
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Virol. , 30 August 2022
Sec. Fundamental Virology
Volume 2 - 2022 | https://doi.org/10.3389/fviro.2022.932187
This article is part of the Research Topic HIV/SIV basic research update View all 10 articles
 Yu Kaku1†
Yu Kaku1† Kaho Matsumoto1†
Kaho Matsumoto1† Takeo Kuwata1
Takeo Kuwata1 Hasan Md Zahid1
Hasan Md Zahid1 Shashwata Biswas1
Shashwata Biswas1 Miroslaw K. Gorny2
Miroslaw K. Gorny2 Shuzo Matsushita1*
Shuzo Matsushita1*The V3 loop of the human immunodeficiency virus type 1 (HIV-1) envelope protein (Env) is one of the conserved immunogenic regions targeted by neutralizing antibodies (nAb). Two different binding modes of anti-V3 abs have been reported in studies using two V3 mimotopes: the ladle-type and cradle-type. We previously isolated a ladle-type nAb, 1C10, that potently and broadly neutralized clade B viruses. Despite its potent neutralization activity, 1C10 possesses no unique features in its amino acid sequence. We hypothesized that the neutralization potency of 1C10 is derived from its antigen-binding characteristics, which are not a consequence of the two previously reported binding modes of anti-V3 nAbs. To analyze epitope-paratope interactions between 1C10 and the V3 loop, we produced five anti-idiotypic antibodies (anti-Id abs) from mice immunized with 1C10 nAb. The idiotopes of the anti-Id Abs on the 1C10 heavy chain were estimated by alanine scanning, germline reversion mutagenesis, and a 1C10 sibling clone. Next-generation sequencing combined with homology modeling revealed contact between R315 at the tip of the V3 loop and 1C10 by D53 of CDRH2 and Phe/Asp of CDRH3. These amino acids were enriched in the anti-Id-ab-reactive B cell receptors encoded by the IGHV3-30 gene. We also found that 20% of HIV-infected individuals had abs specific to the anti-Id abs, as well as both of the V3 mimotopes, that did not respond to the linear V3 peptide. Our findings showed that the anti-Id abs induced by 1C10 recognized a key amino acid formation essential for steric interactions between the ladle-type nAb and the V3 loop. We also revealed the coexistence of anti-V3 ab reactivity to V3 loop mimotopes and to the anti-Id abs in HIV-positive individuals.
Eliciting potent neutralization antibodies (nAbs) is the ultimate goal of the development of vaccines against human immunodeficiency virus type 1 (HIV-1). To date, various vaccines have been developed to induce nAbs targeting the conserved regions of the virus (1–7). The V3 loop of the HIV-1 envelope protein is a target for nAbs, of which glycan epitopes are specifically targeted by broadly neutralizing antibodies (bnAbs) and glycan-independent epitopes are recognized by most anti-V3 antibodies (abs) (8–14). Two types of the latter epitopes have been identified thus far, ladle-like and cradle-like epitopes, which are recognized by V3 mimotopes (15–17). In the binding of anti-V3 Abs to the two epitopes, ladle-type abs recognize the hydrophobic face of the circlet region, in which Gly-Phe-Gly-Arg/Gln (GPGR/Q) at the tip of the V3 loop is found along heavy chain complementarity-determining region 3 (CDRH3) (18–20). Conversely, cradle-type abs attach to the epitope by binding to the band and circlet regions on both sides of the V3 loop. In other words, the former type of binding mode requires a long CDRH3, which forms the handle of the “ladle”, and the latter type requires a binding cavity consisting of both a heavy chain and a light chain, which form the “cradle.” In fact, cradle-type abs are predominantly derived from the IGHV5-51 gene paired with light chain variable region (VL) lambda gene, and are detected in vaccinees’ plasma as non- or weak-nAbs (15, 16, 21, 22). However, it remains unclear if ladle-type abs possess any unique structural features other than the longer-length CDRH3, especially those that are derived from the same immunoglobulin gene segment and can be elicited by vaccination.
From an elite controller, we previously isolated 1C10, which is a potent ladle-type anti-V3 Ab capable of neutralizing 80% of clade B viruses, including 30% of primary isolates of a major subtype in Japan (23). Despite its potent and broad-spectrum neutralization ability, 1C10, which is encoded by the major immunoglobulin gene IGHV3-30, has features common to many antibody types, including an 18-amino-acid-long CDRH3 region. In contrast, bnAbs possess a higher somatic hypermutation (SHM) rate, longer length CDRH3, and novel nucleotide insertions or deletions compared with most antibodies (12, 24–32). Although the details of 1C10’s neutralization potency have not yet been revealed, the induction of 1C10 requires no specific conditions and can be elicited by a vaccine (23, 33, 34). In this study, we generated anti-idiotypic abs that bound to the paratope (or binding cavity) of 1C10 to analyze the key characteristics that make 1C10 a potent anti-V3 nAb. We also aimed to discover the steric structures shared by subgroups of ladle-type abs.
We injected 200 μg of 1C10 into the tail vein of each 6-week-old BALB/c mouse in a prime immunization and the same amount into the peritoneal cavity in further immunizations. After five immunizations at 2-week intervals, the mice were sacrificed and the spleens were extracted. We isolated B220+IgM-IgG+ B cells positive for 1C10 as single cells from the spleens by FACS AriaII (BD, NJ, USA). Following the method reported previously (35), we amplified variable regions of IgG heavy and light chains by RT-PCR and inserted each of the regions into respective expression vectors by means of murine IgG-specific primer pairs.
After incubating a Maxisorp plate (Invitrogen, MA, USA) coated with 0.2 μg NNT-20 JR-FL overnight, which is V3 peptide of HIV-1 clade B strains JR-FL consisting of 20 amino acids (AAs) started from NNT (NNTRKSIHIGPGRAFYTTGE), the plate was blocked by adding 0.1% BSA/PBS for 30 minutes. Anti-Id abs were dispensed into each well at various concentrations from 1 μg to 39 ng and incubated for 30 minutes followed by the addition of 0.1 ng biotinylated 1C10 and 1 hour of incubation. Binding of 1C10 to NNT-20 JR-FL was detected by measuring the OD value (405 nm) following the addition of streptavidin-HRP and 2,2’-azino-bis(3-ethylbenzothiazoline)-6-sulfonic acid (ABTS).
Germline reversion of 1C10 was achieved by AA-replacement of the 1C10 heavy chain variable region (VH) with IGHV3-30 AAs by overlap-extension PCR as we previously described (36). Purified germline 1C10 was dispensed onto a Maxisorp plate (Invitrogen, MA, USA) at 2 μg/well and incubated overnight. After blocking by the addition of 0.1% BSA/PBS, biotinylated anti-Id abs were added to each well as serially diluted concentrations from 1 μg to 39 ng and 1 hour of incubation. Binding of anti-Id abs to germline-reverted 1C10 was detected by measuring the OD value at a wavelength of 405 nm following the addition of streptavidin-HRP (ThermoFisher, MA, USA) and ABTS (Roche, Switzerland). The binding ELISA was also performed to assess the effect of alanine-scanning mutagenesis of the 1C10 CDRH3 contact residues on binding to the anti-Id abs.
Single-cell sorting was performed to isolate abs reactive to the anti-Id ab clones, as previously reported (35), from an HIV-infected patient who was an elite controller and had a plasma viral load constantly below the detection level. Briefly, after CD3-CD8-CD14-cells were negatively sorted from peripheral blood mononuclear cells (PBMCs) using a MojoSort magnet (BioLegend, CA, USA), CD19+CD27+7AAD-IgM-IgG+ B cells that bound to the anti-Id abs were sorted as single cells. The IgG variable regions from the sorted cells were amplified as described above and used for both sequence analysis and cloning into expression vectors for functional analysis. The sequence of the variable region was analyzed by International ImMunoGeneTics (IMGT) and Basic Local Alignment Search Tool (BLAST) searching. Functional analysis was performed using binding ELISA.
CD19+CD27+IgM-IgG+ B cells reactive to anti-Id abs #87 and #102 were isolated by single-cell sorting as a biased group and CD19+CD27+IgM-IgG+B cells were negatively sorted by MojoSort Magnet (BioLegend) as an unbiased group from PBMCs of the elite controller. Single-cell RNA-seq was performed using 10X Chromium (10X Genomics, CA, USA) following the manufacturer’s protocol.
