
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Virol. , 06 April 2023
Sec. Fundamental Virology
Volume 3 - 2023 | https://doi.org/10.3389/fviro.2023.1176840
This article is part of the Research Topic Post-transcriptional regulation of viral protein expression and function View all 5 articles
RNA viruses encode an RNA-dependent RNA polymerase (RdRp), which is essential for transcription and replication of their genome since host cells lack equivalent enzymes. RdRp residues were shown to be phosphorylated by host kinases in several human, animal or plant viruses including flaviviruses, picornaviruses, coronaviruses, influenza viruses and tymoviruses. RdRps can be phosphorylated on several residues by distinct host kinases. Phosphomimetic mutations of identified phosphorylated residues either positively or negatively regulate RNA synthesis or association of RdRps with RNA or other proteins. Interestingly, some RdRps evolved to recruit cellular kinases through direct protein-protein interaction, likely to promote or to tightly control their own phosphorylation. Given the essential nature of RdRps for RNA virus replication, a better knowledge of RdRps’ phosphorylation is expected to facilitate the design of future drugs that strongly affect polymerase activity.
RNA viruses include a wide variety of human, animal and plant viruses. All RNA viruses characterized to date, with the exception of retroviruses, code for an RNA-dependent RNA-polymerase (RdRp) necessary for the replication and the transcription of their RNA genome. RdRps belong to the larger class of enzymes called template-directed nucleic acid polymerases. These polymerases adopt a “right hand” conformation with three subdomains: palm, fingers and thumb (Figure 1). RdRps’ right hand is closed and they share a common core architecture with 7 highly conserved motifs named A to G (2, 3). The detailed structure of RdRps has been largely discussed elsewhere (2, 4–6) and the structural overview presented here only provides the minimal background to discuss the impact of phosphorylation sites.
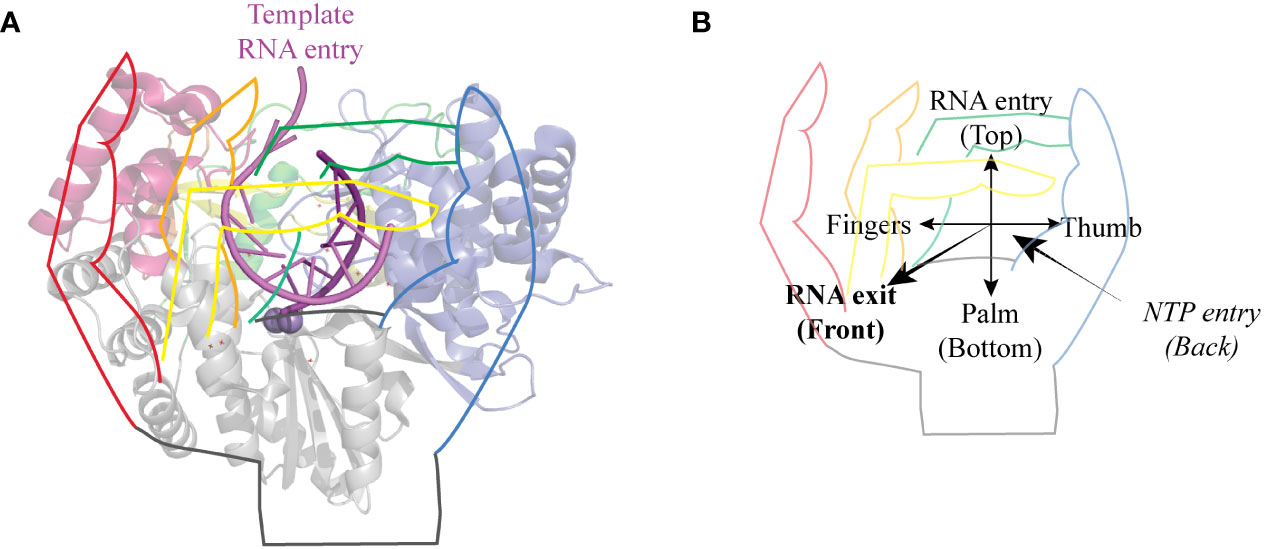
Figure 1 General structure of RdRps. (A) Cartoon representation of HCV genotype 2a protein NS5B seen from the front with a hand drawing on top. The pinky, ring, middle and index fingers are represented in red, yellow, orange, and green, respectively. Thumb is blue and palm is grey. Template RNA is colored in light violet and the newly synthesized strand in purple. Structures were drawn using Pymol (Schrödinger) using PDB entry 4WTI, (1). (B) The same hand representation as in A shows the orientation of the polymerase. The template RNA enters the polymerase from the top and comes out as dsRNA from the front of the polymerase. NTPs enter the polymerase through the back.
The catalytic site in the palm, composed of a three-stranded anti-parallel β-sheet core surrounded by three α-helices, is the most conserved feature of these polymerases. It contains motifs A and C involved in NTP binding and catalysis, motif B important for in NTP recognition and positioning, motif D involved in maintaining polymerase structural integrity and fidelity and motif E that participates in the formation of the NTP entry tunnel. Motifs C and E also participate in locking the primer strand in an optimal position for elongation (2, 3, 7).
On the N-terminal side of the palm are the fingers. In the case of RdRps, fingers and thumb interact to form a closed structure surrounding the active site. This closure is mediated by the index and ring fingertips, which form protrusions that reach the thumb and thereby create a hole, which acts as the template RNA entry channel. This channel contains the conserved motif G (8). RdRps are also characterized by the presence of a positively charged NTP entry channel at the back of the polymerase, of which motif F forms the roof (2, 3, 7).
The C-terminal thumb subdomain is located at the side of the palm. This subdomain is much more diversified than the fingers and palm subdomains. The thumb of picornavirus and calicivirus RdRps is small, leaving an opening to the active site that allows for the accommodation of the protein primer VPg. The thumb of flavivirus RdRp is much larger and protrudes into the active site, thereby stabilizing de novo initiation complexes but imposing more important conformational changes to allow elongation (2, 7).
