- 1Friedrich-Loeffler-Institut, Federal Research Institute for Animal Health, Institute of Novel and Emerging Infectious diseases, Greifswald, Germany
- 2Malopolska Centre of Biotechnology, Laboratory of Virology, Jagiellonian University, Krakow, Poland
Rapid antigen tests (RATs) are used as convenient SARS-CoV-2 tests to minimize infection risks in the private and public domain (e.g., access to shops, concerts, sports, and other social events). RATs are: however, less sensitive than quantitative reverse transcription Polymerase chain reaction (RT-qPCR) assays; hence, samples with low viral loads may be misdiagnosed. Reports on the ability of RATs to detect SARS-CoV-2 variants of concern (VOCs) Delta and Omicron are often only qualitative. We, therefore, examined the analytical sensitivities of four different RATs for the detection of both full virus and recombinant proteins of relevant VOCs. Since most RATs are based on the detection of the SARS-CoV-2 nucleocapsid protein (N-protein), we constructed multiple N-protein mutants (mirroring specific amino acid exchanges of VOC N-proteins) using prokaryotic expression plasmids and site-directed PCR mutagenesis. Testing of recombinant proteins by four RATs revealed amino acid substitutions R203K and R203M, are critical for the sensitivity of some RATs. Interestingly, R203M mutation completely abrogated antigen detection even at high protein concentrations in the Delta variant. As a proof-of-concept study, we show that one or two specific amino acid changes in the N-protein can negatively impact the analytical sensitivity of RATs. Hence, antibodies used in such lateral flow assays should be optimized and target preferentially more conserved regions of N-protein.
Introduction
SARS-CoV-2 is an RNA virus constantly mutating and evolving, with new variants emerging over time. Only a few of those variants are of public health concern in humans because of their high transmission rates, severe pathogenicity, or ability to evade acquired immune responses (1). The five SARS-CoV-2 lineages B.1.1.7 (Alpha), B.1.351 (Beta), P.1 (Gamma), B.1.617.2 (Delta), and B.1.529 (Omicron) constitute variant groups of concern (VOCs) and hence have received vigilant monitoring for their potential impact on SARS-CoV-2 diagnostics, therapeutics, and vaccines (2). In late 2020, the Alpha was first detected in the UK, and was the most predominant cause of new cases worldwide in early 2021 (3). Also, Beta, initially found in South Africa in 2021, is 6-fold less susceptible to vaccine-derived neutralizing antibodies than the SARS-CoV-2 wild-type virus (4). Indeed, as SARS-CoV-2 variants become more contagious or influential on countermeasures, their potential to exacerbate the trajectory of the pandemic via triggering new waves increases. We have seen this with Delta, that has driven the deadly second wave of infections in summer 2021 in India (5), and also the more recent surge of Omicron. Currently, Omicron, with its sublineages, has been identified in more than 150 countries, outcompeting other variants and causing increasing numbers of infections, raising alarms for the need for immediate proactive measures (6). One of these measures is establishing novel platforms for validating the currently available diagnostic tools against the circulating variants.
Quantitative reverse transcriptase-polymerase chain reaction (RT-qPCR) is the gold standard for accurate and reliable COVID-19 diagnostics. However, the time needed for sample collection, transport to the laboratory, the assays’ performance, and patient notification are limitations of RT-qPCR. Besides, the need for specific chemicals, laboratory capacity, and trained staff to accurately execute RT-qPCR assays remains an additional immense challenge. Hence, the development of rapid antigen tests (RAT) for easy detection of SARS-CoV-2 by trained personnel and by ordinary persons was a big step forward to the expeditious testing of a broader scale of the human population. Unfortunately, compared to the RT-qPCRs, the RATs show a lower sensitivity (7), and therefore the confirmation of a negative result in a clinical sample may require confirmation by validated RT-qPCRs in peculiar cases (8).
Recently, several hundred RATs have become available in Germany, and more than 200 different RATs have been licensed for professional use, including 43 tests for self-application. A recent study using the Wuhan SARS-CoV-2 strain, evaluated these tests regarding their diagnostic specificity and sensitivity as listed by the Federal Institute for Drugs and Medical Devices (BfArM) (9, 10). Most of the evaluated RATs have values higher than 95% for both diagnostic criteria, and are therefore suitable for detecting high viral loads in the range of effective viral transmission. Anti-SARS-CoV-2 N-protein antibodies in RATs seem to be the most appropriate, as the nucleoproteins are abundant in infectious virus particles (bound to viral genomic RNA in the nucleocapsid complex) (11). Furthermore, most of the single nucleotide polymorphisms (SNPs) defining specific variants of SARS-CoV-2 are in the spike protein (S) of the viral particle (12, 13) and not in the nucleoprotein. Therefore, detecting a different protein with fewer SNPs should ensure, at least in theory, that RATs also recognize all variants.
In the first part of this study, we used a RAT from BioNote (Nowcheck) to analyze its sensitivity for the detection of non-VOC and VOCs. This particular RAT has been used in previous studies and has been found to detect the non-VOC Strain reliably. In a subsequent study, it was tested whether RATs from other companies yield the same results or whether they produce deviating results, especially for VOCs. For this purpose, RATs were selected from three vendors listed in the BfArM list with good results. Finally, the influence of individual amino acid exchanges typical of certain VOCs on the sensitivity of the RATs used was tested. Taking all parts together, we show that SNPs affect the functionality of RATS and are present not only in S but also in N genes of VOCs.
Materials and methods
Viruses and cells
Infections of Vero E6 (African green monkey (Chlorocebus spec.) kidney cells, Collection of Cell Lines in Veterinary Medicine CCLV, Friedrich-Loeffler-Institut, Greifswald-Insel Riems, Germany) were performed using the following viruses (Non-VOC hCoV-19/Germany/BY-ChVir-929/2020|EPI_ISL_406862|2020-01-28, Alpha hCoV-19/Germany/NW-RKI-I-0026/2020|EPI_ISL_751799|2020-12-07, Beta hCoV-19/Germany/NW-RKI-I-0029/2020|EPI_ISL_803957|2020-12-28, Delta hCoV-19/USA/PHC658/2021, and Omicron hCoV-19/Czech_Republic/KNL_2021-110119140/2021|EPI_ISL_6862005| 2021-11-26) as previously described (14). Cells were maintained in Eagle´s minimal essential medium (Lonza, Germany) with 8% foetal bovine serum (PAA) and antibiotics (100 U/ml penicillin and 100 µg/ml streptomycin) (Sigma, Germany) and incubated at 37°C under 5% CO2 in locked boxes as previously published (15). Virus stocks were preserved at -80°C, and titers were calculated as TCID50/mL in a biosafety-level 3 (BSL-3) laboratory at Friedrich-Loeffler-Institut, Germany, before using viruses for downstream analysis. Due to biosecurity reasons, all experiments using virus have been performed with heat-inactivated material. Heat inactivation of viruses was performed at 95°C for 20 minutes.
