- 1Centre for Virus Research, Kenya Medical Research Institute, Nairobi, Kenya
- 2School of Biomedical Sciences, Jomo Kenyatta University of Agriculture and Technology, Nairobi, Kenya
- 3ILRI Genomics, International Livestock Research Institute, Nairobi, Kenya
- 4Human Health Division, International Centre of Insect Physiology and Ecology, Nairobi, Kenya
Background: Until recently, arbovirus surveillance is mainly focused on mosquito and tick vectors, resulting in the discovery of several mosquito- and tick-borne arboviruses. However, the role of sandflies in arbovirus transmission and disease has remained largely unexplored. This study sought to isolate and characterize arboviruses from phlebotomine sandflies from selected pastoral ecozones in the North Rift region of Kenya.
Methods: Sandflies were collected from selected sites in North Rift Kenya between 2015 and 2018. They were sorted and pooled by sex, site, and collection date. The pools were homogenized and inoculated onto Vero cells for virus isolation. The positive pools were analyzed by polymerase chain reaction targeting different arboviruses. The isolates were further characterized by high-throughput sequencing using Illumina Miseq platform.
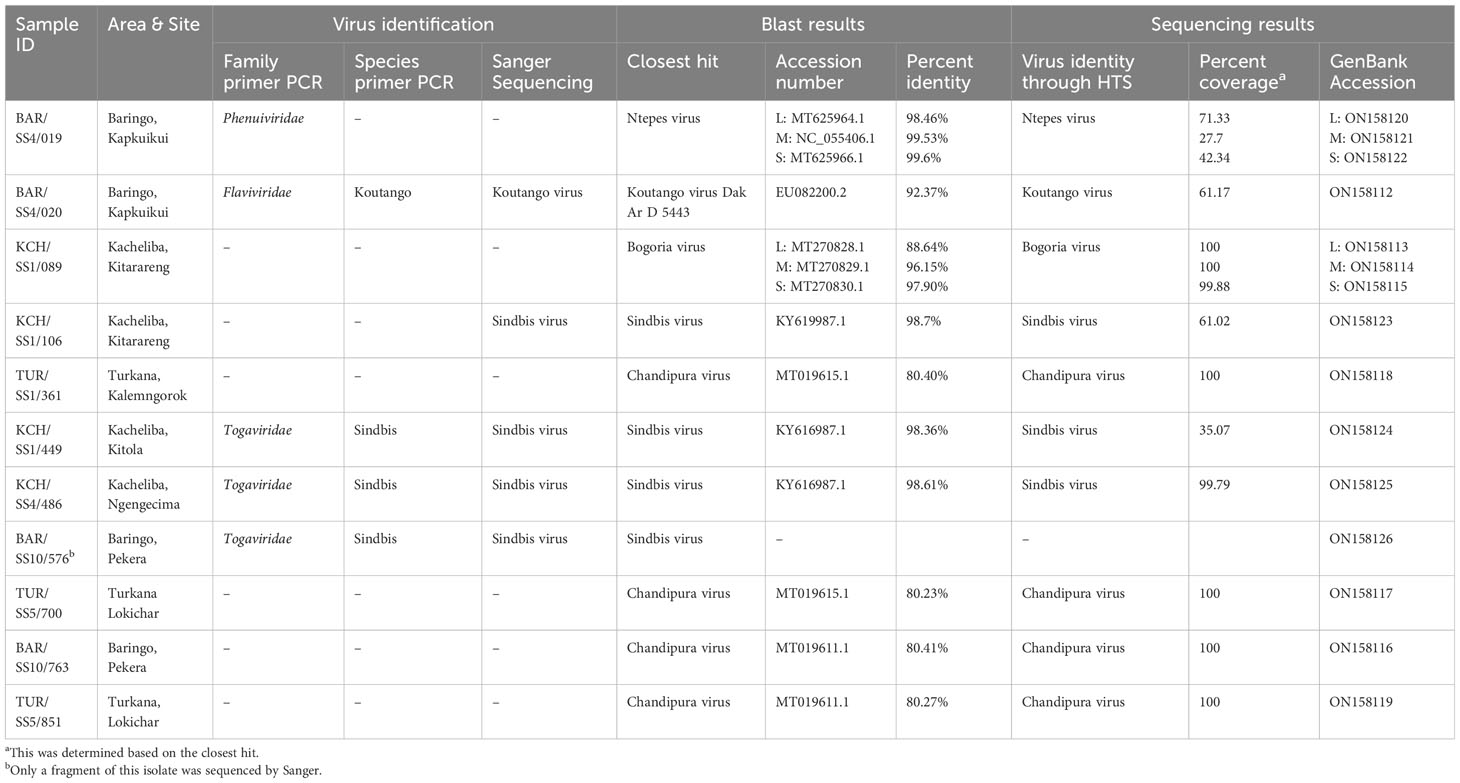
Results: Approximately 28,226 sandflies translating to 824 pools were sampled from the selected regions. A total of 11 showed reproducible cytopathic effects on Vero cells. We identified five arboviruses: sindbis (n = 4) from Kacheliba and Baringo, Chandipura (n = 4) from Turkana and Baringo, Koutango (n = 1) and Ntepes (n = 1) from Baringo, and Bogoria (n = 1) from Kacheliba. The percent identities of the identified viruses were approximately 80% to 98% compared to known viruses in GenBank, suggesting that some of them could be novel viruses.
Conclusion: This study successfully isolated and characterized five arboviruses from sandflies. The findings suggest that sandflies are potential hosts of a wide range of arboviruses and are therefore important vectors to consider in arbovirus surveillance and evaluated for their ability to transmit them. Further studies are needed to determine the public health importance and extent of exposure of these viruses to humans and livestock populations.
Introduction
Phlebotomine sandflies belong to the order Diptera, family Psychididae, subfamily Phlebotominae, and genus Phlebotomus. They include a diverse group of vectors that vary widely in geographic distribution, ecology, and the pathogens that they transmit (1). They are known to transmit both human and animal pathogens, including protozoans that cause leishmaniasis, some bacteria species that cause Carrion’s disease, and a number of phleboviruses that cause sandfly fever (2, 3). They can transmit arboviruses that cause fever, hemorrhage, and even encephalitis, endangering human health and adding to the arbovirus disease burden (3).
Arboviruses (arthropod-borne viruses) are viruses that are transmitted by hematophagous arthropods like mosquitoes, ticks, sandflies, and biting midges to humans and other non-human vertebrates. There are over 500 arboviruses distributed around the world, and approximately 100 are known to cause diseases in humans (4). They include flaviviruses like dengue virus, West Nile virus, and yellow fever virus; alphaviruses like sindbis virus and chikungunya virus; vesiculoviruses like Chandipura virus; and phleboviruses like Rift Valley virus and sandfly fever naples virus.
In the recent past, several sandfly-borne viruses have been on the rise. These viruses are mostly found in the tropics of the New World and semi-arid and temperate parts of the Old World, such as the Mediterranean, North Africa, and central and western Asia, and they can cause short-term febrile illnesses (sandfly fever) or more serious neuroinvasive diseases (5). The past decade has seen many discoveries of new sandfly-borne viruses, some of which have been linked to disease and deaths in different countries, including Turkey, India, and the Balkan region (6–9). Additionally, in recent years, novel viral species have been isolated from sandfly specimens collected all over the world (10).
