- 1GeneOne Life Science, Inc., Seoul, South Korea
- 2Korea Disease Control and Prevention Agency, Center for Vaccine Research, National Institute of Infectious Diseases, National Institute of Health, Cheongju-si, South Korea
- 3Department of Mechanical and Aerospace Engineering, Rutgers, The State University of New Jersey, New Brunswick, NJ, United States
- 4Department of Biomedical and Diagnostic Sciences, University of Tennessee, Knoxville, TN, United States
- 5Department of Biomedical Engineering, Rutgers, The State University of New Jersey, New Brunswick, NJ, United States
- 6PharmaJet, Inc., Golden, CO, United States
- 7Bioqual, Rockville, MD, United States
- 8Vaccine & Immunotherapy Center, The Wistar Institute, Philadelphia, PA, United States
SARS-CoV-2 is the third pathogenic coronavirus to emerge since 2000. Experience from prior outbreaks of SARS-CoV and MERS-CoV has demonstrated the importance of both humoral and cellular immunity to clinical outcome, precepts that have been recapitulated for SARS-CoV-2. Despite the unprecedented rapid development and deployment of vaccines against SARS-CoV-2, more vaccines are needed to meet global demand and to guard against immune evasion by newly emerging SARS-CoV-2 variants. Here we describe the development of pGO-1002, a novel bi-cistronic synthetic DNA vaccine that encodes consensus sequences of two SARS-CoV-2 antigens, Spike and ORF3a. Mice immunized with pGO-1002 developed humoral and cellular responses to both antigens, including antibodies and capable of neutralizing infection by a clinical SARS-CoV-2 isolate. Rats immunized with pGO-1002 by intradermal (ID) injection followed by application of suction with our GeneDerm device also developed humoral responses that included neutralizing antibodies and RBD-ACE2 blocking antibodies as well as robust cellular responses to both antigens. Significantly, in a Syrian hamster vaccination and challenge model, ID+GeneDerm-assisted vaccination prevented viral replication in the lungs and significantly reduced viral replication in the nares of hamsters challenged with either an ancestral SARS-CoV-2 strain or the B.1.351 (Beta) variant of concern. Furthermore, vaccinated immune sera inhibited virus-mediated cytopathic effects in vitro. These data establish the immunogenicity of the SARS-CoV-2 vaccine candidate pGO-1002 which induces potent humoral and cellular responses to the Spike and ORF3a antigens and may provide greater protection against emerging variants.
Introduction
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), the etiologic cause of coronavirus disease 2019 (COVID-19), emerged in late 2019 and quickly spread globally in 2020, causing a pandemic. SARS-CoV-2 is the third recently emergent pathogenic human coronavirus following SARS-CoV in 2002-2003 and the Middle East respiratory syndrome coronavirus (MERS-CoV) first identified in 2012. While SARS-CoV and MERS-CoV infections have higher case-fatality rates than SARS-CoV-2, both are less transmissible with case numbers less than 10,000 for each compared to over 250 million cases and over 5 million deaths caused by SARS-CoV-2. As of December 2021, over 330 SARS-CoV-2 vaccine candidates were in various stages of development or clinical testing, with 22 of these being in active use after receiving regulatory approval or emergency use authorization (EAU) in at least one country (1). Despite the success of these initially approved vaccines at slowing the spread of SARS-CoV-2, more vaccines are needed to meet global demand, specifically in low- and middle-income countries (LMICs), where limited resources and/or lack of medical infrastructure can interfere with obtaining and/or distributing currently approved vaccines (2). Moreover, the emergence of viral variants and recent reports suggesting that vaccine-induced protection against SARS-CoV-2 may wane over time substantiate the continued development of additional SARS-CoV-2 vaccines to meet current and future global demand (3).
Previous efforts in developing vaccines for SARS-CoV and MERS-CoV established that their Spike (S) glycoproteins were potent inducers of humoral and cellular immune responses that could protect animal models from homologous virus challenges (4–6). Most SARS-CoV-2 vaccines in use or development are designed to elicit responses to a single SARS-CoV-2 antigen: the Spike glycoprotein (7). Evaluations of vaccines in animal models have suggested that titers of anti-Spike neutralizing antibodies (nAbs) correlate with vaccine efficacy, but studies of COVID-19 patients suggest that reduced disease severity correlates more with the presence of SARS-CoV-2-specific CD4+ and CD8+ T cells (8, 9). Accumulated evidence from studies of COVID-19 patients suggests that a coordinated adaptive immune response involving rapid induction of SARS-CoV-2-specific CD4+ and CD8+ T cells as well as virus-specific neutralizing antibodies (nAbs) is associated with a milder disease course, with T cells playing a larger role in clearing primary SARS-CoV-2 infection and neutralizing antibodies providing protection from subsequent infections (10). Several other lines of evidence indicate that cellular responses play a significant role in protection from COVID-19. For example, efficacy of the BNT162b mRNA vaccine (Pfizer/BioNTech) has been observed as early as 12 days post-prime immunization, a time when nAbs are largely undetectable but cellular responses are evident (11, 12). Individuals with hematological cancers or those receiving anti-CD20 monoclonal antibody (mAb) therapy (B-cell depleting) have lower antibody responses to SARS-CoV-2 but still develop virus-specific CD8+ T cells which is associated with improvement in survival for this group (13, 14). While antibody responses to SARS-CoV-2 are detectable out to a year post-infection, they have decreased ability to neutralize some variants of SARS-CoV-2, particularly the B.1.351 (Beta) strain (15). However, T cell responses appear to be resistant to mutations that have arisen in SARS-CoV-2 and are thus likely to protect from disease even if breakthrough infections occur due to reduced quantity or activity of nAbs (15, 16). Studies of recovered SARS patients found that while antibody levels dropped after a year post-infection, T cell responses persisted and could be detected out to 17 years later (17, 18).
Here we used the DNA vaccine platform to develop a vaccine candidate, pGO-1002, that expresses two SARS-CoV-2 antigens Spike and ORF3a. The DNA vaccine platform is ideally suited for meeting the challenge of vaccinating residents in LMICs since these vaccines are cost-effective to manufacture in bulk and thermostable with limited cold-chain requirements for distribution. SARS-CoV-2 ORF3a, one of two putative viroporins (along with Envelope) encoded by SARS-CoV-2, forms a Ca2+ permeable cation channel in membranes that is hypothesized to play a role in virus assembly and release (19). Recombinant SARS-CoV-2 with the orf3a gene deleted showed reduced pathogenesis, including decreased cytokine storm and lung tissue damage in a K-18-hACE2 transgenic mouse model (20). The decision to incorporate the SARS-CoV-2 ORF3a into pGO-1002 was made to increase the breadth of T cell immunity induced by our vaccine as preclinical and clinical studies of SARS-CoV and MERS-CoV infection suggested that outcome directly correlated with CD8+ T cell responses (21). ORF3a specifically was found to be an immunodominant T cell antigen for SARS-CoV that induced responses equal in magnitude and frequency to those induced by the SARS-CoV Spike and Nucleocapsid and were detectable to 6 years post-infection (17, 22). Additionally, ORF3a was found to be a target of humoral responses in recovered SARS patients (23, 24). A DNA vaccine encoding SARS-CoV ORF3a induced antigen-specific humoral responses as well as a Th1-based cellular response in mice (25). Several studies of COVID-19 patients have found that SARS-CoV-2 ORF3a is also a target of humoral and cellular immune responses, further underscoring the utility of using ORF3a as a vaccine antigen (26–29). SARS-CoV-ORF3a has been found to activate the NLRP3 inflammasome through TRAF-3 dependent ubiquitination (30) and to inhibit interferon-activated Janus Kinase/signal transducer activator of transcription (JAK/STAT) signaling which suggests that it plays a central role in the pathogenesis of SARS-CoV-2 infection (31).Therefore, ORF3a targeting may provide direct interruption of viral pathogenesis following initial cellular invasion.
Mice immunized with pGO-1002 developed humoral and cellular responses to both Spike and ORF3a that were equivalent to responses induced against each antigen after immunization with Spike- or ORF3a-only vaccines. Full seroconversion to Spike was observed after one immunization with pGO-1002 and humoral responses in pGO-1002-immunized mice blocked binding between the Spike RBD and its cellular receptor ACE2 as well as neutralized infection by a clinical SARS-CoV-2 isolate. Recently we developed GeneDerm, an easy-to-use, cost-effective, and scalable device that applies suction to an intradermal delivery site to enhance in vivo gene expression of DNA plasmids (32). Rats immunized with pGO-1002 by GeneDerm-assisted intradermal (ID) delivery developed cellular responses to Spike and ORF3a as well as Spike-targeting humoral responses that blocked Spike RBD-ACE2 binding and neutralized SARS-CoV-2 infection. GeneDerm-assisted ID immunization with pGO-1002 induced immunity that blocked virus replication in the lungs and reduced virus replication in the nares of Syrian golden hamsters challenged with either wild type SARS-CoV-2 or the B.1.351 (Beta) variant. These data supported the advancement of the SARS-CoV-2 DNA vaccine, GLS-5310 (formulated from pGO-1002), with GeneDerm-enhanced ID delivery into Phase I and Phase 2 clinical trials (NCT04673149).
Results
SARS-CoV-2 DNA Vaccine Design and Characterization
Three mono or bi-cistronic plasmid constructs, pGO-1001, pGO-1002, and pGO-1003, were designed encoding sequence gene cassettes for SARS-CoV-2 Spike, Spike plus ORF3a, and ORF3a, respectively (Figure 1A). The consensus sequences for Spike and ORF3a were generated from publicly available SARS-CoV-2 sequences through mid-February 2020 that were subsequently codon-optimized. For the bi-cistronic construct pGO-1002, a furin cleavage site and a self-cleaving T2A peptide sequence were added between the Spike and ORF3a coding sequences to ensure both proteins were generated after expression and translation of the antigen cassette. An IgE leader sequence was added upstream of the antigen cassettes in each construct. All constructs were made in the pGLS-101 expression vector, including an upstream human cytomegalovirus enhancer/promoter (6).
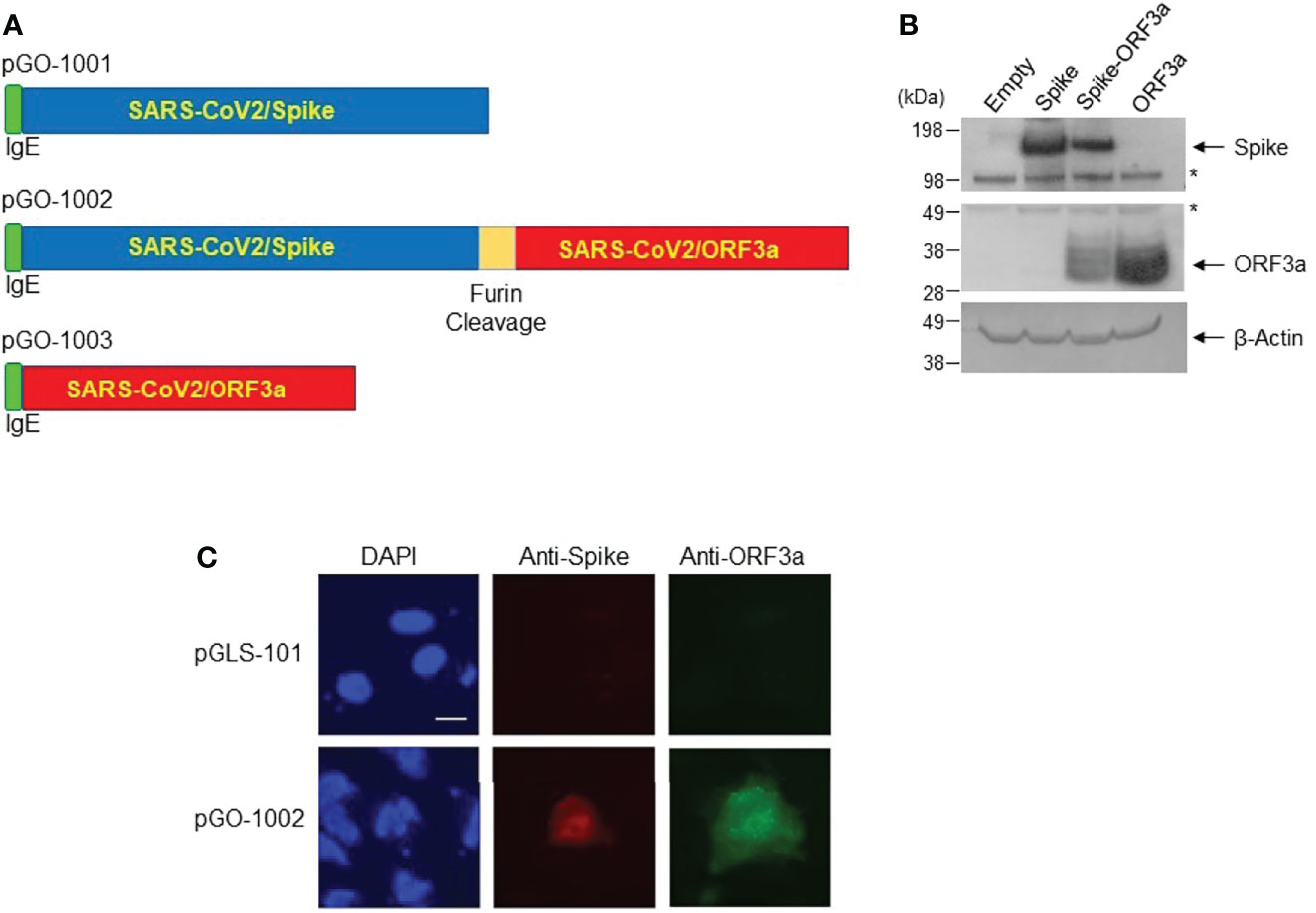
Figure 1 Construction of DNA vaccine constructs and confirmation of expression in eukaryotic cells. (A) Maps of antigen cassettes for plasmids pGO-1001, pGO-1002 and pGO-1003. Antigen cassettes were cloned into the pGLS-101 backbone. (B) SARS-CoV-2 proteins’ expression was determined in cell lysates from 293T cells transfected with pGO-1001, pGO-1002, pGO-1003, or empty pGLS-101. Membranes were blotted sequentially with antibodies to SARS-CoV-2 Spike and ORF3a, as well as β-actin (loading control). Western analysis of cell lysate confirms the presence of SARS-CoV-2 Spike (MW ~140kDa) and/or ORF3a (MW ~31kDa) in transfected cell lysates. The asterisk (*) indicates nonspecific bands. (C) Immunofluorescence labeling of Huh-7 cells transfected with either pGLS-101 or pGO-1002. Cells were fixed and permeabilized 48-hours-post transfection and then incubated sequentially with mouse anti-SARS-CoV-2 Spike or rabbit anti-SARS-CoV-2 ORF3a antibodies followed by incubation with goat anti-mouse IgG-Alexa 568 or goat anti-rabbit IgG-Alexa 488 antibodies, respectively for detection. Nuclei are stained with DAPI. Bar represents 20μm.
