- 1Department of Population Health, College of Veterinary Medicine, University of Georgia, Athens, GA, United States
- 2Virus and Prion Research Unit, National Animal Disease Center, United States Department of Agriculture, Agricultural Research Service (USDA-ARS), Ames, IA, United States
- 3Veterinary Diagnostic and Production Animal Medicine, College of Veterinary Medicine, Iowa State University, Ames, IA, United States
Introduction: The rapid evolution of influenza A viruses (FLUAV) complicates disease control for animal and public health. Although vaccination is an effective way to control influenza, available vaccines for use in swine result in limited protection against the antigenically distinct FLUAV that currently co-circulate in pigs. Vaccines administered parenterally usually stimulate IgG antibodies but not strong mucosal IgA or cell-mediated responses, which are typically more cross-reactive.
Methods: We developed a live attenuated influenza virus (LAIV) vaccine containing IgA-inducing protein (IGIP) as a molecular marker and immunomodulator. This Flu-IGIP vaccine was tested in a bivalent formulation (H1N1 and H3N2) against challenge with antigenically drifted viruses in pigs. Pigs were vaccinated intranasally with either a bivalent Flu-IGIP or a bivalent Flu-att (control without IGIP) and boosted two weeks later. Three weeks post boost, pigs were challenged with antigenically drifted H1N1 or H3N2 virus.
Results: Vaccinated pigs had increased numbers of influenza-specific IgA-secreting cells in PBMC two weeks post boost and higher numbers of total and influenza-specific IgA-secreting cells in bronchoalveolar lavage fluid (BALF) 5 days post inoculation (dpi) compared to naïve pigs. Pigs vaccinated with both Flu-IGIP and Flu-att shed significantly less virus after H1N1 or H3N2 challenge compared to non-vaccinated pigs. Vaccination with Flu-att reduced respiratory transmission, while Flu-IGIP fully blocked transmission regardless of challenge virus. Both Flu-IGIP and Flu-att vaccines reduced virus replication in the lungs and lung lesions after inoculation with either virus. IgG and IgA levels in BALF and nasal wash of vaccinated pigs were boosted after inoculation as soon as 5 dpi and remained high at 14 dpi.
Conclusion: Our results indicate that Flu-IGIP leads to protection from clinical signs, replication and shedding after antigenically drifted influenza virus infection.
1 Introduction
Influenza A viruses (FLUAV) are important respiratory pathogens that impact animal and public health. Clinical signs of influenza in pigs are often mild; however, the high morbidity can result in substantial economic impact to swine producers due to widespread weight loss and reduced weight gain, in addition to increased expenses with vaccines and treatment (1). Pigs are susceptible to infection with avian and human FLUAVs and may serve as intermediary hosts for the generation of novel reassortant viruses (2). Additionally, FLUAV that infect swine can occasionally spillover to humans, and zoonotic infections with swine-origin FLUAV, termed “variant viruses” (or “v”), are reported yearly (3). These zoonotic spillovers are usually associated with close exposure to infected swine and, thus, some individuals might be at increased risk of variant virus infection, such as farmers, meat processing workers, and veterinarians. Further, these variant viruses may sporadically become adapted to humans and acquire the ability to transmit from person-to-person, as was the case of the 2009 pandemic H1N1 (4). This bidirectional transmission of FLUAV between humans and swine can impact human and animal health and has significantly affected virus epidemiology. Hence, controlling influenza infection and transmission in pigs can serve a dual benefit for both animal and public health.
Although only subtypes H1N1, H1N2, and H3N2 are endemic in swine, a diverse range of phylogenetic lineages of FLUAVs co-circulate in swine globally. This diversity is the result of reassortment between different virus lineages leading to antigenic shift and further accumulation of mutations through antigenic drift of the surface genes, hemagglutinin (HA) and neuraminidase (NA) (5, 6). In the U.S., intensified surveillance in the past decade revealed considerable phylogenetic virus diversity that varies regionally, with five major H1 and H3 genetic lineages currently circulating (7). These lineages are further divided into 16 phylogenetic clades based on the HA genetic diversity (7, 8). Substantial antigenic diversity is seen between and within these genetic lineages and clades (9–11). Thus, this diversity complicates diagnosis and control of influenza in pigs more so than the diversity of influenza in humans.
The primary method for controlling FLUAV in swine herds is through vaccination. Current vaccines for use in swine rely primarily on antibody responses to the virus’ HA surface protein. The HA is highly variable and a major site of antigenic drift that can lead to vaccine mismatch. It was demonstrated that changes in only 6 or 7 amino acid positions in the HA (145, 155, 156, 158, 159, 189 and 193) account for major antigenic changes of swine and human H3 influenza viruses, and single amino acid changes can have a significant impact in the emergence of a new antigenic strain (9, 12, 13). Vaccines licensed for use in pigs in the U.S. are adjuvanted, inactivated virus vaccines or alphavirus vectored vaccines (14). These parenterally administered vaccines can elicit protective humoral immunity by inducing the production of high levels of IgG neutralizing antibodies. But they induce limited IgA antibody response and little, if any, T-cell immunity (15) and confer little efficacy in case of antigenic drift or newly emerged viruses. Consequently, there is the need for development of better vaccine strategies that are more broadly cross-protective against a large number (if not all) currently circulating viruses.
In contrast, live attenuated influenza virus (LAIV) vaccines can elicit a combination of humoral and cellular responses by mimicking a natural infection (15, 16). LAIV can also induce mucosal IgA responses in the respiratory tract (17). A bivalent live attenuated vaccine generated by truncation of the non‐structural (NS1) protein became commercially available for use against influenza in pigs in the U.S (18, 19).. However, both vaccine parental viruses were detected recently by routine surveillance in swine herds, with evidence of reassortment with endemic field strains (20), generating concern for its safety. An LAIV vaccine developed by our group containing an attenuated (att) swine-origin backbone was shown to be safe, cross-protective against challenge with antigenically distinct viruses in pigs and significantly more efficacious than inactivated vaccines (15, 21, 22). Here, we generated recombinant H3N2 and H1N1 att LAIVs with modified HA gene segments that express the porcine IgA-inducing protein (IGIP), a highly conserved protein mainly secreted by dendritic cells (DCs) shown to positively regulate IgA expression (23, 24). IGIP was used as a potential immunomodulator that also serves as a molecular marker. A bivalent vaccine using the H1N1/H3N2 Flu-IGIP LAIV strains was tested in pigs and shown to be efficacious against challenge with antigenically distinct viruses.
2 Materials and methods
Generation of LAIV vaccines. The Flu-IGIP H1N1 LAIV was produced and tested previously in mice (25). The Flu-IGIP H3N2 LAIV was produced following the same protocol. Briefly, IGIP-HA plasmids contained the 5’ untranslated region (UTR) and signal peptide sequences of H1 or H3 HA (A/California/04/09 [CA/09; H1N1pdm09] or A/turkey/Ohio/313053/2004 [OH/04; H3N2 C-IV]), followed by the IGIP mature protein sequence, a G4S linker, furin cleavage site, and the Thosea asigna virus (TAV) 2A protease. The Flu-IGIP LAIVs were rescued by reverse genetics using a co-culture of 293T/MDCK cells as previously described (26). The fragments were cloned into the reverse genetics pDP2002 plasmid (22) with the HA of CA/09 or OH/04, respectively. The IGIP-H1 or IGIP-H3 were paired with the CA/09 N1 or OH/04 N2, respectively, and 6 plasmids corresponding to the internal genes of the LAIV att backbone (FLU-att) (22). Virus stocks were generated in 10-day old specific pathogen free (SPF) eggs. The Flu-att (without IGIP) H3N2 and H1N1 LAIV viruses were produced with same gene constellation as the Flu-IGIP LAIVs, as previously described (22). The two strains selected for the vaccines are well-adapted to pigs and have been shown to replicate well in swine (13, 27). All viral stock sequences were confirmed by Sanger sequencing or next-generation sequencing (NGS). Both vaccines (Flu-IGIP and Flu-att) were formulated as bivalent products by mixing equal amounts of the H1 and H3 viruses. Vaccine viruses were propagated once in embryonated chicken eggs.
