- 1Laboratory of Animal Hygiene, Joint Faculty of Veterinary Medicine, Kagoshima University, Kagoshima, Japan
- 2Transboundary Animal Diseases Centre, Joint Faculty of Veterinary Medicine, Kagoshima University, Kagoshima, Japan
- 3Department of Microbiology and Public Health, Faculty of Animal Science and Veterinary Medicine, Patuakhali Science and Technology University, Barishal, Bangladesh
- 4Department of Microbiology and Cell Biology, Tokyo Metropolitan Institute of Medical Science, Tokyo, Japan
- 5Virology Unit, Viral Hepatitis Laboratory, Institut Pasteur du Maroc, Casablanca, Morocco
- 6Department of Virology and Liver Unit, Nagoya City University Graduate School of Medical Sciences, Nagoya, Japan
- 7Faculty of Life Sciences, Kumamoto University, Kumamoto, Japan
Hepatitis B virus (HBV) infection is a global public health problem. The clinical outcomes of HBV infections are influenced by host as well as viral factors, including viral genotypes and subgenotypes. The interplay between HBV and host innate immunity remains unclear because of the lack of a suitable small animal model. Tree shrews (Tupaia belangeri) have been utilized as a useful animal model for hepatitis viruses such as hepatitis B and C viruses. In this study, we characterized acute infections by HBV genotype F (HBV-F) wild type (Wt) and mutant type (Mt) viruses in adult tree shrews. Serum alanine aminotransferase levels were measured before and post- infection 7 and 14 dpi. Both HBV-F-Wt and Mt were detected in the HBV-F-infected tree shrew serum and liver tissue at 7 and 14 dpi. We examined the intrahepatic expression patterns of Toll-like receptors (TLRs) (TLR1–9 mRNAs), cGAS, several transcription factors such as STAT1, STAT2, IRF7, HNF4, PD-L1, and cytokines, including IFN-β, IFN-γ, IL-6, and TNF-α in HBV-F Wt/Mt-infected tree shrews. When compared with uninfected animal group, significant suppression of TLR8 in HBV-F-Wt infected animals and significant suppression of PD-L1 in both HBV-F-Wt and Mt infected animals were observed. Thus, tree shrew can be a useful animal model to characterize HBV-F pathogenesis.
Introduction
Hepatitis B virus (HBV) infection remains a major public health concern worldwide, despite the availability of an effective preventive vaccine (1, 2). HBV infection may cause several hepatic complications, including acute and chronic hepatitis, cirrhosis, and hepatocellular carcinoma (3, 4). According to a recent report by the World Health Organization, 296 million people were infected with chronic HBV infection in 2019, with 1.5 million new infections annually (5).
HBV is an enveloped, circular, and partially double-stranded DNA virus belonging to the Hepadnaviridae family (3). HBV has a genome of approximately 3.2 kb containing four overlapping open reading frames, which encode the following seven proteins: polymerase, core, precore, three envelope/surface proteins (large, middle, and small), and the X protein (6–8). HBV has eight well-known genotypes (A–H), with more than 8% difference in the nucleotide sequence of the HBV genome and distinct geographic distribution (9, 10). Genotype A is highly prevalent in northwestern Europe, Sub-Saharan Africa, India, and North, Central, and South America, whereas genotypes B and C are common in the Asia-Pacific region. Genotype D is prevalent in the Mediterranean area, Central Asia, and South America, genotype E is restricted to West Africa, and genotype F is found in Central and South America. Genotype G has been reported in France, Germany, and the Americas, and genotype H is found in Central America and Mexico (11, 12). In addition to the A–H genotypes, two new genotypes have been described, including genotype I, which was isolated in Vietnam and Laos (13, 14), and genotype J, which was identified in Japan (15). Due to the high genetic variability of HBV, genotypes are further classified into 40 different subgenotypes, distinguished by greater than 4% difference in the nucleotide sequence of the HBV genome (16).
The functional cure, which is the new target for chronic HBV infection treatment, cannot be achieved by the currently available therapies consisting of peg-interferon and nucleos(t)ide analogs (17). Moreover, existing therapies for chronic HBV infection have limitations, such as frequent relapse and emergence of resistant viral variants, particularly in cirrhotic and immunosuppressed patients during short- and long-term treatment, respectively (18, 19). During chronic HBV infection, impairment of the immune response, including the T cell response, is observed (20), highlighting the necessity of developing novel therapies that can improve the immune response. Furthermore, the clinical progression and outcomes of HBV infections are regulated by many factors, including the host’s age and genetic factors, as well as viral factors such as HBV genotypes and subgenotypes (21, 22). All HBV genotypes have not been fully characterized, and HBV-induced immunopathogenesis is not well-understood because of the lack of an appropriate small animal infection model. Moreover, characterization of each HBV genotype is important for the proper understanding of HBV pathogenesis and host immune response for promoting the development of antivirals and vaccines (23). Although the role of adaptive immunity in the control of HBV infection is well-documented, the effects of innate immunity are only partially studied (24).
The innate immune response acts as the first line of immune defense to prevent viral infections (25). Components of viral pathogens, including viral nucleic acids and proteins, can be recognized by different pattern recognition receptors (PRRs), such as Toll-like receptors (TLRs), retinoic acid-inducible gene (RIG)-I-like receptors (RLRs), nucleotide-binding oligomerization domain (NOD)-like receptors (NLRs), and C-type lectin receptors, which may induce the synthesis of interferons (IFNs) and cytokines through distinct signaling pathways, thereby inhibiting infection and enhancing adaptive immune responses (26, 27). TLRs are evolutionarily conserved PRRs, which play an important role in sensing invaders and initiating innate immune responses, thereby limiting the spread of infections and triggering adaptive immune responses (26). Cyclic GMP-AMP (cGAMP) synthase (cGAS), a recently discovered cytosolic DNA sensor, is a key player in inducing the innate immune response against invaders (28, 29). Different TLRs and cGAS can lead to the activation of transcription factors, including signal transducer and activator of transcription (STAT) and IFN regulatory factor (IRF), which may play essential roles in the molecular and cellular signaling pathways of IFN induction (30, 31). Programmed cell death ligand 1 (PD-L1) is a co-inhibitory molecule associated with the immune response, which may inhibit cytokine secretion and induce apoptosis (32). Hepatocyte nuclear factor 4 (HNF4) is considered an important transcription factor for HBV replication, and the control of HNF expression and function might inhibit HBV replication (33, 34).
So far, chimpanzees have been used as natural infection models for HBV; however, high cost and ethical reasons restrict their use in experimental infections. Recently, humanized chimeric mice were developed (35) that can be effectively infected by HBV (36), but limitations including high cost, immunocompromised animal status, and inability to examine chronic HBV infections remained. Therefore, an alternative animal model is essential to study host-virus interactions and develop new drugs or vaccines. In this study, we used Tupaia belangeri (also known as tree shrew or tupaia) to investigate the infectivity of HBV-F and characterize the innate immune responses during an acute HBV infection. Tree shrews are widely distributed in Southeast Asia, including South and Southwest China (37). Tree shrew belongs to the Tupaiidae family, which contains four genera and 19 extant species (37). Whole-genome analysis of tree shrews has revealed that it is more closely related to humans than to rodents (38, 39). Tree shrews have been reported to be susceptible to HBV infection in several studies (40–44). In addition, primary tupaia hepatocytes (PTHs) are being widely used for HBV infection studies, which might provide further insights into HBV pathogenesis (43, 45, 46). It is important to mention that sodium taurocholate co-transporting polypeptide (NTCP), the cellular entry receptor for HBV, was recently identified using PTHs (46). Therefore, tree shrews could be a useful animal model for the experimental analysis of various molecular and clinical aspects of HBV infection, and can be developed as an immunocompetent infection model (47). Furthermore, viral factors such as viral genotypes also influence the clinical outcome and treatment response; therefore, it is essential to study the molecular basis of HBV pathogenesis for each genotype. However, HBV genotype F (HBV-F) has not yet been characterized in the tree shrew model. Notably, it has been reported that in the chimeric mice hepatocytes, HBV-F Mt (BCP/PC/2051 including A1762T/G1764A, G1896A, and A2051C mutation which were observed in HCC patients) can replicate better compared to HBV-F Wt (48). Therefore, in this study, we characterized HBV-F wild- and mutant-type (HBV-F Wt/Mt) infection in tree shrews, including the intrahepatic innate immune response against acute HBV-F Wt/Mt infection to gain insights into the role of host innate immune response in immunopathogenesis during acute HBV-F infection in this model animal.
