
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Vet. Sci., 31 March 2025
Sec. Veterinary Infectious Diseases
Volume 12 - 2025 | https://doi.org/10.3389/fvets.2025.1562444
 Yuwen Shi1,2†
Yuwen Shi1,2† Kaichuang Shi1,3†
Kaichuang Shi1,3† Yan Ma1
Yan Ma1 Yanwen Yin3
Yanwen Yin3 Feng Long3
Feng Long3 Shuping Feng3
Shuping Feng3 Meilan Mo1
Meilan Mo1 Jiakang He1*
Jiakang He1* Zuzhang Wei1*
Zuzhang Wei1*Porcine respiratory coronavirus (PRCoV), porcine reproductive and respiratory syndrome virus (PRRSV), and swine influenza virus (SIV) are important pathogens of significant infectious diseases. They cause similar clinical respiratory symptoms, including fever, cough, runny nose, and respiratory distress, which makes these diseases difficult to distinguish from each other. In this study, three pairs of specific primers and TaqMan probes were designed for the conserved regions of the PRCoV S gene, PRRSV N gene, and SIV M gene, respectively. The annealing temperature, primer and probe concentrations, and reaction cycle were optimized, and a triplex crystal digital PCR (cdPCR) assay was established for the detection of PRCoV, PRRSV, and SIV. According to the test results, the assay was capable of specifically detecting PRCoV, PRRSV, and SIV, and there was no cross-reaction with other control swine viruses. Based on the Poisson distribution analysis, the limits of detection (LODs) for PRCoV, PRRSV, and SIV were 6.00, 5.75 and 6.00 copies/reaction, respectively, and the sensitivity was 26 times higher than those of the corresponding multiplex RT-qPCR. The coefficients of variation (CVs) of the intra-assay and inter-assay ranged from 0.19 to 1.84%. The assay was used to test 1,657 clinical samples, and the positivity rates of PRCoV, PRRSV, and SIV were 1.15, 12.79, and 2.05%, respectively. It showed diagnostic sensitivity and specificity of 100 and 99.82% for PRCoV, 100 and 99.24% for PRRSV, and 100 and 99.69% for SIV, respectively. These results indicated that the triplex cdPCR assay has strong specificity, high sensitivity, and excellent repeatability, which provides a valuable tool for the detection and differentiation of PRCoV, PRRSV, and SIV.
With the development of intensive industrial farms and increasement of feeding density of pig industry, the respiratory infectious diseases have becoming important diseases that seriously endanger pig herds. Of which, porcine respiratory coronavirus (PRCoV), porcine reproductive and respiratory syndrome virus (PRRSV), and swine influenza virus (SIV) are important porcine pathogens which induce similar clinical respiratory symptoms of fever, cough, runny nose, and respiratory distress (1, 2). In serious cases, they can lead to high rates of morbidity and mortality, which cause great economic losses to the pig industry (3, 4).
PRCoV belongs to Alphacoronavirus genus in the coronaviridae family, which is an enveloped, single-stranded, positive-sense RNA virus with a 28 kb genome size (3, 5). PRCoV is considered as a naturally occurring deletion mutant of transmissible gastroenteritis virus (TGEV), with 5′ region deletion (nucleotides 621–681) in the Spike gene (6). After infection, it can lead to mild bronchial interstitial pneumonia and neutrophil infiltration, resulting in respiratory diseases in infected pigs (5, 7). The typical clinical manifestations include dyspnea, wheezing, sneezing, coughing, fever, anorexia, and growth retardation (8, 9). At present, PRCoV distributes in many countries around the world (5, 8, 10, 11), including China (12, 13).
PRRSV is a single-stranded, positive-sense RNA virus that belongs to the Arterivirus genus in the Arteriviridae family, and has a genome size of approximately 15.5 kb (14). According to genetic and antigenic characteristics, PRRSV is divided into European species (PRRSV-1) and American species (PRRSV-2). PRRSV-1 is represented by the Lelystad strain and PRRSV-2 is represented by the VR-2332 strain, and they have only about 60% similarity at nucleotide (15, 16). PRRSV cause immunosuppression, heightening the vulnerability to secondary infections and resulting in increased mortality rates in infected pigs (17, 18). The clinical manifestations include miscarriages, premature deliveries, and stillbirths in sows, as well as respiratory distress, hyperthermia, and fatalities in pigs (13). PRRSV has posed huge economic losses to the pig industry worldwide (19, 20). PRRSV was first reported in 1996 in China, and now has become a significant disease affecting pig herds throughout the country (21, 22).
SIV belongs to Influenza A virus genus in the Orthomyxoviridae family, and is a single-stranded, negative-sense RNA virus with a 13.6 kb genome size (23). It is a respiratory pathogen that is prevalent globally, and causes acute, febrile, and contagious respiratory disease in swine, and exhibits a high incidence of morbidity and low mortality (24). SIV is divided into different subtypes based on antigenic differences in glycoprotein haemagglutinin (HA) and neuraminidase (NA), with 18 subtypes for HA and 11 subtypes for NA (24). Nowadays, SIV is prevalent worldwide, and the mainly prevalent SIV subtypes are H1N1, H1N2, and H3N2, all of which show high diversity (25, 26). SIV was first reported in China in 1918, and to date, SIV has been discovered around the country (27, 28). Furthermore, SIV has been found in humans, and the trans-species transmission and zoonotic potential make serious threat to animals and humans (27, 29).
At present, PRCoV, PRRSV, and SIV are prevalent in many countries around the world (5, 8, 10, 11, 19, 20, 25–28, 30). Even worse, co-infections of PRCoV, PRRSV, and SIV occur in some pig herds, which exacerbates the seriousness of clinical manifestations and pathological damage, and induces more seriously economic losses to pig herds (17, 31–33). PRRSV/PRCoV co-infection was associated with reduced weight, higher incidence of fever, and more serious pneumonia compared with single infection; the infection of PRRSV followed by infection with PRCoV increased the damage of lymph nodes and lungs, and prolonged the persistent period of PRRSV, suggesting the synergistic effect of PRRSV and PRCoV (17). Co-infection with PRCoV and SIV may aggravate clinical signs and lung lesions (31, 32). PRRSV/SIV co-infection cause shortness of breath, dyspnea, fever, and cough, which can lead to more severe illness and growth retardation (33). PRRSV and SIV were found in pig herds (34). The positive antibodies against PRCoV, PRRSV, and SIV were found in German wild boar populations (35), and the positive nucleic acids of PRCoV, PRRSV, and SIV were found in clinical tissue samples in domestic pig herds in China (12). Since PRCoV, PRRSV, and SIV cause similar clinical signs of respiratory disease, the co-infections of two or three of these pathogens increase the complexity and difficulty of accurate diagnosis of these diseases. Therefore, development and application of a rapid, sensitive, and accurate technique for the detection and differentiation of these pathogens is necessary and helpful for early detection of the pathogens, accurate diagnosis of the diseases, and taking timely disposal measures, which is great significance for the prevention and control of these diseases.
The detection of viral nucleic acids is one of the important methods for accurate diagnosis of animal diseases. Of which, conventional RT-PCR/PCR, real-time quantitative RT-PCR/PCR, and digital RT-PCR/PCR are important techniques commonly used in veterinary laboratories (36, 37). Digital PCR (dPCR) is a third-generation PCR detection technology that has recently emerged, and is characterized by the separation of the reaction mixture into thousands to millions of partitions, and then perform amplification of real-time or endpoint detection (38). The target genes are partitioned according to the Poisson distribution model. At the end of the reaction, the target genes can be accurately and absolutely quantified according to the ratio of positive partition to all partitions (39). dPCR has the advantages of the ability to absolute quantification independent of reference gene, standard curve, and Ct value; higher resilience to inhibitors; high sensitivity, specificity, precision, and efficacy (40, 41). At present, dPCR is divided into droplet digital PCR (ddPCR) and crystal digital PCR (cdPCR), and have been used widely and effectively for the detection and quantification of swine pathogens, showing high sensitivity and accuracy (42–44). As for PRCoV, PRRSV, and SIV, the dPCR assays for the detection of PRRSV (45), classical + highly pathogenic + NADC30-like PRRSV (46), and ASFV + CSFV + PRRSV (47) have been reported. However, no multiplex dPCR assay has been reported for the simultaneous detection and differentiation of these three pathogens. In this study, a triplex cdPCR was developed for the detection of PRCoV, PRRSV, and SIV.