To confirm that each antigen was adequately expressed, lysates collected from 293T cells transfected with each of the three plasmids were subjected to Western blot and immunofluorescent analysis (IFA) using antibodies specific to SARS-CoV-2 Spike and ORF3a. Western blot analysis showed that lysates from cells transfected with either mono-cistronic plasmid (pGO-1001 or pGO-1003) contained bands of the expected size for Spike (~140 kDa) or ORF3a (~31 kDa). Lysates from cells transfected with the bi-cistronic pGO-1002 construct had bands of the anticipated sizes of both Spike and ORF3a (Figure 1B). In addition, IFA of 293T cells detected both Spike and ORF3a proteins in pGO-1002 transfected cells (Figure 1C). These data indicate that the production of SARS-CoV-2 Spike and/or ORF3a occurred as expected from each vaccine construct.
Humoral Immune Responses of the pGO-1002 Vaccine in Mice
Safety and immunogenicity of the three candidate vaccines were first evaluated in mice. Groups of five mice (7 weeks of age) were immunized intramuscularly (IM) in the anterior tibialis muscle with 50µg of one of the three vaccine plasmids or empty pGLS-101 (Supplementary Figure S1A). Electroporation (6 pulses at 200V/cm, 50ms, 1Hz) was applied to the injection site using the BTX ECM 830 device. Animals were immunized twice, 14 days apart, and bled at days 0, 14, and 28 to collect sera for immunoassays. Animals were sacrificed on day 28 to obtain spleens for evaluation of cellular responses to the vaccine antigens. All animals remained healthy, without signs of illness and had similar weight gains through the study (Supplementary Figure S1B).
The presence of antibodies to the Spike S1 subunit and to ORF3a in sera were determined by ELISA. A single immunization with pGO-1001 or pGO-1002 induced full seroconversion to Spike in both groups, and equivalent endpoint titers (~104) were seen in both groups by 2 weeks post second immunization (Figure 2A). Two immunizations with either pGO-1003 or pGO-1002 were required for full seroconversion to ORF3a and yielded endpoint titers after the 2nd immunization of 3x103 and ~103, respectively (Figure 2B).
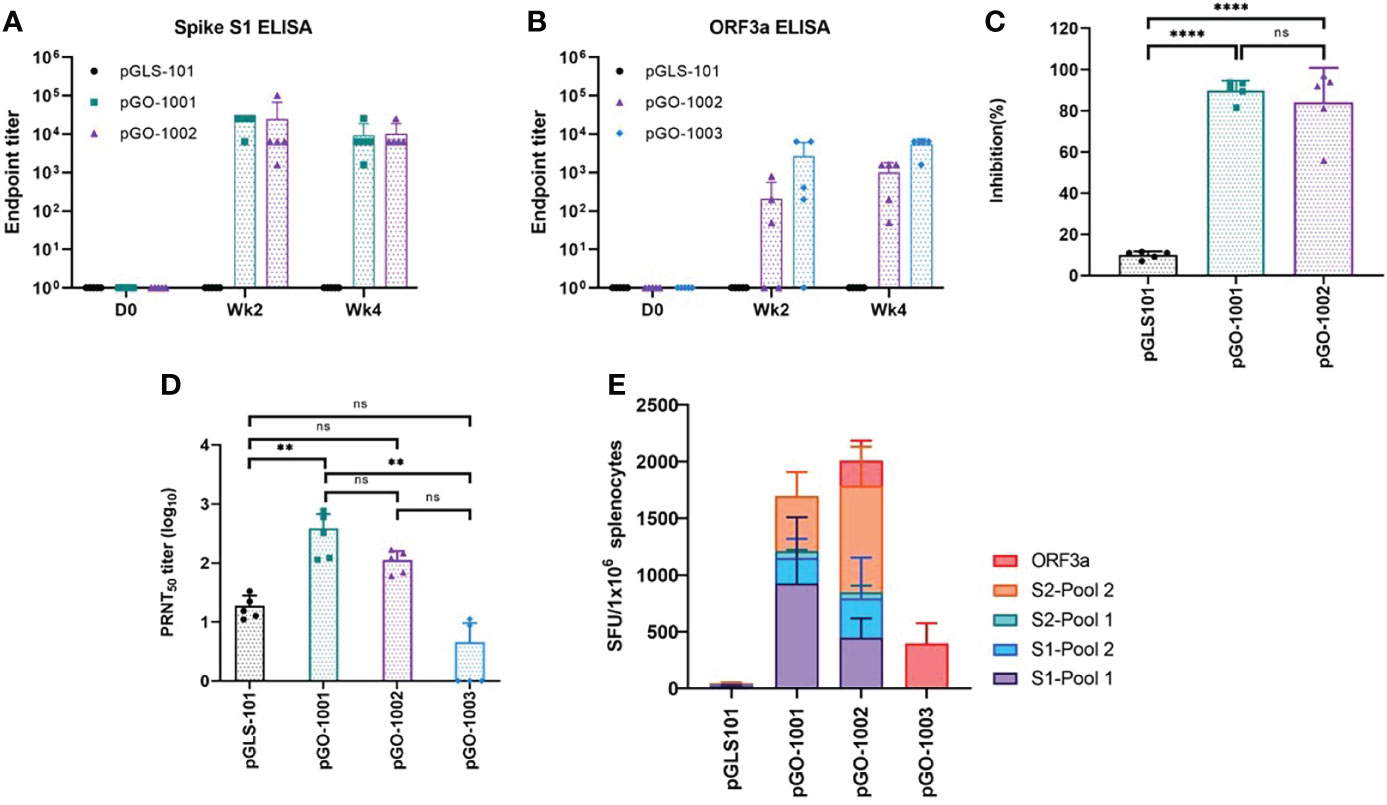
Figure 2 Immunogenicity of SARS-CoV-2 DNA vaccine constructs in mice. All collected serum samples were analyzed by ELISA for binding to SARS-CoV-2 Spike S1 (A) or ORF3a (B). Endpoint titers were calculated as the reciprocal of the highest sera dilution factor whose OD 405nm value was greater than 2.5 standard deviations of the OD 405nm of the day 0 value at that dilution. Each sample was run with three technical replicates on the plate. Day 28 sera from vaccinated mice were evaluated in a surrogate neutralization assay (C) for their ability to block the ACE2-RBD interaction and in a PRNT50 assay (D) for their ability to block infection of βCoV/Korea/KCDC03/2020 SARS-CoV-2 in Vero cells. Data are Mean + SD of percent inhibition of sera from each mouse as compared to inhibition by negative control or mean+ SD of PRNT50 of sera from each mouse. ** = 0.001 to 0.01; *** = 0.0001 to 0.001 (E) Overall splenic T-cell responses. Splenocytes prepared from all mice on day 28 were evaluated by IFN-γ ELISpot assay to characterize cell-mediated responses. Splenocytes were stimulated with each of four linear pools of peptides spanning SARS CoV-2 Spike or a pool of linear peptides spanning SARS-CoV-2 ORF3a. Data represent the mean SFU per 106 splenocytes + SD. Statistical analysis was performed in (C, D) using a one-way ANOVA comparison test where the mean percent inhibition of each group was compared against each other. ns, not significant.
To assess the humoral responses’ functionality, we performed surrogate neutralization assays and/or traditional plaque reduction neutralization titer (PRNT50) assays using sera collected on day 28 (2 weeks post-second injection) from immunized mice. The surrogate neutralization assay measures the ability of sera to inhibit binding between a recombinant SARS-CoV-2 Spike RBD protein and plate-bound ACE2 cellular receptor protein. In this assay, sera from mice receiving two immunizations of either pGO-1001 or pGO-1002 inhibited RBD-ACE2 binding by over 80% (Figure 2C). PRNT50 assays were performed under BSL3 conditions by the Korean Center for Disease Control (KCDC). They measured the ability of each serum to block in vitro infection of Vero cells by the SARS-CoV-2 clinical isolate β-CoV/Korea/KCKC03/2020. In this assay, sera from mice immunized twice with either pGO-1001 or pGO-1002 neutralized infection to an equivalent level (PRNT50 titer ~100) while sera from mice immunized with pGO-1003 were unable to neutralize infection (Figure 2D). These results show that immunization with pGO-1002 induces humoral responses to both encoded SARS-CoV-2 antigens, Spike and ORF3a, and the induced responses to Spike are functional toward blocking binding between the Spike RBD and ACE2 as well as neutralizing infection by a clinical SARS-CoV-2 isolate.
Cellular Immune Responses Induced in Mice Immunized With pGO-1002
Induction of cellular responses to Spike and/or ORF3a was evaluated by performing an IFN-γ ELISpot assay on splenocytes isolated from spleens collected from immunized mice on day 28. Splenocytes were stimulated in vitro with each of four linear pools of peptides spanning the full length of Spike and a pool of linear peptides spanning the full length of ORF3a. Mice immunized with pGO-1001 generated strong cellular responses to each Spike peptide pool, while mice receiving pGO-1003 developed cellular responses to the ORF3a peptide pool. Mice immunized with bi-cistronic construct pGO-1002 generated T cell responses against both the Spike peptide pools and ORF3a peptide pool that mirrored those seen against each peptide pool in mice immunized with pGO-1001 or pGO-1003 (Figure 2E). These results show that the bi-cistronic construct pGO-1002 induced equivalent humoral and cellular responses to Spike and ORF3a as induced by mono-cistronic constructs of each antigen.
These results establish that pGO-1002 induced Spike- and ORF3a-specific humoral and cellular immune responses with similar kinetics and magnitude as induced by monocistronic constructs of each antigen. Based on these results we selected to advance pGO-1002 for further evaluation.
GeneDerm-Assisted Intradermal Immunization of Rats With pGO-1002 Induces Immunity to SARS-CoV-2
In anticipation of future clinical investigation of pGO-1002, we moved to investigate pGO-1002 immunogenicity after intradermal (ID) vaccination as this route is more accessible and more tolerable than intramuscular injection. In a recent study, we developed a new ID delivery protocol for DNA plasmids that involves the application of suction to the ID injection site which they found led to more rapid and robust transgene expression in rat skin (32). Furthermore, they demonstrated that ID immunization with a DNA vaccine followed by suction applied by our GeneDerm device led to more rapid and stronger antibody responses in rats. Here we assessed the immunogenicity of pGO-1002 following GeneDerm-assisted ID delivery in Sprague Dawley (S.D.) rats. Groups of five rats were immunized twice, 2 weeks apart, with either 30µg or 300µg of pGO-1002 or 300µg of empty pGLS-101 by ID injection followed by GeneDerm-applied suction (30 seconds, 65 kPa) at the injection site (Figure 3A). Blood was collected from all animals at days 0, 14 (week 2), and 28 (week 4) for sera to evaluate humoral responses, and animals were sacrificed at day 28 to obtain spleens for evaluation of cellular immune responses.