SARS-CoV-2 rapid antigen assays
For this study, four rapid antigen tests evaluated by the Paul-Ehrlich-Institut have been used; The BioNote NowCheck COVID-19 Ag Test (Sensitivity 89,2%; 95% CI: 81,7% – 93,9%, Specificity 97.6%; 95% CI: 95.1% - 98.8%), the Panbio ™ Covid-19 Ag Rapid Test Device (Nasal) from Abbott (Sensitivity 98.1%; 95% CI: 93.2% - 99.8%; Specificity 99.8%; 95% CI: 98.6% - 100%), the SARS-COV-2 ANTIGEN SCHNELLTEST from Xiamen Boson Biotech Co., Ltd (Sensitivity 96.17%; 95% CI: 94.04% - 98.29%, Specificity 99.16%; 95% CI: 98.49% - 99.83%) and theSARS-CoV-2-Antigen-Schnelltest-Kit from Triplex International Biosciences (China) Co., LTD (Sensitivity 96.17%; 95% CI: 94.04% - 98.29%, Specificity 99.16%; 95% CI: 98.49% - 99.83%). All these assays optimally test human naso/oropharyngeal swabs and are based on SARS-CoV-2 N-protein by using two anti-N-antibodies; a dye-labeled antibody and immobilized fixed antibody. We tested sensitivities of these RATs for the detection of heat-inactivated viruses by following the previously published protocol (16). For the investigations of recombinant proteins, we diluted the proteins in PBS to 8.333 ng/µl (corresponding 1000 ng/120µl). From this amount, we further diluted proteins to 100, 50, 25, 10 and 1 ng/120µl (8.33 pg/µl) in LFD extraction buffer and inoculated these amounts on RATs. Depending on the type of swab used, up to 1450 ng/ml of nucleocapsid were extracted in elution buffer, if 5 x 105 virus particles were loaded experimentally (17). Wolfel et al. showed that 6.76 × 105 genome copies per swab were present until day 5 after symptom onset, and it was concluded that at least 106 viruses per ml were needed for successful cultivation (18). Assuming that 106 viral particles are indeed collected with a swab when sampling a patient with symptoms, this means an estimated amount of 1.5 ng nucleocapsid in approx. 300 µl elution buffer for one RAT, correlating to a concentration of 5 pg nucleocapsid/µl buffer. All concentrations used were above this value. All experiments were carried out in duplicates at least.
Detection of SARS-CoV-2 RNA by real-time RT-PCR
Viruses from diluted cell culture supernatants were subjected to RNA extraction using QIAamp Viral RNA Mini Kit (Qiagen, Hilden, Germany) following their standard protocol. Viral RNA was analyzed by CFX real-time PCR systems (Bio-Rad, Germany) using real-time RT-PCR kits (SuperScript III Reverse Transcriptase, Invitrogen, Germany) and the envelope E gene primers and probes set (19). Dilutions of RNA isolates from standardized samples (provided by INSTAND e.V., Germany) were used to generate standard curves.
Scoring and densitometric analysis of RATs
Immediately after 15 min incubation, RATs were visually assessed (at least four-eyes-principle) as follows: - no test line visible; +: test line weakly visible; ++: test line less intensive than control line; +++: test line as intensive as control line or more intensive. For densitometric analysis, RATs were digitalized using either a Canon flatbed scanner or the BioRad Chemidoc Imaging system. Digitalized images were composed with Adobe Photoshop CS5 software (version 12.0 x64) and densitometrically analyzed using ImageLab 6.0.1 software. Adjusted volumes of control lines and test lines (internal units) were used to normalize the ratio of the test line to the control line.
Cloning of N gene
Sequences of the N-protein open reading frame (ORF) were cloned into the pET19b prokaryotic expression plasmid (Novagen; Cat. No.69677-3) by the use of specific primers (Table 1). The forward primer contains a NdeI recognition site, and the reverse primer harbors a BamHI recognition site for the directed insertion into the expression plasmid. Amplified PCR products were first cloned into the pCR2.1 vector by TA cloning, and an internal NdeI site was silently mutated by site-directed PCR mutagenesis where necessary. Using the NdeI and BamHI (New England Biolabs, MA; USA), the N protein-coding region was inserted with an 8x histidine peptide (His-Tag) into the vector pET19b. Sequence identity and correct in-frame insertion were verified by Sanger sequencing (Eurofins Genomics, Germany) with standard sequencing primer T7 and T7term. Geneious Prime® 2021.0.1 was used to analyze the constructs and sequences.
PCR mutagenesis of N-protein expression plasmids
Complementary mutagenesis primers (Table 2) were designed and used to induce in-frame single nucleotide exchanges in VOCs N genes. These nucleotide exchanges result in substituting one specific amino acid or two neighboring amino acids (Supplementary Figure 3). We used the described primers in Table 2 to amplify the pET19b-N-VOC template by 20 PCR rounds Subsequently, plasmids were digested with DpnI to remove methylated target DNA. Plasmids were further transformed into E. coli strain XL1 and plated onto LA-Amp agar plates. One single colony from a plate was amplified in LB-Broth on a Mini-prep scale. The identity of isolated plasmids and nucleotide exchanges were further proved by Sanger sequencing using the standard T7 and T7term primers.
Production of recombinant SARS-CoV-2 N-proteins
E. coli bacteria of the BL21(DE3) strain were transformed with the non-VOC-N-protein and N-protein-mut expression plasmids. Protein expression was induced by adding IPTG to the LB-Broth growing medium at an OD600nm of 0.6. After 4 hours of expression, bacteria were sedimented, and His-tagged protein was isolated using Ni-NTA (nickel-nitrilotriacetic acid) technology (Thermo ScientificTM, Germany) according to the manufacturer’s instructions and under denaturing conditions described elsewhere. Expression of specific proteins has been confirmed by Coomassie SDS-PAGE and Western blot using His-Tag specific antibody (Supplementary Figure 1). The concentration of the proteins was measured in a microtiter plate using Nanoquant (Carl Roth, Germany), a modified Bradford method (20).
Results
SARS-CoV-2 variants are detected with reduced sensitivity by rapid antigen assays
To test the analytical sensitivity of RATs against SARS-CoV-2 VOCs, equal amounts of heat-inactivated viruses (non-VOC, Alpha, Beta, Delta, and Omicron) were loaded onto the BioNote/NowCheck RATs. These amounts correlated to Ct values (of 24, 27, 30, and 33) known to be in or close to the detectable range of RATs limit of detection (LoD) (16). To confirm the similar amounts of viral antigen in the dilution, Western blot analysis using 20 µl of the Ct 24 dilution has been performed (Supplementary Figure 4). Fifteen minutes post-application, the images of RATs were recorded, and test line intensities were quantified by densitometric analysis. The highest concentrations, corresponding, to Ct of 24 for all strains (non-VOCs and VOCs) were detected by the BioNote RAT (Figures 1A–E). However, cell culture supernatants correlating to Ct values of 27 and 30, showed only positive results in the case of non-VOC (Figure 1A). Visual examination of RATs with Alpha and Beta revealed a negative result, indicating a lower sensitivity of this RAT toward these variants (2 logs, Figures 1B–F).