AlphaScreen (PerkinElmer, Switzerland) was performed to examine the binding between either 1C10 or 3-32 and the looped V3 mimotopes. Mixtures of 2 μg/ml and 20 μg/ml, respectively, were incubated for 40 minutes room temperature. Binding was detected by anti-hIgG acceptor beads and streptavidin donor beads 40 minutes after the addition of the analyte mixture.
Anti-Id-ab Fabs and plasma IgGs obtained from HIV-positive patients’ plasma at 2 μg/ml and a 1:1000 dilution, respectively, were incubated for 40 minutes room temperature. Binding was measured by anti-6x His acceptor beads and protein A donor beads 40 minutes room temperature after the addition of the analyte mixture.
Correlation coefficients (CC) were calculated by Pearson’s correlation. The network map of CC among the contact residues was drawn using NetworkX.
A 1C10 homology model was constructed as described in our previous report (36). In detail, we selected four ladle abs, three unclassified abs, and two cradle abs for which the crystal structures were available as templates for homology modeling of 1C10. Homology models of 1C10 were generated with the SWISS-model (37). First, screening was completed using rigid-body docking simulations with a native V3 loop in zdock (38). Second, flexible-docking simulation by Rossie (39) was performed on the first and second ranked ladle abs and unclassified abs by zdock score. Among the four homology models, we chose the 1C10 homology model using 1334 as the template based on the highest Rossie score of 1369.64. A 1C10-V3 loop complex model was created using 1334 (PDB: 6DB7) and the JRFL V3 loop sequence. Structure models were depicted with Pymol (40), and interactions between the heavy chain and R315 at the tip of the V3 loop in the model were detected using AppA (41). The interface residues of the heavy chain and the V3 loop were identified with the Pymol script, InterfaceResidues.
Using mice immunized with 1C10, we created anti-Id abs against 1C10 as mouse-human chimeric antibodies consisting of murine variable regions and human constant regions. Of these, anti-Id abs specific to the paratope of 1C10 were selected by competitive inhibition against biotinylated anti-V3 abs showing binding to NNT-20 JR-FL (NNTRKSIHIGPGRAFYTTGE) in ELISA. Five different anti-Id ab clones showed interference, not between the V3 peptide and other ladle-type anti-V3 abs, such as KD-247, 5G2, 1D9, 19F8, and 717G2, but between the V3 peptide and 1C10 (Figure 1 and Supplementary Table 1).
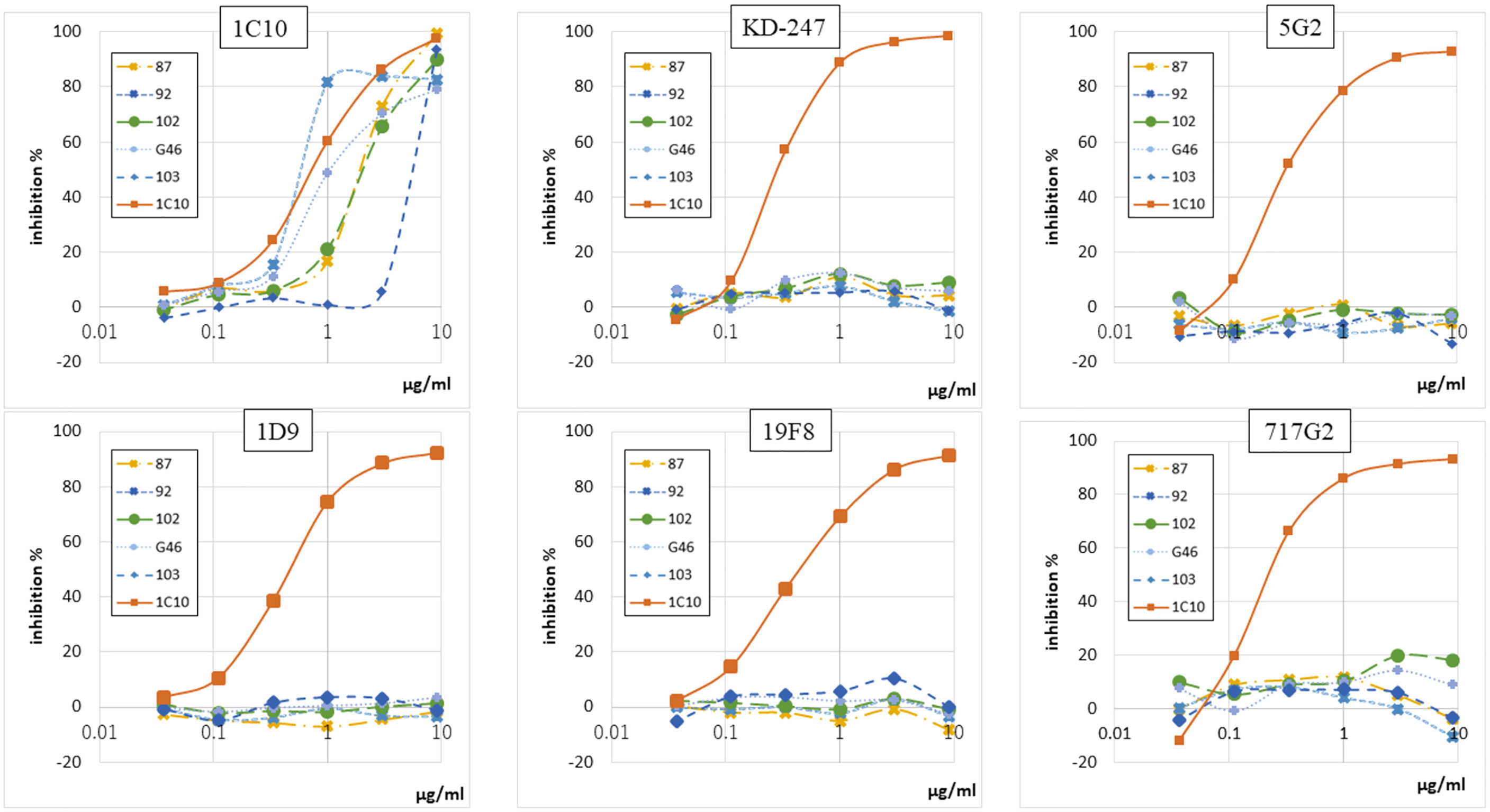
Figure 1 Five anti-Id abs inhibited only 1C10 among six anti-V3 abs. Binding-inhibition ability of five anti-Id abs (87, 92, 102, G46, and 103), which interfere with interactions between anti-V3 abs and NNT-20, was determined by binding-inhibition ELISA using five anti-V3 abs (1C10, KD-247, 5G2, 1D9, 19F8, 717G2) conjugated to biotin. Unconjugated 1C10 was used as a control binding inhibitor for all anti-V3 abs.
We first assessed the region of the 1C10 paratope targeted by the anti-Id abs using 1C10 mutants. For this purpose, we constructed germline-reverted forms of the 1C10 heavy chain VH (Supplementary Figure 1, Figure 2A). We found that all five anti-Id abs bound to the germline-reverted 1C10 mutants in various patterns but did not react to the inferred germline. The result indicated that #87, #102, and #103 had decreased binding potency to the CDR2 mutation, #92 had decreased binding potency to the FR3 mutation, and #G46 had decreased binding potency to the CDR1 mutation (Figure 2B).