Growth, stability, and IgA stimulation of Flu-IGIP vaccines. Each Flu-IGIP virus was inoculated into confluent MDCK monolayers at a multiplicity of infection (MOI) of 0.01 and incubated at 4°C for 15 min, followed by incubation at 35°C for 45 min. After incubation, the inoculum was removed, and cells washed twice with phosphate-buffered saline (PBS). Cells were incubated in Opti-MEM I (Life Technologies, Carlsbad, CA) supplemented with 1 μg/mL of tosylsulfonyl phenylalanyl chloromethyl ketone (TPCK) treated-trypsin (Worthington Biochemicals, Lakewood, NJ) and an antibiotic-antimycotic solution (Sigma-Aldrich, St. Louis, MO) for 72 h. At specific time-points, supernatant was collected, and virus titers determined by real-time reverse transcriptase PCR (RT-qPCR) assay and expressed as log10 TCID50/ml equivalents as previously described (28). The Flu-IGIP viruses were serially passaged at least 5 times in MDCK cells. Briefly, viruses were inoculated at 3 different dilutions in confluent MDCK cells and incubated for 72 h. Supernatant from the highest dilution showing the highest hemagglutination titer was used to inoculate the next passage. RNA from tissue culture supernatants was purified using the RNeasy minikit (Qiagen, Valencia, CA) and used for NGS using the Illumina MiSeq platform as previously described (28). To test the effect of IGIP on IgA expression, naïve swine B cells were cultured with supernatants from Flu-IGIP H3N2 or Flu-att H3N2 infected cells as previously described (23) with modifications. Briefly, total PBMCs were isolated from naïve 5-week-old, cross-bred pigs, and stimulated with B-Poly-S™ reagent (Cellular Technology Ltd, Shaker Hts., OH) in stimulation media (RPMI 1640 media containing 10% fetal bovine serum, 2mM L-Glutamine, antibiotic-antimycotic solution, 8mM HEPES, and 50uM 2-Mercaptomethanol) for 7 days in a 37°C humidified incubator with 5% CO2. Culture supernatants from cells infected with Flu-IGIP H3N2, Flu-att H3N2, or mock supernatant were added at 10% of the total stimulation media volume. Stimulated cells were collected at day 7, and IgA stimulation was measured by ELISPOT as described below.
Animal experimental design. Seventy-two, three-week-old crossbred FLUAV seronegative pigs were obtained from a high health status herd and cared for in compliance with a protocol approved by the Institutional Animal Care and Use Committee at the National Animal Disease Center. Pigs were divided into three groups (n=24/group): non-vaccinated (NV), vaccinated with bivalent Flu-IGIP (H1N1-H3N2), or vaccinated with bivalent Flu-att (H1N1-H3N2). Vaccinated pigs received 2 ml intranasal dose of 1×10^6 TCID50/ml of each vaccine mixture, at 5 and 7 weeks of age. Three weeks post-boost, pigs were challenged (n=12/group) with 2 ml intratracheally and 1 ml intranasally of 1×10^5 TCID50/ml of either A/swine/Iowa/A01778877/2016 (IA/16, 1A.3.3.3 gamma H1N1) or A/swine/Ohio/A01354299/2017 (OH/17, 2010.1 H3N2) under anesthesia as previously described (29). Challenge viruses were antigenically distinct from the OH/04 and CA/09 vaccine strains. At 2-days post inoculation (dpi), 3 naïve pigs were introduced in the same room as challenged pigs but without direct contact to evaluate respiratory transmission. Serum, PBMC, and nasal wash samples were collected at 2 weeks post-vaccination, 3 weeks post-boost, and 2 weeks after inoculation. Nasal swab (Fisherbrand Dacron swabs, Fisher Scientific, Pittsburg, PA) samples were collected pre-challenge (0 dpi), and at 1, 3, and 5 dpi or 3, 5, and 7 days post contact (dpc) as previously described (29). At 5 (n=8/group) and 14 (n=4/group) dpi, pigs were humanely euthanized with a lethal dose of pentobarbital (Fatal Plus, Vortech Pharmaceuticals, Dearborn, MI), and bronchoalveolar lavage fluid (BALF) was collected. Lung and trachea tissues were collected at 5dpi.
Hemagglutination inhibition assay: Sera was heat inactivated at 56°C, treated with receptor-destroying enzyme (RDE; Denka Seiken, VWR) and turkey red blood cells (RBC) using standard techniques. HI assays were performed with serially 2-fold diluted, treated sera obtained from vaccinated pigs using 0.5% turkey RBCs. Viral antigens used included vaccine and challenge strains OH/04, CA/09, IA/16 and OH/17, and a subset of contemporary H1 and H3 antigens that are antigenically different from vaccine and challenge viruses: A/swine/Iowa/A02524480/2020 (IA/20, 1A.3.3.2 H1N1pdm09), A/swine/Iowa/A02245880/2021 (IA/880/21, 1A.3.3.3 gamma H1N1), A/swine/Iowa/A02635890/2021 (IA/890/21, 1990.4.a Cluster-IVA H3N2), and A/swine/Oklahoma/A02479048/2020 (OK/20, 2010.1 H3N2).
ELISA assays. Influenza-specific ELISA assays were performed to measure IgG and IgA in serum (1:2000 dilution), nasal wash and BALF (1:4 dilution each) using OH/04 or CA/09 antigens at 100 HA units, as previously described (30). IgG and IgA were detected using anti-swine IgG and anti-swine IgA (Bethyl Laboratories, Inc., Montgomery, TX) at a 1:1,500 dilution in blocking buffer (Starting Block; Thermo Fisher). Total IgA was quantified using the Pig IgA ELISA Quantitation Set following manufacturer’s recommendation (Bethyl Laboratories, Inc., Montgomery, TX). Antibody levels were reported as the mean O.D. and IgA concentration and were compared between vaccination groups. O.D. levels were shown previously to correlate with antibody titers (31).
ELISPOT assays. Freshly collected PBMC were used to quantify IgA-secreting B cells in vaccinated pigs using the Pig IgA Single-Color ELISPOT kit (Cellular Technology Ltd, Shaker Hts., OH) following manufacturer’s recommendations with a few modifications. Briefly, PBMC was prestimulated with B-Poly-S™ reagent in CTL Test™ B Culture Medium in a 37°C humidified incubator with 7% CO2 for 4 days. ELISPOT plates were pre-coated with either whole OH/04 or CA/09 viruses or Porcine Ig Capture antibody for the comparison of influenza-specific IgA and total IgA levels. Resting B memory cells were pre-stimulated in vitro with B-Poly-STM reagent for 4 days. Pre-stimulated B cells were plated at 2.5×10^5 PBMC/well for total IgA and 1×10^6 PBMC/well for flu-specific IgA. IgA was detected by FITC-labeled Anti-Porcine IgA Antibody. Plates were scanned and analyzed using an automated MultiSpot® Reader system (Autoimmun Diagnostika, Strassberg, Germany). The results are presented in spot-forming units (SFUs).
Virus detection and pathological examination. Nasal swab samples and BALF were titrated in MDCK cells using immunocytochemistry as previously described (32). TCID50/ml titers were calculated for each sample by the Reed and Muench method (33). The percentage of lung surface affected with lesions was calculated based on weighted proportions of each lobe as previously described (34). Formalin-fixed trachea and lung tissue samples were routinely processed, stained with hematoxylin and eosin, and microscopic lesions were scored according to previously described parameters (35). Individual composite scores for each pig were computed for lung and trachea microscopic lesions.
Statistical Analysis. Results are reported as the mean (± the standard error [SE]) of all animals for each treatment group. Analysis of variance (ANOVA) was used to analyze log2 transformed HI reciprocal titers, mean ODs for ELISA assays, log10 transformed virus titers, and SFU counts. Results shown to be significant were subjected to pair-wise comparisons using the Tukey– Kramer test or the Dunn’s test with Bonferroni correction. Statistical analysis and graphs were done using GraphPad Prism software version 9.3.1 (GraphPad Software, La Jolla, CA), with a p-value ≤0.05 considered significant.
3 Results
3.1 Flu-IGIP strains are stable and show efficient replication and IgA stimulation in vitro
Flu-IGIP viruses (H1N1 and H3N2) were successfully rescued in the previously developed Flu-att attenuated backbone (22). The Flu-IGIPs do not need special cell lines to grow and showed similar growth kinetics as the unmodified isogenic LAIVs at 35°C in MDCK cells (Figure S1A). Sequence analysis of the Flu-IGIP strains confirmed that the modified HA segment containing the IGIP cassette was maintained for at least 5 serial passages in SPF eggs or tissue culture cells, and only one mutation was observed in the HA signal peptide of the IGIP-H1 as previously reported (25). Stimulation of porcine naïve B cells with supernatant from Flu-IGIP infected cells in the presence of B-Poly-S™ reagent containing resiquimod and IL-2 resulted in higher number of IgA secreting cells compared to B cells stimulated with supernatant from Flu-att infected cells under the same conditions, although differences were not statistically significant (Figure S1B).
3.2 Flu-IGIP bivalent vaccine is attenuated and stimulates strong immune response in pigs
Twelve pigs per group were vaccinated with bivalent Flu-IGIP or bivalent Flu-att and challenged three weeks post-boost with a 1A.3.3.3 gamma H1N1 (IA/16) or a recent 2010.1 H3N2 (OH/17) virus. Both vaccines were safe in a bivalent formulation and no clinical signs were observed after vaccinations (data not shown). Low HI titers were observed in serum against the vaccine antigens in vaccinated groups before inoculation (Figure 1A) despite high IgG levels (Figure 1B). Low or no HI titers were detected against the challenge viruses or the panel of antigenically different viruses tested (Figure 1A). Levels of IgG and IgA were undetected or low in nasal washes prior to inoculation, but significant difference was observed for IgA levels in Flu-IGIP vaccinated pigs compared to non-vaccinated pigs against CA/09 and OH/04 antigens and compared to Flu-att vaccinated pigs against CA/09 antigen (Figures 1C, D). Numbers of IgA-secreting cells were low before inoculation, particularly for influenza-specific IgA. No differences were observed in the numbers of IgA-secreting cells in PBMC of vaccinated pigs, but a trend for higher counts in total IgA secreting cells was seen in PBMC of vaccinated pigs compared to non-vaccinated pigs prior to inoculation (Figure 1E).
