
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Vet. Sci., 01 April 2025
Sec. Veterinary Infectious Diseases
Volume 12 - 2025 | https://doi.org/10.3389/fvets.2025.1570575
African swine fever is a devastating viral disease of swine causing up to 100% mortality and significant impacts to the swine industry. The causative agent, African swine fever virus (ASFV), is a large, enveloped virus containing a linear, double-stranded DNA genome with 170–190 kb in length. Since its introduction into the Caucasus region in 2007, the genotype II ASFV has continued to spread to Europe, Asia, and Caribbean countries. Early detection is crucial to prevent and control ASF outbreaks for biosecurity purposes, and environmental samples can be used to evaluate the level of biosecurity. Therefore, we evaluated the effect of freeze–thaw cycles and storage at 4°C and room temperature (RT) on ASFV DNA detection in environmental samples. ASFV DNA was stable in environmental samples with no organic contaminants after freeze–thaw and incubation at 4°C and RT. However, incubation at RT negatively affects ASFV detection in swine feces and feed dust samples that were collected using premoistened gauze. There were significant reductions in ASFV detection in environmental samples in the presence of soil and organic mixture after freeze–thaw and incubation at 4°C and RT. These results provide novel insights on the appropriate storage of environmental samples for ASFV detection and contribute to the control and prevention of ASF outbreaks and new introductions.
African swine fever (ASF) is one of the most devastating diseases in the swine industry due to its significant impact on swine health and socioeconomic consequences in affected countries. ASF is a severe, systemic hemorrhagic disease of pigs that produces a wide range of clinical signs and lesions, caused by the African swine fever virus (ASFV). ASFV is a large enveloped, double-stranded DNA virus and belongs to the family Asfarviridae. The virus has strict host specificity capable of infecting animal species in the family Suidae, including domestic pigs and wild boars. The infection in African wild pigs, such as warthogs and bushpigs, is asymptomatic but plays a critical role in ASFV transmission and persistence in Sub-Saharan Africa (1). The recent epidemic is associated with a genotype II virus and originated from an intercontinental transmission from Eastern Africa to Georgia in 2007 (2). The virus subsequently spread to Russia as well as eastern and central European countries. It is assumed that infected wild boar played a critical role in ASFV transmission in these areas (3). In 2018, ASF outbreaks were first reported in China and rapidly spread to nearby Asian countries; human activities were the main contributors to the spread of the virus over long distances (4). The capability of ASFV to cross intercontinental, geographical barriers was indeed supported by ASF outbreaks in the Dominican Republic in 2021 (5).
ASFV transmission occurs through direct contact with infected pigs, wild boar, and soft ticks as well as via indirect routes such as contaminated fomites or pork products. The virus exhibits high stability in pork and pork products, thus contaminated pork and pork products are one of the greatest sources for indirect ASFV transmission (6). In addition, it has been well-documented that the virus is highly resistant in the environment on a wide variety of surface types. The virus retains infectivity for a few weeks to several months in a variety of environmental substrates and surfaces, suggesting the potential role of environmental contamination in ASFV transmission (7–9). For this reason, environmental sampling procedures are useful tools to evaluate the level of biosecurity in pig production system in order to identify shortcomings of biosecurity practices and to determine where enhanced practices should be incorporated to prevent transmission of ASFV or other pathogens. In an ASFV-affected country, a recent investigation utilized environmental sampling techniques in the feed supply chain and found that 0.7% of environmental samples were positive for ASFV; the results were used to implement additional biosecurity procedures (10). Previously, we developed an environmental sampling technique and associated processing methods for detecting ASFV DNA with high sensitivity (11, 12). However, there remained a knowledge gap regarding the storage of samples collected from environmental sampling procedures since scientifically-supported best practices are not currently available. The aim of the present study was to determine the effect of sample storage and handling procedures on detection of ASFV DNA in environmental samples from surfaces contaminated with ASFV and a range of organic materials.
All experiments were approved under the Kansas State University (KSU) Institutional Biosafety Committee (IBC, Protocol #1600) and performed in a biosafety level-3 laboratory in the Biosecurity Research Institute at KSU. Whole blood was collected from Georgia07-infected pigs from a separate animal study and stored at – 80°C until the experiment was conducted. The virus titer was 3.68 × 108 TCID50/mL.