Materials and methods
Animals
Twelve adult male and female tree shrews were used in this study. Among the tree shrews used in this study, #386 and # 395 were female (4 years old) (Figure S1), and the remaining shrews were males (6 years old) (Figure 1; Figure S2). Tree shrew #386 and #395 were immunized with HBs-S antigen (10 μg/dose, intranasally, 6 times, 2017.11.30~2018.02.22). Adult tree shrews were originally purchased from the Laboratory Animal Center at the Kunming Institute of Zoology, Chinese Academy of Sciences (Kunming, China). Tree shrews were reared in separate cages and fed a daily regimen of eggs, fruits, water, and dry mouse food. The animals were handled humanely according to the guidelines of the Institutional Animal Care and Use Committee for Laboratory Animals. This study was carried out in accordance with the ‘Guidelines for Animal Experimentation’ from the Japanese Association for Laboratory Animal Science and the recommendations of the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health.
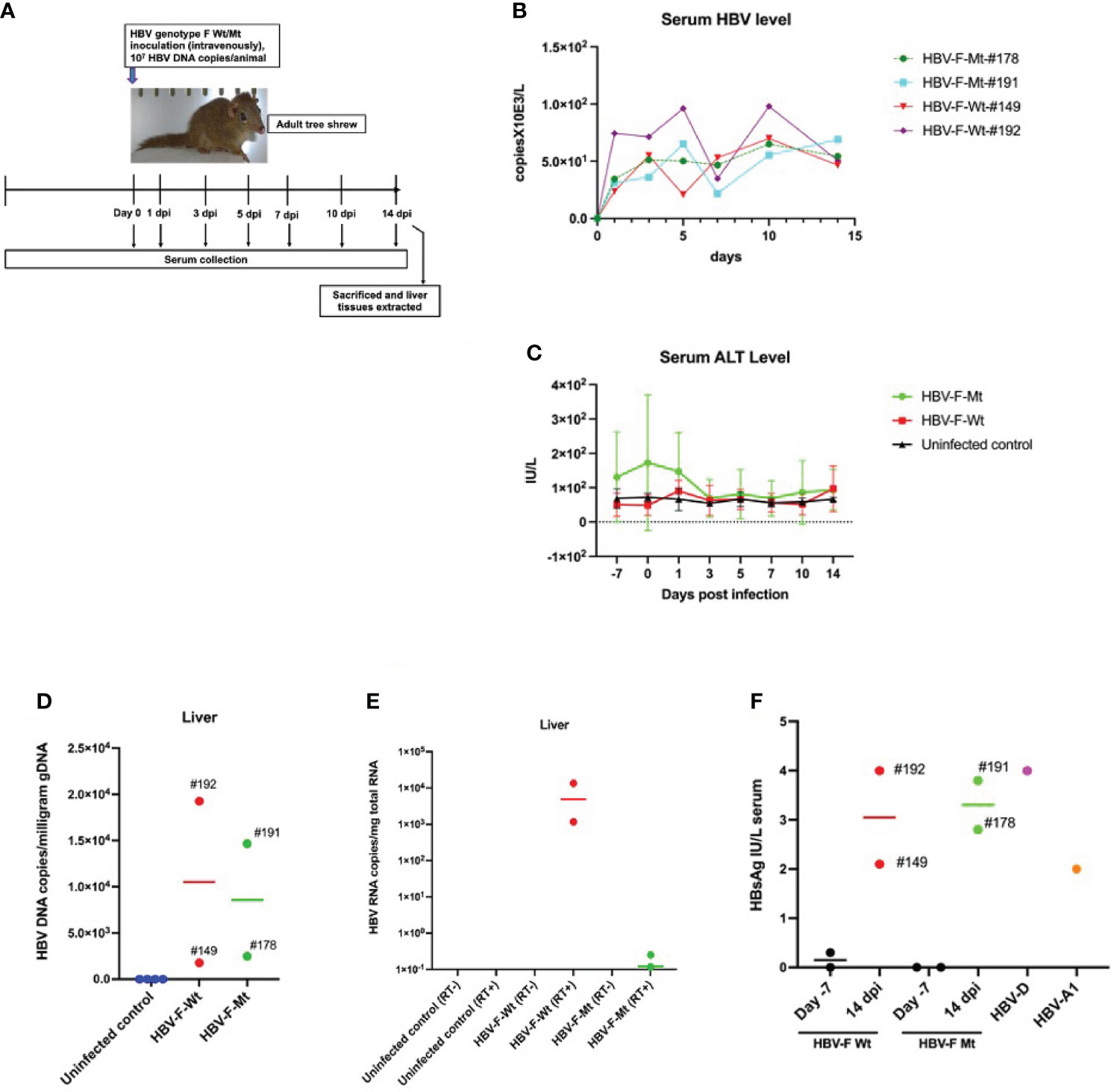
Figure 1 HBV genotype F wild-type or mutant-type (HBV-F Wt#149 and 192/Mt#178 and 191) infections in adult tree shrews. (A) Experimental design of HBV-F infection in tree shrew. (B) Serum HBV levels from day 0 to day 14 in HBV-F Wt/Mt-infected tree shrews. (C) Serum alanine aminotransferase (ALT) levels from day -7 (before infection) to 14 dpi in HBV-F Wt/Mt-infected and uninfected controls. Error bars indicate standard deviation. (D) Intrahepatic HBV DNA loads in HBV-F Wt/Mt-infected tree shrews at 14 dpi. (E) Intrahepatic HBV RNA levels in HBV-F Wt/Mt-infected tree shrews at 14 dpi. (F) Serum hepatitis B surface antigen (HBsAg) levels in HBV-F Wt/Mt-infected tree shrews at 7 days before infection (Day -7) and 14 dpi. Serum HBsAg levels in HBV-D-infected tree shrew after 10 weeks post infection and in HBV-A1-infected tree shrew after 39 weeks post infection were shown (right). Thick horizontal lines indicate arithmetic mean.
Virus infection
All tree shrews used in this study were confirmed to be HBV negative by polymerase chain reaction (PCR), as shown by the HBV DNA measurements, before viral infection. HBV inocula for animal inoculation were derived from humanized chimeric mice but not from humans. Intravenous inoculation of HBV-F Wt (48) (in tree shrews #149, #192, and #395) and HBV-F Mt (48) (in tree shrews number #178, #191, and #386) with 1.0 × 107 HBV DNA copies per animal was performed in the tail vein of tree shrews, and four tree shrews #3, 5, 38, and 53) were used as uninfected controls. The HBV-F Wt/Mt-infected and uninfected tree shrews were bled (0.3 mL) before infection (days -7 and 0), and 1, 3, 5, 7, 10, and 14 days post-infection (dpi). Sera were obtained for the indicated time points and stored at −80°C until further use. At 14 dpi, the shrews were sacrificed, and liver tissues were extracted and stored at −80°C until further use. In addition to the above-mentioned animals, two adults with HBV-Wt (tree shrew #19) and HBV-F Mt (tree shrew #M0-001) were infected and then sampled as previously described (Supplementary Figure S2). Seven days post-infection, the animals were sacrificed, and liver tissues were extracted and stored at −80°C until further use.
Measurement of serum alanine aminotransferase
Serum ALT activity in tree shrews was determined using a commercial kit, Transaminase CII-test Wako (Wako Pure Chemical Industries, Osaka, Japan), and standardized according to the manufacturer’s instructions. ALT activity is presented in international units (IU)/L (L) and the acceptable values for healthy tree shrews were values lower than 100.