While the occurrence and spread of sandfly-borne viruses have been reported in several parts of the world, data for East Africa and Kenya in particular is sparse. Sandflies are abundant in the semi-arid parts of Kenya where the climate is hot and humid, including the North Rift and the Eastern parts of Kenya, where they are linked to outbreaks of cutaneous and visceral leishmaniasis in the human population (1). It is likely that the populations in these areas may also be exposed to unrecognized sandfly-transmitted diseases like sandfly fever. Entomological surveillance of phlebotomine sandflies has led to the detection of arboviruses previously known to be mosquito-borne and tick-borne and even novel viruses which have been detected recently (11–14). In this study, we report the isolation, characterization, and identification of sandfly-borne viruses, including known and/or new sandfly-borne viruses from sandfly populations sampled from selected regions of Baringo, Turkana, and West-Pokot counties in North Rift, Kenya.
Methodology
Study sites
The study sites are located in the North Rift region of Kenya (Figure 1) and were purposely selected due to the high sandfly population in the regions and the endemicity of cutaneous and visceral leishmaniasis. The climate of the North Rift region is generally arid/semi-arid, with hot and dry conditions prevailing throughout much of the year. It is characterized by low and erratic rainfalls of between 500 and 1,000 mm. The region is inhabited by communities practicing agro-pastoralism.
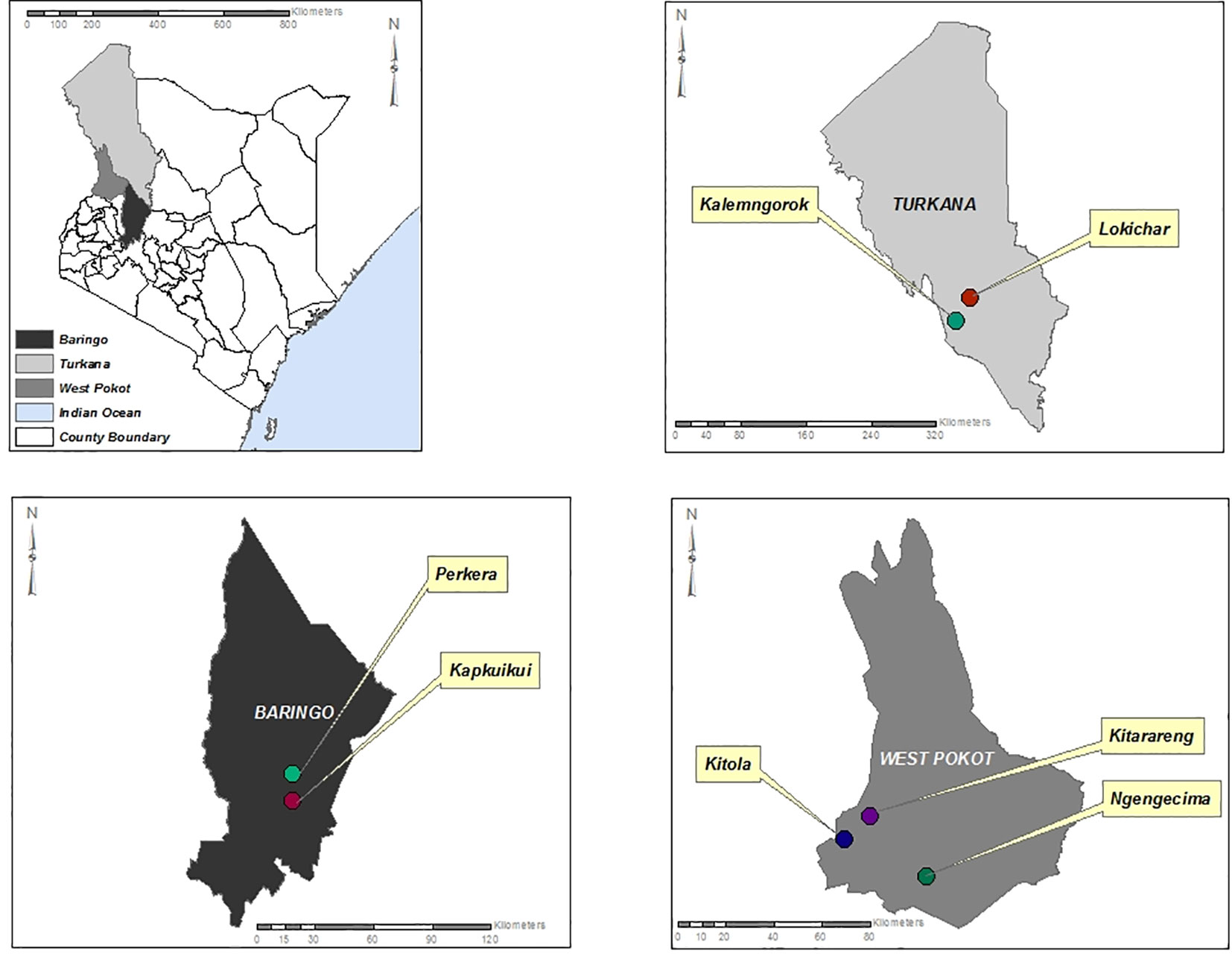
Figure 1 Map of Turkana, Baringo, and West Pokot counties showing the sites where the sandfly sampling was conducted.
Sandfly collection, identification, and pooling
Sandfly collection was carried out in selected sites including Kitola, Kitarareng, and Ngengecima in Kacheliba, West Pokot, Kalemngorok, and Lokichar in Turkana, and Perkera and Kapkuikui in Baringo counties (Figure 1) between October 2015 and July 2018 using the CDC miniature light traps (John W. Hock Company, Gainesville, FL, USA). The collection was done mainly during the wet season (March, August, and October to December) (Table 1) as the numbers of sandflies are high during that time; hence, there is a high vector–vertebrate interaction. The traps were set mostly near live anthills, which serve as breeding habitats for sandflies, 1 m above the ground. A total of 10 traps per night were set overnight beginning from 6 to 7 pm and retrieved between 6 and 7 am of the following morning. The trapped sandflies were knocked down chemically using tri-ethyl amine to immobilize or inactivate them, sorted under a light microscope to remove mosquitoes and other insects that were accidentally trapped during collection, put in cryovials or Eppendorf, and transported to the entomology laboratory at Kenya Medical Research Institute (KEMRI) in liquid nitrogen tanks for further identification and pooling. Mosquitoes accidentally trapped during sandfly collection were put aside and screened in a different project.