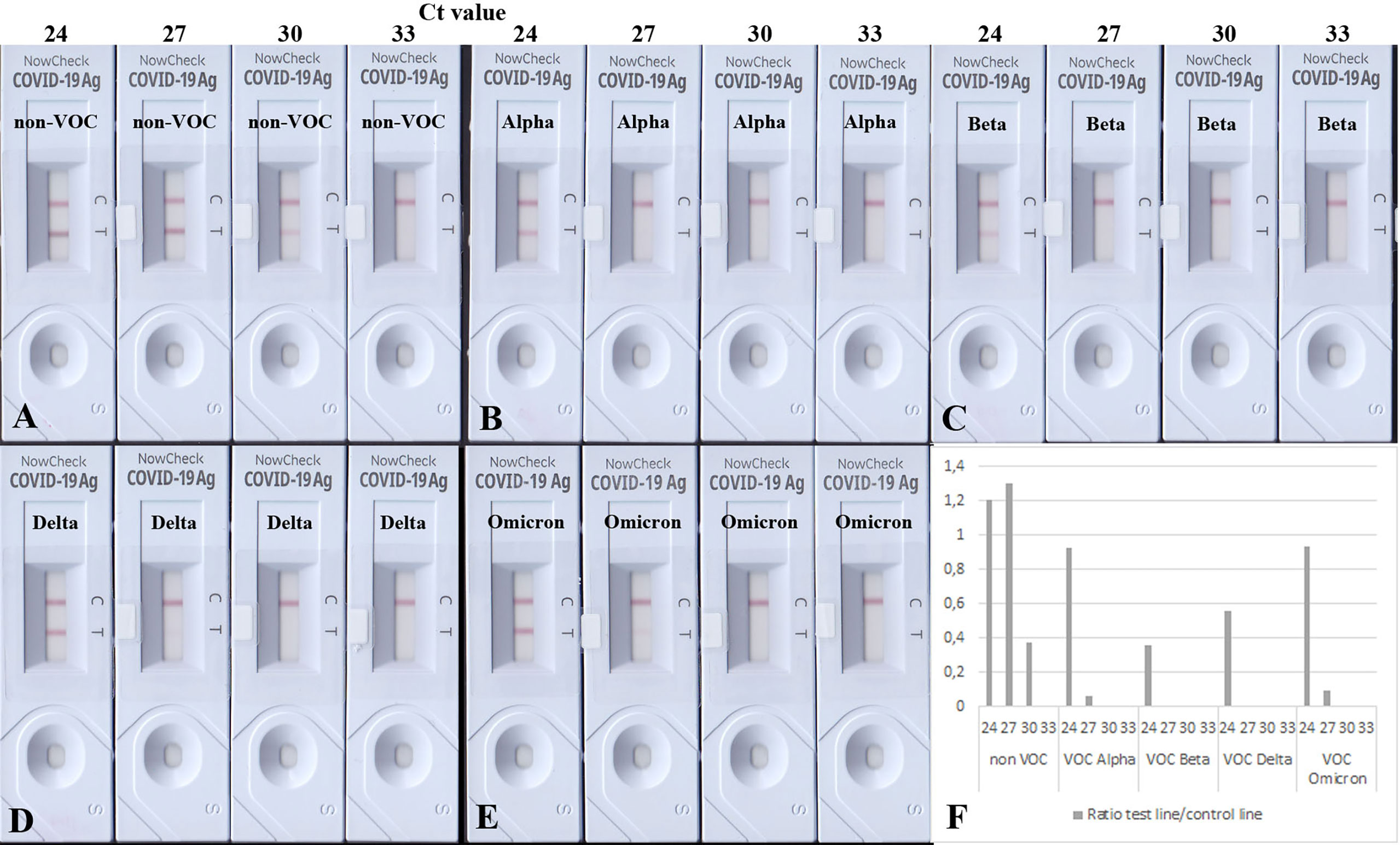
Figure 1 BioNote RATs loaded with samples of heat-inactivated cell culture supernatants. (A) non-VOC, (B) Alpha, (C) Beta, (D) Delta, (E) Omicron. (F) Densitometric analysis of VOCs shown on (A–E). Supernatants were adjusted to comparable viral loads represented by Ct values. All viruses are detected in samples with the highest viral loads but VOCs with less sensitivity (1-2 logs). Densitometric analysis shows comparable detection of VOCs Alpha and Omicron. At Ct = 30, only the non-VOC strain is detected.
Interestingly, Delta and Omicron showed a faint test line in the sample corresponding to Ct 27 (Figures 1D, E), albeit it was also detected with a lower sensitivity by this RAT as shown in Figures 1D–F (1 log lower than the non-VOC strain). Dilutions corresponding to Ct value 33 were not detectable for any viral strains, as they are likely to be beyond the LoD of this particular RAT (16). Together, these data confirm the lowered sensitivity of this RAT to detect Alpha, Beta, Delta, and Omicron (Table 3).

Table 3 Detection of SARS-CoV-2 variants by BioNote RAT: VOCs are detected with less sensitivity compared to non-VOC.
Amino acids sequence analysis of N-proteins from different SARS-CoV-2 variants revealed N-gene mutations
In order to focus the investigations on the target proteins and to avoid side effects, the further investigations on the reasons for the loss of analytical sensitivity of the RATs against VOCs were carried out with recombinant N-protein. Therefore, we amplified N-protein sequences of the non-VOC and VOC viruses and cloned them into prokaryotic expression vectors. Further, we sequenced the constructs to ensure the correct insertion of the coding region into the vector backbone (Supplementary Figure 1). Interestingly, our alignment analysis revealed that the N proteins derived from the non-VOC strain and Alpha and Beta have 14 differences on nucleic acid level, resulting in seven amino acid changes, respectively (Supplementary Figure 2). Moreover, Delta has differences at seven amino acid positions compared to the non-VOC sequences, 13 to Alpha and Beta, and 12 to Omicron (Supplementary Figure 2). VOC Omicron has a deletion of three amino acids and changes at five positions, including R203K, compared to the non-VOC strain. All other nucleotide changes were silent. We observed that the exchange of amino acids 203 and 204 alone or in combination (grey in Table 4) occurs in different VOCs. Primarily at position 203, three different amino acids (R, K, M) are found, making this region interesting for mutagenesis and possible alteration of efficient antibody binding. The amino acid exchanges, including the most VOC defining SNPs (underlined), are listed in Table 4. It should be noted that the detected amino acid exchanges are not necessarily characteristic for the particular VOC in the public SARS-CoV-2 genomic repositories. However, they were found in the isolated viruses, most likely in the context of further viral evolution in-vivo. Mutations due to serial virus passages in cell culture are also possible. To minimize this risk, we used only the third passages in the experiments.
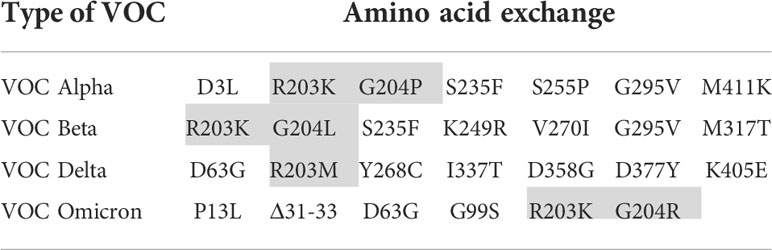
Table 4 Amino acid exchanges in recombinantly expressed N-proteins in comparison to the non-VOC strain (VOC defining SNPs are underlined and investigated amino acid substitutions are shaded).