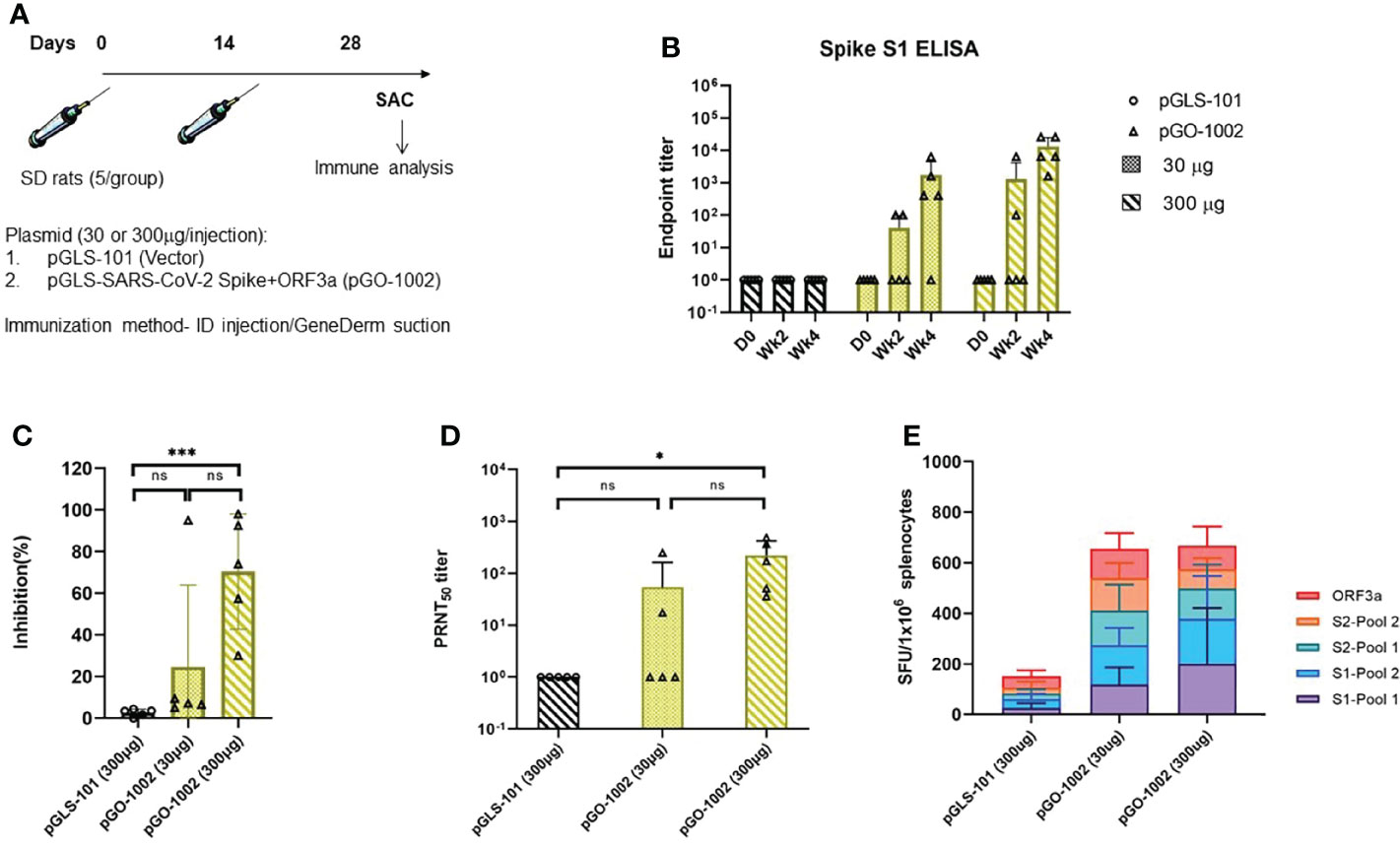
Figure 3 Intradermal DNA immunization followed by application of suction using the GeneDerm device augmented the antigen-specific immune responses in Rats. (A) Schematic representation of rat vaccination schedule. Groups of n = 5 Sprague Dawley (SD) rats were immunized twice with 30 or 300 µg/rat of pGLS-101 or pGO-1002 plasmids at 14-day intervals by intradermal (ID) injection followed by GeneDerm-applied suction (Suction). Rats were bled on days 0, 14, and 28 and sacrificed on day 28 to obtain spleens. ELISA was performed on all collected serum samples for binding to SARS-CoV-2 Spike S1 (B). Endpoint titers were calculated as the reciprocal of the highest sera dilution factor whose OD 405nm value was greater than 2.5 standard deviation of the OD 405nm of the day 0 value at that dilution. Each sample was run with three technical replicates on the plate. A comprehensive evaluation of neutralizing activity in rats. Day 28 sera from vaccinated rats were evaluated in a surrogate neutralization assay (C) for their ability to block the ACE2-RBD interaction and in a PRNT50 assay; *** = 0.0001 to 0.001 (D) for their ability to block infection of βCoV/Korea/KCDC03/2020 SARS-CoV-2 in Vero cells. Data are Mean + SD of percent inhibition of sera from each mouse as compared to inhibition by negative control or mean+SD of PRNT50 of sera from each rat. * = 0.01 to 0.05 (E) Splenocytes prepared from all rats on day 28 were evaluated by IFN-γ ELISpot assay for characterization of cell-mediated responses. Splenocytes were stimulated with each of four linear pools of peptides spanning SARS CoV-2 Spike or a pool of linear peptides spanning SARS-CoV-2 ORF3a. Data represent the mean SFU per 106 splenocytes + SD. Statistical analysis was performed in (C, D) using a one-way ANOVA comparison test where mean percent inhibition of each group was compared against each other.ns, not significant.
Rats with antibodies to SARS-CoV-2 Spike as measured by S1 ELISA were seen after a single immunization of 30μg or 300μg pGO-1002 and the S1 titers were boosted following a second injection of either dose (Figure 3B). All five animals that received two immunizations of 300μg pGO-1002 seroconverted to Spike and had a Spike S1 endpoint titer of ~1.31x104 while four of five animals that received two immunizations of 30μg pGO-1002 seroconverted with a log lower Spike S1 endpoint titer than seen in the 300μg group. The kinetics and Spike antibody endpoint titers in rats receiving pGO-1002 by GeneDerm-assisted ID delivery matched those seen when 300μg pGO-1002 was administered using PharmaJet or EP-enhanced ID delivery (Supplementary Figure S2A). Antibody responses to ORF3a were detected in some animals only in the group receiving two immunizations with 300μg pGO-1002 (data not shown). Sera collected on week 4 from pGO-1002 immunized rats were next evaluated in surrogate neutralization and PRNT50 assays. Sera from rats that received two 30μg doses showed low activity in both the surrogate neutralization and PRNT50 assays (Figures 3C, D). By contrast, sera from rats receiving two 300μg doses of pGO-1002 inhibited RBD-ACE2 binding by 70% in the surrogate neutralization assay, and neutralized in vitro infection by the SARS-CoV-2 clinical isolate β-CoV/Korea/KCKC03/2020 with an average PRNT50 titer ~220 (Figures 3C, D). Surrogate neutralization and PRNT50 titers observed in rats immunized by GeneDerm-assisted ID delivery matched those seen in rats that received pGO-1002 by PharmJet or electroporation enhanced ID delivery (Supplementary Figures S2B, S2C).
An IFN-γ ELISpot was next performed on splenocytes collected from pGO-1002 immunized rats to evaluate the induction of cellular responses to Spike and ORF3a (Figure 3E). Splenocytes isolated on week 4 were stimulated in vitro with each of four linear pools of peptides spanning the full length of Spike and a pool of linear peptides spanning the full length of ORF3a. GeneDerm-assisted ID delivery of pGO-1002 to rats induced robust total cellular responses to S and ORF3a (~650 SFU/10^6 splenocytes). Total cellular responses induced by GeneDerm-assisted ID delivery were higher than cellular responses seen when pGO-1002 was administered ID by Pharmajet or by EP-enhanced ID injection (Supplementary Figure S2D). Notably, T cell responses for animals immunized with 30µg of pGO-1002 by ID+GeneDerm induced an equivalent cellular response as seen in the group receiving 300µg by ID+GeneDerm. Broad reactivity to multiple regions of Spike and ORF3a were seen in pGO-1002 immunized rats. Isotyping of Spike-specific antibodies in rats immunized with pGO-1002 by ID+GeneDerm showed that they were predominantly of the IgG2a/2b subtype consistent with an induction of a Th1 profile of T cell responses by pGO-1002 (Supplementary Figure S3).
These results confirm that GeneDerm-assisted ID delivery of pGO-1002 induces strong cellular responses to Spike and ORF3a as well as humoral responses capable of blocking Spike RBD-ACE2 binding and neutralizing SARS-CoV-2 infection.
Immunization of Syrian Hamsters With GLS-5310 Blocks or Reduces Virus Replication in Lungs and Nares After Challenge With Either Wild Type or B.1.351 Variant SARS-CoV-2 Strains
Based on the positive immunogenicity results in mice and rats, pGO-1002 was reformulated with 1X SSC buffer and designated as GLS-5310. The ability of GLS-5310 to provide protection against SARS-CoV-2 was first evaluated in a Syrian hamster challenge model. Groups of six hamsters were immunized intradermally (ID) twice with 150μg of GLS-5310 or empty pGLS-101 vector on days 0 and 21 (week 3) followed by GeneDerm-applied suction (30 seconds, 65 kPa). Sera were collected from immunized animals on days 0, 21 (week 3), and 41 (week 6). All animals were challenged on day 42 with either a wild type (WT;2019-nCoV/USA-WA1/2020) SARS-CoV-2 strain or the B.1.351 (Beta; 2019-nCoV/South Africa/KRISP-K005325/2020) SARS-CoV-2 variant strain. Challenged animals were sacrificed on day 47 (5 days post-challenge) to collect blood, lung, and nasal turbinate tissues.
Sera collected from immunized animals were evaluated in a modified high-throughput surrogate neutralization assay that uses homogenous time resolved fluorescence (HTRF) technology, where SARS-CoV-2 spike RBD from WT (USA-WA1/2020), Beta (B.1.351), or Delta (B.1.617.2) variants were assessed for ability to interact with ACE2 protein. In this assay, initial experiments showed that 50% human control plasma (i.e., from individuals without SARS-CoV-2 exposure or COVID-19) had no effect on the HTRF signal, while the control inhibitory antibody REGN10933 (casirivimab) blocked HTRF signal due to WT, Beta, or Delta variant RBDs in the presence of human plasma with half-maximal inhibitory concentrations (IC50s) of 47.3, 937.5, and 35.4 ng/mL, consistent with previous reports (33, 34). Serial dilutions of day 0 and 41 sera from hamsters were then evaluated for their ability to inhibit binding between ACE2 and the RBD variants of SARS-CoV-2. Humoral responses in day 41 sera collected from GLS-5310 immunized hamsters were able to inhibit binding between ACE2 and the RBDs of WT or Delta variant SARS-CoV-2 but were unable to block binding between the ACE2 and the RBD from the Beta SARS-CoV-2 variant (Figure 4A). Hamsters immunized with GLS-5310 completely cleared the infectious virus in the lungs and substantially reduced virus levels in the nares 5 days post-challenge with the wt. SARS-CoV-2 strain (Figure 4B). Hamsters immunized with GLS-5310 and challenged with the Beta SARS-CoV-2 variant also had no infectious virus in their lungs 5 days post-challenge (Figure 4C).
