
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Virol., 07 March 2023
Sec. Antivirals and Vaccines
Volume 2 - 2022 | https://doi.org/10.3389/fviro.2022.1064265
 Shuzo Urata1,2*
Shuzo Urata1,2* Jun Takouda3
Jun Takouda3 Yoshihiro Watanabe4
Yoshihiro Watanabe4 Miako Sakaguchi5
Miako Sakaguchi5 Yasuteru Sakurai1,2
Yasuteru Sakurai1,2 Yuki Inahashi4
Yuki Inahashi4 Masato Iwatsuki4
Masato Iwatsuki4 Jiro Yasuda1,2
Jiro Yasuda1,2 Yoshimasa Tanaka6
Yoshimasa Tanaka6 Kohsuke Takeda3
Kohsuke Takeda3Severe fever with thrombocytopenia syndrome virus (SFTSV) is a tick-borne virus first identified in China in 2011 and later reported in other Asian countries. Significant efforts have been made to develop anti-SFTSV compounds; however, there are no approved vaccines or antivirals against SFTSV infections. Marine organisms provide nearly unlimited biological resources to produce therapeutic drugs for the treatment and control of disease. In this study, we aimed to identify anti-SFTSV chemical compounds from the culture broth extracts of marine microbes collected from the coasts of the Nagasaki Prefecture, Japan. Of the 80 extracts, two showed an anti-SFTSV effect. One of them, which exhibited low cell toxicity, was used for further characterization. Chemical analysis combined with the anti-SFTSV effect identified surfactin as one of the main components of the selected extract. Our study showed a proof-of-concept to identify novel antiviral compounds from marine microbes against the virus of interest. Further analysis showed that surfactin affected the integrity of the virion membrane and inhibited SFTSV infection-induced membrane fusion at low pH conditions. Furthermore, surfactin inhibits the post-entry step of viral replication in the cell, which is a novel mode of antiviral action of surfactin. These results indicate that surfactin can target multiple steps of SFTSV replication in cells.
Severe fever with thrombocytopenia syndrome (SFTS) was discovered in China in 2011 and has been reported in several East Asian countries including Japan, South Korea, Taiwan, and Vietnam (1–6). SFTS is an emerging infectious disease caused by the SFTS virus (SFTSV, SFTS phlebovirus, or Huaiyangshan banyangvirus). SFTSV is a tick-borne virus classified under the genus Banyangvirus of the Phenuiviridae family and Bunyavirales order (7). Other phleboviruses that are phylogenetically related to SFTSV are the Heartland and Malsoor viruses, isolated from Missouri, USA, and western India, respectively (8, 9). In addition, a novel virus closely related to the SFTSV and Heartland virus has been identified in China (10). Currently, there is no established prophylactic or treatment for SFTS. Elucidation of the viral replication mechanism could help determine appropriate prophylactic and treatment strategies. Small chemical compound libraries with known structures and functions have been used to identify novel anti-SFTSV compounds. Despite the lack of established treatments against SFTS, several compounds, including Food and Drug Administration (FDA)-approved drugs, have been reported to inhibit SFTSV replication both in vivo and in vitro (11, 12). Apart from this strategy, utilizing living organisms that produce numerous secondary metabolites and natural molecules with almost unlimited structural diversity could be an alternative approach to identify novel anti-SFTSV agents that could be developed into new anti-SFTSV drugs.
A large number of microbes exist in the ocean, and their lives depend on their living environments. These marine microorganisms have been known to provide unlimited biological resources for the production of therapeutic drugs for the treatment and control of diseases, including viral infections (13, 14). In addition, ecology, such as the region, condition, and/or environment of marine microbes, affects their production of metabolites. This implies that marine microbes can produce unique metabolites that are specific to their habitat region and conditions. Among the natural antiviral products identified from marine microbes, alkaloids, quinones, peptides, polyketones, pyrenes, sterols, and terpenoids have been shown to exhibit antiviral effects against several viruses, including enterovirus 71, herpes simplex (HSV)-1, influenza, human immunodeficiency (HIV) type 1, hepatitis C, and dengue virus (13, 14). However, the antiviral effects of these compounds against other viruses remain unclear. Therefore, further research is needed to examine the antiviral effect of the identified compounds against other viruses and to continue the identification of novel antiviral natural products from marine microbes as potential therapeutic drugs.
In this study, we attempted to identify and characterize the effects of the anti-SFTSV compounds extracted from the culture broths of marine microbes collected from the coasts of Nagasaki Prefecture, Japan.
To efficiently collect marine microbes, various sea animals along the coasts of several districts of the Nagasaki Prefecture, Japan, were collected. Marine microbes were mainly isolated from alimentary canals as follows. Alimentary canal samples were streaked onto marine agar plates (Difco Marine Agar 2216; Beckton, Dickinson & Co, NJ, USA) and seawater agar plates containing 8.0 g D(+)-glucose (Fujifilm Wako Chemicals, Osaka, Japan), 4.0 g hipolypepton (Fujifilm Wako), 2.4 g Bacto yeast extract (Beckton, Dickinson & Co), 2.4 g KH2PO4 (Fujifilm Wako), 0.8 mg MgSO4·7H2O (Fujifilm Wako), and Bacto-Agar (Beckton, Dickinson & Co) per liter of natural seawater diluted to 75% of the initial concentration with distilled water, which were incubated at 26 °C for one to seven days. After incubation, colonies were isolated based on their morphology and pigmentation, and marine microbes were grown in 200 mL or 1.6 L marine broth (Difco Marine Broth 2216; Beckton, Dickinson & Co) or seawater broth. This contained 8.0 g D(+)-glucose, 4.0 g hipolypepton, 2.4 g Bacto-yeast extract, 2.4 g KH2PO4, and 0.8 mg MgSO4·7H2O per liter of natural seawater diluted to 75% of the initial concentration with distilled water, in 500 mL or 2 L Erlenmeyer flasks, respectively, at 26 °C for one to two weeks. One-third volume of acetone was added to the marine microbial culture (a mixture of broth and microbes) and sonicated for 5 min. The sonicated samples, including both extracellular and intracellular substances, were filtered through filter paper and the filtrate was subjected to vacuum evaporation to remove acetone. 1/2 volume of ethyl acetate was added to the residue and the mixture was shaken in separatory funnels for 3 min. After the aqueous phase was drained, the organic phase was collected and subjected to vacuum evaporation to remove ethyl acetate. Dimethyl sulfoxide (DMSO) was added to the extract at a concentration of 100 mg/mL, which was dispensed into cryovials and stored at -30 °C.
The human hepatocyte cell line (Huh-7) and the African green monkey kidney cell line (Vero 76) cells were grown in Dulbecco’s modified Eagle’s medium (Fujifilm, 044-29765) containing 10% fetal bovine serum (FBS), 100 IU/mL penicillin, and 100 μg/mL streptomycin. The origin of the SFTSV used in this study has been previously described (15). Sodium surfactant was purchased from Fujifilm (194-12691).
HPLC was performed using a pegasil ODS SP 100 column (4.6 i.d. x 250 mm, Senshu Scientific Co. Ltd., Tokyo, Japan) with a linear gradient from 5% CH3CN aqueous solution to 100% CH3CN for 30 min, followed by 100% CH3CN for 10 min at a flow rate of 1 mL/min, and detected using photodiode array detector. The samples were analyzed by LC-Q-TOF-MS and the data were obtained using an AB Sciex Triple TOF™ 5600+ Liquid Chromatograph (LC)-Mass Spectrometry (MS)/MS Systems (AB Sciex, Framingham, MA, USA) with a reversed-phase HPLC column, Capcell Core C18 (2.7 µm, 3.0 i.d. x 100 mm, Osaka Soda Co. Ltd., Osaka, Japan), with a linear gradient from 5% MeOH aqueous solution containing 0.1% formic acid to 100% MeOH containing 0.1% formic acid at a flow rate of 0.5 mL/min for 13 min. Liquid chromatography-high-resolution electrospray ionization mass spectrometry (LC-MS) spectra were measured using an AB Sciex TripleTOF 5600+ system (AB Sciex, Framingham, MA, USA). All analyses were conducted in positive ion mode. Detailed conditions of the MS analysis were as follows: ion source gas1 20 psi; ion source gas2 15 psi, curtain gas 25 psi, temperature 0°C, ionspray voltage floating 5500 V, declustering potential 100 V, collision energy 45 V, collision energy spread 0 V, ion release delay 67 μs, and ion release delay width 25 μs. LC/MS data were analyzed using the Analyst software (AB Sciex, version 1.7.1).
