
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
OPINION article
Front. Virol. , 20 September 2023
Sec. Emerging and Reemerging Viruses
Volume 3 - 2023 | https://doi.org/10.3389/fviro.2023.1253174
A disease of cotton associated with multiple alternative names [cotton blue disease; (CBD), cotton vein mosaic disease (CVMD), and cotton leafroll dwarf disease (CLRDD)] has been reported from countries throughout Africa, South America, North America and Asia. CBD is believed to have originated from central Africa in 1949 (1–3). The disease has been variably characterized throughout the aforementioned regions. However, the same etiological agent, cotton leafroll dwarf virus (CLRDV), has been associated with all of these diseases reported in different continents, even in the cases where only symptomology has been reported. In many, if not all, recent writings on the subject, the assumption is made that the CBD reported in Africa shares the same etiological agent as the CVMD, later renamed to CBD, reported from South America or CLRDD reported from the United States (US) (1–3). South American CBD and CLRDD from the US have been confirmed biologically and at the molecular level to have the same etiological agent based on numerous independent confirmations from multiple sequence reports (2). Additionally, while CLRDV sequences have been reported from a variety of hosts in different countries, there has not been a reported viral sequence of CLRDV associated with CBD in cotton from Africa, especially from the Central African Republic, where CBD was first documented (Table 1) (2). The main purpose of this article is to highlight these discrepancies among the etiological agents of CBD in cotton reported worldwide to ensure more accurate information is cited in future publications by researchers engaged in research of viral diseases of cotton.
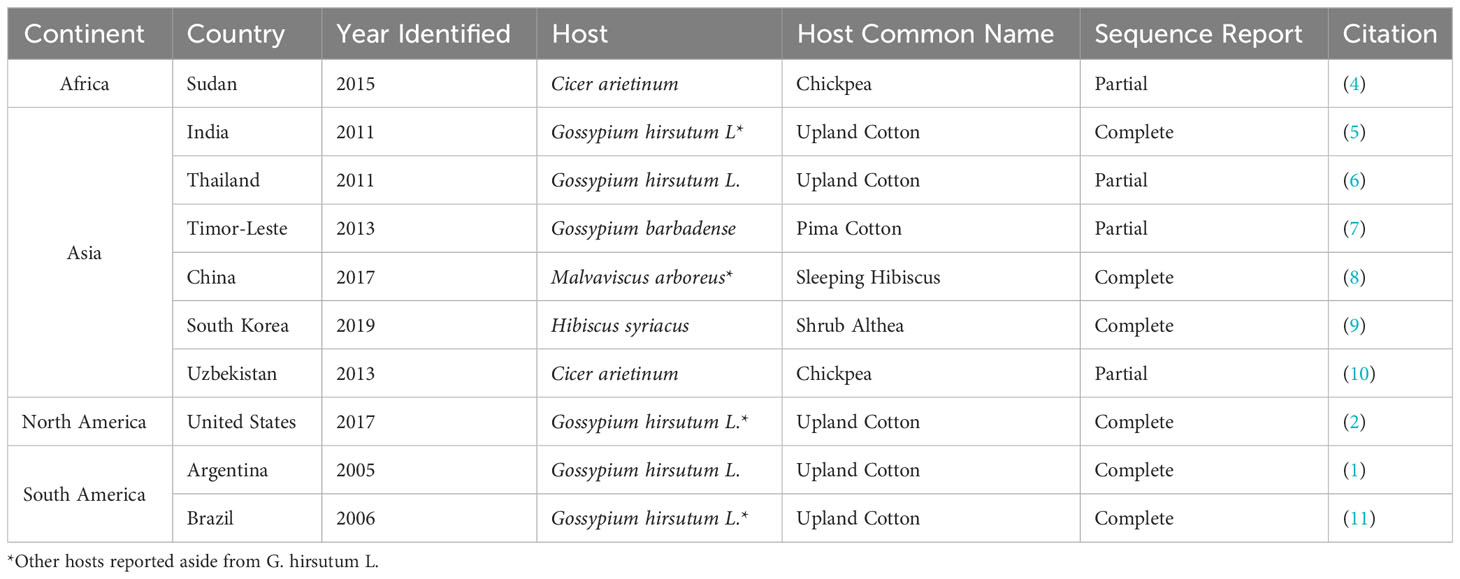
Table 1 All available sequence reports of cotton leafroll dwarf virus (CLRDV) from different countries as of 2022.