The following vaccine strains were purchased from Harbin Harvac Biotechnology Co. Ltd. (Harbin, China): PRRSV (R98, Ch-1R and HuN4-F112 strains), classical swine fever virus (CSFV, CVCC AV1412 strain), TGEV (H strain), porcine epidemic diarrhea virus (PEDV, CV777 strain), and porcine rotavirus (PoRV, G5 type-NX strain). The following vaccine strains were purchased from Huapai Biotechnology Co. Ltd. (Chengdu, China): SIV (TJ strain), foot-and-mouth disease virus (FMDV, O/Mya98/XJ/2010 strain), and porcine circovirus type 2 (PCV2, ZJ/C strain). The clinical positive samples of PRCoV, PRRSV, African swine fever virus (ASFV), and porcine deltacoronavirus (PDCoV) were provided by our laboratory. All vaccine strains and clinical positive samples were stored at −80°C until use.
From July 2023 to March 2024, a total of 1,657 clinical samples from 1,657 pigs (the samples from each pig included trachea, lungs, lymph nodes, tonsils, and spleen) were collected from different pig farms, harmless treatment plants and slaughterhouses in Guangxi Province, China. The tissue samples from each pig were homogenized, and considered as one sample when tested for viral nucleic acids. All samples were stored at −80°C before use.
The genome sequences of 17 strains of PRCoV, 41 strains of PRRSV (16 strains of PRRSV-1, and 25 strains of PRRSV-2), and 40 strains of SIV were downloaded from the NCBI GenBank (https://www.ncbi.nlm.nih.gov/ (accessed on 15 April 2022)), and multiple sequence alignment was compared. Three pairs of primers and TaqMan probes were specifically designed for the conserved regions of the PRCoV S gene, PRRSV N gene, and SIV M gene using Oligo 7.0 primer design software as previous report by Ma et al. (12). The viral strains used for sequence comparison and the locations of the designed primers and probes were described in the supplementary figure S1 of a previous report (12). The information on the specific primers and TaqMan probes is shown in Table 1.
The tissue samples of trachea, lungs, lymph nodes, tonsils, and spleen (about 0.1 g each tissue) were put into 2.0 mL EP tubes, and 1.0 mL of PBS buffer (pH 7.2) was added. The tissues were homogenized using a Retsch MM400 tissue homogenizer (Haan, Germany) with 30 Hz, frozen and thawed three times, and centrifuged at 12,000 rpm g for 5 min at 4°C. Then, 200 μL of the supernatant/vaccine solution was extracted using nucleic acid extractor (TIANGEN, Beijing, China) and Viral DNA/RNA Extraction Kit (TIANGEN, Beijing, China), and stored at −80°C.
The synthesized viral RNAs of PRCoV, PRRSV, and SIV corresponding to the target gene fragments for amplification were provided by Dalian TaKaRa Co. Ltd. (TaKaRa, Dalian, China), which were used for evaluating the sensitivity and repeatability of the developed triplex cdPCR assay.
All standard plasmid constructs were generated referencing the procedure described by Ma et al. (12). PRCoV, PRRSV, and SIV nucleic acids were extracted from positive samples or vaccine solution, reverse transcribed into cDNA using the PrimeScript II 1st Strand cDNA Synthesis Kit (TaKaRa, Dalian, China) according to the manufacturer’s instructions. The target fragments were amplified using three pairs of specific primers in Table 1, using cDNA as templates. After amplification, the target fragments were subjected to agarose gel electrophoresis, purified using MiniBEST DNA Fragment Purification Kit Ver.4.0 (TaKaRa, Dalian, China), cloned into pMD18-T vector (TaKaRa, Dalian, China), and transformed into E. coli DH5α competent cells (TaKaRa, Dalian, China). The positive clones were selected, added into LB solution, and incubated at 37°C for 20–24 h. The plasmid constructs were extracted from cultured bacterial fluids using MiniBEST Plasmid Extraction Kit Ver.5.0 (TaKaRa, Dalian, China), and named p-PRCoV, p-PRRSV, and p-SIV, respectively. The UV absorbance at 260/280 nm were measured using a spectrophotometer (Thermo Fisher, Waltham, MA, USA), and the concentrations of three plasmid constructs were calculated according to the following equation.
The Naica® Sapphire Crystal system (Stilla Technologies, Villejuif, France) was used to determine the optimal reaction conditions for the triplex cdPCR, including primer concentration, probe concentration, and annealing temperature. The total volume of 25 μL of reaction system was as follows: 12.5 μL 2× PerfeCTa Multiplex qPCR Tough Mix (Quanta Biosciences, Gaithersburg, MD, USA), 2.5 μL Fluorescein Sodium Salt (1 μM) (Apexbio Biotechnology, Beijing, China), three pair of primers and TaqMan probes with different final concentrations, 2.5 μL of the mixture of three standard plasmid constructs (final concentration of each plasmid construct was 104 copies/μL), and distilled water to final volume of 25 μL. The annealing temperature of the triplex cdPCR was optimized from 55°C to 60°C, and the concentrations of primers and probes were optimized from 0.1 μM to 0.9 μM. The triplex cdPCR amplification procedure was as follows: 45°C for 5 min, 95°C for 10 s; then, 45 cycles of 95°C for 5 s, and 59°C for 34 s. After reaction, the chips were moved into the Naica™ Prism3 system (Stilla Technologies, Villejuif, France) to obtain images of VIC, FAM, and CY5 detection channels. According to the obtained droplets, the optimal reaction conditions were determined.
The three plasmid constructs of p-PRCoV, p-PRRSV, and p-SIV were mixed at a ratio of 1:1:1, and 10-fold serially diluted from 1 × 105 to 1 × 101 copies/μL (final reaction concentration from 1 × 104 to 1 × 100 copies/μL). The standard curves were generated by performing triplex cdPCR using the optimal reaction conditions.
The extracted nucleic acids (RNA or DNA) of PRCoV, PRRSV, SIV, PRV, TGEV, PEDV, PoRV, ASFV, FMDV, PCV2, PDCoV, and CSFV were used as templates to analyze the specificity of the triplex cdPCR. The mixture of three plasmid constructs, the clinical negative sample, and distilled water were used as controls.
The synthesized viral RNA of PRCoV, PRRSV, and SIV were mixed at a ratio of 1:1:1, and 10-fold serially diluted from 1 × 105 to 1 × 100 copies/μL (final reaction concentration from 1 × 104 to 1 × 10−1 copies/μL). The mixture was used as template to perform triplex cdPCR. The Poisson distribution analysis was used to evaluate the limits of detection (LODs) of the triplex cdPCR.
In addition, to further verify the results of the Poisson distribution analysis, the Probit regression analysis (https://www.ibm.com/cn-zh/spss) was also used to analyze the LODs of the assay. The mixture of PRCoV, PRRSV, and CSFV was 2-flod serially diluted as 31.23, 15.63, 7.81, 3.91, 1.96, 0.98, 0.49 copies/reaction, and used as templates. Each concentration was set for 20 repeats, and the times of positive amplification curve were counted. The Probit regression analysis was used to analyze the relationship between positive hit probability and detection concentration for evaluating the assay’s sensitivity.
The synthesized viral RNA of PRCoV, PRRSV, and SIV were mixed at a ratio of 1:1:1, and diluted to 1 × 105, 1 × 104, and 1 × 103 copies/μL (final reaction concentration of 1 × 104, 1 × 103, and 1 × 102 copies/μL). The mixture was used as template to perform triplex cdPCR. The experiments were performed at three duplicates each time, and repeated at three weeks to assess the intra- and inter-assay variation of the triplex cdPCR.