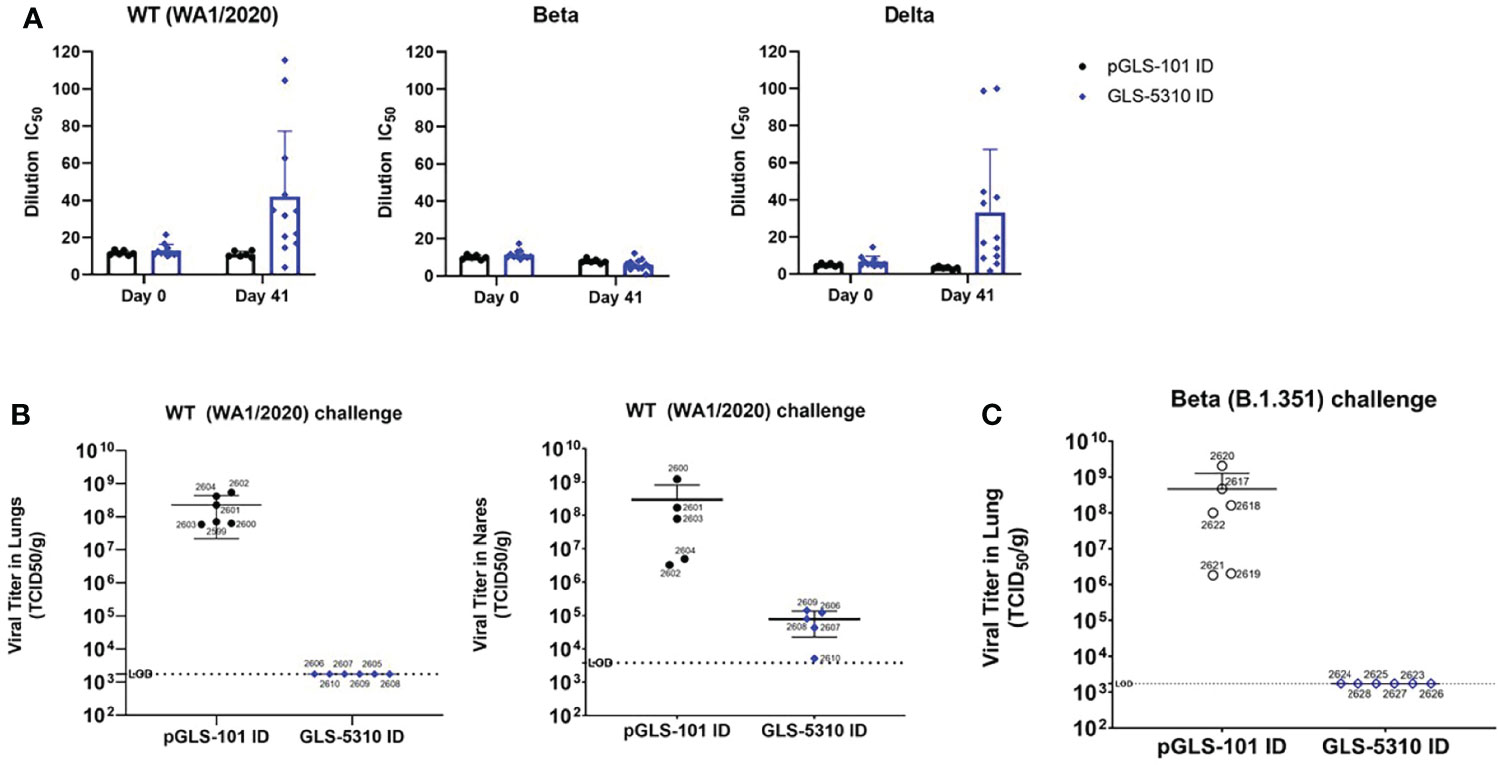
Figure 4 GeneDerm-assisted intradermal immunization of hamsters with GLS-5310 inhibits virus replication in lungs after SARS-CoV-2 challenge. (A) Pre-immune and day 41 sera from Golden Syrian hamsters immunized twice, 3 weeks apart with 150μg pGLS-101 (n = 6) or GLS-5310 (n = 12) by intradermal injection followed by GeneDerm-applied suction were evaluated in (A) modified surrogate neutralization assays for their ability to inhibit interactions between hACE2 and the Spike RBDs of WA1/2020 (Wt.), B.1.351 (Beta) or B.1.617.2 (Delta) SARS-CoV-2 strains. (B) Virus titers in lung and nares tissues collected from animals 5 days-post intranasal challenge (6x103 PFU challenge dose per hamster with the WA1/2020 SARS-CoV-2 strain (Wt.) were evaluated by TCID50 assay. (C) Virus titers in lung tissues collected from animals 5 days-post intranasal challenge with the B.1.351 SARS-CoV-2 strain (Beta) (3.67x102 PFU SARS-CoV-2 dose per hamster) and were evaluated by TCID50 assay.
A separate group of hamsters were immunized twice, 21 days apart with 150μg of GLS-5310 by ID+GeneDerm and 150μg GLS-5310 by intranasal administration and then challenged 3 weeks later (day 42) with wt. SARS-CoV-2. Sera collected on day 41, one day prior to challenge, was able to inhibit binding between ACE2 and the RBDs of wt. or Delta variant SARS-CoV-2, but not between ACE2 and the RBD of the Beta SARS-CoV-2 similar to what was observed in animals immunized with GLS-5310 by ID+GeneDerm alone (Figure 5A). In addition, of five hamsters immunized with GLS-5310 by this ID+GeneDerm/IN route, one demonstrated breakthrough infection in the lungs 5 days post-challenge with wt. SARS-CoV-2, whereas TCID50 for the other four animals were below the limit of detection (Figure 5B).
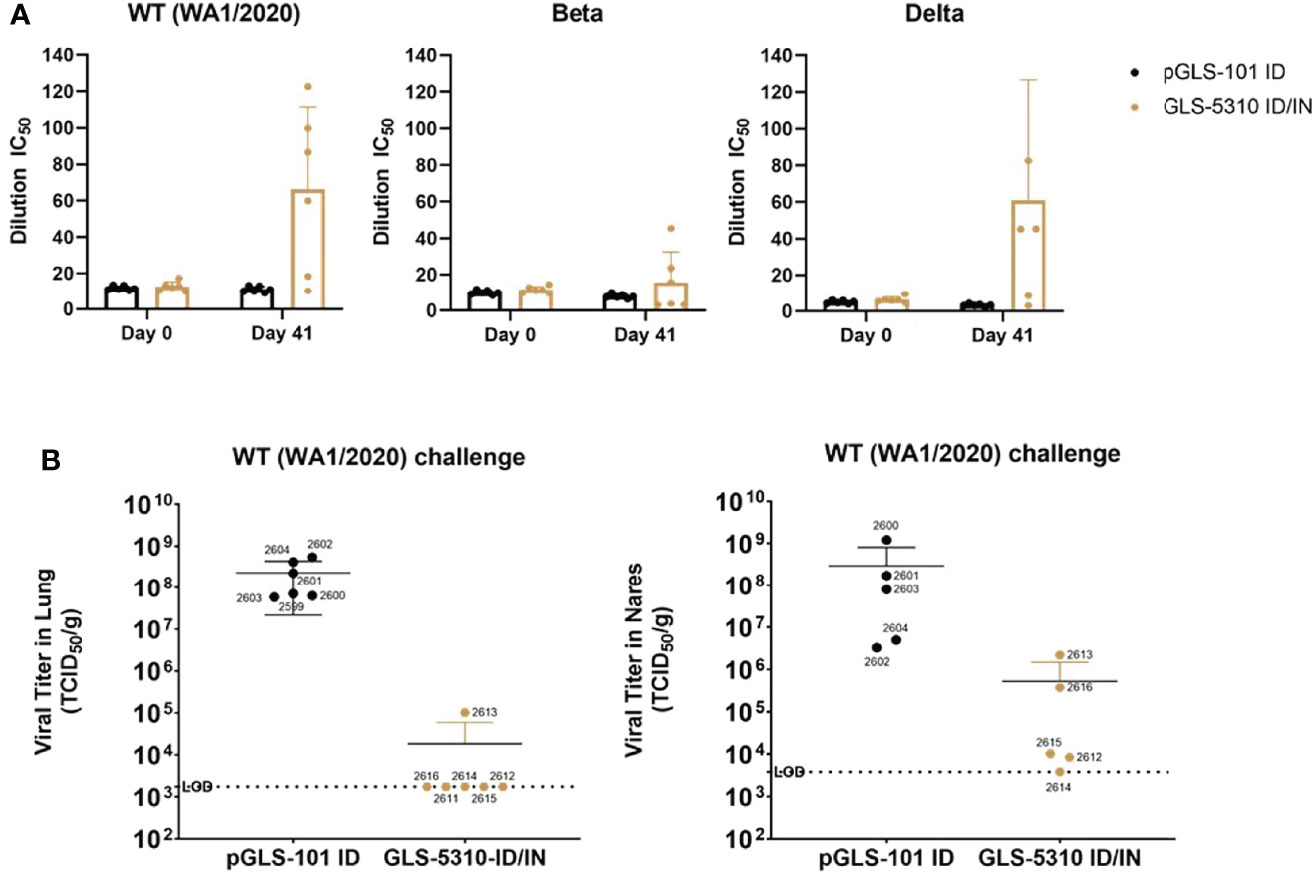
Figure 5 Combined GeneDerm-assisted intradermal and intranasal immunization of hamsters with GLS-5310 reduces virus replication in lungs after the SARS-CoV-2 challenge. (A) Golden Syrian hamsters immunized twice, 3 weeks apart with 150μg pGLS-101 (n = 6) by intradermal injection followed by GeneDerm-applied suction or twice, 3 weeks apart with 150μg GLS-5310 (n = 6) by combined intradermal injection followed by GeneDerm-applied suction and intranasal delivery. Pre-immune and day 41 sera from immunized hamsters were evaluated in modified surrogate neutralization assays for their ability to inhibit interactions between hACE2 and the Spike RBDs of WA1/2020 (Wt.), B.1.351 (Beta), or B.1.617.2 (Delta) SARS-CoV-2 strains. (B) Virus titers in lung and nares tissues collected from animals 5 days-post intranasal challenge (6x103 PFU challenge dose per hamster) with the WA1/2020 SARS-CoV-2 strain (Wt.) and were evaluated by TCID50 assay.
Immune serum was then further assessed for live-virus neutralization and inhibition of cytopathic effect (CPE). Sera collected from a hamster immunized with pGLS-101 and a hamster immunized with GLS-5310 on day 41 were evaluated for their ability to block SARS-CoV-2 replication and cytopathic effect (CPE) in Vero-E6 cells. Dilutions of heat-inactivated immune sera collected from each animal were pre-incubated with wt SARS-CoV-2 and then added to Vero-E6 cell cultures. In these studies, sera from the pGLS-101 injected hamster was unable to block virus-induced CPE and loss of viability, while day 41 immune sera from a hamster immunized with GLS-5310 reduced virus-induced CPE with dose-dependence, as confirmed by direct visual imaging, and maintained or improved infected cell viability, as monitored by resazurin viability dye (Figures 6A, B). These results support that GLS-5310 immunization induces immune responses capable of blocking SARS-CoV-2 infection in hamsters.
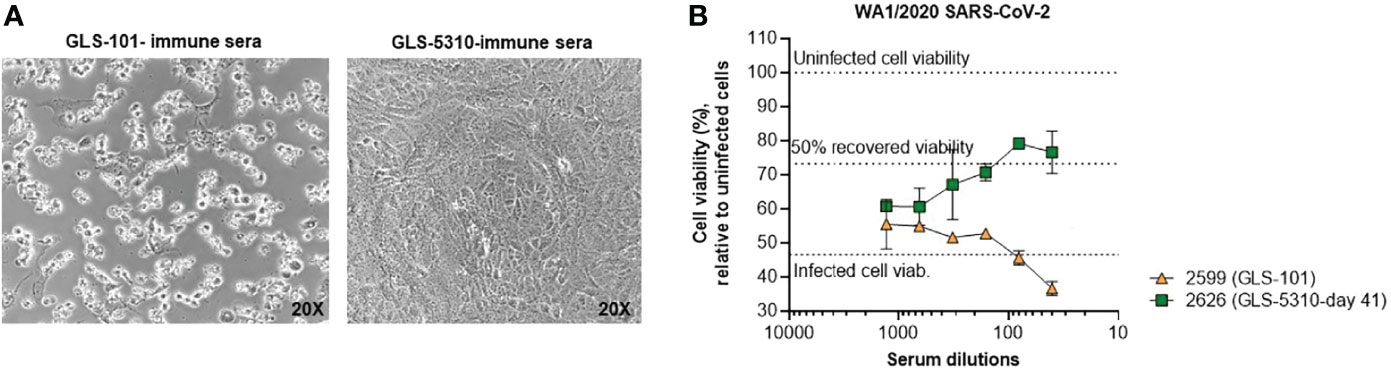
Figure 6 Protective activity of immune sera in live virus in vitro assays. Sera from a GLS-5310 immunized hamster, but not from a pGLS-101 immunized hamster, inhibits SARS-CoV-2-induced CPE. Diluted immune sera from a pGLS-101 and GLS-5310 immunized hamster were pre-incubated with WT SARS-CoV-2 before addition to Vero-E6 cell cultures. (A) Representative images of cell cultures taken at 72 hours post-infection prior to staining for cell viability. (B) Effects of immune sera on WT SARS-CoV-2 infected cells following 72 hours incubation; where 100% cell viability (upper dotted line) denotes viability of uninfected cells, and 46.6% viability (lower dotted line) denotes infected cell viability in the absence of sera.
Discussion
Here we describe the design and preclinical testing of pGO-1002, a novel bi-cistronic SARS-CoV-2 DNA vaccine candidate that encodes consensus sequences of the SARS-CoV-2 Spike and ORF3a antigens. In mice, pGO-1002 generated humoral and cellular responses to Spike and ORF3a that were equivalent in scope and magnitude as seen in mice immunized with mono-cistronic constructs of each antigen. In rats, intradermal immunization of pGO-1002 followed by GeneDerm-applied suction also induced humoral and cellular responses to each antigen. Humoral responses induced by pGO-1002 immunization in both models blocked Spike RBD-ACE2 binding as well as neutralized infection by a clinical SARS-CoV-2 isolate. In a hamster challenge model, GLS-5310 (clinical reformulation of pGO-1002) immunization prevented viral replication in the lungs and significantly reduced viral replication in the nares of animals challenged with the ancestral WA1/2020 SARS-CoV-2 strain. Interestingly, pGO-1002 immunization also prevented viral replication in the lungs of Syrian hamsters challenged with the B.1.351 (Beta) variant despite sera from immunized animals showing no ability to block binding between ACE2 and the recombinant Beta variant Spike RBD. This bi-cistronic SARS-CoV-2 DNA vaccine candidate, pGO-1002, has been formulated for clinical use and advanced into Phase I/IIa human clinical trials as the GLS-5310 DNA vaccine (NCT04673149).
