
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
CORRECTION article
Front. Vet. Sci., 10 April 2025
Sec. Animal Nutrition and Metabolism
Volume 12 - 2025 | https://doi.org/10.3389/fvets.2025.1593695
This article is a correction to:
Probiotic characteristics and whole genome sequencing of Pediococcus pentosaceus SNF15 and its hold a protective effect on mice diarrhea induced by Escherichia coli K99
 Yalan Su†
Yalan Su† Mingque Feng†
Mingque Feng† Jingdi Tong
Jingdi Tong Xiangfu Wen
Xiangfu Wen Meiyi Ren
Meiyi Ren Deyuan Song
Deyuan Song Jinshang Song
Jinshang Song Xiaohan Li
Xiaohan Li Qinna Xie
Qinna Xie Jia Cheng*
Jia Cheng* Mingchao Liu*
Mingchao Liu*by Su, Y., Feng, M., Tong, J., Wen, X., Ren, M., Song, D., Song, J., Li, X., Xie, Q., Cheng, J., and Liu, M. (2025). Front. Vet. Sci. 12:1524658. doi: 10.3389/fvets.2025.1524658
In the published article, there was an error in Figure 1 as published. Labels C and D are incorrect. The corrected Figure 1 and its caption appear below.
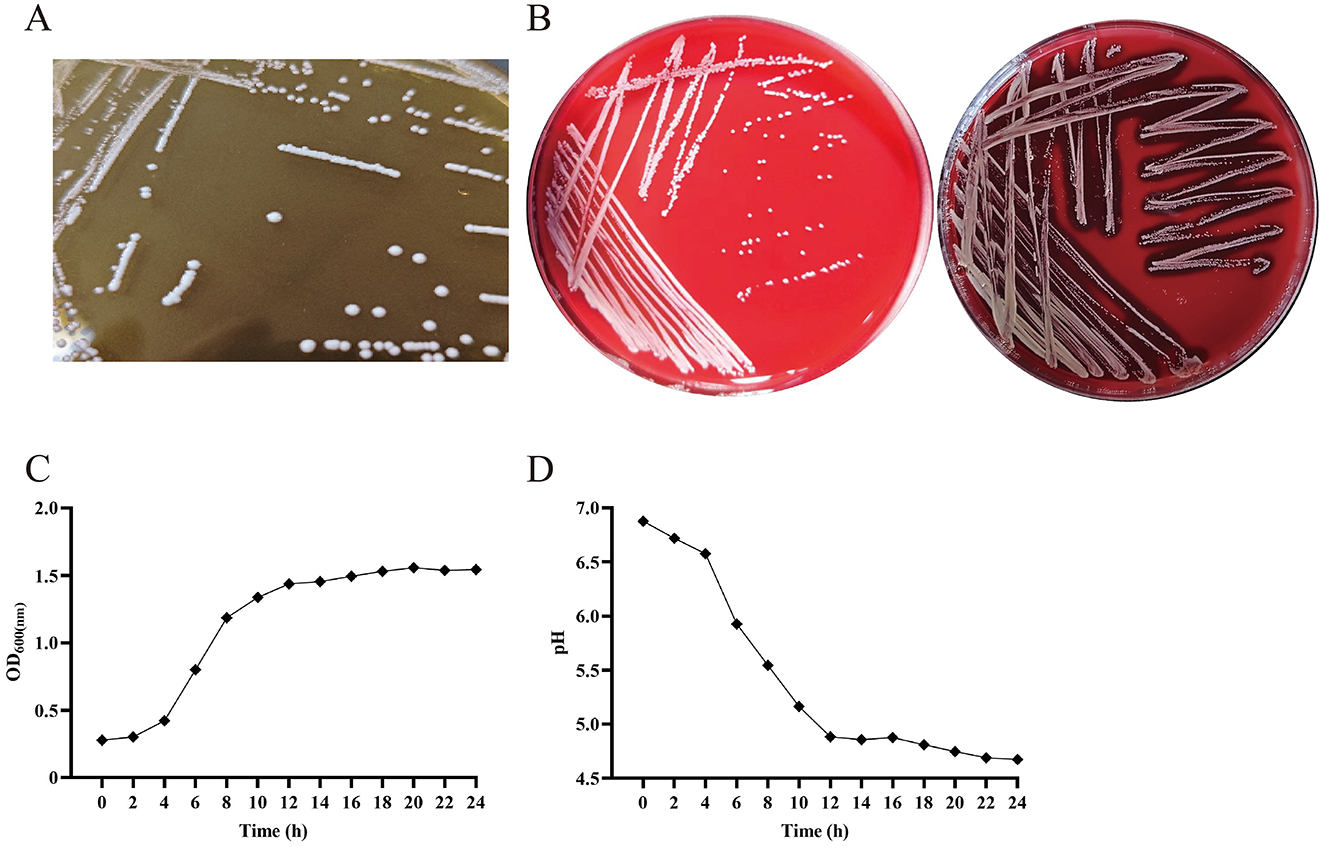
Figure 1. The colony form of P. pentosaceus SNF15 is not hemolytic and has good growth performance and acid production ability. (A) morphology of P. pentosaceus SNF15. (B) γ-hemolysis showed P. pentosaceus SNF15 (left), β -hemolysis showed by S. aureus (right). (C) Growth curve analysis of P. pentosaceus SNF15 was performed by measuring OD600. (D) The acid-producing curve of P. pentosaceus SNF15.