Viral load measurement
HBV DNA from serum samples pre- and post-infection was extracted using SMItest EX-R&D kits (Nippon Genetics Co., Ltd., Tokyo, Japan), according to the manufacturer’s instructions. HBV DNA was measured by real-time PCR, as described previously (49). Genomic DNA (gDNA) from tree shrew liver tissue was extracted using the phenol-chloroform extraction method, and intrahepatic HBV DNA was measured by real-time PCR, as described previously (49). The primers and probes for the S gene consisted of a forward primer HB-166-S21 (nts 166–186): 5′-CACATCAGGATTCCTAGGACC-3′, a reverse primer HB-344-R20 (nts 344-325): 5′-AGGTTGGTGAGTGATTGGAG-3′, and Taq Man probe HB-242-S26FT (nts 242–267): 5′-CAGAGTCTACACTCGTGGTGGACTTC-3′. The limit of detection (LOD) of HBV in sera was 2 copies/mL, and that in genomic DNA was 0.2 copies/μg genomic DNA. The primers and probes for cccDNA were detected using the forward primers HBV-cccDNA 1519-S25; 5’- ACGGGGCGCACCTCTCTTTACGCGG-3’ and the reverse primers 1519-R25; 5’-CCGTGTGCACTTCGCTTCACCTCTGC-3’. The probe used was the HBV-cccDNA 1575 S26FT; 5’-CCGTGTGCACTTCGCTTCACCTCTGC-3’ after treatment of genomic DNA with Plasmid-Safe ATP-dependent DNase (Lucigen) (50). The LOD of HBV cccDNA in the genomic DNA was 0.02 copies/μg genomic DNA. The HBV RNA levels in tree shrew liver tissues were measured using total RNA as a template. After reverse transcription (RT), the template was used for qPCR as described above. Total RNA from liver tissues was extracted using the RNeasy Plus Mini Kit (QIAGEN) according to the manufacturer’s instructions, and genomic DNA was removed using the gDNA Eliminator Spin Column. The concentration and purity of the extracted RNA samples were measured using a NanoDrop ND-1000 spectrophotometer (NanoDrop Technologies, Inc., Waltham, MA, USA). The RNA samples were stored at −80°C until further use. The extracted liver RNA was subsequently used for gene expression analysis.
Measurement of HBsAg
HBsAg levels in the tree shrew sera were measured using the two-step sandwich assay principle with a fully automated chemiluminescent enzyme immunoassay system (Lumipulse G 1200, Fujirebio, Tokyo, Japan), as described previously (51). The LOD for the HBV genotype C HBsAg was 5.0 IU/L and the LOD for the HBV mutant was 1.0 IU/L.
Gene expression analysis by qRT-PCR
To investigate the expression patterns of TLR mRNAs, including TLR1–9 mRNAs, in tree shrews, a one-step qRT-PCR was performed using Brilliant III Ultra-fast SYBR Green QRT-PCR Master Mix (Agilent Technologies, Santa Clara, CA, USA), as described previously (44). The extracted liver RNA was used for gene expression analysis as described above. The expression patterns of immune checkpoint PD-L1 and several transcription factors, including STAT1, STAT2, IRF7, and HNF4, were also measured using one-step qRT-PCR. The cycling conditions included reverse transcription at 50°C for 10 min, primary denaturation at 95°C for 3 min, and 40 cycles at 95°C for 5 s and 60°C for 10 s. IFN-β, IFN-γ, IL-6, and TNF-α mRNA expression were measured by a one-step qRT-PCR, as described previously (44, 52). The sequences of primers used for amplification of the target genes are listed in Table 1. The actual copy number of the gene of interest per microgram of liver RNA was determined by preparing a standard curve from pre-quantified copies of each corresponding gene. Each sample was measured in triplicate and significant difference with uninfected controls was statistically analyzed.
Western blot analysis
Western blot analysis was performed to detect HBc and HBsAg in the liver tissues using a standard protocol. Recombinant HBc and HBs Ag and anti-HBc and HBs rabbit polyclonal or mouse monoclonal antibodies were purchased from Beacle Inc. (Kyoto, Japan).
Histological analysis
For histological analysis, liver tissues were harvested, fixed with paraformaldehyde, and stained with hematoxylin and eosin (H&E).
Statistical analysis
A one-way ANOVA was performed using GraphPad Prism (Version 9) to compare gene expression among uninfected (n=3), HBV-F Wt (n=2) and Mt (n=2) infected groups (Figures 2 and 3). Statistical significance was set at P < 0.05.
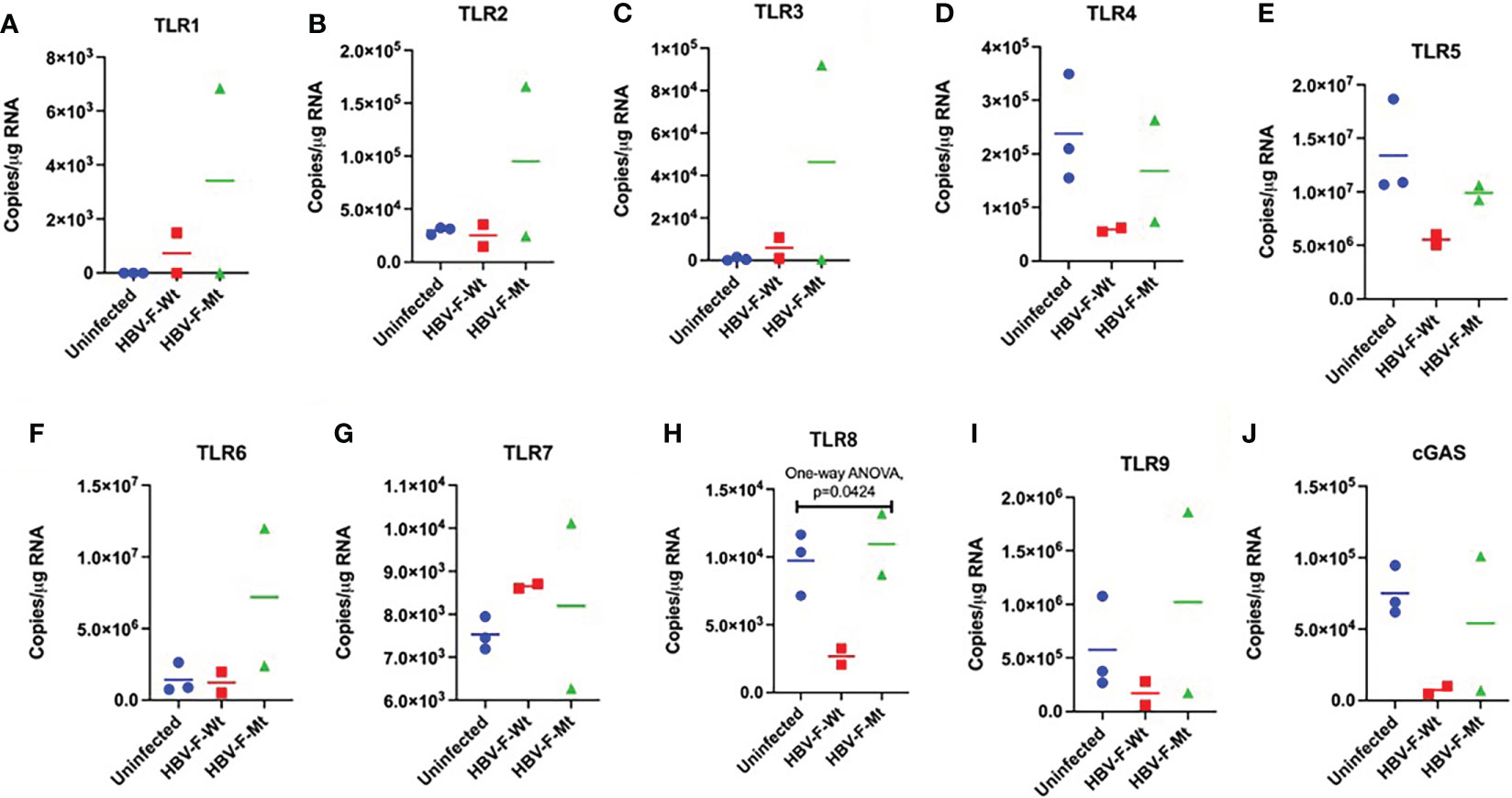
Figure 2 Intrahepatic TLR responses to acute HBV-F Wt/Mt infection in tree shrews. Intrahepatic expression of (A-I) TLR1–9 mRNA in uninfected control (#5, 38 and 53), HBV-F Wt (#149 and 192)/Mt (#178 and 191)-infected tree shrews at 14 dpi. (J) cGAS mRNA expression in uninfected control and HBV-F Wt/Mt-infected tree shrews at 14 dpi. We performed the statistical analysis, and the P values are shown if they are statistically significant. Thick horizontal lines indicate arithmetic mean.