The SFTSV titer was determined using an immunofluorescence assay. Vero 76 cells (3 × 104 cells/well) were seeded in a 96-well plate one day prior to infection. The cells were infected with 1:10 serial dilutions of the virus and incubated for 16 h at 37°C in 5% CO2. The cells were fixed with 4% paraformaldehyde (PFA) for 30 min at 15-25°C, and then incubated in PBS containing 0.1% Tween®20 and 5% FBS for 1 h at 15-25°C. SFTSV N protein was detected using a primary anti-SFTSV N antibody (12, 15), followed by a secondary anti-rabbit IgG-FITC antibody (ab6009, Abcam, Cambridge, UK). SFTSV N-positive cells were counted in a well containing 10–100 SFTSV N-positive cells and expressed as fluorescent focus units (FFU/mL) after appropriate calculation. When counting the N-positive cells, the number below limit of detection (L.O.D.) was considered to be zero. Each viral titer, average, and standard deviation were plotted in the graph.
For the screening, each extract (Figure 1) or fraction (Figure 2A) was mixed with SFTSV (multiplicity of infection (moi) = 0.1), and then infected with Huh-7 cells, which were seeded in 96 -well plates one day prior to infection. For screening of the extracts (Figure 1), infected cells were fixed and stained with anti-SFTSV N antibody, as described in section 2.4. The number of SFTSV N-positive cells and DAPI-positive cells were automatically counted using Cytation 5 (BioTek). For the measurement of the SFTSV titer in NUHE-1 Lots 1 to 3 (Figure 3A) and screening of the fractions (Figure 2A), extracts/fractions were applied throughout the incubation period. The culture supernatants of the infected and treated cells were collected, and the viral titer was examined, as described in section 2.4. To examine the anti-SFTSV effect of surfactin on virus propagation in Huh-7 or Vero 76 cells (Figure 4), both cell lines were infected with SFTSV at an moi of 0.1 for 1 h, in 96-well plates without compounds. After 1 h of incubation, the culture medium was replaced with fresh media containing either DMSO or surfactin. At 48 h post-infection (h p.i.), the culture supernatants were collected and viral titers were measured (Figure 4). The same procedure was followed to examine the effect of surfactin on the post-entry step, except for the fixation and collection time of the supernatant for the effect of a single round of infection (Figures 5A-C). For the assessment of the carryover effect of surfactin in the viral titration, surfactin was applied after virus absorption, incubated for 16 h, then the culture medium was replaced with fresh media that did not contain compounds, incubated further for 6h, then collected for virus titration (Figures 5D, E). For the SFTSV infections with a low cell density, Huh-7 (4 × 103) cells were seeded in 24 well plate, and infected with SFTSV with moi = 1 for 1 h, then treated with either DMSO or surfactin together with 20 mM NH4Cl for 15 h, and then fixed and stained with SFTSV N antibody and DAPI. The number of SFTSV N-positive cells was automatically counted using BZ-X800 (KEYENCE) (Figure 6).
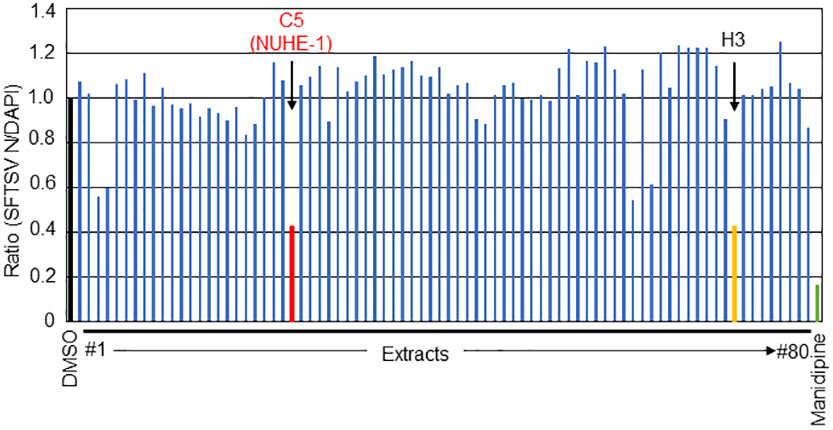
Figure 1 Screening of the culture broth extracts of 80 marine microbes for anti-SFTSV effects. The normalized ratio (SFTSV N positive cell/DAPI positive cell) with DMSO treatment as 1.0. The two hit extracts, C5 (NUHE-1) and H3, are shown in red and orange font, respectively. Manidipine was used as a positive control.
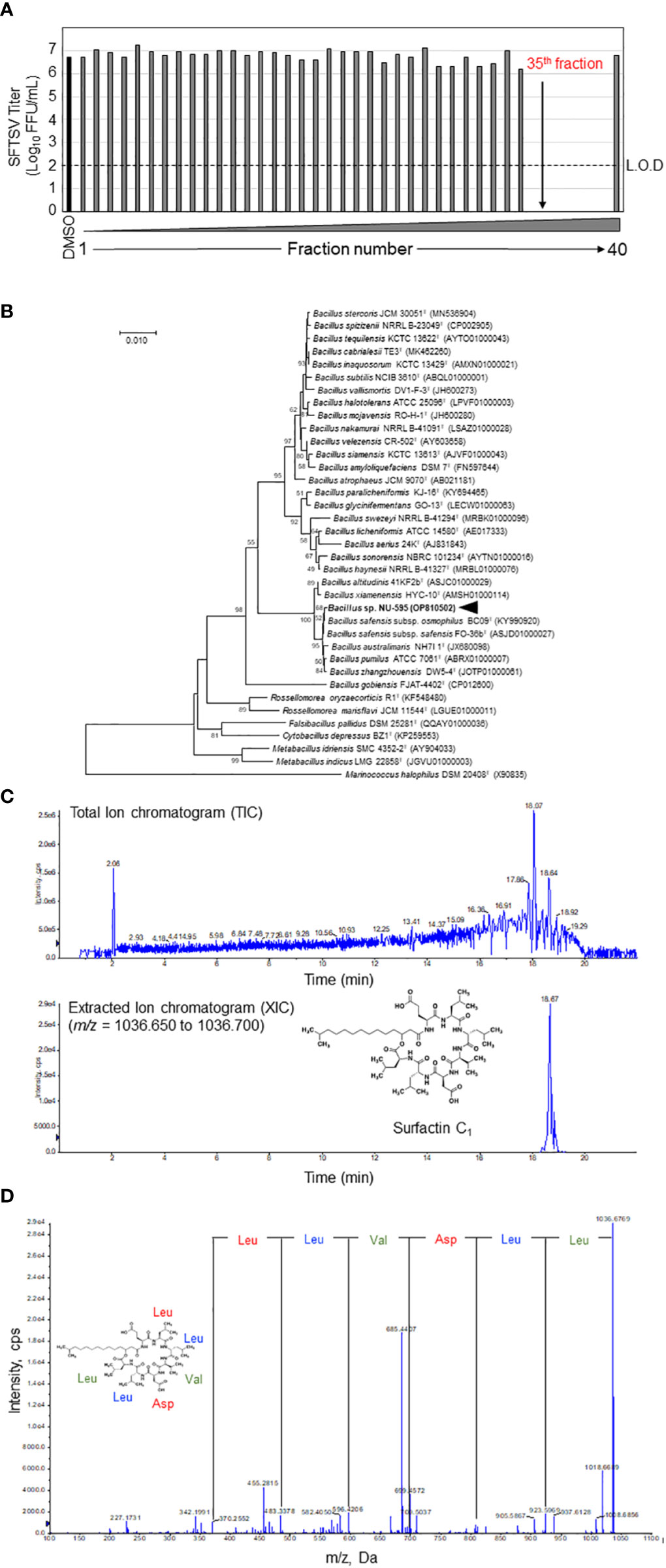
Figure 2 Identification of surfactin as the main anti-SFTSV compound in the NUHE-1. (A) The NUHE-1 was fractionated into 40 fractions and their anti-SFTSV effect was examined in Huh-7 cells. The SFTSV titer was examined using the culture supernatant collected from the SFTSV infected Huh-7 cells supplemented with each fraction or DMSO. (B) Neighbor-joining phylogenetic tree based on 16S rRNA gene sequences. Genetic relatedness between strain NU-595 and type strains in the genus Bacillus and other related genera is shown. Bootstrap values above 50% or higher (percentages of 1000 replicates) are shown. (C) Liquid chromatography quantitative time of flight mass spectrometry (LC-Q-TOF-MS) analysis of the 35th HPLC fraction identified surfactin C1 as the main component of NUHE-1 which exhibited anti-SFTSV effect. Upper panel shows the spectrum of total ion chromatogram (TIC). Lower panel shows the spectrum of extracted ion chromatogram (XIC). (D) Tandem mass spectrometry (MS/MS) analysis of surfactin C1: Characteristic fragment ions were observed at m/z 923.5969, 810.5131, 695.4879, 596.4206, 483.3378, and 370.2552. They are representative of the Leu-Leu-Val-Asp-Leu-Leu sequence of surfactin C1. L.O.D.; limit of detection.