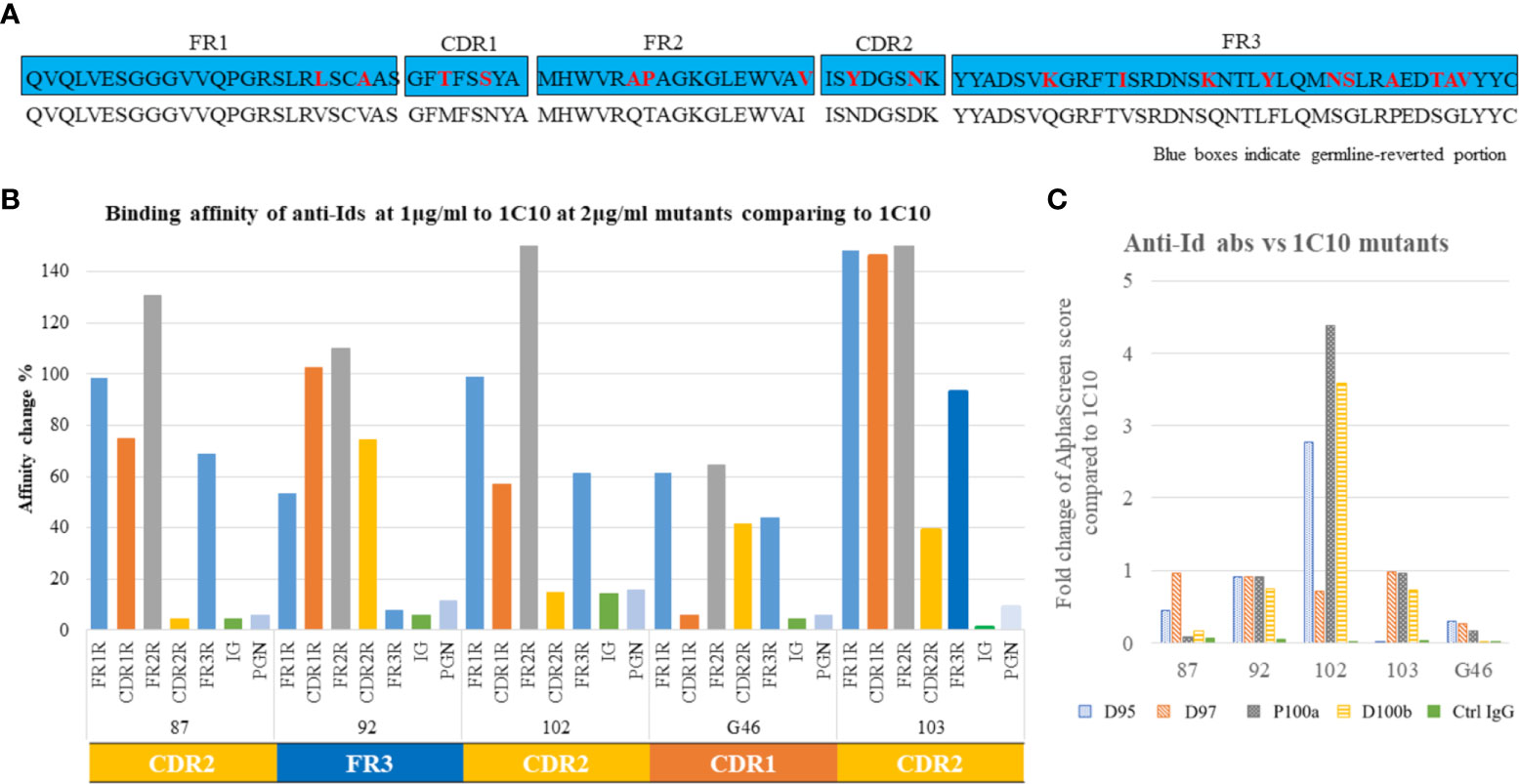
Figure 2 Idiotope classification. Germline reversion of 1C10 was created by overlap PCR to replace five portions of the heavy chain variable region (VH), including framework regions (FR)1-3 and complementarity determining regions (CDR)1-2, with their germline amino acid (AA) sequences. Germline sequences are shown in blue boxes, with replaced AAs as red letters, and the 1C10 sequence is shown below the blue box. (A) Binding of anti-Id abs to germline-reverted 1C10, including inferred germline (IG) and human polyclonal IgG (Ctrl IgG), was tested by binding ELISA and expressed as percentage affinity change, calculated as a relative value based on binding to 1C10. The binding patterns of the anti-Id abs resulted in three types: CDR1-, CDR2-, and FR3-specific types. (B) Alanine scanning mutagenesis was applied to four contact residues of 1C10 CDRH3 (D95, D97, P100a, and D100b). Binding ELISA was used to examine the binding of the anti-Id abs to the 1C10 mutants containing alanine (C).
We next assessed the effects of alanine substitutions at the contact residues of 1C10 CDRH3 that we previously predicted (36) on the anti-Id abs by binding ELISA. Among the contact residues (D95, D97, P100a, and D100b), interactions were observed between D95 and #103, D97 and #102, either or both P100a and D100b, and #87 and #92. While most of the alanine substitutions resulted in a reduced or unchanged interaction strength with anti-Id abs, all substitutions, other than D95, enhanced the affinity of #102 to 1C10 (Figure 2C).
To identify the conformation shared by abs in the 1C10 lineage, antibody-expressing B cells (CD19+CD27+IgM-IgG+) were sorted as single cells using all five anti-Id ab clones from PBMCs of the HIV-1-positive patient, who produced 1C10. From 326 antibody-expressing B cells, we extracted 35 antibody clones, including one 1C10 sibling clone, 3-32. The same AA sequence was shared between 3-32 and 1C10 CDR3, other than 23 AAs of the heavy chain and one AA of the light chain. This also corresponds to the fact that both ab clones are encoded by the same V gene and have the same CDR3 length (Figure 3A). The binding of 3-32 to both looped V3 peptides and the linear V3 peptide (NNT-20) significantly decreased in strength compared with that of 1C10 (Figure 3B).
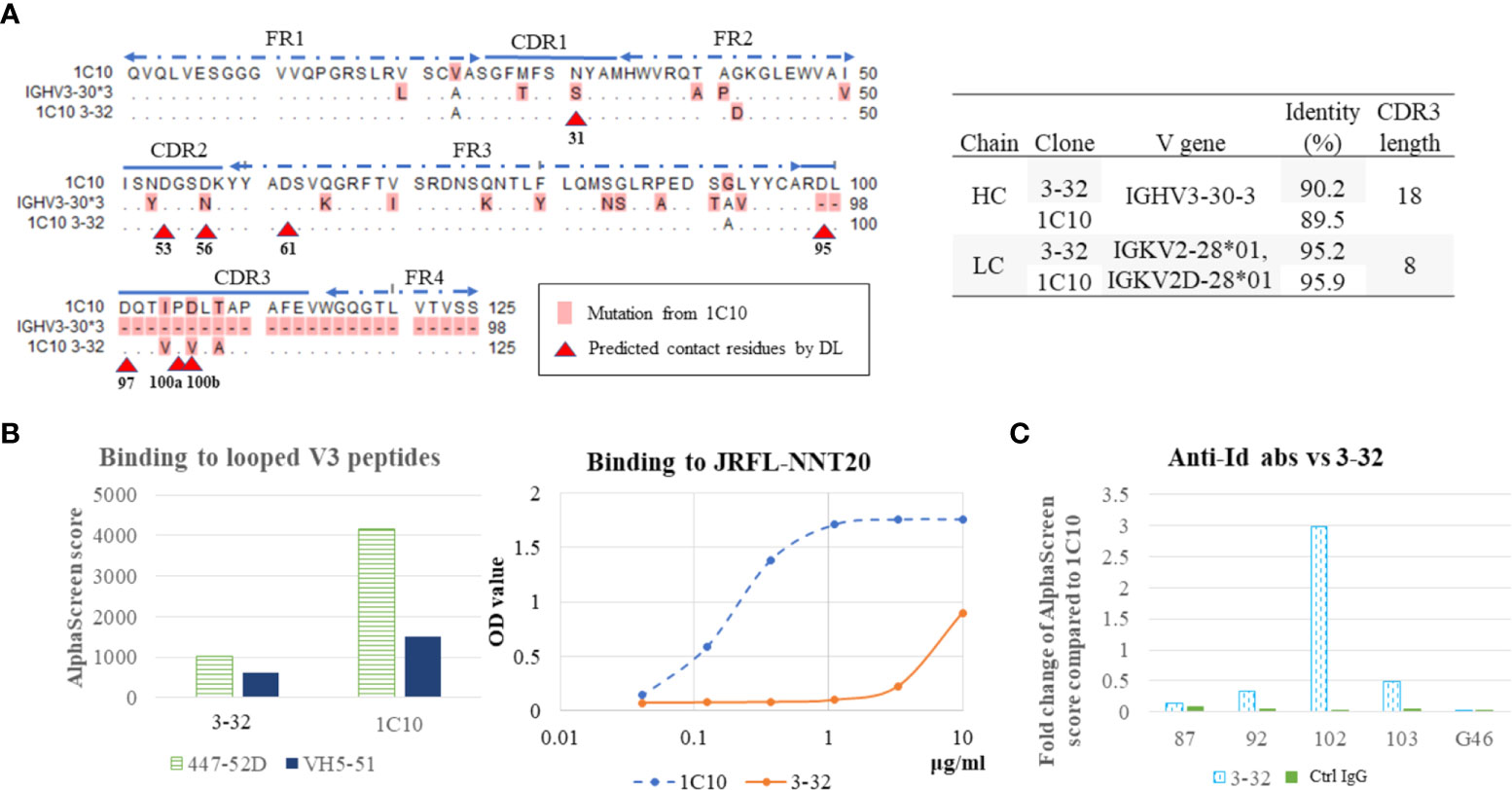
Figure 3 Antibody of 1C10 lineage bound weakly to V3 loops. Amino acid (AA) sequence alignment of heavy chain variable regions of 1C10, germline (IGHV3-30), and a sibling antibody of 1C10 lineage (3-32). The different AAs are highlighted as pink, and predicted contact residues detected by deep learning (DL) are indicated by red arrow heads. Gene usage of the two antibody clones was analyzed with the Basic Local Alignment Search Tool (BLAST) and results are in the table on the right (A). Binding of 1C10 and 3-32 to two looped V3 mimotopes (447-52D and VH5-51) was tested by AlphaScreen, and binding to the linear V3 loop peptide (JRFL-NNT20) was assayed with ELISA (B). Interaction between anti-Id antibodies and 3-32 were examined using AlphaScreen (C).
