, to our knowledge, is the first documented fatal EV-A71 case in Finland. The genetic characteristics of the virus were studied through whole-genome sequencing. Additionally, we describe the public health measures that were implemented.
A seven months old male patient was admitted to intensive care unit (ICU) at Helsinki University Hospital in May 2019, after one-week travel to the Balkan region in Europe. Prior to admission he had fever for four days and vomiting and loose stools for one day. The family had returned to Finland a day before admittance to the hospital. The rest of the family were healthy, but the mother reported that the older sibling had rash around his mouth and poor appetite during their travel. The family had travelled to their destination one week earlier and had sought for medical care one day before returning to Finland. The patient was noted to have no respiratory symptoms but was suspected to have a throat infection. At the airport the child vomited and was vomiting again on their flight back. Back home he received antipyretics and went to sleep. During the night he was breastfed and noticed to be feverish. His temperature was 38.5°C and he was lethargic. He had a transient convulsion, and his eyes were deviating, whereafter he was brought to the local hospital by an ambulance at 5 a.m. His vital signs remained normal during the transfer.
At hospital he was examined and admitted to the ward for follow up at 8 a.m. Leukocytes were slightly elevated 18.5 E9/L and CRP was 38 mg/L. Laboratory results were otherwise within normal variation. Four hours later he rapidly deteriorated before re-assessment. He was pale and cold sweaty, and his blood pressure was 70-85/40-70. His heart rate was 170, he had tachypnea and his consciousness was impaired. His capillary blood-pH was 7.14, base-excess 16 mmol/L. Plasma sodium (Na) was 133 mmol/L and leukocytes 28.1 E6/L. Intravenous fluid resuscitation was initiated, and the patient was transferred to ICU at the University Hospital. In ambulance he was breathing spontaneously (rate 28-36) with O2-supplement and mask ventilation support. His oxygen saturation was 99% but decreased to 71% when arriving to the ICU. At ICU he was immediately resuscitated, and was thereafter dependent on mechanical ventilation and inotrophic drugs to support sufficient circulation. He had convulsions and was treated with anticonvulsants.
Lumbal puncture was performed. Spinal fluid sample showed findings suggestive for viral meningitis: leucocytes 220 E6/L (99% mononuclear), protein 370 mg/L and glucose slightly elevated.
EEG-monitoring demonstrated burst suppression. Magnetic resonance imaging (MRI) of the brain was performed three days after admission to ICU and serious brain damage was demonstrated (Figures 1, 2). The treatment was discontinued the same day due to unfavorable prognosis for recovery. At autopsy hypoxic ischemic encephalopathy was seen and, furthermore, encephalitis lesions in the brain stem could be demonstrated.
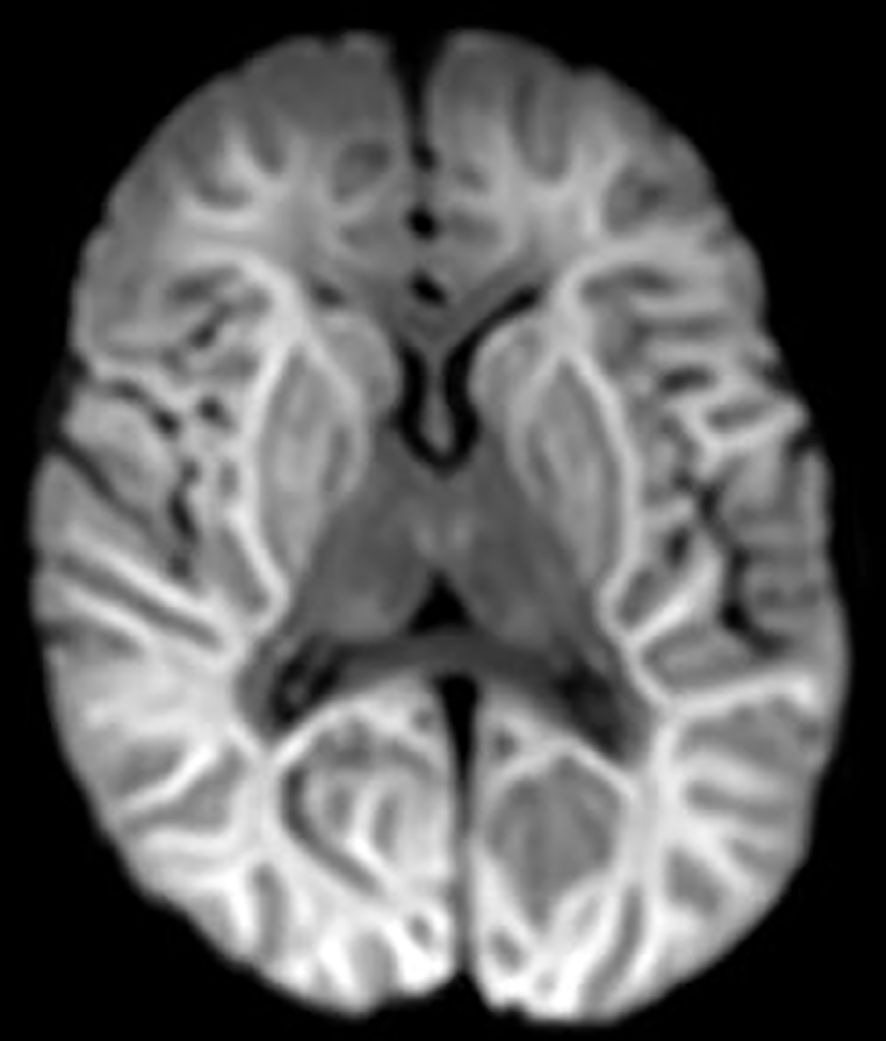
Figure 1 DWI (diffusion weighted imaging) shows diffusion restriction in both hemispheres compatible with hypoxic ischemic encephalopathy. Also, basal ganglia and thalami are involved.