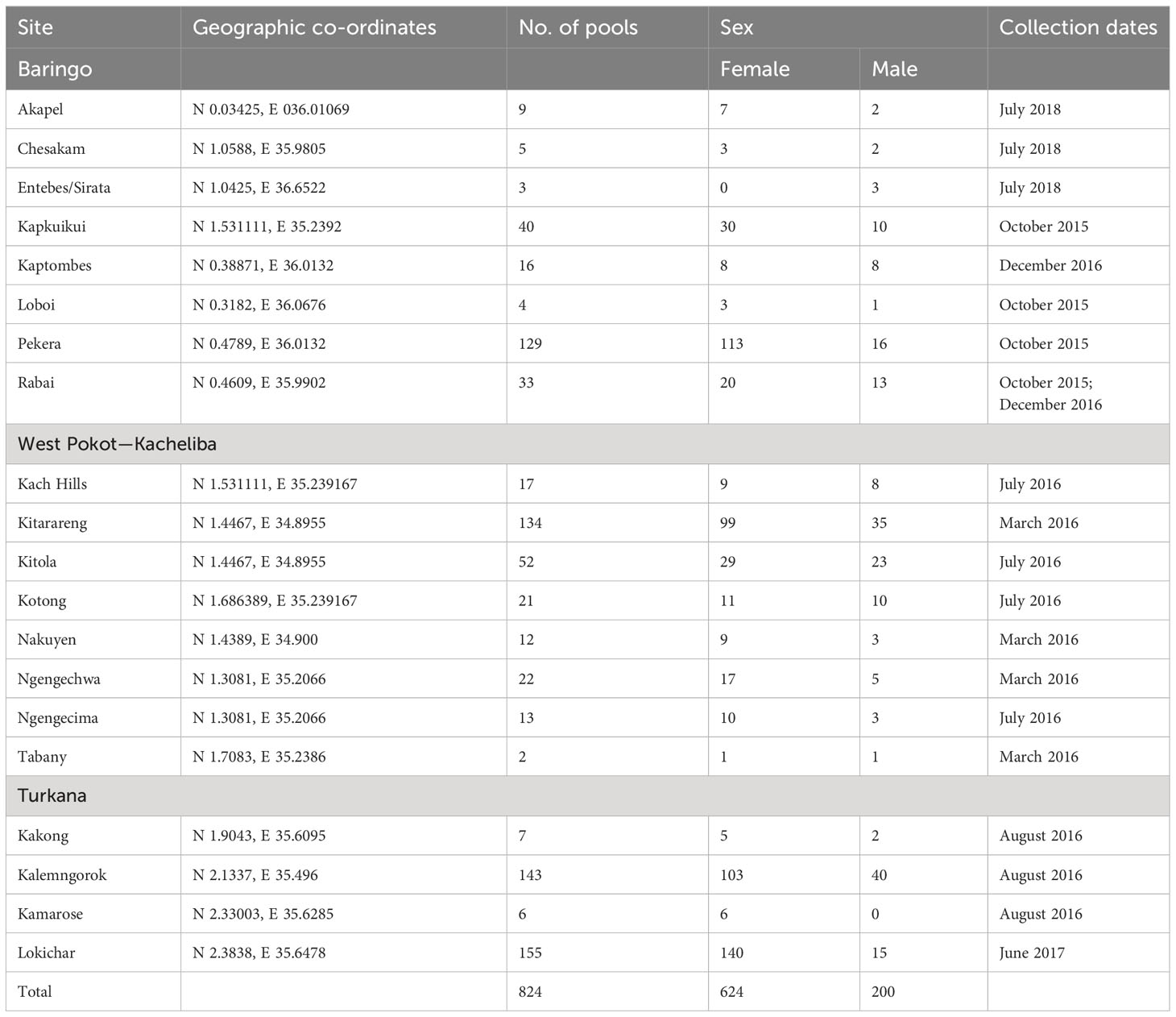
Table 1 Details of sandfly collection from selected sites in the North Rift region of Kenya for virus isolation.
In the lab, the sandflies were identified as to the genus level, separated from other insects like Ceratopogonidae using morphological features (15), and pooled by sex, trapping area, and date of collection with up to 50 sandflies per pool/vial. This is because the sandfly body size is very small, and thus a pool of 50 sandflies will be able to achieve a concentration that will yield viral isolation within the 14-day incubation period in cell culture. The pool numbers also depended on the sandfly abundance, site, and date of collection. They were stored at -80°C while awaiting the virus isolation process.
Cell culture and virus isolation
Virus isolation was done at the Viral Hemorrhagic Fever Laboratory, KEMRI. Vero CCL 81 cells, derived from kidney epithelial cells of an African green monkey, were seeded in 24-well plates using Minimum Essential Media (MEM) containing 10% fetal bovine serum (FBS), 2% L-glutamine, and 2% antibiotic/antimycotic and then incubated at 37°C overnight to enable them to attain at least 80% confluence. The sandfly pools were homogenized in the Omni bead ruptor using 0.45-mm copper beads and homogenizing media (Minimum Essential Media) with 15% (FBS), 2% antibiotic–antimycotic, and 2% L-glutamine at a speed of 1,500 rotations per minute for 30 s and centrifuged at 12,000 rpm for 10 min to separate the supernatant from the debris. The supernatant was then transferred into a sterile cryovial, inoculated in 80% confluent Vero CCL-81 cells, and monitored for 14 days for cytopathic effects (CPE). Pools showing CPE were harvested, frozen, and re-inoculated (passaged) in freshly prepared Vero CCL-81 cells to check for reproducible CPE.
Molecular analysis and sequencing
Viral RNA was extracted from the cell culture isolates using QIAmp® Viral RNA Extraction MiniKit (Qiagen GmbH, Hilden, Germany) according to the manufacturer’s instructions. The extracted RNAs were reverse-transcribed to cDNA using Superscript III cDNA synthesis kit (Invitrogen, Thermo Fisher Scientific, Waltham, MA, USA). Polymerase chain reaction (PCR) was performed using AmpliTaq Gold 360 Master Mix (Invitrogen Thermofisher Scientific, Waltham, MA, USA), together with primers targeting different families of medically important arboviruses (Togaviridae, Flaviviridae, and Phenuiviridae) (Table 2). Any pool that was found to be positive by family primers was further screened using primers specific to the different viruses within the family. Sanger sequencing was done for the sandfly pools that were successfully amplified using a specific set of primers to confirm the identity of the virus isolated from the pool. PCR amplicons were cleaned with ExoCIP (NEB, Frankfurt, Germany), and the cleaned amplicons were submitted for sequencing. Chromatogram files were imported into Chromas v2.6, with the low-quality regions trimmed and the fasta sequences generated for each virus. The sequences generated were compared to publicly available sequences in GenBank using Basic Local Alignment Search Tool (BLAST). Additionally, a subset of the isolates identified by Sanger sequencing as well as those that could not be amplified using the available primers was subjected to high-throughput sequencing (HTS).