Surface contamination was performed as previously described (11, 12). Briefly, 100 μL ASFV-infected blood were mixed with (1) 5 mL of phosphate buffered saline (PBS) or 2.5 g of each organic contaminant and 2.5 mL of phosphate buffered saline (PBS): (2) soil, (3) swine feces collected from finishing pigs, (4) feed dust collected from feed mill, and (5) mixture of soil, swine feces, and feed dust. The mixture was inoculated on a 10 × 10 cm stainless steel surface and dried for 30 min at RT in a biosafety cabinet. The surface was swabbed using either pre-moisten cotton gauze (Dynarex Corporation, Orangeburg, NY, United States) with 5 mL of PBS or a sponge stick (Cat. #SSL100, 3 M, MN, United States) pre-moistened with 10 mL DNA/RNA shield (Zymo Research, Irvine, CA). The cotton gauze was placed in a 50 mL conical tube, and the sponge stick was returned to a plastic bag. Positive control 100 μL ASFV-infected blood in 15 or 20 mL of PBS. These samples were subjected to three different storage conditions: (1) freezing at – 80°C and thawing 0X, 1X, 2X, or 3X, (2) storage at 4°C for 0, 1, 3, or 7 days, and (3) storage at room temperature (RT) for 0, 1, 3, or 7 days. After storage, the gauze samples were processed by adding 5 mL of PBS, vortexing, and incubating for 5 min at RT. The sponge stick was massaged and incubated for 5 min. For the sponge stick samples from feed dust and mix-contaminated surfaces, an additional 5 mL of PBS was added for facilitating the elution step. The supernatant was transferred into a microtube and centrifuged at 700 × g for 5 min. The clarified supernatant was mixed with the equal volume of AL lysis buffer (Qiagen, Germantown, MD, United States) and stored until further analysis. Each treatment combination was replicated three times.
Viral DNA extraction was performed using an automated magnetic bead-based extraction system as previously described (13). Briefly, the AL lysate was heat-inactivated at 70°C for 10 min. A total of 200 μL of AL lysate and 200 μL of isopropanol was added into an extraction plate that was pre-filled with extraction reagents, and extraction was performed in the automated extractor. The eluted DNA was mixed with p72-specific primer and probe set in PCR mastermix. PCR was performed in duplicate using the CFX 96 PCR machine. The Cq value was converted to copy numbers/mL using the standard curve generated from serial dilutions of the known concentration of the plasmid containing the target p72 sequence.
Data were analyzed by a linear model fit using the GLIMMIX function in SAS (version 9.4, Cary, NC) with the response variable for all models being ASFV p72 Log10 copy number/mL. For analysis of storage time at 4°C or RT, sampling device treatment with respective organic matter contamination level, day of storage, and the associated interaction were fit in the model as fixed effects. A similar statistical model was fit to assess the effect of freeze–thaw storage with sampling device treatment with respective organic matter contamination level, number of freeze–thaw cycles, and the associated interaction as fixed effects. In both models, linear and quadratic contrast statements were incorporated to determine the change in ASFV DNA detection by freeze–thaw cycle and over time, respectively. The SLICE function was used to perform an F-test within each sampling device treatment to test the effect of day or number of freeze–thaw cycles. Visual assessment of studentized residual plots was performed to evaluate model assumptions which appeared to be reasonably met. A Tukey multiple comparison adjustment was used to control Type I error rate, and results were considered significant at p < 0.05.
To determine the effect of freeze–thaw cycles on ASFV DNA stability, the environmental samples were subjected to freeze–thaw cycles 0X, 1X, 2X, or 3X. There was no significant reduction of ASFV DNA detection in all environmental samples that were collected using a premoistened gauze (Table 1; p > 0.05). In parallel, there was no effect (p > 0.05) of freeze–thaw on ASFV DNA detection in samples collected from surfaces with no organic material contamination, feces, feed dust, and other organic mixture samples that were collected using the sponge stick with DNA/RNA shield. In contrast, ASFV DNA detection was significantly reduced (p < 0.05 and 0.4 log reduction after incubation for 7 days) after a 3X freeze–thaw cycle compared to the sample with 0X freeze–thaw cycles in samples that were collected using a sponge stick with DNA/RNA shield on surfaces with soil contamination.