Cotton blue disease (CBD), or “maladie bleue,” was first reported in the Central African Republic in 1949 before spreading to neighboring countries such as Sudan, Chad, and Cameroon (3, 12). The disease was characterized by a downward arch of leaves, blistered leaves, shortened terminal internodes, and a typical generalized dark green color causing the coloration to appear blueish (3). The aphid Aphis gossypii Glover was experimentally demonstrated to be a vector associated with the disease (12). However, since the initial studies in the 1970s, further characterization with serological or nucleic acid-based techniques have not been conducted and no etiological agent has been isolated for African CBD in cotton so far.
The only recent report of a single CLRDV sequence from the African continent, SuCp31-15 (MK411565), isolated from chickpea in Sudan, is a partial coat protein sequence (4). When percent identity was determined against the NCBI GenBank (nr) database, the closest related sequence, CLRDV-Pir6 isolate (EU871539) from Brazil, had 89.72% nucleotide identity (13). The amino acid identity of the partial sequence against the highest percent identity match was 87.10% with GA_67 (QWJ75384) from Georgia, US. This 281-nucleotide partial sequence would not be enough criteria for species demarcation for Polerovirus which is defined as below 90% amino acid identity for a whole gene product (14). Additionally, this sequence was not isolated from cotton so, it could represent a new, closely related Polerovirus from Chickpea; however, further genetic information would be required to make that determination (14). The high nucleotide identity with CLRDV-Pir6, a Brazilian CLRDV sequence from 2005, would lend some support with the African CBD and South American CBD possessing a common ancestor. It may also be that the African CLRDV genetic identity has shifted over the decades since its purported initial spread out of Africa.
Cotton vein mosaic disease (CMVD), or “mosaic das nervuras” was first described in South America in Brazil in 1938 with mild mosaic symptoms, and a seemingly more severe form of the disease was observed in 1962 (15, 16). The vector was not identified to be A. gossypii until 1997 (17). The more severe form of CVMD was later termed CBD in 2005 based on the apparent similarities in symptomology as CBD from Africa (1, 15). This more severe form of CVMD was determined to be caused by the virus CLRDV (1). However, this determination raises the issue of the less severe form of CVMD being identified nearly a decade earlier than CBD in Africa (3, 15). Again, the lack of genetic evidence precludes this earlier outbreak from having the same causative agent as the 1962 report of CVMD, especially with the more severe symptoms (16). Nevertheless, CLRDV isolates classified as typical and atypical variants reported in South America have previously demonstrated different symptoms in cotton varieties (18). Therefore, the earlier incidence of CVMD cannot be definitively associated as a precursor to the 1960s incidence (15, 16).
The transmission by the aphid A. gossypii Glover (cotton aphid) in unison with the CBD symptoms are the main factors supporting the hypothesis of the two CBD types possessing the same causative agent. The similarity of symptoms between the African CBD and South American CBD does not preclude different causative agents. This can be exemplified with the issue of diagnosing cotton leaf roll dwarf disease (CLRDD) in the US, which is caused by a variant of CLRDV, due to the symptom similarity with bronze wilt disease (19). Furthermore, the cotton aphid is a known vector of a multitude of other viruses in Polerovirus and other viral genera such as Potyvirus and Mandarivirus (20–22). Therefore, even with the matching broad symptomology and aphid transmission, these factors alone are not definitive in concluding on the shared etiological agent. On a further note, CLRDD and CBD symptomology differs even though the etiological agent has been clearly demonstrated to be CLRDV (19). Therefore, it would be prudent for authors to classify further reports of CLRDV in the US with CLRDD or CBD depending on the symptomology demonstrated by their isolates.
Phylogenetic analysis based on the reported CLRDV P0 gene and complete genome sequences indicates support for CLRDD and South American CBD sharing the same etiological agent as CLRDV (2). However, the same conclusion cannot be reached for African CBD due to lack of genetic information available. The reported CLRDV partial sequence from chickpea in Sudan does not provide enough data to classify the Sudanese isolate as a potential novel species or as the causative agent of African CBD (4).