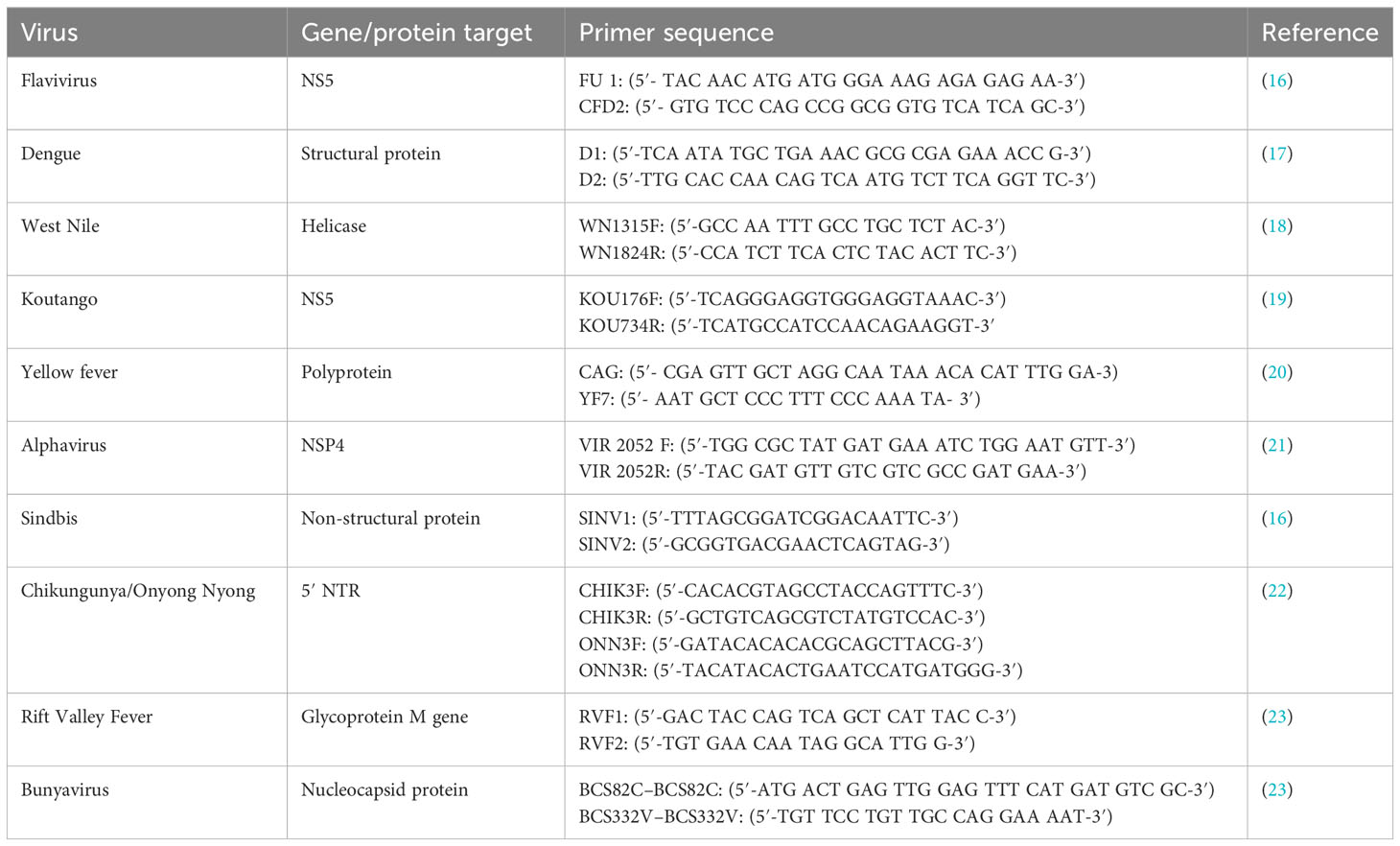
Table 2 Sequences and target regions of the primers used in the identification of Arbovirus isolates.
Library preparation, pooling, and high-throughput sequencing
Library preparation for the CPE-positive samples was performed using the TruSeq Stranded mRNA Library Prep Kit (Illumina, San Diego, CA, USA), according to the manufacturer’s instruction with slight modifications to exclude purification of the PolyA-containing mRNA procedure. Briefly, the RNA was fragmented and primed for cDNA synthesis using Superscript III reverse transcriptase (Invitrogen, Thermofisher Scientific, Waltham, MA, USA) and hexanucleotide primers (Invitrogen, Waltham, MA, USA). Second-strand cDNA synthesis was then performed using DNA Polymerase I and RNase H (Illumina, San Diego, CA, USA). A single adenine (A) nucleotide was then added to the 3′ ends of the blunt fragments. This was to create sticky ends for easy ligation with T-tailed adaptors and to prevent them from ligating to each other during adapter ligation reaction, thus ensuring a low rate of chimera formation. Ligation of the adapters to the ends of the double-stranded cDNA was performed, and the products were purified and enriched by PCR to create the final libraries. Equal volumes of normalized library were then combined, diluted, and denatured using sodium hydroxide (NaOH) in preparation for cluster generation and sequencing. The samples were then loaded onto the MiSeq Sequencing System (Illumina, San Diego, CA, USA). Sequencing was performed using Miseq Reagent V3 reagents (Illumina, San Diego, CA, USA) in a 600-cycle sequencing format.
Sequence analysis
The raw sequence data were cleaned to get high-quality reads for downstream analysis. The raw data were inspected with FastQC. The low-quality reads were subsequently filtered using Prinseq-lite v0.20.4. The high-quality reads were assembled using a de novo approach with Megahit v1.2.9, and the generated contigs were identified by comparing to other publicly available sequences using BlastX and BlastN approaches in GenBank.
Maximum likelihood phylogenetic trees were constructed for each of the identified viruses. Reference sequences were downloaded from GenBank and combined with the generated viral genomes. The combined sets were aligned using Muscle plugin embedded in MEGA7 software. The aligned sequences were manually inspected and edited using Bioedit software. Maximum likelihood phylogenies were inferred in IQTree, where model estimation and tree inference were performed simultaneously for all the viruses using 1,000 bootstrap estimates. The generated trees were visualized in FigTree v1.4.4.
Results
Sandfly collection
A total of 28,226 (24,139 female and 4,087 male) sandflies were collected and identified morphologically. This translated to 824 pools (624 female and 200 male) (Table 1).
Virus isolation and reverse transcription PCR
Out of the 824 sandfly pools processed, 11 gave reproducible CPE after three passages. The pools showed CPE on different days, indicating that some viruses were generating CPE fast while others were generating CPE slowly—for instance, samples BAR/SS4/019 and BAR/SS4/020 from Baringo county and KCH/SS1/089 from West Pokot county started showing CPE between days 2 and 3 post-infection, but it took more than 5 days for the CPE to spread across the cell monolayer and these were harvested between days 5 and 7, indicating that the three isolates are viruses generating CPE slowly. The observed CPE for the viruses generating CPE slowly were characterized by progressive rounding up and detaching of cells from the flask surface, with some of the cells degenerating and floating in the cell culture medium (Figures 2A, B, E). On the other hand, samples KCH/SS1/106, KCH/SS1/449, and KCH/SS4/486, all from West Pokot county, and the isolates from Turkana county (TUR/SS5/700 and TUR/SS5/851) generated CPE in less than 4 days post-inoculation/infection. The CPE generated by these viruses were characterized by very fast progressive rounding and clumping of the cells (Figures 2C, D). The negative control (Figure 2F) retained an intact and confluent monolayer of cells.