Although all RdRps share the same core, a number of polymerases contain additional domains. For example, polymerases of flaviviruses have an N-terminal methyl-transferase (MTase) domain involved in viral mRNA capping (4, 9). Influenza virus polymerase is an heterotrimer formed of polymerase basic 1 and 2 (PB1, PB2) and polymerase acidic (PA) subunits. The RdRp domain is found in the PB1 protein, which is wrapped by PA (10).
This review specifically focuses on core RdRp phosphorylation. It does not address the phosphorylation of regulatory subunits or that of cofactors such as the polymerase basic (PA) subunit of the influenza polymerase, the phosphoprotein (P) of rhabdoviruses or the non-structural 5A (NS5A) protein of hepaciviruses.
Phosphorylation of RdRps has been studied in a variety of viruses, including members of the families Picornaviridae, Coronaviridae, Flaviviridae, Orthomyxoviridae, amongst others. Table 1 provides a snapshot on select RdRp phosphorylated residues shown to impact polymerase activity. Supplemental Table 1 provides a more extensive list of RdRp phosphorylation sites identified to date, including in some plant viruses, and provides some information about the role of phosphorylated residues. As many studies focused on the RdRp of flaviviruses and of hepatitis C virus (HCV) in particular, we used the HCV genotype 1b RdRp as reference for phosphorylation site description and residue numbering (PDB accession: 3MWV).
Fingers interact with the thumb through fingertips protrusions, leaving openings for incoming template RNA and nucleotides. In the polymerase of HCV, non-structural protein 5B (NS5BHCV), Ser27 and Ser29, which can be phosphorylated, are located in the index finger at the back of the polymerase (Figure 2). Ser29 makes van der Waals contacts with thumb residues His428, Pro495, Trp500 and Arg503, thus contributing to the closure of the polymerase (23, 24).
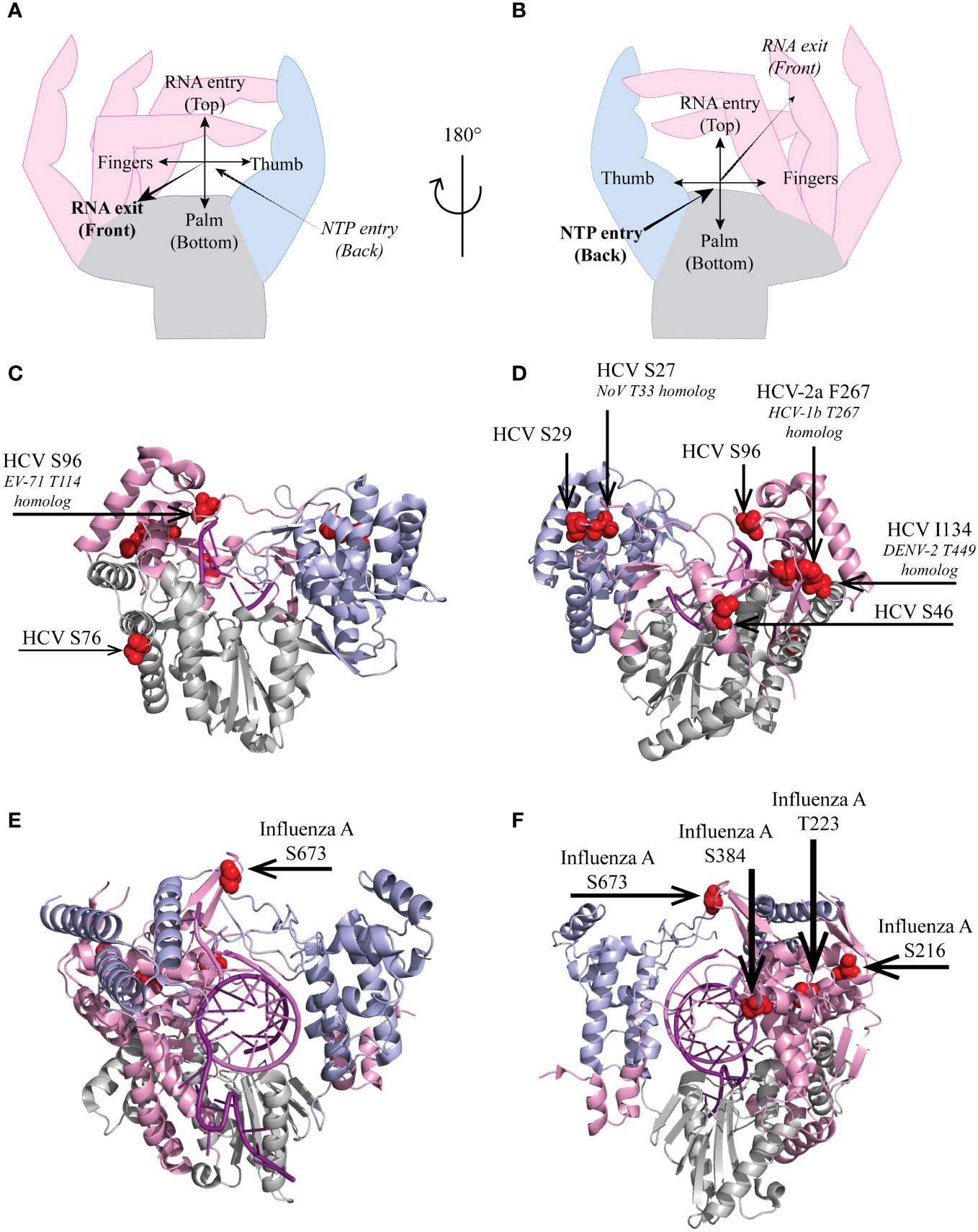
Figure 2 Localization of select HCV and influenza virus RdRp phosphorylated sites. Hand representation of a standard RdRp seen from the front (A) or the back (B). Cartoon representation of (C, D) HCV genotype 2a protein NS5B (PDB 4WTI, (1)) and of (E, F) Influenza A/H7N9 PB1 protein (PDB 7QTL, (22)). Fingers, palm and thumb are colored in pink, grey, light blue, respectively. (C–F), phosphorylated residues presented in this review are represented as red spheres.