In the published article, there was an error in Figure 4 as published. The letters in labels C and E are incorrect. The corrected Figure 4 and its caption appear below.
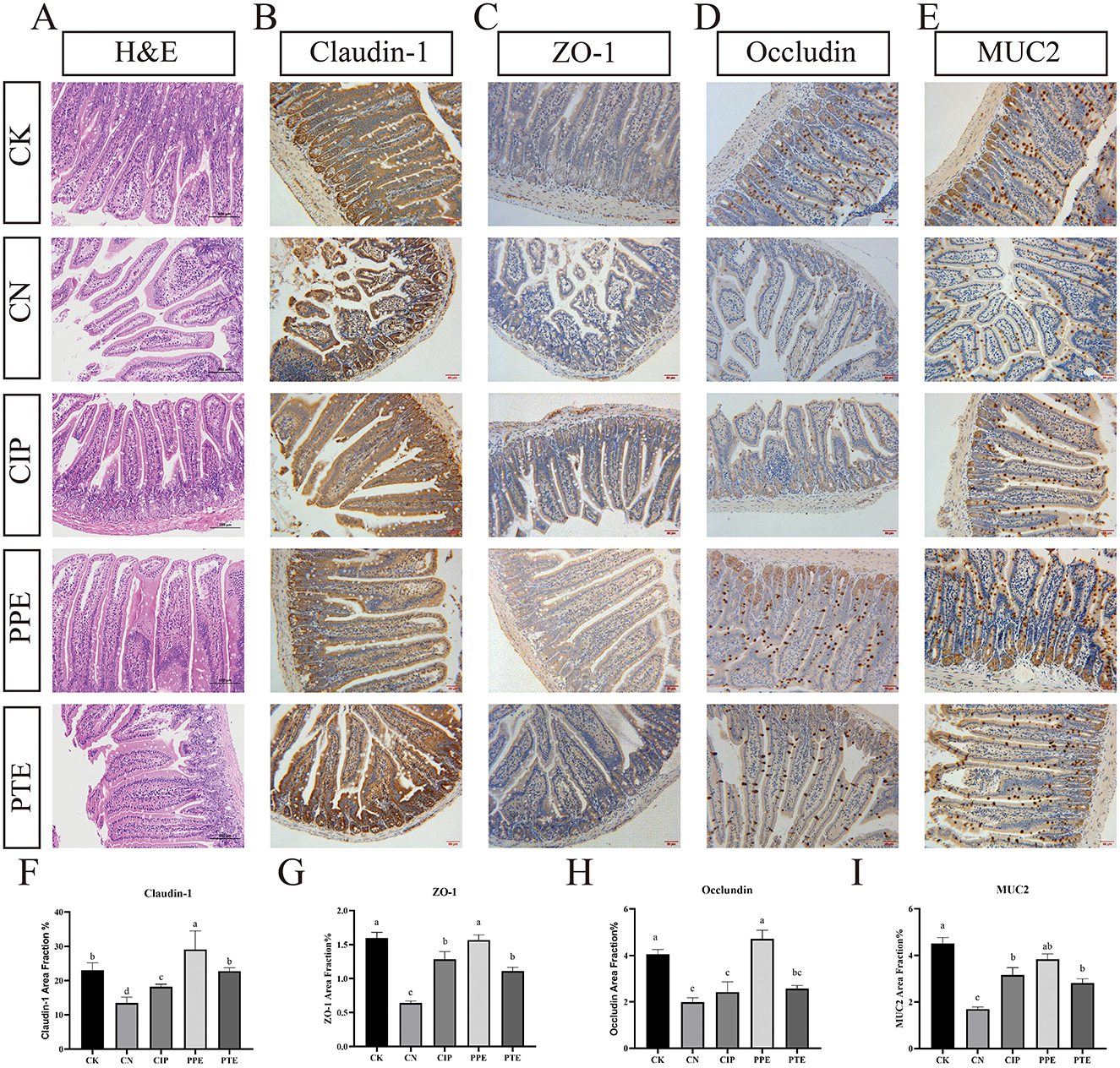
Figure 4. Gavaged P. pentosaceus SNF15 and Ciprofloxacin reduced the damage caused by E. coli K99 stimulation. (A) H&E staining of the jejune tissues. (B) Representative immunohistochemical staining of claudin-1 in jejune tissue for each group. (C) Representative immunohistochemical staining of ZO-1 in jejune tissue for each group. (D) Representative immunohistochemical staining of occludin in jejune tissue for each group. (E) Representative immunohistochemical staining of MUC2 in jejune tissue for each group. (F) The claudin-1 area fraction for each group (G) The ZO-1 area fraction for each group. (H) The occludin area fraction for each group (I) The MUC2 area fraction for each group. Brown is immunopositive, and blue is immunonegative. The database is expressed as the mean ± SEM. Significant differences were considered at P < 0.05. Different letters are marked at bars, without the same superscripts differ significantly (P < 0.05).
In the published article, there was an error. The strain culture condition was wrong, “CO2” was changed to “O2”.
