
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Virol. , 01 November 2022
Sec. Fundamental Virology
Volume 2 - 2022 | https://doi.org/10.3389/fviro.2022.971293
This article is part of the Research Topic HIV/SIV basic research update View all 10 articles
 John J. McAllister1†‡
John J. McAllister1†‡ Satinder Dahiya2,3‡
Satinder Dahiya2,3‡ Rachel Berman2,3
Rachel Berman2,3 Mackenzie Collins2,3
Mackenzie Collins2,3 Michael R. Nonnemacher2,3,4
Michael R. Nonnemacher2,3,4 Tricia H. Burdo5
Tricia H. Burdo5 Brian Wigdahl2,3,4*
Brian Wigdahl2,3,4*Human immunodeficiency virus type 1 (HIV-1) transcription in cells of the monocyte-macrophage lineage is regulated by interactions between the HIV-1 long terminal repeat (LTR) and a variety of host cell and viral proteins. Binding of the Sp family of transcription factors (TFs) to the G/C box array of the LTR governs both basal as well as activated LTR-directed transcriptional activity. The effect of monocytic differentiation on Sp factor binding and transactivation was examined with respect to the HIV-1 LTR. The binding of Sp1, full-length Sp3 and truncated Sp3 to a high affinity HIV-1 Sp element was specifically investigated and results showed that Sp1 binding increased relative to the binding of the sum of full-length and truncated Sp3 binding following chemically-induced monocytic differentiation in monoblastic (U-937, THP-1) and myelomonocytic (HL-60) cells. In addition, Sp binding ratios from PMA-induced cell lines were shown to more closely approximate those derived from primary monocyte-derived macrophages (MDMs) than did ratios derived from uninduced cell lines. The altered Sp binding phenotype associated with changes in the transcriptional activation mediated by the HIV-1 G/C box array. Additionally, analysis of post-translational modifications on Sp1 and Sp3 revealed a loss of phosphorylation on serine and threonine residues with chemically-induced differentiation indicating that the activity of Sp factors is additionally regulated at the level of post-translational modifications (PTMs).
HIV-1 infection within monocyte-macrophage lineage cells represent a crucial cellular reservoir that needs to be targeted and eliminated to achieve complete remission and superior patient outcomes in people living with HIV (PLWH; (1–5). Macrophages are more resistant to cytopathic effects of HIV-1 compared to T cells; as a result, these cells can harbor the virus for a longer period of time and can also traffic across the blood-brain barrier seeding virus into the brain (6–9). Productive infection of the monocyte-macrophage lineage is limited, in comparison to CD4+ T cells, due to the low surface expression of CD4 and HIV-1 co-receptors CCR5 and CXCR4; however, all are displayed on these cell types with the timing and quantity of receptor expression varying depending on the states of stimulation and differentiation of the given cell population (10–13). Circulating monocytes can be infected with HIV-1 and then differentiate into macrophages wherein post-integration latency is established. Once resident in tissues like the brain, these non-proliferative cells are capable of supporting viral gene expression and infectious virus production, albeit to a much lesser extent than activated T cells, with dissemination of virus to surrounding susceptible cells (7, 12, 14, 15). In addition, the production and release of viral proteins such as gp41, gp120, Tat, Vpr, and Nef impact neighboring cellular populations and have been shown to have neurotoxic properties (7, 12, 14, 15).
Monocytic cells are generated in the bone marrow from pluripotent stem cells, which retain the ability to differentiate into either granulocytes or cells of the monocyte-macrophage lineage. The process of differentiation is governed by activity of transcription factors (TFs) that bind cis-acting promoter elements to govern expression of myeloid-specific genes in response to extracellular cues (16). The Sp family of TFs is involved in the regulation of monocyte- and/or myeloid-specific gene expression (17, 18). Sp1 and AP-1 have been shown to act cooperatively in the myeloid-specific regulation of the CD11c leukocyte integrin promoter (19). Similarly, Sp1 and AP-2 have been implicated in regulating lysosomal acid lipase and acid sphingomyelinase during monocytic differentiation (20, 21). Sp1 is also known to interact with C/EBP factors in regulating expression of several genes in myeloid cells (22–24). Other myeloid cell-specific differentiation events have also been attributed to specific activity of Sp factors (25–27). Sp factors have also been implicated in the regulation of chondrocyte, adipocyte, and epithelial cell differentiation (28–30) as well as in neuronal differentiation (31–34).
The Sp family of TFs includes Sp1, Sp2, Sp3, and Sp4 (35, 36). The C-terminus of Sp1 contains three zinc fingers that facilitate DNA binding, while the N-terminus contains the transactivation domains. (17). Sp1, Sp3 and Sp4 share near-equivalent affinity for binding G/C rich sequences, while Sp2 prefers binding to an alternative G/T rich sequence (35–38). Sp1 and Sp3 are ubiquitously expressed, while Sp4 has been shown to be brain-restricted in its pattern of expression (34, 35, 39, 40). Post-translation modifications (PTMs) including phosphorylation, glycosylation, acetylation and sumoylation of the Sp family of TFs have been shown to regulate their DNA binding and transcriptional activity (41). For instance, phosphorylation of Sp1 on Serine 131 by HIV-1 Tat-mediated activation of DNA-dependent protein kinase increases transcription with no effect on DNA binding (42–44) whereas other reports have shown that dephosphorylation of Sp1 mediated increased transcription activity from IL-21R promoter upon T cell receptor stimulation (45). Similarly, dephosphorylation of Sp1 at Serine 59 and Threonine 681 by PP2A resulted in increased association of dephosphorylated Sp1 with chromatin; however, the impact on transcriptional regulation was not studied (46). Additionally, groups have shown that sumoylation of the Sp1 N-terminus and other Sp3 isoforms occurs at basal conditions and can decrease transcriptional activity via chromatin changes promoting local gene silencing (43).
With respect to HIV-1, Sp1 and Sp4 have been shown to function as activators of gene expression through their interactions with the G/C box array of the HIV-1 LTR, while both the full-length Sp3 and truncated Sp3 isoforms have been shown to act as repressors (7, 47, 48). In another report a host factor, interferon γ-inducible protein 16 (IFI16) has also been shown to bind at high levels to Sp1, sequestering it from interacting with the LTR of HIV-1, leading to inefficient transcription. (49, 50). To examine the role of Sp factors in regulating HIV-1 gene expression during the course of monocytic differentiation, a number of immature myeloid cell line models capable of chemically-induced differentiation along with primary monocyte-derived macrophages (MDM) were utilized to model monocytic differentiation (Figure 1). The HL-60 cell line represents a partially committed myeloid cell (51, 52), which retains the ability to differentiate toward either granulocytic or monocytic cell types following treatment with either dimethylsulfoxide (DMSO) or phorbol myristate acetate (PMA), respectively (53, 54). The U-937 and THP-1 cell lines represent committed monoblastic cells (55–58), which may be further differentiated along the monocytic pathway when treated with PMA (59).
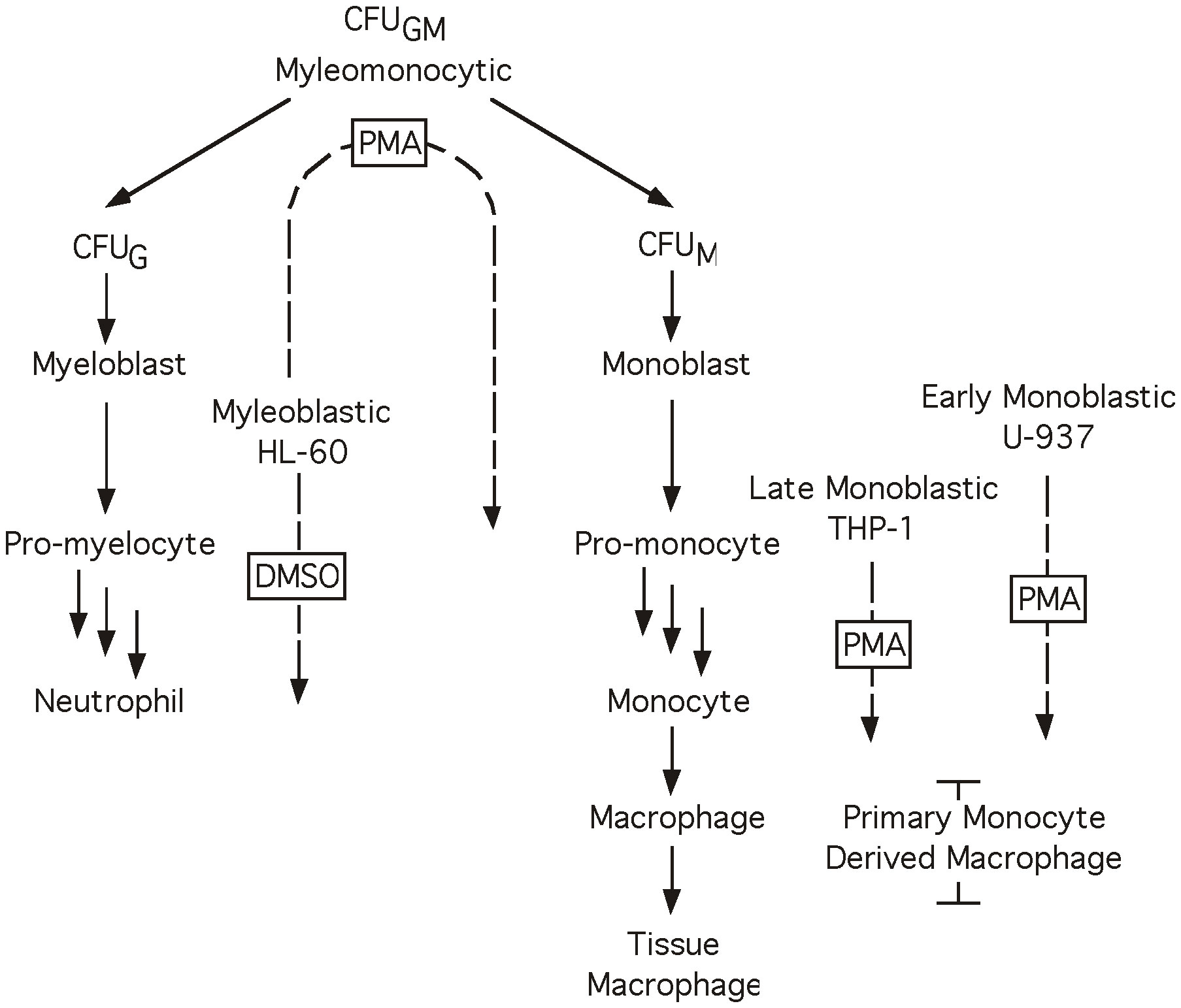
Figure 1 Myeloid differentiation. Diagrammatic representation of the process of myeloid differentiation. Cell lines and primary cell preparations are shown at their approximate stage in the differentiation process, dotted arrows indicate a cell line’s ability to differentiate towards a given cell type in the presence of the boxed chemical inducer. When treated with PMA cell lines HL-60 (48 hrs), THP-1 and U-937 (24 hrs) differentiate toward the monocyte lineage. When treated with DMSO cell line HL-60 (48 hrs) differentiates toward the granulocyte lineage.
Utilizing these in vitro model systems, the binding of the Sp1 activator protein to a high affinity HIV-1 Sp site was shown to increase relative to the binding of the Sp3 repressor isoforms following PMA-induced monocytic differentiation of HL-60, U-937 and THP-1 cell lines. Sp binding ratios from PMA-induced cells lines were also shown to more closely approximate those derived from primary MDMs than did ratios derived from uninduced cell lines. In addition, the altered Sp factor binding phenotype is associated with alterations in the transcriptional activation generated by a series of HIV-1 cis-acting Sp elements. Moreover, the PTM analysis on Sp1 and Sp3 proteins from untreated and PMA-activated U-937 cells demonstrated a loss of phosphorylation on certain residues on Sp1 and Sp3 proteins showing potential functional contribution of these specific PTMs in regulating LTR-directed transcription.
The human monocytic cell lines U-937 and THP-1 were grown in RPMI 1640 media (55, 56). The human myelomonocytic cell line HL-60 was grown in Dulbecco’s media (51, 52). All media were supplemented with 10% heat-inactivated fetal bovine serum, antibiotics (penicillin, streptomycin, and kanamycin at 0.04 mg/ml each), L-glutamine (0.3 mg/ml), and sodium bicarbonate (0.05%). All cell lines were maintained at 37˚C in the presence of 5% CO2 and 90% relative humidity. To induce monocytic differentiation, HL-60 cells were cultured in the presence of PMA (10 μM, Sigma) for 48 hr, while U-937 and THP-1 cells were cultured for 24 hr in the presence of 0.5 μM and 1.5 μM PMA, respectively (20, 60); for review see (59). Granulocytic differentiation of HL-60 cells was induced for 48 hr in the presence of DMSO (1.25%, Sigma) (53, 54).