NS5B Ser29 and the nearby Ser27 were reported to be phosphorylated by both AKT and PRK2 in mammalian Huh7 cell lysates and in in vitro kinase assays (16–18). A Ser-to-Ala mutation, which prevents any phosphorylation (phosphoinhibiting mutation) or a Ser-to-Glu mutation, which mimics phosphorylation (phosphomimetic mutation) in either residue reduced primer extension and de novo synthesis activities of the polymerase as well as replication of a minireplicon (16, 17). It was proposed that contacts between index finger and thumb residues are affected upon Ser29 phosphorylation. The resulting structural perturbation might explain the decreased activity of the RdRp carrying the phosphomimetic mutation. However, the phosphoinhibiting mutation also decreased polymerase activity, suggesting that subtle local structural changes imposed by the Ser-to-Ala mutation might be sufficient to affect the contact between the fingers and the thumb and, hence, the polymerase activity.
In the polymerase of Norovirus (3DNoV) Thr33, which is the homolog of NS5BHCV Ser27, can also be phosphorylated in vitro by AKT. In agreement the above data reported for HCV, the phosphomimetic Thr33-Glu mutation in 3DNoV decreased polymerase speed and affinity for NTPs (12).
Interestingly, two residues (Ser433 or Ser434) structurally occur at a similar position as 3DNoV Thr33 and NS5BHCV Ser27 in the polymerase (non-structural protein 12, nsp12) of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). These residues, which are conserved among β-coronaviruses, were shown to be phosphorylated in infected Vero cells (25). Influence of the phosphorylation of the latter residues was not studied but, given the structural similarity of the polymerases, they likely play similar functions as their homologs in norovirus and HCV.
Several polymerase phosphorylation sites were reported to occur near the RNA template entry channel, including influenza A virus PB1 Thr223 and Ser384 (Figures 2E, F), which are well conserved among influenza viruses (Figure 3). PB1 Thr223 was shown to be phosphorylated in infected A459 cells and when overexpressed in 293T cells (21, 26). An influenza A virus of the strain WSN, carrying the PB1 Thr223-Ala mutation reached lower titers than the parental virus (21). A similar mutant of mouse-adapted influenza strain SC35M displayed however unaffected replication in a reporter replicon assay (27). In contrast, Thr223-Asp phosphomimetic mutants of either strain could not be rescued by reverse genetics and displayed severely affected replication in a luciferase reporter assay (25, 27). This mutation prevented the interaction of PB1 with genomic or complementary viral RNA, thereby impeding nucleocapsid assembly and viral replication (25). Predictions suggest that Thr223 phosphorylation in the polymerase fingers might promote interaction of this residue with the side chain of Arg350 close to the matrix entry channel, thereby negatively affecting RNA template binding and NTP channeling (27).

Figure 3 Conservation of phosphorylatable residues in orthomyxovirus protein PB1. Sequence alignment of orthomyxovirus polymerase subunit PB1. PB1 sequences from a series of Influenza A viruses, influenza B and C, Thogotoviruses and more distantly related viruses was aligned using Clustal Omega. Phosphorylated residues discussed in the text are outlined.
Influenza virus PB1 Ser384 is localized at the back of the polymerase close to the incoming RNA template. It was shown to be phosphorylated in 293T cells overexpressing the polymerase. Mutant influenza PB1 Ser384-Ala virions replicated as the wild type virus in single and multiple round infection. Unlike the Thr223 phosphomimetic mutation which crippled the polymerase, a Ser384-Asp mutation only slightly affected replication. Both Ser384-Ala and -Asp mutant polymerases assembled properly and were as effective as the wild type polymerase in in vitro primer extension assay, suggesting the phosphorylation of this residues has no big impact on PB1 activity although it is localized close to the template entry channel (21).
Only few phosphorylated residues described in the literature mapped to the thumb of RdRps. The best studied is influenza virus PB1 Ser673, located in an unstructured loop close the template RNA (Figures 2E, F). This residue was shown to be phosphorylated in transfected 293T cells as well as in infected A549 cells. A phosphorylatable Ser or Thr residues is very well conserved at the corresponding positition in the PB1 polymerases of influenza and Thogoto viruses (Figure 3). A Ser673-Ala mutant replicated even better than the parental virus in single-round infections but virus titers declined as compared to the parental virus in multiple-round infections. In contrast, a phosphomimetic PB1 Ser673-Asp mutation fully prevented infectious virus rescue although the mutant PB1 kept the capacity to interact with PB2 and PA, to form the heterotrimeric polymerase. Interestingly, in vitro, the mutant polymerase produced wildtype levels of viral genomic RNA but produced very low levels of viral mRNA indicating that phosphorylation of Ser673 probably acts as a switch for the polymerase to favor replication over transcription. The phenotype of the PB1 Ser673-Asp mutant is reminiscent of that of an His510-Ala mutant of the PA subunit of the polymerase, which is located nearby in the trimeric structure (21). That phosphorylation of Ser673 at the level of the RNA entry channel differentially affects transcription and replication suggests that template binding or positioning constraints may be stricter for transcription than for replication.
In the NS5B polymerase of HCV, Ser96 is located in motif G, very close to the template RNA entry channel (Figures 2A-D). Ser96 is highly conserved (Ser or Thr) in HCV, and is predicted to be phosphorylated in genotypes 1a and 1b but such a phosphorylation was not experimentally proven (18, 28). Whereas a phosphoinhibitory Ser96-Ala mutation increased replication levels, a phosphomimetic Ser96-Asp mutation totally abrogated the replication of an HCV minireplicon and in vitro primer extension activity of the mutant polymerase. Phosphorylation of Ser96 is expected to promote the interaction of this residue with Arg168, thereby changing the geometry of the channel needed to accommodate the nascent RNA strand, which might explain the complete loss of polymerase activity (18).
The 3D polymerase of enterovirus 71 carries a well-conserved Thr114 – Ser115 doublet at a location structurally close to NS5BHCV Ser96 (Figure 4). Mutagenesis of these residues suggested that they critically constraint the RNA template and incoming nucleotide positioning thus regulating polymerase translocation during elongation (8).