A total of 1,657 clinical samples collected between July 2023 and March 2024 in Guangxi Province of China were analyzed using the established triplex cdPCR. Meanwhile, these samples were also evaluated using the quadruplex RT-qPCR established by Ma et al. (12), which was used as a reference method for the simultaneous detection of PRCoV, PRRSV, and SIV. The diagnostic sensitivity and specificity of the developed triplex cdPCR were calculated. The consistency of the developed triplex cdPCR and the reference quadruplex RT-qPCR was assessed using SPSS 27.0 software.
After PCR amplification of the PRCoV S gene, PRRSV N gene, and SIV M gene using the specific primers in Table 1, the target fragments were purified, and connected to pMD18-T vector, then transformed into E. coli DH5α competent cells. The positive clones were screened and cultured, and the recombinant plasmid constructs were extracted and confirmed by sequence analysis. Three obtained plasmid constructs were named p-PRCoV, p-PRRSV, and p-SIV, respectively, and their concentrations were determined. The results showed that the initial concentrations of the plasmid constructs p-PRCoV, p-PRRSV, and p-SIV were 7.85 × 1010, 7.65 × 1010, and 6.35 × 1010 copies/μL, respectively. All standard plasmid constructs were diluted to 1.0 × 1010 copies/μL, and stored at −80°C until use.
To obtain the optimal reaction conditions, the annealing temperatures, primer and probe concentrations, and amplification cycles were adjusted to perform triplex cdPCR. The optimal reaction conditions were determined according to the following states of the reaction results: the largest number of total and positive droplets, the highest fluorescence signal values, the best droplet density, obvious distinction between positive and negative droplet clusters, and the least number of dispersed droplets between negative and positive droplets. Finally, the optimal reaction system of the triplex cdPCR is shown in Table 2. The amplification parameters were as follows: 45°C for 5 min, 95°C for 10 s, and then 45 cycles of 95°C for 5 s, and 60°C for 30 s. The triplex cdPCR achieved the best states of reaction results under the above optimal reaction conditions (Figure 1).
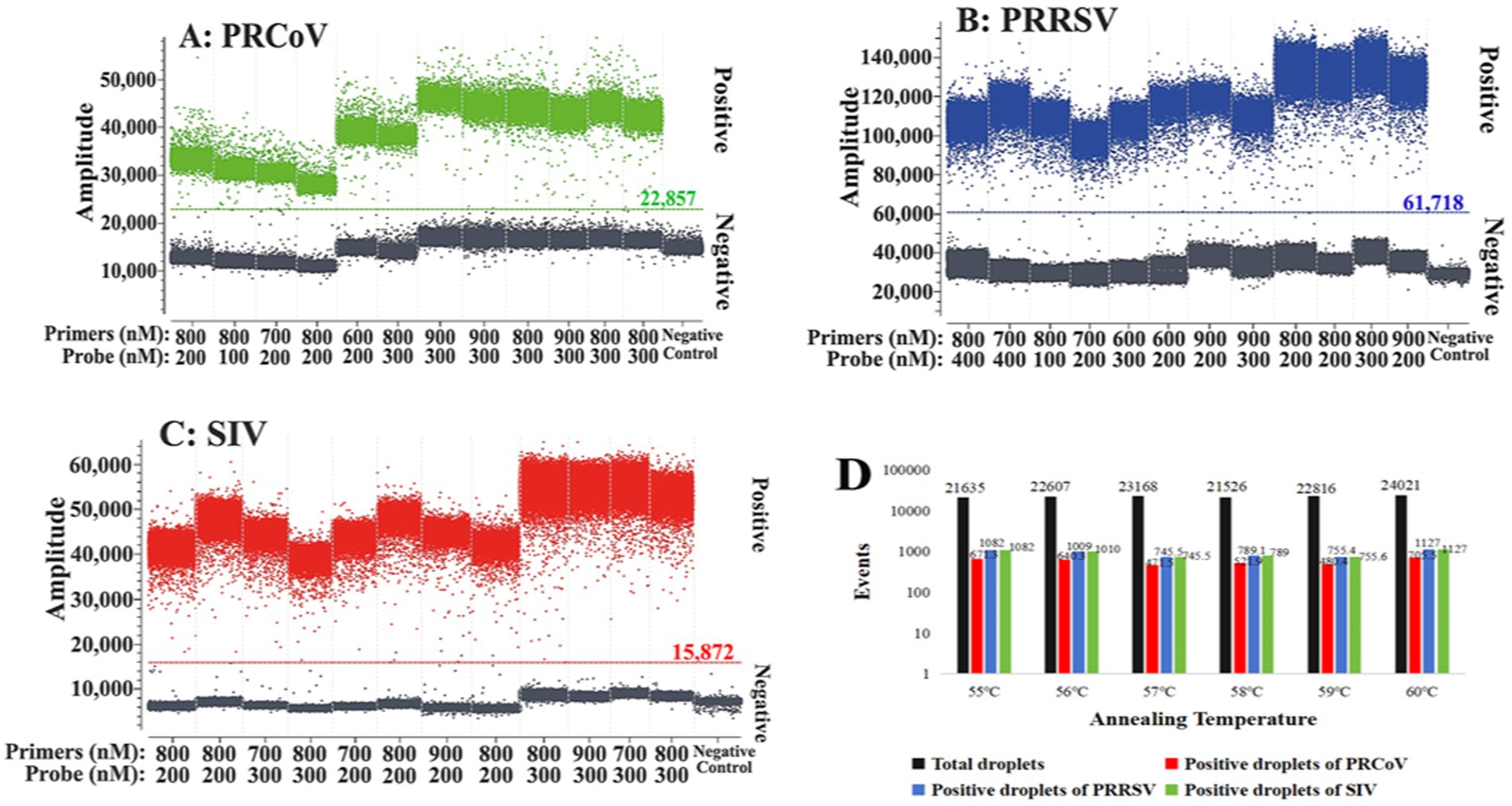
Figure 1. Optimization of the optimal reaction conditions for the triplex cdPCR. (A–C) Optimization of primer and probe concentration for PRCoV (A), PRRSV (B), and SIV (C). (D) Optimization of annealing temperature.
The standard curves of the triplex cdPCR were generated using the mixture of three standard plasmid constructs from 1 × 105 to 1 × 101 copies/μL (final reaction concentration: from 1 × 104 to 1 × 100 copies/μL) as templates. The results showed that the slopes and the correlation coefficients (R2) of PRCoV, PRRSV, and SIV were 0.999 and 0.9993, 0.993 and 0.9986, and 1.004 and 0.9990, respectively (Figure 2), indicating excellent linear relationship between templates and positive droplets.