Beta and Delta are the most poorly recognized VOCs by some RATs
Next, we aimed to test the performance of the RATs by using 100 ng purified recombinant N-protein variants per RAT. All analyzed RATs (BioNote, Abbott, Boson, TIB) detected the N-protein of the non-VOC SARS-CoV-2 at this amount (100 ng). Nonetheless, the Beta and Delta recombinant N-proteins were only detected by Boson and TIB RATs and faintly detected by BioNote and Abbott (Figures 2, 3 and Table 5). Densitometric analysis of the BioNote RAT loaded with VOCs showed that the intensity of the test line produced by the N protein of Beta is less than 20% compared to the control line (Figure 2B). The N protein of Delta produced a test line with higher intensity, which was also under 40% of the intensity of the control line (Figure 2C). In contrast, Omicron was easily detected, and the test line was even more intense than the control line. Except for Alpha, these findings are consistent with the results of applying cell culture supernatants to the BioNote RAT (Figure 1). Unsurprisingly, though, although VOCs are primarily defined by differences of the amino acid sequence in the spike (S) protein, changes in other viral proteins also contribute to the definition of the PANGOLIN lineages. Therefore, it is very likely, that amino acid exchanges in N-protein might impair the binding of a monoclonal antibody to its epitope. For this, we tested the function of the RAT by adding purified recombinant N-protein to the extraction buffer at defined amounts.

Figure 2 (A) RATs (BioNote, Abbott, Boson, and TIB) loaded with 100 ng recombinant N-protein of the non-VOC strain. All used RATs detected this amount of protein. (B) The exact amount of protein produced a comparable test line in the case of Alpha and Omicron, but the test lines using protein from Beta and Delta appeared much weaker. (C) Densitometric analysis of RATs shown in (B). The N protein from all variants was detected, but for Beta and Delta, the sensitivity seems much lower.
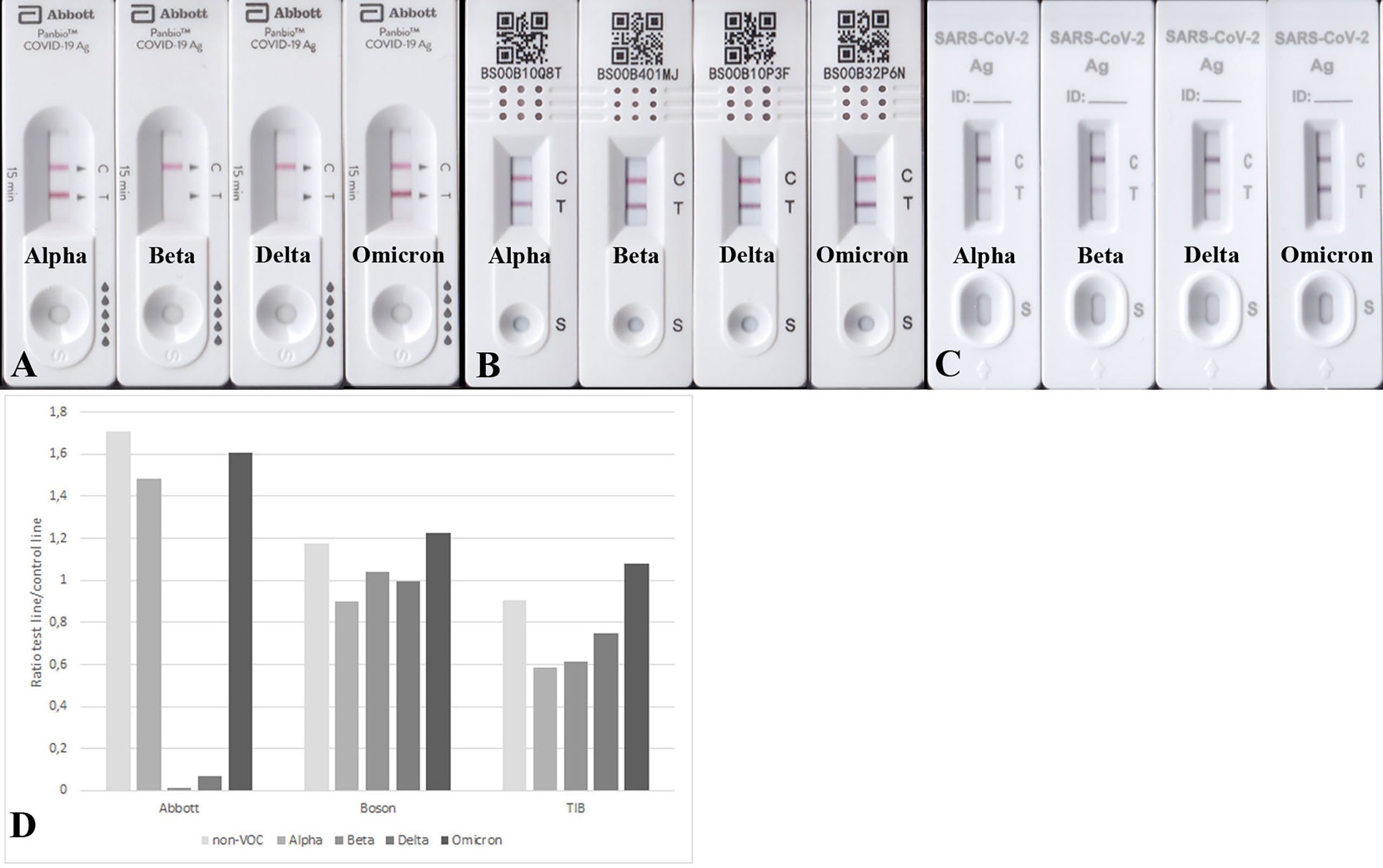
Figure 3 RATs detecting 100 ng N protein of Alpha, Beta, Delta, and Omicron. RATs were from different companies: Abbott (A), Boson (B), and TIB (C). (D) Densitometric analysis of RATs shown in (A-C). Corresponding RATs loaded with non-VOC strain are shown in Figure 2A.
The reduced detection of VOC Beta and Delta is due to the amino acid exchange at position 203 of the N-protein
We tried to understand the reason for such lowered sensitivities for VOCs in some RATs. Therefore, we decided to introduce specific mutations in the N-protein using PCR-based mutagenesis (Table 6). This allowed us to analyze whether the band intensity reduction results from the described amino acid exchange/s. Interestingly, we found that applying the mutated N proteins R203K and G204L onto BioNote lateral flows did not decrease the detection capability of this particular RAT. In contrast, an R203M mutation (a defining SNP of VOC Delta) completely impaired the detection (Figure 4A). This evidence is a clear proof of concept that even one amino acid exchange can affect protein epitopes or stability, allowing abrogated detection by RATs. Interestingly, the R203K mutation on VOC Omicron does not alter the detection of this variant by RATs.