Edula et al. (2023) reported a BEAST analysis to estimate the age of the CLRDV population using available P0 sequences. Their analysis indicated 1945 to be the most likely origin year with 1938 to1962 being the interval (2). Even without considering the posterior probability spanning 1914 to 1971, the interval range also includes the 1938 CVMD original outbreak (2). Again, while the evidence does lean towards the conclusion of African CBD sharing CLRDV as its etiological agent, there is still a lack of direct evidence to support that conclusion as well as confounding variables.
CLRDV research has grown at a rapid rate due in part to the spread of the virus to new regions including the US; however, the base assumption on the causative agent of CBD in Africa being CLRDV needs to be addressed (19). The lack of genetic evidence from CBD presenting cotton samples from Africa as well as potential conflicting reports concerning the origin between South America and Brazil raises reasonable doubt on the etiological connection between South American and African CBD (3, 4, 12, 15). Furthermore, while the broad symptomology and aphid transmission similarities support the idea of a shared causative agent, these factors alone are not definitive for classifying the etiological agent (1).
In order to better support or entirely validate the assumption that is the premise of this work, cotton samples from Africa with symptoms of CBD must be tested for CLRDV. Additionally, given the low amino acid identity from the Sudanese isolate, the complete sequence of CLRDV from CBD in Africa should be collected with associated phylogenetic and chronogram analyses performed with existing data to provide a clear representation of the potential origins of CBD. Until this data is collected and analyzed, the causative agent of CBD in Africa should not be listed as CLRDV. We would recommend that further work pertaining to CLRDV could still use the CBD and CLRDD terminology, but due to lack of genetic evidence, the connection to CVMD and African CBD cannot be made. Any references to CBD should clarify the reference is to South American CBD and not African CBD until further characterization is performed.
AA conceived the study, rewrote partially, edited and proofread the manuscripts several times. CF wrote the first draft and final version of the manuscript. All authors contributed to the article and approved the submitted version
This work was supported by Cotton Incorporated (award ID, 2023 project 19-112). We thank the Department of Biological Science and the Office of Research and Sponsored Programs at the University of Tulsa for supporting the publication charges for this work.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
1. Corrêa RL, Silva TF, Simões-Araújo JL, Barroso PAV, Vidal MS, Vaslin MFS. Molecular characterization of a virus from the family Luteoviridae associated with cotton blue disease. Arch Virol (2005) 150:1357–67. doi: 10.1007/s00705-004-0475-8
2. Edula SR, Bag S, Milner H, Kumar M, Suassuna ND, Chee PW, et al. Cotton leafroll dwarf disease: An enigmatic viral disease in cotton. Mol Plant Pathol (2023) 24:513–26. doi: 10.1111/mpp.13335
3. Cauquil J. Etudes sur une maladie d’origine virale du cotonnier: la maladie bleue. Coton Fibres (1977) 32:259–78.
4. Moukahel A, Kumari SG, Hamed AA, Sharman M, Ahmed S. Distribution and identification of luteovirids affecting chickpea in Sudan. Phytopathol Mediterr (2021) 60:199–214. doi: 10.36253/phyto-12135
5. Mukherjee AK, Mukherjee PK, Kranthi S. Genetic similarity between cotton leafroll dwarf virus and chickpea stunt disease virus in India. Plant Pathol J (2016) 32(6):580–3. doi: 10.5423/PPJ.NT.09.2015.0197
6. Sharman M, Lapbanjob S, Sebunruang P, Belot J-L, Galbieri R, Giband M, et al. First report of cotton leafroll dwarf virus in Thailand using a species-specific PCR validated with isolates from Brazil. Aust Plant Dis Notes (2015) 10:24. doi: 10.1007/s13314-015-0174-1
7. Ray JD, Sharman M, Quintao V, Rossel B, Westaway J, Gambley C. Cotton leafroll dwarf virus detected in Timor-Leste. Aust Plant Dis Notes (2016) 11:29. doi: 10.1007/s13314-016-0217-2
8. Wang L, Fang Y, Yang Y, Qing L, Li M. First report of cotton leafroll dwarf virus infection of Malvaviscus arboreus in China. Plant Dis (2023). doi: 10.1094/PDIS-12-22-2909-PDN
9. Igori D, Shin AY, Kim SE, Kwon SY, Moon JS. First report of cotton leafroll dwarf virus infecting Hibiscus Syriacus in South Korea. Plant Dis (2022) 106(11):1798. doi: 10.1094/PDIS-11-21-2559-PDN
10. Kumari SG, Sharman M, Moukahel A, Ziyaev Z, Ahmed S. First report of cotton leafroll dwarf virus affecting chickpea (Cicer arietinum) in Uzbekistan. Plant Dis (2022) 104(9):2532. doi: 10.1094/PDIS-01-20-0085-PDN
11. Ramos-Sobrinho R, Ferro MMM, Lima GSA, Nagata T. First report of cotton leafroll dwarf virus infecting cacao (Theobroma cacao) trees in Brazil. Plant Dis (2023) 107(4):1251. doi: 10.1094/PDIS-07-22-1570-PDN
12. Cauquil J, Vaissayre M. La “maladie bleue” du cotonnier en Afrique: transmission de cottonier à cotonnier par Aphis gossypii glover. Coton Fibres Tropicales (1971) 26:436–66.