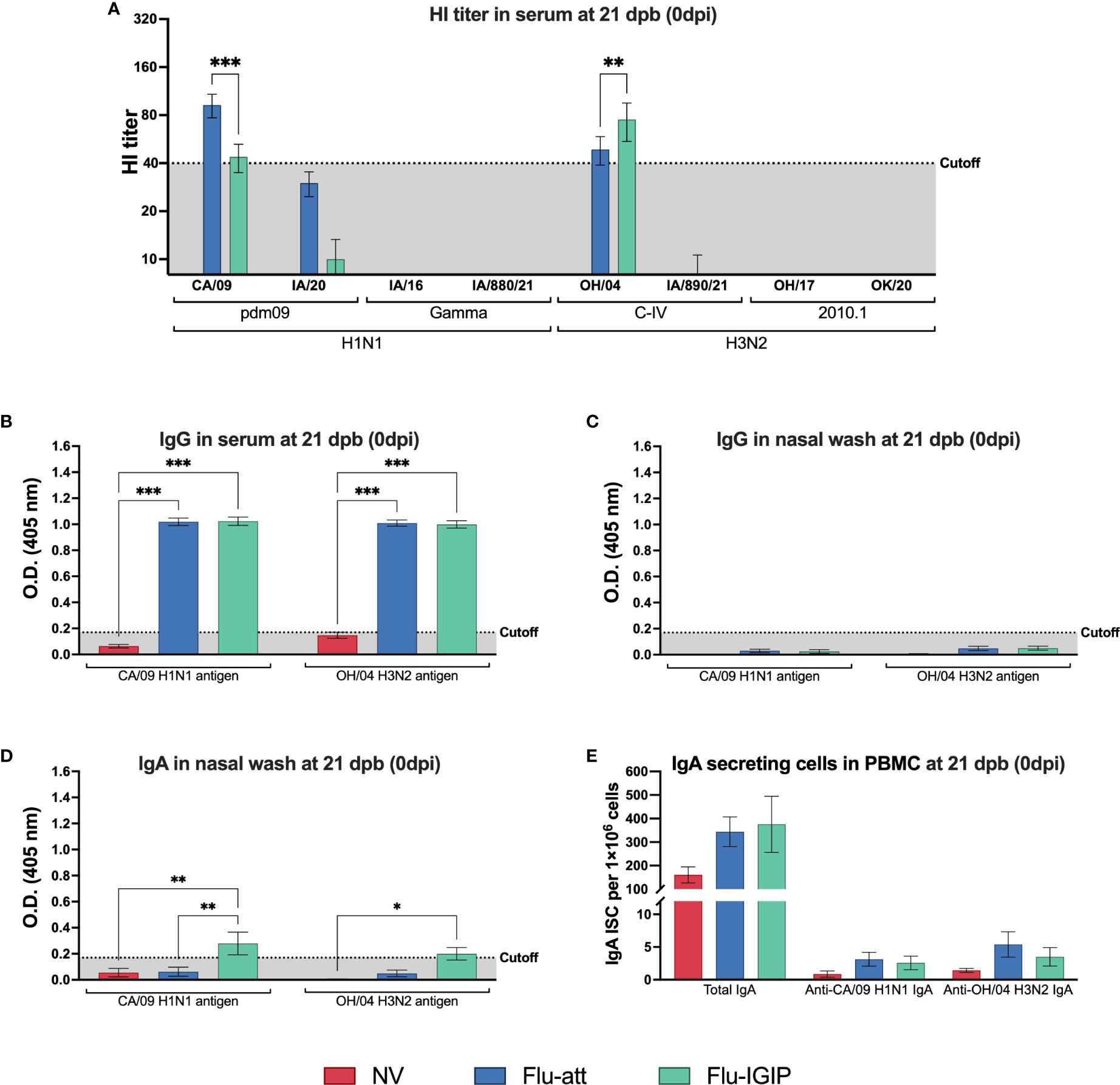
Figure 1 Influenza-specific immune response prior to inoculation. Pigs were not vaccinated (NV, red bars) or vaccinated with bivalent Flu-att (blue bars) or Flu-IGIP (green bars) containing CA/09 H1N1 and OH/04 H3N2 viruses. Hemagglutination inhibition titers were measured (A) 21 days post-boost (dpb). IgG levels in sera (B) and nasal wash (C), and IgA levels in nasal wash (D) were measured by ELISA at 21 dpb. Number of IgA secreting B cells (ISC) in peripheral blood mononuclear cells (PBMC) following B cell stimulation was determined by ELISPOT for total and influenza-specific IgA (E). Viruses on the x-axis represent antigens used in each assay: CA/09, A/California/04/09 (H1N1pdm09); IA/20, A/swine/Iowa/A02524480/2020 (1A.3.3.2 H1N1pdm09); IA/16, A/swine/Iowa/A01778877/2016 (1A.3.3.3 gamma H1N1); IA/880/21, A/swine/Iowa/A02245880/2021 (1A.3.3.3 gamma H1N1); OH/04, A/turkey/Ohio/313053/2004 (H3N2 C-IV); IA/890/21, A/swine/Iowa/A02635890/2021 (H3N2 C-IVA); OH/17, A/swine/Ohio/A01354299/2017 (H3N2 2010.1); and OK/20, A/swine/Oklahoma/A02479048/2020 (2010.1 H3N2). Results displayed as means ± standard error of the mean. Dotted shaded areas indicate assay cutoff values known to be protective for HI assays or calculated as the average of negative pigs + 3 times the standard error of the mean for ELISA assays. *p ≤ 0.05; **p ≤ 0.01; ***p ≤ 0.001.
HI titers against the vaccine antigens were boosted in the serum of vaccinated pigs after inoculation, with a stronger boost for the antigen with the same subtype as the challenge virus (e.g., IA/16 H1N1 challenge strongly boosted CA/09-specific antibodies but not as strongly for OH/04 antibodies; Figure 2A). Infection stimulated high HI antibody titers specific to each challenge virus in all groups, including non-vaccinated pigs, in addition to cross-reactive response to the antigens of the same clade as the challenge virus (e.g., IA/16 H1N1 challenge resulted in HI antibody titers to IA/16 and IA/880/21 of the gamma clade). However, vaccinated pigs showed cross-reactive response to all viruses of the same subtype of the challenge virus (e.g., vaccinated pigs challenged with IA/16 H1N1 showed cross-reactive response against CA/09, IA/20, IA/16, and IA/880/21 H1N1 viruses). Cross-reactive IgG levels to the vaccine antigens in serum remained high post-inoculation in vaccinated groups (Figure 2B) and reached similar levels in non-vaccinated pigs 2 weeks post inoculation (Figure 2C).