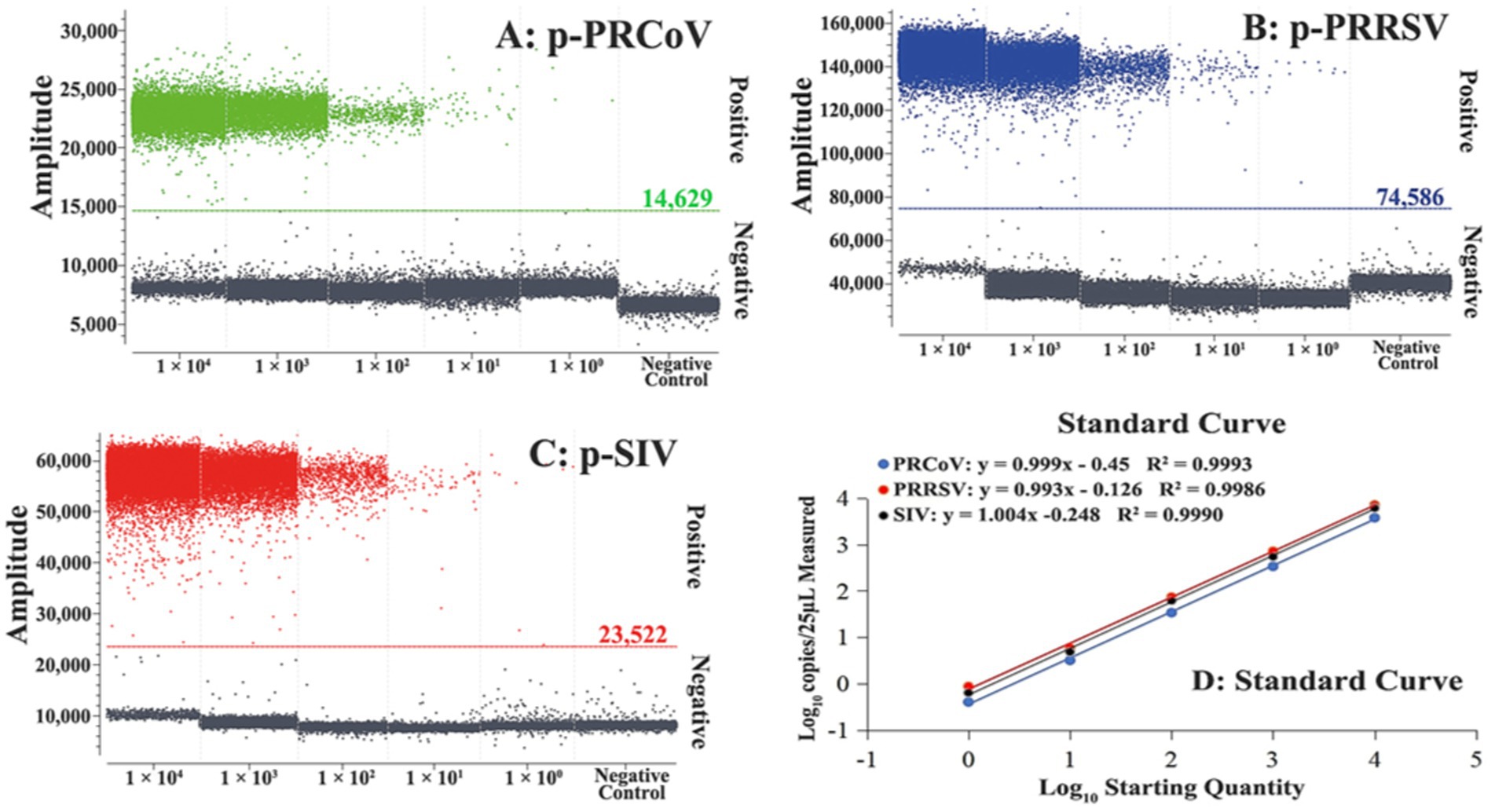
Figure 2. Generation of standard curves of the triplex cdPCR. The amplification plots of different final reaction concentrations (from 1 × 104 to 1 × 100 copies/μL) of plasmid constructs p-PRCoV (A), p-PRRSV (B), and p-SIV (C) and their standard curves (D) are shown.
The specificity of the triplex cdPCR was assessed using the extracted nucleic acids (RNA or DNA) of PRCoV, PRRSV, SIV, TGEV, PEDV, PoRV, ASFV, FMDV, PCV2, PDCoV, and CSFV as templates. The results showed that the triplex cdPCR could specifically detect PRCoV, PRRSV, and SIV, and there was no cross-reaction with other control swine viruses (Figure 3), indicating excellent specificity of the developed triplex cdPCR.
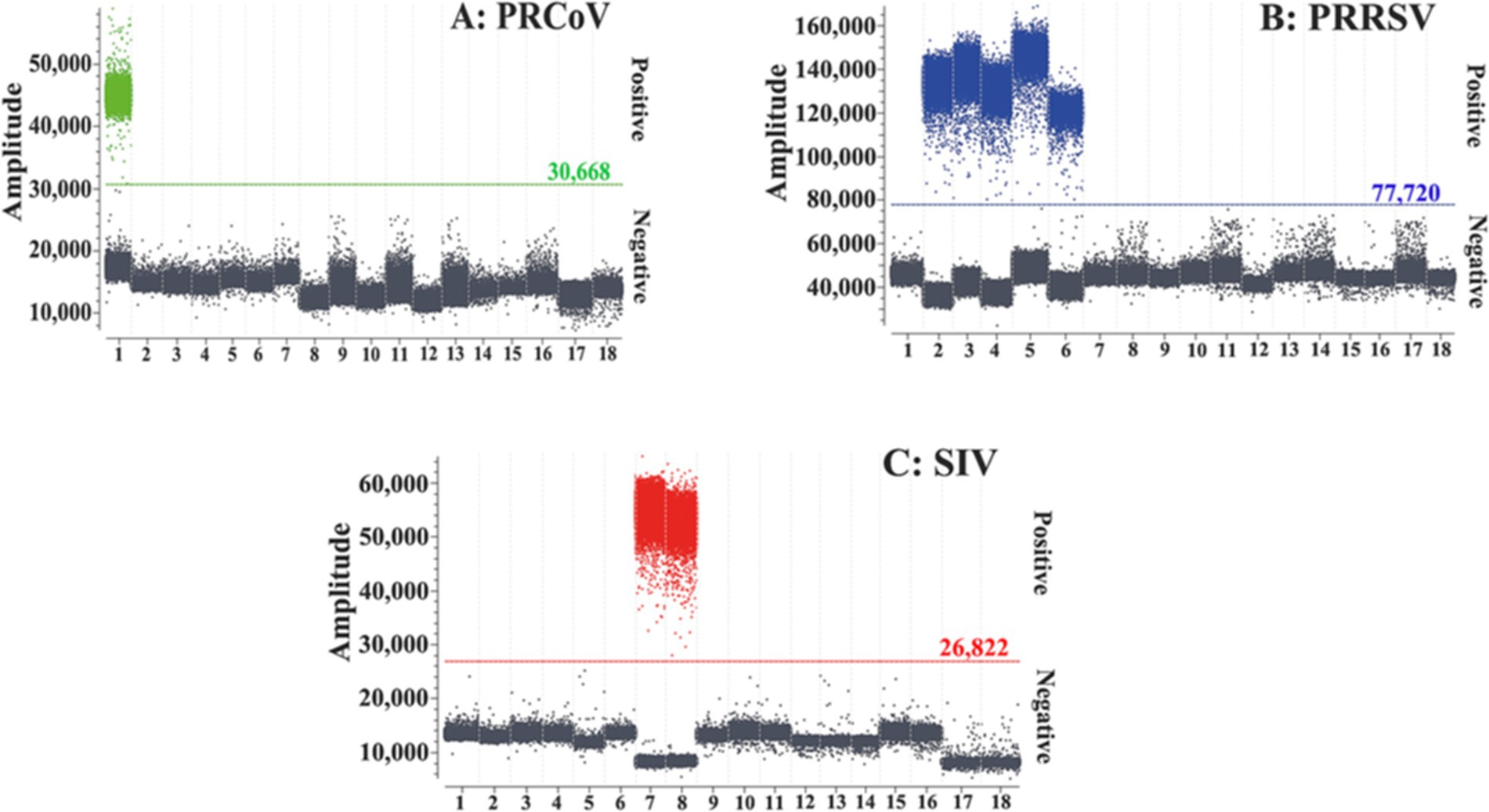
Figure 3. Specificity of the triplex cdPCR for PRCoV (A), PRRSV (B), and SIV (C). 1: p-PRCoV; 2: p-PRRSV; 3: PRRSV; 4: PRRSV CH-1R strain; 5: PRRSV R98 strain; 6: PRRSV HuN4-F112 strain; 7: p-SIV; 8: SIV TJ strain; 9: TGEV H strain; 10: PEDV CV777 strain; 11: PoRV G5 type-NX strain; 12: FMDV O/Mya98/XJ/2010 strain; 13: PCV2 ZJ/C strain; 14: CSFV CVCC AV1412 strain; 15: ASFV; 16: PDCoV; 17: Clinical negative sample; 18: Nuclease-free distilled water.
The mixtures of three synthesized viral RNA of PRCoV, PRRSV, and SIV from 1 × 105 to 1 × 100 copies/μL (final reaction concentration: from 1 × 104 to 1 × 10−1 copies/μL) were used as templates for evaluation of the sensitivity of the triplex cdPCR. The results basing on Poisson distribution analysis showed that the LODs of PRCoV, PRRSV, and SIV were 6.00, 5.75, and 6.00 copies/reaction, respectively (Figure 4).