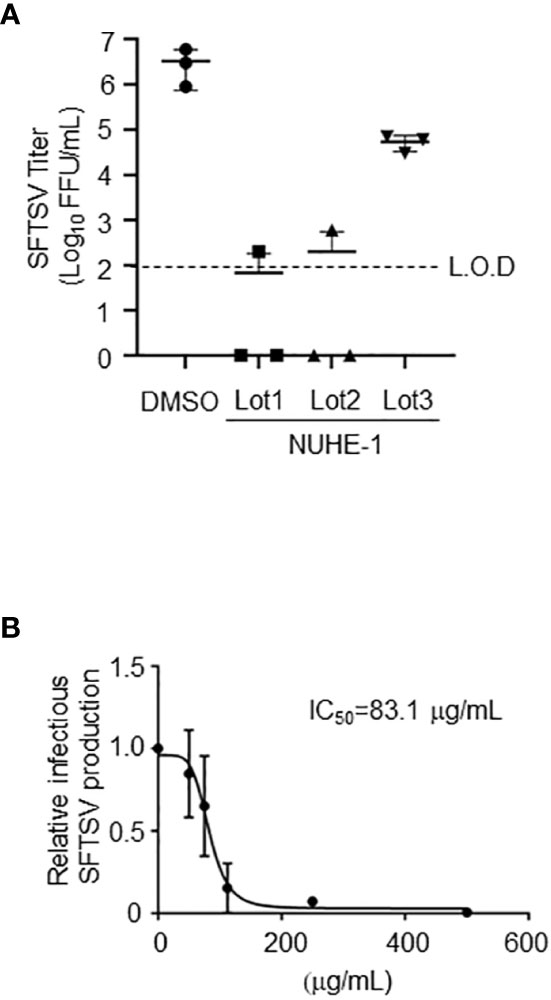
Figure 3 Anti-SFTSV effects of the NUHE-1. (A) NUHE-1 Lot 1, Lot 2, and Lot 3 were examined for their anti-SFTSV effects on Huh-7 cells. At 48 h post-infection (h p.i.), culture media which contains infectious SFTSV was collected, and examined for its infectivity. (B) The half maximal inhibitory concentration (IC50) was calculated with the condition described above and in the Materials and Methods section, using 31.25, 62.5, 125, 250, and 500 μg/mL of NUHE-1 Lot 3 (IC50 = 83.1 μg/mL). L.O.D.; limit of detection.
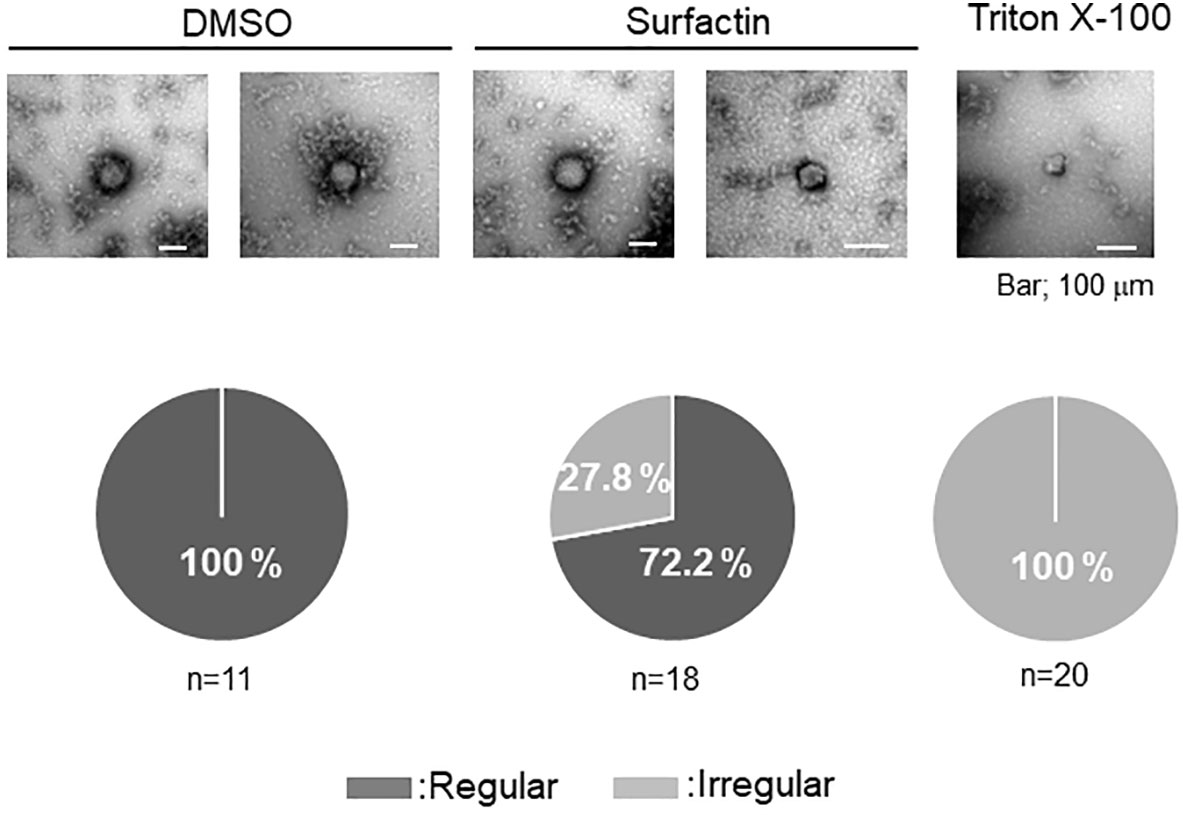
Figure 4 Effect of surfactin on SFTSV integrity. Electron microscope analysis was performed to examine the effect of surfactin on the virion membrane integrity. Infectious SFTSV was mixed with DMSO, surfactin (final 100 μM), or Triton X-100 (final 0.01%), and incubated for 30 min at 37°C. Samples were prepared as described in the Materials and Methods section. Eleven pictures from the DMSO treatment (two were shown), eighteen pictures from the surfactin treatment (two were shown), and twenty pictures from the Triton X-100 treatment (only one was shown) were captured, and the ratio of the particle number with the regular/irregular membrane integrity was shown.
The viability of Huh-7 and Vero 76 cells after surfactin treatment was assessed using the CellTiter-Glo Luminescent Cell Viability Assay (Promega), which determines the number of viable cells in a culture based on ATP levels. Huh-7 and Vero 76 cells (2 × 104 cells/well) were seeded in 96 well plates to form a monolayer. After seeding, the cells were treated with surfactin (100 μM) or DMSO (1%) as a control. At 48 h post-treatment, the culture supernatant was removed, and CellTiter-Glo reagent was added. The assay was performed according to the manufacturer’s recommendations using a SpectraMAX iD5 microplate reader (Molecular Devices, San Jose, CA, USA). The viability of DMSO-treated control cells was set to 1.0.
The culture media which contains infectious virions (2 × 106 FFU) were mixed with DMSO, surfactin (final 100 μM), or Triton X-100 (final 0.01%) for 30 min at 37°C (16). Then, samples were fixed with 2% glutaraldehyde (Nacalai Tesque, Kyoto, Japan) in 0.1 M sodium cacodylate buffer containing 1 mM CaCl2 and 1 mM MgCl2 (cacodylate buffer, pH 7.4) at 4°C for 60 min. Each sample was loaded on a 200-mesh copper grid with carbon-coated plastic film (Nisshin EM, Tokyo, Japan) immediately following glow discharge and negatively stained with uranyl acetate solution (1%, w/v) for 15 s. The morphology of each sample was observed using JEM-1400Flash (JEOL, Tokyo, Japan) with an acceleration voltage of 80 kV.
Syncytium formation was induced by SFTSV infection of cells at low pH (17). Huh-7 cells were infected with SFTSV for 1 h at a moi of 1, and then replaced with DMSO- or surfactin-containing media. At 24 h post-infection, the culture supernatant was collected to measure the viral titer. The cells were rinsed once with PBS (-) and then incubated with citrate-phosphate buffers (0.1 M citric acid and 0.2 M sodium dihydrogen orthophosphate) adjusted to pH 5.0 or 7.0 for 3 min. The citrate-phosphate buffers were then replaced with culture media, and cell fusion within the monolayers was observed under a phase-contrast microscope after 4-6 h.