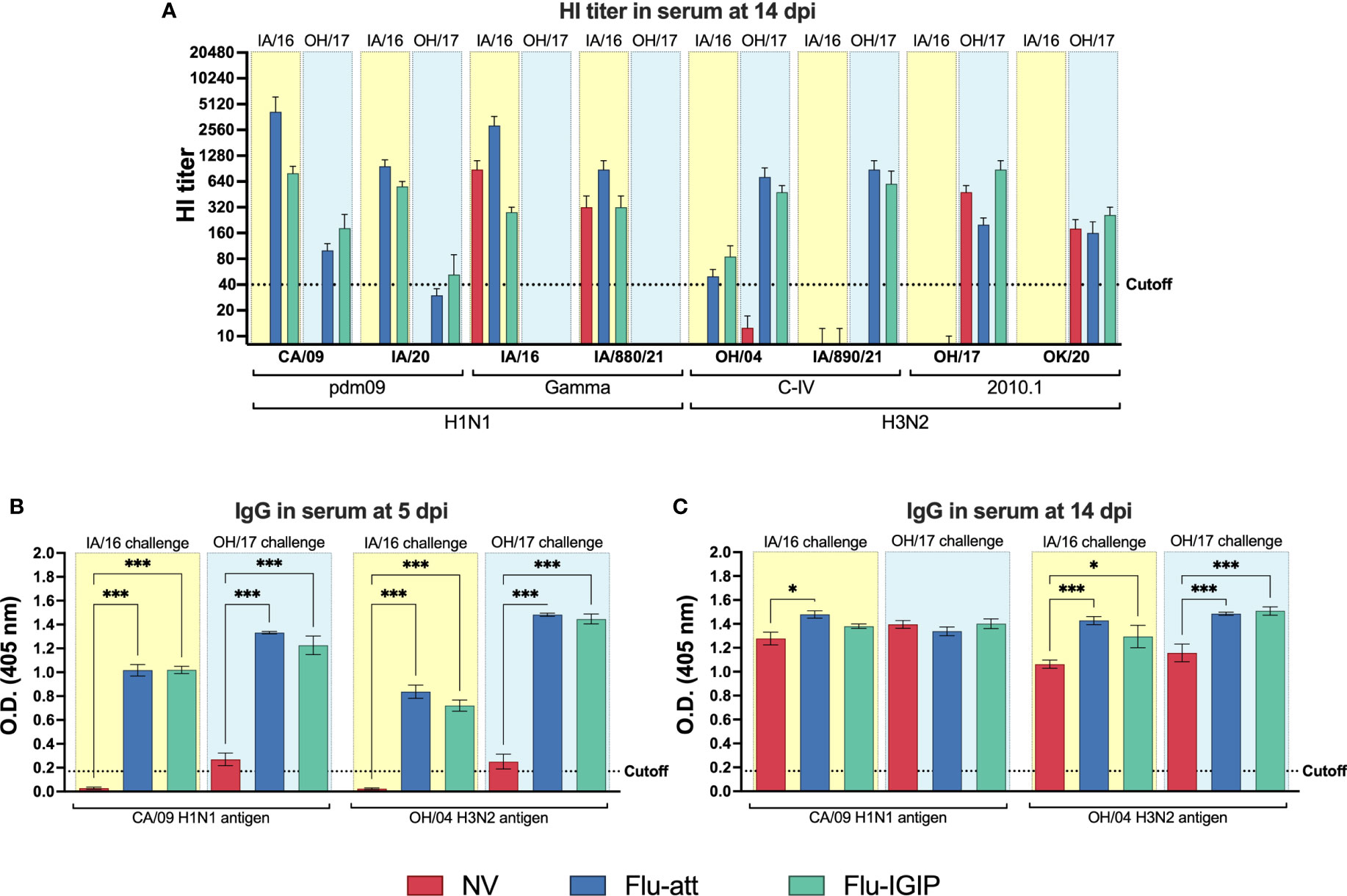
Figure 2 Influenza-specific antibody response in serum after inoculation. Pigs were not vaccinated (NV, red bars) or vaccinated with bivalent Flu-att (blue bars) or Flu-IGIP (green bars) containing CA/09 H1N1 and OH/04 H3N2 viruses. Pigs were challenged with IA/16 H1N1 (light-yellow boxes) or OH/17 H3N2 (light-blue boxes) viruses. Hemagglutination inhibition titers were measured (A) 14 days post inoculation (dpi). IgG levels were measured in serum by ELISA at (B) 5 and (C) 14 dpi. Viruses on the x-axis represent antigens used in each assay: CA/09, A/California/04/09 (H1N1pdm09); IA/20, A/swine/Iowa/A02524480/2020 (1A.3.3.2 H1N1pdm09); IA/16, A/swine/Iowa/A01778877/2016 (1A.3.3.3 gamma H1N1); IA/880/21, A/swine/Iowa/A02245880/2021 (1A.3.3.3 gamma H1N1); OH/04, A/turkey/Ohio/313053/2004 (H3N2 C-IV); IA/890/21, A/swine/Iowa/A02635890/2021 (H3N2 C-IVA); OH/17, A/swine/Ohio/A01354299/2017 (H3N2 2010.1); and OK/20, A/swine/Oklahoma/A02479048/2020 (2010.1 H3N2). Results displayed as means ± standard error of the mean. Vaccine and challenge viruses and additional antigenically distinct viruses were used as HI antigens and vaccine viruses as ELISA antigens. Results displayed as means ± standard error of the mean. Dotted lines indicate assay cutoff values known to be protective for HI assays or calculated as the average of negative pigs + 3 times the standard error of the mean for ELISA assays. *p ≤ 0.05; ***p ≤ 0.001 within groups challenged with the same virus.
IgG and IgA levels in BALF were detected only in vaccinated pigs soon after inoculation (5dpi) compared to non-vaccinated pigs (Figures 3A, B), suggesting that the infection boosted a vaccine-specific memory response. IgG and IgA levels remained high in vaccinated pigs 2 weeks post infection, with similar levels in non-vaccinated pigs challenged with the H1N1 IA/16, but levels were overall lower in non-vaccinated pigs challenged with the H3N2 OH/17 (Figures 3C, D). IgG levels were overall low in nasal washes (Figure 3E). IgA levels in nasal wash were boosted after inoculation in vaccinated groups in a similar pattern as observed for the HI response, with a stronger boost for the antigen with the same subtype as the challenge virus (Figure 3F). Cross-reactive IgA for the vaccine antigen with the same subtype as the challenge virus was also stimulated in non-vaccinated pigs, but levels remained lower than vaccinated animals (Figure 3F).
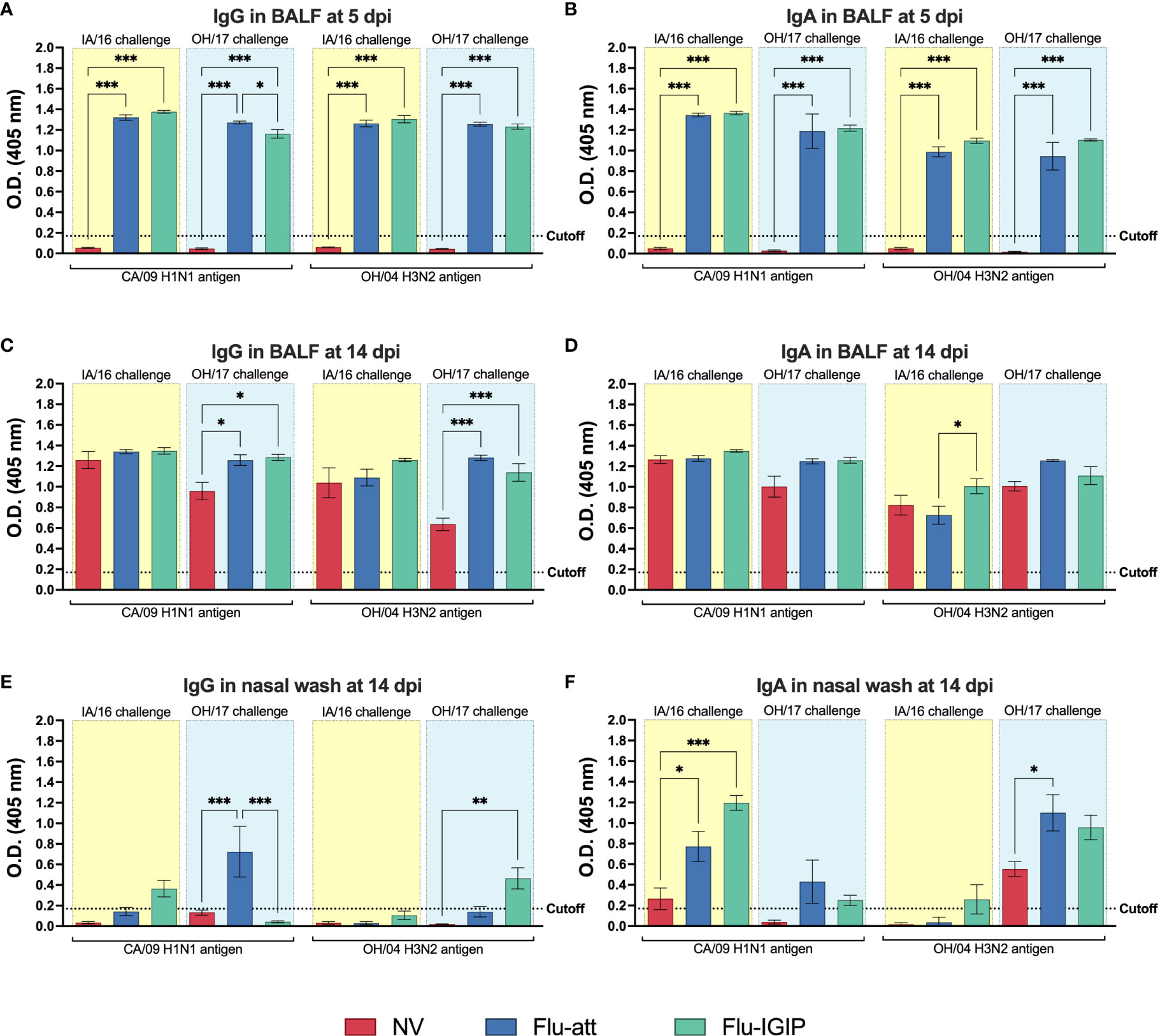
Figure 3 Influenza-specific antibody response in mucosal surfaces after inoculation. Pigs were not vaccinated (NV, red bars) or vaccinated with bivalent Flu-att (blue bars) or Flu-IGIP (green bars) containing CA/09 H1N1 and OH/04 H3N2 viruses. Pigs were challenged with IA/16 H1N1 (light-yellow boxes) or OH/17 H3N2 (light-blue boxes) viruses. IgG levels were measured in bronchoalveolar lavage fluid (BALF) at (A) 5 and (C) 14 days post inoculation (dpi) and in nasal wash at (E) 14 dpi. IgA levels were measured by ELISA in BALF at (B) 5 and (D) 14 dpi and in nasal wash at (F) 14 dpi. Viruses on the x-axis represent antigens used in the ELISA. Results displayed as means ± standard error of the mean. Dotted lines indicate assay cutoff values calculated as the average of negative pigs + 3 times the standard error of the mean for ELISA. *, p ≤ 0.05; **, p ≤ 0.01; ***, p ≤ 0.001 within groups challenged with same virus. CA/09, A/California/04/09; OH/04, A/turkey/Ohio/313053/2004; IA/16, A/swine/Iowa/A01778877/2016; OH/17, A/swine/Ohio/A01354299/2017.