A correction has been made to 2.1. Isolation, culture, and safety identification of probiotics, Paragraph Number 1. This sentence previously stated:
The feces were fully homogenized, spread on MRS agar (Qingdao hopebiol, China), and hermetically cultured at 37°C without CO2 for 24 h, MRS plate was sealed with a sealing membrane.
The corrected sentence appears below:
The feces were fully homogenized, spread on MRS agar (Qingdao hopebiol, China), and hermetically cultured at 37°C without O2 for 24 h, MRS plate was sealed with a sealing membrane.
A correction has been made to 2.1. Isolation, culture, and safety identification of probiotics, Paragraph Number 1. This sentence previously stated:
“The purified bacteria were inoculated in MRS broth and static cultured at 37°C without CO2 for 24 h, and the centrifugal tube is sealed with a sealing membrane to isolate oxygen.”
The corrected sentence appears below:
“The purified bacteria were inoculated in MRS broth and static cultured at 37°C without O2 for 24 h, and the centrifugal tube is sealed with a sealing membrane to isolate oxygen.”
In the published article, there was an error. A unit was misrepresented, and “(1% w/v)” was changed to “(1% v/v)”.
A correction has been made to 2.2.1. Growth and acid-producing curve curves, Paragraph Number 1. This sentence previously stated:
“The cultured solutions of isolated strains were added to 80 mL MRS broth (1% w/v) and hermetically incubated at 37°C.”