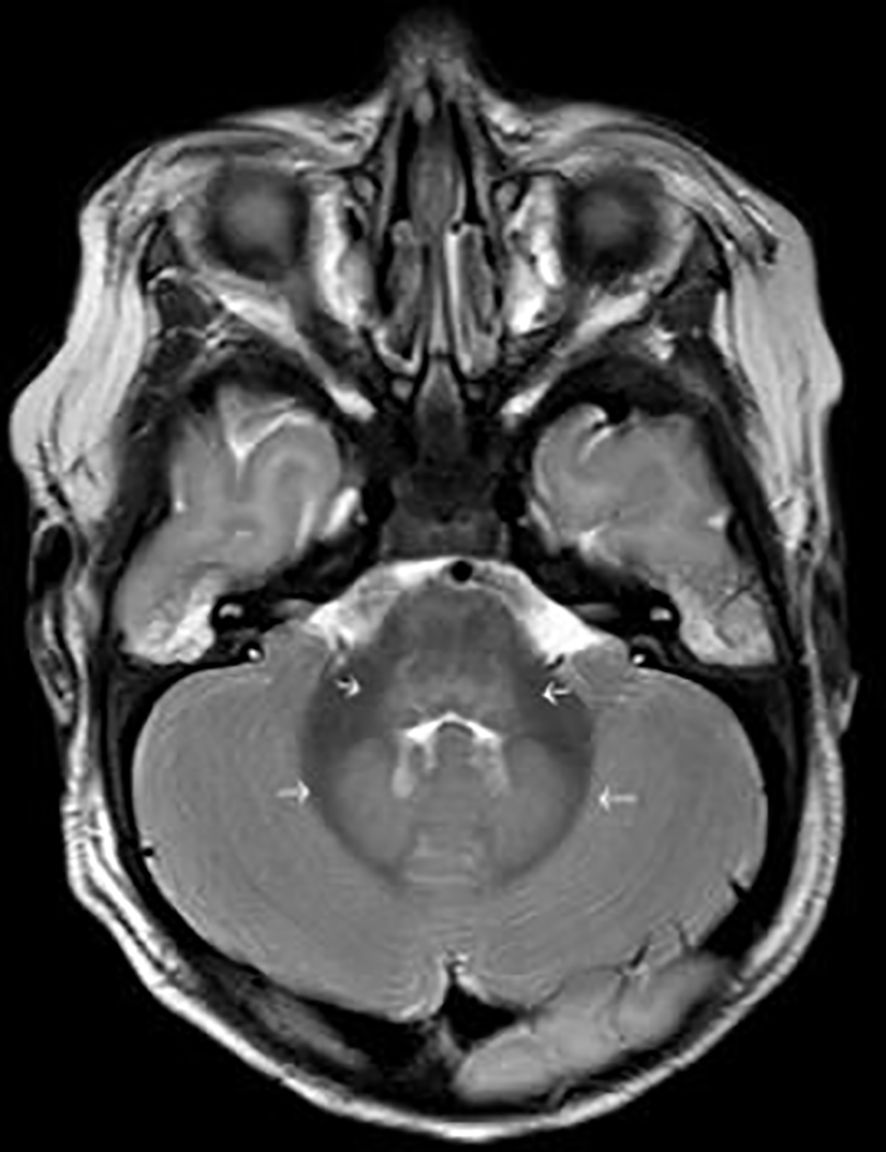
Figure 2 T2 weighted image shows oedema in dorsal part of brain stem and in dentate nuclei, common sites of involvement in enterovirus infection (short arrows in the middle). There is also general oedema compatible with hypoxic ischemic encephalopathy (thin arrows around the brain).