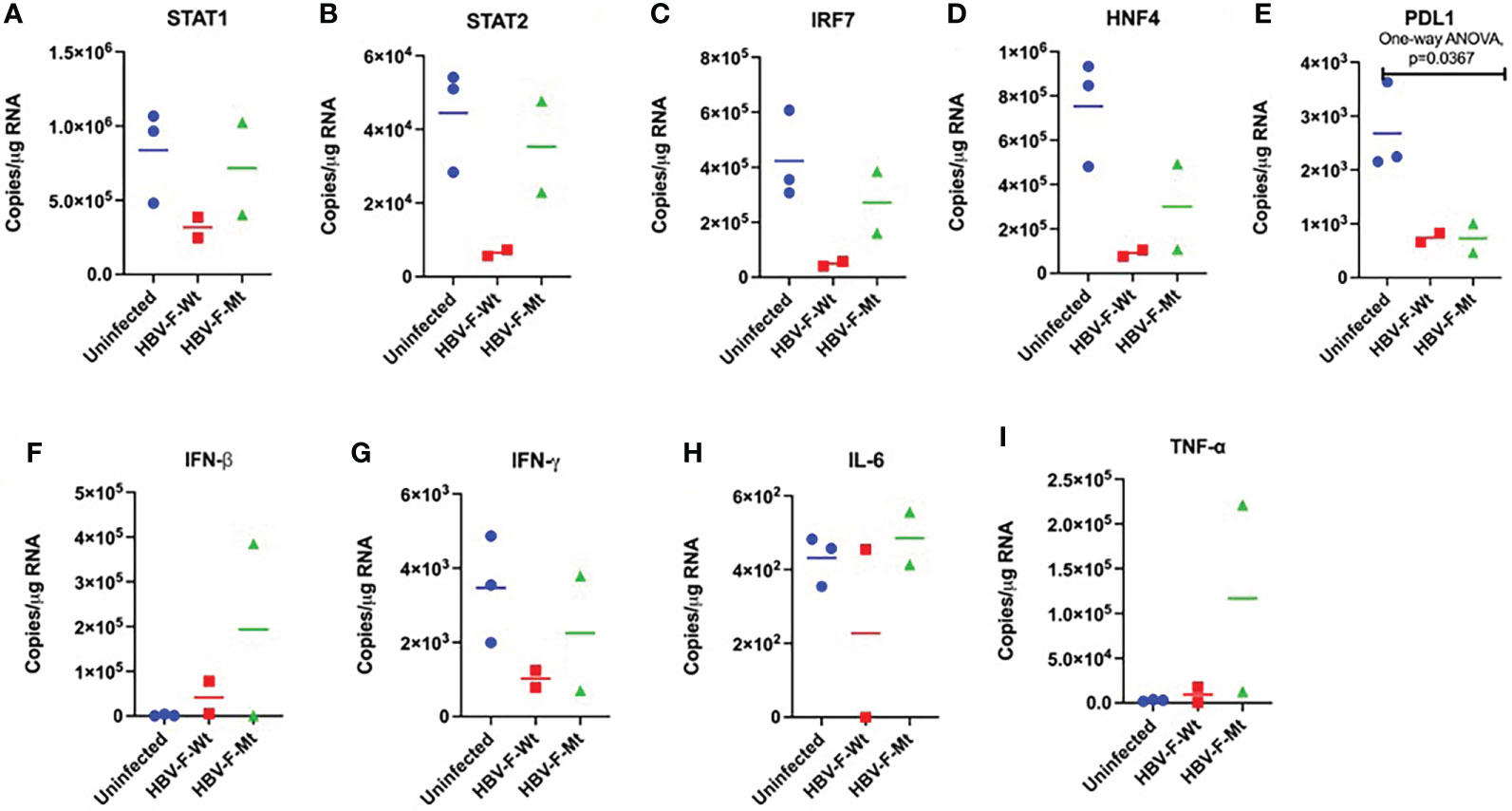
Figure 3 Changes in the intrahepatic transcription factors and cytokines in acute HBV-F Wt/Mt infection in tree shrews. The mRNA expression of transcription factors, including (A) STAT1 (B) STAT2 (C) IRF7 (D) HNF4, and immune checkpoint (E) PDL1, and cytokines, including (F) IFN-β (G) IFN-γ, (H) IL-6, and (I) TNF-α in HBV-F Wt/Mt-infected tree shrews at 14 dpi. We performed the statistical analysis, and the P values are shown if they are statistically significant. Thick horizontal lines indicate arithmetic mean.
Results
Infection of tree shrews with HBV-F Wt/Mt
To investigate the infectivity of HBV-F Wt and Mt in adult tree shrews, the animals were inoculated with HBV-F Wt (n=2) or HBV-F Mt (n=2) and investigated for 14 days post infection (dpi) (Figure 1A). We also investigated the infectivity of HBV-F Wt/Mt in two immunized adult tree shrews for 14 dpi (Figure S1). Serum HBV and ALT levels were monitored for 14 days (Figures 1B, C; Figures S1A, B). At 14 dpi, HBV DNA was detected in the sera of all HBV-F Wt/Mt-infected tree shrews (Figure 1B; Figure S1A). HBV DNA quantification in the genomic DNA (gDNA) isolated from HBV-F Wt/Mt-infected tree shrew liver tissues was performed by real-time PCR at 14 dpi. HBV DNA was detected in the liver tissues of all HBV-F Wt/Mt-infected tree shrews (Figure 1D; Figure S1C). The mean intrahepatic viral loads in HBV-F Wt- and HBV-F Mt-infected tree shrews were 10510 and 8570 copies HBV DNA/milligram gDNA, respectively (Figure 1D). The intrahepatic viral loads in immunized tree shrews infected with HBV-F Wt- and HBV-F Mt were 1165 and 4630 copies HBV DNA/milligram gDNA, respectively at 14 dpi (Figure S1C). Although the titer was low, HBV RNA was detected in the liver tissues of HBV-F Wt- and HBV-F Mt-infected tree shrews (Figure 1E). Also, HBsAg was measured in the serum of HBV-F Wt and HBV-F Mt-infected tree shrews at 7 days before infection (Day -7) and 14 dpi (Figure 1F; Figure S1D). HBV-D and HBV-A1 infected tree shrew sera were also pleasured as positive controls. Two additional animals were infected with HBV-F Wt (n=1) or HBV-F Mt (n=1) and then monitored for 7 dpi (Figure S2A). Serum HBV and ALT levels were monitored 4 days before infection to 7 dpi in HBV-F Wt/Mt-infected tree shrews (Figure S2B, C). HBV DNA was detected in HBV-F Wt/Mt-infected tree shrew liver gDNA at 7 dpi (Figure S2D). HBV cccDNA was only detected in HBV-F Mt-infected tree shrew liver genomic DNA (Figure S2E). Western blot analysis detected intrahepatic HBc and HBsAg at 14 dpi (Figure S2F).
Intrahepatic TLR response in tree shrews to HBV-F infection
In this study, we investigated the innate immune response, including expression of TLRs (TLR1–9) and cytokine responses in HBV-F Wt/Mt-infected adult tree shrews in order to have comparable results with other HBV subtypes (39). We have performed statistical analysis on each value comparing with uninfected control group (Tree shrew No. 5, 38 and 53) (Figure 2). In addition, significant suppression of TLR8 was observed in HBV-F Wt-infected tree shrew liver tissue as compared to the uninfected controls (Figure 2H). The intrahepatic TLRs and cGAS response were also characterized in immunized tree shrews infected with HBV-F Wt- and HBV-F Mt at 14 dpi (Figure S3A-J). TLR1-9 mRNAs were upregulated in immunized tree shrew liver tissues infected with HBV-F Wt as compared to uninfected controls (Figure S3A-I). cGAS mRNAs were found to be upregulated in immunized tree shrew liver tissues infected with HBV-F Wt/Mt at 14 dpi (Figure S3J). The TLR response was also characterized in HBV-F Wt/Mt-infected tree shrew liver tissues at 7 dpi, and TLR response was mostly downregulated except for TLR7 in HBV-F Mt infected animals (Figure S4).
Effects of HBV-F infection on expression levels of transcription factors and cytokines in tree shrew livers
We measured the changes in expression patterns of several transcription factors and cytokines, including STAT1, STAT2, IRF7, HNF4, PDL1, IFN-β, IFN-γ, IL-6 and TNF-α in the livers of HBV-F Wt/Mt-infected tree shrews (Figure 3). We observed significant decrease of PDL1 (Figure 3E) in HBV-F Wt-infected tree shrew livers. We also measured the intrahepatic changes in expression patterns of several transcription factors and cytokines, including STAT1, STAT2, IRF7, HNF4, PDL1, IFN-β, IFN-γ, IL-6 and TNF-α in immunized tree shrew infected with HBV-F Wt/Mt at 14 dpi (Figure S5A-I). Compared with the controls, STAT2, IFN-β and TNF-α mRNAs were found to be upregulated in immunized tree shrew livers infected with HBV-F Wt (Figures S5B, F, I). The intrahepatic cytokine expression in HBV-F Wt/Mt-infected tree shrew liver tissue at 7 dpi was also measured (Figure S6).