Numbers of IgA-secreting cells (total IgA and influenza-specific) were higher in BALF of vaccinated pigs early after inoculation (5dpi) (Figure 4A), but total IgA-secreting cell numbers increased in non-vaccinated pigs at 14dpi (Figures 4B, C). Nevertheless, no major differences were observed between the two vaccines (IGIP vs non-IGIP LAIVs) for IgA stimulation.
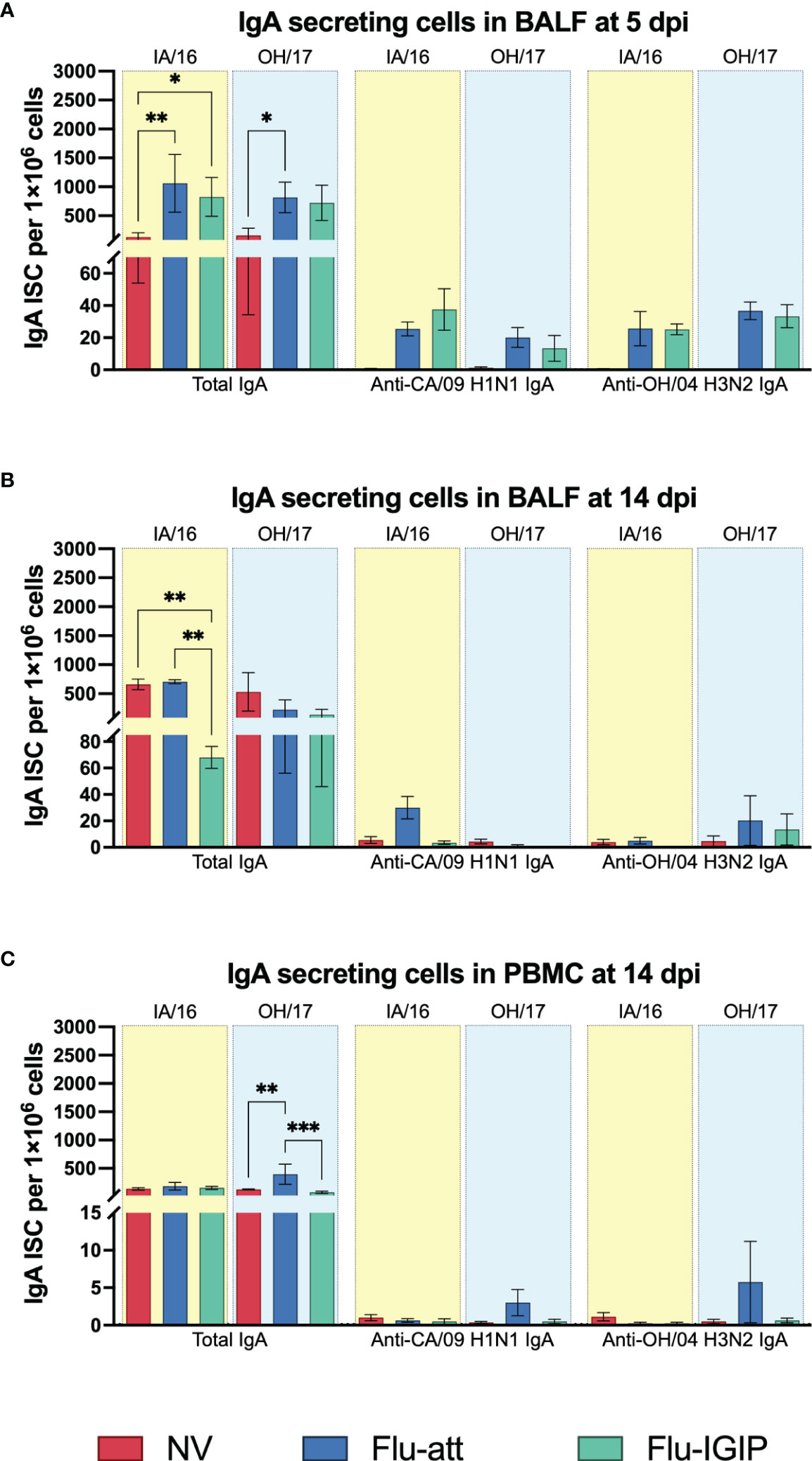
Figure 4 Total and influenza-specific systemic and local IgA secreting B cells after inoculation. Pigs were not vaccinated (NV, red bars) or vaccinated with bivalent Flu-att (blue bars) or Flu-IGIP (green bars) containing A/California/04/09 (CA/09 H1N1) and A/turkey/Ohio/313053/2004 (OH/04 H3N2) viruses. Pigs were challenged with A/swine/Iowa/A01778877/2016 (IA/16 H1N1, light-yellow boxes) or A/swine/Ohio/A01354299/2017 (OH/17 H3N2, light-blue boxes). Number of total and influenza-specific IgA secreting B cells (ISC) were determined by ELISpot in bronchoalveolar lavage fluid (BALF) at (A) 5 and (B) 14 days post inoculation (dpi) and in peripheral blood mononuclear cells (PBMC) at 14 dpi (C) following B cell stimulation. Results displayed as mean spot counts ± standard error of the mean. *, p ≤ 0.05; **, p ≤ 0.01; ***, p ≤ 0.001 within groups challenged with the same virus.
3.3 Flu-IGIP bivalent vaccine protected pigs against infection with antigenically distinct viruses
Following inoculation with antigenically distinct viruses (IA/16 and OH/17), Flu-IGIP and Flu-att vaccinated pigs were completely protected and had macroscopic lung lesions not different from non- challenged pigs (Figure 5A). In contrast, non-vaccinated pigs had higher percentage of lungs affected with lesions. Microscopic lesions in vaccinated pigs were lower than non-vaccinated pigs, particularly for H1N1 challenged pigs (Figure 5B). Additionally, little to no virus was detected in BALF or nasal swabs of vaccinated pigs, but titers were high in BALF and nasal swabs of non-vaccinated pigs (Figures 5C, D). Titers were higher in non-vaccinated pigs challenged with the H3N2 OH/17 virus despite the lower percentage of lung lesions observed (Figures 5A, C, D).
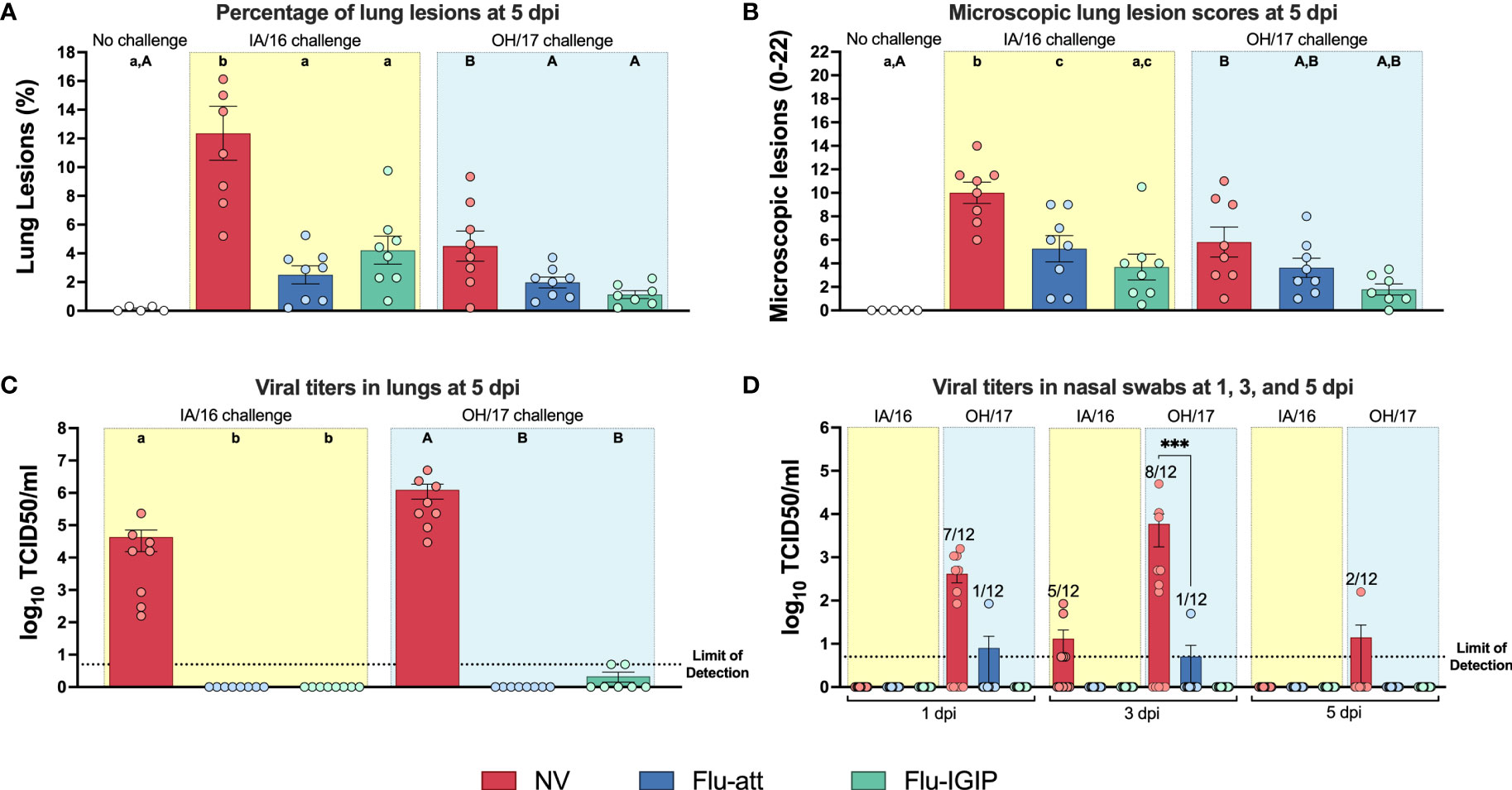
Figure 5 Lung lesions and virus titers in challenged pigs. Non-vaccinated pigs (NV, red bars) or pigs vaccinated with bivalent Flu-att (blue bars) or Flu-IGIP (green bars) were challenged with A/swine/Iowa/A01778877/2016 (IA/16 H1N1, light-yellow boxes) or A/swine/Ohio/A01354299/2017 (OH/17 H3N2, light-blue boxes). (A) Macroscopic and (B) microscopic pneumonia. Results displayed as mean percentage macroscopic consolidation or microscopic scores of lungs ± standard error of the mean. Viral titers in (C) bronchoalveolar lavage fluid (BALF) collected at 5 days post inoculation (dpi) or (D) nasal swabs collected at 1, 3, and 5 dpi. Results displayed as mean viral titers ± standard error of the mean. Different uppercase (OH/17 challenge) or lowercase (IA/16 challenge) letters indicate statistical differences within groups challenged with the same virus (only groups challenged with same virus were compared between each other). ***, p ≤ 0.001 within groups challenged with the same virus. Dotted lines indicate limit of detection.