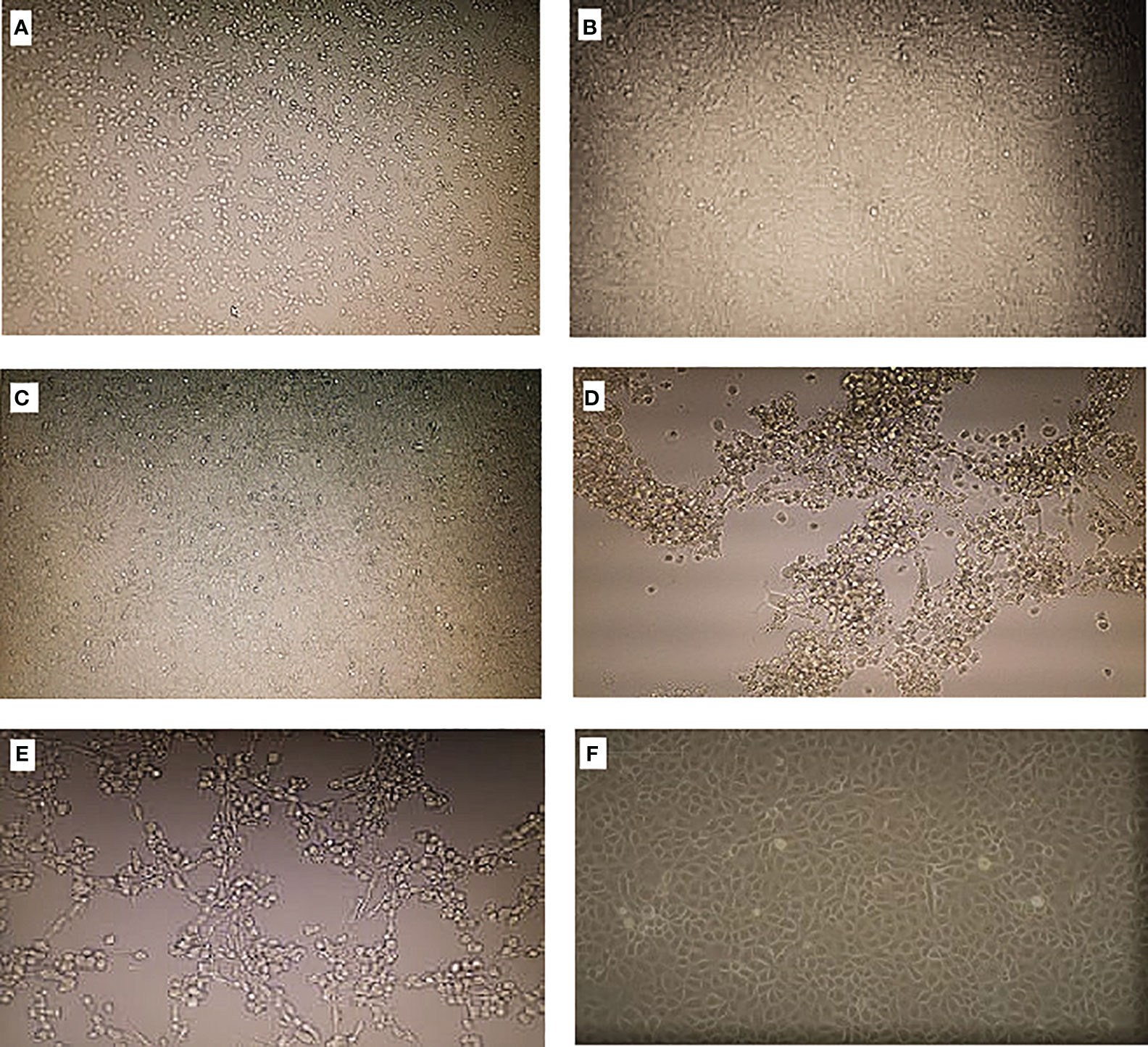
Figure 2 Cytopathic effects (CPE) on Vero cells when inoculated with the isolates. (A–C) Viruses generating CPE slowly. (D, E) Viruses generating CPE fast. (F) Negative control.
The isolates were harvested by freezing and thawing of the contents of the cell culture flask in ultra-low freezer (-65°C to -85°C) and thereafter centrifuging using a refrigerated centrifuge at 1,2000 rpms for 10 min to get the supernatant. The harvested samples were characterized using RT-PCR, partial sequencing, and HTS.
One sample BAR/SS4/020, which was positive using the Flaviviridae family primers, was further analyzed by RT-PCR using the virus-specific primers for West Nile virus, Koutango virus, dengue virus, and yellow fever virus and was positive for Koutango virus using the Koutango primers. Three samples—KCH/SS1/449, KCH/SS4/486, and BAR/SS10/576—tested positive using Togaviridae family primers and, when tested using alphavirus-specific primers, tested positive for sindbis virus. Sanger sequencing performed on the four identified isolates confirmed the RT-PCR results.
High-throughput sequencing and phylogenetic analysis
The 11 isolates were obtained and characterized using Sanger sequencing and HTS for further identification and characterization. Five isolates were sequenced by Sanger (one Koutango and four sindbis isolates), and out of these, one Koutango and three sindbis isolates, together with six unidentified isolates, were sequenced by HTS. The HTS results for the four virus isolates identified by Sanger matched. A total of five viruses were detected. Sindbis virus, which is a known mosquito-borne virus, was identified from sandfly pools from Kacheliba (n = 3). Chandipura virus was identified from sandfly pools from Turkana (n = 4) and Baringo (n = 4). Koutango virus, which is a known tick-borne virus, was identified in a sandfly pool from Baringo (n = 1). Ntepes virus (n = 1) and Bogoria virus (n = 1), which are both novel viruses, were identified in sandfly pools from Baringo and Kacheliba, respectively. BLAST search analysis for the genome sequences obtained showed very close relatedness to the known viruses (Table 3).
The phylogenetic analysis showed that isolate BAR/SS4/019 from Kapkuikui in Baringo clustered with Ntepes virus (Figures 3A–C), which is a phlebovirus recently described in Marigat within the same county of Baringo. The nucleotide identity of this isolate to known Ntepes virus was 98.46%, 99.53%, and 99.6% at L, M, and S segments, respectively (Table 3). Another isolate, KCH/SS1/089, from Kitarareng in Kacheliba, West Pokot, was also classified as a phlebovirus, and it clustered with a recently described Bogoria virus (Figures 3A–C). Compared with the known Bogoria virus, it had a nucleotide identity of 88.64%, 96.15%, and 97.90% at large, medium, and small segments, respectively (Table 3). The isolate BAR/SS4/020, also from Kapkuikui in Baringo, was identified as a Koutango virus, and it clustered with Koutango viruses from West Africa (Figure 3F). The nucleotide identity of the identified virus to a Koutango virus from GenBank (EU082200.2) was found to be 92.3% (Table 3). Three isolates from different regions of Kacheliba and one from Pekera in Baringo—KCH/SS1/106, KCH/SS1/449, KCH/SS4/486, and BAR/SS10/576—were identified as sindbis viruses, and they clustered under genotype 1 with other sindbis viruses from Kenya (Figure 3D). The percentage nucleotide identity scores to this previously reported sindbis virus were 98.7%, 98.36%, and 98.61%, respectively (Table 3). Four isolates—three from Turkana and one from Baringo (TUR/SS1/361, TUR/SS5/700, BAR/SS10/763, and TUR/SS5/851)—were all identified as Chandipura virus. These isolates formed a single cluster which was unique compared to the isolates from Senegal and India (Figure 3E). The nucleotide percent identity of the Chandipura isolates from Kenya to those from Senegal in West Africa was 80.4%, 80.23%, 80.41%, and 80.27%, respectively. The sequences of all the isolates from the study have been deposited in GenBank (Table 3).