Histological analysis of liver tissues from HBV-F-infected tree shrews
Histological analysis showed lymphocyte infiltration into the liver tissues of HBV-F Wt/Mt-infected tree shrews (Figure 4; Figure S7). A comparison with the liver tissues of uninfected control tree shrew (#38) showed abnormal architecture of liver cell cords, as indicated by destruction and necrosis of hepatocytes in HBV-F Wt/Mt-infected tree shrew liver tissues (Figure 4; Figure S7). At 7 dpi, abnormal architecture of the liver cell cords was also observed in the liver tissues of HBV-F Wt/Mt-infected tree shrews (Figure S7B).
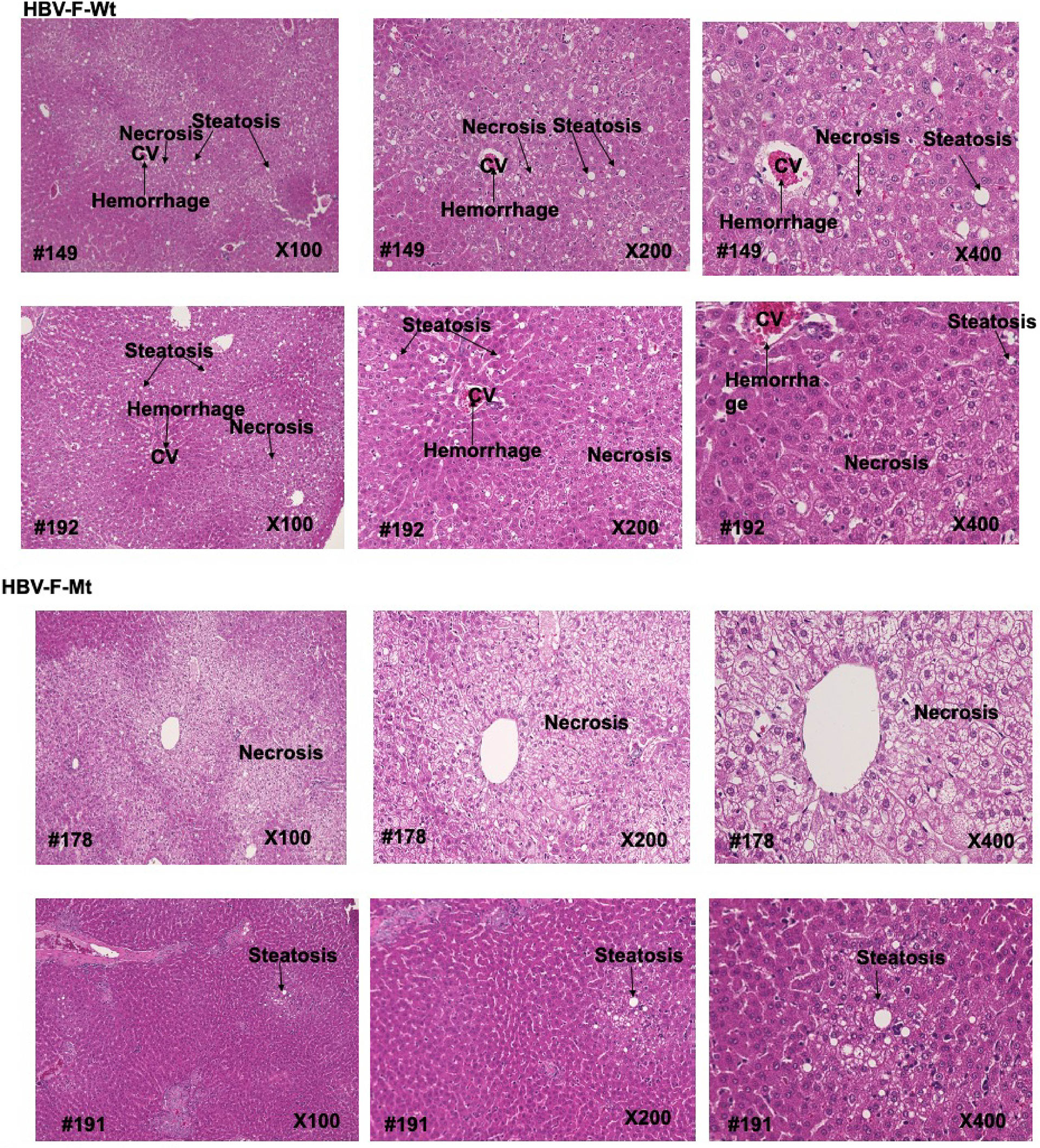
Figure 4 Histopathological analysis of liver tissues from uninfected control and HBV-F Wt/Mt-infected tree shrews at 14 dpi. Histopathological analysis of liver tissues from uninfected control and HBV-F Wt (#149, 192)/Mt (#178, #191)-infected tree shrews at 14 dpi. Histopathological analysis was performed with H&E. The images were taken using a BZ-X700 fluorescence microscope (Keyence Co., Japan) at 100x, 200x and 400× magnification.
Discussion
Tree shrews are a promising animal model for studying human viral infections, including HBV (53). In this study, we investigated the infectivity of HBV-F Wt/Mt in tree shrews and characterized the innate immune response against HBV infection in this model. We observed that HBV-F Wt/Mt could establish an acute infection in adult tree shrews, as demonstrated by the increased level of ALT in the sera and the detection of HBV DNA in the liver tissues of HBV-F Wt/Mt-infected tree shrews at both 7 and 14 dpi. Also, intrahepatic HBV RNA and serum HBsAg could be detected at 14 dpi, indicating the replication of HBV-F in tree shrew liver. Although HBV-F Mt (BCP/PC/2051) showed a higher replication compared to HBV-F Wt in the chimeric mice hepatocytes (48); it was not reflected in the tree shrew model, which could be due to differences in animal models and host immune response. Induction of IFN-β and upstream TLR1, 3, 7, 8, and 9 was only observed in HBV-F-Wt-infected tree shrew liver tissues (#395), suggesting a stronger stealth nature of HBV-F-Wt infection. Tree shrews (#395 and #386) were immunized, intranasally, with HBs-S antigen four years ago, therefore, lymphocyte infiltration was more pronounced than in other Tupaia tissues (Figure 4A). Characterization of TLR response showed some different response between HBV-F-Wt and Mt, such as down regulation of TLR8 (#178, #386) in HBV-F-Wt infected Tupaia liver tissue. Thus, these differences might link to the higher replication activity and pathogenesis of HBV-F-Mt than Wt, but further investigation using tree shrew model is required. Suppression of PDL1 was observed in both HBV-F Wt and Mt infected tree shrew liver tissues at 14 dpi, which may link to pathogenesis of HBV infection (54).
Several previous studies have also highlighted the stealth nature of HBV, where the induction of innate immune response, including IFNs and IFN-stimulated genes (ISGs), was not observed in acute HBV infection in chimpanzee (55) and woodchuck models (56). An early innate intrahepatic response was determined to be partially effective in controlling woodchuck hepatitis virus (WHV) replication in a woodchuck model of HBV infection (57). It has also been reported that in patients with acute HBV infection, type I IFN induction is either lacking or there is negligible induction (58, 59). However, in our previous study, during acute infection at 28 dpi, a significantly upregulated expression of TLR2, 3, 8, and 9 was observed in HBV A2_JP1-infected tree shrews (44), which suggest the effects of different conditions, including genotype difference and days after infection. Moreover, the sensing of HBV by PRRs, including TLRs, has been reported in several studies (24, 60, 61). An earlier study showed that a single intravenous injection of TLR3, 4, 5, 7, and 9 agonists induced an IFN-α/β production that inhibited HBV replication in the liver of HBV-transgenic mice (62). The role of TLR4 in inhibiting HBV replication has also been demonstrated in an in vitro study (63). TLR signaling is involved in anti-HBV response, while HBV has devised strategies to avoid the host TLR response to establish infection (64, 65). A recent study demonstrated that TLR-mediated signaling pathways play a significant role in eliciting functional HBV-specific CD8+ T cell responses in vivo (66). HBV reportedly suppresses TLR-mediated antiviral response in hepatic cells by inhibiting the activation of IRF-3, nuclear factor kappa B (NF-κB), and extracellular signal-regulated kinase 1/2 (ERK1/2) (67). HBV polymerase can inhibit IRF activation by interfering with the interaction between IκB kinase-ϵ and DEAD box RNA helicase, which may inhibit IFN production (68).