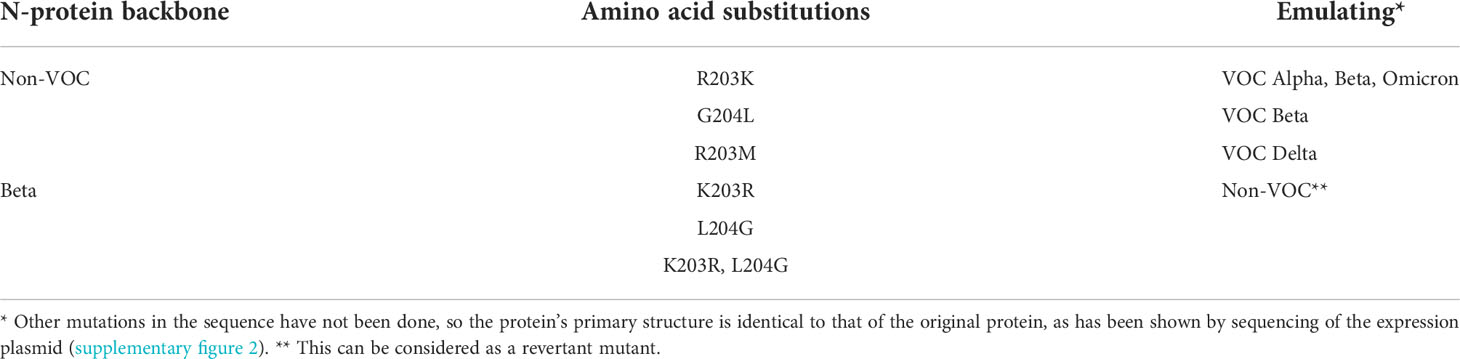
Table 6 Overview of the induced amino acid exchanges (PCR-based mutagenesis) in SARS-CoV-2 recombinant N-protein.
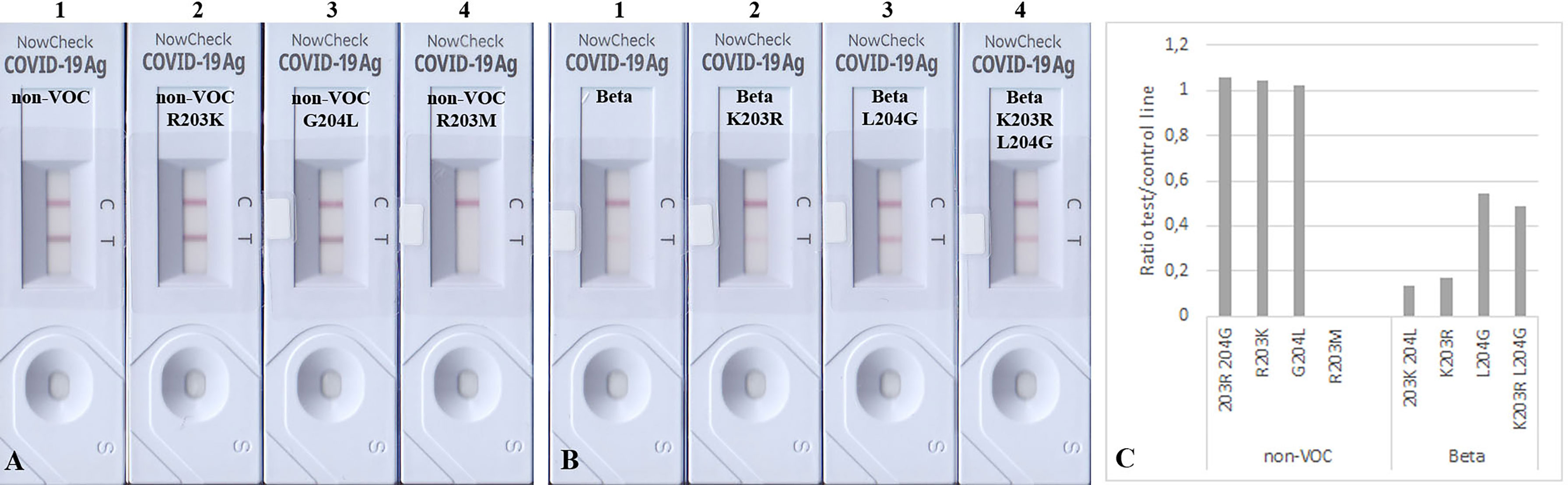
Figure 4 Detection of 100 ng mutated N proteins by RATs from BioNote. Numbers label specific RATs in (A) and (B) for easier orientation. Changes at positions 203 (A2) and 204 (A3) of the non-VOC N protein to the amino acids found in VOC Beta at these positions do not alter the detection of the N protein by the BioNote RAT as seen by densitometric analysis (C). In contrast, R203M mutation (A4) impairs the detection ultimately. Whereas the K203R mutation (B2) in the Beta N protein does not alter the test line intensity, the L204G (B3, alone and B4 in combination with K203R) mutation enhanced the detection of Beta compared to Beta (B1).
Since we observed a significant reduction in BioNote RAT analytical sensitivity for detecting Beta, we also introduced single aa changes reflecting the Beta in an attempt to recover the band intensity of this protein variant, as these amino acids exist in the non-VOC protein. An exchange of amino acid 203 alone in the VOC Beta from Lysin to Arginin did not alter the detection, but a Leucine to Glycine exchange at position 204 enhanced the detection (Figures 4B, C). The latter was also true when both amino acids (203, 204) were exchanged (Figure 4B).
The mutated N proteins were also used in other RATs. Boson and TIB RATs that detected all non-VOC and VOCs (Figure 2) were not affected by the introduced mutations (Figures 3B–D). However, RATs produced by Abbott did not detect the N protein mutants on the Beta background, or the R203M mutation (Figure 5). We could not explain why the K203R/L204G mutations retrieved the band intensity in one RAT (BioNote) and not in the other (Abbott), but both tests are presumable to use monoclonal antibodies whose target site is not bona fide identical. Hence, we concluded that the mutation at position 203 (R203K or R203M) on non-VOC background seems to be responsible for a decreased intensity of the test line.