Next, we investigated the effect of storage at 4°C on ASFV DNA detection. Samples were incubated at 4°C for 0, 1, 3, and 7 days after sample collection. There was a significant interaction between treatment and the time (days) of storage (Table 2; p < 0.0001). The effect of storage time was dependent upon the sampling device and level of surface organic matter contamination. In non-organic, swine feces and feed dust samples using either sampling device, no ASFV DNA degradation was observed. ASFV DNA concentrations significantly decreased over time for samples collected from surfaces contaminated with soil using the premoistened gauze sampling device (Linear, p < 0.0001 and 1.2 log reduction after incubation for 7 days). In addition, ASFV DNA detection was significantly reduced in samples collected from surfaces contaminated with the organic mixture using the sponge stick with DNA/RNA shield, with the greatest reduction in detection occurring within 1 day of incubation at 4°C with no subsequent reduction when stored beyond 1 day (Quadratic, p < 0.0001 and 0.5 log reduction after incubation for 7 days). There was no evidence of a difference in ASFV DNA detection based on day of storage for any of the other combinations of sampling device and organic matter contamination level (p > 0.05).
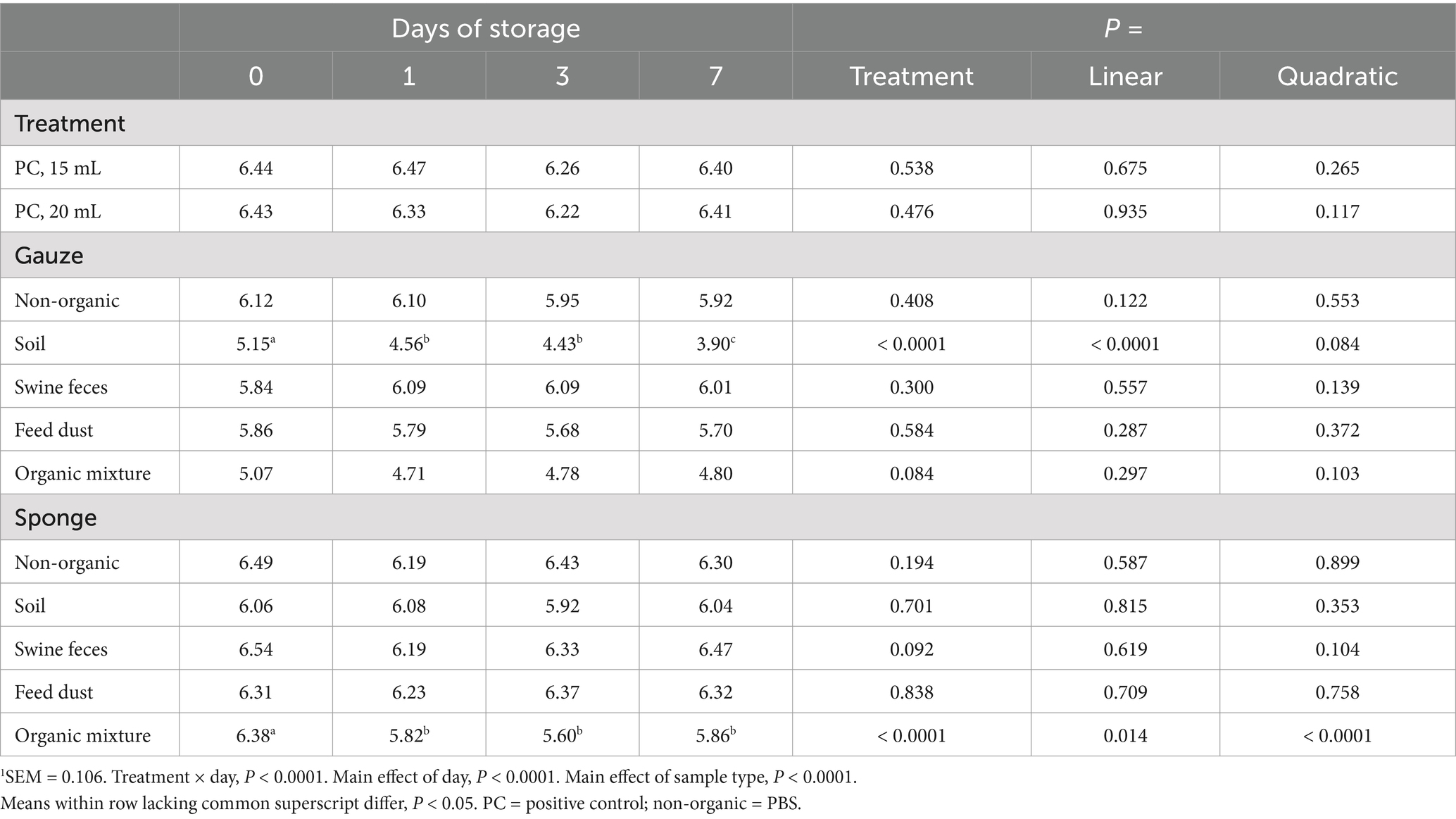
Table 2. Effect of storage at 4°C on detection of ASFV p72 log10 copy number/mL1.