According to our previous prediction (36), there is one contact residue in CDRH1 (N31), two contact residues in CDRH2 (D53 and D56), and one contact residue in FR3 (D61), in addition to the four contact residues in CDRH3. Of these positions, 3-32 possessed N31 and D56, which are mutated from germline AAs, as in 1C10, and V100b, which is different from the D100b of 1C10 (Figure 3A). Furthermore, 3-32 enhanced the binding of #102, despite the decreased interactions with the other four anti-Id ab clones (Figure 3C).
We analyzed the idiotopes that were shared by abs produced by an individual who produced 1C10 that were targeted by the anti-Id abs. On the basis of idiotopic analysis of 1C10 VH, we selected two CDRH2-specific anti-Id abs for NGS analysis, #87 and #102, with opposing affinities to mutations of the contact residues. We isolated CD2-CD14-CD16-CD36-CD43-Cd235a-B cells (unbiased B cells) as total B cells with magnet beads for analysis of the whole BCR repertoire of the individual. We also sorted B cells (CD19+CD27+IgM-IgG+) expressing idiotypic abs capable of binding to #87 and #102 to analyze the BCRs expressed on the responder B cells (biased B cells). Single-cell RNA-seq of BCRs was performed on 21,179 BCRs of unbiased B cells and 2,164 BCRs of biased B cells by 10X chromium, and we found 1,008 CDRH3s and 1,253 CDRH3s from each BCR group. In the IGHV3 BCR, we focused on five Asp residues and one Asn residue at the same positions as the predicted contact residues of 1C10 shown in Figure 3A. Of interest, Phe/Asp (PD) at any place in CDRH3 were analyzed as residues corresponding to the P100a and D100b of the 1C10 CDRH3. We obtained BCRs containing 35 and 64 N31, 165 and 212 D53, 4 and 5 D56, 138 and 209 D95, and 9 and 19 PD from the unbiased and biased groups, respectively (Figure 4A). In addition to CDRH3 length and the mutation ratio of the IGHV gene, IGHV3 usage was also increased in the biased group compared to the unbiased group (55.7% vs 32.8%) (Figure 4B).
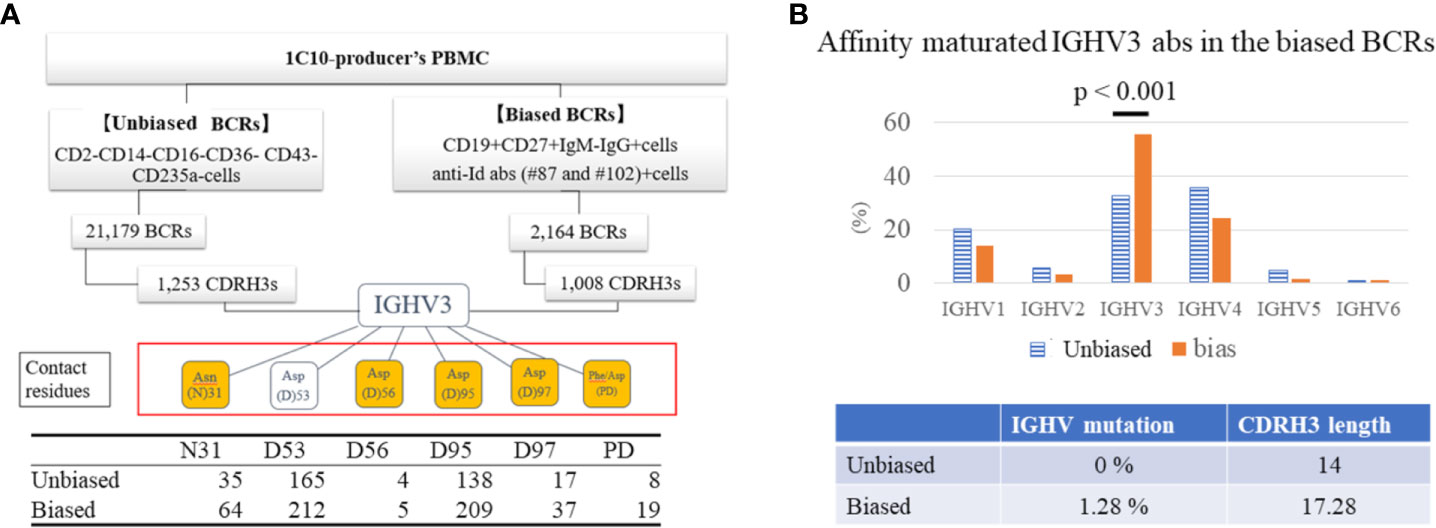
Figure 4 IGHV3 BCR responded to anti-Id abs. BCRs were analyzed in CD2-CD14-CD16-CD36-CD43-Cd235a B cells (unbiased BCRs) as total BCRs and in CD19+CD27+IgM-IgG+ B cells with response to anti-Id ab #87 and #102 (biased BCRs) respectively sorted from an elite controller. We obtained 1,253 CDRH3s from unbiased BCRs and 1,008 CDRH3s from biased BCRs. Finally, we analyzed five aspartic acid (Asp) residues and one asparagine (Asn) residue at the same positions as contact residues of 1C10 in IGHV3 abs. Amino acids (AAs) in orange boxes are mutated AAs from germline, of which N31 is located in CDR2, and sequential phenylalanine (Phe) and Asp residues (PD) are considered mutated AAs corresponding to P100a and D100b. The table at the bottom shows the number of IGHV3 CDRH3 regions containing each residue (A). Unbiased and biased BCRs were compared for genes encoding immunoglobulin heavy chain variable (IGHV) regions, mutation ratio of AAs of IGHV, and length of CDRH3 (B).
Although there were no correlations among the contact residues, once both BCR groups were classified by the possession of a single contact residue, several correlations appeared among the other contact residues. In the unbiased group, D56 and D97, and D53 and D95 correlated with the possession of N31 and D95, respectively. In the biased group, D53 and D97 were associated with PD when one of the two residues existed. Furthermore, in biased BCRs containing either D56 or PD in their CDRH3 region, N31 and D95 were correlated (Figure 5, Supplementary Figure 2). The results indicate the coexistence of these AAs in individual BCRs of each group; the AA pairs in the biased group, especially, implied the presence of epitopes for the anti-Id abs.