Buffy coat preparations obtained from normal healthy individuals were centrifuged over a Ficoll gradient (300 x g) for 45 min (61) and the mononuclear fraction was collected and washed in Hank’s balanced salt solution containing EDTA (0.5 mM final concentration). The cells were resuspended in Dulbecco’s phosphate-buffered saline containing 10% human AB serum (US Biological) and HEPES buffer (2.5 mM). This preparation was subsequently centrifuged (550 x g) for 45 min over a 46% Percol solution (Pharmacia), and mononuclear cells were harvested at the resulting interface (62). Cells were washed with PBS and plated into 100-mm tissue culture plates (Falcon) at a density of 1 x 106 cell per ml. Following a 1 hr incubation at 37˚C in the presence of 5% CO2 and 90% relative humidity, nonadherent T cells were removed by washing with RPMI 1640. Adherent T cells were subsequently maintained in RPMI 1640 supplemented with 10% heat-inactivated human AB serum (US biological), M-CSF (100 U/ml; R&D Systems), GM-CSF (40 U/ml; R&D Systems), antibiotics (penicillin, streptomycin, and kanamycin at 0.04 mg/ml each), L-glutamine (0.3 mg/ml), and sodium bicarbonate (0.05%). Cells were cultured at 37˚C in the presence of 5% CO2 and 90% relative humidity for 5 to 10 days before use. The purity of monocytic preparations was assessed three days after isolation. Purity was determined to be >95% by flow cytometry. Cells reactive with antibodies directed against either CD45, CD4 (dim) and CD14; or CD45, CD4 (dim) and CD15 (dim) were considered monocytic by previous definition (63). Assessment of the major contaminating cell populations within the monocytic cell preparations was assessed by flow cytometry utilizing antibodies directed against CD45, CD20, and CD15 (bright) for identification of B cells and granulocytes; and antibodies directed against CD45, CD4, CD3 for the identification of T cells. All antibodies were purchased from Becton Dickenson.
Small-scale nuclear extracts were prepared from low-passage, exponentially-growing cells as previously described (3). Briefly, cells (1 × 107) were collected by centrifugation, washed once with ice-cold 1× Dulbecco phosphate-buffered saline (Mediatech), and lysed in ice-cold lysis buffer [HEPES (10 mM) pH 7.9, KCl (10 mM), EDTA (0.1 mM), EGTA (0.1 mM), octylphenoxypolyethoxyethanol (IGEPAL; Rhodia) (0.4%), dithiothreitol (DTT; 1 mM), phenylmethylsulfonyl fluoride (PMSF; 0.5 mM), and HALT protease inhibitor cocktail (1:100; Thermo Scientific, Rockford, IL)]. After centrifugation (1000 × g), the supernatant (cytoplasmic extract) was discarded. The pelleted nuclei were gently resuspended in nuclear extract buffer [HEPES (20 mM), NaCl (0.4 M), EDTA (1 mM), EGTA (1 mM), DTT (1 mM), PMSF (1 mM), and HALT protease inhibitor cocktail (1:100)], shaken vigorously on a rocker for 30 min at 4°C, and subjected to centrifugation for 10 min (14,000 × g). The supernatant (nuclear extract) was then dialyzed two times for 45 minutes using mini dialysis units (Thermo Scientific, 2000 molecular weight cutoff) in dialysis buffer [HEPES (20 mM), KCl (100 mM), EDTA (0.2 mM), glycerol (20%), DTT (1 mM), and PMSF (1 mM)]. The nuclear extract was recovered from the dialysis units and transferred to an Eppendorf tube and stored at -80°C in small aliquots. The protein concentration was determined by Bradford assay as described by the manufacturer (Bio-Rad, Hercules, CA).
Double-stranded, gel-purified oligonucleotides for EMS analysis were prepared as previously described (64). The sequence of the probe used in these studies was derived from the HIV-1 subtype B consensus Sp site III, 5’-CCAGGGAGGCGTGGCCTGGG-3’. Oligonucleotides were synthesized by Integrated DNA Technologies (Coralville, IA) as previously described (64). For antibody supershift/abrogation analysis, the indicated antibody (2 μg per reaction) (Santa Cruz Biotechnologies, Santa Cruz, CA) was added to the reaction and incubated for 30 min at room temperature prior to addition of the labeled probe. Complexes were separated on a 4% native TGE gel.
Complementary single-stranded oligonucleotides representing the subtype B consensus HIV-1 core LTR sequence (-83 to +2, relative to the transcriptional start site) were synthesized and annealed. In addition, an HIV-1 natural variant Sp element that fails to recruit Sp family factors (GAAGCGTGGC; (65) was substituted for 1, 2 or all 3 Sp elements in three additional synthetic oligonucleotides. Substitution of variant Sp elements in place of subtype B consensus binding elements began at the NF-κB-proximal Sp site and sequentially moved toward the TATA element. The four resulting oligonucleotides were cloned into pGL3-Basic (Promega) to generate Sp(3x)TATA-Luc, Sp(2x)TATA-Luc, Sp(1x)TATA-Luc, and Sp(0x)TATA-Luc (Figure 8A). Sp(3x)TATA-Luc contains three subtype B consensus Sp binding elements. Sp(2x)TATA-Luc contains a low affinity Sp element at position III, and subtype B consensus elements at positions II and I. Sp(1x)TATA-Luc contains two low affinity Sp elements at positions III and II, and a subtype B consensus element at position I. Sp(0x)TATA-Luc contains three low affinity Sp elements (Figure 8). Each construct contains a functional HIV-1 LTR TATA box as well as additional sequence out to the start site of transcription. The sequence of each construct was confirmed prior to experimentation.
Reporter plasmids were transfected in the U-937 and THP-1 monoblastic cell lines using Fugene 6 transfection system (Promega, Madison, WI) while electroporation was utilized in the HL-60 myelomonocytic cell line. U-937 or THP-1 cells were seeded from exponentially growing, low passage cultures at 5 x 105 cells per ml growth medium and transfected with Fugene 6 in 35-mm plates (Falcon). Premixed Fugene (94 μl serum free RPMI with 6 μl Fugene) was added dropwise to 2.5 μg of the experimental luciferase vector (THP-1 and U-937) and either 200 ng pRL-CMV, or 100 ng pRL-TK Renilla internal control vector (THP-1 and U-937, respectively). After a 15-min incubation, the Fugene/DNA mixture was added directly to the cells. Cultures were incubated at 37˚C for 24 hr. Cell extracts were harvested and assayed for luminescence using the Dual Luciferase Assay (Promega). HL-60 cells were seeded from exponentially growing, low passage cultures. The experimental luciferase vector (15 μg) and the pRL-CMV Renilla internal control vector (50 ng) were added to 5 x 106 cells in 250 μl of culture medium. Cells were subsequently electroporated (250 V and 960 μF) and transferred to 35-mm plates (Falcon) in 2 ml growth media. Cultures were incubated for 24 hr at 37°C. Cell extracts were prepared, and luminescence was measured using the Dual Luciferase Assay (Promega). Chemical differentiating agents, DMSO (1.25%) or PMA (0.5 μM U-937, 1.0 μM HL-60 or 1.5 μM THP-1); were diluted in culture media and added to transfected cells 30 min after the addition of the DNA/Fugene mixture. In each case, firefly luminescence was normalized to Renilla luminescence to control for variability in transfection efficiency. Activity of the constructs is presented as the fold-increase in normalized firefly activity over the activity of the Sp(0x)TATA-Luc construct. Each transfection was performed in duplicate and repeated at least three separate times. Error bars indicate the standard deviation among data obtained from independent experiments.
Total protein was isolated from U-937 and U-937 PMA-treated cell pellets. The isolated proteins were then digested with trypsin in solution and enriched for phosphopeptides with Titanium dioxide (TiO2) column. The digested peptides were then extracted with 50% acetonitrile containing 5% formic acid, dried in a speedvac and resuspended in the appropriate mobile phase for LC-MS/MS analysis. A linear acetonitrile gradient was used to separate the tryptic peptides based on their hydrophobicity prior to MS analysis on a Thermo Scientific LTQ XL mass spectrometer and with a total run time of 90 min per sample. The samples were then analyzed separately by nano-LC-MS/MS and the PTM analyses for phosphorylation were performed. A data-dependent Top 5 method was used where a full MS scan from m/z 400-1500 was followed by MS/MS scans of the five most abundant ions. Each ion was subjected to CID (Collision Induced Dissociation) for fragmentation and peptide identification. Raw data files from each sample were then searched using Proteome Discoverer 1.4 (Thermo Scientific) and SEQUEST algorithm against FASTA databases for Sp1 (P08047.3) and Sp3 (Q02447) from UniProt. PhosphoRS 3.0 algorithm (integrated into the Proteome Discoverer) was used for phosphorylation site localization, whereas fixed value PSM Validator was used for PSM validation in database searches for Sp1 and Sp3. pRS site probabilities above 75% were used as a cutoff to obtain good evidence that a responsive site was truly phosphorylated.
The quantitative two-tailed student t-test was utilized and p < 0.05 was considered significant.
To investigate the relative amounts of the different Sp family TFs present during the course of monocytic differentiation it was necessary to identify the Sp family members present in each of the cell types examined. EMS analyses were performed utilizing a 20-bp oligonucleotide probe centered on the subtype B consensus HIV-1 Sp site III in conjunction with HL-60, U-937, THP-1 and primary MDM nuclear extracts. The U-937 and THP-1 nuclear extracts each generated four distinct DNA-protein complexes (Figure 2, lanes 1 and 6). Addition of antibody directed against Sp1 resulted in both the partial abrogation of complex A and the generation of a lower mobility complex (Figure 2, compare lanes 1 and 6 to lanes 2 and 7). Complexes B and C were shown to be reactive with antibody directed against Sp3 by a combination of abrogation (complexes B and C) and supershift (either complex B or complex C) (Figure 2, compare lanes 1 and 6 to lanes 3 and 8). Consistent with the analysis of individual antibodies, the combined use of both Sp1 and Sp3 antibodies resulted in both the abrogation of complexes A, B and C and the generation of multiple low mobility complexes (Figure 2, lanes 4 and 9). Sp4 antibody did not react significantly with any of the complexes in reactions containing U-937 or THP-1 nuclear extract. Complex D was unaffected by the presence of any of the Sp antibodies examined and has been shown to be nonspecific by competition with homologous and non-homologous competitors (data not shown). EMS reactions containing HL-60 nuclear extract generated DNA-protein complexes consistent with complexes A through D in the U-937 and THP-1 extracts (Figure 2, compare lane 11 to lanes 1 and 6). Complex A and B was found to contain Sp1 while complexes B and C were found to contain Sp3 (Figure 2, lanes 11-14). In addition to these four complexes, a fifth high mobility complex was observed (Figure 2, lane 11, complex E). This complex was shown to be partially abrogated in the presence of either Sp1 or Sp3 antibody. Use of both Sp1 and Sp3 antibody resulted in nearly complete abrogation of complex E, suggesting that this high mobility complex may represent proteolytic Sp factor fragments, which retain their DNA-binding activity (27). Analysis of the primary MDM extract also yielded complexes similar to those observed in the U-937, THP-1 and HL-60 analysis. Complex A was shown to contain Sp1 (Figure 2, compare lane 16 and 17). Although complexes B and C appeared in low abundance, supershift analysis with Sp3 antibody yielded a faint low mobility complex consistent with the presence of Sp3 containing DNA-protein complexes (Figure 2, lane 18). As before, complex D was shown to be nonspecific by competition analysis (data not shown). A fifth complex (complex E), similar to that observed in EMS reactions utilizing HL-60 nuclear extract (Figure 2C), again demonstrated partial abrogation in the presence of either Sp1 or Sp3 antisera (Figure 2, lanes 17 and 18). Use of both Sp1 and Sp3 antibodies resulted in nearly complete abrogation of the complex (Figure 2, lane 19).