Serum, cerebrospinal fluid (CSF), tracheal aspirate and fecal samples were collected for microbiological testing on day 5 after the onset of symptoms. The CSF sample was negative for herpes simplex virus (HSV), Varicella-Zoster virus (VZV), human herpesvirus 6 (HHV6), enterovirus, and bacterial nucleic acids, and there were no antibodies against HSV, VZV, HHV6 or Mycoplasma pneumoniae in the CSF or serum. In addition, the serum was negative for antibodies against cytomegalovirus (CMV) and Epstein-Barr virus (EBV). No respiratory syncytial virus, influenza-, adeno-, parainfluenza-, picorna-, human metapneumo-, corona- or human bocavirus nucleic acids were detected from tracheal aspirate. Virus cultivation was carried out from a fecal sample, and enterovirus was isolated and detected in Vero cells (strain FIN/2019/1589). No other microbial pathogens were detected from the sample. A second fecal sample was taken post-mortem, and shown to be positive for enterovirus by PCR.
The virus strain FIN/2019/1589 was typed as EV-A71 by partial VP1 sequencing (19) using a previously described procedure (20). Phylogenetic analysis using Maximum Likelihood analysis (21) with the Kimura 2-parameter substitution model (22) showed that the EV-A71 strain was closely related to subgenotype C1-like viruses first detected in Germany in 2015 (7) (Figure 3A).
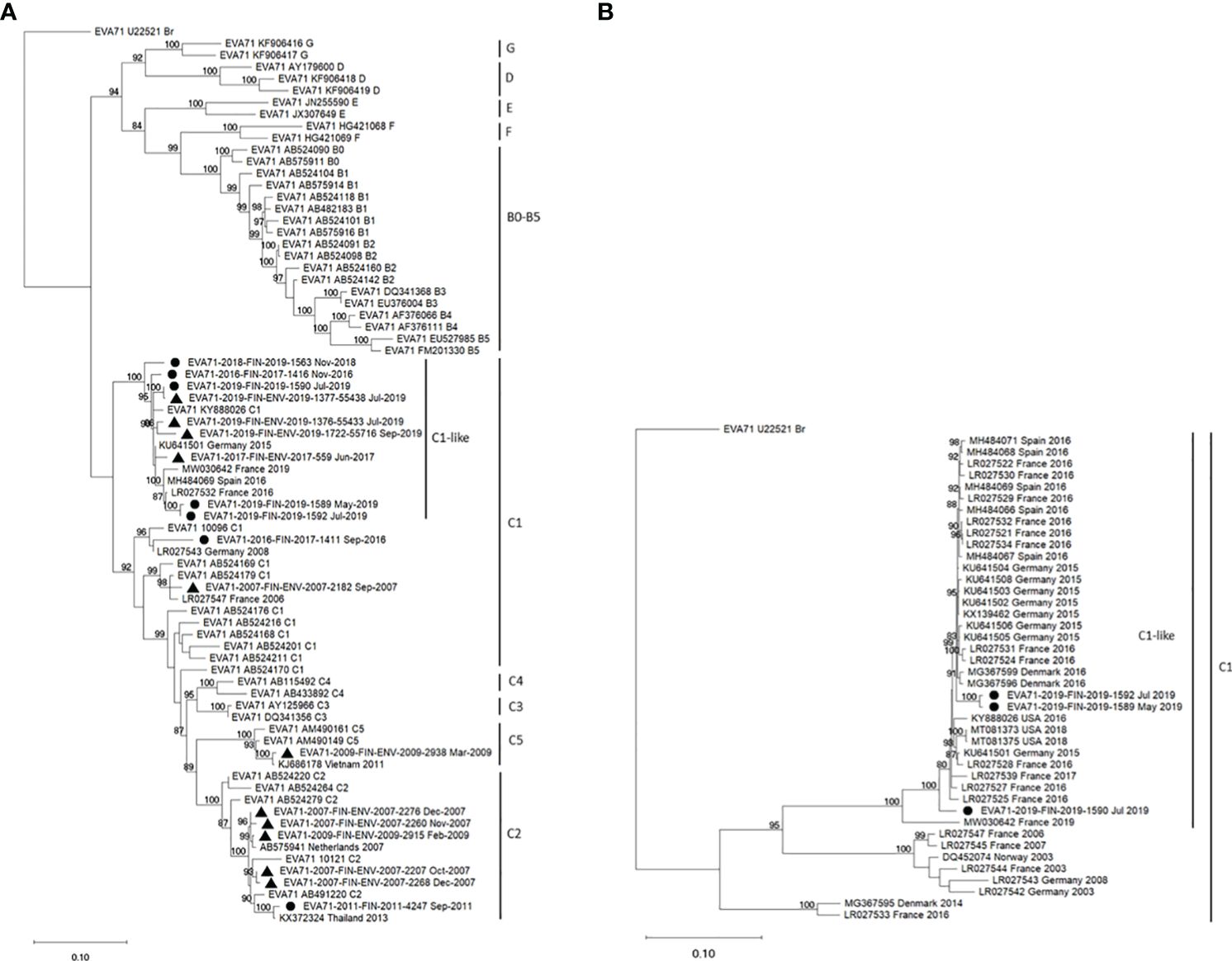
Figure 3 Phylogenetic analysis of EV-A71 strains isolated in Finland from 2007 to 2019 was conducted. Maximum Likelihood trees (21) were generated using the Kimura 2-parameter method (22) in W-IQTree (23, 24) with ultrafast bootstrap approximation (25), visualized in iTOL [Interactive tree of life, (26), https://itol.embl.de/] and finalized in MEGA11 (27). Bootstrap values ≥80% are indicated next to the branches (28). (A) The phylogenetic tree was constructed using 453 nucleotides from the 3’ end of the VP1 capsid protein encoding region. The analysis comprised 76 nucleotide sequences, including 7 clinical isolates (●) and 11 environmental isolates (▲) from Finland, 9 selected representative sequences from GenBank, and all 49 reference EV-A71 sequences from the RIVM database used for subtyping of EV-A71 strains (https://www.rivm.nl/mpf/typingtool/enterovirus/). (B) Phylogenetic analysis involved 1386 nucleotides encoding the complete 3D polymerase encoding region. The dataset included 3 clinical strains from Finland, 39 C1-subgenotype sequences from the USA or Europe selected from GenBank, and the prototype strain (accession number U22521).