3.4 Flu-IGIP bivalent vaccine reduced viral shedding and blocked viral transmission to naïve pigs
Protection against respiratory transmission was tested by placing 3 naïve pigs in the same room with each challenged group. While pigs vaccinated with the bivalent Flu-att LAIV challenged with both viruses showed little shedding (Figure 5D), transmission was confirmed in all pigs exposed to H3N2-challenged pigs and in 1 pig exposed to H1N1-challenged pigs either by detection of viral titers or antibodies (Figure 6). In contrast, bivalent Flu-IGIP-vaccinated pigs did not show evidence of transmission; neither H1N1 nor H3N2 viruses were detected in respiratory contacts in these groups (Figure 6A) and very low levels of antibodies (potentially background) were detected in only one pig (Figure 6B).
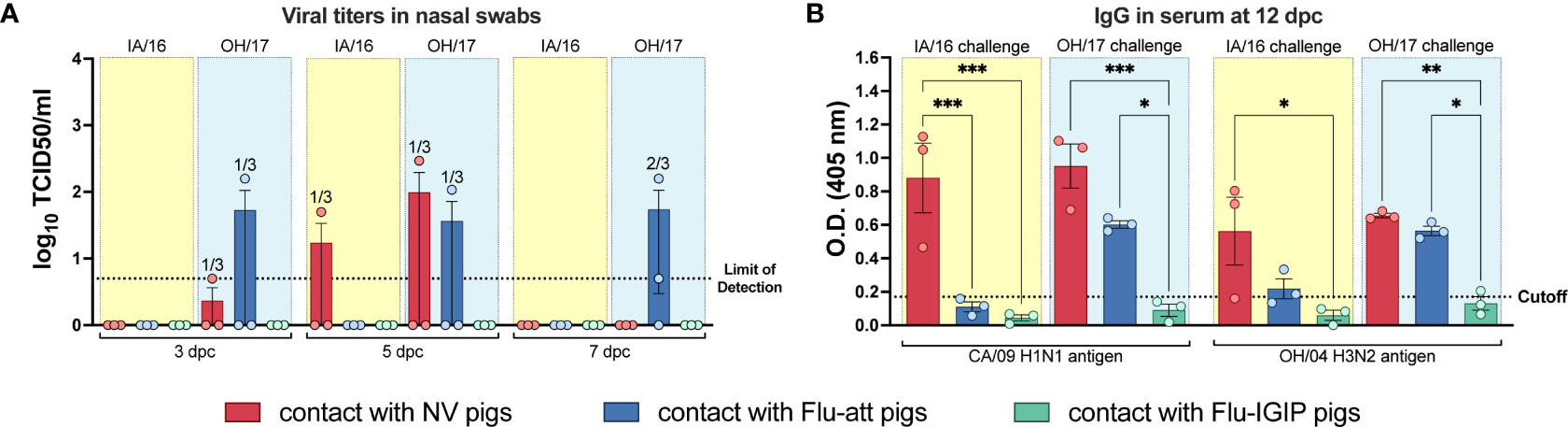
Figure 6 Respiratory transmission from challenged pigs to naïve contacts. Non-vaccinated pigs (NV, red bars) or pigs vaccinated with bivalent Flu-att (blue bars) or Flu-IGIP (green bars) were challenged with A/swine/Iowa/A01778877/2016 (IA/16 H1N1, light-yellow boxes) or A/swine/Ohio/A01354299/2017 (OH/17 H3N2, light-blue boxes). Naïve pigs (n=3/group) were placed in the same room as challenged pigs but with no direct contact at 2 days post inoculation (dpi). (A) Viral titers in nasal swabs were measured by TCID50 at 3, 5 and 7 days post contact (dpc) and (B) IgG antibody titers were measured in serum by ELISA at 12 dpc. Viruses on the x-axis of (B) represent antigens used in the ELISA: CA/09, A/California/04/09 H1N1; OH/04, A/turkey/Ohio/313053/2004 H3N2. *, p ≤ 0.05; **, p ≤ 0.01; ***, p ≤ 0.001 within groups challenged with the same virus. Dotted lines indicate limit of detection for titration or ELISA cutoff values calculated as the average of negative pigs + 3 times the standard error of the mean.
4 Discussion
Although vaccination is an effective tool for the prevention and control against FLUAV, the diversity of strains that co-circulate in North American swine continues to challenge veterinarians and producers. Most vaccines available for use in pigs are inactivated products that only stimulate protective immunity against closely related viruses. Although LAIVs have the potential to provide more multidimensional cellular and humoral responses, an LAIV that was recently introduced to the market was withdrawn due to safety concerns regarding reassortment with circulating influenza viruses (20). In this research, we sought to improve the safety and efficacy of previously generated LAIVs for swine (22). Thus, the HA segment of an H1N1 strain (CA/09) and an H3N2 strain (OH/04) were modified to carry the IGIP mature peptide with additional modifications. The IGIP was chosen due to its potential as a natural vaccine adjuvant and to stimulate IgA class switch (24). The IGIP was also introduced as a molecular marker with the potential to reduce the reassortment of the HA gene. Further, our previous study in mice showed that the inclusion of the IGIP on the HA resulted in a more attenuated strain (25). Since there is no evidence of inflammatory diseases associated with overexpression of IGIP, it has the potential to be a safe vaccine adjuvant that promotes IgA upregulation without side effects.
The Flu-IGIP bivalent vaccine was tested in 5 week-old pigs in comparison to an LAIV (Flu-att) that has been repeatedly shown to be safe, stimulate a strong humoral and cellular response, and be protective against influenza in pigs (22, 36, 37). Since immunization with this Flu-att has consistently shown to stimulate sterilizing immunity against homologous viruses, we selected antigenically mismatched viruses for infection, to compare the two vaccines and be able to detect any differences in protection. The Flu-IGIP bivalent vaccine was as efficient as the bivalent Flu-att in protecting pigs against antigenically distinct H1N1 and H3N2 viruses. Lung lesions were significantly reduced, as was viral replication in the lungs and nasal cavity. This was particularly true for the H1N1 IA/16 infection, a representative of current gamma viruses (1A.3.3.3 clade) circulating in pigs, which showed higher virulence in pigs compared to the H3N2 OH/17 virus from the 2010.1 lineage. Levels of neutralizing antibodies measured by HI assay were low in both vaccinated groups before infection, as previously observed for these and other LAIV vaccines (19, 36). This is expected since systemic HI responses stimulated by LAIVs are at lower detectable levels compared to inactivated vaccines (17). Hence, HI titers are typically not considered an ideal correlate of protection for LAIVs as is the standard for inactivated influenza vaccines (38). Following challenge, vaccinated pigs showed higher cross-reactivity against all viruses tested in our panel that were of the same subtype of the challenge viruses, while non-vaccinated pigs only showed cross-reactivity to viruses of the same clade of the challenge viruses. LAIVs cell-mediated and mucosal immune responses have been associated with the protection against infection in the absence of neutralizing antibodies (15, 17). In contrast to the low HI antibody titers, high levels of IgG were detected in serum of vaccinated animals prior to and a few days after infection. These antibody levels were measured against the whole viruses in an in-house ELISA and represent antibodies against all viral proteins, including more conserved ones such as the nucleoprotein (NP). Non-neutralizing, anti-NP humoral responses have been associated with protection against heterologous influenza infection (39).