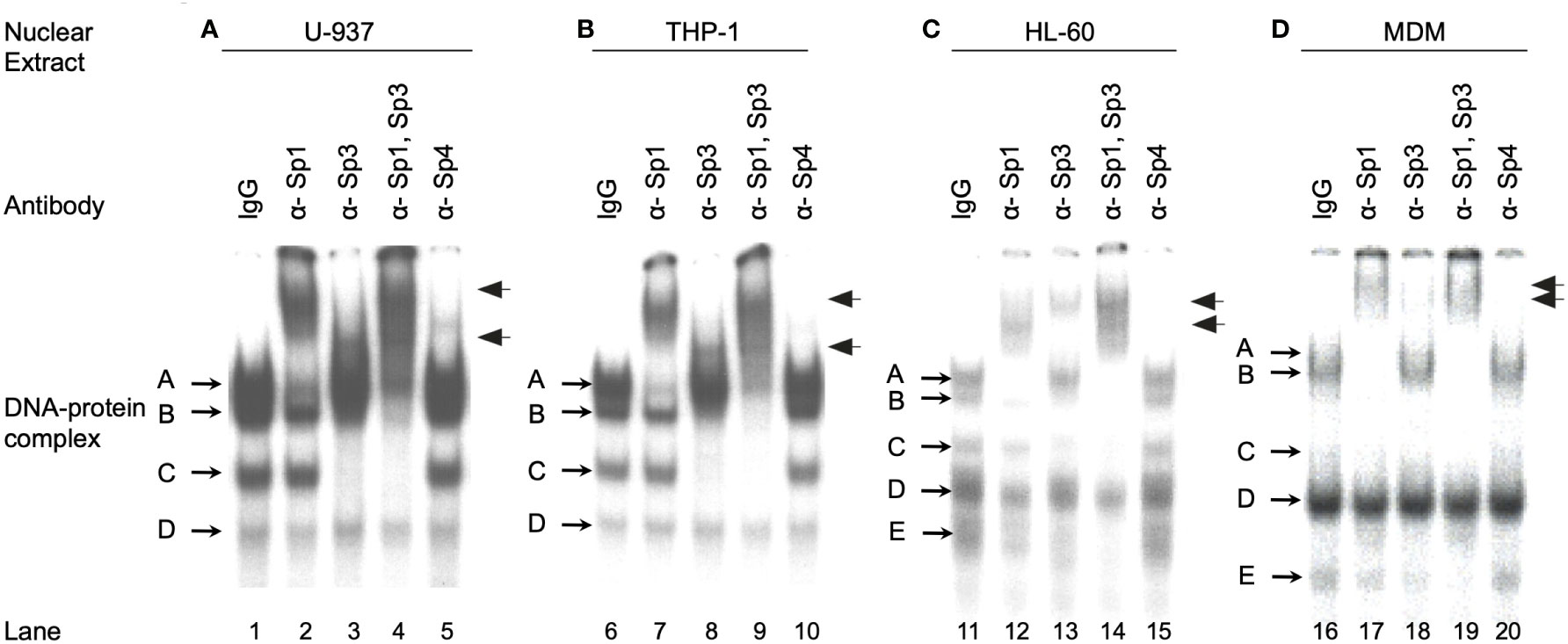
Figure 2 Monocytic Sp TFs bind a high affinity HIV-1 Sp binding site. EMS analyses using the HIV-1 subtype B consensus Sp site III were performed as described in the Materials and Methods. Binding reactions included 6 μg of U-937 (A), THP-1 (B), or HL-60 (C) nuclear extract with 60,000 cpm radiolabled probe (approximately 1 ng). MDM nuclear extract (20 μg) (D) was also reacted with 60,000 cpm of radiolabled probe. Antisera (1 μl) was added as indicated. DNA-protein complexes are labeled A through E, while supershifted DNA-protein complexes are indicated with arrows. This was performed as three independent experiments with a representative image presented.
It was observed in initial studies that the ratio of Sp1 to Sp3 might vary among nuclear extracts prepared from primary MDM, monoblastic (U-937 and THP-1) and myelomonoblastic (HL-60) cell populations. To determine whether alterations in the relative binding of Sp family activators (Sp1) and Sp family repressors (full-length and truncated Sp3) occurred during the course of monocytic differentiation, the binding of Sp factors was examined in a probe titration EMS analysis under conditions where each of the three major Sp complexes could be quantitated. Increasing amounts of a 20-bp probe centered on the HIV-1 subtype B consensus Sp site III was reacted with a constant amount of nuclear extract. When the probe is saturated with respect to the available binding protein, the relative abundance of each complex will primarily reflect the relative amounts of protein in the extract capable of binding probe. Under these conditions, a comparison of unstimulated, DMSO-stimulated, and PMA-stimulated HL-60 nuclear extracts (Figure 3A) provides an indication of the relative levels of Sp factors present in myelomonocytic cells (unstimulated), granulocytic cells (DMSO-treated), and monocytic cells (PMA-treated).
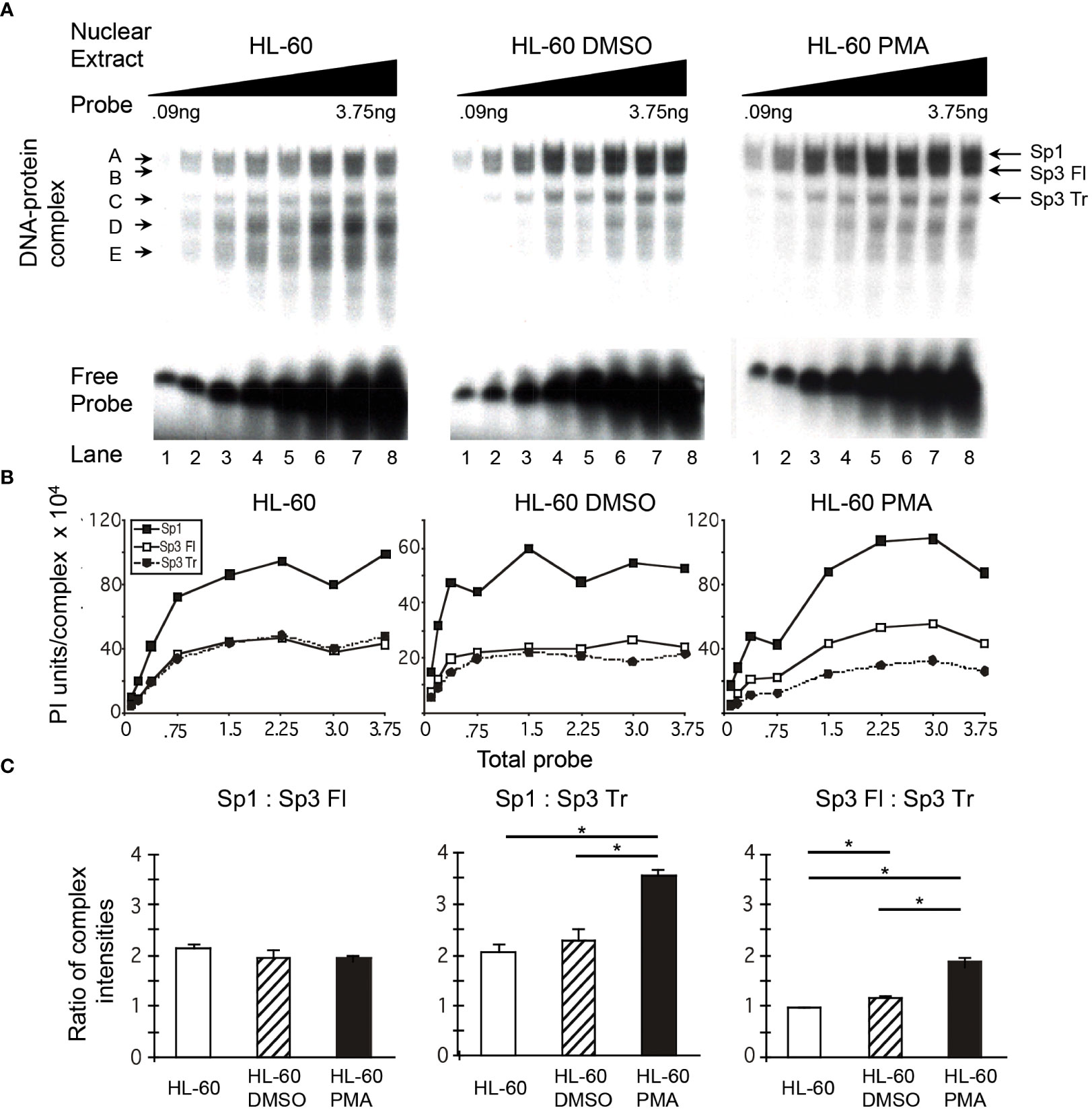
Figure 3 Relative levels of Sp factor binding in the myleomonocytic HL-60 cell line are altered with the addition of the chemical differentiating agent, PMA. Relative Sp factor binding was examined by EMS probe titration analyses. (A) Untreated, DMSO-treated, and PMA-treated HL-60 nuclear extracts (6 μg) were reacted with increasing amounts of labeled HIV-1 Sp site III probe (12,500 to 500,000 cpm; 0.09 to 3.75 ng). Complexes were separated utilizing a 2-hr electrophoresis interval through a 4% acrylamide gel to retain unbound probe and facilitate subsequent analysis. DNA-protein complexes are labeled A through E; arrows indicate the position of low mobility Sp-related complexes. Representative image shown. (B) Untreated, DMSO-treated, and PMA-treated HL-60 nuclear extracts (6 μg) were reacted with increasing amounts of labeled HIV-1 Sp site III probe (12,500 to 500,000 cpm; 0.09 to 3.75 ng). Complexes were subjected to a 3-hr electrophoresis interval utilizing 4% acrylamide to allow for greater separation and a more accurate subsequent quantitation of the resulting DNA-protein complexes. Unbound probe was not retained. DNA-protein complexes were quantitated and graphed as phosphorimager units per complex. Representative graph shown. (C) DNA-protein complexes intensities (B) were averaged at saturation and their relative levels compared within a given gel. This process was repeated three times and the average of the three relative intensities was graphed for each pair of DNA-protein complexes examined: Sp1 relative to full-length Sp3 (Sp3 Fl), Sp1 relative to truncated Sp3 (Sp3 Tr), and Sp3 Fl relative to Sp3 Tr. Comparisons between all were evaluated by the quantitative two-tailed student t-test and p < 0.05 was considered significant (*).
Quantitation of the resulting DNA-protein complexes and subsequent comparison of the complex intensities at saturation (Figure 3B) allows identification of the relative levels of each DNA-protein complex. The relative levels of Sp1 and Sp3 demonstrated little or no difference when nuclear extracts of untreated and DMSO-treated HL-60 cells were examined. In each case, the level of Sp1 relative to full-length Sp3 was approximately 2:1 (Figure 3C). Similarly, the level of Sp1 relative to truncated Sp3 was also approximately 2:1, while the level of full-length Sp3 relative to truncated Sp3 was nearly 1:1 in both extracts (Figure 3C). In contrast to the similarity among the Sp1- and Sp3-containing complexes, the intensity of the nonspecific complex D and the Sp1-Sp3 reactive low mobility complex E (Figure 3A) were notably reduced following DMSO treatment. Overall, it appears that DMSO treatment, which approximates myeloid differentiation (53), has little impact on the relative binding levels of Sp1, full-length Sp3, or truncated Sp3 to the HIV-1 subtype B consensus Sp site III.
In contrast to the Sp ratios observed between untreated and DMSO-treated HL-60 nuclear extracts, a comparison of untreated and PMA-treated HL-60 nuclear extracts yielded a different result. The level of Sp1 relative to full-length Sp3 was approximately 2:1 in each case (Figure 3C). However, the level of Sp1 relative to truncated Sp3 increased from 2:1 in untreated HL-60 nuclear extracts to nearly 3.6:1.0 in PMA-treated extracts (Figure 3C, middle panel). Similarly, the ratio of full-length Sp3 relative to truncated Sp3 increased from just under 1:1 in untreated extracts to nearly 2:1 in PMA-treated nuclear extracts. As with the DMSO-treated extracts, complexes D and E (Figure 3A) were noticeably reduced following differentiation. Overall, it appears that PMA treatment, which approximates monocytic differentiation (54), causes an increase in both Sp1 and full-length Sp3 relative to truncated Sp3.
To determine if the changes in relative Sp factor binding observed during the chemical differentiation of HL-60 cells was cell type-specific or related to a more general physiologic process, similar experiments were performed using the U-937 and THP-1 monoblastic cell lines. Nuclear extracts from untreated and PMA-treated cells were utilized in probe titration analyses under conditions in which each of the three Sp complexes could be separately quantitated (U-937, Figure 4A; THP-1, Figure 5A). Subsequent quantitation and comparison of complex intensities at saturation allowed for comparisons of relative factor binding profiles (U-937, Figure 4B; THP-1, Figure 5B).

Figure 4 Relative levels of Sp factor binding in the monoblastic U-937 cell line varies with the addition of the chemical differentiating agent, PMA. Relative Sp factor binding was examined by EMS probe titration analyses. (A) Untreated and PMA-treated U-937 nuclear extracts (6 μg) were reacted with increasing amounts of labeled HIV-1 Sp site III probe (12,500 to 500,000 cpm; 0.09 to 3.75 ng). Complexes were separated by a 2-hr electrophoresis interval through a 4% acrylamide gel to retain unbound probe to facilitate subsequent analysis. DNA-protein complexes are labeled A through D; arrows indicate the position of low mobility Sp-related complexes. Representative image shown. (B) Untreated and PMA-treated U-937 nuclear extracts (6 μg) were reacted with increasing amounts of labeled HIV-1 Sp site III probe (12,500 to 500,000 cpm; 0.09 to 3.75 ng). Complexes were subjected to a 3-hr electrophoresis interval utilizing 4% acrylamide to allow for greater separation and a more accurate subsequent quantitation of the resulting DNA-protein complexes. Unbound probe was not retained. DNA-protein complexes were quantitated and graphed as phosphorimager units per complex. Representative graph shown. (C) DNA-protein complex intensities (B) were averaged at saturation and their relative levels compared within a given gel. This process was repeated three times and the average of the three relative intensities was graphed for each pair of DNA-protein complexes examined: Sp1 relative to Sp3 Fl, Sp1 relative to Sp3 Tr, and Sp3 Fl relative to Sp3 Tr. Comparisons between all were evaluated by the quantitative two-tailed student t-test and p < 0.05 was considered significant (*).