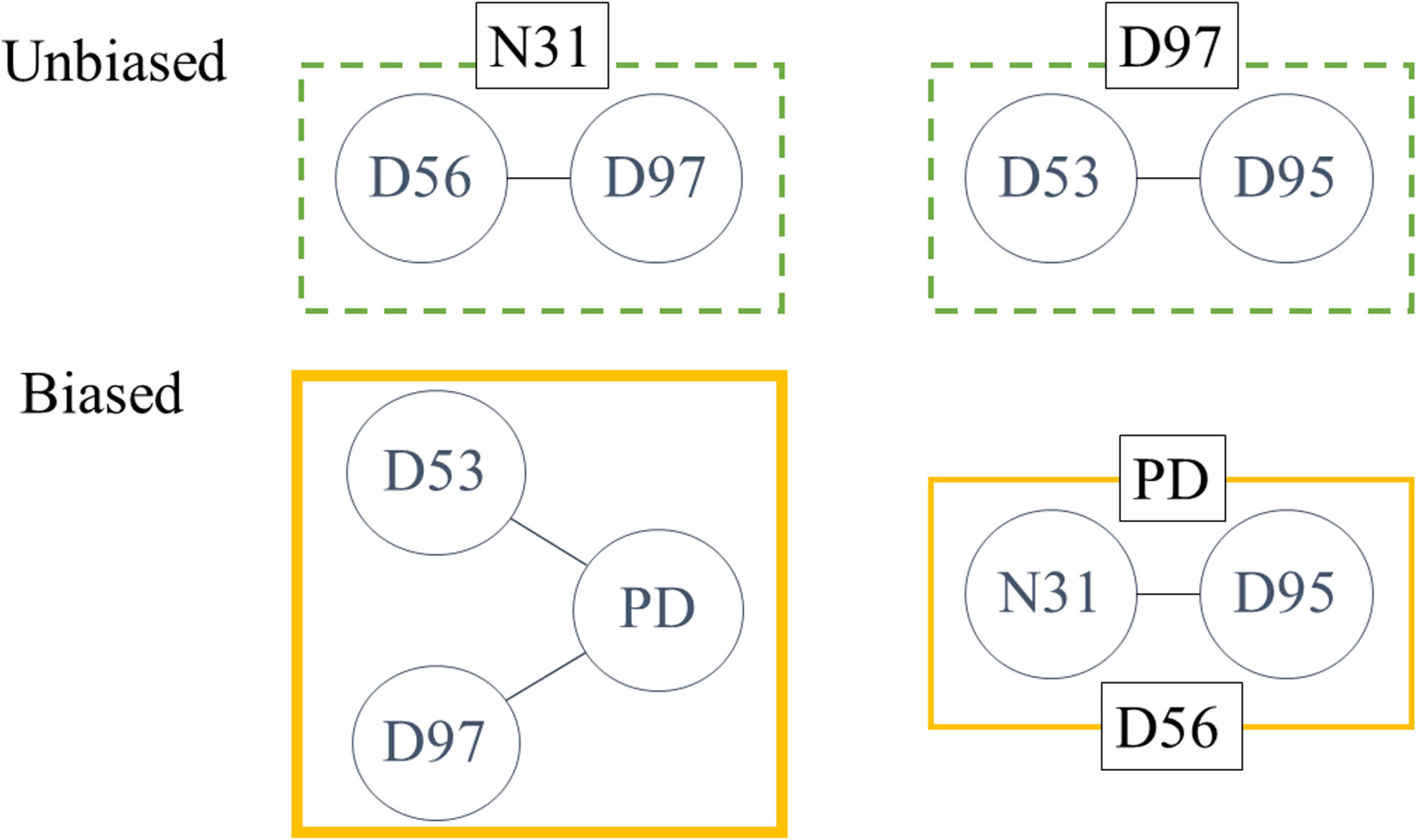
Figure 5 Correlated AAs in both samples. Within six subgroups of both unbiased and biased BCRs classified using one of the contact residues (N31, D53, D56, D95, D97, and PD), correlations among other contact residues were calculated by Pearson’s correlation. Two correlations were detected in subgroups containing either N31 or D97 in unbiased BCRs. In biased BCRs, D53 and D97 were correlated to PD when the subgroup was chosen by the other residue. The correlation between N31 and D95 was seen in both PD and D56 subgroups. Amino acids are annotated by Kabat numbering.
We selected a ladle-type ab, 1334, as a template for homology modeling based on its zdock score and Rossie score. The V and D segments of 1C10 and 1334 are encoded by IGHV3-30 and IGHV1-3, and IGHD2-21 and IGHD3-9, respectively, while the J segments of both antibodies are encoded by IGHJ4. The root mean square deviation of the 1C10 homology model and 1334 was 0.315, which was the lowest value among ladle-type abs, though it was higher than the unclassified and cradle-type abs. This difference was attributed to the longer CDRH3 of the ladle-type abs compared with other ab types (Supplementary Table 2, Figure 6A).
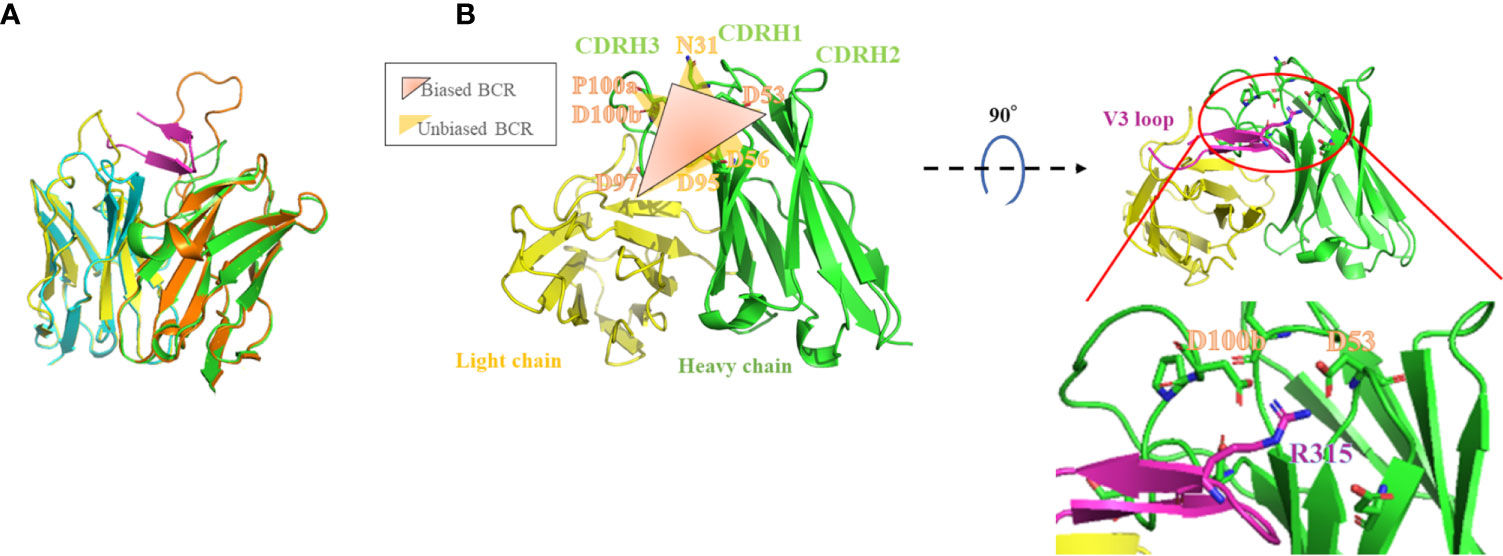
Figure 6 1C10 homology model illustrates contact residues on the adapted structure located close to the V3 tip region. The crystal structure of 1334 (PDB: 6db7) is superposed onto the 1C10 homology model. Heavy and light chains of 1334 are drawn as orange and light blue ribbons, respectively, and those of 1C10 are light green and light blue, respectively, and the V3 loop is magenta (A). The structures composed by the correlated amino acids (AAs) in biased BCRs and unbiased BCRs are drawn in the 1C10 homology model as graduated dark orange triangles consisting of D53, D97, and PD, and yellow triangles consisting of N31, D56, and D97, and D53, D95, and D97, respectively (left). The positions of D53 of 1C10 CDRH2, D100b of 1C10 CDRH3, and R315 of the V3 loop are also depicted in the homology model (right) (B). AAs are annotated by Kabat numbering.
The 1C10-homology model constructed using SWISS-model exhibited a negatively charged binding cavity consisting of both heavy and light chains. More than half (57.0%) of this binding pocket is formed by the heavy chain, especially by CDRH3 (23.9%), and the remaining area (42.9%) is formed mainly by CDRL1 (23.2%) (data not shown).
In this model, correlated contact residues detected by NGS analysis formed a part of the 1C10 binding pocket. In the complex model with the V3 loop, the R315 at the tip of the V3 loop was placed close to D53 or D100b of PD, which were correlated AAs in the biased group (Figure 6B). Interactive contact analysis of these close AAs by AppA also suggested the formation of strong interactions comprising hydrogen bonds, VdW interactions, and ionic interactions. This is comparable to the binding of 1334 to the V3 loop, which uses CDRH2, including N52, D53, and D54, and CDRH3, including D99. Intriguingly, the D99 of 1334 CDRH3 comprises a PD sequence with P98 (Figure 7, Table 1).

Figure 7 Contact residues between the V3 loop and 1C10 and 1334. Contact residues of the 1C10 heavy chain are numbered with down forward arrow heads and those of 1334 are indicated by asterisks. Different amino acids are highlighted in pink, and the same residues are shown by dots. Amino acids are annotated by Kabat numbering. The single asterisk and the double asterisk indicate the elite controller who produced 1C10 and a healthy donor, respectively.