The near-complete genome sequence of the FIN/2019/1589 strain, as well as two other clinical EV-A71 strains isolated in Finland in 2019 (FIN/2019/1590, FIN/2019/1592), were determined through amplification of the genome with pan-EV- and HEV-A specific primers (29). Library construction was carried out using the Rapid Barcoding Kit 96 (SQK-RBK110.96, Oxford Nanopore Technologies, ONT), followed by sequencing on the MinION (ONT) device. The raw sequence data were utilized to generate consensus sequences with Geneious Prime software (version 2023.0.2, Biomatters Ltd.), employing the MiniMap2 assembler (30) and a set of reference EV-A71 sequences sourced from the GenBank (https://www.ncbi.nlm.nih.gov/genbank). The resulting sequences (nucleotides 102-7393, following the numbering of EV-A71 prototype strain BrCr, GenBank accession number U22521) were classified as subtype C1 using both the Enterovirus Genotyping Tool Version 1.0 (https://www.rivm.nl/mpf/typingtool/enterovirus/31) and MegaBLAST in BLAST®, National Library of Medicine, National Center for Biotechnology Information.
The GenBank sequences with the highest similarities to FIN/2019/1589 were EV-A71 C1-like subcluster sequences from Spain and France, collected in 2016 (according to Blast search as of 17.11.2023). Near-complete genome nucleotide identities with MH484069 (Spain 2016) and LR027532 (France 2016) were 97.86% and 97.71%, respectively. In the complete 2193 amino acid long protein coding region, there were 9 and 8 substitutions compared to MH484069 and LR027532, respectively. The only amino acid difference in the capsid coding region was at VP1 position 13, where the Spanish and French strains had Met instead of Val, as observed in FIN/2019/1589 and the prototype strain BrCr.
Recombination was analyzed by constructing a maximum likelihood tree based on the complete 3D polymerase encoding region (Figure 3B). The Finnish strains FIN/2019/1589, FIN/2019/1590 and FIN/2019/1592 clustered together with the C1-like strains from Europe and the USA. Notably, strain MW030642 from Marseilles, France, 2019, previously reported as a novel recombinant form (32), displayed an exception. Further investigation of recombination between complete EV-A71 genomes was conducted using SimPlot++ software (33), employing a Jukes-Cantor distance model with a sliding window of 200 nucleotides moving in 20-nucleotide steps. As illustrated in Figure 4, high levels of nucleotide similarity were observed throughout the genome between the Finnish strains and C1-like strains from Europe during 2015-2016, indicating no additional recombination during the circulation of these subgenotype C1 strains between 2016 and 2019. However, strain MW030642 from Marseilles in 2019, exhibited distinct differences from the other strains.