Lastly, the effect of storage at RT on ASFV DNA detection was studied in environmental samples. Similar to storage at 4°C, there was a treatment × day effect (Table 3; p < 0.0001). ASFV DNA was stable over time in samples collected from surfaces with no organic matter contamination using either sampling device, but significant reductions of ASFV DNA were observed in soil, feces, and feed dust samples collected with the premoistened gauze (p ≤ 0.043 and 1.5, 0.2, and 1.2 log reduction, respectively, after incubation for 7 days). Using the sponge stick with DNA/RNA shield samples, ASFV DNA was stable in swine feces and feed dust, however detection of ASFV DNA gradually decreased in the presence of soil and organic mixture (Quadratic, p ≤ 0.003 and 0.6 log reduction, respectively, after incubation for 7 days). There was no evidence of a reduction in ASFV DNA when environmental samples were collected using the sponge stick with DNA/RNA shield from surfaces contaminated with swine feces or feed dust (p > 0.05).
Table 3. Effect of storage at room temperature on detection of ASFV p72 log10 copy number/mL1.
Due to the lack of an effective and safe vaccine to control and prevent ASFV infection, current approaches rely on enhanced biosecurity to prevent the introduction of ASFV into ASFV-free areas and to confine and destroy the source of infections. Early detection is one of the most important factors to determine successful implementation of biosecurity. The detection of ASFV DNA is a gold standard for agent identification, and most diagnostic laboratories utilize automated DNA extraction and real-time PCR for ASFV detection (14). In addition, novel assays, such as point-of-care detection, have been developed for fast turnaround time and increased sensitivity (15). Clinical samples from pigs or wild boar are the most popular sample type for detecting ASFV DNA, thus these types of samples, such as blood and tissue, are validated for molecular diagnostics. In contrast, a few manuscripts attempted to validate and improve the use of environmental samples for ASFV detection (11, 12). The successful ASFV DNA detection in environmental samples is affected by several factors: choice of sampling devices, appropriate sample collection from the targeted area, rapid transportation of samples to the diagnostic laboratory, and the validated assays for ASFV DNA detection. In modern swine production systems, samples are usually shipped to diagnostic laboratory in a few days. Although overnight shipping of samples is common in the United States, some countries experience delayed sample transportation, and subsequently diagnostic sensitivity could be greatly compromised. In particular, most environmental samples contain a variety of organic contaminants, in which different types of proteinases, DNases, and RNases can potentially degrade nucleic acids and/or inhibit subsequent molecular analysis. Therefore, this study aimed to evaluate the effect of sample storage conditions on ASFV DNA detection in environmental samples.