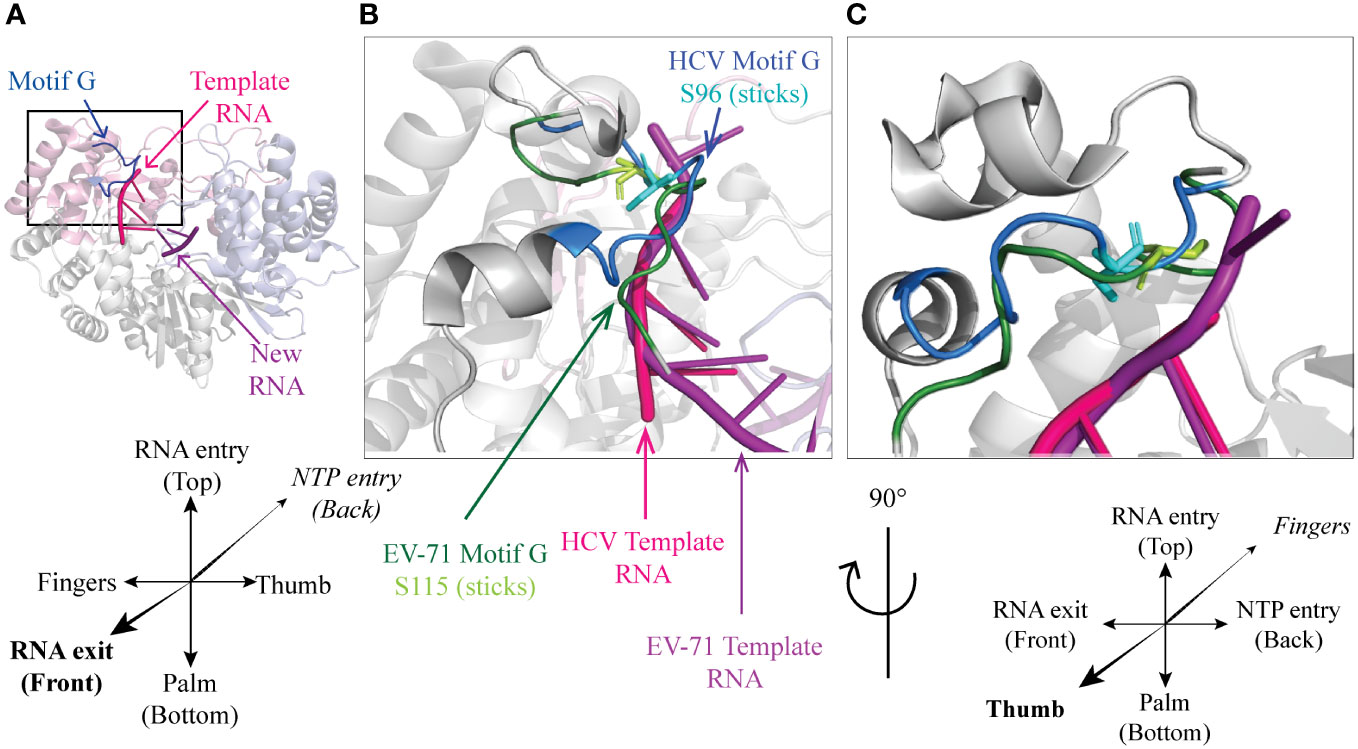
Figure 4 A conserved Ser residue in motif G (A) Cartoon representation of HCV genotype 2a protein NS5B (PDB 4WTI, (1)). Fingers, palm and thumb are colored in pink, grey, light blue respectively. Conserved motif G is represented in blue, template RNA in magenta and the new RNA strand in purple. Front (B) and side (C) close-up on HCV NS5B motif G (PDB 4WTI) with EV-71 3D motif G and RNA aligned (PDB 6LSE, (8)). HCV polymerase is colored in grey with motif G in blue, template RNA in magenta and within motif G, Ser96 is represented as cyan sticks. From EV-71 only motif G (dark green) and template RNA (purple) are represented, with Ser115 shown as Light green sticks.
Phosphorylation of these residue was however documented for neither NSB5HCV Ser96 nor 3DEV71 Thr114-Ser115 and, although these residues critically regulate the elongation process, there is no evidence so far that this process is regulated by phosphorylation in physiological conditions.
Mass spectrometry analysis of 293T cells transfected with NS5 expression constructs identified a common phosphorylation site in the NS5 polymerases of dengue virus type 2 (DENV-2) (Thr449) and of yellow fever virus (YFV) (Ser450). NS5DENV-2 Thr449 was confirmed to be phosphorylated in both mammalian and insect (Aedes aegypti) cells infected with DENV-2. In agreement with in silico predictions, this residue, which is contained in the consensus CXT/SC motif, was phosphorylated by the human cyclic nucleotide-dependent protein kinase G (PKG or PRKG1) in an in vitro kinase assay. It is noteworthy that PKG can be expressed in both mammalian and insect cells (14, 15, 29).
Whereas the CXT/SC motif is highly conserved among mosquito-borne flaviviruses, thick-borne flaviviruses harbor a His or a Gln residue at the corresponding position. Interestingly, treatment of HEK293T cells with a PKG activator increased DENV-2 viral titers up to 4 times and treatment with a PKG inhibitor or with a siRNA targeting PKG decreased viral titers by up to 10 times whereas none of the treatments significantly affected the replication of the thick-borne Langat virus (LGTV) (14). Taken together, these data suggest that phosphorylation of NS5 Thr449 by PKG can activate mosquito-borne but not thick-borne flavivirus replication in both mammalian and insect cells.
A phosphomimetic Thr449-Glu mutation however unexpectedly decreased DENV-2 replication suggesting that either very subtle conformation changes at this position heavily affect polymerase activity or that Thr449 phosphorylation only allows polymerase activity when transient or partial (14).