13. Silva T, Corrêa R, Castilho Y, Silvie P, Bélot J-L, Vaslin M. Widespread distribution and a new recombinant species of Brazilian virus associated with cotton blue disease. Virol J (2008) 5:123. doi: 10.1186/1743-422X-5-123
14. Lefkowitz EJ, Dempsey DM, Hendrickson RC, Orton RJ, Siddell SG, Smith DB, et al. (2018). Virus taxonomy: the database of the International Committee on Taxonomy of Viruses (ICTV). Nucleic Acids Res. 46, D708–D717. doi: 10.1093/nar/gkx932
15. Costa AS, Forster R. Nota preliminar sobre uma nova moléstia de vírus do algodoeiro - mosaico das nervuras. Boletim Técnico do Instituto Agronômico Campinas (1938) 51:11.
17. Michelotto MD, Busoli AC. Efficiency of nymphs and adults of Aphis Gossypii Glov. To transmit cotton vein mosaic virus. Bragantia (2002) 62(2):255–9. doi: 10.1590/S0006-87052003000200010
18. Agrofoglio YC, Delfosse VC, Casse MF, Hopp HE, Kresic IB, Distéfano AJ. Identification of a new cotton disease caused by an atypical cotton leafroll dwarf virus in Argentina. Phytopathology (2017) (2017) 107:369–76. doi: 10.1007/s00705-004-0475-8
19. Parkash V, Sharma DB, Snider J, Bag S, Roberts P, Tabassum A, et al. Effect of cotton leafroll dwarf virus on physiological processes and yield of individual cotton plants. Front Plant Sci (2021) 12:734386. doi: 10.3389/fpls.2021.734386
20. Ellis MH, Silva TF, Stiller WN, Wilson LJ, Vaslin MFS, Sharman M, et al. Identification of a new Polerovirus (family Luteoviridae) associated with cotton bunchy top disease in Australia. Aust Plant Pathol (2012) 42:261–9. doi: 10.1007/s13313-012-0177-8
21. Gadhave KR, Dutta B, Coolong T, Srinivasan R. A non-persistent aphid-transmitted Potyvirus differentially alters the vector and non-vector biology through host plant quality manipulation. Sci Rep (2019) 9:2503. doi: 10.1038/s41598-019-39256-5
Keywords: cotton leafroll dwarf virus (CLRDV), cotton blue disease (CBD), etiology, polerovirus, cotton vein mosaic disease (CVMD)
Citation: Ferguson C and Ali A (2023) Cotton blue disease from Africa and its de facto relationship with cotton leafroll dwarf virus: a misleading etiological discrepancy. Front. Virol. 3:1253174. doi: 10.3389/fviro.2023.1253174
Received: 05 July 2023; Accepted: 07 September 2023;
Published: 20 September 2023.
Edited by:
Ashish Srivastava, Amity University, IndiaReviewed by:
Lava Kumar, International Institute of Tropical Agriculture (IITA), NigeriaCopyright © 2023 Ferguson and Ali. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Akhtar Ali, QWtodGFyLWFsaUB1dHVsc2EuZWR1
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.