The efforts of the swine industry in the past focused on preserving the integrity of RNA molecules of RNA viruses in diagnostic specimens because of the economic impact of endemic RNA viruses, such as porcine reproductive and respiratory syndrome virus (PRRSV) (16). A previous study showed that PRRSV detection was maintained for up to 168 h in serum that was stored at 4, 10 or 20°C; however, incubation at 30°C had a negative effect on PRRSV RNA stability (17). In contrast, PRRSV RNA detection decreased in oral fluid and fecal samples over time at 4, 10, 20, or 30°C. The reduced detection of porcine epidemic diarrhea virus (PEDV) and transmissible gastroenteritis virus (TGEV) RNA in fecal samples was observed after storage at 4, 21, 36, and 45°C (18). DNA in general is more stable than RNA, however, prolonged storage of ASFV-infected tissues at 4 and 23°C led to reduction of ASFV DNA detection (8). Our data supports the high stability of ASFV DNA in environmental samples with no contamination with organic materials such as feed dust, fecal material, or soil. However, we observed significant reduction in the stability of ASFV DNA in environmental samples with organic contaminants, in particular in the presence of soil, suggesting that ASFV DNA stability is dependent on the type of organic contaminant. Bacteria and fungi play an important role in building the microenvironment of soils and produce a variety of extracellular DNases which could affect the stability of DNA viruses. The optimal temperature of soil DNase activity is between 30 to 40°C, but DNase activity can be sufficient for DNA degradation even at 4°C (19, 20). It could be plausible that soil DNases in environmental samples contaminated with soil could result in ASFV DNA degradation in this study. In addition, the pH level may also affect ASFV DNA stability. Soil could have an acidic or alkaline pH level which may also affect DNAse activity (21). Furthermore, we also observed reduced ASFV DNA concentrations in some samples collected with the sponge stick with DNA/RNA shield after freeze–thaw cycles and incubation at 4°C and RT. The role of DNA/RNA shield in preserving nucleic acids has been well-documented, and our study confirmed better sensitivity with the sponge stick plus DNA/RNA shield than the gauze plus PBS. Importantly, the volume of DNA/RNA shield (10 mL) may not be sufficient to preserve ASFV DNA in environmental samples containing excessive amounts of organic contaminants (2.5 g) as the manufacturer’s instruction indicates an optimal ratio of 10% (v/v and w/v), since the present study stimulated the field situation where organic matters are heavily contaminating environment samples.
Environmental samples have been used to monitor disease status and the distribution of infectious agents in swine herds after a disease outbreak. For example, following a PRRS outbreak, viral RNA remained detectable up to 14 weeks post-outbreak in environmental wipes and the sensitivity of environmental samples was similar to blood samples (22). In addition, environmental sampling revealed the extensive contamination and distribution of PRRSV RNA on contaminated farms, such as exhaust fan cones, door knobs, anteroom floors, mortality carts/sleds (23). Similarly, long-term persistence and extensive contamination of PEDV RNA was found after an outbreak and the results of environmental samples were useful to implement a modified biosecurity protocol (24). In contrast to endemic viruses, it is impossible to monitor the disease status in ASFV-affected farms over time because the affected herd is readily culled for control purposes. Rather, the environmental samples can be used to evaluate the level of cleaning and disinfecting process for the re-introduction of animals after the ASF outbreak. In addition, they are valuable sample types to detect ASFV on equipment and transportation vehicles in affected areas, in order to reduce ASFV transmission between farms. In this scenario, the environmental samples should be properly collected and immediately shipped to the diagnostic laboratory at 4°C. If not possible, our data support and recommend freezing the environmental samples before shipping. We found when using different sampling devices with differing organic contamination levels, there were minimal reductions in ASFV DNA concentrations; even after 3 freeze–thaw cycles, the amount of ASFV DNA recovered still is likely to generate positive PCR results.
One of limitations of our study is the high virus titer for surface contamination, approximately 3.7 × 107 TCID50 per sample type. This amount of virus is certainly present in the blood of infected animals. However, in reality, ASFV DNA is present at a variable level in environmental samples and therefore, ASFV DNA decay on various surfaces may be more variable, compared to that of high level of contamination used in this study. Secondly, we tested the viral DNA stability in certain types of organic contaminants on a single surface type, stainless steel. Others surfaces such as cement, plastic and cardboard present on farms were not tested. Therefore, further exploration using a variety of infectious loads, organic contaminants and surfaces would provide additional understanding of viral DNA decay in environmental samples. Lastly, due to the limitations at the BSL-3 facility, the experiments were performed for relatively short durations at certain temperatures. In real-world conditions, samples may be stored for extended periods of time at −20°C before processing, thus it may be valuable to assess ASFV DNA degradation under longer storage conditions.
Nevertheless, the data provided in this study are highly valuable to provide a baseline understanding of handling of environmental samples for the detection of ASFV DNA which can be used to establish the appropriate biosecurity procedures for ASFV preparedness and response.
The original contributions presented in the study are included in the article/supplementary material; further inquiries can be directed to the corresponding authors.
TK: Conceptualization, Formal analysis, Investigation, Methodology, Writing – review & editing. JG: Conceptualization, Funding acquisition, Methodology, Project administration, Writing – review & editing, Supervision. EL: Investigation, Writing – review & editing, Formal analysis. NG: Funding acquisition, Methodology, Writing – review & editing. JT: Methodology, Writing – review & editing. JR: Conceptualization, Funding acquisition, Methodology, Supervision, Writing – review & editing.