To identify the strain NU-595 that produced an anti-SFTSV compound, 16S rRNA gene sequencing was conducted as described below. DNA was extracted using MightyPrep reagent for DNA (9182, TaKaRa) according to the manufacturer’s protocol. The 16S rRNA gene was amplified using Q5 High-Fidelity DNA polymerase (M0491A, New England BioLabs) and primers (11F: AGTTTGATCATGGCTCAG, 520F: CAGCAGCCGCGGTAATAC, 925R: CGTCAATTCATTTGAGTT, and 1540R: AAGGAGGTGATCCAGCCGCA). Amplifications were performed in a GeneAmp PCR system 9700 (Thermo Fisher Scientific) with an initial incubation for 1 min at 94°C, followed by 35 cycles of 20 s at 94°C, 30 s at 50°C, and 1 min at 72°C, followed by 2 min final extension at 72°C. PCR products were purified using NucleoSpin Gel and PCR Clean-up (U0609A, TaKaRa). Sequence analysis was performed using a DNA sequencer (3730xI DNA Analyzer, Thermo Fisher Scientific) using a BigDye Terminator v3.1 Cycle Sequencing Kit (Thermo Fisher Scientific). The sequence was subjected to BLAST search using the EzBioCloud server (https://www.ezbiocloud.net/). For the phylogenetic analysis, the sequence of strain NU-595 was aligned with sequences of related species using CLUSTAL W (18), and the phylogenetic tree was constructed using the neighbor-joining method (19) in MEGA X (20).
To detect and analyze the expression of SFTSV N in the infected cells, cell lysate was prepared with Lysis A buffer (1% NP-40, 50 mM Tris-HCl [pH 8.0], 62.5 mM EDTA, and 0.4% sodium deoxycholate), and WES (ProteinSimple, Tokyo, Japan) was used to detect SFTSV N with Ez standard Pack 1 (PS-ST01EZ-8, ProteinSimple), anti-rabbit chemiluminescence detection module (DM-001, ProteinSimple), together with anti-SFTSV N antibody as described above (section 2.4) (12, 15).
To quantify the SFTSV S segment RNA in the cell, total RNA was collected from the infected cell using RNeasy mini kit (74106, QIAGEN). Reverse Transcription was performed using equal amount of RNA to obtain cDNA with a primer (5’- AGCTGAAGGAGACAGGTGGA -3’) and PrimeScript™ II 1st strand cDNA synthesis kit (6210A, TaKaRa). Then, the cDNA was used as a template for the quantification with the primer set (5’-GCTGAAGGAGACAGGTGGAG-3’ and 5’-GCAACCCTCACAGGAGTGAT-3’), TB Green Premix Ex Taq II (Tli RNaseH Plus) (RR820A, TaKaRa) and StepOnePlus™ real-time PCR system (Thermo Fisher Scientific). A plasmid containing the whole SFTSV S segment was diluted and used to obtain a standard curve.
Statistically significant differences between groups were determined by the Student’s t‐test using Excel software (Microsoft). Quantitative data were presented as the mean ± SD from at least three independent experiments. For all calculations, p < 0.05 (*), p < 0.01 (**), p < 0.001 (***) were considered significant and represented using asterisks.
To identify compounds that exhibit anti-SFTSV effects from 80 culture broth extracts of marine microbes, each extract was mixed with SFTSV, followed by application to Huh-7 cells, as described in the Materials and Methods section. At 48 h post infection (h p.i.), the infected cells were fixed and stained with anti-SFTSV N antibody and DAPI to monitor viral propagation and cell number. The relative number of SFTSV N-positive cells (SFTSV N-positive cells/DAPI positive cells normalized to DMSO-treated cells as 1.0) is shown in Figure 1. Manidipine, an L-type calcium channel inhibitor (12, 21) was used as a positive control. Among the 80 extracts tested in this experiment, the relative number of SFTSV N-positive cells/DAPI-positive cells from the two extracts (C5 and H3) was less than 0.5, as shown in red and orange, respectively (Figure 1). Although both extracts reduced SFTSV N-positive cell numbers to similar extents, H3 exhibited higher cell toxicity than C5, suggesting potential cytotoxic effects. Based on this result, we decided to focus on the C5 extract, which exhibits anti-SFTSV activity with lower cell toxicity, and renamed it Nagasaki University hit extract (NUHE)-1.
To examine the reproducibility of the anti-SFTSV effect of NUHE-1, the microbial strain NU-595 (from which NUHE-1 was prepared) was cultured again in 200 mL and 1.6 L media, and different lots of extracts, Lots 2 and 3, respectively, were prepared from the culture broth (Figure 3A). The culture conditions for Lot 2 were the same as those for Lot 1, which were used in the initial screening. The culture scale for Lot 3 was set based on the subsequent purification of the compound responsible for exerting the anti-SFTSV effect of the extract. Lots 1 and 2 exhibited equivalent anti-SFTSV activities (4-logs reduction compared to DMSO treatment). In the case of Lot 3, it only reduced up to 2-logs compared to the DMSO treatment. Although the efficacy of anti-SFTSV by Lot 3 was reduced compared to that of Lot 1 and Lot 2, we concluded that this reduction was enough to isolate the responsible compounds in NUHE-1. 50% anti-SFTSV inhibitory concentration (IC50) of NUHE-1 Lot 3 was calculated as 83.1 ug/mL (Figure 3B).
NUHE-1 Lot 3 was fractionated every minute using HPLC for 40 min, and the anti-SFTSV effect was evaluated for each fraction to identify the fraction that exhibited the strongest anti-SFTSV effect (Figure 2A). Fractions 35–39 significantly inhibited SFTSV production. A taxonomic study of the strain NU-595 was also conducted. The 16S rRNA sequence of strain NU-595 (GenBank accession number: OP810502) showed the highest similarity value (99.93%) to those of Bacillus safensis subsp. safensis FO-36bT and B. safensis subsp. osmophilus BC09T. Phylogenetic analysis indicated that the strain NU-595 clustered with Bacillus species (Figure 2B). Therefore, strain NU-595 was identified as belonging to the genus Bacillus. Surfactin is an antiviral agent produced by members of the genus Bacillus (16, 22, 23). Based on this knowledge, fraction 35 in Figure 2A was analyzed using LC-Q-TOF-MS to determine the presence of surfactin in this fraction. As expected, surfactin was the main component of fraction 35 (Figure 2C). MS/MS analysis confirmed that fraction 35 contained surfactin C1 with a molecular weight of 1036, a surfactin congener (Figure 2D). Analysis of fractions 36-39 by MS/MS revealed that these fractions also contained other surfactin congeners with molecular weights of 1050, 1064, and 1078 (Supplemental Figure 1).
Owing to its ability to disrupt the viral lipid membrane surfactin has been reported to possess strong antiviral activity against envelope viruses, including Semliki Forest (SFV), herpes simplex (HSV-1, HSV-2), suid herpes (SHV-1), vesicular stomatitis (VSV), simian immunodeficiency (SIV) (22), influenza, Ebola, Zika, Nipah, Chikungunya, Una, Mayaro, Dugbe, and Crimean-Congo hemorrhagic fever viruses (23), as well as severe acute respiratory syndrome (SARS-CoV), and Middle East respiratory syndrome coronaviruses (MERS-CoV). To examine whether surfactin affects virion integrity as reported with other enveloped viruses, infectious SFTSV was treated with DMSO or surfactin. The viruses were treated with Triton X-100 as a positive control. The treated virions were observed by electron microscopy. Virions treated with DMSO were pleomorphic, but mostly round in shape (n=11). In contrast, the integrity of all the virions treated with Triton X-100 was affected (n=20). Approximately 30% of the virion integrity was affected when treated with surfactin (n=18) (Figure 4).
To examine the anti-SFTSV effect of surfactin, two cell lines, Huh-7 (Figure 7A) and Vero 76 (Figure 7B), were infected with SFTSV at a multiplicity of infection (moi) of 0.1, and treated with surfactin for 48 h. At 48 h p.i., the culture supernatant was collected and the viral titer was measured. The production of SFTSV in both Huh-7 and Vero 76 cells was reduced to below the limit of detection (L.O.D.). The cell viability following surfactin treatment was also evaluated. While surfactin treatment reduced the viable cell number to 60% in Huh-7 cells compared to DMSO treatment (Figure 4A, right), the same treatment in Vero 76 cells did not significantly affect the cell viability compared to the DMSO treatment (Figure 7B, right). In addition to its ability to disrupt the viral lipid membrane, surfactin was reported to reduce the infectivity of viruses, including porcine epidemic diarrhea and transmissible gastroenteritis virus (16), by inhibiting the viral envelope-cell membrane fusion event. Based on these reports, the same inhibitory effect of surfactin on syncytium formation at low pH due to virion-cell membrane fusion induced by SFTSV glycoprotein was examined in Huh-7 cells (Figure 8) and Vero 76 cells (Supplemental Figure 2). Both Huh-7 and Vero 76 cells were infected with SFTSV at moi =1.0 and treated with surfactin. At 24 h p.i., infected cells were treated with citrate-phosphate buffers (pH 7.0 or 5.0) for 3 min and then the buffer was replaced with fresh media, followed by a 4 h incubation period. The cells were fixed and stained with an anti-SFTSV N antibody and DAPI. When cells were treated with DMSO and citrate-phosphate buffers at pH 5.0, syncytium formation was observed, as indicated by the white dotted lines (Figure 8 for Huh-7 cells and Supplemental Figure 2 for Vero 76 cells). SFTSV N-positive cells were also observed in the fused cells. These effects were not observed in cells treated with DMSO-citrate-phosphate buffer (pH 7.0). When the infected cells were treated with surfactin, either treatment with pH 7.0 nor 5.0, exhibited syncytium formation.