Although mucosal IgA responses were low prior to inoculation in the nasal cavity of vaccinated pigs, they were boosted after infection, reaching significantly higher levels compared to non-vaccinated challenged animals, suggesting that the vaccines stimulated a strong memory IgA response. Both IgG and IgA responses were detected in the lungs (BALF) of vaccinated pigs soon after inoculation but only reached similar levels in non-vaccinated pigs after 2 weeks post-infection. Similarly, numbers of flu-specific IgA-secreting cells showed a trend to be higher in vaccinated pigs prior to inoculation and became significantly higher than non-vaccinated pigs shortly after inoculation, reaching similar numbers in all groups 2 weeks post-inoculation. Although no major differences were observed between vaccine groups, these results highlight that the IGIP modification did not alter the immunity and efficacy against antigenically different viruses compared to the control vaccine.
Importantly, the Flu-IGIP bivalent vaccine completely blocked transmission to respiratory contacts, more so than the Flu-att control vaccine. This is of particular importance as it suggests that the Flu-IGIP vaccine is more protective against shedding and transmission than the Flu-att, which could be a result of improved mucosal IgA response. It is possible that the levels of IgA and IgA-secreting cells were enhanced in Flu-IGIP vaccinated pigs but not detected in the timepoints of sample collection selected in the present study. IgA has been shown to have a shorter circulating half-life (~4-6 days) than IgG in multiple species (40). Although IgA-producing plasma cells may be long-lived, these memory cells are usually confined to the bone marrow in mice and humans (29), which may be similar in pigs. This may be the reason why a stronger IgA response was observed 5 days post infection in vaccinated pigs but numbers of IgA-secreting cells in lungs or circulation were low at 14 dpi. Further work is needed to dissect the mechanisms for stimulation of memory IgA response post-vaccination in pigs. Although little is known of the IGIP’s function in the respiratory tract, our combined results suggest that IGIP may act as an adjuvant to stimulate enhanced humoral responses in the respiratory tract.
Protection elicited by live attenuated vaccines results from the induction of multiple branches of the immune system. The combination of multiple correlates of protection needs to be considered when evaluating such response, including levels of mucosal IgA antibodies, which are essential for broader cross-protection (41). Here, we showed that the incorporation of IGIP to an LAIV platform previously shown to be highly efficacious in swine resulted in protection against influenza infection. Although both vaccines resulted in similar protection against clinical signs and viral replication, the vaccine expressing IGIP resulted in improved protection against transmission, and it is tempting to speculate that it is likely the result of IgA upregulation. Although we showed a trend for higher stimulation of IgA-secreting cells by the Flu-IGIP virus in vitro, further studies are further studies are needed to truly appreciate the effect of IGIP as an immune stimulant of B cells, either for IgA response but also IgG response. Nevertheless, this new attenuation strategy is expected to further reduce virus fitness, possibly generating more attenuated LAIVs with increased safety profiles. Overall, our results suggest that the Flu-IGIP is safe and more efficacious against antigenically distinct viruses.
Data availability statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation upon request.
Ethics statement
The animal study was reviewed and approved by Institutional Animal Care and Use Committee of the National Animal Disease Center, USDA-ARS.
Author contributions
DSR, TKA, ALVB, and DRP contributed to conception and design of the study. DSR, GCZ, MWB, SK, MEM, LMF, CJC, SCG, CKS, PCG performed experiments and data analysis. DSR performed the statistical analysis. DSR wrote the first draft of the manuscript. All authorscontributed to the article and approved the submitted version.
Funding
Funding was provided by The National Pork Board (Project #19-009) and in part by USDA-ARS and by Diseases of Agricultural Animals Program [grant no. 2022-67015-37205/project accession no. 1028058] from the USDA National Institute of Food and Agriculture.
Acknowledgments
We thank the animal care personnel from the National Animal Disease Center-USDA for assistance with animals and Michelle Harland for technical assistance with laboratory techniques. This research was supported in part by an appointment to the Agricultural Research Service (ARS) Research Participation Program administered by the Oak Ridge Institute for Science and Education (ORISE) through an interagency agreement between the U.S. Department of Energy (DOE) and the U.S. Department of Agriculture (USDA). ORISE is managed by ORAU under DOE contract number DE-SC0014664. All opinions expressed in this paper are the author’s and do not necessarily reflect the policies and views of USDA, DOE, or ORAU/ORISE. Mention of trade names or commercial products in this article is solely for the purpose of providing specific information and does not imply recommendation or endorsement by UGA, USDA, DOE, or ORISE. USDA is an equal opportunity provider and employer.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fviro.2023.1042724/full#supplementary-material
Supplementary Figure 1 | In vitro growth kinetics profiles of Flu-IGIP viruses. Growth kinetics of Flu-IGIP H1N1 and Flu-IGIP H3N2 in MDCK compared to isogenic non-IGIP Flu-att H1N1 and H3N2. Results for H3N2 viruses are from 2 independent experiments in triplicates and titers are expressed as log10 TCID50 equivalents. Results for H1N1 viruses were previously reported in (25) and are presented here for comparison.
References
1. Rajao DS, Anderson TK, Gauger PC, Vincent AL. Pathogenesis and vaccination of influenza a virus in swine. In: Compans RW, Oldstone MBA, editors. Influenza pathogenesis and control. New York, NY: Springer (2014). p. 307–26. Curr Top Microbiol Immunol. 1.
2. Ito T, Couceiro JN, Kelm S, Baum LG, Krauss S, Castrucci MR, et al. Molecular basis for the generation in pigs of influenza a viruses with pandemic potential. J Virol (1998) 72(9):7367–73. doi: 10.1128/JVI.72.9.7367-7373.1998
3. Centers for Disease Control and Prevention. Key facts about human infections with variant viruses (2016). Available at: https://www.cdc.gov/flu/swineflu/keyfacts-variant.htm.
4. Dawood FS, Jain S, Finelli L, Shaw MW, Lindstrom S, Garten RJ, et al. Emergence of a novel swine-origin influenza a (H1N1) virus in humans. N Engl J Med (2009) 360(25):2605–15. doi: 10.1056/NEJMoa0903810
5. Shao W, Li X, Goraya MU, Wang S, Chen JL. Evolution of influenza a virus by mutation and re-assortment. Int J Mol Sci (2017) 18(8):1650. doi: 10.3390/ijms18081650
6. Anderson TK, Nelson MI, Kitikoon P, Swenson SL, Korslund JA, Vincent AL. Population dynamics of cocirculating swine influenza a viruses in the united states from 2009 to 2012. Influenza Other Respir Viruses (2013) 7(Suppl 4):42–51. doi: 10.1111/irv.12193
7. Walia RR, Anderson TK, Vincent AL. Regional patterns of genetic diversity in swine influenza a viruses in the united states from 2010 to 2016. Influenza Other Respir Viruses (2018) 13(3):262–73. doi: 10.1111/irv.12559
8. Anderson TK, Macken CA, Lewis NS, Scheuermann RH, Van Reeth K, Brown IH, et al. A phylogeny-based global nomenclature system and automated annotation tool for H1 hemagglutinin genes from swine influenza a viruses. mSphere (2016) 1(6):e00275–16. doi: 10.1128/mSphere.00275-16
9. Lewis NS, Anderson TK, Kitikoon P, Skepner E, Burke DF, Vincent AL. Substitutions near the hemagglutinin receptor-binding site determine the antigenic evolution of influenza a H3N2 viruses in U.S. swine. J Virol (2014) 88(9):4752–63. doi: 10.1128/JVI.03805-13
10. Lorusso A, Vincent AL, Harland ML, Alt D, Bayles DO, Swenson SL, et al. Genetic and antigenic characterization of H1 influenza viruses from united states swine from 2008. J Gen Virol (2011) 92(Pt 4):919–30. doi: 10.1099/vir.0.027557-0
11. Rajao DS, Anderson TK, Kitikoon P, Stratton J, Lewis NS, Vincent AL. Antigenic and genetic evolution of contemporary swine H1 influenza viruses in the united states. Virology (2018) 518:45–54. doi: 10.1016/j.virol.2018.02.006
12. Koel BF, Burke DF, Bestebroer TM, van der Vliet S, Zondag GC, Vervaet G, et al. Substitutions near the receptor binding site determine major antigenic change during influenza virus evolution. Science (2013) 342(6161):976–9. doi: 10.1126/science.1244730