Several of the studied phosphomimetic mutations do decrease polymerase activity (see Table 1 and Supplemental Table 1). Such decreases can be due to steric hindrance caused by the phosphate group or by the phosphomimetic lateral chain, which alter the local RdRp structure, or prevent interactions between residues. Interestingly, other phosphomimetic mutations do positively impact polymerase activity whereas the corresponding phosphoinhibitory mutations (usually to Ala) hardly impacted activity if at all. Phosphorylation of these residues likely occurs to boost polymerase activity in specific physiological conditions or cell types.
Such activating phosphomimetic mutations were reported in the case of NS5BHCV Ser46, Ser76 and Thr267 and in the case of influenza virus polymerase PB1 residue Ser216.
NS5BHCV Ser46 is located in an α-helix of the index finger close to the NTP entry channel. This residue was however not documented to be phosphorylated in cells and is poorly conserved among hepacivirus strains hepacivirus strains. The physiological relevance of the phosphomimetic mutant phenotype can thus be questionned.
Ser76, which is well-conserved among hepaciviruses, is also located in the fingers but rather close to the exit site of the RNA (Figures 2A-D). This residue was phosphorylated in vitro, after incubation with Huh7 cell extracts (18). Phosphomimetic mutants produced more colonies than wild type replicons. Such a phosphorylation thus likely promotes polymerase activity.
NS5BHCV Thr267 can be phosphorylated in vitro by AKT/PKB. While mutation of either Thr267 or Ser269 into the non-phosphorylatable Ala residue yielded a polymerase with less activity than WT for primer extension and de novo synthesis, the Thr267-Asp mutant displayed an increased activity in both assays. This poorly conserved residue is located at the back of the polymerase and the reason for enhanced polymerase activity of the phosphomimetic mutant is unclear (17).
A number of viruses have their core RNA-dependent RNA-polymerase domain extended with N-terminal domains important for RNA replication, RNA protection or interaction with other proteins. This is the case of flavivirus (WNV, YFV, ZIKA, DENV) NS5 polymerases which have their RdRp domain N-terminally fused to an S-adenosyl-L-methionine-dependent methyltransferase (MTase) domain responsible for viral mRNA caping (9, 30, 31). MTase domains of flaviviruses have been shown to be phosphorylated. In particular, NS5DENV Thr39, as its homolog Ser38 in West Nile virus (WNV), were reported to be phosphorylated in infected mosquito as well as mammalian cells. Interestingly, Ser or Thr residues are well-conserved at this position in the polymerases of flaviviruses and, when not conserved, they are often replaced by phosphomimetic Asp or Glu residues. In vitro, Ser38/Thr39 residues can be phosphorylated by PKGα. The impact of the phosphorylation of these residues by PKG is unknown but the overall impact of PKG-mediated phosphorylation on viral replication was shown to be positive (15, 20).
SARS-CoV-2 nsp12 contains a N-terminal nidovirus RdRp-associated nucleotidyltransferase domain (NiRAN) likely involved in cap formation (32). Another characteristic of nsp12 is that it needs an nsp8 monomer and an nsp7/nsp8 heterodimer to function, and has additional structures for binding other non-structural proteins involved in RNA replication (33). nsp12SARS-CoV-2 Thr20, located in the NiRAN domain of the protein, was convincingly shown to be phosphorylated by cyclin-dependent kinase 2 (CDK2) (13). A phosphoinhibiting Thr20-Ala mutation decreased replication of a reporter construct by 60% in CDK2-positive cells but not in CDK2 knock down cells, whereas the phosphomimetic Thr20-Glu mutation slightly increased replication as compared to the wild type. Inhibition of RdRp by the Thr20-Ala mutation turned out to be due to the inefficient association of nsp12 with nsp8 and nsp7 to form a functional polymerase. Interestingly, pharmacological inhibition of CDK2 reduced SARS-CoV-2 replication in Vero cells (13). Thus, phosphorylation of the NiRAN domain by CDK2 promotes viral replication by promoting the assembly of polymerase with the co-factors nsp7 and nsp8.
Taken together, these data illustrate that phosphorylation of residues located outside of the core RdRp can affect polymerase activity.
Strikingly, several host protein kinases were shown to be recruited by RdRps, through direct protein-protein contact. For instance, in the case of SARS-CoV-2, nsp12 was shown to recruit CDK2 through its NiRAN domain (13). The polymerase of infectious bursal disease virus (IBDV) can interact with the CDK1-cyclinB1 complex (11). Polymerases of several flaviviruses including West Nile virus, Usutu virus, Zika virus and HCV were all shown to interact with AKT/PKB and to be phosphorylated by these kinases in vitro (19, 34). In the case of NS5BHCV, interaction of the polymerase with AKT/PKB was shown to modify the cellular localization of this kinase from the cytoplasm to the perinuclear region in infected cells (35).
Some RdRps likely recruit more than one host kinase. Phage display, co-immunoprecipitation, immunolabeling and in vitro kinase experiments converged to show that NS5BHCV can also interact with PRK2 (28). In the case of WNV, in addition to AKT/PKB, NS5 was shown to interact with PKG in transfected cells. This interaction is mediated by an α-helix located in the MTase domain (19, 20).
Recruited kinases were shown to phosphorylate RdRp residues and/or to impact viral replication.
In SARS-CoV-2 nsp12, CDK2 was found to be responsible for Thr20 phosphorylation, which activates replication by facilitating the interaction of nsp12 with nsp8 and nsp7 (13).
In the case of HCV, siRNA-mediated silencing of PRK2 but not of related kinases, reduced NS5B phosphorylation (16, 28), an effect that was also achieved with the PRK2 inhibitor HA1077 (36). Interestingly, PRK2 knock down reduced HCV RNA copy numbers in infected mice (37). In contrast, AKT reduced in vitro polymerase activity of NS5BHCV(35), showing that phosphorylation by recruited kinases can either positively or negatively impact RdRp activity.