To explore the effect of surfactin on viral replication and production, in addition to the cell entry procedure, the time course of SFTSV replication in Huh-7 cells was examined. Huh-7 cells were infected with SFTSV (moi = 1), and the production of virions in the culture supernatant and the expression of SFTSV N protein at different time points (0, 6, 8, 10, 12, 16, and 24 h p.i.) were examined (Supplemental Figure 3). The production of infectious virions was detected at 12 h p.i., and the expression of SFTSV N was detected at 16 h p.i. Based on these results, 16 h p.i. was considered to reflect a single round of infection. Next, the cells were infected with SFTSV without treatment for 1 h, the media was replaced with fresh media containing either DMSO or surfactin, and incubated for 15 h (Figure 5A). Total RNA was isolated at 1 h p.i. and 16 h p.i. (+/- surfactin treatment). The reverse transcription (RT) followed by the quantitative PCR (qPCR) was performed to quantify the SFTSV RNA (S segment) replication in the cell. Without the surfactin treatment, the SFTSV RNA amount was increased approximately 15 times from the 1 h p.i. to the 16 h p.i. When the infected cells were treated with surfactin after the infection, approximately 6.8-fold reduction of the SFTSV S segment was observed compared to the DMSO control treatment (Figure 5B). Accordingly, the SFTSV N expression in the cell was reduced (five-fold reduction) with the surfactin post-infection treatment (Figure 5C). Infected cells were also fixed and stained with anti-SFTSV N to count the SFTSV N-positive cells. SFTSV N-positive cells were significantly reduced under surfactin treatment compared to the control DMSO treatment (2-fold reduction, Figure 5D). To exclude the possibility that our conclusion about surfactin inhibiting the post-entry step and virion production was not due to a carryover of surfactin in the supernatant collected during the titration experiment, the following experiment was conducted. After virus infection and either DMSO or surfactin treatment and incubation for 16 h, the culture medium was replaced with fresh media without the compound, incubated for a further 6 h without any compounds, and the viral titer in the supernatant was examined (Figure 5D). As a result, virus production after media replacement was reduced by 1-2 logs with surfactin treatment compared to DMSO treatment (Figure 5E). Furthermore, to exclude the possible cell-to-cell transmission of SFTSV, cells were seeded at a low density, infected with SFTSV (moi = 1) for 1 h, then the medium was replaced with fresh media containing 20 mM NH4Cl to prevent re-infection and either DMSO or surfactin, and incubated for a further 15 h. At 16 h p.i., the infected cells were fixed and stained with an anti-SFTSV N antibody to count the SFTSV N-positive cells (Figure 6A). SFTSV N positive cell number was reduced to 57% compared with the DMSO treatment (Figure 6B).
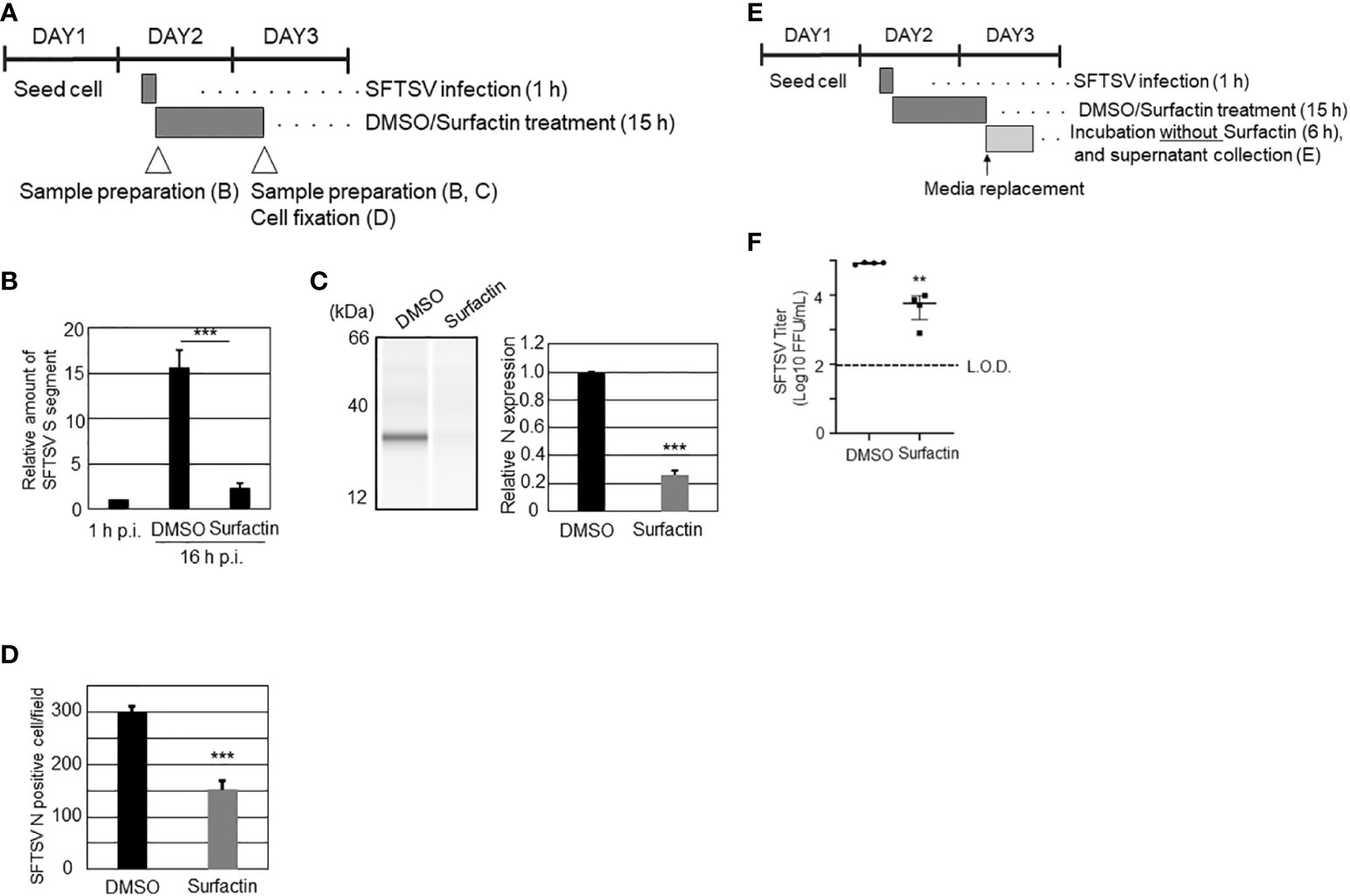
Figure 5 The effects of surfactin on SFTSV replication. (A) Scheme of the experiment for (B–D). On day 1, Huh-7 cells were seeded on 24 well plate (B, C) or 96 well plates (D). On day 2, cells were infected with SFTSV for 1 h (moi = 1), following which, the medium is replaced with media containing either DMSO or surfactin, and incubated for 15 h. On day 3 (16 h p.i.), infected cells were either collected cell lysate for quantifying SFTSV S segment (B), detecting SFTSV N by western blotting analysis (C), or fixed for staining SFTSV N to count N positive cells (D). Cell lysate was also collected at 1 h p.i. for the normalization (B). (E) Scheme of the experiment for (F). Until the 16 h p.i., the procedure was same as above. At 16 h p.i., media were replaced with fresh media, and further incubated for 6 h. The culture media collected at 22 h p.i. was collected for viral titration (F). Statistically significant differences between groups were determined by the Student’s t‐test (**p < 0.01, ***p < 0.001).