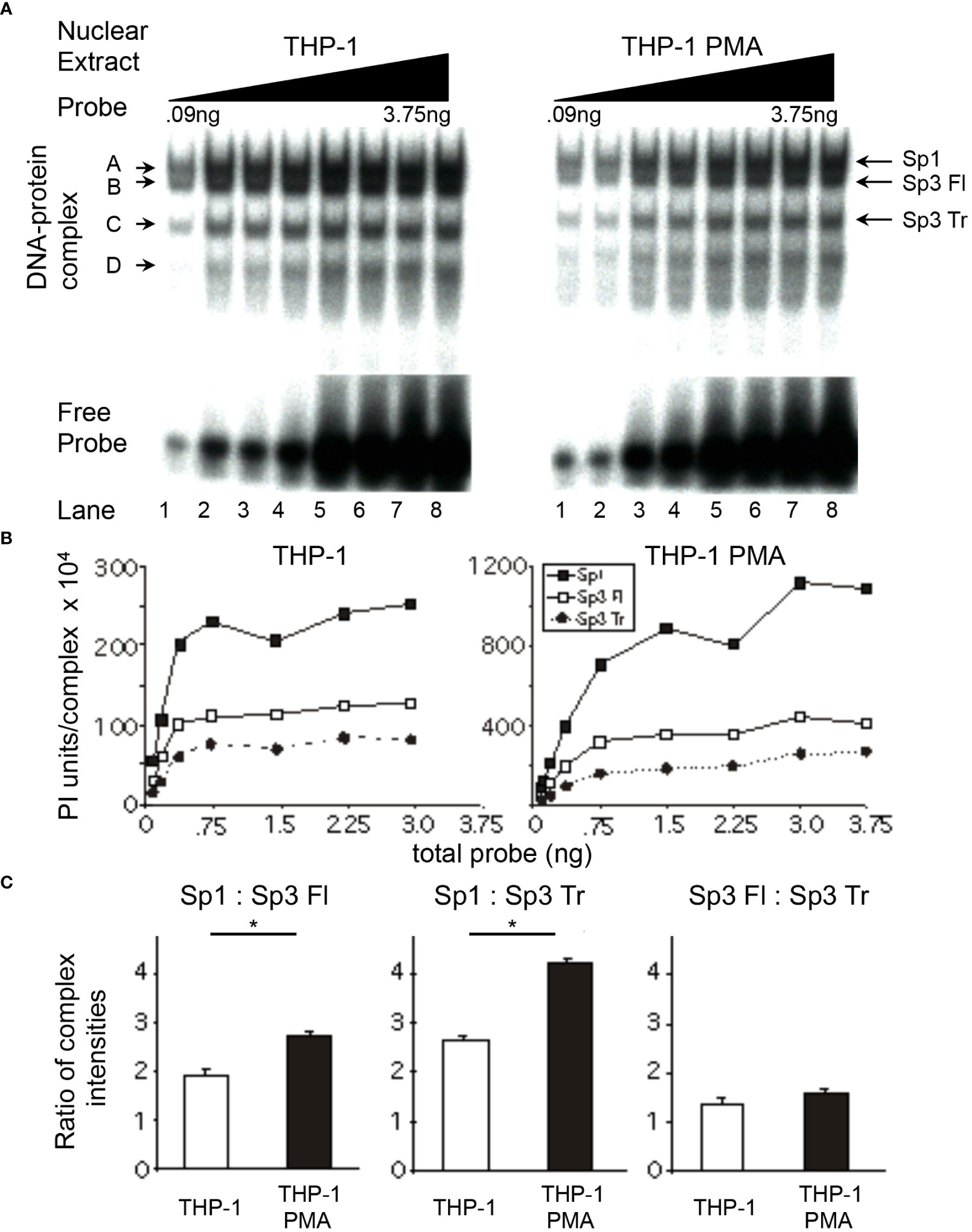
Figure 5 Relative levels of Sp factor binding in the monoblastic THP-1 cell line varies with the addition of the chemical differentiating agent, PMA. Relative Sp factor binding was examined by EMS probe titration analyses. (A) Untreated and PMA-treated THP-1 nuclear extracts (6 μg) were reacted with increasing amounts of labeled HIV-1 Sp site III probe (12,500 to 500,000 cpm; 0.09 to 3.75 ng). Complexes were separated by a 2-hr electrophoresis interval utilizing 4% acrylamide gel to retain unbound probe to facilitate subsequent analysis. DNA-protein complexes are labeled A through D; arrows indicate the position of low mobility Sp-related complexes. Representative image shown. (B) Untreated and PMA-treated THP-1 nuclear extracts (6 μg) were reacted with increasing amounts of labeled HIV-1 Sp site III probe (12,500 to 500,000 cpm; 0.09 to 3.75 ng). Complexes were subjected to a 3-hr electrophoresis interval utilizing 4% acrylamide to allow for greater separation and a more accurate subsequent quantitation of the DNA-protein complexes. Unbound probe was not retained. DNA-protein complexes were quantitated and graphed as phosphorimager units per complex. Representative graph shown. (C) DNA-protein complexes intensities (B) were averaged at plateau and their relative levels compared within a given gel. This process was repeated three times and the average of the three relative intensities was graphed for each pair of DNA-protein complexes examined: Sp1 relative to full-length Sp3, Sp1 relative to Sp3 Tr, and Sp3 Fl relative to Sp3 Tr. Comparisons between all were evaluated by the quantitative two-tailed student t-test and p < 0.05 was considered significant (*).
Significant alterations in relative factor binding were observed with chemical differentiation of U-937 cells with PMA. The level of Sp1 relative to full-length Sp3 in untreated U-937 cells was similar to that seen in untreated HL-60 cells, nearly 2:1 in each case (compare Figures 3C, 4C). This ratio increased to 3:1 in PMA-treated U-937 nuclear extracts (Figure 4C). The level of Sp1 relative to truncated Sp3 in untreated U-937 cells was again similar to that observed in nuclear extract prepared from untreated HL-60 cells (compare Figure 3C, 4C). However, this ratio increased from 1.8:1.0 in untreated U-937 nuclear extract to approximately 5.6:1.0 in nuclear extract prepared from PMA-treated U-937 cells (Figure 4C). The level of full-length Sp3 relative to truncated Sp3 was again near equivalent in nuclear extracts derived from untreated U-937 and HL-60 cells, approximately 1:1 in each case (compare Figures 3C, 4C). Examination of nuclear extracts from PMA-treated U-937 cells yielded a 2:1 ratio between full-length Sp3 and truncated Sp3 (Figure 4C). In summary, the Sp factor ratios observed in nuclear extracts prepared from the U-937 cell line were consistent with those observed in extracts prepared from the HL-60 cell line, in that a relative increase in Sp1 over both full-length and truncated Sp3 was observed following PMA-induced monocytic differentiation.
Relative Sp factor ratios were also examined in THP-1 monoblastic nuclear extracts. The level of Sp1 relative to full-length Sp3 in untreated THP-1 nuclear extracts (Figure 5C) was similar to those observed in untreated U-937 (Figure 4C) and HL-60 extracts (Figure 3C). Nuclear extracts prepared from PMA-treated THP-1 cells exhibited Sp1 to full-length Sp3 ratios of 2.7:1.0, a value above that of untreated U-937 and THP-1 extracts, but below that of PMA-treated U-937 extracts (Figure 5C). The level of Sp1 relative to truncated Sp3 in untreated THP-1 extracts (2.7:1.0) was above that observed with U-937 (2:1) and HL-60 (2:1) nuclear extracts (THP-1, Figure 5C; U-937, Figure 4C; HL-60, Figure 3C). Similar to previous results, nuclear extracts prepared from PMA-treated THP-1 cells exhibited an increase in the Sp1 to truncated Sp3 ratio as compared to the ratio observed with extracts prepared from untreated THP-1 cells (compare 4.3:1.0 to 2.7:1.0, respectively; Figure 5C). The full-length Sp3 to truncated Sp3 ratio obtained with nuclear extracts prepared from untreated THP-1 cells (Figure 5C) was slightly above those ratios generated from U-937 (Figure 4C) and HL-60 (Figure 3C) nuclear extract. However, nuclear extracts prepared from PMA-treated THP-1 cells exhibited little or no increase in the full-length Sp3 to truncated Sp3 ratio compared to extracts prepared from untreated THP-1 cells (Figure 5C). In summary, the use of the THP-1 monoblastic cell line as a model of PMA-induced monocytic differentiation yielded results similar to those observed in the U-937 monoblastic and HL-60 myleomonoblastic cell lines. In each case, the activating Sp1 was found to increase relative to the repressor Sp3 isoforms.
To determine whether the increase in relative Sp1 binding observed during chemical differentiation of monoblastic and myelomonocytic cell lines was a direct result of a monocytic differentiation process or a process specific to monocytic cell lines and thus unrelated to the differentiation process that occurs in primary monocytic cell populations in vivo, the relative binding of Sp factors in primary MDMs was examined. MDMs are known to differentiate during the process of isolation from peripheral blood. Hence, it was hypothesized that Sp factor binding ratios derived from MDM nuclear extracts would be similar to those observed in chemically differentiated nuclear extracts, if the alterations in Sp factor ratios were dependent on the process of monocytic differentiation.
MDM nuclear extracts were utilized in probe titration analyses (Figure 6A) under conditions in which each of the major Sp complexes could be quantitated (Figure 6B). The ratio of Sp1 relative to full-length Sp3 was 3:1 (MDM, Figure 6C), a value similar to ratios derived from PMA-treated U-937 (Figure 4C) and THP-1 (Figure 5C) nuclear extracts (3.0:1.0 and 2.8:1.0, respectively) and greater than the value obtained with PMA-treated HL-60 nuclear extracts (Figure 3C, 2.1:1.0). The level of Sp1 relative to truncated Sp3 was 3.7:1.0 (MDM, Figure 6C), a level similar to PMA-treated HL-60 (Figure 3C) and THP-1 (Figure 5C) cells (3.6:1.0 and 4.3:1.0, respectively), but somewhat below the level derived from PMA-treated U-937 cells (Figure 4C, 5.6:1.0). The level of full-length Sp3 relative to truncated Sp3 was 1.3:1.0 in primary MDM nuclear extracts (Figure 6C). When compared to chemically-differentiated cell lines exhibiting ratios between 1.6:1.0 and 1.9:1.0, it was apparent that MDM nuclear extracts exhibit less binding of full-length Sp3 relative to truncated Sp3 (MDM, Figure 6C; U-937, Figure 4C; THP-1, Figure 5C; HL-60, Figure 3C).