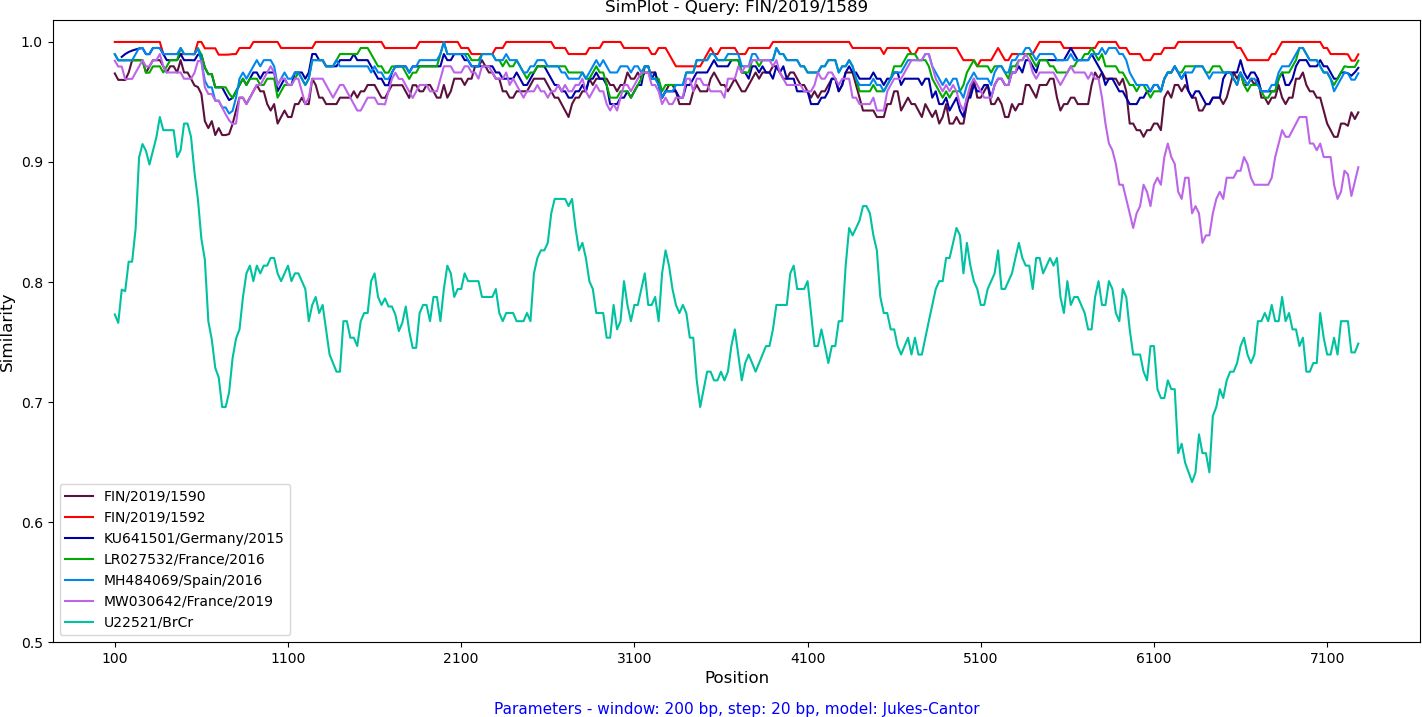
Figure 4 Analysis of the near-complete genome nucleotide sequence of the FIN/2019/1589 EV-A71 strain was conducted through a similarity plot. The Jukes-Cantor distance model was applied using a sliding window of 200 nucleotides, moving in 20 nucleotide steps. The FIN/2019/1589 EV-A71 strain served as the query sequence and was compared to the Finnish clinical strains, 2019, EV-A71 prototype strain BrCr (U22521) as well as representative sequences from EV-A71 outbreaks in Germany, 2015 (KU641501), Catalonia, Spain, 2016 (MH484069), and France, 2016 (LR027532) and 2019 (MW030642).
In Finland, clinical microbiology laboratories report all EV detections to the National Infectious Disease Register maintained by the Finnish Institute for Health and Welfare (THL). The total number of EV cases was 151 in 2019, significantly lower than in the previous years (357 cases in 2018, 283 in 2017). However, EV findings are not systematically genotyped. Although the notification of HFMD or EV-A71 cases to THL is encouraged, it is not mandatory. THL had previously been notified of clusters of EV-A71 cases in 2000 and 2007. Typing data from environmental samples collected in 2007 suggest the circulation of both subgenotypes C1 and C2 (Figure 3A). In other years, detections have typically involved singular cases (C2 in 2011, C1 in 2016) or rare instances found in sewage samples (C2 and C5 in 2009).
The first C1-like subgroup virus infection was reported to THL in the autumn of 2016, and the same virus type was identified once in a sewage sample in 2017. The second patient was notified in November 2018, preceding the case described here, which occurred in May 2019. Two additional cases from the Helsinki-Uusimaa region were detected in July 2019; these cases involved a 1-month-old male (FIN/2019/1590) and a 3-month-old female (FIN/2019/1592), respectively, with no known severe symptoms. The near-complete genome sequences of these cases exhibited 95.83% and 99.49% nucleotide identities with the index case FIN/2019/1589, respectively.
To investigate the prevalence of EV-A71 in Finland, sewage samples collected in 2019 for environmental surveillance of polio and other enteroviruses were subjected to screening for EV-A71 using a specific real-time RT-PCR assay (34). The assay was retrospectively conducted on cytopathic effect (CPE) positive RD isolates from samples collected between January and June 2019. Starting from July, RD isolates were additionally inoculated into Vero cells, known for efficiently supporting EV-A71 growth. CPE positive Vero cells were then utilized in the EV-A71 specific real-time RT-PCR assay.
In 2019, the initial detection of EV-A71 in sewage occurred in early May 2019. By July, the virus was identified at three of the five collection sites, including two sites in the Helsinki-Uusimaa region and one in Lapland. Notably, EV-A71 continued to be detected in Helsinki sewage through November. Although the screening was performed by a rapid and specific real-time assay, three of the positive findings were confirmed by partial VP1 sequencing. These environmental strains clustered in VP1 region together with the C1-like strains from Finland and other Europe (Figure 3A).