Other studies examined the impact of human and insect PKG on NS5YFV and NS5DENV-2 phosphorylation and activity. As indicated above, a PKG activator increased DENV-2 viral titers up to 4 times whereas PKG inhibition decreased viral titers by up to 10 times, similarly to the positive effect of AKT/PKB, observed for the related HCV (14, 35, 38). In addition to PKG, NS5DENV-2 can be phosphorylated in vitro by PKC, although this kinase was not shown to interact with NS5. While the PKC inhibitor increased viral copy numbers, PKC induction by PMA reduced it (39). Thus, the RdRp of DENV-2 can be phosphorylated by at least 3 host kinases (PKG, AKT and PKC), which play partly antagonist effects on polymerase activity.
Due to technical limitations, providing a global mechanistic interpretation of available data is still challenging for several reasons: i) The identification of specific residues that are phosphorylated by a kinase often stems from in silico predictions and in vitro kinase assays, which are prone to false positive results. ii) Another common way of identifying phosphorylated residues is the overexpression of the viral polymerase by transfection in cells, a model that lacks the complexity due to the many molecular changes induced by a viral infection. iii) Phenotypic data provided by phosphomimetic and phosphoinhibitory mutations must be interpreted with caution since these mutations, in addition to mimicking or preventing phosphorylation, may affect the local conformation of the protein and thereby the interaction landscape of the mutated residue. iv) The use of kinase activators or inhibitors, which provides a more global view on the effect of a specific kinase on viral replication can be subjected to off-target effects.
RdRps also appear to be phosphorylated by a set of different kinases, which can have antagonistic effects, and expression of these kinases has a cell type-dependent pattern. Moreover, some phosphorylation events were proposed to be transient or partial because both phosphomimetic and phosphoinhibitory mutations negatively affected polymerase activity.
Phosphorylation of viral proteins may have quite diverse effects (40). It may affect protein stability, turnover, subcellular localization and assembly with other proteins to form functional complexes. It may also impact the local structure of RdRps and thereby affect the interaction with RNA and nucleotides and fine-tune the polymerase catalytic activity. In the case of cellular gene transcription by RNA polymerase II, phosphorylation of the C-terminal domain of the polymerase occurs as a well-orchestrated mechanism to control stepwise transcription initiation, pausing, and elongation (41).
Given the impressive development of mass spectrometry techniques, major developments are expected in the understanding of mechanistic impacts of RdRp phosphorylation in the near future but much is still to be done to get a better picture of the physiological impact of RdRp phosphorylation.
RdRps are among the best targets for antiviral drugs as they are essential to the viral cycle and reasonably diverge from host polymerases to allow specific targeting. For example, nucleoside analogs targeting the nucleotide binding pocket of viral polymerases have proven important in the treatment of viral infections (42, 43). However, the use of inhibitor cocktails may be needed to counteract the emergence of resistant mutants, which easily arise with RNA viruses given the error-prone nature of RdRps.
On the one hand, one can examine the influence of specific kinase inhibitors on viral replication in vitro or in vivo in search of approved drugs that may be repurposed to globally decrease viral replication. These empiric approaches can lead to important therapeutic tools, even if the inhibitor may impact many other processes than RdRp phosphorylation and RdRp-dependent viral replication. On the other hand, with the exception of phosphorylation sites that are buried in the polymerase (e.g. Thr223 of influenza PB1) and might be phosphorylated before folding or complex assembly (21), RdRp phosphorylation sites usually correspond to amino acids that are accessible to small molecules. Thus, the knowledge of phosphorylation sites that strongly impact polymerase activity may prove interesting in the design of molecules, which target polymerase activity.
It is worth noting that a screening for molecules that inhibit enterovirus replication yielded a broad-spectrum inhibitor, which inhibits RdRp activity by binding to the RNP entry channel, next to EV71 3D Thr114-Ser115 doublet that was shown to block replication when mutated into phosphomimetics (44). The knowledge of RdRp structures at the level of phosphoresidues may thus provide structural bases for the improvement or the design of molecules, which block RdRp function.
In conclusion, although much remains to be done, the expected progresses in identifying RdRp phosphorylated residues, the kinases responsible for these phosphorylations and the mechanistic impact of such phosphorylations on RdRp activity should help the understanding of basic virus biology as well as the development of new antiviral drugs.
CD prepared the draft and the figures of the manuscript. CD and TM wrote and edited the manuscript. All authors contributed to the article and approved the submitted version.
CD was the recipient of a fellowship of the FRIA (fonds pour la recherche dans l’industrie et l’agriculture). Work on RdRp was funded by the EOS joint programme of Fonds de la recherche scientifique-FNRS and Fonds wetenschappelijk onderzoek-Vlaanderen-FWO (EOS ID: 30981113 and 40007527), as well as by a grant from FNRS (CDR n° 40008453), Actions de Recherche concertées (ARC), Loterie Nationale through support to the de Duve Institute.