Most SARS-CoV-2 vaccines developed to date endeavor to induce a strong neutralizing antibody response to Spike. In mice, full seroconversion to Spike was seen after one immunization with pGO-1002, while two injections of pGO-1002 were needed to get maximal responses to Spike in rats. While further characterization of the antibodies induced by pGO-1002 is needed, pGO-1002 immunization induced antibodies targeting the Spike RBD in all animal models tested that blocked its interaction with its cellular receptor, ACE2. Studies in nonhuman primate (NHP) models of SARS-CoV-2 infection showed that antibodies recovered from convalescent rhesus macaques could protect naïve macaques from infection, and in vaccinated macaques, anti-Spike antibodies correlated with the protective efficacy of mRNA vaccines (9, 35). Levels of SARS-CoV-2 neutralizing antibodies in convalescent COVID-19 patients or SARS-CoV-2 vaccinated individuals is highly predictive of protection from symptomatic SARS-CoV-2 infection (36). While antibody levels induced by natural SARS-CoV-2 infection or vaccination appear to persist out to a year, antibody levels decline post-exposure, which may compromise immune protection and allow for reinfection.
Another concern for antibodies is their ability to recognize new variants of SARS-CoV-2 as mutations are concentrated primarily in the Spike proteins, and in particular the receptor binding domain, that mediates virus entry and is critical immune cell recognition (37). Five of these variants, B.1.1.7 (Alpha), B.1.351 (Beta), P.1 (Gamma), B.1.617.2 (Delta), and B.1.1.529 (Omicron), have been designated variants of concern (VOCs) due to observed or suspected increases in transmissibility and/or their ability to cause breakthrough infections in individuals recovered from COVID-19 or vaccinated with first-generation COVID vaccines (that all present a Spike antigen similar to that of the original ancestral strains). Studies have found that sera from recovered COVID-19 patients or those immunized with mRNA-based vaccines have detectable binding antibodies to both ancestral and variant SARS-CoV-2 strains over 7 months post-infection or vaccination, but there is a decrease in the ability of these antibodies to neutralize variants, particularly the B.1.351 variant, compared to ancestral strains (15, 38, 39). In our experiments, sera from GLS-5310 immunized hamsters were able to block binding between the Spike RBD of the Delta variant SARS-CoV-2 and hACE2 to a similar extent as it blocked binding between the Spike RBD of the wild type (wt.) WA1/2020 strain and hACE2, but it was less able to block binding between the Spike RBD of the Beta variant SARS-CoV-2 and hACE2.
Early induction of SARS-CoV-2-specific T cells, particularly SARS-CoV-2-specific CD4+ T cells, correlates with reduced COVID-19 severity more so than nAb titers (8, 40, 41). At least 1,434 unique, non-redundant epitopes spanning the whole SARS-CoV-2 proteome, including 1052 epitopes targeted by CD8+ T cells and 382 targeted by CD4+ T cells, have been identified in studies of COVID-19 patients (42). Most of the epitopes are in the structural proteins Spike, Nucleocapsid (N), and Membrane (M), which appears to correlate with both the size of these antigens and their expression levels though some bias may be inferred because investigators often focused on these antigens only (27, 42). However, several studies have identified dominant T cell targets in other SARS-CoV-2 proteins including nsp3, nsp4, nsp12, ORF3a, and ORF8 (26, 42–44). A study of convalescent COVID-19 patients found that on average, 19 CD4+T cell and 17 CD8+ T cell epitopes were recognized per donor (44). The significant breadth and diversity of T cell reactivity seen in recovered COVID-19 patients has been associated with milder COVID-19 symptoms and likely provides additional protection against immune evasion by new variants of SARS-CoV-2 (45). A follow-up study by Tarke et al. found that over 93% of CD4+ T cell and 97% of CD8+ T cell epitopes were 100% conserved between variants (B.1.1.7, B.1.351, P.1, and CAL.20C) and the original Wuhan strain, and that total CD4+ and CD8+ T cell responses resultant from natural infection or vaccination with either the BNT162b2 or mRNA-1273 RNA vaccines had similar reactivity against the four variants as found against the original isolates (16).
ORF3a was added as a second antigen in pGO-1002 to provide greater breadth and persistence of T cell immune responses. Our data show that immunization with pGO-1002 induces broad-based T cell responses spanning the entire S protein and robust cellular response against ORF3a. Cellular responses to SARS-CoV ORF3a were detected in recovered SARS patients 6 years after recovery (22). Several groups have found that SARS-CoV-2 ORF3a is an immunodominant CD4+ and CD8+ T cell antigen with responses prevalent in 90%-95% of recovered patients against multiple HLA-restricted epitopes (26, 27, 43, 44). SARS-CoV-2 ORF3a appears to be a particularly strong target of CD8 T cells, particularly in proportion to its size (29). Importantly, with regard to vaccine development, while mutations of ORF3a have been described, a comprehensive analysis of non-synonymous mutations of ORF3a of 2782 strains showed that detected mutations were relatively uncommon, and many mutations were considered deleterious (46). Immunization with pGO-1002 also induced humoral immune responses to SARS-CoV-2 ORF3a in mice and rats, but the contribution of these responses to protection from infection and/or disease needs further investigation. Antibodies induced in mice by the ORF3a-only construct pGO-1003 were unable to neutralize SARS-CoV-2 infection. ORF3a-targeting antibodies have been detected in COVID-19 patients (28, 47, 48).
Viral challenge of Syrian hamsters with SARS-CoV-2 results in mild-to-moderate disease characterized by progressive weight loss, lethargy, ruffled fur, and signs of respiratory distress, including labored breathing (49). High levels of virus replication occur in the lungs of challenged hamsters and histopathological analysis of lung tissues shows evidence of moderate- to severe-lung pathology including features seen in the lungs of human COVID-19 patients such as ground-glass opacities (50). However, Syrian hamsters do not succumb to SARS-CoV-2 challenge, but the mild disease phenotype it manifests after the challenge makes it a suitable model for evaluating the efficacy of vaccines and therapeutics (51). A study by Tostanoski et al. found that infection of Syrian hamsters with the WA1/2020 strain provided robust protection against lung viral load and clinical disease when the animals were re-challenged with WA1/2020 or with either the B.1.1.7 (Alpha) or B.1.351 (Beta) variants of SARS-CoV-2 (52). They observed complete suppression of replicating virus in the lungs of infected animals re-challenged with either of the three SARS-CoV-2 strains even though RBD-binding and pseudovirus neutralizing antibody titers to B.1.351 induced by WA1/2020 infection were significantly lower than titers to WA1/2020 and B.1.1.7. This group also investigated the protection of Syrian hamsters from challenge with either the WA1/2020 or B.1.351 SARS-CoV-2 strains after vaccination with the Ad26.COV2.S vaccine (Janssen) which encodes a Spike antigen similar to that in the WA1/2020 strain (52). They found that vaccination provided complete protection against weight loss induced by virus challenge, and they observed a reduction in replicating virus in the lungs of vaccinated animals challenged with either strain, but not complete suppression of virus replication seen when they re-challenged WA1/2020-infected animals. Overall binding and pseudovirus neutralizing antibody titers observed in vaccinated animals were nearly a log lower than those induced by natural infection, but even in this context, there were significantly decreased titers of binding antibodies and nAb titers to B.1.351 compared to WA1/2020. These data suggest that immunity to the Spike antigen of an early pandemic strain alone may not be able to control or prevent virus replication in the lung in this model and that immunity to other SARS-CoV-2 proteins may be needed. In a separate study, Syrian hamsters immunized with a DNA vaccine encoding only Spike antigens of the original ancestral strain or the B.1.351 strains similarly reduced but did not eliminate virus titers in the lungs after challenge with the B.1.351 SARS-CoV-2 strain (53). Here we found Syrian hamsters immunized with the bi-cistronic pGO-1002 DNA vaccine exhibited complete suppression of virus replication in the lungs after challenged with either an ancestral or B.1.351 SARS-CoV-2 strains suggesting that additional immunity provided by inclusion of the ORF3a antigen is beneficial in this model.
Efficient DNA vaccine delivery into cells in vivo, has required a device for administration such as electroporation (EP) or jet delivery. In studies using ID vaccine delivery plus EP, a significant proportion of study participants experienced pain, swelling, erythema, and pruritis following vaccination (54). Moreover, EP causes significant tissue damage, though this damage has been cited as being required to elicit APC migration into region of vaccine administration to accentuate the immune response (55). Human skin tissue damage caused by EP was observed in clinical trials as scabbing at the injection site (54). Because of the pain and discomfort associated with EP, the level of expertise required for either EP or other delivery modalities, and questions of mass production and use in resource-limited regions, we sought to develop an alternative means for DNA vaccine delivery to enhance in vivo transfection.
The concept of applying suction to the skin using the GeneDerm device borrows loosely from the alternative medicine practice of cupping where suction is applied to acupuncture points on the skin as a treatment for a variety of diseases (56). The application of suction to the skin has been found to increase vasodilation and blood flow to the application site and to cause tensile stresses that may stimulate recruitment of immune cells, both of which would be expected to contribute to better immunogenicity after ID vaccine delivery (57, 58). Design, modeling, and optimization of the GeneDerm suction devise was recently described (32). In this prior study, application of suction to rat skin with GeneDerm was atraumatic by histologic exam and induced gene expression within an hour after injection. Here, we demonstrate that GeneDerm-assisted ID immunization of pGO-1002 into rats resulted in the development of antigen-specific humoral and cellular responses. The Spike S1-specific antibodies induced after GeneDerm-assisted immunization of rats had similar kinetics and reached similar magnitude as seen in rats that were immunized ID with pGO-1002 using the Pharmajet Tropis device or rats immunized ID followed by electroporation. Cellular responses induced by pGO-1002 after ID+GeneDerm immunization were higher than those induced when pGO-1002 was administered with either comparator modalities even when one tenth of the dose was administered. To our knowledge this is the first demonstration of the ability of suction to enhance DNA vaccine immunogenicity.
As a vaccine platform, DNA plasmids offer many advantages such as (1) they can be rapidly designed and constructed, (2) there are well-established and scalable plasmid manufacturing processes in place, and (3) they exhibit thermostability at both 4°C and ambient temperature for prolonged periods of time. Numerous clinical studies have established the safety of DNA vaccines and their ability to induce robust cellular as well as humoral responses to a variety of infectious disease antigens (59, 60). The GeneDerm device is easily mass producible, portable, inexpensive, and requires minimal training in use. Combined, these factors significantly alleviate vaccine supply and distribution challenges for underserved populations in developed countries and in Low/Middle Income Countries (LMICs), where access to life-saving therapies and vaccines are limited by poor public health infrastructures.
In summary, pGO-1002 is highly immunogenic in multiple small animal models and induces humoral responses that can neutralize SARS-CoV-2 infection. Further, we show that pGO-1002 is highly immunogenic when delivered intradermally followed by application of suction at the injection site using our GeneDerm device. Immunization of Syrian hamsters with GLS-5310 (formulated from pGO-1002) significantly reduced virus levels in the lungs and substantially dropped virus levels in the nares of animals challenged with a Wt. or Beta variant SARS-CoV-2. The results presented here support advancement of the GLS-5310 SARS-CoV-2 DNA vaccine with ID+GeneDerm delivery into clinical trials to evaluate the ability of this platform to contribute to the challenge of vaccinating enough of the world’s population to end the SARS-CoV-2 pandemic.
Materials and Methods
Cell Lines and Antibodies
Human embryonic kidney cells 293T (HEK 293T; ATCC, cat#CRL-3216) were obtained from ATCC and cultured in Dulbecco’s Modified Eagle Media (DMEM; Welgene, cat#LM001-05) supplemented with 10% fetal bovine serum (FBS; Welgene, cat#S101-07). Antibody to SARS-CoV-2 Spike was obtained from Sino Biological (cat#40591-MM42). Human hepatoma cell line Huh-7 (ATCC, CVCL-0336) were grown in Dulbecco’s modified Eagle medium (DMEM) supplemented with 10% fetal bovine serum (FBS) and 1% antibiotics at a 37°C incubator, respectively (6). Antibody to SARS-CoV-2 ORF3a obtained from immunized mice serum. Antibody to β-actin was obtained from Abcam (cat#ab8226).