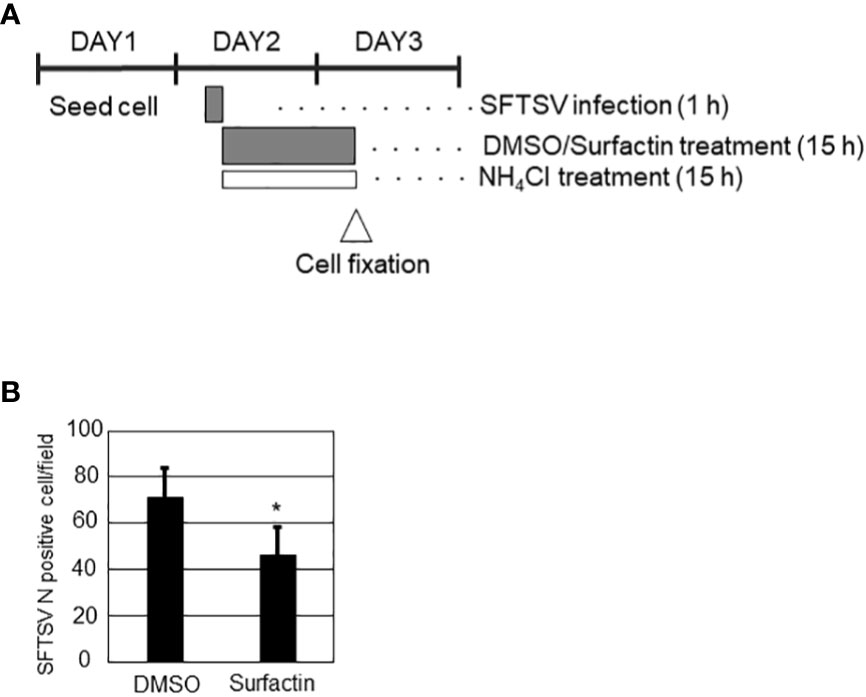
Figure 6 (A) Schematic representation of experiments. On day 1, Huh-7 cells were seeded in 24 well plate at a low cell density to avoid cell-cell contact. On day 2, cells were infected with SFTSV for 1 h, following which the medium was replaced with media containing 20 mM NH4Cl to avoid re-infection and either DMSO or surfactin, and incubated for a further 15 h. On day 3 (16 h p.i.), the infected cells were fixed for staining SFTSV N to count N-positive cells (B). Statistically significant differences between groups were determined by the Student’s t‐test (*p < 0.05).
Although significant efforts have been made to develop effective antiviral agents against multiple viruses, only few have been approved for clinical use. It is difficult to predict whether a specific compound has a strong antiviral effect without any significant side effects at the screening point. Therefore, it is important to identify as many candidate compounds as possible to evaluate their antiviral effects and safety.
In this study, considering there are no approved vaccines nor antivirals to treat SFTS patients, we attempted to identify anti-SFTSV compounds extracted from the culture broth of marine microbes. A small library of 80 marine microbial extracts prepared from the culture media of marine microbes collected from the coast of the Nagasaki prefecture, Japan, was used for screening. Two extracts were selected as the hit extracts, and one of them (NUHE-1) was used in this study because of its higher cell toxicity compared to the other one.
While the same culture volume of NU-595 (200 mL Lots 1 and 2) exhibited similar anti-SFTSV activities, Lot 3, which was prepared from the 1.6 L culture scale required for the subsequent HPLC analysis, showed reduced anti-SFTSV activity. In some cases, changing the culture conditions, including increasing the microbial culture volume, affected the final composition of the extracts. As described above, it is possible that 1.6 L culture is not the best condition for extracting hit compounds compared to smaller culture volumes in case of NUHE-1. Nonetheless, we concluded that the anti-SFTSV effect by NUHE-1 Lot 3 was enough to isolate the responsible compound and selected for further examination.
To reduce the number of steps required to identify the responsible anti-SFTSV compound in the extracts, NUHE-1 Lot 3 was fractionated every minute using HPLC and the anti-SFTSV effect was examined for each fraction (Figure 2A). Surfactin and other surfactin congeners were identified as the main compounds that exhibited significant anti-SFTSV activity in fractions 35-39 (Figures 2A, B, and Supplemental Figure 1). These results provide a clear proof of concept for the identification of antiviral compounds from the microbial extracts.
Surfactins are cyclic lipopeptides with amphiphilic properties. Surfactin is known to be produced by the gram-positive endospore-forming bacterium Bacillus subtilis (24), as well as by B. natto, B. pumilus, and B. slichenifomis. Surfactin congeners have also been isolated from Bacillus subtilis (25). Furthermore, it has been reported that surfactin congeners with varied amino acid alignments were produced from different strains of B. subtilis because of the high flexibility of nonribosomal peptide synthase (26). Since the first identification and characterization of surfactin (27), a series of studies have reported the function of surfactin, including antibacterial, antiviral, antifungal, antimycoplasma, and hemolytic activities (28). So far, surfactin’s antiviral effect has been considered to be based on the detergent effect, which leads to the disruption of the lipid membrane on the virion (29) as well as an inhibitory effect on viral-host membrane fusion at the virus entry step (16). In terms of the surfactin’s antiviral effect, enveloped viruses, as listed in the introduction section as well as non-enveloped viruses such as feline calicivirus (Caliciviridae) and murine encephalomyocarditis virus (Picornaviridae) (22) have been shown to be affected by the Surfactin treatment. The inactivation of enveloped viruses is much more efficient than that of non-enveloped viruses (22). It is worth noting that the antiviral effect of surfactin against Semliki Forest virus (Togaviridae) was significantly lower than that on the other enveloped viruses and almost equal to that on the non-enveloped viruses (22), implying that the antiviral effect of surfactin is not common to all enveloped viruses. Therefore, it is important to examine the effect of surfactin against all the viruses that are of interest.
In the current study, the significant anti-SFTSV effect of surfactin was observed in two cell lines: Huh-7 and Vero 76. As far as we know, the antiviral effect of surfactin is dependent on the surfactant effect, which affects the virion membrane and inhibits the virus-host membrane fusion event, as described above. The former effect has been observed in a wide range of envelope viruses (16, 22). Therefore, the effect of surfactin on SFTSV envelope was examined using an electron microscope. Our results showed that surfactin partially affected the SFTSV envelope (Figure 7). Next, to examine the effect of surfactin on virion-membrane fusion events, the syncytium formation in cells induced by SFTSV G was examined. As expected, surfactin treatment inhibited syncytium formation in both cell lines (Figure 8), strongly suggesting that one of its anti-SFTSV actions is to inhibit the SFTSV G-mediated membrane fusion event at the entry step. These results indicate that surfactin affects both the virion envelope and virion-host membrane fusion events, which have been reported as the mechanisms of action of surfactin against other viruses.
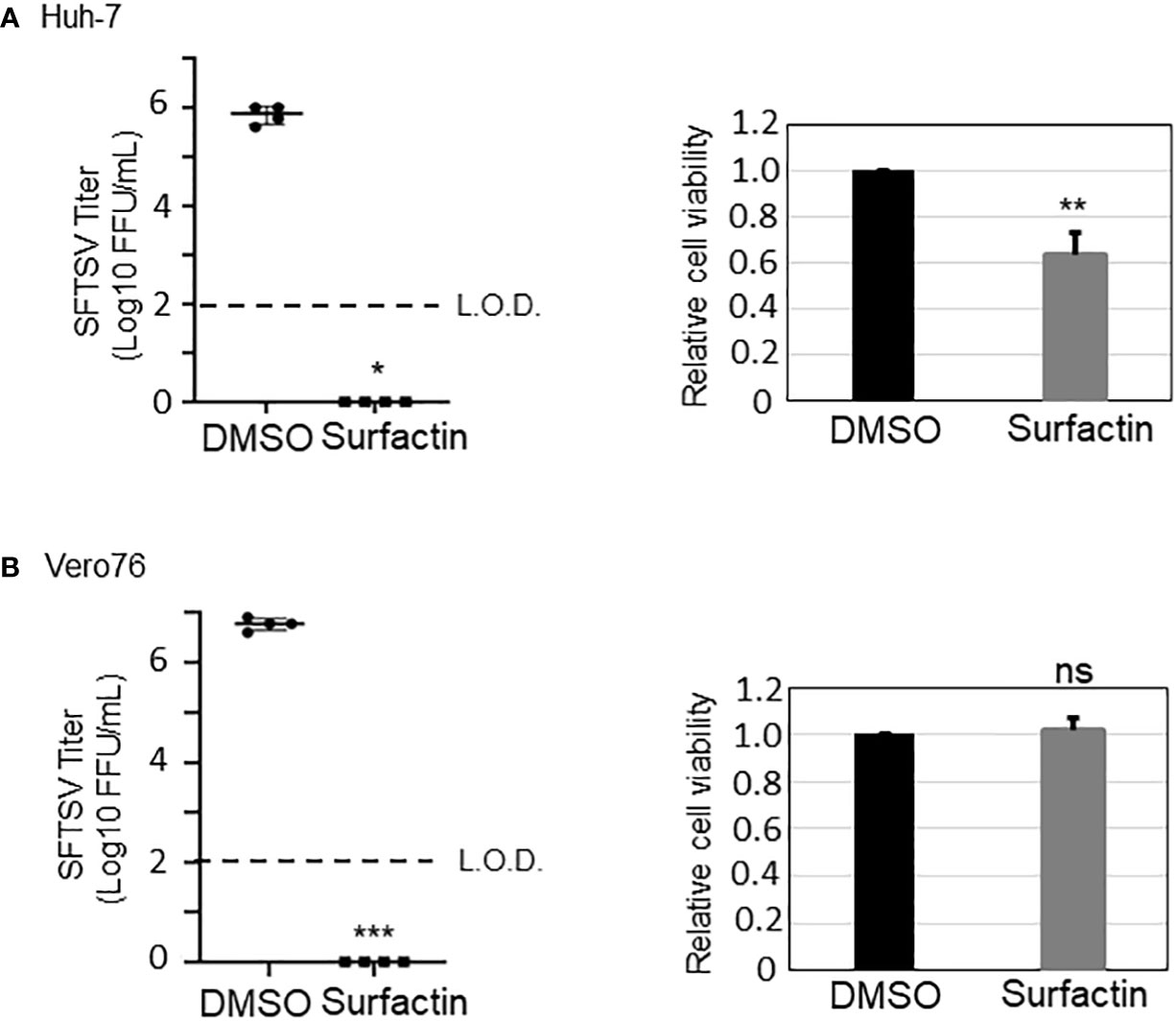
Figure 7 Surfactin inhibits SFTSV propagation. The effect of surfactin (100 μM) on SFTSV propagation (48 h post infection (h p.i.)) in Huh-7 cells (A) and in Vero76 cells (B). The cell viability of Huh-7 cells and Vero76 cells treated with 100 μM surfactin was shown on the right side of both figures. L.O.D.; limit of detection. Statistically significant differences between groups were determined by the Student’s t‐test (*p < 0.05, **p < 0.01, ***p < 0.001). ns, not significant.