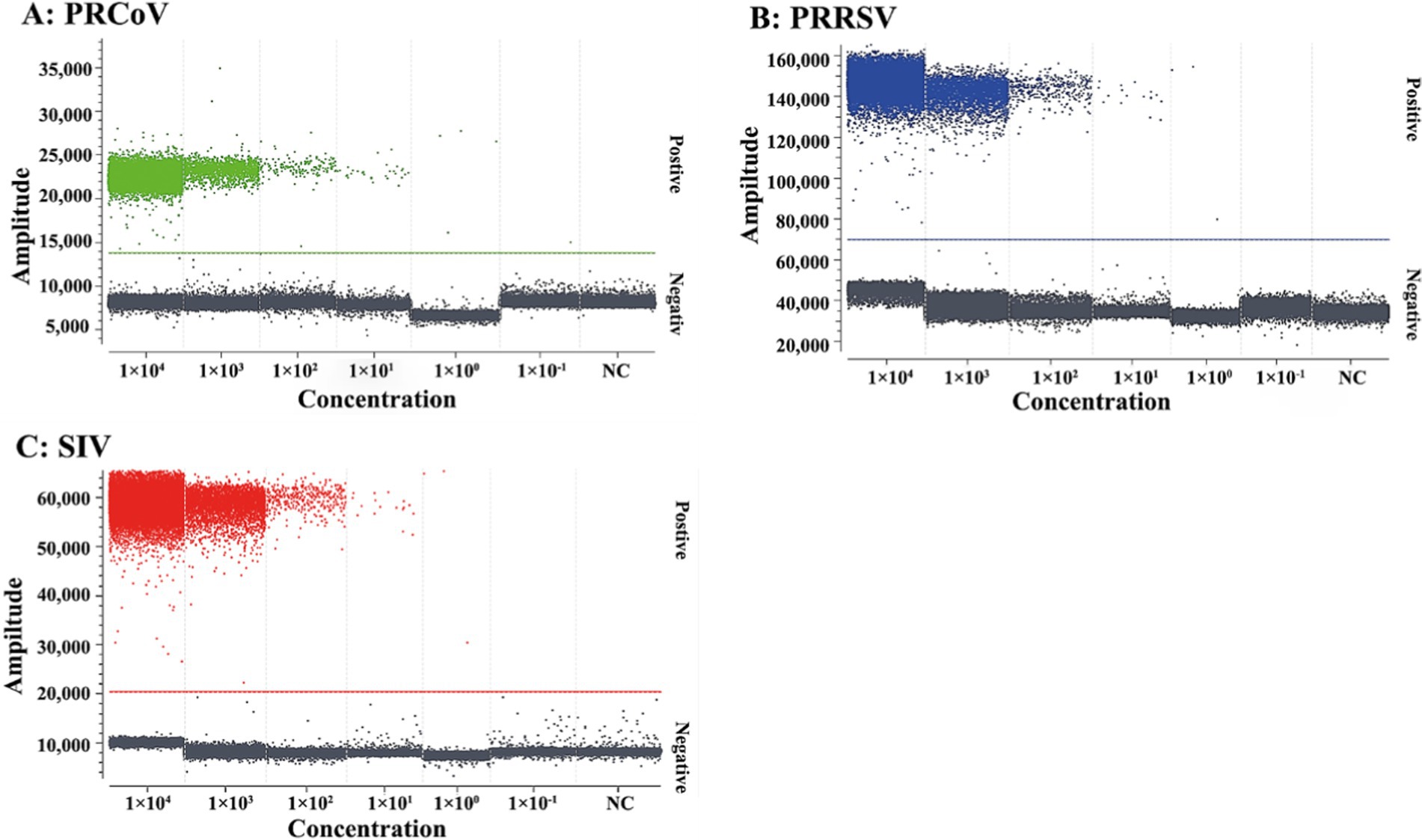
Figure 4. Sensitivity of the triplex cdPCR for PRCoV (A), PRRSV (B), and SIV (C). The final reaction concentrations of the mixtures of synthesized viral RNA of PRCoV, PRRSV, and SIV ranged from 1 × 104 to 1 × 10−1 copies/μL. NC, negative control.
In addition, the mixtures of three synthesized viral RNA of PRCoV, PRRSV, and SIV of 31.23, 15.63, 7.81, 3.91, 1.96, 0.98, 0.49 copies/reaction were used to performed triplex cdPCR, and the LODs were assessed using the Probit regression analysis. The number of positive droplets and hit rates were detected and recorded (Table 3). The LODs obtained for PRCoV, PRRSV, and SIV were 5.384, 4.822 and 5.029 copies/reaction, respectively (Figure 5).
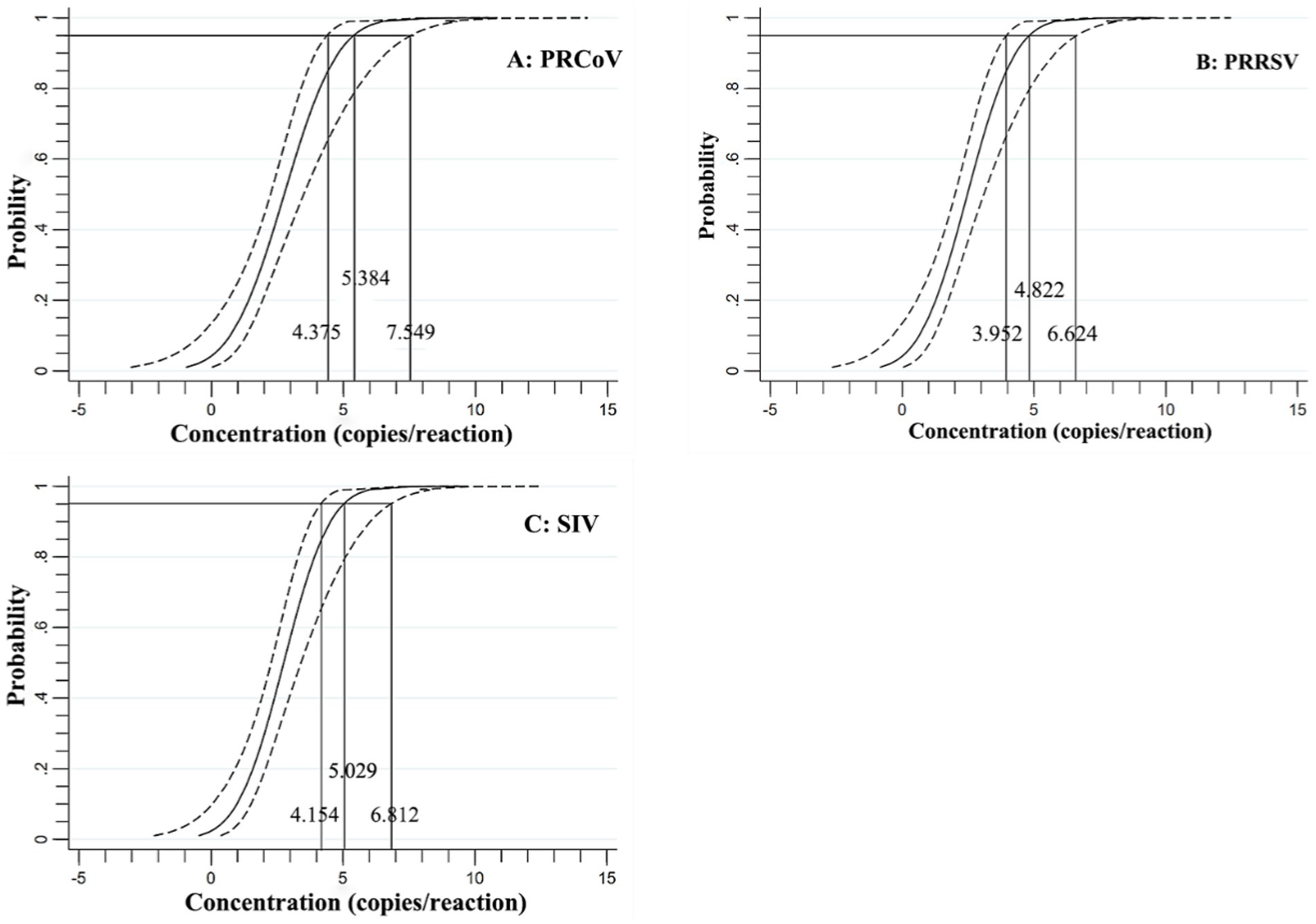
Figure 5. Specificity of the triplex cdPCR using Probit regression analysis for PRCoV (A), PRRSV (B), and SIV (C).