Animals and Vaccine Immunizations
Male, 7-week-old BALB/c mice were purchased from RaonBio (Seoul, Korea). Female, 6- to 8-week-old Sprague Dawley rats were purchased from OrientBio (Seoul Korea). Male and female Syrian golden hamsters (6– to 8-week-old) were acquired from Charles River Laboratories and housed at Bioqual, Inc., for the duration of the study. All animal testing and research complied with relevant ethical regulations and studies received approval from each institution’s IACUC. For mouse studies, 50µg empty vector or vaccine plasmids were administered intramuscularly twice, 2 weeks apart, by needle into the tibialis anterior (T.A.) muscle and then in vivo electroporation (EP) was applied to the injection site using BTX ECM 830 series device BTX, MA, USA. The machine was set to deliver six pulses at 200V/cm, 50ms, 1Hz. Mice were weighed starting 1 week prior to first immunization and then every 3 to 4 days until mice were sacrificed on day 28 post injection to obtain spleens for analysis of cellular immune responses. On days 0, 14, and 28, blood was collected from mice by tail vein sampling.
For intradermal (ID) immunizations, rat skin was first prepared by gentle clipping hair in a spot on the backs of animals and then a depilatory cream was applied to spot to remove remaining hair. Control (pGLS-101; 300µg) or pGO-1002 (30 or 300µg) plasmid was administered using one of three methods; (1) plasmid (100µl) was injected to the shallow intradermal space using a tuberculin syringe (BD, Cat.328820) followed by application of 65 kPa suction for 30 sec using GeneDerm device such that the bleb induced by injection was contained within the opening of the cap of the device (32); (2) plasmid (100µl) was injected using PharmaJet Tropis® needle-free device following manufacturer’s protocol; or (3) plasmid (100µl) was injected to the shallow intradermal space using a tuberculin syringe and electroporation 200V/cm, 50ms, 1 Hz, 6 pulses, 1s interval (BTX) was applied with a EP device.
Hamsters were vaccinated in a manner similar to rats with either pGLS-101 or GLS-5310. GLS-5310 is a formulated drug product of pGO-1002 at a concentration of 6 mg/mL in 1X saline sodium citrate (SSC) buffer for human use. Prior to use, vaccine was diluted with 1X SSC. For animals vaccinated ID only, 150 µg of pGLS-101 or GLS-5310 vaccine in a 50µl volume was administered whereas for animals vaccinated ID and IN 150 µg of vaccine in a 50 µl volume was administered to both sites for a total dose of 300µg.
Western Blot Analysis
Cultures of HEK293T cells were transfected using PEI MAX transfection reagent (Polysciences, Inc., cat# 24765-1, following manufacturer’s protocol) with 100µg empty pGLS-101 vector or one of the three vaccine candidate plasmids (pGO-1001, pGO-1002, pGO-1003). Each cell culture was lysed 48 hours later in 1% Triton X-100 buffer. Proteins in lysates were separated by SDS-PAGE (Bio-Rad, Mini-PROTEAN Tetra System) and then transferred to PVDF membrane. Membrane was blotted with antibodies to SARS-CoV-2 Spike (1:5000 dilution); ORF3a (1:200 dilution), and β-actin (1:1000 Dilution) followed by blotting with horseradish peroxidase (HRP)-conjugated anti-mouse IgG. Blots were developed using ECL system (Biomax, cat# BWD0100).
Intracellular Fluorescence Assay (IFA)
Cultures of Huh7 cells were transfected with pGLS-101 or pGO-1002 plasmids as described above. Forty-eight hours post transfection cells were washed twice in 1×PBS and fixed in 4% v/v formaldehyde (Fisher) supplemented with 0.0075% v/v glutaraldehyde (Sigma) in PBS for 30 minutes at room temperature. Cell membranes were permeabilized in 0.1% Triton X-100 in PBS for 30 minutes. Autofluorescence was quenched using 0.1 M glycine in PBS for 15 minutes Blocking was performed with 3% w/v bovine serum albumin (BSA) for at least 1h at room temperature, were incubated with primary antibodies for 90 minutes at room temperature or overnight at 4°C, Immunofluorescence labeling was performed by incubation with the mouse anti-SARS-CoV-2 Spike antibody or anti-SARS-CoV-2-ORF3a antibody followed incubation with goat anti-rabbit IgG-Alexa 488 and goat anti-mouse IgG-Alexa 568 antibodies. DAPI panels show control staining of cell nuclei (6).
Enzyme-Linked Immunosorbent Assay
Corning ELISA plates (cat #3366) were coated with recombinant protein antigens in phosphate-buffered saline (1x PBS) overnight at 4°C. Plates were blocked with blocking buffer (Seracare, Cat.5140-0011) for 2 hours at 37°C. Plates were then washed and incubated for 2 hours at 37°C with 50µl of 4-fold serial dilutions of mouse or rat sera made in blocking buffer. Plates were again washed and then incubated for 1 hour at room temperature (R.T.) with horse radish peroxidase (HRP)-conjugated anti-mouse IgG or anti-rat IgG secondary antibody diluted in blocking buffer. After final washes, plates were developed using ABTS® Peroxidase Substrate System (KPL, cat. 5120-0032) and the reaction stopped with ABTS® Peroxidase Stop Solution (KPL, cat. 5150-0017). Plates were read at 405 nm wavelength within 30 minutes using a Synergy HTX (Bio Tek Instruments, Highland Park, VT). Calculation of cutoffs for endpoint titer determined using formula: Day 0 of each group; (each value - blank) Avg +(1.923 x SD) as previously described (61).
For mouse sera ELISA, wash buffer was 1x PBS with 0.05% Tween 20 (diluted Phosphate-Buffered Saline (10X) 1:10 (v/v) and Tween 20 1:20 (v/v) with deionized water), binding antigens tested included recombinant S1 subunit of SARS-CoV-2 Spike (1µg/ml; Sino Biological, 40591-V02H) or internally purified recombinant SARS-CoV-2 ORF3a protein (1µg/ml), and secondary antibody was HRP-goat anti-mouse IgG (H+L) (diluted 1:2500; Invitrogen, Cat# 31430). For rat sera ELISA, wash buffer was 1x PBS with 5% Tween 20, binding antigens tested included recombinant S1 subunit of SARS-CoV-2 Spike (4µg/ml) or recombinant SARS-CoV-2 ORF3 protein (5µg/ml; Bioworld technology, NCP0026P), and secondary antibody was HRP-goat anti-rat IgG (H+L) (diluted 1:2500 for S1 ELISA and 1:1000 for ORF3 ELISA; Sigma, Cat. A9037).
ELISpot Assay
Spleens collected from mice and rats were collected individually and processed into single cell suspensions in R10 media: RPMI1640 media (Gibco, Cat 11875135) supplemented with 10% FBS (Welgene, Cat S101-07) and (1%) penicillin/streptomycin (Gibco, cat 15140-122). Cell pellets were re-suspended in 5 mL of ACK lysis buffer for 8 minutes at R.T. and then R10 was added to stop the reaction. The samples were again centrifuged at 1400 rpm for 5 minutes and cell pellets were re-suspended in R10 and counted. ELISpot assays for mice and rat splenocytes were performed using Mouse IFN-γ or Rat IFN-γ ELISpotPLUS plates (MABTECH, cat. 3221-4APW-10 and 3220-4APW-10respectively) following manufacturer’s protocol. Basically, 96-well ELISpot plates were precoated with INF-α capture antibody and blocked with R10 medium for 1hr at room temperature. 200,000 mouse or rat splenocytes were plated into each well and stimulated for 14 hours with: (1) one of 5 pools of 15-mer peptides overlapping by ten amino acids spanning the SARS CoV-2 Spike (Genscript); or (2) ORF3a (Mimotopes). Cells were stimulated with a final concentration of 1µg/ml of each peptide per well in R10 media. The spots were developed based on manufacturer’s instructions. R10+5% DMSO (Sigma, # D2650) and Concanavalin A (Con-A; Sigma, cat#: C5275) were used for negative and positive controls, respectively. Spots were scanned and quantified by ImmunoSpot CTL reader. Spot-forming unit (SFU) per million cells was calculated by subtracting the R10+5% DMSO control wells. The cutoff values were determined as previously described using the following equation: Cutoff = Mean of Spot numbers in overlapping peptides (OLPs) + (Standard deviation of spot numbers in OLPs * 2 with the sample used for cut off coming from splenocytes of unvaccinated mice or rats (6, 61).
Plaque Reduction Neutralization Titer 50 (PRNT50)
SARS-CoV-2/South Korea/2020 isolate neutralization assays were performed at Korea Centers for Disease Control and Prevention. Neutralizing virus titers were measured for mice or rat sera that had been heat-inactivated at 56°C for 30 minutes SARS-CoV-2 (βCoV/Korea/KCDC03/2020) was diluted to a concentration of 60 pfu/ml and mixed 50:50 in Dulbecco’s modified Eagle’s medium (DMEM, Invitrogen, Carlsbad, USA) supplemented with 10% fetal bovine serum (FBS), 100 IU/ml penicillin, and 100μg/ml streptomycin, with serial serum dilutions from 1:4 to 1:2040 in a 96-well U-bottomed plate. The plate was incubated at 37°C in a humidified box for 1 hour before the virus was transferred into the wells of 12-well plate that had been seeded the previous day at 2.5 × 10^5 Vero cells (ATCC, CCL-81) per well in Dulbecco’s Modified Eagle’s Medium (DMEM, Invitrogen, Carlsbad, USA) supplemented with 10% fetal bovine serum (FBS), 100 IU/ml penicillin, and 100μg/ml streptomycin. Virus was allowed to adsorb at 37°C for 1 hour and then plaque assay overlay media (2× MEM/1.5% Agar/4% FBS final) was added to cells. After 3 days incubation at 37°C in a humidified box, the plates were fixed, stained and plaques counted. Cytopathic effect (CPE) of each well was recorded under microscopes and the neutralizing titer was calculated by the dilution number of 50% protective condition (inverse of the dilution of sera where plaques first appeared). PRNT50 values were determined by the following equation: PRNT50=DL + ((PRNT50-PL)(DH-DL)/(PH-PL))-DL: plaque is the reciprocal of the lower dilution bracketing the 50% endpoint.-DH: is the reciprocal of the higher dilution bracketing the 50% endpoint; -P50: is the number of plaques at the 50% endpoint; PL.: is the number of plaques at the lower dilution bracketing the 50% endpoint; PH.: is the number of plaques at the higher dilution bracketing the 50% endpoint.
Surrogate Neutralization Assay
A surrogate neutralization assay was performed on immune sera from mice and rats to detect whether circulating antibodies could block the interaction between the receptor binding domain (Genscript, Cat #: L00847) of the SARS-CoV-2 Spike glycoprotein with the ACE2 cell surface receptor (Genscript, cat#: L00847 or Ray Biotech, Cat# CoV-ACE2S2). In separate tubes, diluted positive control (Genscript, Cat#L00847), diluted negative control (Genscript, Cat#L00847), and the sera samples were mixed with diluted HRP-RBD solution (concentration 1:999) at a volume ratio of 1:1. Mixtures were incubated at 37°C for 30 minutes, and then 100µl each of the positive control mixture, the negative control mixture, and the sera sample mixture was added to the corresponding wells that were pre-coated with recombinant ACE2 protein. Plate was sealed and incubated at 37°C for 15 minutes. Plate seal was removed, and plate washed four times with 260µl of 1x Wash solution and then blotted dry. Then 100µl of TMB Solution (Genscript, Cat#L00847) was added to each well of plate which was then incubated in the dark at 20-25°C for 15 minutes before 50µl of Stop solution added to wells to quench the reaction. Absorbance at 450nm for wells of plate read immediately using the microtiter plate reader (Biotek, Synergy™ HTX). Cutoff values were determined by the following equation: Inhibition = (1-O.D. Value of sample/OD value of Negative Control) * 100%. The cut off value is based on validation with Genscript panel of confirmed COVID-19 patient sera and healthy control sera. Values over 20% constituted a positive result meaning SARS-CoV-2 neutralizing antibody was present in the sera sample while vales under 20% constituted a negative result meaning no detectable SARS-CoV-2 neutralizing antibody was present.
HTRF Assay for Testing Plasma Samples for Inhibition of CoV-Spike Binding to ACE2
Plasma samples were serially diluted 1:2 in assay buffer (25 mM Tris, pH 7.4, 150 mM KCl, 0.05% CHAPS, 0.1% BSA), and dilutions were pre-incubated with 2 nM HIS-CoV-Spike RBD (either wild-type, Beta, or Delta variants, Sino Biological) prebound to 600 ng/mL anti-HIS-d2 HTRF acceptor (PerkinElmer) in a total volume of 10μL of assay buffer in white, low-volume 384 well plates. After 1 hour, assays were initiated by adding 5μL of 6 nM biotin-ACE2 (2 nM final concentration, Acros BioSciences) prebound to 50 ng/mL streptavidin-terbium HTRF donor (PerkinElmer). After an additional 2 hours incubation, HTRF signals were measured using a ClarioStar plate reader (BMG LabTech) at 320 nm excitation and 620/665 nm emission with a 50μs delay and window time of 200μs. The raw data at each wavelength were then converted to the HTRF ratio by RFU 665/RFU 620 x 10000. Ratio values were then converted to percent inhibition, where 0% was equal to the HTRF value in the absence of plasma and 100% was equal to the HTRF value in the absence of HIS-CoV-Spike RBD. To calculate IC50s, percent inhibition values were then fit to a 4-parameter dose-response curves using the dilution factor as the X coordinate, the top parameter fixed to 100% and the slope constrained between 1-2. REGN10933 was obtained from excess aliquot volumes at the Perelman School of Medicine, University of Pennsylvania, PA, USA which could not be used for patients (a gift from Dr. Pablo Tebas).