We are grateful to Belén Lizcano-Perret and Frédéric Sorgeloos for critical reading of the manuscript. We are also indebted to Frédéric Sorgeloos for valuable help in bioinformatics and sequence alignments.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fviro.2023.1176840/full#supplementary-material
1. Appleby TC, Perry JK, Murakami E, Barauskas O, Feng J, Cho A, et al. Viral replication. structural basis for RNA replication by the hepatitis c virus polymerase. Science (2015) 347:771–5. doi: 10.1126/science.1259210
2. Ferrero D, Ferrer-Orta C, Verdaguer N. Viral RNA-dependent RNA polymerases: A structural overview. Subcell Biochem (2018) 88:39–71. doi: 10.1007/978-981-10-8456-0_3
3. Smertina E, Urakova N, Strive T, Frese M. Calicivirus RNA-dependent RNA polymerases: Evolution, structure, protein dynamics, and function. Front Microbiol (2019) 10:1280. doi: 10.3389/fmicb.2019.01280
4. Lu G, Gong P. A structural view of the RNA-dependent RNA polymerases from the flavivirus genus. Virus Res (2017) 234:34–43. doi: 10.1016/j.virusres.2017.01.020
5. Peersen OB. A comprehensive superposition of viral polymerase structures. Viruses (2019) 11:745. doi: 10.3390/v11080745
6. Ramaswamy K, Rashid M, Ramasamy S, Jayavelu T, Venkataraman S. Revisiting viral RNA-dependent RNA polymerases: Insights from recent structural studies. Viruses (2022) 14:2200. doi: 10.3390/v14102200
7. Ferrer-Orta C, Arias A, Escarmís C, Verdaguer N. A comparison of viral RNA-dependent RNA polymerases. Curr Opin Struct Biol (2006) 16:27–34. doi: 10.1016/j.sbi.2005.12.002
8. Wang M, Li R, Shu B, Jing X, Ye HQ, Gong P. Stringent control of the RNA-dependent RNA polymerase translocation revealed by multiple intermediate structures. Nat Commun (2020) 11:2605. doi: 10.1038/s41467-020-16234-4
9. Egloff M-P, Benarroch D, Selisko B, Romette J-L, Canard B. An RNA cap (nucleoside-2′-O-)-methyltransferase in the flavivirus RNA polymerase NS5: crystal structure and functional characterization. EMBO J (2002) 21:2757–68. doi: 10.1093/emboj/21.11.2757
10. Te Velthuis AJW, Grimes JM, Fodor E. Structural insights into RNA polymerases of negative-sense RNA viruses. Nat Rev Microbiol (2021) 19:303–18. doi: 10.1038/s41579-020-00501-8
11. Hu X, Chen Z, Wu X, Ding Z, Huang Y, Fu Q, et al. Phosphorylation of VP1 mediated by CDK1-cyclin B1 facilitates infectious bursal disease virus replication. J Virol (2023) 97:e0194122. doi: 10.1128/jvi.01941-22
12. Eden JS, Sharpe LJ, White PA, Brown AJ. Norovirus RNA-dependent RNA polymerase is phosphorylated by an important survival kinase, akt. J Virol (2011) 85:10894–8. doi: 10.1128/JVI.05562-11
13. Guo S, Lei X, Chang Y, Zhao J, Wang J, Dong X, et al. SARS-CoV-2 hijacks cellular kinase CDK2 to promote viral RNA synthesis. Signal Transduct Target Ther (2022) 7:400. doi: 10.1038/s41392-022-01239-w
14. Bhattacharya D, Mayuri, Best SM, Perera R, Kuhn RJ, Striker R. Protein kinase G phosphorylates mosquito-borne flavivirus NS5. J Virol (2009) 83:9195–205. doi: 10.1128/JVI.00271-09
15. Keating JA, Bhattacharya D, Rund SS, Hoover S, Dasgupta R, Lee SJ, et al. Mosquito protein kinase G phosphorylates flavivirus NS5 and alters flight behavior in aedes aegypti and anopheles gambiae. Vector Borne Zoonotic Dis (2013) 13:590–600. doi: 10.1089/vbz.2012.1110
16. Han SH, Kim SJ, Kim EJ, Kim TE, Moon JS, Kim GW, et al. Phosphorylation of hepatitis c virus RNA polymerases ser29 and ser42 by protein kinase c-related kinase 2 regulates viral RNA replication. J Virol (2014) 88:11240–52. doi: 10.1128/JVI.01826-14
17. Sabariegos R, Albentosa-González L, Palmero B, Clemente-Casares P, Ramírez E, García-Crespo C, et al. Akt phosphorylation of hepatitis c virus NS5B regulates polymerase activity and hepatitis c virus infection. Front Microbiol (2021) 12:754664. doi: 10.3389/fmicb.2021.754664
18. Hernández S, Figueroa D, Correa S, Díaz A, Aguayo D, Villanueva RA. Phosphorylation at the n-terminal finger subdomain of a viral RNA-dependent RNA polymerase. Biochem Biophys Res Commun (2015) 466:21–7. doi: 10.1016/j.bbrc.2015.08.082
19. Albentosa-González L, Jimenez De Oya N, Arias A, Clemente-Casares P, Martin-Acebes M, Saiz JC, et al. Akt kinase intervenes in flavivirus replication by interacting with viral protein NS5. Viruses (2021) 13:896. doi: 10.3390/v13050896
20. Keating JA, Bhattacharya D, Lim PY, Falk S, Weisblum B, Bernard KA, et al. West Nile Virus methyltransferase domain interacts with protein kinase G. Virol J (2013) 10:242. doi: 10.1186/1743-422X-10-242
21. Dawson AR, Wilson GM, Freiberger EC, Mondal A, Coon JJ, Mehle A. Phosphorylation controls RNA binding and transcription by the influenza virus polymerase. PloS Pathog (2020) 16:e1008841. doi: 10.1371/journal.ppat.1008841
22. Kouba T, Dubankova A, Drncova P, Donati E, Vidossich P, Speranzini V, et al. Direct observation of backtracking by influenza a and b polymerases upon consecutive incorporation of the nucleoside analog T1106. Cell Rep (2023) 42:111901. doi: 10.1016/j.celrep.2022.111901
23. Biswal BK, Cherney MM, Wang M, Chan L, Yannopoulos CG, Bilimoria D, et al. Crystal structures of the RNA-dependent RNA polymerase genotype 2a of hepatitis c virus reveal two conformations and suggest mechanisms of inhibition by non-nucleoside inhibitors. J Biol Chem (2005) 280:18202–10. doi: 10.1074/jbc.M413410200
24. Chinnaswamy S, Yarbrough I, Palaninathan S, Kumar CT, Vijayaraghavan V, Demeler B, et al. A locking mechanism regulates RNA synthesis and host protein interaction by the hepatitis c virus polymerase. J Biol Chem (2008) 283:20535–46. doi: 10.1074/jbc.M801490200