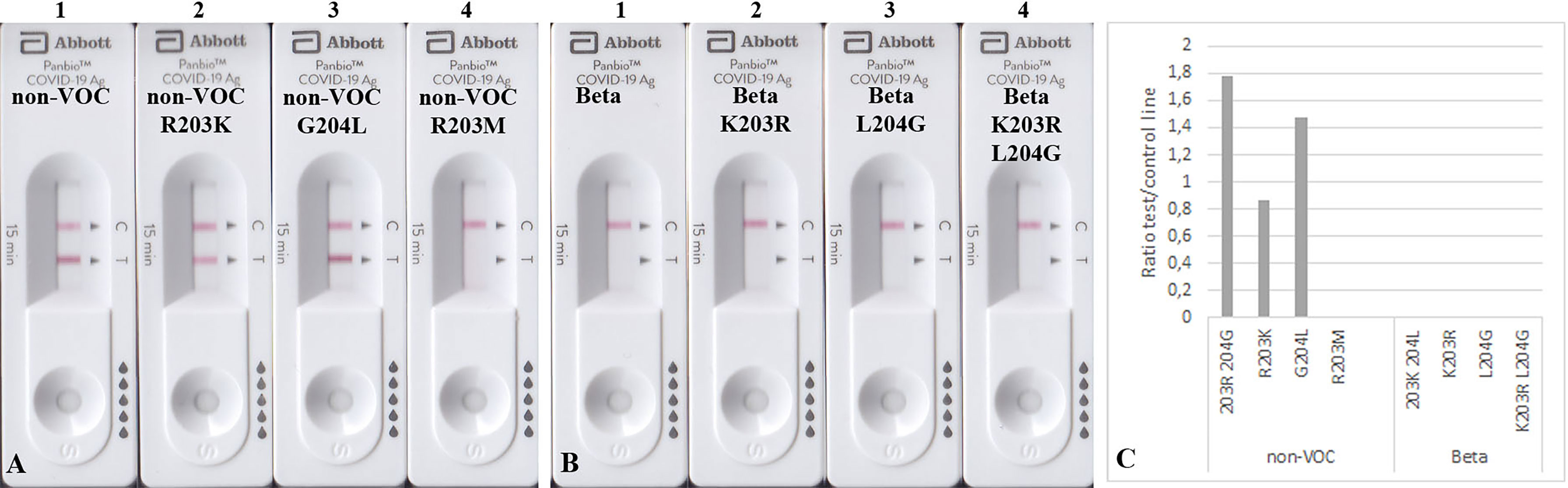
Figure 5 Detection of 100 ng mutated N proteins by RATs from Abbott. Numbers label specific RATs in (A) and (B) for easier orientation Amino acid change at position 203 (A2) decreases the test line intensity, whereas the change at position 204 (A3) of the non-VOC N protein does not have an effect. R203M mutation (A4) also impairs the detection by these RATs. (B) All N proteins on the Beta background were not detected. (C) Densitometric analysis of RATs shown in (A, B).
Limits of detection (LOD) for VOC delta N protein and non-VOC R203M
As the R203M mutation was one of the decisive SNPs for Delta, we further aimed to determine the limits of detection of different RATs for N proteins of the non-VOC strain, Delta, and the non-VOC protein mutant with R203M exchange. Therefore, 50, 25, 10, and 1 ng of recombinant proteins were applied to the RATs from BioNote, Boson, and TIB. RATs from BioNote and Boson detected 1 ng of the non-VOC N protein but the same amount is not detected by TIB RAT. BioNote RAT detected the Delta protein until 50 ng, whereas the R203M protein variant was not detected at all by this RAT (Table 7 and Figure 6). This can be explained by the fact that Delta harbors additional mutations in the N-protein and not only the R203M that exist in the mutated protein (Table 7 and Supplementary Figure 2). These mutations might retain protein stability and/or epitopes. We found that TIB RATs detected 10 ng of Delta and non-VOC R203M but not the lowest amount of 1 ng, raising questions about the LoD of this particular RAT. Strikingly, Boson RATs showed the best performance in this study since all proteins (VOC and non-VOC) were recognized even at 1 ng, and the test to control lines had the best ratios in all experiments (Figure 6).

Table 7 Summary of RAT sensitivity for N-protein from non-VOC, Delta, and non-VOC carrying the R203M amino acid exchange.
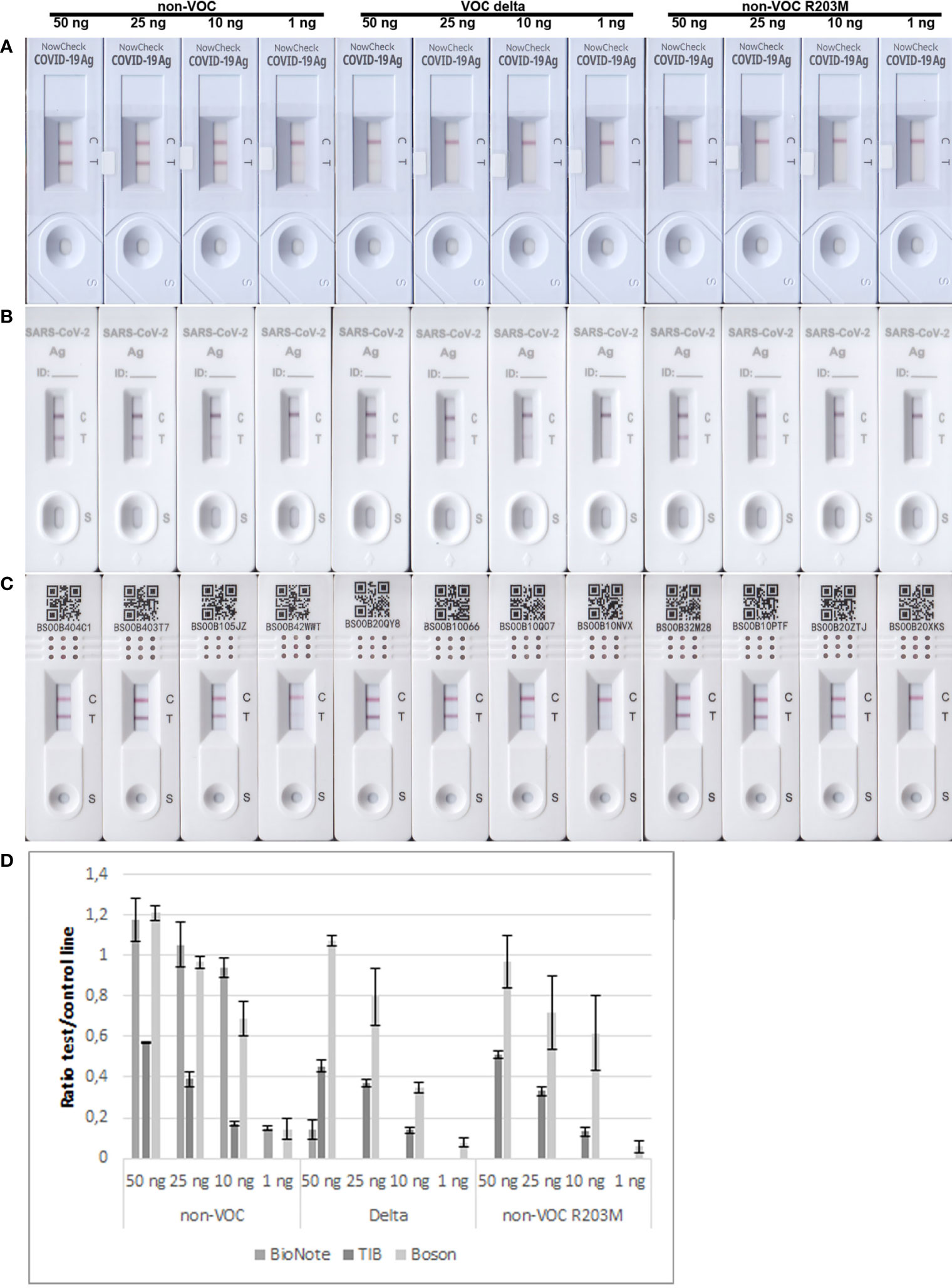
Figure 6 Limits of detection of BioNote, TIB, and Boson RATs for detection of non-VOC, Delta, and non-VOC R203M recombinant proteins. (A) BioNote, (B) TIB, (C) Boson, using amounts of 50 ng, 25 ng, 10 ng, 1 ng respectively. (D) Means of densitometric analysis of RATs is shown in (A–C) (n=2).