Cell Viability Protection Assay
Day 41 sera from a pGLS-101 and GLS-5310 immunized hamster were heat-inactivated at 56°C for 30 minutes, serially diluted 2-fold from 1:20-1:1280 in DMEM+2% FBS, and then incubated with 100x TCID50 of USA-WA1/2020 SARS-CoV-2 for 1 hour before addition to cultures of 20,000 Vero-E cells plated 24 hours previously in 96 well plates. For each experiment, all culture conditions were performed in duplicate. After 72 hours, cell cultures were imaged at 20X magnification and then treated with resazurin (Sigma-Aldrich) to a final concentration of 20μg/ml. Cells were incubated for an additional 4 hours before fluorescence intensity was measured using a ClarioStar plate reader (BMG Labtech). Background fluorescence was subtracted from wells containing resazurin and media but no cells.
Syrian Hamster Challenge Experiments and Quantification of Viral Titers in Lung Tissues
Syrian hamsters were immunized on study days 0 and 21 via ID/suction route (or IN depending on the groups being used) and challenged on Study Day 42, via intranasal (IN) route, with 6x103 PFU of SARS-CoV-2 (2019-nCoV/USA-WA1/2020, LOT #12152020-1235, derived from BEI seed stock Cat # NR-52281 Lot # 70036318) or 3.67x102 PFU of SARS-CoV-2-RSA (2019-nCoV/South Africa/KRISP-K005325/2020 in Calu-3, LOT 030621-750, derived from BEI seed stock cat # NR-54974, Lot # 70041987) challenge dose per hamster. Prior to challenge procedure, the animals were anesthetized by injecting 80 mg/kg ketamine and 5 mg/kg xylazine via the intramuscular (IM) route. The animals were injected with an antisedan, at 1 mg/kg via IM route, 20 minutes post challenge. All animals were monitored until completely recovered. Post-challenge, animals were observed for weight loss and clinical symptoms of disease until the terminal day, study day 5 post-challenge, when hamsters were euthanized, and tissues collected for viral RNA quantification and histopathology analysis. Differences viral load between experimental groups of animals were analyzed statistically using t-test.
Statistical Analyses
Statistical analyses were conducted using Student’s t test in Prism (version 9) software (GraphPad). Adjusted probability p values (p) of smaller than 0.05 was considered statistically significant.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding authors.
Ethics Statement
The animal study was reviewed and approved by BALB/c mice were purchased from RaonBio (Seoul, Korea). Sprague Dawley rats were purchased from OrientBio (Seoul Korea). Male and female Syrian golden hamsters (6–8-week-old) were acquired from Charles River Laboratories and housed at Bioqual, Inc., for the duration of the study. All animal testing and research complied with relevant ethical regulations and studies received approval from each institution’s IACUC.
Author Contributions
KM, CR, and JNM conceived and designed the experiments for this study. MJ, SBK, AG, BJ, GP, YC, HeL, MC, WK, Y-HH, J-AL, HyL, MK, EL, TB, NJ, LP, MP, AR, and DN performed the experiments. SAK, JS, JZ, DS, JPS, HaL, ES, SK, HA, IT, JC, JMS, and LM provided resources. SBK, KM, and JNM wrote the original concept and manuscript, and KM, CR, and JM reviewed and edited it. KM and JNM provided supervision. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the Research Program funded by the Korea Disease Control and Prevention Agency (Fund code#:2020-ER5507-00) and by a grant of the Korea Health Technology R&D Project through the Korea Health Industry Development Institute (KHIDI), funded by the Ministry of Health & Welfare, Republic of Korea (grant number : HQ20C0040). It was also supported by GeneOne Life Science (to LM) and by the following grants to LM: the Robert I. Jacobs Fund of the Philadelphia Foundation and the Herbert Kean, M.D., Family Professorship.
Conflict of Interest
MJ, SBK, AG, BJ, GP, YC, HyL, MC, WK, YKP, KM, CR, and JNM are employees of GeneOne Life Science, Inc. and, as such, receive salary and benefits that may include ownership of stock and stock options. GeneOne is developing the GLS-5310 vaccine. ES is employed by Pharmajet and, as such, receives salary and benefits from the company. LP, MP, AR, DN, SK, and HA are employees of Bioqual Inc. and, as such, receive salary and benefits from the company.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors. No writing assistance was utilized in the production of this manuscript.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We would like to thank the Animal Facility staff at the Korea Preclinical Center (KPC), for providing house and care to the animals. We would like to thank the Bioqual Inc staff members Zackery Flinchbaugh, Brandon Narvaez and Brittany Spence for providing care to the animals and experiments and for their technical assistance. We thank the Center for Vaccine Research, Korea National Institute of Health, KDCPA for their neutralizing assay assistance, as well as Dr. Pablo Tebas for the gift of REGN10987 control antibody.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fviro.2022.891540/full#supplementary-material
Supplementary Figure 1 | (A) Schematic representation of mice vaccination schedule. Groups of n=5 BALB/c mice were immunized twice with 50 µg/mouse of pGLS-101, pGO-1001, pGO-1002, or pGO-1003 plasmids at 14-day intervals by intramuscular (IM) injection followed by BTX-EP. Mice were bled on days 0, 14, and 28 and sacrificed on day 28 to obtain spleens. (B) Evaluation of toxicity induced by expressed SARS-CoV2 protein by measuring weight changes in mice vaccinated with Vaccines.
Supplementary Figure 2 | Groups of n = 5 Sprague Dawley (SD) rats were immunized twice with 30 or 300 µg/rat of pGLS-101 or pGO-1002 plasmids at 14-day intervals intradermally by PharmaJet Tropis® device (PharmaJet), or intradermal injection followed by EP (EP). Rats were bled on days 0, 14, and 28 and sacrificed on day 28 to obtain spleens. ELISA analyzed all collected serum samples for binding to SARS-CoV-2 Spike S1 (A). Endpoint titers were calculated as the reciprocal of the highest sera dilution factor whose OD 405nm value was greater than 2.5 standard deviation of the OD 405nm of the day 0 value at that dilution. Each sample was run with three technical replicates on the plate. A comprehensive evaluation of neutralizing activity in rats. Day 28 sera from vaccinated rats were evaluated in a surrogate neutralization assay (B) for their ability to block the ACE2-RBD interaction and in a PRNT50 assay (C) for their ability to block infection of βCoV/Korea/KCDC03/2020 SARS-CoV-2 in Vero cells. Data are Mean + SD of percent inhibition of sera from each mouse as compared to inhibition by negative control or mean+SD of PRNT50 of sera from each rat. (D) Splenocytes prepared from all rats on day 28 were evaluated by IFN-γ ELISpot assay for characterization of cell-mediated responses. Splenocytes were stimulated with each of four linear pools of peptides spanning SARS CoV-2 Spike or a pool of linear peptides spanning SARS-CoV-2 ORF3a. Data represent the mean SFU per 106 splenocytes + SD. Statistical analysis for Pharmajet was performed in (B) using a one-way ANOVA comparison test where mean percent inhibition of each groups were compared against each other. Statistical analysis for EP was performed in (B, C) using an unpaired t-test.
Supplementary Figure 3 | SD rats (n = 5) were immunized twice, 2 weeks apart with 50μg of pGO-1002 by intradermal injection followed by GeneDerm-applied suction. Day 29 sera (2 weeks post second injection) were evaluated by ELISA to determine the IgG subclass of anti-Spike S1 antibodies in sera (A). Data for each subtype were normalized to the level of total anti-Spike S1 IgG. (B) The ratio of subclasses IgG2a and IgG2b to subclass IgG1 for S1-specific antibodies is reported.
References
1. Shrotri M, Swinnen T, Kampmann B, Parker EPK. An Interactive Website Tracking COVID-19 Vaccine Development. Lancet Glob Health (2021) 9:e590–2.
2. Bollyky TJ, Gostin LO, Hamburg MA. The Equitable Distribution of COVID-19 Therapeutics and Vaccines. JAMA (2020) 323:2462–3.
3. Kim JH, Marks F, Clemens JD. Looking Beyond COVID-19 Vaccine Phase 3 Trials. Nat Med (2021) 27:205–11.
4. Yang ZY, Kong WP, Huang Y, Roberts A, Murphy BR, Subbarao K, et al. A DNA Vaccine Induces SARS Coronavirus Neutralization and Protective Immunity in Mice. Nature (2004) 428:561–4.
5. Martin JE, Louder MK, Holman LA, Gordon IJ, Enama ME, Larkin BD, et al. A SARS DNA Vaccine Induces Neutralizing Antibody and Cellular Immune Responses in Healthy Adults in a Phase I Clinical Trial. Vaccine (2008) 26:6338–43.
6. Muthumani K, Falzarano D, Reuschel EL, Tingey C, Flingai S, Villarreal DO, et al. A Synthetic Consensus Anti-Spike Protein DNA Vaccine Induces Protective Immunity Against Middle East Respiratory Syndrome Coronavirus in Nonhuman Primates. Sci Transl Med (2015) 7:301ra132.
7. Heinz FX, Stiasny K. Distinguishing Features of Current COVID-19 Vaccines: Knowns and Unknowns of Antigen Presentation and Modes of Action. NPJ Vaccines (2021) 6:104.
8. Rydyznski Moderbacher C, Ramirez SI, Dan JM, Grifoni A, Hastie KM, Weiskopf D, et al. Antigen-Specific Adaptive Immunity to SARS-CoV-2 in Acute COVID-19 and Associations With Age and Disease Severity. Cell (2020) 183:996–1012e1019.
9. Corbett KS, Nason MC, Flach B, Gagne M, O’connell S, Johnston TS, et al. Immune Correlates of Protection by mRNA-1273 Vaccine Against SARS-CoV-2 in Nonhuman Primates. Science (2021) 373:eabj0299.
11. Kalimuddin S, Tham CYL, Qui M, De Alwis R, Sim JXY, Lim JME, et al. Early T Cell and Binding Antibody Responses are Associated With COVID-19 RNA Vaccine Efficacy Onset. Med (N Y) (2021) 2:682–688e684.
12. Sahin U, Muik A, Vogler I, Derhovanessian E, Kranz LM, Vormehr M, et al. BNT162b2 Vaccine Induces Neutralizing Antibodies and Poly-Specific T Cells in Humans. Nature (2021) 595:572–7.
13. Bange EM, Han NA, Wileyto P, Kim JY, Gouma S, Robinson J, et al. CD8(+) T Cells Contribute to Survival in Patients With COVID-19 and Hematologic Cancer. Nat Med (2021) 27:1280–9.
14. Marasco V, Carniti C, Guidetti A, Farina L, Magni M, Miceli R, et al. T-Cell Immune Response After mRNA SARS-CoV-2 Vaccines is Frequently Detected Also in the Absence of Seroconversion in Patients With Lymphoid Malignancies. Br J Haematol (2021) 196(3)548–58.
15. Geers D, Shamier MC, Bogers S, Den Hartog G, Gommers L, Nieuwkoop NN, et al. SARS-CoV-2 Variants of Concern Partially Escape Humoral But Not T-Cell Responses in COVID-19 Convalescent Donors and Vaccinees. Sci Immunol (2021) 6(59):eabj1750.
16. Tarke A, Sidney J, Methot N, Yu ED, Zhang Y, Dan JM, et al. Impact of SARS-CoV-2 Variants on the Total CD4(+) and CD8(+) T Cell Reactivity in Infected or Vaccinated Individuals. Cell Rep Med (2021) 2:100355.
17. Li CK, Wu H, Yan H, Ma S, Wang L, Zhang M, et al. T Cell Responses to Whole SARS Coronavirus in Humans. J Immunol (2008) 181:5490–500.
18. Le Bert N, Tan AT, Kunasegaran K, Tham CYL, Hafezi M, Chia A, et al. SARS-CoV-2-Specific T Cell Immunity in Cases of COVID-19 and SARS, and Uninfected Controls. Nature (2020) 584(7821):457–62.
19. Kern DM, Sorum B, Mali SS, Hoel CM, Sridharan S, Remis JP, et al. Cryo-EM Structure of SARS-CoV-2 ORF3a in Lipid Nanodiscs. Nat Struct Mol Biol (2021) 28:573–82.
20. Silvas JA, Vasquez DM, Park JG, Chiem K, Allue-Guardia A, Garcia-Vilanova A, et al. Contribution of SARS-CoV-2 Accessory Proteins to Viral Pathogenicity in K18 Human ACE2 Transgenic Mice. J Virol (2021) 95:e0040221.
21. Zhao J, Alshukairi AN, Baharoon SA, Ahmed WA, Bokhari AA, Nehdi AM, et al. Recovery From the Middle East Respiratory Syndrome Is Associated With Antibody and T-Cell Responses. Sci Immunol (2017) 2(124):eaan5393.