The author(s) declare that financial support was received for the research and/or publication of this article. This material is based upon work supported by the U.S. Department of Homeland Security under grant award number 18STCBT00001-03-00 through the Cross-Border Threat Screening and Supply Chain Defense Center of Excellence. Additional funding was made available through the National Bio and Agro-Defense Facility (NBAF) Transition Fund from the State of Kansas, and the MCB Core of the Center on Emerging and Zoonotic Infectious Diseases (CEZID) of the National Institutes of General Medical Sciences under award number P20GM130448.
We gratefully thank the lab members of Richt’s lab for collecting blood samples in the animal study.
The JR laboratory received support from Tonix Pharmaceuticals, Xing Technologies, and Zoetis, outside of the reported work. JR is an inventor on patents and patent applications on the use of antivirals and vaccines for the treatment and prevention of virus infections, owned by Kansas State University.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declare that no Gen AI was used in the creation of this manuscript.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The views and conclusions contained in this document are those of the authors and should not be interpreted as necessarily representing the official policies, either expressed or implied, of the U.S. Department of Homeland Security.
1. Thomson, GR. The epidemiology of African swine fever: the role of free-living hosts in Africa. Onderstepoort J Vet Res. (1985) 52:201–9.
2. Rowlands, RJ, Michaud, V, Heath, L, Hutchings, G, Oura, C, Vosloo, W, et al. African swine fever virus isolate, Georgia, 2007. Emerg Infect Dis. (2008) 14:1870–4. doi: 10.3201/eid1412.080591
3. Guberti, V, Khomenko, S, Masiulis, M, and Kerba, S. African swine fever in wild boar – ecology and biosecurity. Second ed FAO; World Organisation for Animal Health (WOAH) (founded as OIE); European Commission Paris, France. (2022).
4. Tao, D, Sun, D, Liu, Y, Wei, S, Yang, Z, An, T, et al. One year of African swine fever outbreak in China. Acta Trop. (2020) 211:105602. doi: 10.1016/j.actatropica.2020.105602
5. Ramirez-Medina, E, O'Donnell, V, Silva, E, Espinoza, N, Velazquez-Salinas, L, Moran, K, et al. Experimental infection of domestic pigs with an African swine fever virus field strain isolated in 2021 from the Dominican Republic. Viruses. (2022) 14:1090. doi: 10.3390/v14051090
6. Mazur-Panasiuk, N, Zmudzki, J, and Wozniakowski, G. African swine fever virus—persistence in different environmental conditions and the possibility of its indirect transmission. J Vet Res. (2019) 63:303–10. doi: 10.2478/jvetres-2019-0058
7. Prodelalova, J, Kavanova, L, Salat, J, Moutelikova, R, Kobzova, S, Krasna, M, et al. Experimental evidence of the long-term survival of infective African swine fever virus strain Ba71V in soil under different conditions. Pathogens. (2022) 11:648. doi: 10.3390/pathogens11060648
8. Mazur-Panasiuk, N, and Wozniakowski, G. Natural inactivation of African swine fever virus in tissues: influence of temperature and environmental conditions on virus survival. Vet Microbiol. (2020) 242:108609. doi: 10.1016/j.vetmic.2020.108609
9. Nuanualsuwan, S, Songkasupa, T, Boonpornprasert, P, Suwankitwat, N, Lohlamoh, W, and Nuengjamnong, C. Persistence of African swine fever virus on porous and non-porous fomites at environmental temperatures. Porcine Health Manag. (2022) 8:34. doi: 10.1186/s40813-022-00277-8
10. Gebhardt, JT, Dritz, SS, Elijah, CG, Jones, CK, Paulk, CB, and Woodworth, JC. Sampling and detection of African swine fever virus within a feed manufacturing and swine production system. Transbound Emerg Dis. (2022) 69:103–14. doi: 10.1111/tbed.14335
11. Kwon, T, Gebhardt, JT, Lyoo, EL, Gaudreault, NN, Trujillo, JD, Woodworth, JC, et al. Development and optimization of sampling techniques for environmental samples from African swine fever virus-contaminated surfaces with no organic contaminants. Front Vet Sci. (2024) 11:1425928. doi: 10.3389/fvets.2024.1425928
12. Kwon, T, Gebhardt, JT, Lyoo, EL, Gaudreault, NN, Trujillo, JD, Woodworth, JC, et al. Improved African swine fever detection for environmental samples in the presence of organic contaminants. Transbound Emerg Dis. (2024) 2024:8841168. doi: 10.1155/tbed/8841168
13. Houston, GE, Trujillo, JD, Jones, CK, Kwon, T, Stark, CR, Cool, K, et al. Detection of African swine fever virus in feed and feed mill environment following extended storage. Transbound Emerg Dis. (2023) 2023:1–7. doi: 10.1155/2023/3455128
14. World Organization for Animal Health. Chapter 3.9.1. African swine fever (infection with African swine fever virus). Paris, France: WOAH Terrestrial Manual (2024).