We tested the polyclonal ab responses of HIV-positive patients’ plasma samples and anti-Id ab binding to V3 peptides with AlphaScreen. We used the linear V3 peptide and cyclic V3 mimotopes (447-D and VH5-51 type peptides), which were reported to be involved in the differentiation into the two binding modes of anti-V3 abs (cradle-type and ladle-type) (15). Among the 44 plasma samples, there were 12 (27.2%) with reactivity to the anti-Id abs mixture, 10 (22.7%) with reactivity to the cyclic V3 peptides, and 5 (11.3%) with reactivity to the linear peptide. No correlation in reactivity was found between anti-Id abs and the linear peptide (NNT-20 JR-FL). Responders to the anti-Id abs also bound to the cyclic peptides (Figure 8). Interestingly, the plasma IgG responses of the responders to each V3 mimotope were equally strong, other than the response of the elite controller who produced 1C10. This result suggests that the anti-Id abs form a structure similar to the steric structure shared by both cyclic V3 mimotopes but not by the linear V3 peptide.
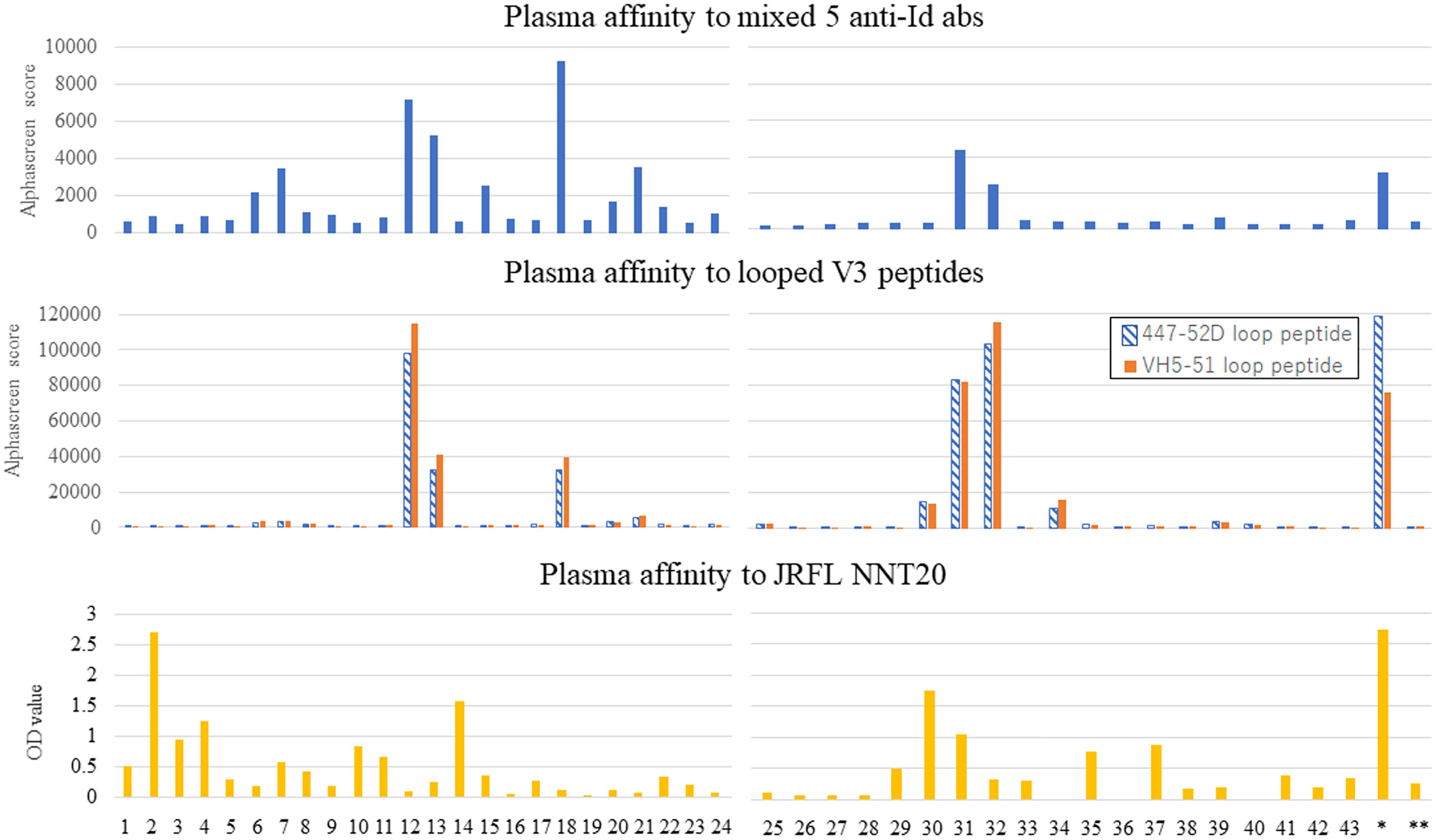
Figure 8 Affinity of 44 patients’ plasma to anti-Id abs correlated to that to looped V3 peptides. Plasma samples from 44 HIV-positive patients were collected to investigate the binding of IgG in sera to a mixture of the five anti-Id abs, the looped V3 peptides (447-52D and VH5-51), and a linear V3 peptide (JRFL NNT20). The former two affinities were examined by AlphaScreen, and the latter affinity was shown by ELISA.
The purpose of this study was using anti-Id abs to investigate the structural features of 1C10 as a potent ladle-type anti-V3 nAb without a long CDRH3 and as an ab originated from the IGHV3 gene. Our data demonstrated that components of the 1C10 paratope shared among IGHV3 abs corresponded to the response to the V3 loop. For this purpose, we generated five anti-Id antibodies specific for the 1C10 paratope without cross-reactivity against other anti-V3 abs. The results of binding to germline-reverted 1C10s revealed that the five anti-Id ab clones should have unique steric structures that consist of at least two epitopes: an epitope bound by 1C10 contact-residue-like AAs on CDRH2 and CDRH3 and an epitope shared with the cyclic V3 loop of a mimotope. The anti-Id abs were specific to 1C10 and predominantly recognized the idiotopes on CDRH2. In addition, alanine replacement at contact residues also showed the existence of the idiotopes of 1C10 CDRH3 based on changes to the anti-Id ab binding potency. More evidence was provided by a 1C10 sibling clone showing many different AAs of CDR3 in 3-32, especially N31S, D56N, and D100bD, at contact residues, leading to a loss of affinity to the V3 loop and alterations to anti-Id ab recognition (Figures 4A, B). Our NGS data suggested that the anti-Id abs recognized the same AAs at the same positions or order as the contact residues of 1C10 CDRH3. In particular, D53 of CDRH2 correlated to D97 and PD of CDRH3 in the biased IGHV3 BCRs. Other combinations were found in D56 of CDRH2 or PD, N31 of CDRH1, and D95 of CDRH3. The homology model postulated strong interactions between D53 or D100b and R315, which is known to be a conserved immunogenic AA on the tip of the clade B virus V3 loop (11). The crystal structure determined for 1334 yielded evidence for the involvement of D53 and PD in contact between the V3 loop and ladle-type abs. This also gave rise to our interest in the interaction between 1334 and the anti-Id ab clones, which should be explored in more detail.
In the binding assay, 27.2% (12/44) of HIV-infected patients were shown to possess plasma IgGs reactive to the anti-Id abs, of which 75% (9/12) also bound to both types of V3 mimotopes equally. Interestingly, only the elite controller, who produced 1C10, had plasma IgG that bound to the NNT-20 JRFL peptide, namely the linear peptide, and preferentially bound to the 447-52D mimotope rather than the VH5-51 mimotope. The relatively low plasma response, namely 22% (2/9, to the linear peptide in the anti-Id abs and V3 mimotope responders was considered to be a specific immune response in the moderately inflammatory chronic phase of HIV-1 infection, as opposed to a non-specific immune response in the acute phase. This plasma response specific to the V3 mimotopes consisted of both a response to the 447-52D (ladle-type) mimotope and a response to the VH5-51 (cradle-type) mimotope. However, cradle-type abs specific to the VH5-51 mimotope have been reported to be vaccine-inducible abs with cross-neutralizing activities due to binding to conserved residues. Conversely, the ladle-type abs may be potent but are more specific to viruses with R315 at the tip of the V3 region (15, 21, 42–44).
Despite the current study’s lack of crystal structural analyses, bioinformatic analysis using the anti-Id abs asserted that the anti-Id abs specifically recognize the D53 and D100b that form solid bonds between the 1C10 heavy chain and R315 of the V3 tip region. This contributed to results showing that the anti-Id abs targeted IGHV3-30 abs with higher SHM rates and a longer CDRH3 length, and the anti-Id abs derived a synchronized response to cyclic V3 mimotopes from HIV-infected individuals. These findings concur with the idea that, in addition to cradle-type abs, subgroups of ladle-type abs lacking the typical feature of a long CDRH3 are induced by vaccines.