We found no induction of STAT1, but STAT2 in HBV-F-infected tree shrew livers, compared to uninfected controls. A previous study reported that HBV polymerase can impair IFN-α-induced STAT activation (69). Poor induction of early intracellular innate responses are characteristics of HBV infection (70). Ruan et al. reported similar histopathological changes in chronically infected tree shrew livers as observed in CHB (71). Our study had similar results to a previous study whereby abnormal architecture of the liver cell cords was also observed (44).
Thus, HBV-F Wt/Mt acute infection model in tree shrews can become a suitable animal model for investigation of early immune response. The mechanisms by which HBV-F modifies the host innate immune response remain to be investigated. Although further investigations are required, with a larger number of tree shrews, to unravel the mechanisms implicated in the immunopathogenesis during HBV-F acute infection in the tree shrew model, this study highlights the tree shrew as a suitable small animal model for acute HBV-F infection and the host-virus interaction study.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Materials. Further inquiries can be directed to the corresponding author.
Ethics statement
This study was reviewed and approved by Laboratory Animal Research committee in Kagoshima University.
Author contributions
Conceptualization, MEHK, MK, and KT-K. Performed the experiments, MEHK, MH, TS, BK, MR, LA, SE, SM, SO, YT, MK, KT-K. Original draft preparation, MEHK and KT-K. Review and editing, MEHK, MK, and KT-K. Supervision, KT-K. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by grants from the Japan Agency for Medical Research and Development grant number, 20fk0310111h0004 KT-K and 22fk0310518h0001 YT.
Acknowledgments
The authors would like to thank Drs. Tatsuro Hifumi and Noriaki Miyoshi for their support in the pathological analysis.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fviro.2022.926831/full#supplementary-material
Supplementary Figure 1 | HBV genotype F wild-type or mutant-type (HBV-F Wt #386/Mt #395) infections in HBs-S immunized adult tree shrews. (A) Serum HBV levels from day 0 to day 14 in HBs-S immunized adult tree shrews infected with HBV-F Wt/Mt (B) Serum alanine aminotransferase (ALT) levels from day -7 (before infection) to 14 dpi in immunized adult tree shrews infected with HBV-F Wt/Mt-infected and uninfected controls. (C) Intrahepatic HBV DNA loads in HBs-S immunized adult tree shrews infected with HBV-F Wt/Mt at 14 dpi. (D) Serum HBsAg levels in HBs-S immunized adult tree shrews infected with HBV-F Wt/Mt at 7 days before infection (Day -7) and 14 dpi. Serum HBsAg levels in HBV-D-infected tree shrew after 10 weeks post infection and in HBV-A1-infected tree shrew after 39 weeks post infection were shown (right).
Supplementary Figure 2 | HBV genotype F wild-type or mutant-type (HBV-F Wt/Mt) infections in adult tree shrews up until 7 days post infection (dpi). (A) Experimental design of HBV-F infection in tree shrews. Serum ALT and HBV-DNA levels in HBV-F Wt (Tupaia #19) (B) or HBV-F Mt (Tupaia #M0-001) (C) infected Tupaia from 4 days before infection (dbi) to 7 dpi. Intrahepatic HBV DNA (D) or cccDNA (E) loads in HBV-F Wt/Mt-infected tree shrews at 7 dpi. (F) Detection of HBs-S antigen and HBc antigen in the livers from HBV-F Mt infected (#191) or HBV-F Wt infected (#149, #192) Tupaia at 14 dpi. Recombinant HBs-S and HBc protein were also indicated (controls).
Supplementary Figure 3 | Intrahepatic TLR responses to acute HBV-F Wt/Mt infection in HBs-S immunized adult tree shrews. Intrahepatic expression of (A-I) TLR1–9 mRNA in uninfected control (#5, 38 and 53), and HBs-S immunized and HBV-F Wt (#395) and Mt (#386)-infected tree shrews at 14 dpi. (J) cGAS mRNA expression in uninfected control and HBs-S immunized and HBV-F Wt (#395) and Mt (#386)-infected tree shrews at 14 dpi.
Supplementary Figure 4 | Intrahepatic TLR responses to acute HBV-F Wt/Mt infection in tree shrews. TLR and cGAS mRNA response in uninfected control (#5, 38, and 53) and HBV-F Wt (#19)/Mt (#M0-001) infected tree shrews at 7 dpi have been indicated.
Supplementary Figure 5 | Changes in the intrahepatic transcription factors and cytokines in acute HBV-F Wt/Mt infection in HBs-S immunized adult tree shrews (#395 and #386). The mRNA expression of transcription factors, including (A) STAT1 (B) STAT2 (C) IRF7 (D) HNF4, and immune checkpoint (E) PDL1, and cytokines, including (F) IFN-β (G) IFN-γ, (H) IL-6, and (I) TNF-α in HBs-S immunized adult tree shrews infected with HBV-F Wt/Mt at 14 dpi.
Supplementary Figure 6 | Changes in the intrahepatic transcription factors and cytokines in acute HBV-F Wt/Mt infection in tree shrews. The mRNAs were measured in uninfected controls (#5, 38, and 53) and HBV-F Wt (#19)/Mt (#M0-001) infected tree shrews at 7 dpi.
Supplementary Figure 7 | (A) Histopathological analysis was performed with H&E. H&E staining images of liver tissues from uninfected control (#38), HBs-S immunized and HBV-F Wt-infected (#395), and HBV-F Mt-infected (#386) tree shrews are shown. The images were taken using a BZ-X700 fluorescence microscope (Keyence Co., Japan) at 100x, 200x and 400× magnification. (B) Histopathological analysis of liver tissues from uninfected controls and HBV-F Wt/Mt-infected tree shrews at 7 dpi (x400). In HBV-F Wt (#19) liver tissues, abnormal architecture, necrosis of hepatocyte and inflammatory cells were observed. In HBV-F Mt (#M0-001) liver tissues, abnormal architecture and necrosis (N) of hepatocytes were observed.
References
1. Seeger C, Mason WS. Molecular biology of hepatitis b virus infection. Virology (2015) 479-480:672–86. doi: 10.1016/j.virol.2015.02.031
2. Mastrodomenico M, Muselli M, Provvidenti L, Scatigna M, Bianchi S, Fabiani L. Long-term immune protection against HBV: associated factors and determinants. Hum Vaccin Immunother (2021) 17:2268–72. doi: 10.1080/21645515.2020.1852869
3. Lee WM. Hepatitis b virus infection. N Engl J Med (1997) 337:1733–45. doi: 10.1056/NEJM199712113372406
4. Kappus MR, Sterling RK. Extrahepatic manifestations of acute hepatitis b virus infection. Gastroenterol Hepatol (N Y) (2013) 9:123–6.
5. World Health Organization. Hepatitis b (2021). Available at: https://www.who.int/news-room/fact-sheets/detail/hepatitis-b (Accessed 17 May, 2022).