Repeatability of the triplex cdPCR was analyzed using the mixtures of three synthesized viral RNA of PRCoV, PRRSV, and SIV with final reaction concentration of 1 × 104, 1 × 103, and 1 × 102 copies/μL as templates. The results showed that the coefficients of variation (CVs) were 0.44–1.46% for intra-assay and 0.41–1.67% for inter-assay (Table 4), indicating excellent repeatability.
The 1,657 clinical samples collected from Guangxi Province in China from July 2023 to March 2024 were tested using the developed triplex cdPCR. The results showed that the positivity rates of PRCoV, PRRSV, and SIV were 1.15% (19/1,657), 12.79% (212/1,657), and 2.05% (34/1,657), respectively. The co-infection positivity rates of PRCoV + PRRSV + SIV, PRCoV + PRRSV, and PRRSV + SIV were 0.18% (3/1,657), 0.18% (3/1,657), and 0.12% (2/1,657), respectively. The positive samples were shown in a 3D dot-plots to display the data using three-dimensional scatterplots for direct visualization (Figure 6). The samples collected from pig farms, slaughterhouses, and harmless treatment plants showed positivity rates of 2.76, 0.56, and 3.06% for PRCoV, 8.76, 11.58, and 25.00% for PRRSV, and 3.23, 0.96, and 7.65% for SIV (Table 5).
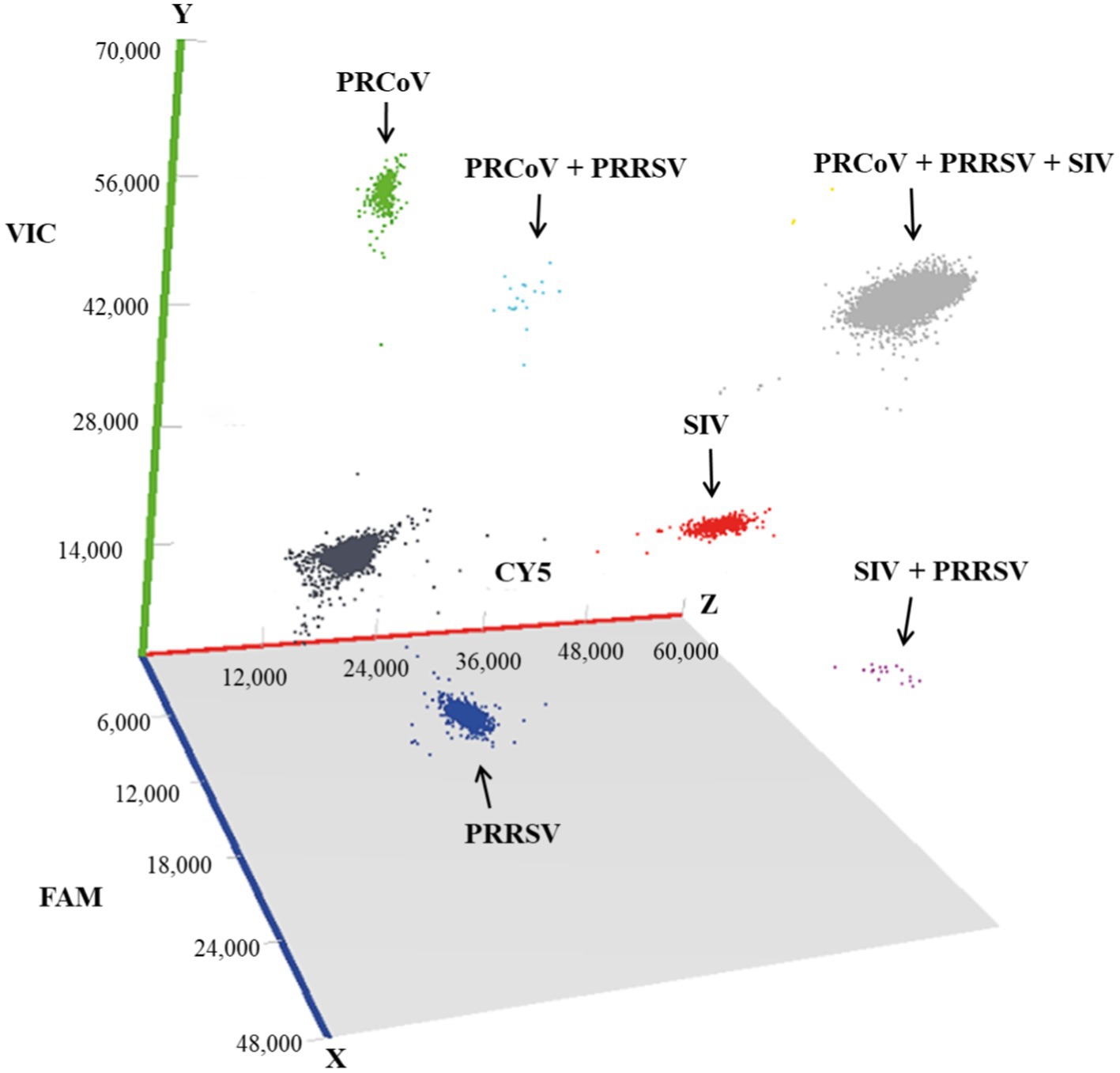
Figure 6. Test results of the clinical samples shown with 3D scatterplots. The fluorescence intensities were acquired in the Blue (x axis), Green (y axis) and Red (z axis) acquisition channels.
The 1,657 samples were also tested using the reference multiplex RT-qPCR reported by Ma et al. (12). The results showed that the positivity rates of PRCoV, PRRSV, and SIV were 0.97% (16/1,657), 12.13% (201/1,657), and 1.75% (29/1,657), respectively. Compared with the reference multiplex RT-qPCR, the diagnostic sensitivity and specificity of the developed triplex cdPCR were 100 and 99.82% for PRCoV, 100 and 99.24% for PRRSV, and 100 and 99.69% for SIV, respectively (Table 6). The compliance rates of these two methods were higher than 99.34%, and the Kappa values of PRCoV, PRRSV, and SIV were 0.913, 0.970, and 0.919, respectively (Table 7).
Digital PCR (dPCR) is a highly sensitive detection technology that can detect low concentration of target template (40, 41). The samples to be tested are diluted to form millions of droplet reaction chambers, where either one or no target template is distributed. Subsequently, PCR amplification is conducted and fluorescence signals are obtained. Finally, the absolute quantification of nucleic acid is achieved through Poisson distribution correction to obtain the copy number of nucleic acid (38, 39, 42–44). Compared with other assays, this method is able to accurately detect and quantify viral nucleic acids in samples without relying on standard curves, Ct values and amplification efficiencies. It is able to detect nucleic acids in samples with high sensitivity and accuracy, and exhibit low LOD (38–41). Therefore, dPCR has been gradually applicated in different veterinary laboratories for the detection of a variety of pathogens, which ensures the exact diagnosis of swine diseases. Unfortunately, the cdRT-PCR has the limitation of relatively high cost while compared with the RT-qPCR, but the development of multiplex cdPCR has decreased dramatically the average cost of each sample, so it has been applied in more and more laboratories now (47–49).
PRCoV, PRRSV, and SIV are important respiratory pathogens. Nowadays, these swine viruses circulate in many countries around the world (5, 8, 10, 11, 19, 20, 25–28, 30). Recently, these pathogens have been reported in many provinces in China, indicating that these pathogens are still circulating in many pig herds (12, 13, 21, 22, 27, 28, 30). They cause similar clinical respiratory symptoms of fever, cough, runny nose, and respiratory distress, and similar pathological damage of pneumonia. It is hard to differentially diagnose these diseases depending only on clinical signs and pathological changes. Therefore, it is necessary to establish specific, sensitive, and accurate method to detect and differentiate PRCoV, PRRSV, and SIV. To date, two dPCR assays for the detection of PRRSV (45, 46), and a triplex cdPCR assay for the detection of ASFV, CSFV, and PRRSV (47) have been reported. However, no multiplex cdPCR has been reported for the simultaneous detection and differentiation of PRCoV, PRRSV, and SIV.