In this report, we first described the remarkable features of mouse anti-Id abs raised against the potent cross-neutralizing human anti-V3 mAb 1C10. Our data indicate that the anti-Id abs target not only the distinctive AAs shared among 1C10-like abs but also the conformational epitope corresponding to epitopes of anti-V3 ab specific to cyclic V3 mimotopes. Of note, 20% of individuals chronically infected with HIV-1 produced anti-V3 abs cross-reactive to the anti-Id abs. These results help to confirm our hypothesis that subgroups of ladle-type abs are derived from a certain immunoglobulin gene and can be elicited by vaccination.
The findings may also lead to a strategy to induce 1C10-like anti-V3 nAbs in HIV-1-infected individuals using anti-Id abs, based on the idea that anti-Id abs mimic an antigen of their target antibody, which first appeared in the “idiotypic network theory” (45–47). By way of illustration, a recent study demonstrated bnAb precursor B cells can be activated by BCR cross-links with anti-Id abs (48). Further studies are needed to complete our comprehension of the effects of these anti-Id abs on responsive B cells; such studies may include structural analyses to investigate the interfaces within complexes of anti-Id abs and 1C10 as well as the V3 loop and 1C10, in vivo experiments to examine the abs induced by the anti-Id abs, and experiments to test the neutralization ability of 1C10-like abs.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: GenBank, accession numbers OP006668-OP006677.
The studies involving human participants were reviewed and approved by the ethics committee for Clinical Research and Advanced Medical Technology at the Kumamoto University Medical School. The patients/participants provided their written informed consent to participate in this study. The animal study was reviewed and approved by Institutional Animal Care and Use Committee at Kumamoto University.
YK, KM, TK, and SM designed the experiments. YK generated anti-idiotypic antibodies, germline-reverted mutants and isolated B cells. YK, KM, HZ, and SB performed antibody cloning. YK performed binding and inhibition assay. MG produced V3 mimotopes. YK did homology modeling and next generation sequencing. YK, KM, TK, and SM interpreted data and prepared the figures and wrote the first draft and MG revised the draft. SM designed the study and SM and MG coordinated the study. All authors contributed to manuscript revision, read, and approved the submitted version.
This work was supported in part by Global Education and Research Center Aiming at the Control of AIDS, by JSPS KAKENHI Grant Number18H0285400 and by a grant for Research Program on HIV/AIDS from the Japan Agency for Medical Research and Development (JP19fk0410025h0001, JP20fk0410025h0002, JP21fk0410025h0003, JP22fk0410054h0001).
The looped V3 mimotopes were kindly gifted by MG. We thank Ms. Noriko Kuramoto, Yoko Kawanami, and Mikiko Shimizu for their assistance in the experiments, and Chie Hirai for her kind administrative assistance and Suzanne Leech, PhD, from Edanz (https://jp.edanz.com/ac) for editing a draft of this manuscript.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fviro.2022.932187/full#supplementary-material
1. Zagury D, Léonard R, Fouchard M, Réveil B, Bernard J, Ittelé D, et al. Immunization against AIDS in humans. Nature (1987) 326:249–50. doi: 10.1038/326249a0
2. Esparza J. A brief history of the global effort to develop a preventive HIV vaccine. Vaccine (2013) 31:3502–18. doi: 10.1016/j.vaccine.2013.05.018
3. Dolin R, Graham BS, Greenberg SB, Tacket CO, Belshe RB, Midthun K, et al. The safety and immunogenicity of a human immunodeficiency virus type 1 (HIV-1) recombinant gp160 candidate vaccine in humans. NIAID AIDS vaccine clinical trials network. Ann Intern Med (1991) 114:119–27. doi: 10.7326/0003-4819-114-2-119
4. Marovich MA. ALVAC-HIV vaccines: clinical trial experience focusing on progress in vaccine development. Expert Rev Vaccines (2004) 3(4 Suppl):S99–S104. doi: 10.1586/14760584.3.4.S99
5. Sanders RW, Derking R, Cupo A, Julien JP, Yasmeen A, de Val N, et al. A next-generation cleaved, soluble HIV-1 env trimer, BG505 SOSIP.664 gp140, expresses multiple epitopes for broadly neutralizing but not non-neutralizing antibodies. PloS Pathog (2013) 9:e1003618. doi: 10.1371/journal.ppat.1003618
6. Pauthner MG, Nkolola JP, Havenar-Daughton C, Murrell B, Reiss SM, Bastidas R, et al. Vaccine-induced protection from homologous tier 2 SHIV challenge in nonhuman primates depends on serum-neutralizing antibody titers. Immunity (2019) 50:241–52.e6. doi: 10.1016/j.immuni.2018.11.011
7. Rerks-Ngarm S, Pitisuttithum P, Nitayaphan S, Kaewkungwal J, Chiu J, Paris R, et al. Vaccination with ALVAC and AIDSVAX to prevent HIV-1 infection in Thailand. N Engl J Med (2009) 361:2209–20. doi: 10.1056/NEJMoa0908492
8. Burton DR, Ahmed R, Barouch DH, Butera ST, Crotty S, Godzik A, et al. A blueprint for HIV vaccine discovery. Cell Host Microbe (2012) 12:396–407. doi: 10.1016/j.chom.2012.09.008
9. Ringe RP, Ozorowski G, Rantalainen K, Struwe WB, Matthews K, Torres JL, et al. Reducing V3 antigenicity and immunogenicity on soluble, native-like HIV-1 env SOSIP trimers. J Virol (2017) 91:e00677–17. doi: 10.1128/JVI.00677-17
10. Escolano A, Gristick HB, Abernathy ME, Merkenschlager J, Gautam R, Oliveira TY, et al. Immunization expands b cells specific to HIV-1 V3 glycan in mice and macaques. Nature (2019) 570:468–73. doi: 10.1038/s41586-019-1250-z
11. Zolla-Pazner S. Improving on nature: focusing the immune response on the V3 loop. Hum Antibodies (2005) 14:69–72. doi: 10.3233/HAB-2005-143-403
12. Zolla-Pazner S, Edlefsen PT, Rolland M, Kong XP, deCamp A, Gottardo R, et al. Vaccine-induced human antibodies specific for the third variable region of HIV-1 gp120 impose immune pressure on infecting viruses. EBioMedicine (2014) 1:37–45. doi: 10.1016/j.ebiom.2014.10.022
13. Gottardo R, Bailer RT, Korber BT, Gnanakaran S, Phillips J, Shen X, et al. Plasma IgG to linear epitopes in the V2 and V3 regions of HIV-1 gp120 correlate with a reduced risk of infection in the RV144 vaccine efficacy trial. PloS One (2013) 8:e75665. doi: 10.1371/journal.pone.0075665
14. Gorny MK, Williams C, Volsky B, Revesz K, Cohen S, Polonis VR, et al. Human monoclonal antibodies specific for conformation-sensitive epitopes of V3 neutralize human immunodeficiency virus type 1 primary isolates from various clades. J Virol (2002) 76:9035–45. doi: 10.1128/JVI.76.18.9035-9045.2002
15. Gorny MK, Sampson J, Li H, Jiang X, Totrov M, Wang XH, et al. Human anti-V3 HIV-1 monoclonal antibodies encoded by the VH5-51/VL lambda genes define a conserved antigenic structure. PloS One (2011) 6:e27780. doi: 10.1371/journal.pone.0027780
16. Balasubramanian P, Kumar R, Williams C, Itri V, Wang S, Lu S, et al. Differential induction of anti-V3 crown antibodies with cradle- and ladle-binding modes in response to HIV-1 envelope vaccination. Vaccine (2017) 35:1464–73. doi: 10.1016/j.vaccine.2016.11.107
17. Kumar R, Pan R, Upadhyay C, Mayr L, Cohen S, Wang XH, et al. Functional and structural characterization of human V3-specific monoclonal antibody 2424 with neutralizing activity against HIV-1 JRFL. J Virol (2015) 89:9090–102. doi: 10.1128/JVI.01280-15
