
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
BRIEF RESEARCH REPORT article
Front. Virol. , 17 November 2022
Sec. Emerging and Reemerging Viruses
Volume 2 - 2022 | https://doi.org/10.3389/fviro.2022.1050880
 Idrissa Dieng1*
Idrissa Dieng1* Samba Niang Sagne2
Samba Niang Sagne2 Mignane Ndiaye1
Mignane Ndiaye1 Mamadou Aliou Barry2
Mamadou Aliou Barry2 Cheikh Talla2
Cheikh Talla2 Moufid Mhamadi1
Moufid Mhamadi1 Diamilatou Balde1
Diamilatou Balde1 Cheikh Talibouya Toure1
Cheikh Talibouya Toure1 Boly Diop3
Boly Diop3 Amadou Alpha Sall1
Amadou Alpha Sall1 Gamou Fall1
Gamou Fall1 Cheikh Loucoubar2
Cheikh Loucoubar2 Oumar Faye1
Oumar Faye1 Ousmane Faye1
Ousmane Faye1Dengue virus 2 (DENV-2) was detected in a febrile patient living in Saré Yoba in the Kolda region of southern Senegal. Phylogenetic analysis based on the full coding region revealed that the virus belongs to the DENV-2 sylvatic genotype and is closely related to a strain (JF260983/99.66% identity) detected in Spain in a tourist who traveled to Guinea-Bissau (which borders the Kolda region) in 2009. This highlights a potential recent under-reported circulation of sylvatic dengue in the southern part of Senegal and calls for reinforced integrated surveillance among humans, non-human primates, and arboreal mosquitoes through a one-health approach.
Dengue is the most prevalent arboviral disease in tropical and subtropical areas. Dengue is caused by the dengue virus (DENV) and the etiological agents exist in four antigenically and phylogenetically distinct serotypes (DENV 1–4) (1). Infection with any DENV serotypes causes diseases ranging from flu-like illness (i.e., dengue fever) to a life-threatening disease known as severe dengue (2). The WHO estimates that one-third of the world’s population is at risk of dengue infection (2). Each of the existing dengue serotypes is maintained in two different ecologically and evolutionary distinct transmission cycles, namely the human cycle and the sylvatic cycle. The human cycle, in which only humans act as a reservoir, is maintained between domestic and peridomestic mosquitoes; in contrast, the sylvatic cycle involves non-human primates and arboreal mosquitoes (3). Despite the central and basal role that sylvatic strains of DENV play in evolution and emergence, there is no report of continuous and sustained transmission (4).
In Senegal, mainly in the south of the country (i.e., the Kédougou area), the landscape of DENV circulation was long dominated by the occurrence and maintenance of sylvatic cycles (5). In 2009, a shift occurred, with the first reported urban DENV epidemic in Dakar, and this was followed by the recurrent and yearly multifocal and multiserotype circulation of human cycle strains (6). Here, we report a case of dengue virus 2 (DENV-2) infection. A phylogenetic analysis, based on full coding region, revealed that the DENV-2 infection was closely related to a strain circulating in Guinea-Bissau in 2009.