22. Oh HL, Chia A, Chang CX, Leong HN, Ling KL, Grotenbreg GM, et al. Engineering T Cells Specific for a Dominant Severe Acute Respiratory Syndrome Coronavirus CD8 T Cell Epitope. J Virol (2011) 85:10464–71.
23. Tan YJ, Goh PY, Fielding BC, Shen S, Chou CF, Fu JL, et al. Profiles of Antibody Responses Against Severe Acute Respiratory Syndrome Coronavirus Recombinant Proteins and Their Potential Use as Diagnostic Markers. Clin Diagn Lab Immunol (2004) 11:362–71.
24. Qiu M, Shi Y, Guo Z, Chen Z, He R, Chen R, et al. Antibody Responses to Individual Proteins of SARS Coronavirus and Their Neutralization Activities. Microbes Infect (2005) 7:882–9.
25. Lu B, Tao L, Wang T, Zheng Z, Li B, Chen Z, et al. Humoral and Cellular Immune Responses Induced by 3a DNA Vaccines Against Severe Acute Respiratory Syndrome (SARS) or SARS-Like Coronavirus in Mice. Clin Vaccine Immunol (2009) 16:73–7.
26. Ferretti AP, Kula T, Wang Y, Nguyen DMV, Weinheimer A, Dunlap GS, Xu Q, Nabilsi N, et al. Immunity (2020) 53:1095–107.
27. Grifoni A, Weiskopf D, Ramirez SI, Mateus J, Dan JM, Moderbacher CR, et al. Targets of T Cell Responses to SARS-CoV-2 Coronavirus in Humans With COVID-19 Disease and Unexposed Individuals. Cell (2020) 181:1489–1501e1415.
28. Hachim A, Kavian N, Cohen CA, Chin AWH, Chu DKW, Mok CKP, et al. ORF8 and ORF3b Antibodies Are Accurate Serological Markers of Early and Late SARS-CoV-2 Infection. Nat Immunol (2020) 21:1293–301.
29. Saini SK, Hersby DS, Tamhane T, Povlsen HR, Amaya Hernandez SP, Nielsen M, et al. SARS-CoV-2 Genome-Wide T Cell Epitope Mapping Reveals Immunodominance and Substantial CD8(+) T Cell Activation in COVID-19 Patients. Sci Immunol (2021) 6(58):eabf7550.
30. Siu KL, Yuen KS, Castano-Rodriguez C, Ye ZW, Yeung ML, Fung SY, et al. Severe Acute Respiratory Syndrome Coronavirus ORF3a Protein Activates the NLRP3 Inflammasome by Promoting TRAF3-Dependent Ubiquitination of ASC. FASEB J (2019) 33:8865–77.
31. Xia H, Cao Z, Xie X, Zhang X, Chen JY, Wang H, et al. Evasion of Type I Interferon by SARS-CoV-2. Cell Rep (2020) 33:108234.
32. Lallow EO, Jhumur NC, Ahmed I, Kudchodkar SB, Roberts CC, Jeong M, et al. Novel Suction-Based In Vivo Cutaneous DNA Transfection Platform. Sci Adv (2021) 7:eabj0611.
33. Tietjen I, Cassel J, Register ET, Zhou XY, Messick TE, Keeney F, et al. The Natural Stilbenoid (-)-Hopeaphenol Inhibits Cellular Entry of SARS-CoV-2 USA-WA1/2020, B.1.1.7 and B.1.351 Variants. Antimicrob Agents Chemother (2021) 65:e0077221.
34. Wang P, Nair MS, Liu L, Iketani S, Luo Y, Guo Y, et al. Antibody Resistance of SARS-CoV-2 Variants B.1.351 and B.1.1.7. Nature (2021) 593:130–5.
35. Mcmahan K, Yu J, Mercado NB, Loos C, Tostanoski LH, Chandrashekar A, et al. Correlates of Protection Against SARS-CoV-2 in Rhesus Macaques. Nature (2021) 590:630–4.
36. Khoury DS, Cromer D, Reynaldi A, Schlub TE, Wheatley AK, Juno JA, et al. Neutralizing Antibody Levels Are Highly Predictive of Immune Protection From Symptomatic SARS-CoV-2 Infection. Nat Med (2021) 27:1205–11.
37. Tao K, Tzou PL, Nouhin J, Gupta RK, De Oliveira T, Kosakovsky Pond SL, et al. The Biological and Clinical Significance of Emerging SARS-CoV-2 Variants. Nat Rev Genet (2021) 22:757–73.
38. Dupont L, Snell LB, Graham C, Seow J, Merrick B, Lechmere T, et al. Neutralizing Antibody Activity in Convalescent Sera From Infection in Humans With SARS-CoV-2 and Variants of Concern. Nat Microbiol (2021) 6:1433–42.
39. Pegu A, O’connell SE, Schmidt SD, O’dell S, Talana CA, Lai L, et al. Durability of mRNA-1273 Vaccine-Induced Antibodies Against SARS-CoV-2 Variants. Science (2021) 373:1372–7.
40. Bonifacius A, Tischer-Zimmermann S, Dragon AC, Gussarow D, Vogel A, Krettek U, et al. COVID-19 Immune Signatures Reveal Stable Antiviral T Cell Function Despite Declining Humoral Responses. Immunity (2021) 54:340–354e346.
41. Tan AT, Linster M, Tan CW, Le Bert N, Chia WN, Kunasegaran K, et al. Early Induction of Functional SARS-CoV-2-Specific T Cells Associates With Rapid Viral Clearance and Mild Disease in COVID-19 Patients. Cell Rep (2021) 34:108728.
42. Grifoni A, Sidney J, Vita R, Peters B, Crotty S, Weiskopf D, et al. SARS-CoV-2 Human T Cell Epitopes: Adaptive Immune Response Against COVID-19. Cell Host Microbe (2021) 29:1076–92.
43. Peng Y, Mentzer AJ, Liu G, Yao X, Yin Z, Dong D, et al. Broad and Strong Memory CD4(+) and CD8(+) T Cells Induced by SARS-CoV-2 in UK Convalescent Individuals Following COVID-19. Nat Immunol (2020) 21:1336–45. Oxford Immunology Network Covid-19 Response, ISARIC4C Investigators
44. Tarke A, Sidney J, Kidd CK, Dan JM, Ramirez SI, Yu ED, et al. Comprehensive Analysis of T Cell Immunodominance and Immunoprevalence of SARS-CoV-2 Epitopes in COVID-19 Cases. Cell Rep Med (2021) 2:100204.
45. Nelde A, Bilich T, Heitmann JS, Maringer Y, Salih HR, Roerden M, et al. SARS-CoV-2-Derived Peptides Define Heterologous and COVID-19-Induced T Cell Recognition. Nat Immunol (2021) 22:74–85.
46. Issa E, Merhi G, Panossian B, Salloum T, Tokajian S. SARS-CoV-2 and ORF3a: Nonsynonymous Mutations, Functional Domains, and Viral Pathogenesis. mSystems (2020) 5(3):e00266–20.
47. Wang H, Wu X, Zhang X, Hou X, Liang T, Wang D, et al. SARS-CoV-2 Proteome Microarray for Mapping COVID-19 Antibody Interactions at Amino Acid Resolution. ACS Cent Sci (2020) 6:2238–49.
48. Zhang XY, Guo J, Wan X, Zhou JG, Jin WP, Lu J, et al. Biochemical and Antigenic Characterization of the Structural Proteins and Their Post-Translational Modifications in Purified SARS-CoV-2 Virions of an Inactivated Vaccine Candidate. Emerg Microbes Infect (2020) 9:2653–62.
49. Munoz-Fontela C, Dowling WE, Funnell SGP, Gsell PS, Riveros-Balta AX, Albrecht RA, et al. Animal Models for COVID-19. Nature (2020) 586:509–15.
50. Imai M, Iwatsuki-Horimoto K, Hatta M, Loeber S, Halfmann PJ, Nakajima N, et al. Syrian Hamsters as a Small Animal Model for SARS-CoV-2 Infection and Countermeasure Development. Proc Natl Acad Sci U.S.A. (2020) 117:16587–95.
51. Rosenke K, Meade-White K, Letko M, Clancy C, Hansen F, Liu Y, et al. Defining the Syrian Hamster as a Highly Susceptible Preclinical Model for SARS-CoV-2 Infection. Emerg Microbes Infect (2020) 9:2673–84.
52. Tostanoski LH, Al. E. Immunity Elicited by Natural Infection or Ad26.COV2.S Vaccination Protects Hamsters Against SARS-CoV-2 Variants of Concern. Sci Transl Med (2021) 13:eabj3789.
53. Reed CC, Schultheis K, Andrade VM, Kalia R, Tur J, Schouest B, et al. Design, Immunogenicity and Efficacy of a Pan-SARS-CoV-2 Synthetic DNA Vaccine. bioRxiv (2021). 2021.2005.2011.443592.
54. Tebas P, Roberts CC, Muthumani K, Reuschel EL, Kudchodkar SB, Zaidi FI, et al. Safety and Immunogenicity of an Anti-Zika Virus DNA Vaccine - Preliminary Report. N Engl J Med (2021) 385(12):e35.
55. Schultheis K, Smith TRF, Kiosses WB, Kraynyak KA, Wong A, Oh J, et al. Delineating the Cellular Mechanisms Associated With Skin Electroporation. Hum Gene Ther Methods (2018) 29:177–88.
56. Cao H, Li X, Liu J. An Updated Review of the Efficacy of Cupping Therapy. PloS One (2012) 7:e31793.
58. Mehta P, Dhapte V. Cupping Therapy: A Prudent Remedy for a Plethora of Medical Ailments. J Tradit Complement Med (2015) 5:127–34.
59. Modjarrad K, Roberts CC, Mills KT, Castellano AR, Paolino K, Muthumani K, et al. Safety and Immunogenicity of an Anti-Middle East Respiratory Syndrome Coronavirus DNA Vaccine: A Phase 1, Open-Label, Single-Arm, Dose-Escalation Trial. Lancet Infect Dis (2019) 19:1013–22.
60. Tebas P, Roberts CC, Muthumani K, Reuschel EL, Kudchodkar SB, Zaidi FI, et al. Safety and Immunogenicity of an Anti-Zika Virus DNA Vaccine. N Engl J Med (2021) 385:e35.
Keywords: SARS-CoV-2, Bi-cistronic DNA vaccine, T cell, antibody, SARS-CoV-2 variant of concern, viral challenge
Citation: Jeong M, Kudchodkar SB, Gil A, Jeon B, Park GH, Cho Y, Lee H, Cheong MS, Kim W, Hwang Y-H, Lee J-A, Lim H, Kim MY, Lallow EO, Brahmbhatt T, Kania SA, Jhumur NC, Shan JW, Zahn JD, Shreiber DI, Singer JP, Lin H, Spiegel EK, Pessaint L, Porto M, Van Ry A, Nase D, Kar S, Andersen H, Tietjen I, Cassel J, Salvino JM, Montaner LJ, Park YK, Muthumani K, Roberts CC and Maslow JN (2022) Immune Responses of a Novel Bi-Cistronic SARS-CoV-2 DNA Vaccine Following Intradermal Immunization With Suction Delivery. Front.Virol. 2:891540. doi: 10.3389/fviro.2022.891540
Received: 07 March 2022; Accepted: 06 April 2022;
Published: 24 May 2022.
Edited by:
Xin Yin, Chinese Academy of Agricultural Sciences (CAAS), ChinaReviewed by:
Chao Zhang, Fifth Medical Center of the PLA General Hospital, ChinaLonghuan Ma, University of Florida, United States
Copyright © 2022 Jeong, Kudchodkar, Gil, Jeon, Park, Cho, Lee, Cheong, Kim, Hwang, Lee, Lim, Kim, Lallow, Brahmbhatt, Kania, Jhumur, Shan, Zahn, Shreiber, Singer, Lin, Spiegel, Pessaint, Porto, Van Ry, Nase, Kar, Andersen, Tietjen, Cassel, Salvino, Montaner, Park, Muthumani, Roberts and Maslow. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Kar Muthumani, a211dGh1bWFuaUBnZW5lbHMudXM=; Joel N. Maslow, am1hc2xvd0BnZW5lbHMudXM=
†These authors have contributed equally to this work
 Moonsup Jeong
Moonsup Jeong Sagar B. Kudchodkar
Sagar B. Kudchodkar Areum Gil1
Areum Gil1 Gee Ho Park
Gee Ho Park Mi Sun Cheong
Mi Sun Cheong Wonil Kim
Wonil Kim Emran O. Lallow
Emran O. Lallow Stephen A. Kania
Stephen A. Kania David I. Shreiber
David I. Shreiber Jonathan P. Singer
Jonathan P. Singer Laurent Pessaint
Laurent Pessaint Danielle Nase
Danielle Nase Swagata Kar
Swagata Kar Ian Tietjen
Ian Tietjen Joseph M. Salvino
Joseph M. Salvino Luis J. Montaner
Luis J. Montaner Kar Muthumani
Kar Muthumani Christine C. Roberts
Christine C. Roberts Joel N. Maslow
Joel N. Maslow