18. Jiang X, Burke V, Totrov M, Williams C, Cardozo T, Gorny MK, et al. Conserved structural elements in the V3 crown of HIV-1 gp120. Nat Struct Mol Biol (2010) 17:955–61. doi: 10.1038/nsmb.1861
19. Burton DR, Hangartner L. Broadly neutralizing antibodies to HIV and their role in vaccine design. Annu Rev Immunol (2016) 34:635–59. doi: 10.1146/annurev-immunol-041015-055515
20. Zolla-Pazner S, Cohen SS, Boyd D, Kong XP, Seaman M, Nussenzweig M, et al. Structure/Function studies involving the V3 region of the HIV-1 envelope delineate multiple factors that affect neutralization sensitivity. J Virol (2015) 90:636–49. doi: 10.1128/JVI.01645-15
21. Hessell AJ, McBurney S, Pandey S, Sutton W, Liu L, Li L, et al. Induction of neutralizing antibodies in rhesus macaques using V3 mimotope peptides. Vaccine (2016) 34:2713–21. doi: 10.1016/j.vaccine.2016.04.027
22. Balasubramanian P, Williams C, Shapiro MB, Sinangil F, Higgins K, Nádas A, et al. Functional antibody response against V1V2 and V3 of HIV gp120 in the VAX003 and VAX004 vaccine trials. Sci Rep (2018) 8:542. C. E. doi: 10.1038/s41598-017-18863-0
23. Ramirez Valdez KP, Kuwata T, Maruta Y, Tanaka K, Alam M, Yoshimura K, et al. Complementary and synergistic activities of anti-V3, CD4bs and CD4i antibodies derived from a single individual can cover a wide range of HIV-1 strains. Virology (2015) 475:187–203. doi: 10.1016/j.virol.2014.11.011
24. Roskin KM, Jackson K, Lee JY, Hoh RA, Joshi SA, Hwang KK, et al. Aberrant b cell repertoire selection associated with HIV neutralizing antibody breadth. Nat Immunol (2020) 21:199–209. doi: 10.1038/s41590-019-0581-0
25. Wu X, Yang ZY, Li Y, Hogerkorp CM, Schief WR, Seaman MS, et al. Rational design of envelope identifies broadly neutralizing human monoclonal antibodies to HIV-1. Sci (New York NY) (2010) 329:856–61. doi: 10.1126/science.1187659
26. Scheid JF, Mouquet H, Ueberheide B, Diskin R, Klein F, Oliveira TY, et al. Sequence and structural convergence of broad and potent HIV antibodies that mimic CD4 binding. Sci (New York NY) (2011) 333:1633–7. doi: 10.1126/science.1207227
27. Walker LM, Phogat SK, Chan-Hui PY, Wagner D, Phung P, Goss JL, et al. Broad and potent neutralizing antibodies from an African donor reveal a new HIV-1 vaccine target. Sci (New York NY) (2009) 326:285–9. doi: 10.1126/science.1178746
28. Walker LM, Huber M, Doores KJ, Falkowska E, Pejchal R, Julien JP, et al. Broad neutralization coverage of HIV by multiple highly potent antibodies. Nature (2011) 477:466–70. doi: 10.1038/nature10373
29. Bonsignori M, Hwang KK, Chen X, Tsao CY, Morris L, Gray E, et al. Analysis of a clonal lineage of HIV-1 envelope V2/V3 conformational epitope-specific broadly neutralizing antibodies and their inferred unmutated common ancestors. J Virol (2011) 85:9998–10009. doi: 10.1128/JVI.05045-11
30. Pejchal R, Doores KJ, Walker LM, Khayat R, Huang PS, Wang SK, et al. A potent and broad neutralizing antibody recognizes and penetrates the HIV glycan shield. Sci (New York NY) (2011) 334:1097–103. doi: 10.1126/science.1213256
31. Huang J, Ofek G, Laub L, Louder MK, Doria-Rose NA, Longo NS, et al. Broad and potent neutralization of HIV-1 by a gp41-specific human antibody. Nature (2012) 491:406–12. doi: 10.1038/nature11544
32. Doria-Rose NA, Schramm CA, Gorman J, Moore PL, Bhiman JN, DeKosky BJ, et al. Developmental pathway for potent V1V2-directed HIV-neutralizing antibodies. Nature (2014) 509:55–62. doi: 10.1038/nature13036
33. Maruta Y, Kuwata T, Tanaka K, Alam M, Valdez KP, Egami Y, et al. Cross-neutralization activity of single-chain variable fragment (scFv) derived from anti-V3 monoclonal antibodies mediated by post-attachment binding. Jpn J Infect Dis (2016) 69:395–404. doi: 10.7883/yoken.JJID.2015.667
34. Tanaka K, Kuwata T, Alam M, Kaplan G, Takahama S, Valdez K, et al. Unique binding modes for the broad neutralizing activity of single-chain variable fragments (scFv) targeting CD4-induced epitopes. Retrovirology (2017) 14:44. doi: 10.1186/s12977-017-0369-y
35. Kaku Y, Kuwata T, Zahid HM, Hashiguchi T, Noda T, Kuramoto N, et al. Resistance of SARS-CoV-2 variants to neutralization by antibodies induced in convalescent patients with COVID-19. Cell Rep (2021) 36:109385. doi: 10.1016/j.celrep.2021.109385
36. Kaku Y, Kuwata T, Gorny MK, Matsushita S. Prediction of contact residues in anti-HIV neutralizing antibody by deep learning. Jpn J Infect Dis (2020) 73:235–41. doi: 10.7883/yoken.JJID.2019.496
37. Waterhouse A, Bertoni M, Bienert S, Studer G, Tauriello G, Gumienny R, et al. SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res (2018) 46(W1):W296–303. doi: 10.1093/nar/gky427
38. Pierce BG, Hourai Y, Weng Z. Accelerating protein docking in ZDOCK using an advanced 3D convolution library. PloS One (2011) 6:e24657. doi: 10.1371/journal.pone.0024657
39. Lyskov S, Chou FC, Conchúir SÓ, Der BS, Drew K, Kuroda D, et al. Serverification of molecular modeling applications: The Rosetta online server that includes everyone (ROSIE). PloS One (2013) 8:e63906. doi: 10.1371/journal.pone.0063906
40. Schrödinger L, DeLano W. PyMOL (2020). Available at: http://www.pymol.org/pymol.
41. Nguyen MN, Verma CS, Zhong P. AppA: a web server for analysis, comparison, and visualization of contact residues and interfacial waters of antibody-antigen structures and models. Nucleic Acids Res (2019) 47(W1):W482–9. doi: 10.1093/nar/gkz358
42. Gorny MK, Wang XH, Williams C, Volsky B, Revesz K, Witover B, et al. Preferential use of the VH5-51 gene segment by the human immune response to code for antibodies against the V3 domain of HIV-1. Mol Immunol (2009) 46:917–26. doi: 10.1016/j.molimm.2008.09.005
43. Pan R, Qin Y, Banasik M, Lees W, Shepherd AJ, Cho MW, et al. Increased epitope complexity correlated with antibody affinity maturation and a novel binding mode revealed by structures of rabbit antibodies against the third variable loop (V3) of HIV-1 gp120. J Virol (2018) 92:e01894–17. doi: 10.1128/JVI.01894-17
44. Spencer DA, Malherbe DC, Vázquez Bernat N, Ádori M, Goldberg B, Dambrauskas N, et al. Polyfunctional tier 2-neutralizing antibodies cloned following HIV-1 env macaque immunization mirror native antibodies in a human donor. J Immunol (Baltimore Md 1950) (2021) 206:999–1012. doi: 10.4049/jimmunol.2001082
47. Kohler H, Pashov A, Kieber-Emmons T. The promise of anti-idiotype revisited. Front Immunol (2019) 10:808. doi: 10.3389/fimmu.2019.00808
Keywords: HIV-1, neutralizing antibody, vaccine, anti-idiotypic antibody, anti-V3 antibody
Citation: Kaku Y, Matsumoto K, Kuwata T, Zahid HM, Biswas S, Gorny MK and Matsushita S (2022) Development and characterization of a panel of anti-idiotype antibodies to 1C10 that cross-neutralize HIV-1 subtype B viruses. Front. Virol. 2:932187. doi: 10.3389/fviro.2022.932187
Received: 29 April 2022; Accepted: 08 August 2022;
Published: 30 August 2022.
Edited by:
Akio Adachi, Kansai Medical University, JapanReviewed by:
George N. Pavlakis, National Cancer Institute (NIH), United StatesCopyright © 2022 Kaku, Matsumoto, Kuwata, Zahid, Biswas, Gorny and Matsushita. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shuzo Matsushita, c2h1em9Aa3VtYW1vdG8tdS5hYy5qcA==
†These authors have contributed equally to this work and share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.