In this study, three pairs of specific primers and TaqMan probes were designed for the conserved regions of PRCoV S gene, PRRSV N genes, and SIV M gene. After optimization of the optimal conditions of primer and probe concentrations, annealing temperatures, and reaction cycles, one triplex cdPCR for the simultaneous detection of PRCoV, PRRSV, and SIV was successfully developed. The assay showed strong specificity to detect PRCoV, PRRSV, and SIV, and high sensitivity to have very low LODs with about 5 copies/reaction for all three viruses, which is about 26 times higher sensitive than the corresponding multiplex RT-qPCR assay (about 130 copies/reaction) (12). In addition, basing on Poisson distribution analysis, the LODs of the individual cdPCR for PRCoV, PRRSV, and SIV in this study were 3.25, 3.25, and 3.25 copies/reaction, respectively, which were slightly higher sensitive than those of the corresponding triplex cdPCR. The assay had excellent repeatability with intra- and inter-assay CVs less than 2% (Table 4). The assay was used to test 1,657 clinical samples to evaluate its applicability, and showed diagnostic sensitivity and specificity higher than 99.24% for three viruses (Table 6). The assay had coincidence rate with the reference multiplex RT-qPCR higher than 99.34%, and the Kappa values were higher than 0.913 (Table 7). The results showed that the detection rates of PRCoV, PRRSV, and SIV were 1.15, 12.79, and 2.05% by the triplex cdPCR, and 0.97, 12.13, and 1.75% by the multiplex RT-qPCR. These results showed that the triplex cdPCR has a higher detection rate and it can be effectively applied in the clinical samples with low concentration of nucleic acid. These results indicated that a specific, sensitive, accurate, and reliable triplex cdPCR was established for the simultaneous and differential detection of PRCoV, PRRSV, and SIV.
The 1,657 clinical samples from Guangxi Province during 2023–2024 were tested using the developed triplex cdPCR, and the positivity rates of PRCoV, PRRSV, and SIV were 1.15, 12.79, and 2.05%, respectively. The results indicated that PRCoV, PRRSV, and SIV were still circulating in some pig herds in Guangxi Province, southern China. According to our previous surveillance, the positivity rates of PRRSV in Guangxi province from 2018 to 2023 ranged from 8.64 to 22.13% (unpublished data). The overall positivity rates of PRRSV in South China during 2017–2021 were 18.82% (1,279/6,795), ranging from 4.92 to 25% positivity rates (50). The suspected samples collected from Sichuan Province of China from 2021 to 2023 showed a prevalence rate of 39.74% (643/1,618) for PRRSV, with PRRSV-2 as dominant (95.65%, 615/643) and PRRSV-1 as minor (4.35%, 28/643) (51). The 7,518 samples collected from 100 intensive pig farms in 21 provinces in China during 2021–2022 showed a positivity rate of 32.1% (2,416/7,518) for PRRSV (52). At present, PRRSV-1 (European species) and PRRSV-2 (American species) are circulating in China (21, 51, 53), which increases the complexity and difficulty of prevention and control of PRRS. The surveillance of SIV in several regions of China during 2016–2021 found the highest positivity rate of Eurasian Avian-like (EA) H1N1 in pig farms, indicating that EA H1N1 has become an endemic subtype in the farm (54), and the EA H1N1 SIV was undergoing mutations and some of its genes were rearranged (55). In addition, SIV infection in humans has been reported (25, 29, 56, 57), which demonstrates the zoonotic potential of SIV strains and the importance of SIV surveillance. Compared with PRRSV and SIV, there has fewer reports on PRCoV at home and abroad until now. The 4,909 clinical specimens collected from Guangxi Province in China during 2022–2023 showed a positivity rate of 1.36% (67/4,909) for PRCoV (12). Two PRCV positive samples were found in the 7,645 samples from clinically healthy pigs at abattoirs in 13 provinces of China in 2017 (13). Further surveillance of PRCoV is necessary to take sufficient data on the endemic of PRCoV in China. Since the common prevalence of PRCoV, PRRSV, and SIV worldwide, the developed triplex cdPCR can be used as an important usefully tool to perform investigation and epidemiology of these pathogens, especially for the samples with very low viral loads.
Especially, the results showed the co-infection positivity rates of PRCoV + PRRSV + SIV, PRCoV + PRRSV, and PRRSV + SIV in these 1,657 samples were 0.18, 0.18, and 0.12%, respectively, which indicated that there were co-infections of two or three pathogens in pig herds. Co-infections of these viruses exacerbate the diseases (17, 31–35), which may be one of the reasons for the increased morbidity rate of pig herd. So, it is necessary to strengthen the surveillance of PRCoV, PRRSV, and SIV, to evaluate the epidemic situation of these pathogens, and to prevent and control these diseases effectively.
At present, PRCoV, PRRSV, and SIV are common prevalent in pig herds in China and other countries, and they induce similar clinical signs and pathological changes. Multiplex cdPCR assay is an important technical method for the detection of several pathogens in one reaction. The multiplex cdPCR assay established in this study provides a new technical tool for the clinical detection and differentiation of PRCoV, PRRSV, and SIV, which is beneficial to specially detect the existence of pathogens with very low concentration, and exactly diagnose the diseases. In addition, the specific primers and probes used in this study were designed for the conserved regions basing on the sequence alignment of the viral strains from different countries around the world, so the developed triplex cdPCR is suit for detection of the viral strains of PRCoV, PRRSV, and SIV from different countries.
A triplex cdPCR assay that can simultaneously detect and differentiate PRCoV, PRRSV, and SIV was successfully established for the first time in this study, which showed high sensitivity, strong specificity, and excellent repeatability. This assay can detect PRCoV, PRRSV, and SIV in the clinical samples with very low concentration in one reaction in a short time, and provide a powerful technical tool for rapid, and accurate detection of PRCoV, PRRSV, and SIV.
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
The animal studies were approved by the Guangxi Center for Animal Disease Control and Prevention. The studies were conducted in accordance with the local legislation and institutional requirements. Written informed consent was obtained from the owners for the participation of their animals in this study.
YS: Writing – original draft, Data curation, Methodology. KS: Writing – original draft, Funding acquisition, Writing – review & editing. YM: Methodology, Writing – original draft. YY: Investigation, Validation, Writing – original draft. FL: Data curation, Methodology, Supervision, Writing – original draft. SF: Software, Supervision, Validation, Writing – original draft. MM: Investigation, Validation, Writing – original draft. JH: Methodology, Project administration, Writing – original draft. ZW: Funding acquisition, Writing – original draft, Writing – review & editing.
The author(s) declare that financial support was received for the research and/or publication of this article. This work was supported by the Guangxi Key Laboratory of Animal Breeding, Disease Control and Prevention (Grant No. ABDC-b202304), and the Key Research and Development Program (Grant No. AB21238003) of Guangxi Science and Technology Bureau, China.