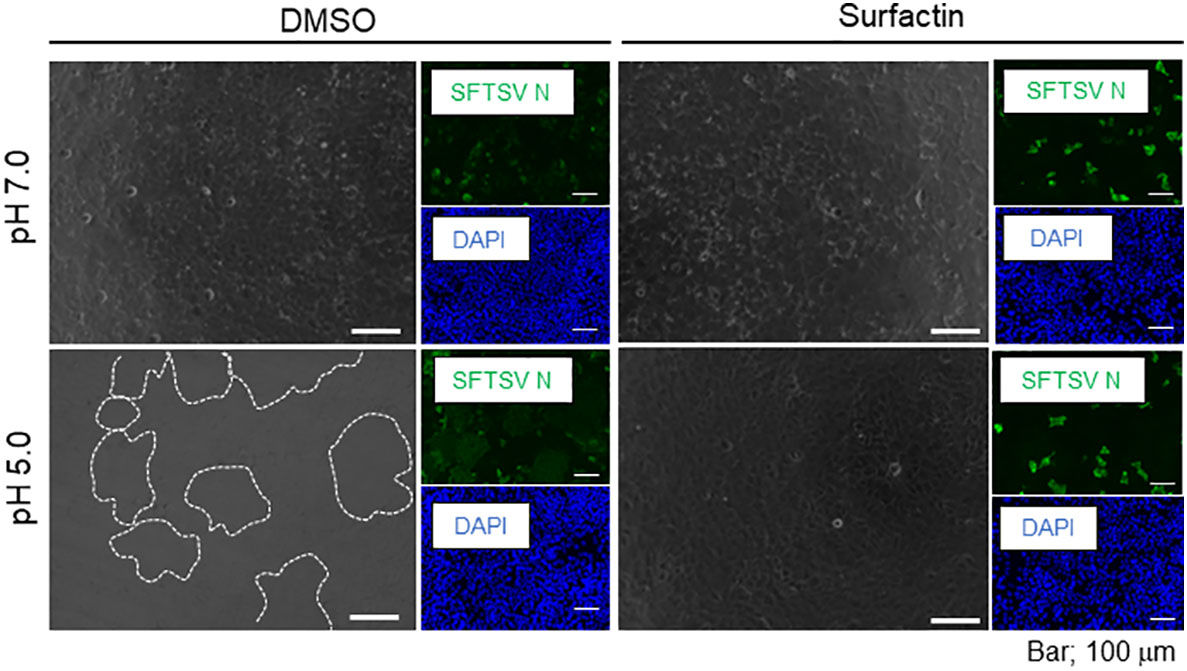
Figure 8 Cell-cell membrane fusion in low pH. Huh-7 cells were infected with SFTSV at multiplicity of infection = 1.0, replaced with either DMSO- or surfactin-containing media, and incubated for 24 h. Infected cells were treated with citrate phosphate buffer adjusted with either pH 7.0 or 5.0 for 2 min, replaced with fresh media, further incubated for 4 h, and fixed with 4% paraformaldehyde (PFA). Fixed cells were then stained with anti-SFTSV N antibody and DAPI. Captures of fluorescence and phase contrast images are shown. Syncytia are highlighted with a white dotted line.
To further investigate whether Surfactin affects viral life cycle steps other than the virion itself and/or the entry step, surfactin was applied after virus internalization into the cell (Figure 5). The reduction in the number of SFTSV N-positive cells at 16 h p.i., which was considered to reflect a single round of infection based on our results shown in Supplemental Figure 3, suggested that surfactin affected SFTSV genome replication until viral protein (N) translation. To exclude the possibility that the reduction in the virus titer was not due to the carryover of surfactin, the culture medium was replaced with fresh media that did not contain surfactin and was incubated for an additional 6 h. Then, the virus titer in the culture supernatant was compared between the DMSO- and surfactin-treated (before media replacement) cells. The result showed more than one log reduction in the virus titer after 16 h of surfactin treatment compared to that of control DMSO-treated cells (Figure 5E). This result suggested that surfactin also reduced virus production from 17 to 23 h p.i. However, the reduction in virus production at this point might be partially due to the reduction in viral replication, such as the N expression level, rather than the assembly and/or budding procedure.
Inhibition of NF-κB signaling by surfactin might be one of the mechanisms of its anti-SFTSV function (30), since an NF-kB inhibitor was reported to reduce SFTSV replication (31). However, others have reported that surfactin activates NF-κB in macrophages (32). It is important to reveal the molecular mechanisms underlying the anti-SFTSV activity of surfactin, including its effect on NF-κB in the cell lines used in this study.
Here, surfactin was shown to target more than two steps, membrane fusion at the entry and post-entry steps, in SFTSV infection, and a direct effect on the virion envelope, making the emergence of a surfactin-resistant mutant virus difficult. It might also be possible that surfactin could exhibit additive and/or synergetic anti-SFTSV effects with lower amounts than the ones used in this study when applied with other anti-SFTSV compounds. Through this study, it became clear that compounds extracted from the culture broth of marine organisms could serve as potential antiviral agents, such as surfactin. We also demonstrated that the strategy of utilizing bioassay-guided HPLC before large-scale cultivation is an effective approach to identify the target compound.
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
SU and KT designed the project. SU, JT, YW, MS and YI conducted the experiments. SU, YS, and KT analyzed the results. SU and KT prepared the original draft of the manuscript, and JT, YW, MS, YS, YI, MI, JY, YT, and KT supported the project and modified the original draft of the manuscript. All authors contributed to the article and approved the submitted version.
This study was partially supported by the Platform Project for Supporting Drug Discovery and Life Science Research (Basis for Supporting Innovative Drug Discovery and Life Science Research (BINDS)) of the Japan Agency for Medical Research and Development (AMED) under grant numbers JP21am0101088 and JP21am0101096.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fviro.2022.1064265/full#supplementary-material
Supplementary Figure 1 | MS/MS analysis of fractions 36 (A), 37 (B), 38 (C), and 39 (D). Analysis showed that surfactin congeners were included in the fractions.
Supplementary Figure 2 | Cell-cell membrane fusion in low pH in Vero 76 cells. The experiment was performed in the same manner as for Huh-7 cells ().
Supplementary Figure 3 | Initiation of SFTSV N expression and viral production. Huh-7 cells were infected with SFTSV at 0, 6, 8, 10, 12, 16, and 24 h p.i. Culture supernatant was collected to measure virus titer and cell lysates were collected to detect SFTSV N and actin by SDS-PAGE and western blot analysis. A panel of low- and high-intensity was prepared for the same outcome.