13. Abente EJ, Santos J, Lewis NS, Gauger PC, Stratton J, Skepner E, et al. The molecular determinants of antibody recognition and antigenic drift in the H3 hemagglutinin of swine influenza a virus. J Virol (2016) 90(18):8266–80. doi: 10.1128/JVI.01002-16
14. Vander Veen RL, Loynachan AT, Mogler MA, Russell BJ, Harris DL, Kamrud KI. Safety, immunogenicity, and efficacy of an alphavirus replicon-based swine influenza virus hemagglutinin vaccine. Vaccine (2012) 30(11):1944–50. doi: 10.1016/j.vaccine.2012.01.030
15. Loving CL, Vincent AL, Pena L, Perez DR. Heightened adaptive immune responses following vaccination with a temperature-sensitive, live-attenuated influenza virus compared to adjuvanted, whole-inactivated virus in pigs. Vaccine (2012) 30(40):5830–8. doi: 10.1016/j.vaccine.2012.07.033
16. Gorse GJ, Belshe RB, Munn NJ. Superiority of live attenuated compared with inactivated influenza a virus vaccines in older, chronically ill adults. Chest (1991) 100(4):977–84. doi: 10.1378/chest.100.4.977
17. Hoft DF, Lottenbach KR, Blazevic A, Turan A, Blevins TP, Pacatte TP, et al. Comparisons of the humoral and cellular immune responses induced by live attenuated influenza vaccine and inactivated influenza vaccine in adults. Clin Vaccine Immunol (2017) 24(1):e00414–16. doi: 10.1128/CVI.00414-16
18. Richt JA, Garcia-Sastre A. Attenuated influenza virus vaccines with modified NS1 proteins. Curr Top Microbiol Immunol (2009) 333:177–95. doi: 10.1007/978-3-540-92165-3_9
19. Richt JA, Lekcharoensuk P, Lager KM, Vincent AL, Loiacono CM, Janke BH, et al. Vaccination of pigs against swine influenza viruses by using an NS1-truncated modified live-virus vaccine. J Virol (2006) 80(22):11009–18. doi: 10.1128/JVI.00787-06
20. Sharma A, Zeller MA, Li G, Harmon KM, Zhang J, Hoang H, et al. Detection of live attenuated influenza vaccine virus and evidence of reassortment in the U.S. swine population. J Vet Diagn Invest (2020) 32(2):301–11. doi: 10.1177/1040638720907918
21. Gauger PC, Loving CL, Khurana S, Lorusso A, Perez DR, Kehrli ME Jr., et al. Live attenuated influenza a virus vaccine protects against A(H1N1)pdm09 heterologous challenge without vaccine associated enhanced respiratory disease. Virology (2014) 471-473:93–104. doi: 10.1016/j.virol.2014.10.003
22. Pena L, Vincent AL, Ye J, Ciacci-Zanella JR, Angel M, Lorusso A, et al. Modifications in the polymerase genes of a swine-like triple-reassortant influenza virus to generate live attenuated vaccines against 2009 pandemic H1N1 viruses. J Virol (2011) 85(1):456–69. doi: 10.1128/JVI.01503-10
23. Austin AS, Haas KM, Naugler SM, Bajer AA, Garcia-Tapia D, Estes DM. Identification and characterization of a novel regulatory factor: IgA-inducing protein. J Immunol (2003) 171(3):1336–42. doi: 10.4049/jimmunol.171.3.1336
24. Estes DM. Regulation of IgA responses in cattle, humans and mice. Vet Immunol Immunopathol (2010) 138(4):312–7. doi: 10.1016/j.vetimm.2010.10.009
25. Cáceres CJ, Cardenas-Garcia S, Jain A, Gay LC, Carnaccini S, Seibert B, et al. Development of a novel live attenuated influenza a virus vaccine encoding the IgA-inducing protein. Vaccines (2021) 9(7):703. doi: 10.3390/vaccines9070703
26. Perez DR, Angel M, Gonzalez-Reiche AS, Santos J, Obadan A, Martinez-Sobrido L. Plasmid-based reverse genetics of influenza a virus. Methods Mol Biol (2017) 1602:251–73. doi: 10.1007/978-1-4939-6964-7_16
27. Henningson JN, Rajao DS, Kitikoon P, Lorusso A, Culhane MR, Lewis NS, et al. Comparative virulence of wild-type H1N1pdm09 influenza a isolates in swine. Vet Microbiol (2015) 176(1-2):40–9. doi: 10.1016/j.vetmic.2014.12.021
28. Santos JJS, Abente EJ, Obadan AO, Thompson AJ, Ferreri L, Geiger G, et al. Plasticity of amino acid residue 145 near the receptor binding site of H3 swine influenza a viruses and its impact on receptor binding and antibody recognition. J Virol (2019) 93(2):e01413–18. doi: 10.1128/JVI.01413-18
29. Khodadadi L, Cheng Q, Radbruch A, Hiepe F. The maintenance of memory plasma cells. Front Immunol (2019) 10(721). doi: 10.3389/fimmu.2019.00721
30. Gauger PC, Loving CL, Vincent AL. Enzyme-linked immunosorbent assay for detection of serum or mucosal isotype-specific IgG and IgA whole-virus antibody to influenza a virus in swine. Methods Mol Biol (2014) 1161:303–12. doi: 10.1007/978-1-4939-0758-8_25
31. Souza CK, Rajao DS, Loving CL, Gauger PC, Perez DR, Vincent AL. Age at vaccination and timing of infection do not alter vaccine-associated enhanced respiratory disease in influenza a virus-infected pigs. Clin Vaccine Immunol (2016) 23(6):470–82. doi: 10.1128/CVI.00563-15
32. Vincent AL, Ma W, Lager KM, Richt JA, Janke BH, Sandbulte MR, et al. Live attenuated influenza vaccine provides superior protection from heterologous infection in pigs with maternal antibodies without inducing vaccine-associated enhanced respiratory disease. J Virol (2012) 86(19):10597–605. doi: 10.1128/JVI.01439-12
33. Reed LJ, Muench H. A simple method of estimating fifty per cent endpoints. Am J Epidemiol (1938) 27(3):493–7. doi: 10.1093/oxfordjournals.aje.a118408
34. Gauger PC, Vincent AL, Loving CL, Lager KM, Janke BH, Kehrli ME Jr., et al. Enhanced pneumonia and disease in pigs vaccinated with an inactivated human-like (delta-cluster) H1N2 vaccine and challenged with pandemic 2009 H1N1 influenza virus. Vaccine (2011) 29(15):2712–9. doi: 10.1016/j.vaccine.2011.01.082
35. Gauger PC, Vincent AL, Loving CL, Henningson JN, Lager KM, Janke BH, et al. Kinetics of lung lesion development and pro-inflammatory cytokine response in pigs with vaccine-associated enhanced respiratory disease induced by challenge with pandemic (2009) A/H1N1 influenza virus. Vet Pathol (2012) 49(6):900–12. doi: 10.1177/0300985812439724
36. Abente EJ, Rajao DS, Santos J, Kaplan BS, Nicholson TL, Brockmeier SL, et al. Comparison of adjuvanted-whole inactivated virus and live-attenuated virus vaccines against challenge with contemporary, antigenically distinct swine H3N2 influenza a viruses. J Virol (2018) 92(22):e01323–18. doi: 10.1128/JVI.01323-18
37. Loving CL, Lager KM, Vincent AL, Brockmeier SL, Gauger PC, Anderson TK, et al. Efficacy of inactivated and live-attenuated influenza virus vaccines in pigs against infection and transmission of emerging H3N2 similar to the 2011-2012 H3N2v. J Virol (2013) 87:9895–903. doi: 10.1128/JVI.01038-13
38. Jang YH, Seong BL. Call for a paradigm shift in the design of universal influenza vaccines by harnessing multiple correlates of protection. Expert Opin Drug Discovery (2020) 15(12):1441–55. doi: 10.1080/17460441.2020.1801629
39. Isakova-Sivak I, Matyushenko V, Kotomina T, Kiseleva I, Krutikova E, Donina S, et al. Sequential immunization with universal live attenuated influenza vaccine candidates protects ferrets against a high-dose heterologous virus challenge. Vaccines (Basel) (2019) 7(3):61. doi: 10.3390/vaccines7030061
40. Rifai A, Fadden K, Morrison SL, Chintalacharuvu KR. The n-glycans determine the differential blood clearance and hepatic uptake of human immunoglobulin (Ig)a1 and Iga2 isotypes. J Exp Med (2000) 191(12):2171–82. doi: 10.1084/jem.191.12.2171
Keywords: influenza, swine, vaccine, LAIV, IgA, molecular marker
Citation: Rajao DS, Zanella GC, Wymore Brand M, Khan S, Miller ME, Ferreri LM, Caceres CJ, Cadernas-Garcia S, Souza CK, Anderson TK, Gauger PC, Vincent Baker AL and Perez DR (2023) Live attenuated influenza A virus vaccine expressing an IgA-inducing protein protects pigs against replication and transmission. Front. Virol. 3:1042724. doi: 10.3389/fviro.2023.1042724
Received: 12 September 2022; Accepted: 23 January 2023;
Published: 14 February 2023.
Edited by:
Rosa M. Del Angel, Centro de Investigaciones y Estudios Avanzados, Instituto Politécnico Nacional de México (CINVESTAV), MexicoReviewed by:
Adolfo García-Sastre, Icahn School of Medicine at Mount Sinai, United StatesFernando Roger Esquivel-Guadarrama, Autonomous University of the State of Morelos, Mexico
Copyright © 2023 Rajao, Zanella, Wymore Brand, Khan, Miller, Ferreri, Caceres, Cadernas-Garcia, Souza, Anderson, Gauger, Vincent Baker and Perez. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Daniela S. Rajao, ZGFuaWVsYS5yYWphb0B1Z2EuZWR1
 Daniela S. Rajao
Daniela S. Rajao Giovana C. Zanella
Giovana C. Zanella Meghan Wymore Brand2
Meghan Wymore Brand2 C. Joaquin Caceres
C. Joaquin Caceres Stivalis Cadernas-Garcia
Stivalis Cadernas-Garcia Carine K. Souza
Carine K. Souza Tavis K. Anderson
Tavis K. Anderson Phillip C. Gauger
Phillip C. Gauger Daniel R. Perez
Daniel R. Perez