In collaboration with the Ministry of Health and Social Action (Dakar, Senegal) and the Unit of Epidemiology Clinical Research and Data Sciences at the Institut Pasteur de Dakar (IPD) (Dakar, Senegal), our laboratory (i.e., the Virology Unit at IPD) is conducting syndromic surveillance of fever around the country through a program named the 4S (Syndromic Sentinel Surveillance in Senegal) Network (7). As part of this nationwide surveillance project, samples from febrile patients are collected and shipped on a weekly basis to the WHO collaborating center for suspected arbovirus infection diagnosis. In November 2021, a patient suspected of arbovirus infection presented in the Saré Yoba health district, located in the Kolda region (southern Senegal). The patient was male, aged 28 years, and presented with symptoms that included headaches, myalgia, asthenia, arthralgia, and chills. A malaria rapid diagnostic test yielded a negative result. Following 2 days of fever, a venous blood sample was collected from the patient and shipped to the Virology Unit at IPD for diagnosis. At IPD, the blood sample was centrifuged at 2000 rpm for 5 minutes, and serum was harvested and aliquoted into 2-ml tubes. RNA extraction was performed using the QIAGEN viral RNA kit (QIAGEN, Hilden, Germany), using 140 µl of input serum, in accordance with the manufacturer’s recommendations. Extracted nucleic acid was subjected to screening for seven arboviruses, as previously mentioned by Dieng and colleagues (8), of which only DENV gave a positive result. To define the incriminating serotype, DENV-positive RNA was subjected to a multiplex quantitative reverse transcription PCR assay using a TIB Molbiol Modular Dx Dengue typing kit (cat. no. 40-0700-24; TIB Molbiol, Berlin, Germany) (5). Surprisingly, the multiplex quantitative reverse transcription (qRT) PCR assay failed to define the dengue serotype. To define the virus serotype/genotype, we successfully amplified a partial NS5 gene sequence using FU1/FD3 (9), and the obtained amplicon was approximately ≈ 1 kb. The amplicon was purified using AMPure (Beckman Coulter Inc., Brea, CA, USA) beads at a ratio of 1:0.8. A sequencing library was prepared for the Oxford Nanopore MinION (Oxford Nanopore Technologies plc, Oxford, UK) using the rapid barcoding kit (SQK RBQ110.96), loaded onto a R9 flow cell and sequenced using a MinION MK1C device.
Raw data were collected and basecalled using guppy (https://community.nanoporetech.com). Adapters were trimmed using NanoFilt (10) (options -headcrop 50 and -tailcrop 50) and reads were mapped to a DENV reference genome (NC_001474.2) using Minimap2 (11). A National Center for Biotechnology Information (NCBI) BLASTn search of the obtained sequence matched with sylvatic DENV-2 (JF260983).
Based on results from the NCBI BLASTn (12) search, we downloaded full genome sequences of closely related sylvatic DENV-2 sequences and designed a tilling PCR primal scheme, generating amplicons of around 900 bp and covering the coding region of sylvatic DENV-2 strains. PCR amplification was performed using Q5® High-Fidelity 2X Master Mix (New England Biolabs, Ipswich, MA, USA) in accordance with a protocol previously described by Dieng and colleagues (13). The sequencing and data analysis were the same as previously used for NS5 gene sequencing.
To determine the evolutionary history, we download representative sequences of described DENV-2 genotypes. The obtained dataset was aligned using MAFFT (14) and a maximum likelihood (ML) tree was constructed using IQ-TREE (15).
The obtained ML (Figure 1) tree clearly shows that, based on a nearly full genome sequence, our strain fell within the West African DENV-2 sylvatic genotype and is closely related to a strain linked to hemorrhagic DENV detected in Spain from a tourist who traveled to Guinea-Bissau through Senegal in 2009 (16), and not to strains of the DENV-2 cosmopolitan genotype, which was responsible for the latest DENV-2 epidemic in Senegal (6, 13). This is the first identification of circulating sylvatic DENV-2 in Senegal since 2000 (17). Interestingly, the Kolda region borders the Niokolo-Koba National Park, which is home to monkey species (Papio papio, Erythrocebus patas) known to be a reservoir of sylvatic DENV-2 (3, 18). In addition, experimental findings from the surrogate human models of infection and from cultured cells suggest that there is little or no adaptive barrier for the emergence of sylvatic DENV in human populations, possibly reflecting the evolution of DENV as an opportunistic virus that is capable of infecting a wide range of primate species (3).