The corrected sentence appears below:
“The cultured solutions of isolated strains were added to 80 mL MRS broth (1% v/v) and hermetically incubated at 37°C.”
In the published article, there was an error. The incorrect test method, “2.2.1.” was changed to “2.2.2.”.
A correction has been made to 2.2.3. Bacteriostatic substance, Paragraph Number 1. This sentence previously stated:
“Use the same way as in 2.2.1 to get P. pentosaceus SNF15 supernatant. Catalase or protease K or NaOH (Beijing Solarbio Science & Technology Co., Ltd., China) was added to the supernatant of P. pentosaceus SNF15.”
The corrected sentence appears below:
“Use the same way as in 2.2.2. to get P. pentosaceus SNF15 supernatant. Catalase or protease K or NaOH (Beijing Solarbio Science & Technology Co., Ltd., China) was added to the supernatant of P. pentosaceus SNF15.”
A correction has been made to 2.2.3. Bacteriostatic substance, Paragraph Number 1. This sentence previously stated:
“The same method was used as 2.2.1 to determine the inhibitory effect of the treated supernatant on E. coli K99.”
The corrected sentence appears below:
“The same method was used as 2.2.2 to determine the inhibitory effect of the treated supernatant on E. coli K99.”
In the published article, there was an error. In the experimental group of mice, the experimental treatment of writing errors, “stroke-physiological saline solution” was changed to “E. coli K99 (1 × 108 CFU/mL)”.
A correction has been made to 2.4.1. Animal management and experimental procedures, Paragraph Number 3. This sentence previously stated:
“(III) CIP group received 200 μL stroke-physiological saline solution at 1-7 days and 200 μL of 50mg/kg Ciprofloxacin solution at 8-14 days.”
The corrected sentence appears below:
“(III) CIP group received 200 μL E. coli K99 (1 × 108 CFU/mL) at 1-7 days and 200 μL of 50mg/kg Ciprofloxacin solution at 8-14 days.”
A correction has been made to 2.4.2. RNA extraction and Quantitative real-time polymerase chain reaction, Paragraph Number 1. This sentence previously stated:
“qCR assays were performed to assess the relative expression levels of the IL-6, IL-1β, and TNF-α. The expression level of GAPDH was used as the reference gene.”
The corrected sentence appears below:
“qPCR assays were performed to assess the relative expression levels of the IL-6, IL-1β, and TNF-α. The expression level of GAPDH was used as the reference gene.”
A correction has been made to 2.4.2. RNA extraction and Quantitative real-time polymerase chain reaction, Paragraph Number 1. Due to an oversight in writing, the primer sequence was not written. This sentence previously stated:
“All primers were synthesized by Sangon Biotech Co., Ltd. (Shanghai, China).”
The corrected sentence appears below:
“All primers were synthesized by Sangon Biotech Co., Ltd. (Shanghai, China). The specific primers for interleukin were IL-6 (5′-CTTCTTGGGACTGATGCTGGTGAC-3′; 5′-TCTGTTGGGAGTGGTATCCTCTGTG-3′), IL-1β (5′-CCTGGGCTGTCCTGATGAGAG-3′; 5′-TCCACGGGAAAGACACAGGTA-3′), TNF-α (5′-GGACTAGCCAGGAGGGAGAACAG-3′; 5′-CAATGTGTCCGTCGTGGATCT-3′), GAPDH (5′-CAATGTGTCCGTCGTGGATCT-3′; 5′-GTCCTCAGTGTAGCCCAAGATG-3′).”
A correction has been made to 3.2.3. Adhesion to Caco-2 cells of P. pentosaceus SNF15, Paragraph Number 1, as the test results were incorrectly described. This sentence previously stated:
“As shown in Table 3, the adhesion rate of P. pentosaceus SNF15 on Caco-2 cells is 13.91%.”
The corrected sentence appears below:
“As shown in Table 3, the adhesion rate of P. pentosaceus SNF15 on Caco-2 cells is 13.93%.”