Discussion
Negative RAT results can be used to minimize SARS-CoV-2 exposure risks, especially when combined with the user’s immune status (vaccinated, recovered, versus non-immune) (21). Therefore, RATs must work for most, if not all, circulating VOCs at an analytical sensitivity equaling one infectious dose for humans (conservatively estimated: ~106 to ~107 copies/mL) (16, 18, 22).
Previously, we have shown that the limit of detection for the non-VOC strain was up to the level of Ct value 32,25 and gene copy number of 103-104 copies/mL using randomly selected RATs (16). In the present study, we in assessed the ability of RATs to detect formerly and currently circulating VOCs by using heat-inactivated cell culture supernatant and recombinant proteins. Differences between the data of this study and previous studies regarding the sensitivity of RATs against the different VOCs presented are most probably due to different study designs. Standardized procedures, such as those used in large-scale series before (23, 24), would make the results of different studies more comparable. However, this was not the primary goal of the study, but rather to show how much the analytical sensitivity of the RATs can vary depending on amino acid variations in the target regions of the VOCs.
Alpha and Beta VOCs were not well-detected by one particular RAT (Abbott) as already published (16). Here, we also included the most recently circulating VOCs, Delta, and Omicron. Initially, Ct values of RNA isolations from native (Ct 14.35) and heat-inactivated (Ct 20.4) cell culture supernatants have been compared to quantify the loss of sensitivity due to inactivation. Data illustrate that heat-inactivation of SARS-CoV-2 results in a loss of sensitivity of up to 2 logs in RT-qPCR, which is in good accordance with published data (25). To increase the comparability of the assays, recombinant proteins were used in the present study in addition to heat-inactivated cell culture supernatants. This excludes the possibility that the lower sensitivity of the RATs is solely due to the denaturation of the antigen.
This reduced sensitivity to VOCs is due, at least in part, to a mutation at position 203 (R203K, R203M) in the N protein of SARS-CoV-2 variants. Since many RATs are based on the less-variable N-protein, they are thought to be able to detect all the variants. However, this study clearly shows that the performance of RATs depends heavily on the anti-N-antibodies used for detection. RATs rely on two anti-N-antibodies; one in the sample pad and another immobilized capture antibody at the test line (16). Both antibodies are not necessarily the same, but they may be. If one of the antibodies cannot bind N-protein (e.g., due to an amino acid exchange), the test will not or only partially detect the SARS-CoV-2 antigens in the sample.
In both VOCs Beta and Delta, amino acid 203 (Arginine) of the N-protein has been changed to Lysine in Beta and Methionine in Delta, respectively. Using PCR-mediated mutagenesis, it was possible to mimic this amino acid exchange in the non-VOC N-gene in vitro, and use it to evaluate different RATs. While the R203K mutation only reduced the intensity at the test line, an R203M mutation (as in the Delta) completely impaired the antigen detection. Therefore, antibodies binding to a more conserved part of the N-protein or polyclonal antibodies should be used preferentially.
Interestingly, the R203K mutation seems not to influence the RAT sensitivity in Omicron. Reasons for this could be the replacement of other single amino acids in other domains, especially the three amino acid deletions in the N-terminal domain of the protein. The possible interaction of different secondary structures due to the changes in the primary structure may expose or mask epitopes in the tertiary structure of the proteins, which may result in different binding properties of the antibodies. This has to be further evaluated in future studies using correspondingly mutated N-protein.
Several studies have been conducted using RATs for special cohort screening (26–29), and several assays have been recommended to be useful for the early detection of infection with SARS-CoV-2. This study is not intended to warn users about specific products. The selected sample size of RATs from only 4 manufacturers is too small and not representative for this purpose, but the aim of this article is to raise awareness in terms of a VOC specific evaluation of RATs.
Data availability statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Author contributions
ITH, MG, MK conceived and designed the experiments. ITH, MK performed experiments and analyzed data. ITH, SW, KP, AB-B, MK contributed reagents/materials/analysis tools. MHG attained funding. ITH, MHG, MK wrote the paper. All authors reviewed, edited, and approved the final version.
Funding
This work was made possible by FLI intramural funding, and was partially supported by funding from the COVRIN project of the European Union’s Horizon 2020 Research and Innovation programme under grant agreement No 773830: One Health European Joint Programme.
Acknowledgments
Non-VOC SARS-CoV-2 isolate hCoV-19/Germany/BY-ChVir-929/2020|EPI_ISL_406862| 2020-01-28 was kindly provided by Bundeswehr Institute of Microbiology, Munich, Germany. SARS-CoV-2 VOC Omicron virus (hCoV-19/Czech_Republic/KNL_2021-110119140/2021| EPI_ISL_6862005|2021-11-26) has been kindly provided by Prof. Dr hab. KP, Laboratory of Virology, Malopolska Centre of Biotechnology, Jagiellonian University Krakow. We thank Katrin Schwabe for her excellent technical assistance.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fviro.2022.971862/full#supplementary-material
References
1. Walensky RP, Walke HT, Fauci AS. Sars-Cov-2 variants of concern in the united states–challenges and opportunities. Jama (2021) 325(11):1037–8. doi: 10.1001/jama.2021.2294
2. World Health Organization. Tracking sars-Cov-2 variants (2021). Available at: https://www.who.int/en/activities/tracking-SARS-CoV-2-variants/.
3. Kim Y, Kim E-J, Lee S-W, Kwon D. Review of the early reports of the epidemiological characteristics of the b. 1.1. 7 variant of sars-Cov-2 and its spread worldwide. Osong Public Health and Research Perspectives (2021) 12(9):139–48. doi: 10.24171/j.phrp.2021.0037
4. Wibmer CK, Ayres F, Hermanus T, Madzivhandila M, Kgagudi P, Oosthuysen B, et al. Sars-Cov-2 501y. V2 escapes neutralization by south African covid-19 donor plasma. Nat Med (2021) 27(4):622–5. doi: 10.1038/s41591-021-01285-x
5. Roy B, Dhillon J, Habib N, Pugazhandhi B. Global variants of covid-19: Current understanding. J Biomed Sci (2021) 8(1):8–11. doi: 10.3126/jbs.v8i1.38453
6. World Health Organization. Weekly epidemiological update on covid-19, edition 80. New York : World Health Organization (2022).
7. Treggiari D, Piubelli C, Caldrer S, Mistretta M, Ragusa A, Orza P, et al. Sars-Cov-2 rapid antigen test in comparison to rt-pcr targeting different genes: A real-life evaluation among unselected patients in a regional hospital of Italy. J Med Virol (2021) 94(3):1190–5. doi: 10.1002/jmv.27378
8. Stromer A, Rose R, Schafer M, Schon F, Vollersen A, Lorentz T, et al. Performance of a point-of-Care test for the rapid detection of sars-Cov-2 antigen. Microorganisms (2020) 9(1):58–68. doi: 10.3390/microorganisms9010058
9. Federal Institute for Drugs and Medical Devices. Antigen-tests zum direkten erregernachweis des coronavirus sars-Cov-2 (2020). Available at: https://www.bfarm.de/DE/Medizinprodukte/Aufgaben/Spezialthemen/Antigentests/_node.html;jsessionid=4A724075BCB2578FD0EC0D6E020DC338.intranet241.