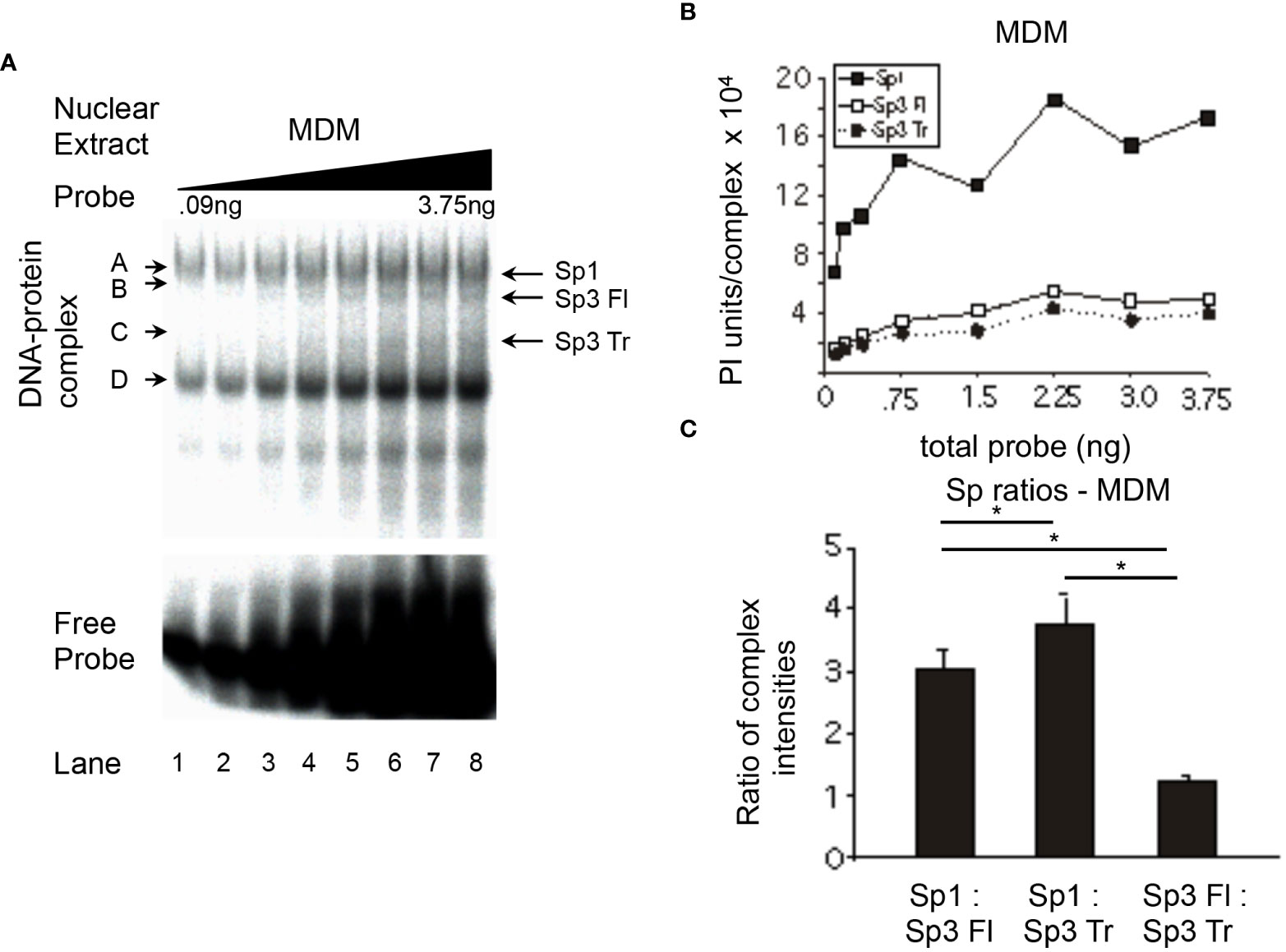
Figure 6 Relative levels of Sp factor binding in primary MDMs. Relative Sp factor binding was examined by EMS probe titration analyses. (A) Primary MDM nuclear extracts (20 μg) were reacted with increasing amounts of labeled HIV-1 Sp site III probe (12,500 to 500,000 cpm; 0.09 to 3.75 ng). Complexes were separated by a 2-hr electrophoresis interval utilizing 4% acrylamide gel to retain unbound probe for subsequent analysis. DNA-protein complexes are labeled A through D; arrows indicate the position of low mobility Sp-related complexes. Representative image shown. (B) Primary MDM nuclear extracts (6 μg) were reacted with increasing amounts of labeled HIV-1 Sp site III probe (12,500 to 500,000 cpm.; 0.09 to 3.75 ng). Complexes were subjected to a 3-hr electrophoresis interval utilizing 4% acrylamide to allow for greater separation and a more accurate subsequent quantitation of the DNA-protein complexes. Unbound probe was not retained. DNA-protein complexes were quantitated and graphed as phosphorimager units per complex. (C) DNA-protein complexes intensities Representative graph shown. (B) were averaged at saturation and their relative levels compared within a given gel. This process was repeated three times and the average of the three relative intensities was graphed for each pair of DNA-protein complexes examined: Sp1 relative to Sp3 Fl, Sp1 relative to Sp3 Tr, and Sp3 Fl relative to Sp3 Tr. Comparisons between all were evaluated by the quantitative two-tailed student t-test and p < 0.05 was considered significant (*).
Because Sp1 activates transcription from the HIV-1 LTR while both Sp3 isoforms act as transcriptional repressors (47), a comparison of activator to repressor ratios [Sp1/(Full length Sp3 + Truncated Sp3)], may allow for a more succinct view of the functional impact of different levels of Sp factor binding (Figure 7). Nuclear extracts from untreated cell lines, modeling undifferentiated monoblastic cells, each exhibited lower activator/repressor ratios than their PMA differentiated counterparts, indicating a potential increase in transcriptional activation from HIV-1 promoters containing active cis-acting Sp elements following monocytic differentiation. In the case of the HL-60 and U-937 nuclear extracts, each exhibited a nearly 1:1 ratio of activating Sp factors to repressing Sp factors. Following chemical differentiation with PMA, these ratios increased to 1.9:1.0 and 2.0:1.0, respectively (Figure 7). However, DMSO-induced granulocytic differentiation of the HL-60 myelomonocytic cell line generated a much smaller change in the activator/repressor ratio (compare 1.0:1.0 untreated to 1.2:1.0 treated) implying that the change in Sp factor recruitment observed with PMA-treatment were more a function of monocytic differentiation, than granulocytic differentiation. Nuclear extracts prepared from untreated cells exhibited similar levels of activating and repressing Sp factors (compare THP-1, 1.1:1.0; U-937, 0.9:1.0; and HL-60, 1.0:1.0; Figure 7). Interestingly, nuclear extracts from PMA-differentiated THP-1 cells, exhibited a smaller increase in the activator/repressor ratio (compare 1.1:1.0 untreated to 1.7:1.0 PMA-treated, Figure 7) than did nuclear extracts from either PMA-differentiated HL-60 or U-937 cells (compare 1.0:1.0 untreated to 1.9:1.0 PMA-treated, HL-60; and 0.9:1.0 untreated to 2.0:1.0 PMA-treated U-937; Figure 7). When the activator/repressor ratio of primary MDM (1.7:1.0) was compared to that of the cell lines examined, it was clear that primary MDMs have Sp factor binding profiles which more closely resemble profiles from chemically-differentiated monoblastic and myelomonoblastic cell lines as compared to their untreated counterparts (Figure 7).
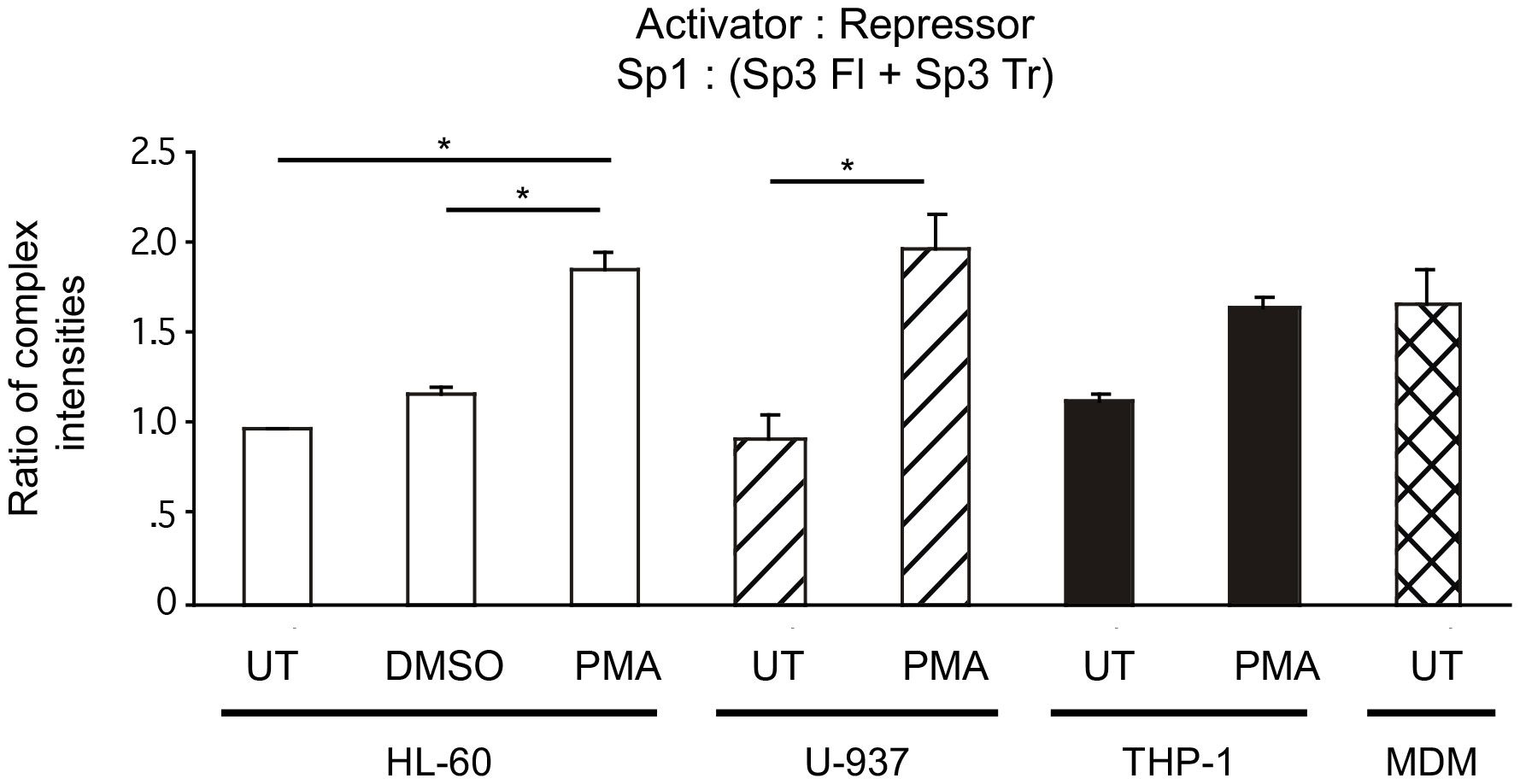
Figure 7 Comparisons of relative Sp family activator to repressor levels in untreated and chemically differentiated cell lines as well as primary MDM. To facilitate an examination of the effect of changing Sp factor binding on Sp-mediated activation of the HIV-1 LTR, results from Figures 3–6 have been coalesced to illustrate Sp1 (total activator) to the sum of both Sp3 isoforms (total repressor). Results from untreated and chemically differentiated cell lines are presented along with results from primary MDM. Statistical analysis between the three basal cells lines show no statistical difference. PMA treated cells cell lines were all statistically significantly higher than their basal comparator. Most important is that there was not a statistical difference between the PMA-treated cell lines and primary MDMs in this ratio. Comparisons between all were evaluated by the quantitative two-tailed student t-test and p < 0.05 was considered significant (*).
To examine the effect of the altered Sp factor ratios on HIV-1 LTR-directed gene expression in the absence of any confounding effects from non-Sp related changes in the transcriptional factor milieu which might be induced following monocyte differentiation, analyses were performed with a series of constructs containing only the TATA box and Sp elements. Specifically, transient transfection-reporter gene analyses were performed on truncated HIV-1 subtype B LTR-luciferase constructs containing three, two, one or no active cis-acting Sp elements (Figure 8A). To test the impact of altered Sp factor binding ratios on the activity of these constructs, fold-activation of the Sp(0x)TATA construct following the addition of one, two or three active Sp elements was examined in the HL-60, U-937 and THP-1 cell lines both in their undifferentiated and chemical-differentiated states. When these studies were performed in the HL-60 cell line, the addition of each active Sp element caused a sequential increase in promoter activity [Sp(0x)TATA ≤ Sp(1x)TATA < Sp(2x)TATA < Sp(3x)TATA] in both untreated and PMA-treated HL-60 cells (Figure 8B). The activity of each individual construct relative to that of the Sp(0x)TATA construct was then compared in untreated and chemically differentiated cells. Similar increases in activity were observed when both the Sp(1x)TATA and the Sp(2x)TATA constructs were examined (Figure 8B). However, the Sp(3x)TATA construct generated a small increase in promoter activity in PMA-treated cells than in untreated cells (compare 17-fold in untreated to 27-fold in PMA-treated; Figure 8B). This increase in fold-activation of the Sp(0x)TATA construct in the presence of PMA was qualitatively consistent with the 92% elevation in Sp activator binding observed in PMA-treated HL-60 nuclear extracts (compare untreated and PMA-treated HL-60 cells; Figure 7).