25. Davidson AD, Williamson MK, Lewis S, Shoemark D, Carroll MW, Heesom KJ, et al. Characterisation of the transcriptome and proteome of SARS-CoV-2 reveals a cell passage induced in-frame deletion of the furin-like cleavage site from the spike glycoprotein. Genome Med (2020) 12:68. doi: 10.1186/s13073-020-00763-0
26. Hutchinson EC, Denham EM, Thomas B, Trudgian DC, Hester SS, Ridlova G, et al. Mapping the phosphoproteome of influenza a and b viruses by mass spectrometry. PloS Pathog (2012) 8:e1002993. doi: 10.1371/journal.ppat.1002993
27. Weber A, Dam S, Saul VV, Kuznetsova I, Müller C, Fritz-Wolf K, et al. Phosphoproteome analysis of cells infected with adapted and nonadapted influenza a virus reveals novel pro- and antiviral signaling networks. J Virol (2019) 93:e00528–19. doi: 10.1128/JVI.00528-19
28. Kim SJ, Kim JH, Kim YG, Lim HS, Oh JW. Protein kinase c-related kinase 2 regulates hepatitis c virus RNA polymerase function by phosphorylation. J Biol Chem (2004) 279:50031–41. doi: 10.1074/jbc.M408617200
29. Bhattacharya D, Hoover S, Falk SP, Weisblum B, Vestling M, Striker R. Phosphorylation of yellow fever virus NS5 alters methyltransferase activity. Virology (2008) 380:276–84. doi: 10.1016/j.virol.2008.07.013
30. Klema VJ, Ye M, Hindupur A, Teramoto T, Gottipati K, Padmanabhan R, et al. Dengue virus nonstructural protein 5 (NS5) assembles into a dimer with a unique methyltransferase and polymerase interface. PloS Pathog (2016) 12:e1005451. doi: 10.1371/journal.ppat.1005451
31. Ferrero DS, Ruiz-Arroyo VM, Soler N, Usón I, Guarné A, Verdaguer N. Supramolecular arrangement of the full-length zika virus NS5. PloS Pathog (2019) 15:e1007656. doi: 10.1371/journal.ppat.1007656
32. Yan L, Ge J, Zheng L, Zhang Y, Gao Y, Wang T, et al. Cryo-EM structure of an extended SARS-CoV-2 replication and transcription complex reveals an intermediate state in cap synthesis. Cell (2021) 184:184–193.e10. doi: 10.1016/j.cell.2020.11.016
33. Campagnola G, Govindarajan V, Pelletier A, Canard B, Peersen OB. The SARS-CoV nsp12 polymerase active site is tuned for Large-genome replication. J Virol (2022) 96:e0067122. doi: 10.1128/jvi.00671-22
34. Albentosa-González L, Sabariegos R, Arias A, Clemente-Casares P, Mas A. Akt interacts with usutu virus polymerase, and its activity modulates viral replication. Pathogens (2021) 10:244. doi: 10.3390/pathogens10020244
35. Valero ML, Sabariegos R, Cimas FJ, Perales C, Domingo E, Sánchez-Prieto R, et al. Hepatitis c virus RNA-dependent RNA polymerase interacts with the Akt/PKB kinase and induces its subcellular relocalization. Antimicrob Agents Chemother (2016) 60:3540–50. doi: 10.1128/AAC.03019-15
36. Kim SJ, Kim JH, Sun JM, Kim MG, Oh JW. Suppression of hepatitis c virus replication by protein kinase c-related kinase 2 inhibitors that block phosphorylation of viral RNA polymerase. J Viral Hepat (2009) 16:697–704. doi: 10.1111/j.1365-2893.2009.01108.x
37. Moon JS, Lee SH, Han SH, Kim EJ, Cho H, Lee W, et al. Inhibition of hepatitis c virus in mouse models by lipidoid nanoparticle-mediated systemic delivery of siRNA against PRK2. Nanomedicine (2016) 12:1489–98. doi: 10.1016/j.nano.2016.02.015
38. Liu Z, Tian Y, Machida K, Lai MM, Luo G, Foung SK, et al. Transient activation of the PI3K-AKT pathway by hepatitis c virus to enhance viral entry. J Biol Chem (2012) 287:41922–30. doi: 10.1074/jbc.M112.414789
39. Noppakunmongkolchai W, Poyomtip T, Jittawuttipoka T, Luplertlop N, Sakuntabhai A, Chimnaronk S, et al. Inhibition of protein kinase c promotes dengue virus replication. Virol J (2016) 13:35. doi: 10.1186/s12985-016-0494-6
40. Jakubiec A, Jupin I. Regulation of positive-strand RNA virus replication: the emerging role of phosphorylation. Virus Res (2007) 129:73–9. doi: 10.1016/j.virusres.2007.07.012
41. Gulyas L, Glaunsinger BA. RNA Polymerase II subunit modulation during viral infection and cellular stress. Curr Opin Virol (2022) 56:101259. doi: 10.1016/j.coviro.2022.101259
42. Barik S. Inhibition of viral RNA-dependent RNA polymerases by nucleoside inhibitors: An illustration of the unity and diversity of mechanisms. Int J Mol Sci (2022) 23:12649. doi: 10.3390/ijms232012649
43. Stevaert A, Groaz E, Naesens L. Nucleoside analogs for management of respiratory virus infections: mechanism of action and clinical efficacy. Curr Opin Virol (2022) 57:101279. doi: 10.1016/j.coviro.2022.101279
44. Van Der Linden L, Vives-Adrian L, Selisko B, Ferrer-Orta C, Liu X, Lanke K, et al. The RNA template channel of the RNA-dependent RNA polymerase as a target for development of antiviral therapy of multiple genera within a virus family. PloS Pathog (2015) 11:e1004733. doi: 10.1371/journal.ppat.1004733
Keywords: RNA-dependent RNA polymerase, virus, phosphorylation, flavivirus, coronavirus, influenza virus, protein kinase, RdRp
Citation: Duflos C and Michiels T (2023) Regulation of viral RNA-dependent RNA polymerases by phosphorylation. Front. Virol. 3:1176840. doi: 10.3389/fviro.2023.1176840
Received: 28 February 2023; Accepted: 23 March 2023;
Published: 06 April 2023.
Edited by:
Sonia Zuñiga, National Center for Biotechnology (CSIC), SpainReviewed by:
Antonio Mas, University of Castilla-La Mancha, SpainCopyright © 2023 Duflos and Michiels. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Thomas Michiels, dGhvbWFzLm1pY2hpZWxzQHVjbG91dmFpbi5iZQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.