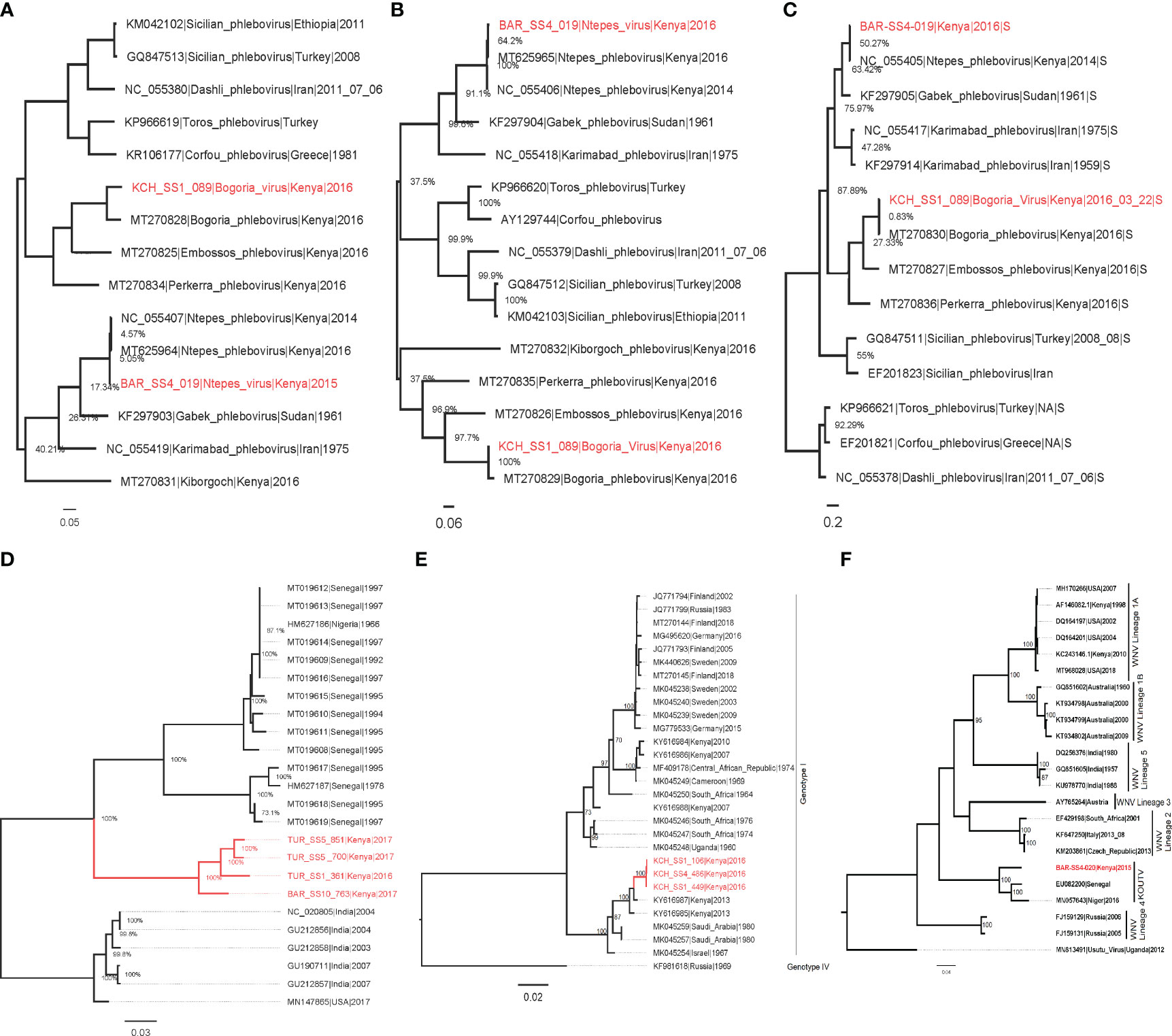
Figure 3 Maximum likelihood phylogenies of Ntepes and Bogoria virus segments L (A), M (B), and S (C), Chandipura virus (D), sindbis virus (E), and Koutango virus (F). The virus strains isolated from the study are shown in red. The trees were inferred based on 1,000 bootstrap replicates.
Discussion
Targeting of sandflies during arbovirus surveillance in Kenya has led to the discovery of known and novel phleboviruses, including arboviruses of both known and unknown medical importance. This study successfully isolated for the first time from sandflies sindbis virus, a known mosquito-borne virus, and Koutango virus, a known tick-borne arbovirus. There has been a recent increase in reports of sandfly-borne virus circulation across the world, including Europe, Middle East, America, and Africa (6, 24), where these viruses have been isolated from humans, rodents, and birds, which are potential reservoirs or incidental hosts (25–27). Some of the viruses isolated have low percent identity thresholds to previously identified viruses, as low as 80%, suggesting that phlebotomine sandflies are hosts to many potentially pathogenic viruses that remain unidentified.
In this study, we also successfully isolated Chandipura virus from sandflies sampled from neighboring Turkana and Baringo counties. This constitutes the first time that this virus has been isolated in Kenya from sandflies. The isolates obtained from this study form a single monophyletic clade, which is different from the other Chandipura virus strains from Africa and South Asia. However, the Chandipura virus identified from this study share a common ancestor with Chandipura virus strains previously isolated in West Africa though distantly related (Figure 3D). Chandipura virus, a Rhabdoviridae, was first isolated from sandflies and also human serum during an outbreak in Maharashtra in India, where it has been associated with an epidemic that killed thousands of children (28). After this initial detection, there have been subsequent isolations of this virus from sandflies and humans in various parts of India (9). In Africa, CHPV was previously isolated from wild-caught sandflies and a rodent, hedgehog, in a couple of West African countries (Senegal and Nigeria) (29). The strain of Chandipura virus isolated in this study can be considered as a unique lineage. From the phylogenetic analysis, the long branch length leading to the isolates obtained in this study indicates a relatively high evolutionary distance of the Kenyan Chandipura virus strain compared to the West African strain. This finding suggests that the Kenyan Chandipura virus strain may have been circulating undetected in Kenya for a long time. The detection of multiple isolates from the two different and distant sampling sites also implies the widespread circulation of the virus in parts of the country that goes on undetected.
A new strain of West Nile, Koutango virus lineage (WN-KOUTV), was also isolated and identified in this study. Koutango virus, which was first isolated in 1968 from a wild rodent in Tatera Kempi in Senegal and from a rodent mastomys in Central African Republic (30), was isolated from gebrils in Somalia later in 1974 (31) and recently from sandflies in Niger (12). This WN-KOUTV strain isolated from sandflies from Baringo county shares 92.37% identity with Dak Ar D-5443 Koutango strain from Senegal and forms a monophyletic clade with the WN-KOUTV strains from Senegal and Niger (Figure 3F). Koutango virus is mainly associated with ticks but is also known to be transmitted by Aedes spp. mosquitoes (32). This is also the first time that this virus is being isolated in Kenya, although the closely related West Nile virus has been commonly found circulating among birds, ticks, and mosquitoes in Kenya (33–36).
Sindbis virus (Family Togaviridae, genus Alphavirus) is a known mosquito-borne virus pathogen often transmitted by Culex species and occasionally by Aedes species mosquitoes. Different species of birds, including migratory as well as game birds, serve as amplifying hosts of sindbis virus. Sindbis virus was first isolated from Culex mosquitoes in 1952 in the Nile river delta in Egypt (37) and has subsequently been isolated over the years from mosquitoes and the amplifying hosts, i.e., birds and humans (38, 39). From this study, we isolated it from sandflies circulating in parts of North Rift, Kenya. This finding constitutes the first time that this virus has been isolated from these potential hosts (phlebotomine sandflies). Three sindbis virus isolates were identified in this study, with the phylogenetic analyses showing that the isolates share a high similarity among themselves, possibly because they were isolated from the same region. The isolates form a single cluster with two Kenyan sindbis virus isolates which were recently isolated from mosquitoes (40) (Figure 3E). The isolates share a high similarity with the sindbis virus isolates from mosquitoes with about 98% identity, suggesting that the same strain that infects mosquitoes also infects and is possibly transmitted by sandflies. The close relatedness of the isolated virus to the one previously isolated from mosquitoes in Boni, in the coastal region, is an indication that the virus could be circulating across the country among migratory birds. Furthermore, the isolation from this new vector suggests that these sandfly-associated sindbis virus originates from a common pool of virus quasi species circulating among mosquitoes, birds, and sandflies in Kenya.