1. Yu XJ, Liang MF, Zhang SY, Liu Y, Li JD, Sun YL, et al. Fever with thrombocytopenia associated with a novel bunyavirus in China. N Engl J Med (2011) 364(16):1523–32. doi: 10.1056/NEJMoa1010095
2. Tran XC, Yun Y, Van An L, Kim SH, Thao NTP, Man PKC, et al. Endemic severe fever with thrombocytopenia syndrome, Vietnam. Emerg Infect Dis (2019) 25(5):1029–31. doi: 10.3201/eid2505.181463
3. Xu B, Liu L, Huang X, Ma H, Zhang Y, Du Y, et al. Metagenomic analysis of fever, thrombocytopenia and leukopenia syndrome (FTLS) in henan province, China: Discovery of a new bunyavirus. PloS Pathog (2011) 7(11):e1002369. doi: 10.1371/journal.ppat.1002369
4. Kim KH, Yi J, Kim G, Choi SJ, Jun KI, Kim NH, et al. Severe fever with thrombocytopenia syndrome, south Korea, 2012. Emerg Infect Dis (2013) 19(11):1892–4. doi: 10.3201/eid1911.130792
5. Takahashi T, Maeda K, Suzuki T, Ishido A, Shigeoka T, Tominaga T, et al. The first identification and retrospective study of severe fever with thrombocytopenia syndrome in Japan. J Infect Dis (2014) 209(6):816–27. doi: 10.1093/infdis/jit603
6. Lin TL, Ou SC, Maeda K, Shimoda H, Chan JP, Tu WC, et al. The first discovery of severe fever with thrombocytopenia syndrome virus in Taiwan. Emerg Microbes Infect (2020) 9(1):148–51. doi: 10.1080/22221751.2019.1710436
7. Abudurexiti A, Adkins S, Alioto D, Alkhovsky SV, Avsic-Zupanc T, Ballinger MJ, et al. Taxonomy of the order bunyavirales: Update 2019. Arch Virol (2019) 164(7):1949–65. doi: 10.1007/s00705-019-04253-6
8. McMullan LK, Folk SM, Kelly AJ, MacNeil A, Goldsmith CS, Metcalfe MG, et al. A new phlebovirus associated with severe febrile illness in Missouri. N Engl J Med (2012) 367(9):834–41. doi: 10.1056/NEJMoa1203378
9. Mourya DT, Yadav PD, Basu A, Shete A, Patil DY, Zawar D, et al. Malsoor virus, a novel bat phlebovirus, is closely related to severe fever with thrombocytopenia syndrome virus and heartland virus. J Virol (2014) 88(6):3605–9. doi: 10.1128/JVI.02617-13
10. Shen S, Duan X, Wang B, Zhu L, Zhang Y, Zhang J, et al. A novel tick-borne phlebovirus, closely related to severe fever with thrombocytopenia syndrome virus and heartland virus, is a potential pathogen. Emerg Microbes Infect (2018) 7(1):95. doi: 10.1038/s41426-018-0093-2
11. Takayama-Ito M, Saijo M. Antiviral drugs against severe fever with thrombocytopenia syndrome virus infection. Front Microbiol (2020) 11:150. doi: 10.3389/fmicb.2020.00150
12. Urata S, Yasuda J, Iwasaki M. Loperamide inhibits replication of severe fever with thrombocytopenia syndrome virus. Viruses (2021) 13(5):869. doi: 10.3390/v13050869
13. Teng YF, Xu L, Wei MY, Wang CY, Gu YC, Shao CL. Recent progresses in marine microbial-derived antiviral natural products. Arch Pharm Res (2020) 43(12):1215–29. doi: 10.1007/s12272-020-01286-3
14. Yi M, Lin S, Zhang B, Jin H, Ding L. Antiviral potential of natural products from marine microbes. Eur J Med Chem (2020) 207:112790. doi: 10.1016/j.ejmech.2020.112790
15. Urata S, Uno Y, Kurosaki Y, Yasuda J. The cholesterol, fatty acid and triglyceride synthesis pathways regulated by site 1 protease (S1P) are required for efficient replication of severe fever with thrombocytopenia syndrome virus. Biochem Biophys Res Commun (2018) 503(2):631–6. doi: 10.1016/j.bbrc.2018.06.053
16. Yuan L, Zhang S, Wang Y, Li Y, Wang X, Yang Q. Surfactin inhibits membrane fusion during invasion of epithelial cells by enveloped viruses. J Virol (2018) 92(21):e00809–18. doi: 10.1128/JVI.00809-18
17. Tani H, Shimojima M, Fukushi S, Yoshikawa T, Fukuma A, Taniguchi S, et al. Characterization of glycoprotein-mediated entry of severe fever with thrombocytopenia syndrome virus. J Virol (2016) 90(11):5292–301. doi: 10.1128/JVI.00110-16
18. Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res (1994) 22(22):4673–80. doi: 10.1093/nar/22.22.4673
19. Saitou N, Nei M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol Biol Evol (1987) 4(4):406–25. doi: 10.1093/oxfordjournals.molbev.a040454
20. Kumar S, Stecher G, Li M, Knyaz C, Tamura K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol (2018) 35(6):1547–9. doi: 10.1093/molbev/msy096
21. Li H, Zhang LK, Li SF, Zhang SF, Wan WW, Zhang YL, et al. Calcium channel blockers reduce severe fever with thrombocytopenia syndrome virus (SFTSV) related fatality. Cell Res (2019) 29(9):739–53. doi: 10.1038/s41422-019-0214-z
22. Vollenbroich D, Ozel M, Vater J, Kamp RM, Pauli G. Mechanism of inactivation of enveloped viruses by the biosurfactant surfactin from bacillus subtilis. Biologicals (1997) 25(3):289–97. doi: 10.1006/biol.1997.0099
23. Johnson BA, Hage A, Kalveram B, Mears M, Plante JA, Rodriguez SE, et al. Peptidoglycan-associated cyclic lipopeptide disrupts viral infectivity. J Virol (2019) 93(22):e01282–19. doi: 10.1128/JVI.01282-19
24. Peypoux F, Bonmatin JM, Wallach J. Recent trends in the biochemistry of surfactin. Appl Microbiol Biotechnol (1999) 51(5):553–63. doi: 10.1007/s002530051432
25. Kracht M, Rokos H, Ozel M, Kowall M, Pauli G, Vater J. Antiviral and hemolytic activities of surfactin isoforms and their methyl ester derivatives. J Antibiot (1999) 52(7):613–9. doi: 10.7164/antibiotics.52.613
26. Kaspar F, Neubauer P, Gimpel M. Bioactive secondary metabolites from bacillus subtilis: A comprehensive review. J Nat Prod (2019) 82(7):2038–53. doi: 10.1021/acs.jnatprod.9b00110
27. Arima K, Kakinuma A, Tamura G. Surfactin, a crystalline peptidelipid surfactant produced by bacillus subtilis: Isolation, characterization and its inhibition of fibrin clot formation. Biochem Biophys Res Commun (1968) 31(3):488–94. doi: 10.1016/0006-291X(68)90503-2
28. Singh P, Cameotra SS. Potential applications of microbial surfactants in biomedical sciences. Trends Biotechnol (2004) 22(3):142–6. doi: 10.1016/j.tibtech.2004.01.010
29. Kragh-Hansen U, le Maire M, Moller JV. The mechanism of detergent solubilization of liposomes and protein-containing membranes. Biophys J (1998) 75(6):2932–46. doi: 10.1016/S0006-3495(98)77735-5
30. Park SY, Kim Y. Surfactin inhibits immunostimulatory function of macrophages through blocking NK-kappaB, MAPK and akt pathway. Int Immunopharmacol (2009) 9(7-8):886–93. doi: 10.1016/j.intimp.2009.03.013
31. Mendoza CA, Yamaoka S, Tsuda Y, Matsuno K, Weisend CM, Ebihara H. The NF-kappaB inhibitor, SC75741, is a novel antiviral against emerging tick-borne bandaviruses. Antiviral Res (2021) 185:104993. doi: 10.1016/j.antiviral.2020.104993
Keywords: severe fever with thrombocytopenia syndrome virus (SFTSV), anti-SFTSV compound, marine microbes, surfactin, screening
Citation: Urata S, Takouda J, Watanabe Y, Sakaguchi M, Sakurai Y, Inahashi Y, Iwatsuki M, Yasuda J, Tanaka Y and Takeda K (2023) Identification of surfactin as an anti-severe fever with thrombocytopenia syndrome virus multi-target compound extracted from the culture broth of marine microbes. Front. Virol. 2:1064265. doi: 10.3389/fviro.2022.1064265
Received: 08 October 2022; Accepted: 28 December 2022;
Published: 07 March 2023.
Edited by:
Siew Pheng Lim, Denka Life Innovation Research (DLIR), SingaporeReviewed by:
Xufang Deng, Oklahoma State University, United StatesCopyright © 2023 Urata, Takouda, Watanabe, Sakaguchi, Sakurai, Inahashi, Iwatsuki, Yasuda, Tanaka and Takeda. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shuzo Urata, c2h1em91cmF0YUBuYWdhc2FraS11LmFjLmpw
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.