6. Huang HL, Jeng KS, Hu CP, Tsai CH, Lo SJ, Chang C. Identification and characterization of a structural protein of hepatitis b virus: a polymerase and surface fusion protein encoded by a spliced RNA. Virology (2000) 275:398–410. doi: 10.1006/viro.2000.0478
7. Locarnini S. Molecular virology of hepatitis b virus. Semin Liver Dis (2004) 24 Suppl 1:3–10. doi: 10.1055/s-2004-828672
8. Hu J, Seeger C. Hepadnavirus genome replication and persistence. Cold Spring Harb Perspect Med (2015) 5:a021386. doi: 10.1101/cshperspect.a021386
9. Okamoto H, Tsuda F, Sakugawa H, Sastrosoewignjo RI, Imai M, Miyakawa Y, et al. Typing hepatitis b virus by homology in nucleotide sequence: comparison of surface antigen subtypes. J Gen Virol (1988) 69(Pt 10):2575–83. doi: 10.1099/0022-1317-69-10-2575
10. Arauz-Ruiz P, Norder H, Robertson BH, Magnius LO. Genotype h: a new Amerindian genotype of hepatitis b virus revealed in central America. J Gen Virol (2002) 83:2059–73. doi: 10.1099/0022-1317-83-8-2059
11. Miyakawa Y, Mizokami M. Classifying hepatitis b virus genotypes. Intervirology (2003) 46:329–38. doi: 10.1159/000074988
12. Lin CL, Kao JH. Hepatitis b virus genotypes and variants. Cold Spring Harb Perspect Med (2015) 5:a021436. doi: 10.1101/cshperspect.a021436
13. Tran TT, Trinh TN, Abe K. New complex recombinant genotype of hepatitis b virus identified in Vietnam. J Virol (2008) 82:5657–63. doi: 10.1128/JVI.02556-07
14. Phung TB, Alestig E, Nguyen TL, Hannoun C, Lindh M. Genotype X/C recombinant (putative genotype I) of hepatitis b virus is rare in Hanoi, Vietnam–genotypes B4 and C1 predominate. J Med Virol (2010) 82:1327–33. doi: 10.1002/jmv.21775
15. Tatematsu K, Tanaka Y, Kurbanov F, Sugauchi F, Mano S, Maeshiro T, et al. A genetic variant of hepatitis b virus divergent from known human and ape genotypes isolated from a Japanese patient and provisionally assigned to new genotype J. J Virol (2009) 83:10538–47. doi: 10.1128/JVI.00462-09
16. Pourkarim MR, Amini-Bavil-Olyaee S, Kurbanov F, Van Ranst M, Tacke F. Molecular identification of hepatitis b virus genotypes/subgenotypes: revised classification hurdles and updated resolutions. World J Gastroenterol (2014) 20:7152–68. doi: 10.3748/wjg.v20.i23.7152
17. Ezzikouri S, Kayesh MEH, Benjelloun S, Kohara M, Tsukiyama-Kohara K. Targeting host innate and adaptive immunity to achieve the functional cure of chronic hepatitis b. Vaccines (Basel) (2020) 8:216. doi: 10.3390/vaccines8020216
18. Lok AS, Lai CL, Leung N, Yao GB, Cui ZY, Schiff ER, et al. Long-term safety of lamivudine treatment in patients with chronic hepatitis b. Gastroenterology (2003) 125:1714–22. doi: 10.1053/j.gastro.2003.09.033
19. Minami M. Future therapy for hepatitis b virus infection. Clin J Gastroenterol (2015) 8:167–71. doi: 10.1007/s12328-015-0590-y
20. Ye B, Liu X, Li X, Kong H, Tian L, Chen Y. T-Cell exhaustion in chronic hepatitis b infection: current knowledge and clinical significance. Cell Death Dis (2015) 6:e1694. doi: 10.1038/cddis.2015.42
21. Chen YC, Chu CM, Liaw YF. Age-specific prognosis following spontaneous hepatitis b e antigen seroconversion in chronic hepatitis b. Hepatology (2010) 51:435–44. doi: 10.1002/hep.23348
22. Akcay IM, Katrinli S, Ozdil K, Doganay GD, Doganay L. Host genetic factors affecting hepatitis b infection outcomes: Insights from genome-wide association studies. World J Gastroenterol (2018) 24:3347–60. doi: 10.3748/wjg.v24.i30.3347
23. Keating GM, Noble S. Recombinant hepatitis b vaccine (Engerix-b): a review of its immunogenicity and protective efficacy against hepatitis b. Drugs (2003) 63:1021–51. doi: 10.2165/00003495-200363100-00006
24. Kayesh MEH, Kohara M, Tsukiyama-Kohara K. Toll-like receptor response to hepatitis b virus infection and potential of tlr agonists as immunomodulators for treating chronic hepatitis b: an overview. Int J Mol Sci (2021) 22:10462. doi: 10.3390/ijms221910462
25. Zuniga EI, Macal M, Lewis GM, Harker JA. Innate and adaptive immune regulation during chronic viral infections. Annu Rev Virol (2015) 2:573–97. doi: 10.1146/annurev-virology-100114-055226
26. Takeuchi O, Akira S. Pattern recognition receptors and inflammation. Cell (2010) 140:805–20. doi: 10.1016/j.cell.2010.01.022
27. Kawai T, Akira S. Toll-like receptors and their crosstalk with other innate receptors in infection and immunity. Immunity (2011) 34:637–50. doi: 10.1016/j.immuni.2011.05.006
28. Civril F, Deimling T, De Oliveira Mann CC, Ablasser A, Moldt M, Witte G, et al. Structural mechanism of cytosolic DNA sensing by cGAS. Nature (2013) 498:332–7. doi: 10.1038/nature12305
29. Gao P, Ascano M, Zillinger T, Wang W, Dai P, Serganov AA, et al. Structure-function analysis of STING activation by c[G(2’,5’)pA(3’,5’)p] and targeting by antiviral DMXAA. Cell (2013) 154:748–62. doi: 10.1016/j.cell.2013.07.023
30. Mogensen TH. IRF and STAT transcription factors - from basic biology to roles in infection, protective immunity, and primary immunodeficiencies. Front Immunol (2018) 9:3047. doi: 10.3389/fimmu.2018.03047
31. Gehring AJ, Protzer U. Targeting innate and adaptive immune responses to cure chronic HBV infection. Gastroenterology (2019) 156:325–37. doi: 10.1053/j.gastro.2018.10.032
32. Han Y, Liu D, Li L. PD-1/PD-L1 pathway: current researches in cancer. Am J Cancer Res (2020) 10:727–42.
33. Tang H, McLachlan A. Transcriptional regulation of hepatitis b virus by nuclear hormone receptors is a critical determinant of viral tropism. Proc Natl Acad Sci USA (2001) 98:1841–6. doi 10.1073/pnas.98.4.184
34. Kim DH, Kang HS, Kim KH. Roles of hepatocyte nuclear factors in hepatitis b virus infection. World J Gastroenterol (2016) 22:7017–29. doi: 10.3748/wjg.v22.i31.7017
35. Mercer DF, Schiller DE, Elliott JF, Douglas DN, Hao C, Rinfret A, et al. Hepatitis c virus replication in mice with chimeric human livers. Nat Med (2001) 7:927–33. doi: 10.1038/90968
36. Nakagawa S, Hirata Y, Kameyama T, Tokunaga Y, Nishito Y, Hirabayashi K, et al. Targeted induction of interferon-lambda in humanized chimeric mouse liver abrogates hepatotropic virus infection. PloS One (2013) 8:e59611. doi 10.1371/journal.pone.0059611
37. Tsukiyama-Kohara K, Kohara M. Tupaia belangeri as an experimental animal model for viral infection. Exp Anim (2014) 63:367–74. doi: 10.1538/expanim.63.367
38. Fan Y, Huang ZY, Cao CC, Chen CS, Chen YX, Fan DD, et al. Genome of the Chinese tree shrew. Nat Commun (2013) 4:1426. doi: 10.1038/ncomms2416
39. Sanada T, Tsukiyama-Kohara K, Shin IT, Yamamoto N, Kayesh MEH, Yamane D, et al. Construction of complete tupaia belangeri transcriptome database by whole-genome and comprehensive RNA sequencing. Sci Rep (2019) 9:12372. doi: 10.1038/s41598-019-48867-x
40. Walter E, Keist R, Niederost B, Pult I, Blum HE. Hepatitis b virus infection of tupaia hepatocytes in vitro and in vivo. Hepatology (1996) 24:1–5. doi: 10.1053/jhep.1996.v24.pm0008707245
41. Wang Q, Schwarzenberger P, Yang F, Zhang J, Su J, Yang C, et al. Experimental chronic hepatitis b infection of neonatal tree shrews (Tupaia belangeri chinensis): a model to study molecular causes for susceptibility and disease progression to chronic hepatitis in humans. Virol J (2012) 9:170. doi: 10.1186/1743-422X-9-170
42. Yang C, Ruan P, Ou C, Su J, Cao J, Luo C, et al. Chronic hepatitis b virus infection and occurrence of hepatocellular carcinoma in tree shrews (Tupaia belangeri chinensis). Virol J (2015) 12:26. doi: 10.1186/s12985-015-0256-x