Sandfly-associated phleboviruses were earlier limited to the Old World, which included the countries around the semi-arid Mediterranean basin. Over the past decade, a wide range of novel phleboviruses is being detected within the Mediterranean basin and even in other parts of the world (New World) (14, 24, 41, 42). In this study, two phleboviruses were isolated and identified and found to show a high similarity to two novel viruses which were recently identified in Kenya from sandflies: Ntepes virus and Bogoria virus (13). The Ntepes virus isolated in the study has over 98% identity to novel Ntepes virus recently isolated from sandflies collected in Marigat in Kenya in 2014 (Figures 3C). The short branches suggest a short evolutionary distance and limited diversity between the isolates. The Bogoria virus isolated in this study was identified from sandflies sampled from Kacheliba in West Pokot county. It has over 95% identity to the novel Bogoria virus recently identified in sandflies from Baringo county. The Bogoria virus isolates share a common ancestor and cluster with three other novel phleboviruses also isolated from the same locality: Embossos virus, Pekerra virus, and Kiborgoch virus (Figures 3C). The identification of these two phleboviruses with a high similarity to previously isolated novel viruses indicates that they are widely distributed in the North Rift region of Kenya. The implication of the circulation of all these viruses to public health should be explored as they may be contributing to frequently undiagnosed febrile illnesses that are reported in these areas.
Conclusion
The successful isolation and identification of sandfly-associated arboviruses indicate the presence and circulation of these viruses, some of which are known to be of public health importance, e.g., sindbis virus, Chandipura virus, and Koutango virus. The findings suggest that sandflies are potential hosts of a wide range of arboviruses, some of which are yet to be identified and/or characterized. Apart from mosquitoes and ticks, phlebotomine sandflies should be treated as important hosts to consider in arbovirus disease transmission ecology and epidemiology. They should therefore be targeted during arbovirus surveillance and whenever there are outbreaks of unidentified febrile illnesses. Potential novel viruses were also isolated, suggesting that phlebotomine sandflies are hosts to many circulating, potentially pathogenic viruses that remain unidentified. More studies are therefore recommended to determine the host and epidemic range of the viruses, the public health importance, and the impact and the level of exposure of these viruses to humans, livestock, and other animal populations in the study areas.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: NCBI GenBank, ON158112–ON158126. The raw sequence data used to generate genomes have been deposited in the NCBI SRA, PRJNA1023525.
Ethics statement
Approval for the study was granted by the Scientific and Ethical Review Unit of the Kenya Medical Research Institute, SSC No. 3690.
Author contributions
EK: Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Writing – original draft, Writing – review & editing. SL: Data curation, Formal analysis, Software, Visualization, Writing – review & editing. JM: Methodology, Writing – review & editing. FM: Methodology, Writing – review & editing. JL: Methodology, Resources, Writing – review & editing. HK: Methodology, Writing – review & editing. SO: Methodology, Resources, Writing – review & editing. RW: Supervision, Writing – review & editing. SM: Writing – original draft, Writing – review & editing, Methodology, Supervision. RS: Conceptualization, Project administration, Resources, Supervision, Writing – review & editing.
Funding
The author(s) declare that no financial support was received for the research, authorship, and/or publication of this article.
Acknowledgments
We thank Betty Chelangat, Samuel Owaka, John Gachoya, Dunstone Beti, and Reuben Lugalia for their technical assistance in the field and laboratory.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Anjili CO, Ngumbi PM, Kaburi JC, Irungu LW. The phlebotomine sandfly fauna (Diptera: Psychodidae) of Kenya. J Vector Borne Dis. (2011) 48:183.
2. Maroli M, Feliciangeli MD, Bichaud L, Charrel RN, Gradoni L. Phlebotomine sandflies and the spreading of leishmaniases and other diseases of public health concern. Med Vet Entomol. (2013) 27:123–47. doi: 10.1111/j.1365-2915.2012.01034.x
3. Depaquit J, Grandadam M, Fouque F, Andry PE, Peyrefitte C. Arthropod-borne viruses transmitted by Phlebotomine sandflies in Europe: a review. Euro surveillance: Bull Europeen sur les maladies transmissibles = Eur communicable Dis bulletin. (2010) 15:19507. doi: 10.2807/ese.15.10.19507-en
4. Quah SR, Cockerham W. International encyclopedia of public health. (2017) (Amsterdam: Academic Press, Elsevier).
5. Marklewitz M, Tchouassi DP, Hieke C, Heyde V, Torto B, Sang Ret , et al. Insights into the evolutionary origin of mediterranean sandfly fever viruses. mSphere. (2020) 5(5):10–1128. doi: 10.1128/mSphere.00598-20
6. Ayhan N, Charrel RN. Emergent sand fly–borne Phleboviruses in the Balkan region. Emerg Infect Dis. (2018) 24:2324. doi: 10.3201/eid2412.171626
7. Temocin F, Sari Tb, Tulek N. Sandfly fever with skin lesions: A case series from Turkey. J Arthropod Borne Dis. (2016) 10:608–612.
8. Torun Edis C, Yağçı Çağlayık D, Uyar Y, Korukluoğlu G, Ertek M. [Sandfly fever outbreak in a province at Central Anatolia, Turkey]. Mikrobiyol Bul. (2010) 44:431–9.
9. Sudeep AB, Gurav YK, Bondre VP. Changing clinical scenario in Chandipura virus infection. Indian J Med Res. (2016) 143:712–21. doi: 10.4103/0971-5916.191929
10. Elliott RM, Brennan B. Emerging phleboviruses. Curr Opin Virol. (2014) 5:50–7. doi: 10.1016/j.coviro.2014.01.011
11. Wang Q, Fu S, Cheng J, Xu X, Wang J, Wu B, et al. Re-isolation of wuxiang virus from wild sandflies collected from Yangquan County, China. Virologica Sinica. (2021) 36:1177–86. doi: 10.1007/s12250-021-00398-4
12. Fall G, Diallo D, Soumaila H, Ndiaye EH, Lagare A, Sadio BD, et al. First detection of the west nile virus koutango lineage in sandflies in Niger. Pathogens. (2021) 10(3):257. doi: 10.3390/pathogens10030257
13. Tchouassi DP, Marklewitz M, Chepkorir E, Zirkel F, Agha SB, Tigoi CC, et al. Sand fly-associated phlebovirus with evidence of neutralizing antibodies in humans, Kenya. Emerg Infect Dis. (2019) 25:681–90. doi: 10.3201/eid2504.180750
14. Marklewitz M, Dutari LC, Paraskevopoulou S, Page RA, Loaiza JR, Junglen S. Diverse novel phleboviruses in sandflies from the Panama Canal area, Central Panama. J Gen Virol. (2019) 100:938–49. doi: 10.1099/jgv.0.001260
15. Abonnenc E, Minter D. Bilingual keys for the identification of the sandflies of the Ethiopian region. Entomologie Medicale. (1965) 5.