A correction has been made to 3.3.1. Genome assembly result, Paragraph Number 1, as the test results were incorrectly described. This sentence previously stated:
“It contains 1,855 protein-coding sequences, 1,668,747 bp, accounting for 88.03% of the total length; the average gene length is 899 bp, the max gene length is 8,601 bp, and the min gene length is 90bp. The total repetitive sequence length is 4,435 bp, and the repetitive sequence content is 0.23%. The genome contains 15 rRNA genes and 59 tRNA genes.”
The corrected sentence appears below:
“It contains 1,855 protein-coding sequences, 1,668,747 bp, accounting for 88.02% of the total length; the average gene length is 899 bp, the max gene length is 8,601 bp, and the min gene length is 90 bp. The total repetitive sequence length is 4,435 bp, and the repetitive sequence content is 0.23%. The genome contains 15 rRNA genes and 56 tRNA genes.”
A correction has been made to 3.4.2. P. pentosaceus SNF15 attenuated Damages in the mechanical intestinal barrier of E. coli K99-infection mice, Paragraph Number 1, as the test results were incorrectly described. This sentence previously stated:
“There was no edema within the lamina propria. Jejunal intestinal villi in the CK group were severely damaged, and there was prominent edema within the lamina propria of the intestinal villi. Compared with the CK group, the intervention of P. pentosaceus SNF15 and ciprofloxacin can significantly alleviate the intestinal damage caused by E. coli K99.”
The corrected sentence appears below:
“There was no edema within the lamina propria. Jejunal intestinal villi in the CN group were severely damaged, and there was prominent edema within the lamina propria of the intestinal villi. Compared with the CN group, the intervention of P. pentosaceus SNF15 and ciprofloxacin can significantly alleviate the intestinal damage caused by E. coli K99.”
A correction has been made to 3.4.3. P. pentosaceus SNF15 attenuated damages in the inflammatory properties' intestinal barrier of E. coli K99-infection mice, Paragraph Number 2. This sentence previously stated:
“The CIP group also has the same trends with expression mRNA level, compared with secretion levels of IL-6, IL-1β, and TNF-α significantly decreased with the CK group (p < 0.05). The secretion levels of IL-6, IL-1β, and TNF-α in the CIP, PPE, and PTE groups were significantly reduced compared with the CN group (p < 0.05) and had no significant difference with the CK group (p > 0.05).”
The corrected sentence appears below:
“The secretion levels of IL-6, IL-1β, and TNF-α in the CIP, PPE, and PTE groups were significantly reduced compared with the CN group (p < 0.05). The secretion levels of IL-6 and TNF-α in PPE group, secretion levels of IL-1β in CIP group had no significant difference with the CK group (p > 0.05).”
The authors apologize for this error and state that this does not change the scientific conclusions of the article in any way. The original article has been updated.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Keywords: Pediococcus pentosaceus SNF15, Escherichia coli, diarrhea, probiotic properties, whole genome sequencing, gut microbiota
Citation: Su Y, Feng M, Tong J, Wen X, Ren M, Song D, Song J, Li X, Xie Q, Cheng J and Liu M (2025) Corrigendum: Probiotic characteristics and whole genome sequencing of Pediococcus pentosaceus SNF15 and its protective effect on mice diarrhea induced by Escherichia coli K99. Front. Vet. Sci. 12:1593695. doi: 10.3389/fvets.2025.1593695
Received: 14 March 2025; Accepted: 18 March 2025;
Published: 10 April 2025.
Approved by:
Frontiers Editorial Office, Frontiers Media SA, SwitzerlandCopyright © 2025 Su, Feng, Tong, Wen, Ren, Song, Song, Li, Xie, Cheng and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jia Cheng, eWluZ3Rhb2NoZW5nMjAxMEAxNjMuY29t; Mingchao Liu, bGl1bWluZ2NoYW9AMTYzLmNvbQ==
†These authors share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.