This is the first established severe pediatric EV-A71 case in Finland. The extraordinarily rapid progression of the disease was distracting for all health care personnel participating in care of the patient, but a neuropathogenic virus was highly suspected as a causative etiology at early stage of his care. EV-A71 is rarely detected in sterile sites (35), as CSF or blood, as was the case with our patient. However, shedding EV-A71 in stool is frequent, which was the means the virus was detected in our patient. The source of the infection remains unresolved. The incubation period of enteroviruses varies, and it is possible that the case acquired the infection in Finland before traveling abroad for one week.
Continuous evolution and recombination events contribute to the emergence of novel EV-A71 strains, which can lead to variations in clinical outcomes. The availability of complete genome sequence data is essential for understanding the genetic diversity of EV-A71, which is crucial for effective epidemiological surveillance, vaccine development, and the design of antiviral strategies. The genomic results presented in this study confirm that the new C1-like recombinant EV-A71 genetic lineage was the causative agent of the fatal case described. Additionally, a similar EV-A71 strain was detected in two non-severe neonatal cases in Finland in 2019. This C1-like lineage emerged in 2015 and has since circulated in Europe in various recombinant forms (32), leading to severe cases among children in several European countries.
EV-A71 strains, which clustered in the VP1 region together with the new C1-like viruses, had been detected in Finland through both clinical and environmental surveillance already before 2019. However, the absence of complete genome sequences for all EV-A71 strains included in this study limits our ability to interpret the spread of the C1-like strains to Finland. Despite the similarity in clustering within the VP1 region, identifying the specific recombinant forms is not feasible. This underscores the significance of ongoing research on the molecular epidemiology of complete EV-A71 genomes to deepen our comprehension of the virus and enhance public health responses to outbreaks.
In Finland, as in other European countries, EV infections usually reach their peak in the autumn, with the number of cases varying from year to year. In 2019, the confirmed number of EV cases was notably lower than in previous years, and the reason for this variation is not known. Due to the discovery of an EV-A71 case in May 2019, efforts to enhance EV communication, surveillance, and typing were implemented, but the overall case count remained low. Only three EV-A71 cases were confirmed during the season. However, the broader circulation of EV-A71 was confirmed through environmental surveillance.
Public health responses to enterovirus outbreaks aim to prevent the spread of the virus, mitigate the impact on affected individuals, and reduce the severity of cases. These responses involve collaboration with international health organizations and neighboring countries to share information and coordinate responses, particularly if cross-border outbreaks are suspected. Upon identifying EV-A71 as the causative agent of a fatal case, the case was promptly reported through the ECDC EPIS system and subsequently through a confidential information system to Finnish health authorities, including chief pediatricians. Clinical microbiology laboratories were encouraged to report and type enteroviruses in a timely manner. Public awareness was raised in August, coinciding with the typical start of the enterovirus season in Finland. The primary public message emphasized the importance of good hygiene practices, particularly frequent handwashing, especially in settings with a high risk of transmission.
The sequence datasets presented in this study can be found in the online repository GenBank https://www.ncbi.nlm.nih.gov/genbank/ under accession numbers PP417828–PP417845.
Ethical approval was not required for the study involving humans in accordance with the local legislation and institutional requirements. Written informed consent to participate in this study was not required from the participants or the participants’ legal guardians/next of kin in accordance with the national legislation and the institutional requirements. Written informed consent was obtained from the minor(s)’ legal guardian/next of kin for the publication of any potentially identifiable images or data included in this article.
TN: Writing – original draft, Writing – review & editing. AJ: Methodology, Writing – original draft, Writing – review & editing. EL: Methodology, Writing – review & editing. SB: Methodology, Writing – original draft, Writing – review & editing. CS: Supervision, Writing – review & editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was financially supported by HUS Diagnostic Center (Helsinki University Hospital, Helsinki, Finland; TYH2019263 and TYH2023102).