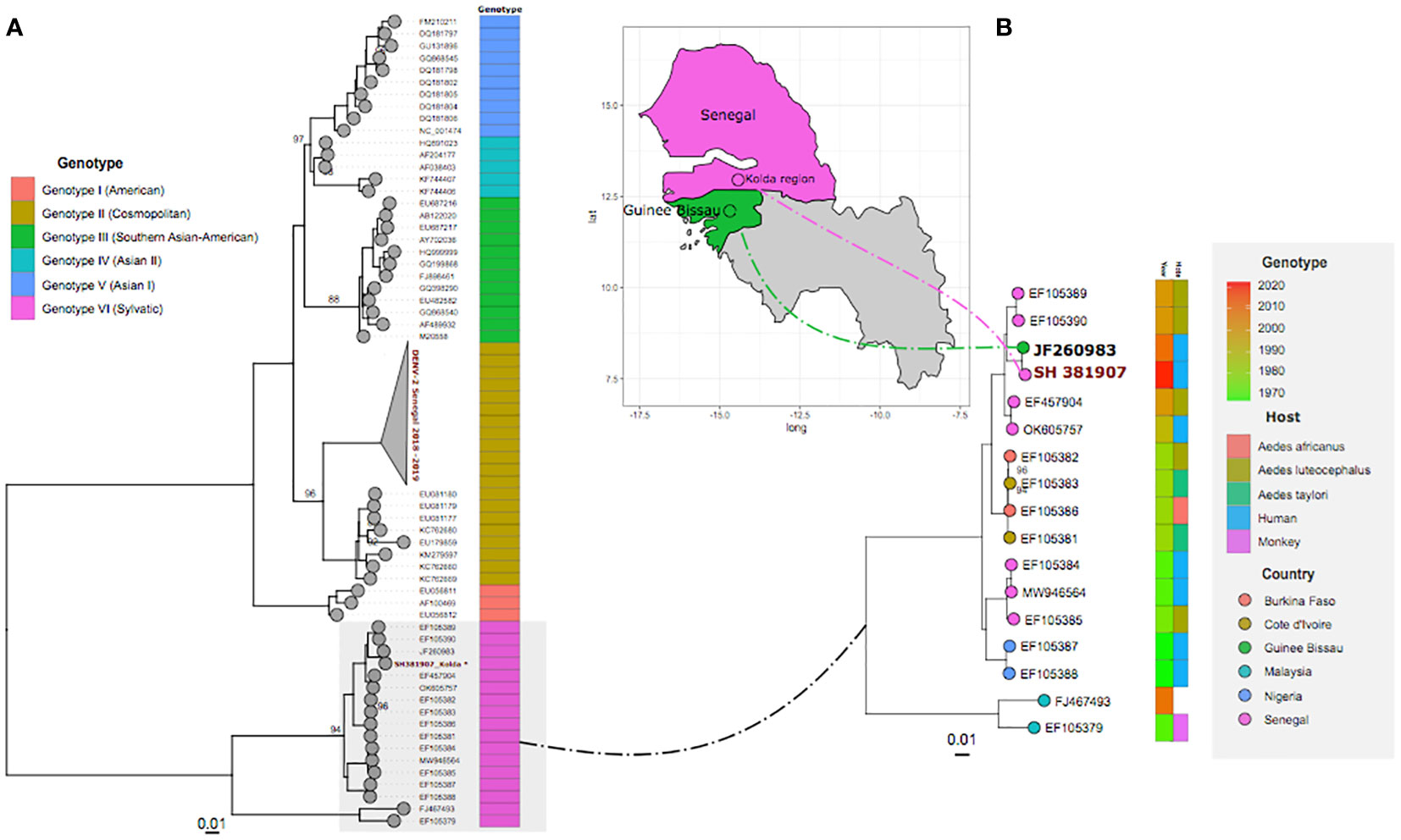
Figure 1 A maximum likelihood (ML) phylogenetic tree based on nearly complete genome sequences. Panel (A) shows that the obtained sequence used in this work belongs to the sylvatic genotype of dengue virus 2 (DENV-2). Panel (B) is an expansion of the sylvatic genotype (highlighted in grey on panel A). The heatmap shows, respectively, the year of isolation and the host of used sylvatic DENV-2 strains during phylogenetic analysis. Tips are colored according to the country or provenance. The arrow shows the itinerary of the patient linked to sequence JF260983, who traveled to Guinea-Bissau through Senegal before returning to Spain where he was diagnosed.
This finding, in addition to the present detection of this sylvatic genotype in the Kolda area (which shares a border with Guinea-Bissau), highlights a potential unnoticed circulation of sylvatic DENV-2 in the south of Senegal, and this is corroborated by the fact that the patient was not traveling, confirming that the case was autochthonous.
In 2021, a national seroprevalence study conducted in 14 administrative regions in Senegal led to the detection of five DENV immunoglobulin M-positive samples in the Kolda region (unpublished data). No samples have been reported to be qRT PCR positive prior to this case.