16. Bryant JE, Crabtree MB, Nam VS, Yen NT, Duc HM, Miller BR. Isolation of arboviruses from mosquitoes collected in northern Vietnam. Am J Trop Med Hyg. (2005) 73:470–3. doi: 10.4269/ajtmh.2005.73.470
17. Lanciotti RS, Calisher CH, Gubler DJ, Chang GJ, Vorndam AV. Rapid detection and typing of dengue viruses from clinical samples by using reverse transcriptase-polymerase chain reaction. J Clin Microbiol. (1992) 30:545–51. doi: 10.1128/jcm.30.3.545-551.1992
18. Turell MJ, Dohm DJ, Sardelis MR, Oguinn ML, Andreadis TG, Blow JA. An update on the potential of north American mosquitoes (Diptera: Culicidae) to transmit West Nile Virus. J Med Entomol. (2005) 42:57–62. doi: 10.1093/jmedent/42.1.57
19. Sang R, Onyango C, Gachoya J, Mabinda E, Konongoi S, Ofula V, et al. Tickborne arbovirus surveillance in market livestock, Nairobi, Kenya. Emerg Infect Dis. (2006) 12:1074–80. doi: 10.3201/eid07.060253
20. Ayers M, Adachi D, Johnson G, Andonova M, Drebot M, Tellier R. A single tube RT-PCR assay for the detection of mosquito-borne flaviviruses. J Virol Methods. (2006) 135:235–9. doi: 10.1016/j.jviromet.2006.03.009
21. Eshoo MW, Whitehouse CA, Zoll ST, Massire C, Pennella TT, Blyn LB, et al. Direct broad-range detection of alphaviruses in mosquito extracts. Virology. (2007) 368:286–95. doi: 10.1016/j.virol.2007.06.016
22. Smith DR, Lee JS, Jahrling J, Kulesh DA, Turell MJ, Groebner JL, et al. Development of field-based real-time reverse transcription-polymerase chain reaction assays for detection of Chikungunya and O’nyong-nyong viruses in mosquitoes. Am J Trop Med Hyg. (2009) 81:679–84. doi: 10.4269/ajtmh.2009.09-0138
23. Kuno G, Mitchell CJ, Chang GJ, Smith GC. Detecting bunyaviruses of the Bunyamwera and California serogroups by a PCR technique. J Clin Microbiol. (1996) 34:1184–8. doi: 10.1128/jcm.34.5.1184-1188.1996
24. Calzolari M, Chiapponi C, Bellini R, Bonilauri P, Lelli D, Moreno A, et al. Isolation of three novel reassortant phleboviruses, Ponticelli I, II, III, and of Toscana virus from field-collected sand flies in Italy. Parasites Vectors. (2018) 11:84. doi: 10.1186/s13071-018-2668-0
25. Moriconi M, Rugna G, Calzolari M, Bellini R, Albieri A, Angelini P, et al. Phlebotomine sand fly-borne pathogens in the Mediterranean Basin: Human leishmaniasis and phlebovirus infections. PLoS Negl Trop Dis (2017) 11(8):. doi: 10.1371/journal.pntd.0005660
26. Becker N, Jöst H, Ziegler U, Eiden M, Höper D, Emmerich P, et al. Epizootic emergence of Usutu virus in wild and captive birds in Germany. PloS One. (2012) 7:e32604. doi: 10.1371/journal.pone.0032604
27. Hall RA, Scherret JH, Mackenzie JS. Kunjin virus: an Australian variant of West Nile? Ann New York Acad Sci. (2001) 951:153–60. doi: 10.1111/j.1749-6632.2001.tb02693.x
28. Dhanda V, Rodrigues FM, Ghosh SN. Isolation of Chandipura virus from sandflies in Aurangabad. Indian J Med Res. (1970) 58:179–80.
29. Ba Y, Trouillet J, Thonnon J, Fontenille D. [Phlebotomus of Senegal: survey of the fauna in the region of Kedougou. Isolation of arbovirus]. Bull la Societe pathologie exotique (1990). (1999) 92:131–5.
30. de Araujo Lobo JMDA. Koutango: Under Reported Arboviral Disease in West Africa. (Doctoral dissertation, Louisiana State University and Agricultural & Mechanical College) (2012) p. 32–6.
31. Butenko AM, Semashko IV, Skvortsova TM, Gromashevskiĭ VL, Kondrashina NG. [Detection of the koutango virus (Flavivirus, togaviridae) in Somalia]. Meditsinskaia parazitologiia i parazitarnye bolezni. (1986) 3):65–8.
32. Lobo JMDA, Christofferson RC, Mores CN. Investigations of koutango virus infectivity and dissemination dynamics in Aedes aEgypti mosquitoes. Environ Health Insights. (2014) 8:EHI-S16005. doi: 10.4137/EHI.S16005
33. Crabtree M, Sang R, Lutomiah J, Richardson J, Miller B. Arbovirus surveillance of mosquitoes collected at sites of active Rift Valley fever virus transmission: Kenya, 2006–2007. J Med Entomol. (2009) 46:961–4. doi: 10.1603/033.046.0431
34. LaBeaud AD, Sutherland LJ, Muiruri S, Muchiri EM, Gray LR, Zimmerman PA, et al. Arbovirus prevalence in mosquitoes, Kenya. Emerg Infect Dis. (2011) 17:233. doi: 10.3201/eid1702.091666
35. Lwande OW, Venter M, Lutomiah J, Michuki G, Rumberia C, Gakuya F, et al. Whole genome phylogenetic investigation of a West Nile virus strain isolated from a tick sampled from livestock in north eastern Kenya. Parasites Vectors. (2014) 7:1–10. doi: 10.1186/s13071-014-0542-2
36. Nyamwaya D, Wang’ondu V, Amimo J, Michuki G, Ogugo M, Ontiri E, et al. Detection of West Nile virus in wild birds in Tana river and Garissa Counties, Kenya. BMC Infect Diseases. (2016) 16:1–10. doi: 10.1186/s12879-016-2019-8
38. Eiden M, Ziegler U, Keller M, Müller K, Granzow H, Jöst H, et al. Isolation of sindbis virus from a hooded crow in Germany. Vector Borne Zoonotic Dis (Larchmont NY). (2014) 14:220–2. doi: 10.1089/vbz.2013.1354
39. Ziegler U, Fischer D, Eiden M, Müller K, Granzow H, Jöst H, et al. Sindbis virus- a wild bird associated zoonotic arbovirus circulates in Germany. Veterinary Microbiol. (2019) 239:108453. doi: 10.1016/j.vetmic.2019.108453
40. Sigei F, Nindo F, Mukunzi S, Ng’ang’a Z, Sang R. Evolutionary analyses of Sindbis virus strains isolated from mosquitoes in Kenya. Arch Virol. (2018) 163:2465–9. doi: 10.1007/s00705-018-3869-8
41. Xu Z, Fan N, Hou X, Wang J, Fu S, Song J, et al. Isolation and identification of a novel phlebovirus, Hedi Virus, from sandflies collected in China. Viruses. (2021) 13(5):772. doi: 10.3390/v13050772
Keywords: sandflies, Koutango virus, Chandipura virus, sindbis virus, Ntepes virus, Bogoria virus
Citation: Koskei E, Langat S, Mutisya J, Mulwa F, Lutomiah J, Koka H, Oyola SO, Waihenya R, Mabeya SN and Sang R (2024) Isolation and phylogenetic characterization of arboviruses circulating among phlebotomine sandflies in parts of North Rift, Kenya. Front. Virol. 4:1289258. doi: 10.3389/fviro.2024.1289258
Received: 05 September 2023; Accepted: 09 April 2024;
Published: 01 May 2024.
Edited by:
Shengzhang Dong, Johns Hopkins University, United StatesReviewed by:
Rafael Kroon Campos, University of Texas Medical Branch at Galveston, United StatesEdgar Simulundu, University of Zambia, Zambia
Copyright © 2024 Koskei, Langat, Mutisya, Mulwa, Lutomiah, Koka, Oyola, Waihenya, Mabeya and Sang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Edith Koskei, ZWNoZXBraXJ1aUBrZW1yaS5nby5rZQ==
 Edith Koskei
Edith Koskei Solomon Langat
Solomon Langat James Mutisya
James Mutisya Francis Mulwa
Francis Mulwa Joel Lutomiah
Joel Lutomiah Hellen Koka
Hellen Koka Samuel O. Oyola
Samuel O. Oyola Rebecca Waihenya2
Rebecca Waihenya2