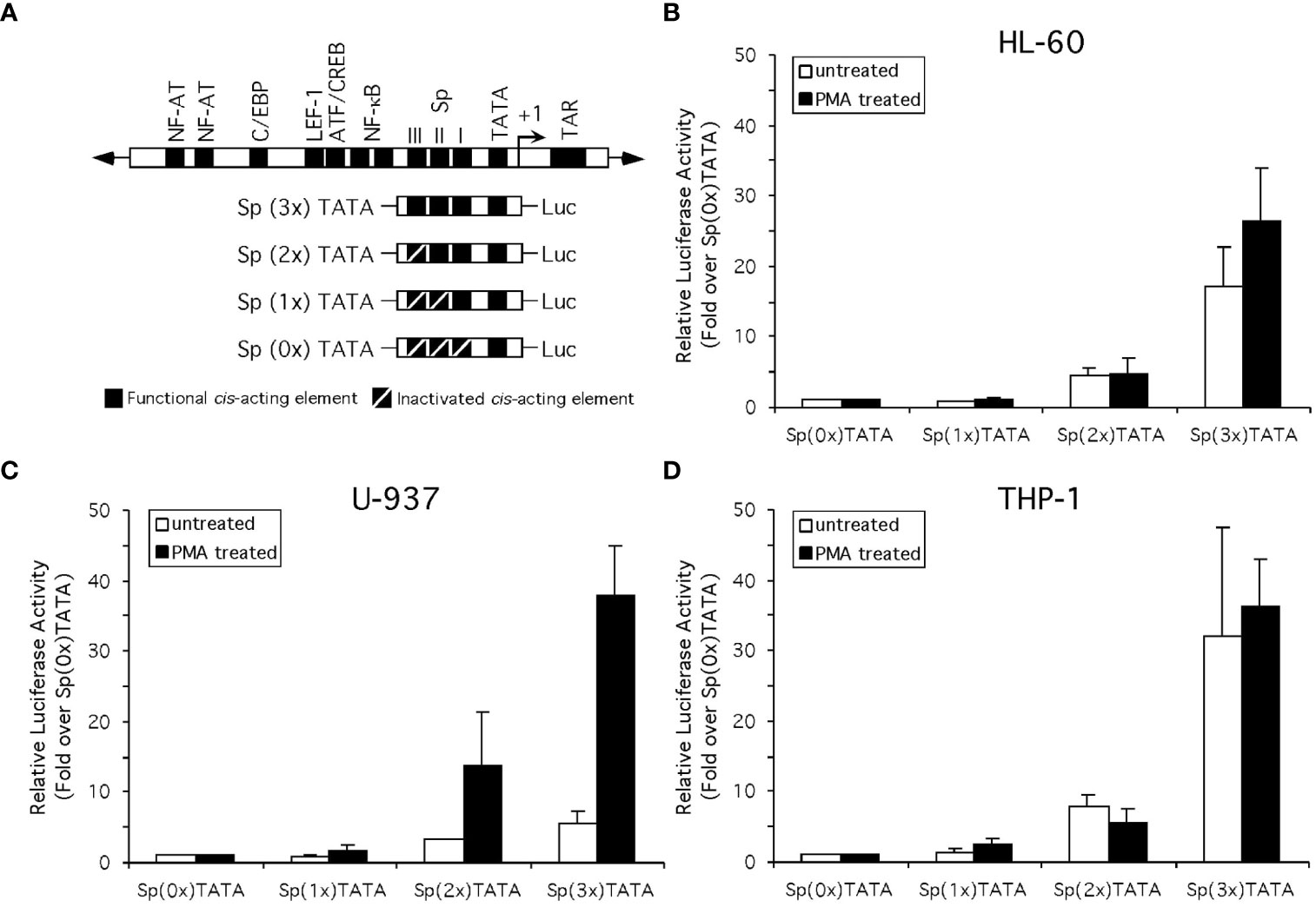
Figure 8 Altered Sp factor binding correlated with chemical differentiation alters the activity of the Sp elements of the HIV-1 LTR. (A) Diagrammatic depiction of the truncated LTR-Luciferase constructs examined. (B) Standard reporter gene analysis utilizing the dual luciferase reporter gene system (see Materials and Methods for details) was employed to determine the effect of chemically-induced monocytic differentiation on the activity of constructs containing three, two, one or no active Sp cis-acting elements. Results compare the activity of constructs in the HL-60 (B), U-937 (C), and THP-1 (D) cells in the absence or presence of PMA treatment. Activity is shown as fold-increase over the Sp(0x)TATA construct. Individual experiments were performed in duplicate and repeated at least three times. Error bars represent the standard deviation between the replicate experiments.
When these constructs were examined in U-937 cells, the addition of each active Sp element again caused a sequential increase in promoter activity [Sp(0x)TATA ≤ Sp(1x)TATA < Sp(2x)TATA < Sp(3x)TATA] in both untreated and PMA-treated U-937 cells (Figure 8C). When the activity of each individual construct relative to that of the Sp(0x)TATA construct was compared in untreated and chemically differentiated cells, significant differences were observed. When examined in untreated U-937 cells the Sp(1x)TATA construct resulted in no increase in activity over the Sp(0x)TATA construct, however a 1.8-fold increase in activity was observed when the constructs were tested in PMA-treated U-937 cells (Figure 8C). Similarly, the untreated Sp(2x)TATA construct generated a 3.3-fold increase in activity relative the Sp(0x)TATA construct in untreated cells, and a 14-fold increase in this activity in PMA-treated cells (Figure 8C). The Sp(3x)TATA construct, containing three active Sp elements, generated activity 5.7-fold over that of the Sp(0x)TATA construct in untreated cells, while the same comparison in PMA-treated cells yielded a 38-fold increase in promoter activity (Figure 8C). These results are consistent with results observed in HL-60 cells, in that a 116% increase in the relative levels of Sp family activators observed in PMA-treated extracts (compare untreated and PMA-treated U-937 cells; Figure 8) qualitatively correlates with the increased levels of LTR-luciferase activity observed in the presence of PMA.
Similar to observations in HL-60 and U-937 cells, the addition of each active Sp element caused a sequential increase in promoter activity [Sp(0x)TATA ≤ Sp(1x)TATA < Sp(2x)TATA < Sp(3x)TATA] in both untreated and PMA-treated THP-1 cells (Figure 8D). As before, the activity of each individual construct relative to that of the Sp(0x)TATA construct was compared in untreated and chemically differentiated cells. Unlike the results obtained with HL-60 or U-937 cells, a comparison of construct activity in untreated and PMA-treated THP-1 cells generated little or no difference in activity relative to the Sp(0x)TATA construct (Figure 8D). The lack of a PMA-induced increase in the activation of the Sp-containing LTR-Luciferase constructs may be related to the smaller (47%) increase in the binding of Sp family activators relative to repressors observed with THP-1 nuclear extracts following PMA treatment (compare untreated and PMA-treated THP-1 cells; Figure 8). Cumulatively, results from the HL-60, U-937 and THP-1 cell lines indicate that enhancement of Sp1 factor binding relative to Sp3 isoform binding may influence, but not fully predict, the transcriptional activity generated from the cis-acting Sp elements of the HIV-1 LTR.
It is well established that activity and stability of cellular TFs is regulated by PTMs including phosphorylation, glycosylation, acetylation, ubiquitination, methylation and sumoylation [reviewed in (66)]. For the Sp family of TFs, PTMs have been shown to influence several functional aspects including protein-protein interactions, sequence-specific DNA binding, and transcriptional activity (41). To evaluate the changes in PTMs on Sp1 and Sp3 in the process of monocytic differentiation, mass spectroscopy analysis was performed as described above. U-937 promocytic cells were utilized in this analysis as these cells demonstrated a good separation of transcriptional activity between untreated and PMA-treated conditions (see Figure 8C). Untreated and 0.5 μM PMA-treated U-937 cells were lysed and subjected to LC MS/MS analysis after trypsin digestion followed by PTM analyses as described above. Results identified loss of phosphorylation on S126, S137, S360, S365, T375, S376, S379, S765, S777, and S781 positions for Sp1 (Table 1). There was also a gain of phosphorylation at S127. Similarly, for Sp3, loss of phosphorylation was observed but only on the T574 residue (Table 2). The PTM analysis did not detect the well-studied amino acid residues in the transactivation domains of Sp1 and Sp3 including Ser131, T355 and others (43). It may be due to inefficient digestion with trypsin (data not shown) or uniqueness of the cell types and culture conditions utilized in this study. Nevertheless, the results identified additional novel sites where Sp1 and Sp3 were post-translationally modified upon activation in monocytic-lineage cells. Is the specific loss of phosphorylation on these residues a result of other competitive PTMs and what is their functional relevance to LTR-directed transcription? These questions are highly relevant and would require additional studies focused on mutating these residues and evaluating the impact on Sp1/Sp3 binding to cognate sites in the HIV-1 LTR and downstream transcriptional activation.
Cells of the monocyte-macrophage lineage play critical roles in the function of immune system during infection by pathogens such as HIV-1. The process of monocytic differentiation induces a shift in the relative levels of Sp TFs with a favorable binding of the activating Sp1 over the two Sp3 isoforms. These alterations in Sp TFs appear to increase the transcriptional activity provided by the cis-acting Sp elements of the HIV-1 subtype B consensus LTR.
A comparison of Sp factor binding ratios derived from MDMs, chemically-differentiated cell lines, and their undifferentiated counterparts, illustrated that ratios derived from MDMs and chemically-differentiated cells more closely resemble each other than did ratios from undifferentiated cells (compare Figure 6C to Figures 3C, 4C, 5C). These results suggest that Sp factor binding ratios derived from chemically differentiated cells lines may act as an appropriate proxy for Sp factor binding in primary MDMs.
After demonstrating an alteration in the binding profiles of Sp factors following monocytic differentiation, it was of interest to examine the mechanism by which their binding profiles were altered. To this end, the level of both Sp1 and Sp3 were examined by western immunoblot analyses. Interestingly, no significant differences in the levels of Sp factors were detected between nuclear extracts derived from untreated and chemically differentiated U-937 or THP-1 cells (data not shown). Although a small increase in Sp factors was observed in HL-60 cells upon PMA-induced monocytic differentiation, a similar increase was also observed following DMSO-induced granulocytic differentiation (data not shown) indicating that the increased protein levels were not the likely cause of the altered factor binding. Hence, changes in Sp factor binding do not appear to be driven by changes in the nuclear concentrations of the Sp factors, though this observation could also be validated through chromatin immunoprecipitation and qPCR. Thus, it was hypothesized that Sp factor binding may be regulated by a post-translational mechanism. The PTM analysis on Sp1 and Sp3 upon differentiation with PMA revealed loss of phosphorylation on several residues on Sp1 and one such modification was seen on Sp3 (Tables 1, 2). The PTM data did not result in the detection of some of the reported phosphorylated residues in Sp1 and Sp3. This may be due a variety of factors including inefficient digestion with trypsin prior to MS/MS or it may be due to unique patterns of phosphorylation of Sp factors in the promonocytic U-937 cells. Additional studies will need to be performed that will involve the use of a combination of proteases (Chymotrypsin and trypsin) to generate superior digestion to yield smaller peptides and better coverage. In addition, in future studies in this regard, we would also want to perform these studies in other myeloid cells to see if there is differential phosphorylation in myeloid cell type populations. These studies are extensive and beyond the scope of this report. Additionally, the functional contribution of these newly-identified phosphorylation sites on Sp1 and Sp3 will be evaluated in context of LTR-directed transcription by sequentially mutating these residues coupled with additional functional studies to pinpoint the role of these residues with respect to the impact of Sp phosphorylation on regulating HIV-1 gene expression during differentiation and activation of this important lineage of cells.
After observing an increase in Sp1 binding relative to the two Sp3 isoforms, the mechanisms guiding altered Sp factor binding profiles and how they impact the activity of subtype B consensus HIV-1 LTR cis-acting Sp elements was examined. Because Sp1 has been shown to activate the HIV-1 LTR while both Sp3 isoforms have been shown to repress it (47), we hypothesized that an increase in Sp1 binding relative the Sp3 isoforms would correlate with increased LTR activity. To examine this hypothesis a series of truncated HIV-1 LTR constructs (-83 to +2 relative the transcriptional start site) containing a TATA box and either three, two, one or no active Sp binding elements were utilized in transient expression analyses in HL-60, U-937, and THP-1 cells in the absence or presence of PMA (Figure 8A). The activity of each construct was then compared to the activity of the construct containing no active Sp elements. This measure of relative activity facilitated a comparison of each construct activity in a given cell line in the absence or presence of PMA. Furthermore, one might anticipate a greater increase in the relative activity of a construct containing Sp elements in a cellular environment, which favors binding of the activating Sp1 over the repressing Sp3 isoforms. In general, assuming that the activation potential for the Sp factors themselves remains unchanged during differentiation, the increase in LTR activity observed with each additional Sp element would be greater after differentiation.
Results from two of the three cell lines examined qualitatively support this hypothesis. The HL-60 and U-937 cell lines both exhibit increases in their activator to repressor ratio (Figure 7) and an increased relative activity of cis-acting Sp elements (Figures 8B, C) following chemically induced monocytic differentiation. However, the qualitative relationship between the activator to repressor ratio and the increase in relative activity of cis-acting Sp elements is significantly different. The HL-60 cell line exhibited a 92% increase in the activator to repressor ratio (Figure 7) and a 55% increase in the relative activity of the Sp(3x)TATA LTR-Luciferase construct (Figure 8B) with PMA treatment. While the U-937 cell line exhibited a 116% increase in the activator to repressor ratio (Figure 7) and a 5.6-fold increase in the relative activity of the Sp(3x)TATA LTR-Luciferase construct (Figure 8C) with PMA treatment. In addition, PMA treatment causes the Sp(1x)TATA and Sp(2x)TATA constructs to exhibit little increase in relative activity in HL-60 cells while a greater increase was observed in U-937 cells (compare Figure 8B to Figure 8C). Conversely, the THP-1 cell line did not appear to qualitatively or quantitatively adhere to our aforementioned hypothesis. THP-1 cells exhibited a 47% increase in the activator to repressor ratio (Figure 7) and either a minor or no increase in the relative activity of the Sp(3x)TATA LTR-Luciferase construct (Figure 8D) upon PMA treatment. Taken together, these relationships imply that although Sp factor binding may influence the activity of cis-acting Sp elements it does not directly predict this activity, thus implying that factors or processes other than the ratio of Sp factor binding may also modulate the activation of cis-acting Sp elements. To further explore these findings, knockdowns of Sp1 and Sp3 may also be used to support our hypotheses however it has previously been shown that loss of Sp1 significantly dysregulates gene expression during differentiation (67).