10. EUROPEAN COMMISSION DIRECTORATE-GENERAL FOR HEALTH AND FOOD SAFETY. Eu health preparedness: A common list of covid-19 rapid antigen tests and a common standardised set of data to be included in covid-19 test result certificates. European Commission (2021). Available at: https://ec.europa.eu/health/sites/default/files/preparedness_response/docs/covid-19_rat_common-list_en.pdf.
11. Wang S, Trilling M, Sutter K, Dittmer U, Lu M, Zheng X, et al. A crowned killer’s résumé: Genome, structure, receptors, and origin of sars-Cov-2. Virol Sin (2020) 35(6):673–84. doi: 10.1007/s12250-020-00298-z
12. Harvey WT, Carabelli AM, Jackson B, Gupta RK, Thomson EC, Harrison EM, et al. Sars-Cov-2 variants, spike mutations and immune escape. Nat Rev Microbiol (2021) 19(7):409–24. doi: 10.1038/s41579-021-00573-0
13. Centers for Disease Control and Prevention. Sars-Cov-2 variant classifications and definitions (2021). Available at: https://www.cdc.gov/coronavirus/2019-ncov/variants/variant-info.html.
14. van den Worm SH, Eriksson KK, Zevenhoven JC, Weber F, Zust R, Kuri T, et al. Reverse genetics of sars-related coronavirus using vaccinia virus-based recombination. PloS One (2012) 7(3):e32857. doi: 10.1371/journal.pone.0032857
15. de Wilde AH, Raj VS, Oudshoorn D, Bestebroer TM, van Nieuwkoop S, Limpens R, et al. Mers-coronavirus replication induces severe in vitro cytopathology and is strongly inhibited by cyclosporin a or interferon-alpha treatment. J Gen Virol (2013) 94(Pt 8):1749–60. doi: 10.1099/vir.0.052910-0
16. Hagag IT, Weber S, Sadeghi B, Groschup MH, Keller M. Impact of animal saliva on the performance of rapid antigen tests for detection of sars-Cov-2 (Wildtype and variants B.1.1.7 and B.1.351). Vet Microbiol (2021) 262:109243. doi: 10.1016/j.vetmic.2021.109243
17. Grant BD, Anderson CE, Alonzo LF, Garing SH, Williford JR, Baughman TA, et al. A sars-Cov-2 coronavirus nucleocapsid protein antigen-detecting lateral flow assay. PloS One (2021) 16(11):e0258819. doi: 10.1371/journal.pone.0258819
18. Wolfel R, Corman VM, Guggemos W, Seilmaier M, Zange S, Muller MA, et al. Virological assessment of hospitalized patients with covid-2019. Nature (2020) 581(7809):465–9. doi: 10.1038/s41586-020-2196-x
19. Corman VM, Landt O, Kaiser M, Molenkamp R, Meijer A, Chu DKW, et al. Detection of 2019 novel coronavirus (2019-ncov) by real-time rt-pcr. Euro Surveill Bull Euro Sur Maladies Transmissibles = Eur Commun Dis Bull (2020) 25(3):23–30. doi: 10.2807/1560-7917.ES.2020.25.3.2000045
20. Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem (1976) 72:248–54. doi: 10.1006/abio.1976.9999
21. Deutscher Bundestag - Wissenschaftliche qDienste. Verfassungsrechtliche bewertung der beschränkungen ungeimpfter durch die sogenannten 3g- und 2g-regeln (2021). Available at: https://www.bundestag.de/resource/blob/863844/70947388a152d82f48ed6013fe3f1d7a/WD-3-149-21-pdf-data.pdf.
22. Vierbaum L, Wojtalewicz N, Grunert HP, Lindig V, Duehring U, Drosten C, et al. Rna reference materials with defined viral rna loads of sars-Cov-2-a useful tool towards a better pcr assay harmonization. PloS One (2022) 17(1):e0262656. doi: 10.1371/journal.pone.0262656
23. Paul Ehrlich Institut. Mindestkriterien für sars-Cov-2 antigentests im sinne Von § 1 abs. 1 satz 1 testvo: Antigenschnelltests (2020). Available at: https://www.pei.de/SharedDocs/Downloads/DE/newsroom/dossiers/mindestkriterien-sars-cov-2-antigentests-01-12-2020.pdf?:blob=publicationFile&v=6
24. Paul Ehrlich Institut. Statement - many rapid tests detect omicron (2022). Available at: https://www.pei.de/SharedDocs/Downloads/EN/newsroom-en/positions/antigen-test-omicron.pdf?:blob=publicationFile&v=5.
25. Pastorino B, Touret F, Gilles M, de Lamballerie X, Charrel RN. Heat inactivation of different types of sars-Cov-2 samples: What protocols for biosafety, molecular detection and serological diagnostics? Viruses (2020) 12(7):735–42. doi: 10.3390/v12070735
26. Winkel B, Schram E, Gremmels H, Debast S, Schuurman R, Wensing A, et al. Screening for sars-Cov-2 infection in asymptomatic individuals using the panbio covid-19 antigen rapid test (Abbott) compared with rt-pcr: A prospective cohort study. BMJ Open (2021) 11(10):e048206. doi: 10.1136/bmjopen-2020-048206
27. Garcia-Finana M, Hughes DM, Cheyne CP, Burnside G, Stockbridge M, Fowler TA, et al. Performance of the innova sars-Cov-2 antigen rapid lateral flow test in the Liverpool asymptomatic testing pilot: Population based cohort study. BMJ (2021) 374:n1637. doi: 10.1136/bmj.n1637
28. Landaas ET, Storm ML, Tollanes MC, Barlinn R, Kran AB, Bragstad K, et al. Diagnostic performance of a sars-Cov-2 rapid antigen test in a Large, Norwegian cohort. J Clin Virol (2021) 137:104789. doi: 10.1016/j.jcv.2021.104789
Keywords: SARS-CoV-2, rapid antigen test, nucleocapsid mutation, variants of concern, sensitivity loss
Citation: Hagag IT, Pyrc K, Weber S, Balkema-Buschmann A, Groschup MH and Keller M (2022) Mutations in SARS-CoV-2 nucleocapsid in variants of concern impair the sensitivity of SARS-CoV-2 detection by rapid antigen tests. Front. Virol. 2:971862. doi: 10.3389/fviro.2022.971862
Received: 17 June 2022; Accepted: 11 August 2022;
Published: 09 September 2022.
Edited by:
Yoon-Seok Chung, Korea Disease Control and Prevention Agency, South KoreaReviewed by:
Jason Kindrachuk, University of Manitoba, CanadaAnan Jongkaewwattana, National Center for Genetic Engineering and Biotechnology (BIOTEC), Thailand
Copyright © 2022 Hagag, Pyrc, Weber, Balkema-Buschmann, Groschup and Keller. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Martin H. Groschup, bWFydGluLmdyb3NjaHVwQGZsaS5kZQ==
 Ibrahim T. Hagag1
Ibrahim T. Hagag1 Krzysztof Pyrc
Krzysztof Pyrc Martin H. Groschup
Martin H. Groschup Markus Keller
Markus Keller