Interestingly, clinical infection with sylvatic strains is indistinguishable from human transmission cycle strains, and this can lead to under-reporting of sylvatic DENV (16) because genomic surveillance of the circulating DENV strain in Senegal, as well as in Africa, is limited (19). All parameters mentioned above support the high likelihood of spillover of sylvatic DENV from Africa or Asia in the human transmission cycle. In addition, many studies report that people with African ancestry confer some level of protection against severe dengue infection (20), and this, in addition to increased tourism in Africa (21), raises concerns about the risk of increased sylvatic dengue infection in Africa among non-indigenous people in near future.
This calls for determining the genetic diversity of circulating DENV strains, which is crucial before any vaccination policy can be implemented. Indeed, determining which contemporary genotypes are in circulation in a given area is crucial for ensuring effective diagnostics and for developing preventive and therapeutic countermeasures.
DENV is now hyperendemic in Senegal, with the co-circulation of DENV 1–3 belonging to human transmission cycles, and it is marked by yearly epidemics that may constrain the identification of sylvatic DENV. Nevertheless, as human DENV has the potential to enter human cells and cause hemorrhagic disease, an integrated one-health approach between humans, mosquitoes, and non-human primates is urgently needed in regions, other than the Kédougou area, located in the south of Senegal. A one-health approach could improve dengue fever surveillance through active, existing human malaria-like illness surveillance within the 4S Network.
Finally, to be able the discriminate between sylvatic and epidemic DENV strains, real-time genomic surveillance of DENV could play a key role in virus surveillance around the country. This real-time genomic surveillance will help us to better understand the evolutionary history, transmission, and spread, with complex transmission dynamics involving both urban and sylvatic DENV cycles.
The original contributions presented in the study are included in the article/supplementary material. Further inquiries can be directed to the corresponding author.
In this study, we used samples collected as part of approved ongoing surveillance conducted by the IPD (i.e., a WHO collaborating center for arboviruses and hemorrhagic fever reference and research). The Senegalese National Ethical Committee approved the protocol as a less than minimal risk research, and written consent forms were not required. All samples from humans were deidentified before we performed virus detection, characterization, and analysis.
Conceptualization: OusF, AS, BD, MB, CL, CT, and OumF; methodology: ID and MN; software: ID; original draft: ID; final draft and review: all authors. All authors contributed to the article and approved the submitted version.
This work was supported by IPD proper funds, Senegal.
We would like to acknowledge the Ministry of Health and Social Action for its support and all the sentinel sites’ healthcare workers. We thank Diogop Camara, Cherif Sylla, and Mame Diarra Sall for their excellent technical assistance during laboratory diagnosis. Warm thanks to Dr Joseph Fauver, Assistant Professor, Department of Epidemiology, University of Nebraska Medical Center, Omaha, NE, USA, for their English proofreading during manuscript pre-submission.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
2. WHO. Global strategy for dengue prevention and control, 2012-2020 (2012). Geneva, Switzerland: World Health Organization. Available at: http://apps.who.int/iris/bitstream/10665/75303/1/9789241504034_eng.pdf (Accessed cité 12 sept 2020).
3. Vasilakis N, Cardosa J, Hanley KA, Holmes EC, Weaver SC. Fever from the forest: prospects for the continued emergence of sylvatic dengue virus and its impact on public health. Nat Rev Microbiol (2011) 9(7):532−41. doi: 10.1038/nrmicro2595
4. Liu W, Pickering P, Duchêne S, Holmes EC, Aaskov JG. Highly divergent dengue virus type 2 in traveler returning from Borneo to Australia. Emerg Infect Dis (2016) 22(12):2146−8. doi: 10.3201/eid2212.160813
5. Dieng I, Diarra M, Diagne MM, Faye M, Dior Ndione MH, Ba Y, et al. Field deployment of a mobile biosafety laboratory reveals the Co-circulation of dengue viruses serotype 1 and serotype 2 in louga city, Senegal, 2017. J Trop Med (2021) 2021:8817987. doi: 10.1155/2021/8817987
6. Dieng I, Ndione MHD, Fall C, Diagne MM, Diop M, Gaye A, et al. Multifoci and multiserotypes circulation of dengue virus in Senegal between 2017 and 2018. BMC Infect Dis (2021) 21(1):867. doi: 10.1186/s12879-021-06580-z
7. Dia N, Diene Sarr F, Thiam D, Faye Sarr T, Espié E, OmarBa I, et al. Influenza-like illnesses in Senegal: Not only focus on influenza viruses. PLoS one (2014). Available at: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3968133/ (Accessed cité 24 oct 2019).