One possible inference from these results is that the activation potential of the Sp factors themselves may be altered during the course of monocytic differentiation. An increase in the activation potential of the Sp factors following monocytic differentiation could explain how the small difference in increased activator to repressor ratios observed between HL-60 and U-937 cells following PMA treatment [compare a 92% increase (HL-60) to a 116% increase (U-937); Figure 7), could be coupled to such notable differences in increased relative activity of cis-acting Sp elements following PMA treatment [compare a 55% increase (HL-60) to a 5.6-fold increase (U-937); Figures 8B, C]. Similarly, in the THP-1 cell line a marginal change in the activating potential of Sp factors might explain the lack of a significant increase in the relative activity of the Sp(3x)TATA construct following (Figure 8D) the induction of a 42% increase in the activator to repressor ratio (Figure 7) following PMA treatment. Another potential explanation for these findings would hypothesize the existence of significant synergism between activating Sp family members. Such a synergism could be used to explain the large changes in transcriptional activity observed after smaller quantitative alterations in Sp factor recruitment in U-937s. Furthermore, the 42% increase in activating Sp factor binding coupled with only minor alterations in transcriptional activity observed following PMA treatment of THP-1 cells, could be explained if a certain threshold level of Sp activator binding was required before noticeable synergism is achieved. In addition to these, the PTMs on Sp factors showed some significant differences between the untreated and PMA-stimulated U-937 cells (Tables 1, 2). Additional studies in HIV-infected monocytic cell lines with induced differentiation, demonstrating altered PTMs of Sp isoforms would support the significance of this observation.
With respect to the HIV-1 gene regulation system, the alterations in Sp factor binding observed may have significance beyond that presently illustrated. HIV-1 LTR activity has been shown to be activated in the presence of either the viral transactivating protein, Tat, (68) or the viral accessory protein Vpr (69). Moreover, it has been shown that both Tat- and Vpr-mediated LTR activation is modulated through interactions with the G/C box array of the HIV-1 LTR and recruited Sp factors. Specifically, the loss of Sp elements in the HIV-1 LTR dramatically reduces Tat-mediated LTR activity in HeLa cells but only when all three are mutated but not basal activity (57). In Figure 8, we show that individual Sp binding site mutations in Sp binding site III contribute the most to basal and PMA-induced activity in various monocytic cells. However, previous studies from our group have shown that in HIV-1 replication experiments when Sp binding site III alone is mutated to prevent Sp1 and/or 3 binding, viral replication is reduced in Jurkat T cells and not U-937 monocytic cells (70). However, in this scenario Tat and Vpr would be present. This is interesting as the Tat inducibility of certain synthetic promoters has been shown to be Sp1-dependent (71) and Tat has been shown to induce the phosphorylation of Sp factors (42), a modification which might directly influence the DNA binding affinity of the Sp factors themselves. Evidence that Vpr-mediated LTR activation is influenced by an interaction between Vpr and the Sp1 HIV-1 LTR complex has also been reported (72, 73). In light of these studies, one might hypothesize that the altered recruitment of Sp factors during monocytic differentiation illustrated herein may likely impact Tat- and/or Vpr-mediated LTR activation or that Tat and/or Vpr may aid in stabilizing lower affinity Sp binding sites to help induce replication.
In summary, we have illustrated that the process of monocytic differentiation can induce a shift in the relative levels of Sp factor binding to an HIV-1 Sp element. This shift favors binding of the activated Sp1 over the two Sp3 isoforms. Such an alteration in Sp factor binding may have significance across a variety of Sp-dependent cellular and viral promoters present in cells of the monocyte-macrophage lineage during differentiation. Furthermore, the alteration in Sp factor binding was shown to alter the transcriptional activity provided by the cis-acting Sp elements of the HIV-1 subtype B consensus LTR. These observations may be attributed to the changes in PTMs on Sp isoforms upon receiving differentiation and activation stimuli as observed in U-937 cells. Finally, HIV-1 transcription and replication is driven through the LTR and cell phenotype and differentiation status impacts this. In fact, we have previously shown that all three Sp binding sites explored here have significantly different mean genetic differences between CXCR4-utilizing and CCR5-utilizing virus (proxy for T cell versus myeloid infection) and that these differential genetic signatures are predicted to impact the Sp binding phenotype at each Sp site (65, 70, 74). Taken together, this study adds to the importance of the Sp binding sites for viral replication and shows the isoforms of the proteins that can regulate the LTR in various differentiation states of myeloid lineages.
All relevant data is contained within the article: The original contributions presented in the study are included in the article/supplementary material. Further inquiries can be directed to the corresponding author.
Conceptualization, JM., SD, BW; methodology, JM., SD, BW; formal analysis, SD, JM, MN, TB, BW; investigation, SD, JM, MN, TB, BW; resources, BW; data curation, SD, JM., MN, TB, BW; writing—original draft preparation, SD, JM, MN, TB, BW; writing—review and editing, SD, JM, RB, MC, MN, TB, BW; visualization, SD, JM, MN, TB, BW; supervision, MN, BW; project administration, MN, BW; funding acquisition, BW. All authors contributed to the article and approved the submitted version.
The authors were funded in part by the Public Health Service, National Institutes of Health, through grants from the National Institute of Mental Health (NIMH) R01 MH110360 (Contact PI, BW), the NIMH Comprehensive NeuroAIDS Center (CNAC) P30 MH092177 (KK, PI; BW, PI of the Drexel subcontract involving the Clinical and Translational Research Support Core), the Ruth L. Kirschstein National Research Service Award T32 MH079785 (PI, TB; BW, Principal Investigator of the Drexel University College of Medicine component). The contents of the paper are solely the responsibility of the authors and do not necessarily reflect the official views of the NIH.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
1. Le Douce V, Herbein G, Rohr O, Schwartz C. Molecular mechanisms of HIV-1 persistence in the monocyte-macrophage lineage. Retrovirology (2010) 7:32. doi: 10.1186/1742-4690-7-32
2. Burdo TH, Lackner A, Williams KC. Monocyte/macrophages and their role in HIV neuropathogenesis. Immunol Rev (2013) 254(1):102–13. doi: 10.1111/imr.12068
3. Dahiya S, Liu Y, Nonnemacher MR, Dampier W, Wigdahl B. CCAAT enhancer binding protein and nuclear factor of activated T cells regulate HIV-1 LTR via a novel conserved downstream site in cells of the monocyte-macrophage lineage. PloS One (2014) 9(2):e88116. doi: 10.1371/journal.pone.0088116
4. Kumar A, Abbas W, Herbein G. HIV-1 latency in monocytes/macrophages. Viruses (2014) 6(4):1837–60. doi: 10.3390/v6041837
5. Avalos CR, Price SL, Forsyth ER, Pin JN, Shirk EN, Bullock BT, et al. Quantitation of productively infected monocytes and macrophages of SIV-infected macaques. J Virol (2016) 5643–56. doi: 10.1128/JVI.00290-16
6. Carter CA, Ehrlich LS. Cell biology of HIV-1 infection of macrophages. Annu Rev Microbiol (2008) 62:425–43. doi: 10.1146/annurev.micro.62.081307.162758
7. Kilareski EM, Shah S, Nonnemacher MR, Wigdahl B. Regulation of HIV-1 transcription in cells of the monocyte-macrophage lineage. Retrovirology (2009) 6:118. doi: 10.1186/1742-4690-6-118
8. Honeycutt JB, Wahl A, Baker C, Spagnuolo RA, Foster J, Zakharova O, et al. Macrophages sustain HIV replication in vivo independently of T cells. J Clin Invest (2016) 126(4):1353–66. doi: 10.1172/JCI84456
9. Sattentau QJ, Stevenson M. Macrophages and HIV-1: An unhealthy constellation. Cell Host Microbe (2016) 19(3):304–10. doi: 10.1016/j.chom.2016.02.013
10. Lee B, Sharron M, Montaner LJ, Weissman D, Doms RW. Quantification of CD4, CCR5, and CXCR4 levels on lymphocyte subsets, dendritic cells, and differentially conditioned monocyte-derived macrophages. Proc Natl Acad Sci U.S.A. (1999) 96(9):5215–20. doi: 10.1073/pnas.96.9.5215
11. Joly M, Pinto JM. CXCR4 and CCR5 regulation and expression patterns on T- and monocyte-macrophage cell lineages: implications for susceptibility to infection by HIV-1. Math Biosci (2005) 195(1):92–126. doi: 10.1016/j.mbs.2005.01.002
12. Kruize Z, Kootstra NA. The role of macrophages in HIV-1 persistence and pathogenesis. Front Microbiol (20192828) 10:2828. doi: 10.3389/fmicb.2019.02828
13. Wong ME, Jaworowski A, Hearps AC. The HIV reservoir in monocytes and macrophages. Front Immunol (2019) 10:1435. doi: 10.3389/fimmu.2019.01435
14. Hendricks CM, Cordeiro T, Gomes AP, Stevenson M. The interplay of HIV-1 and macrophages in viral persistence. Front Microbiol (2021) 12:646447. doi: 10.3389/fmicb.2021.646447
15. Veenhuis RT, Abreu CM, Shirk EN, Gama L, Clements JE. HIV Replication and latency in monocytes and macrophages. Semin Immunol (2021) 51:101472. doi: 10.1016/j.smim.2021.101472
16. Valledor AF, Borras FE, Cullell-Young M, Celada A. Transcription factors that regulate monocyte/macrophage differentiation. J Leukoc Biol (1998) 63(4):405–17. doi: 10.1002/jlb.63.4.405
17. Li L, He S, Sun JM, Davie JR. Gene regulation by Sp1 and Sp3. Biochem Cell Biol (2004) 82(4):460–71. doi: 10.1139/o04-045
18. Resendes KK, Rosmarin AG. Sp1 control of gene expression in myeloid cells. Crit Rev Eukaryot Gene Expr (2004) 14(3):171–81. doi: 10.1615/CritRevEukaryotGeneExpr.v14.i3.20
19. Noti JD, Reinemann BC, Petrus MN. Sp1 binds two sites in the CD11c promoter in vivo specifically in myeloid cells and cooperates with AP1 to activate transcription. Mol Cell Biol (1996) 16(6):2940–50. doi: 10.1128/MCB.16.6.2940
20. Ries S, Buchler C, Langmann T, Fehringer P, Aslanidis C, Schmitz G. Transcriptional regulation of lysosomal acid lipase in differentiating monocytes is mediated by transcription factors Sp1 and AP-2. J Lipid Res (1998) 39(11):2125–34. doi: 10.1016/S0022-2275(20)32467-6
21. Langmann T, Buechler C, Ries S, Schaeffler A, Aslanidis C, Schuierer M, et al. Transcription factors Sp1 and AP-2 mediate induction of acid sphingomyelinase during monocytic differentiation. J Lipid Res (1999) 40(5):870–80. doi: 10.1016/S0022-2275(20)32122-2
22. Friedman AD. Transcriptional regulation of granulocyte and monocyte development. Oncogene (2002) 21(21):3377–90. doi: 10.1038/sj.onc.1205324
23. Payton SG, Whetstine JR, Ge Y, Matherly LH. Transcriptional regulation of the human reduced folate carrier promoter c: synergistic transactivation by Sp1 and C/EBP beta and identification of a downstream repressor. Biochim Biophys Acta (2005) 1727(1):45–57. doi: 10.1016/j.bbaexp.2004.11.006
24. Richer E, Campion CG, Dabbas B, White JH, Cellier MF. Transcription factors Sp1 and C/EBP regulate NRAMP1 gene expression. FEBS J (2008) 275(20):5074–89. doi: 10.1111/j.1742-4658.2008.06640.x
25. Kao WY, Briggs JA, Kinney MC, Jensen RA, Briggs RC. Structure and function analysis of the human myeloid cell nuclear differentiation antigen promoter: evidence for the role of Sp1 and not of c-myb or PU.1 in myelomonocytic lineage-specific expression. J Cell Biochem (1997) 65(2):231–44. doi: 10.1002/(sici)1097-4644(199705)65:2<231::aid-jcb8>3.0.co;2-v