43. Sanada T, Tsukiyama-Kohara K, Yamamoto N, Ezzikouri S, Benjelloun S, Murakami S, et al. Property of hepatitis b virus replication in tupaia belangeri hepatocytes. Biochem Biophys Res Commun (2016) 469:229–35. doi: 10.1016/j.bbrc.2015.11.121
44. Kayesh MEH, Ezzikouri S, Chi H, Sanada T, Yamamoto N, Kitab B, et al. Interferon-beta response is impaired by hepatitis b virus infection in tupaia belangeri. Virus Res (2017) 237:47–57. doi: 10.1016/j.virusres.2017.05.013
45. Kock J, Nassal M, Macnelly S, Baumert TF, Blum HE, Von Weizsacker F. Efficient infection of primary tupaia hepatocytes with purified human and woolly monkey hepatitis b virus. J Virol (2001) 75:5084–9. doi: 10.1128/JVI.75.11.5084-5089.2001
46. Yan H, Zhong G, Xu G, He W, Jing Z, Gao Z, et al. Sodium taurocholate cotransporting polypeptide is a functional receptor for human hepatitis b and d virus. Elife (2012) 1:e00049. doi: 10.7554/eLife.00049
47. Kayesh MEH, Hashem MA, Kitab B, Tsukiyama-Kohara K. Pathogenesis and immune response caused by vector-borne and other viral infections in a tupaia model. Microorganisms (2019) 7:686. doi: 10.3390/microorganisms7120686
48. Hayashi S, Khan A, Simons BC, Homan C, Matsui T, Ogawa K, et al. An association between core mutations in hepatitis b virus genotype f1b and hepatocellular carcinoma in alaskan native people. Hepatology (2019) 69:19–33. doi 10.1002/hep.30111
49. Tanaka T, Inoue K, Hayashi Y, Abe A, Tsukiyama-Kohara K, Nuriya H, et al. Virological significance of low-level hepatitis b virus infection in patients with hepatitis c virus associated liver disease. J Med Virol (2004) 72:223–9. doi: 10.1002/jmv.10566
50. Yamamoto N, Sato Y, Munakata T, Kakuni M, Tateno C, Sanada T, et al. Novel pH-sensitive multifunctional envelope-type nanodevice for siRNA-based treatments for chronic HBV infection. J Hepatol (2016) 64:547–55. doi: 10.1016/j.jhep.2015.10.014
51. Shinkai N, Matsuura K, Sugauchi F, Watanabe T, Murakami S, Iio E, et al. Application of a newly developed high-sensitivity HBsAg chemiluminescent enzyme immunoassay for hepatitis b patients with HBsAg seroclearance. J Clin Microbiol (2013) 51:3484–91. doi: 10.1128/JCM.00726-13
52. Sanada T, Yasui F, Honda T, Kayesh MEH, Takano JI, Shiogama Y, et al. Avian H5N1 influenza virus infection causes severe pneumonia in the northern tree shrew (Tupaia belangeri). Virology (2019) 529:101–10. doi: 10.1016/j.virol.2019.01.015
53. Kayesh MEH, Sanada T, Kohara M, Tsukiyama-Kohara K. Tree shrew as an emerging small animal model for human viral infection: A recent overview. Viruses (2021) 13:1641. doi: 10.3390/v13081641
54. Schonrich G, Raftery MJ. The PD-1/PD-L1 axis and virus infections: a delicate balance. Front Cell Infect Microbiol (2019) 9:207. doi: 10.3389/fcimb.2019.00207
55. Wieland S, Thimme R, Purcell RH, Chisari FV. Genomic analysis of the host response to hepatitis b virus infection. Proc Natl Acad Sci USA (2004) 101:6669–74. doi 10.1073/pnas.040177110
56. Fletcher SP, Chin DJ, Cheng DT, Ravindran P, Bitter H, Gruenbaum L, et al. Identification of an intrahepatic transcriptional signature associated with self-limiting infection in the woodchuck model of hepatitis b. Hepatology (2013) 57:13–22. doi: 10.1002/hep.25954
57. Guy CS, Mulrooney-Cousins PM, Churchill ND, Michalak TI. Intrahepatic expression of genes affiliated with innate and adaptive immune responses immediately after invasion and during acute infection with woodchuck hepadnavirus. J Virol (2008) 82:8579–91. doi: 10.1128/JVI.01022-08
58. Dunn C, Peppa D, Khanna P, Nebbia G, Jones M, Brendish N, et al. Temporal analysis of early immune responses in patients with acute hepatitis b virus infection. Gastroenterology (2009) 137:1289–300. doi: 10.1053/j.gastro.2009.06.054
59. Fisicaro P, Valdatta C, Boni C, Massari M, Mori C, Zerbini A, et al. Early kinetics of innate and adaptive immune responses during hepatitis b virus infection. Gut (2009) 58:974–82. doi: 10.1136/gut.2008.163600
60. Liu HY, Zhang XY. Innate immune recognition of hepatitis b virus. World J Hepatol (2015) 7:2319–22. doi: 10.4254/wjh.v7.i21.2319
61. Zhang Z, Trippler M, Real CI, Werner M, Luo X, Schefczyk S, et al. Hepatitis b virus particles activate toll-like receptor 2 signaling initially upon infection of primary human hepatocytes. Hepatology (2020) 72:829–44. doi: 10.1002/hep.31112
62. Isogawa M, Robek MD, Furuichi Y, Chisari FV. Toll-like receptor signaling inhibits hepatitis b virus replication in vivo. J Virol (2005) 79:7269–72. doi: 10.1128/JVI.79.11.7269-7272.2005
63. Das D, Sarkar N, Sengupta I, Pal A, Saha D, Bandopadhyay M, et al. Anti-viral role of toll like receptor 4 in hepatitis b virus infection: An in vitro study. World J Gastroenterol (2016) 22:10341–52. doi: 10.3748/wjg.v22.i47.10341
64. Zhang E, Lu M. Toll-like receptor (TLR)-mediated innate immune responses in the control of hepatitis b virus (HBV) infection. Med Microbiol Immunol (2015) 204:11–20. doi: 10.1007/s00430-014-0370-1
65. Ma Z, Cao Q, Xiong Y, Zhang E, Lu M. Interaction between hepatitis b virus and toll-like receptors: Current status and potential therapeutic use for chronic hepatitis b. Vaccines (Basel) (2018) 6:6. doi: 10.3390/vaccines6010006
66. Ma Z, Liu J, Wu W, Zhang E, Zhang X, Li Q, et al. The IL-1R/TLR signaling pathway is essential for efficient CD8(+) T-cell responses against hepatitis b virus in the hydrodynamic injection mouse model. Cell Mol Immunol (2017) 14:997–1008. doi: 10.1038/cmi.2017.43
67. Wu J, Meng Z, Jiang M, Pei R, Trippler M, Broering R, et al. Hepatitis b virus suppresses toll-like receptor-mediated innate immune responses in murine parenchymal and nonparenchymal liver cells. Hepatology (2009) 49:1132–40. doi: 10.1002/hep.22751
68. Wang H, Ryu WS. Hepatitis b virus polymerase blocks pattern recognition receptor signaling via interaction with DDX3: implications for immune evasion. PloS Pathog (2010) 6:e1000986. doi: 10.1371/journal.ppat.1000986
69. Chen J, Wu M, Zhang X, Zhang W, Zhang Z, Chen L, et al. Hepatitis b virus polymerase impairs interferon-alpha-induced STA T activation through inhibition of importin-alpha5 and protein kinase c-delta. Hepatology (2013) 57:470–82. doi: 10.1002/hep.26064
70. Ferrari C. HBV and the immune response. Liver Int (2015) 35 Suppl 1:121–8. doi: 10.1111/liv.12749
Keywords: hepatitis B virus, tree shrew, toll-like receptors, transcription factors, cytokines
Citation: Kayesh MEH, Hashem MA, Sanada T, Kitab B, Rashid MHO, Akter L, Ezzikouri S, Murakami S, Ogawa S, Tanaka Y, Kohara M and Tsukiyama-Kohara K (2022) Characterization of innate immune response to hepatitis B virus genotype F acute infection in tree shrew (Tupaia belangeri) model. Front. Virol. 2:926831. doi: 10.3389/fviro.2022.926831
Received: 23 April 2022; Accepted: 29 July 2022;
Published: 18 August 2022.
Edited by:
Ishan Patro, Jiwaji University, IndiaReviewed by:
Tomasz I Michalak, Memorial University of Newfoundland, CanadaYong-Tang Zheng, Kunming Institute of Zoology, Chinese Academy of Sciences (CAS), China
Ye Zhang, Tangdu Hospital, China
Copyright © 2022 Kayesh, Hashem, Sanada, Kitab, Rashid, Akter, Ezzikouri, Murakami, Ogawa, Tanaka, Kohara and Tsukiyama-Kohara. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Kyoko Tsukiyama-Kohara, a2tvaGFyYUB2ZXQua2Fnb3NoaW1hLXUuYWMuanA=
 Mohammad Enamul Hoque Kayesh
Mohammad Enamul Hoque Kayesh Md Abul Hashem
Md Abul Hashem Takahiro Sanada
Takahiro Sanada Bouchra Kitab1,2
Bouchra Kitab1,2 Sayeh Ezzikouri
Sayeh Ezzikouri Shuko Murakami
Shuko Murakami Michinori Kohara
Michinori Kohara Kyoko Tsukiyama-Kohara
Kyoko Tsukiyama-Kohara