We thank Paula Väre (HUS, Diagnostic Center, Helsinki, Finland; virus cultivation and PCR) for her excellent technical assistance and the staff of the National Poliovirus Laboratory (THL), especially Katri Keino for screening of EV-A71 from environmental samples and Anna Kolodiazieva for the ONT sequencing.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
1. Solomon T, Lewthwaite P, Perera D, Cardosa MJ, McMinn P, Ooi MH. Virology, epidemiology, pathogenesis, and control of enterovirus 71. Lancet Infect Dis. (2010) 10:778–90. doi: 10.1016/S1473-3099(10)70194-8
2. Savolainen-Kopra C, Blomqvist S, Susi P. Enteroviruses (Picornaviridae). In: Bamford DH, Zuckerman M, editors. Encyclopedia of Virology (Fourth Edition). Oxford: Academic Press (2021). p. 245–55. doi: 10.1016/B978-0-12-809633-8.21544-6
3. Puenpa J, Wanlapakorn N, Vongpunsawad S, Poovorawan Y. The history of enterovirus A71 outbreaks and molecular epidemiology in the Asia-Pacific region. J BioMed Sci. (2019) 26(1):75. doi: 10.1186/s12929-019-0573-2
4. Ooi MH, Wong SC, Lewthwaite P, Cardosa MJ, Solomon T. Clinical features, diagnosis, and management of enterovirus 71. Lancet Neurol. (2010) 9:1097–105. doi: 10.1016/S1474-4422(10)70209-X
5. Jones E, Pillay TD, Liu F, Luo L, Bazo-Alvarez JC, Yuan C, et al. Outcomes following severe hand foot and mouth disease: A systematic review and meta-analysis. Eur J Paediatr Neurol. (2018) 22:763–73. doi: 10.1016/j.ejpn.2018.04.007
6. Bessaud M, Razadindratsimandresy R, Nougairède A, Joffret ML, Deshpande JM, Dubot-Pérès A. Molecular comparison and evolutionary analyses of VP1 nucleotide sequences of new African human enterovirus 71 isolates reveal a wide genetic diversity. PloS One. (2014) 9:e90624. doi: 10.1371/journal.pone.0090624
7. Böttcher S, Obermeier PE, Neubauer K, Diedrich S. Laboratory network for enterovirus diagnostics. Recombinant enterovirus A71 subgenogroup C1 strains, Germany 2015. Emerg Infect Dis. (2016) 22:1843–6. doi: 10.3201/eid2210.160357
8. Karrasch M, Fischer E, Scholten M, Sauerbrei A, Henke A, Renz DM, et al. A severe pediatric infection with a novel enterovirus A71 strain, Thuringia, Germany. J Clin Virol. (2016) 84:90–5. doi: 10.1016/j.jcv.2016.09.007
9. Böttcher S, Diedrich S, Keeren K. The Laboratory Network For Enterovirus Diagnostic LaNED. Increased detection of enterovirus A71 infections, Germany 2019. Euro Surveill. (2019) 24(39):pii=1900556. doi: 10.2807/1560-7917.ES.2019.24.39.1900556
10. Akinnurun OM, Narvaez Encalada M, Orth J, Petzold M, Böttcher S, Diedrich S, et al. Enterovirus A71-associated acute flaccid paralysis in a pediatric patient: a case report. J Med Case Rep. (2023) 17:310. doi: 10.1186/s13256-023-04041-6
11. Antona D, Kossorotoff M, Schuffenecker I, Mirand A, Leruez-Ville M, Bassi C, et al. Severe paediatric conditions linked with EV-A71 and EV-D68, France, May to October 2016. Euro Surveill. (2016) 21:30402. doi: 10.2807/1560-7917.ES.2016.21.46.30402
12. Ngangas ST, Lukashev A, Jugie G, Ivanova O, Mansuy JM, Mengelle C, et al. Multirecombinant enterovirus A71 subgenogroup C1 isolates associated with neurologic disease, France 2016-2017. Emerg Infect Dis. (2019) 25:1204–8. doi: 10.3201/eid2506.181460
13. Casas-Alba D, de Sevilla MF, Valero-Rello A, Fortuny C, García-García JJ, Ortez C, et al. Outbreak of brainstem encephalitis associated with enterovirus-A71 in Catalonia, Spain, (2016): a clinical observational study in a children’s reference centre in Catalonia. Clin Microbiol Infect. (2017) 23:874–81. doi: 10.1016/j.cmi.2017.03.016
14. González-Sanz R, Casas-Alba D, Launes C, Muñoz-Almagro C, Ruiz-García MM, Alonso M, et al. Molecular epidemiology of an enterovirus A71 outbreak associated with severe neurological disease, Spain 2016. Euro Surveill. (2019) 24:1800089. doi: 10.2807/1560-7917.ES.2019.24.7.1800089
15. Taravilla CN, Pérez-Sebastián I, Salido AG, Serrano CV, Extremera VC, Rodríguez AD, et al. Enterovirus A71 infection and neurologic disease, Madrid, Spain 2016. Emerg Infect Dis. (2019) 25:25–32. doi: 10.3201/eid2501.181089
16. Midgley SE, Nielsen AG, Trebbien R, Poulsen MW, Andersen PH, Fischer TK. Co-circulation of multiple subtypes of enterovirus A71 (EV- A71) genotype C, including novel recombinants characterised by use of whole genome sequencing (WGS), Denmark 2016. Euro Surveill. (2017) 22:30565. doi: 10.2807/1560-7917.ES.2017.22.26.30565
17. Foli-Andersen PJ, Munkholm A, Rønde G, Børresen ML, Nielsen JEK, Midgley S, et al. Acute flaccid rhombencephalomyelitis with radiculitis in a child with an enterovirus A71 infection seen for the first time in Denmark: a case report. J Med Case Rep. (2022) 16:32. doi: 10.1186/s13256-021-03246-x