26. Hauses M, Tonjes RR, Grez M. The transcription factor Sp1 regulates the myeloid-specific expression of the human hematopoietic cell kinase (HCK) gene through binding to two adjacent GC boxes within the HCK promoter-proximal region. J Biol Chem (1998) 273(48):31844–52. doi: 10.1074/jbc.273.48.31844
27. Rao J, Zhang F, Donnelly RJ, Spector NL, Studzinski GP. Truncation of Sp1 transcription factor by myeloblastin in undifferentiated HL60 cells. J Cell Physiol (1998) 175(2):121–8. doi: 10.1002/(SICI)1097-4652(199805)175:2<121::AID-JCP1>3.0.CO;2-Q
28. Apt D, Watts RM, Suske G, Bernard HU. High Sp1/Sp3 ratios in epithelial cells during epithelial differentiation and cellular transformation correlate with the activation of the HPV-16 promoter. Virology (1996) 224(1):281–91. doi: 10.1006/viro.1996.0530
29. Dharmavaram RM, Liu G, Mowers SD, Jimenez SA. Detection and characterization of Sp1 binding activity in human chondrocytes and its alterations during chondrocyte dedifferentiation. J Biol Chem (1997) 272(43):26918–25. doi: 10.1074/jbc.272.43.26918
30. Tang QQ, Jiang MS, Lane MD. Repressive effect of Sp1 on the C/EBPalpha gene promoter: role in adipocyte differentiation. Mol Cell Biol (1999) 19(7):4855–65. doi: 10.1128/MCB.19.7.4855
31. Billon N, Carlisi D, Datto MB, van Grunsven LA, Watt A, Wang XF, et al. Cooperation of Sp1 and p300 in the induction of the CDK inhibitor p21WAF1/CIP1 during NGF-mediated neuronal differentiation. Oncogene (1999) 18(18):2872–82. doi: 10.1038/sj.onc.1202712
32. Yoo J, Jeong MJ, Kwon BM, Hur MW, Park YM, Han MY. Activation of dynamin I gene expression by Sp1 and Sp3 is required for neuronal differentiation of N1E-115 cells. J Biol Chem (2002) 277(14):11904–9. doi: 10.1074/jbc.M111788200
33. Milagre I, Nunes MJ, Castro-Caldas M, Moutinho M, Gama MJ, Rodrigues E. Neuronal differentiation alters the ratio of sp transcription factors recruited to the CYP46A1 promoter. J Neurochem (2012) 120(2):220–9. doi: 10.1111/j.1471-4159.2011.07577.x
34. Guo H, Cao C, Chi X, Zhao J, Liu X, Zhou N, et al. Specificity protein 1 regulates topoisomerase IIbeta expression in SH-SY5Y cells during neuronal differentiation. J Neurosci Res (2014) 1374–83. doi: 10.1002/jnr.23403
35. Hagen G, Muller S, Beato M, Suske G. Cloning by recognition site screening of two novel GT box binding proteins: A family of Sp1 related genes. Nucleic Acids Res (1992) 20(21):5519–25. doi: 10.1093/nar/20.21.5519
36. Kingsley C, Winoto A. Cloning of GT box-binding proteins: A novel Sp1 multigene family regulating T-cell receptor gene expression. Mol Cell Biol (1992) 12(10):4251–61. doi: 10.1128/mcb.12.10.4251-4261.1992
37. Hagen G, Muller S, Beato M, Suske G. Sp1-mediated transcriptional activation is repressed by Sp3. EMBO J (1994) 13(16):3843–51. doi: 10.1002/j.1460-2075.1994.tb06695.x
38. Terrados G, Finkernagel F, Stielow B, Sadic D, Neubert J, Herdt O, et al. Genome-wide localization and expression profiling establish Sp2 as a sequence-specific transcription factor regulating vitally important genes. Nucleic Acids Res (2012) 40(16):7844–57. doi: 10.1093/nar/gks544
39. Lalonde J, Saia G, Gill G. Store-operated calcium entry promotes the degradation of the transcription factor Sp4 in resting neurons. Sci Signal (2014) 7(328):ra51. doi: 10.1126/scisignal.2005242
40. Saia G, Lalonde J, Sun X, Ramos B, Gill G. Phosphorylation of the transcription factor Sp4 is reduced by NMDA receptor signaling. J Neurochem (2014) 129(4):743–52. doi: 10.1111/jnc.12657
41. Waby JS, Bingle CD, Corfe BM. Post-translational control of sp-family transcription factors. Curr Genomics (2008) 9(5):301–11. doi: 10.2174/138920208785133244
42. Chun RF, Semmes OJ, Neuveut C, Jeang KT. Modulation of Sp1 phosphorylation by human immunodeficiency virus type 1 tat. J Virol (1998) 72(4):2615–29. doi: 10.1128/JVI.72.4.2615-2629.1998
43. Tan NY, Khachigian LM. Sp1 phosphorylation and its regulation of gene transcription. Mol Cell Biol (2009) 29(10):2483–8. doi: 10.1128/MCB.01828-08
44. Chu S. Transcriptional regulation by post-transcriptional modification–role of phosphorylation in Sp1 transcriptional activity. Gene (2012) 508(1):1–8. doi: 10.1016/j.gene.2012.07.022
45. Wu Z, Kim HP, Xue HH, Liu H, Zhao K, Leonard WJ. Interleukin-21 receptor gene induction in human T cells is mediated by T-cell receptor-induced Sp1 activity. Mol Cell Biol (2005) 25(22):9741–52. doi: 10.1128/MCB.25.22.9741-9752.2005
46. Vicart A, Lefebvre T, Imbert J, Fernandez A, Kahn-Perles B. Increased chromatin association of Sp1 in interphase cells by PP2A-mediated dephosphorylations. J Mol Biol (2006) 364(5):897–908. doi: 10.1016/j.jmb.2006.09.036
47. Majello B, De Luca P, Hagen G, Suske G, Lania L. Different members of the Sp1 multigene family exert opposite transcriptional regulation of the long terminal repeat of HIV-1. Nucleic Acids Res (1994) 22(23):4914–21. doi: 10.1093/nar/22.23.4914
48. Rohr O, Marban C, Aunis D, Schaeffer E. Regulation of HIV-1 gene transcription: From lymphocytes to microglial cells. J Leukoc Biol (2003) 74(5):736–49. doi: 10.1189/jlb.0403180
49. Hotter D, Bosso M, Jonsson KL, Krapp C, Sturzel CM, Das A, et al. IFI16 targets the transcription factor Sp1 to suppress HIV-1 transcription and latency reactivation. Cell Host Microbe (2019) 25(6):858–872 e813. doi: 10.1016/j.chom.2019.05.002
50. Bosso M, Prelli Bozzo C, Hotter D, Volcic M, Sturzel CM, Rammelt A, et al. Nuclear PYHIN proteins target the host transcription factor Sp1 thereby restricting HIV-1 in human macrophages and CD4+ T cells. PloS Pathog (2020) 16(8):e1008752. doi: 10.1371/journal.ppat.1008752
51. Collins SJ, Gallo RC, Gallagher RE. Continuous growth and differentiation of human myeloid leukaemic cells in suspension culture. Nature (1977) 270(5635):347–9. doi: 10.1038/270347a0
52. Gallagher R, Collins S, Trujillo J, McCredie K, Ahearn M, Tsai S, et al. Characterization of the continuous, differentiating myeloid cell line (HL-60) from a patient with acute promyelocytic leukemia. Blood (1979) 54(3):713–33. doi: 10.1182/blood.V54.3.713.713
53. Collins SJ, Ruscetti FW, Gallagher RE, Gallo RC. Terminal differentiation of human promyelocytic leukemia cells induced by dimethyl sulfoxide and other polar compounds. Proc Natl Acad Sci U.S.A. (1978) 75(5):2458–62. doi: 10.1073/pnas.75.5.2458
54. Lotem J, Sachs L. Regulation of normal differentiation in mouse and human myeloid leukemic cells by phorbol esters and the mechanism of tumor promotion. Proc Natl Acad Sci U.S.A. (1979) 76(10):5158–62. doi: 10.1073/pnas.76.10.5158
55. Sundstrom C, Nilsson K. Establishment and characterization of a human histiocytic lymphoma cell line (U-937). Int J Cancer (1976) 17(5):565–77. doi: 10.1002/ijc.2910170504
56. Tsuchiya S, Yamabe M, Yamaguchi Y, Kobayashi Y, Konno T, Tada K. Establishment and characterization of a human acute monocytic leukemia cell line (THP-1). Int J Cancer (1980) 26(2):171–6. doi: 10.1002/ijc.2910260208
57. Harrich D, Garcia J, Wu F, Mitsuyasu R, Gonazalez J, Gaynor R. Role of SP1-binding domains in in vivo transcriptional regulation of the human immunodeficiency virus type 1 long terminal repeat. J Virol (1989) 63(6):2585–91. doi: 10.1128/jvi.63.6.2585-2591.1989
58. Auwerx J. The human leukemia cell line, THP-1: A multifacetted model for the study of monocyte-macrophage differentiation. Experientia (1991) 47(1):22–31. doi: 10.1007/BF02041244
59. Koeffler HP. Induction of differentiation of human acute myelogenous leukemia cells: Therapeutic implications. Blood (1983) 62(4):709–21. doi: 10.1182/blood.V62.4.709.709
60. Lopez-Rodriguez C, Zubiaur M, Sancho J, Concha A, Corbi AL. An octamer element functions as a regulatory element in the differentiation-responsive CD11c integrin gene promoter: OCT-2 inducibility during myelomonocytic differentiation. J Immunol (1997) 158(12):5833–40.
61. Boyum A. Isolation of mononuclear cells and granulocytes from human blood. isolation of monuclear cells by one centrifugation, and of granulocytes by combining centrifugation and sedimentation at 1 g. Scand J Clin Lab Invest Suppl (1968) 97:77–89.
62. Gmelig-Meyling F, Waldmann TA. Separation of human blood monocytes and lymphocytes on a continuous percoll gradient. J Immunol Methods (1980) 33(1):1–9. doi: 10.1016/0022-1759(80)90077-0
63. Graziani-Bowering GM, Graham JM, Filion LG. A quick, easy and inexpensive method for the isolation of human peripheral blood monocytes. J Immunol Methods (1997) 207(2):157–68. doi: 10.1016/S0022-1759(97)00114-2
64. Grant C, Nonnemacher M, Jain P, Pandya D, Irish B, Williams SC, et al. CCAAT/enhancer-binding proteins modulate human T cell leukemia virus type 1 long terminal repeat activation. Virology (2006) 348(2):354–69. doi: 10.1016/j.virol.2005.12.024
65. Millhouse S, Krebs FC, Yao J, McAllister JJ, Conner J, Ross H, et al. Sp1 and related factors fail to interact with the NF-kappaB-proximal G/C box in the LTR of a replication competent, brain-derived strain of HIV-1 (YU-2). J Neurovirol (1998) 4(3):312–23. doi: 10.3109/13550289809114532
66. Filtz TM, Vogel WK, Leid M. Regulation of transcription factor activity by interconnected post-translational modifications. Trends Pharmacol Sci (2014) 35(2):76–85. doi: 10.1016/j.tips.2013.11.005
67. Gilmour J, O'Connor L, Middleton CP, Keane P, Gillemans N, Cazier JB, et al. Robust hematopoietic specification requires the ubiquitous Sp1 and Sp3 transcription factors. Epigenet Chromatin (2019) 12(1):33. doi: 10.1186/s13072-019-0282-9
68. Arya SK, Guo C, Josephs SF, Wong-Staal F. Trans-activator gene of human T-lymphotropic virus type III (HTLV-III). Science (1985) 229(4708):69–73. doi: 10.1126/science.2990040
69. Cohen EA, Terwilliger EF, Jalinoos Y, Proulx J, Sodroski JG, Haseltine WA. Identification of HIV-1 vpr product and function. J Acquir Immune Defic Syndr (1990) 3(1):11–8.
70. McAllister JJ, Phillips D, Millhouse S, Conner J, Hogan T, Ross HL, et al. Analysis of the HIV-1 LTR NF-kappaB-proximal sp site III: Evidence for cell type-specific gene regulation and viral replication. Virology (2000) 274(2):262–77. doi: 10.1006/viro.2000.0476
71. Kamine J, Subramanian T, Chinnadurai G. Sp1-dependent activation of a synthetic promoter by human immunodeficiency virus type 1 tat protein. Proc Natl Acad Sci U.S.A. (1991) 88(19):8510–4. doi: 10.1073/pnas.88.19.8510
72. Wang L, Mukherjee S, Jia F, Narayan O, Zhao LJ. Interaction of virion protein vpr of human immunodeficiency virus type 1 with cellular transcription factor Sp1 and trans-activation of viral long terminal repeat. J Biol Chem (1995) 270(43):25564–9. doi: 10.1074/jbc.270.43.25564
73. Sawaya BE, Khalili K, Mercer WE, Denisova L, Amini S. Cooperative actions of HIV-1 vpr and p53 modulate viral gene transcription. J Biol Chem (1998) 273(32):20052–7. doi: 10.1074/jbc.273.32.20052
Keywords: HIV-1 LTR, Sp, G/C box array, transcription, monocytes
Citation: McAllister JJ, Dahiya S, Berman R, Collins M, Nonnemacher MR, Burdo TH and Wigdahl B (2022) Altered recruitment of Sp isoforms to HIV-1 long terminal repeat between differentiated monoblastic cell lines and primary monocyte-derived macrophages. Front. Virol. 2:971293. doi: 10.3389/fviro.2022.971293
Received: 16 June 2022; Accepted: 14 October 2022;
Published: 01 November 2022.
Edited by:
Akio Adachi, Kansai Medical University, JapanReviewed by:
Carine M. C. Van Lint, Université libre de Bruxelles, BelgiumCopyright © 2022 McAllister, Dahiya, Berman, Collins, Nonnemacher, Burdo and Wigdahl. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Brian Wigdahl, Ync0NUBkcmV4ZWwuZWR1
†Present address: John J. McAllister, Anesthesia Company LLC, Luminis Health, Annapolis, MD, United States
‡These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.