15. Mee, PT, Wong, S, O'Riley, KJ, da Conceicao, F, da Costa, B, Jong, J, et al. Field verification of an African swine fever virus loop-mediated isothermal amplification (LAMP) assay during an outbreak in Timor-Leste. Viruses. (2020) 12:1444. doi: 10.3390/v12121444
16. Munguia-Ramirez, B, Gimenez-Lirola, L, and Zimmerman, J. Assessment of strategies for preserving swine viral RNA targets in diagnostic specimens. Microorganisms. (2024) 12:410. doi: 10.3390/microorganisms12020410
17. Munguia-Ramirez, B, Armenta-Leyva, B, Henao-Diaz, A, Cheng, TY, Zhang, J, Rawal, G, et al. Effect of extrinsic factors on the detection of PRRSV and a porcine-specific internal sample control in serum, oral fluid, and fecal specimens tested by RT-rtPCR. J Vet Diagn Invest. (2023) 35:375–84. doi: 10.1177/10406387231174556
18. Jung, K, and Chae, C. Effect of temperature on the detection of porcine epidemic diarrhea virus and transmissible gastroenteritis virus in fecal samples by reverse transcription-polymerase chain reaction. J Vet Diagn Invest. (2004) 16:237–9. doi: 10.1177/104063870401600312
19. Kamino, LN, and Gulden, RH. The effect of crop species on DNase-producing bacteria in two soils. Ann Microbiol. (2021) 71:14. doi: 10.1186/s13213-021-01624-w
20. Ahrenholtz, I, Lorenz, MG, and Wackernagel, W. The extracellular nuclease of Serratia marcescens: studies on the activity in vitro and effect on transforming DNA in a groundwater aquifer microcosm. Arch Microbiol. (1994) 161:176–83. doi: 10.1007/BF00276480
21. Frankenberger, WT, and Johanson, JB. Effect of pH on enzyme stability in soils. Soil Biol Biochem. (1982) 14:433–7. doi: 10.1016/0038-0717(82)90101-8
22. Vilalta, C, Sanhueza, J, Garrido, J, Murray, D, Morrison, R, Corzo, CA, et al. Indirect assessment of porcine reproductive and respiratory syndrome virus status in pigs prior to weaning by sampling sows and the environment. Vet Microbiol. (2019) 237:108406. doi: 10.1016/j.vetmic.2019.108406
23. Melini, CM, Kikuti, M, Bruner, L, Allerson, M, O'Brien, K, Stahl, C, et al. Assessment of porcine reproductive and respiratory syndrome virus (PRRSV) farm surface contamination through environmental sampling. Porcine Health Manag. (2024) 10:34. doi: 10.1186/s40813-024-00387-5
Keywords: African swine fever, ASFV, environmental sample, freeze–thaw, fomite, surface, storage
Citation: Kwon T, Gebhardt JT, Lyoo EL, Gaudreault NN, Trujillo JD and Richt JA (2025) The effect of freeze–thaw and storage on African swine fever virus detection in environmental samples. Front. Vet. Sci. 12:1570575. doi: 10.3389/fvets.2025.1570575
Received: 03 February 2025; Accepted: 17 March 2025;
Published: 01 April 2025.
Edited by:
Aleksandra Kosowska, University of Castilla La Mancha, SpainReviewed by:
Saurabh Gupta, GLA University, IndiaCopyright © 2025 Kwon, Gebhardt, Lyoo, Gaudreault, Trujillo and Richt. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jordan T. Gebhardt, amdlYmhhcmR0QHZldC5rLXN0YXRlLmVkdQ==; Juergen A. Richt, anJpY2h0QHZldC5rLXN0YXRlLmVkdQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.