18. Wieczorek M, Purzyńska M, Krzysztoszek A, Ciąćka A, Figas A, Szenborn L. Genetic characterization of enterovirus A71 isolates from severe neurological cases in Poland. J Med Virol. (2018) 90:372–6. doi: 10.1002/jmv.24958
19. Oberste MS, Nix WA, Maher K, Pallansch MA. Improved molecular identification of enteroviruses by RT-PCR and amplicon sequencing. J Clin Virol. (2003) 26:375–7. doi: 10.1016/s1386-6532(03)00004-0
20. Blomqvist S, Paananen A, Savolainen-Kopra C, Hovi T, Roivainen M. Eight years of experience with molecular identification of human enteroviruses. J Clin Microbiol. (2008) 46:2410–3. doi: 10.1128/JCM.00313-08
21. Felsenstein J. Evolutionary trees from DNA sequences: A maximum likelihood approach. J Mol Evol. (1981) 17:368–76. doi: 10.1007/BF01734359
22. Kimura M. A simple method for estimating evolutionary rate of base substitutions through comparative studies of nucleotide sequences. J Mol Evol. (1980) 16:111–20. doi: 10.1007/BF01731581
23. Nguyen L-T, Schmidt HA, von Haeseler A, Minh BQ. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum likelihood phylogenies. Mol Biol Evol. (2015) 32:268–74. doi: 10.1093/molbev/msu300
24. Trifinopoulos J, Nguyen L-T, von Haeseler A, Minh BQ. W-IQ-TREE: a fast online phylogenetic tool for maximum likelihood analysis. Nucleic Acids Res. (2016) 44:W232–5. doi: 10.1093/nar/gkw256
25. Hoang DT, Chernomor O, von Haeseler A, Minh BQ, Vinh LS. UFBoot2: Improving the ultrafast bootstrap approximation. Mol Biol Evol. (2017) 35:518–22. doi: 10.1093/molbev/msx281
26. Letunic I, Bork P. Interactive Tree Of Life (iTOL) v5: an online tool for phylogenetic tree display and annotation. Nucleic Acids Res. (2021) 49(W1):W293–6. doi: 10.1093/nar/gkab301
27. Tamura K, Stecher G, Kumar S. MEGA 11: molecular evolutionary genetics analysis version 11. Mol Biol Evol. (2021) 38(7):3022–7. doi: 10.1093/molbev/msab120
28. Felsenstein J. Confidence limits on phylogenies: An approach using the bootstrap. Evolution. (1985) 39:783–91. doi: 10.1111/j.1558-5646.1985.tb00420.x
29. Joffret ML, Polston PM, Razafindratsimandresy R, Bessaud M, Heraud JM, Delpeyroux F. Whole genome sequencing of enteroviruses species A to D by high-throughput sequencing: application for viral mixtures. Front Microbiol. (2018) 9:2339. doi: 10.3389/fmicb.2018.02339
30. Li H. Minimap2: pairwise alignment for nucleotide sequences. Bioinformatics. (2018) 34:3094–100. doi: 10.1093/bioinformatics/bty191
31. Kroneman A, Vennema H, Deforche K, Avoort HV, Penaranda S, Oberste MS, et al. Koopmans. An automated genotyping tool for enteroviruses and noroviruses. J Clin Virol. (2011) 51:121–5. doi: 10.1016/j.jcv.2011.03.006
32. Luciani L, Morand A, Zandotti C, Piorkowski G, Boutin A, Mazenq J, et al. Circulation of enterovirus A71 during 2019-2020, Marseille, France. J Med Virol. (2021) 93:5163–6. doi: 10.1002/jmv.26893
33. Samson S, Lord É, Makarenkov V. SimPlot++: a Python application for representing sequence similarity and detecting recombination. Bioinformatics. (2022) 38:3118–20. doi: 10.1093/bioinformatics/btac287
34. Thanh TT, Anh NT, Tham NT, Van HM, Sabanathan S, Qui PT, et al. Validation and utilization of an internally controlled multiplex Real-time RT-PCR assay for simultaneous detection of enteroviruses and enterovirus A71 associated with hand foot and mouth disease. Virol J. (2015) 12:85. doi: 10.1186/s12985-015-0316-2
35. World Health Organization, Regional Office for the Western Pacific. A guide to clinical management and public health response for hand, foot and mouth disease (HFMD). WHO Regional Office for the Western Pacific (2011). Available at: https://iris.who.int/handle/10665/207490.
Keywords: enterovirus A71, enterovirus, whole-genome sequencing, viral meningitis, molecular epidemiology
Citation: Nieminen T, Jääskeläinen AJ, Lindh E, Blomqvist S and Savolainen-Kopra C (2024) A fatal pediatric infection with a C1-like subgenogroup enterovirus A71: case study and enterovirus A71 epidemiology in Finland. Front. Virol. 4:1381001. doi: 10.3389/fviro.2024.1381001
Received: 02 February 2024; Accepted: 25 March 2024;
Published: 15 April 2024.
Edited by:
Thea Kølsen Fischer, Nordsjællands Hospital, DenmarkReviewed by:
Andi Krumbholz, University of Kiel, GermanyCopyright © 2024 Nieminen, Jääskeläinen, Lindh, Blomqvist and Savolainen-Kopra. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Soile Blomqvist, c29pbGUuYmxvbXF2aXN0QHRobC5maQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.