8. Dieng I, Barry M, Diagne M, Diop B, Ndiaye M, Faye M, et al. Detection of Crimean Congo haemorrhagic fever virus in north-eastern Senegal, bokidiawé 2019. Emerging Microbes Infections (2020) 9:2485–7. doi: 10.1080/22221751.2020.1847605
9. Kuno G, Chang GJ, Tsuchiya KR, Karabatsos N, Cropp CB. Phylogeny of the genus flavivirus. J Virol (1998) 72(1):73−83. doi: 10.1128/JVI.72.1.73-83.1998
10. De Coster W, D’Hert S, Schultz DT, Cruts M, Van Broeckhoven C. NanoPack: visualizing and processing long-read sequencing data. In: Berger B, editor. Bioinformatics, vol. 34 (2018). p. 2666−9.
11. Li H. Minimap2: pairwise alignment for nucleotide sequences. In: Birol I, editor. Bioinformatics, vol. 34 (2018). p. 3094−100.
12. Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol (1990) 215(3):403−10. doi: 10.1016/S0022-2836(05)80360-2
13. Dieng I, Diallo A, Ndiaye M, Mhamadi M, Diagne MM, Sankhe S, et al. Full genome analysis of circulating DENV-2 in Senegal reveals a regional diversification into separate clades. J Med Virol (2022) 94(11):5593–600. doi: 10.20944/preprints202201.0057.v1
14. Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol (2013) 30(4):772−80. doi: 10.1093/molbev/mst010
15. Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol (2015) 32(1):268−74. doi: 10.1093/molbev/msu300
16. Franco L, Palacios G, Martinez JA, Vázquez A, Savji N, De Ory F, et al. First report of sylvatic DENV-2-Associated dengue hemorrhagic fever. In: Powers AM, editor. PLoS negl trop dis, vol. 5 . West Africa (2011).
17. Diallo M, Ba Y, Sall AA, Diop OM, Ndione JA, Mondo M, et al. Amplification of the sylvatic cycle of dengue virus type 2, Senegal, 1999–2000: Entomologic findings and epidemiologic considerations. Emerg Infect Dis (2003) 9(3):362−7. doi: 10.3201/eid0903.020219
18. Patzelt A, Zinner D, Fickenscher G, Diedhiou S, Camara B, Stahl D, et al. Group composition of Guinea baboons (Papio papio) at a water place suggests a fluid social organization. Int J Primatol (2011) 32(3):652−68. doi: 10.1007/s10764-011-9493-z
19. Letizia AG, Pratt CB, Wiley MR, Fox AT, Mosore M, Agbodzi B, et al. Retrospective genomic characterization of a 2017 dengue virus outbreak, Burkina faso. emerg infect dis (2022). Available at: https://wwwnc.cdc.gov/eid/article/28/6/21-2491_article.htm (Accessed 15 sept 2022).
20. Gainor EM, Harris E, LaBeaud AD. Uncovering the burden of dengue in Africa: Considerations on magnitude, misdiagnosis, and ancestry. Viruses (2022) 14(2):233. doi: 10.3390/v14020233
Keywords: fever, DENV 2, sylvatic, Senegal, one health
Citation: Dieng I, Sagne SN, Ndiaye M, Barry MA, Talla C, Mhamadi M, Balde D, Toure CT, Diop B, Sall AA, Fall G, Loucoubar C, Faye O and Faye O (2022) Detection of human case of dengue virus 2 belonging to sylvatic genotype during routine surveillance of fever in Senegal, Kolda 2021. Front. Virol. 2:1050880. doi: 10.3389/fviro.2022.1050880
Received: 22 September 2022; Accepted: 26 October 2022;
Published: 17 November 2022.
Edited by:
John H.-O. Pettersson, Uppsala University, SwedenReviewed by:
Ernest Gould, Aix Marseille Université, FranceCopyright © 2022 Dieng, Sagne, Ndiaye, Barry, Talla, Mhamadi, Balde, Toure, Diop, Sall, Fall, Loucoubar, Faye and Faye. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Idrissa Dieng, SWRyaXNzYS5ESUVOR0BwYXN0ZXVyLnNu
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.