YS was employed by the Nanning Kedi Biotechnology Co., Ltd.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The authors declare that no Gen AI was used in the creation of this manuscript.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
1. Oba, P, Wieland, B, Mwiine, FN, Erume, J, Gertzell, E, Jacobson, M, et al. Status and gaps of research on respiratory disease pathogens of swine in Africa. Porcine Health Manag. (2020) 6:5. doi: 10.1186/s40813-020-0144-7
2. Jiang, C, Li, L, Xue, M, Zhao, L, Liu, X, Wang, W, et al. Long-term expanding porcine airway organoids provide insights into the pathogenesis and innate immunity of porcine respiratory coronavirus infection. J Virol. (2022) 96:e0073822. doi: 10.1128/jvi.00738-22
3. Turlewicz-Podbielska, H, and Pomorska-Mól, M. Porcine coronaviruses: overview of the state of the art. Virol Sin. (2021) 36:833–51. doi: 10.1007/s12250-021-00364-0
4. Boeters, M, Garcia-Morante, B, van Schaik, G, Segalés, J, Rushton, J, and Steeneveld, W. The economic impact of endemic respiratory disease in pigs and related interventions - a systematic review. Porcine Health Manag. (2023) 9:45. doi: 10.1186/s40813-023-00342-w
5. Woo, PCY, de Groot, RJ, Haagmans, B, Lau, SKP, Neuman, BW, Perlman, S, et al. ICTV virus taxonomy profile: Coronaviridae 2023. J Gen Virol. (2023) 104:001843. doi: 10.1099/jgv.0.001843
6. Pensaert, M, Callebaut, P, and Vergote, J. Isolation of a porcine respiratory, non-enteric coronavirus related to transmissible gastroenteritis. J Vet Q. (1986) 8:257–61. doi: 10.1080/01652176.1986.9694050
7. Jung, K, Alekseev, KP, Zhang, X, Cheon, DS, Vlasova, AN, and Saif, LJ. Altered pathogenesis of porcine respiratory coronavirus in pigs due to immunosuppressive effects of dexamethasone: implications for corticosteroid use in treatment of severe acute respiratory syndrome coronavirus. J Virol. (2007) 81:13681–93. doi: 10.1128/JVI.01702-07
8. Halbur, PG, Pallarés, FJ, Opriessnig, T, Vaughn, EM, and Paul, PS. Pathogenicity of three isolates of porcine respiratory coronavirus in the USA. Vet Rec. (2003) 152:358–61. doi: 10.1136/vr.152.12.358
9. Vannier, P. Disorders induced by the experimental infection of pigs with the porcine respiratory coronavirus (P.R.C.V.). Zentralbl Veterinarmed B. (1990) 37:177–80. doi: 10.1111/j.1439-0450.1990.tb01043.x
10. Bedsted, AE, Rasmussen, TB, Martinenghi, LD, Bøtner, A, Nauwynck, H, and Belsham, GJ. Porcine respiratory coronavirus genome sequences; comparisons and relationships to transmissible gastroenteritis viruses. Virology. (2024) 595:110072. doi: 10.1016/j.virol.2024.110072
11. Antas, M, and Olech, M. First report of transmissible gastroenteritis virus (TGEV) and porcine respiratory coronavirus (PRCV) in pigs from Poland. BMC Vet Res. (2024) 20:517. doi: 10.1186/s12917-024-04364-6
12. Ma, Y, Shi, K, Chen, Z, Shi, Y, Zhou, Q, Mo, S, et al. Simultaneous detection of porcine respiratory coronavirus, porcine reproductive and respiratory syndrome virus, swine influenza virus, and pseudorabies virus via quadruplex one-step RT-qPCR. Pathogens. (2024) 13:341. doi: 10.3390/pathogens13040341
13. Sun, W, Shi, Z, Wang, P, Zhao, B, Li, J, Wei, X, et al. Metavirome analysis reveals a high prevalence of porcine hemagglutination encephalomyelitis virus in clinically healthy pigs in China. Pathogens. (2023) 12:510. doi: 10.3390/pathogens12040510
14. Brinton, MA, Gulyaeva, AA, Balasuriya, UBR, Dunowska, M, Faaberg, KS, Goldberg, T, et al. ICTV virus taxonomy profile: Arteriviridae 2021. J Gen Virol. (2021) 102:001632. doi: 10.1099/jgv.0.001632
15. Nelsen, CJ, Murtaugh, MP, and Faaberg, KS. Porcine reproductive and respiratory syndrome virus comparison: divergent evolution on two continents. J Virol. (1999) 73:270–80. doi: 10.1128/JVI.73.1.270-280.1999
16. Forsberg, R, Storgaard, T, Nielsen, HS, Oleksiewicz, MB, Cordioli, P, Sala, G, et al. The genetic diversity of European type PRRSV is similar to that of the north American type but is geographically skewed within Europe. Virology. (2002) 299:38–47. doi: 10.1006/viro.2002.1450
17. Jung, K, Renukaradhya, GJ, Alekseev, KP, Fang, Y, Tang, Y, and Saif, LJ. Porcine reproductive and respiratory syndrome virus modifies innate immunity and alters disease outcome in pigs subsequently infected with porcine respiratory coronavirus: implications for respiratory viral co-infections. J Gen Virol. (2009) 90:2713–23. doi: 10.1099/vir.0.014001-0
18. Chen, XX, Qiao, S, Li, R, Wang, J, Li, X, and Zhang, G. Evasion strategies of porcine reproductive and respiratory syndrome virus. Front Microbiol. (2023) 14:1140449. doi: 10.3389/fmicb.2023.1140449
19. Neumann, EJ, Kliebenstein, JB, Johnson, CD, Mabry, JW, Bush, EJ, Seitzinger, AH, et al. Assessment of the economic impact of porcine reproductive and respiratory syndrome on swine production in the United States. J Am Vet Med Assoc. (2005) 227:385–92. doi: 10.2460/javma.2005.227.385
20. Han, J, Zhou, L, Ge, X, Guo, X, and Yang, H. Pathogenesis and control of the Chinese highly pathogenic porcine reproductive and respiratory syndrome virus. Vet Microbiol. (2017) 209:30–47. doi: 10.1016/j.vetmic.2017.02.020
21. Sun, Q, Xu, H, An, T, Cai, X, Tian, Z, and Zhang, H. Recent progress in studies of porcine reproductive and respiratory syndrome virus 1 in China. Viruses. (2023) 15:1528. doi: 10.3390/v15071528
22. Zhou, L, Han, J, and Yang, H. The evolution and diversity of porcine reproductive and respiratory syndrome virus in China. Vet Microbiol. (2024) 298:110252. doi: 10.1016/j.vetmic.2024.110252
23. Kuhn, JH, Abe, J, Adkins, S, Alkhovsky, SV, Avšič-Županc, T, Ayllón, MA, et al. Annual (2023) taxonomic update of RNA-directed RNA polymerase-encoding negative-sense RNA viruses (realm Riboviria: kingdom Orthornavirae: phylum Negarnaviricota). J Gen Virol. (2023) 104:001864. doi: 10.1099/jgv.0.001864
24. Ma, W. Swine influenza virus: current status and challenge. Virus Res. (2020) 288:198118. doi: 10.1016/j.virusres.2020.198118
25. Kuntz-Simon, G, and Madec, F. Genetic and antigenic evolution of swine influenza viruses in Europe and evaluation of their zoonotic potential. Zoonoses Public Health. (2009) 56:310–25. doi: 10.1111/j.1863-2378.2009.01236.x
26. Chauhan, RP, and Gordon, ML. A systematic review analyzing the prevalence and circulation of influenza viruses in swine population worldwide. Pathogens. (2020) 9:355. doi: 10.3390/pathogens9050355
27. Kong, W, Ye, J, Guan, S, Liu, J, and Pu, J. Epidemic status of swine influenza virus in China. Indian J Microbiol. (2014) 54:3–11. doi: 10.1007/s12088-013-0419-7
28. Cai, M, Wei, Z, Hu, X, Ji, Y, Li, S, Huang, J, et al. The evolution, complexity, and diversity of swine influenza viruses in China: a hidden public health threat. Virology. (2024) 598:110167. doi: 10.1016/j.virol.2024.110167
29. Cui, X, Ma, J, Pang, Z, Chi, L, Mai, C, Liu, H, et al. The evolution, pathogenicity and transmissibility of quadruple reassortant H1N2 swine influenza virus in China: a potential threat to public health. Virol Sin. (2024) 39:205–17. doi: 10.1016/j.virs.2024.02.002
30. Zhao, Y, Han, L, Sang, H, Yang, P, Hou, Y, and Xiao, Y. Two genotypes of H3N2 swine influenza viruses identified in pigs from Shandong Province, China. Front Cell Infect Microbiol. (2024) 14:1517023. doi: 10.3389/fcimb.2024.1517023
31. Peng, JY, Shin, DL, Li, G, Wu, NH, and Herrler, G. Time-dependent viral interference between influenza virus and coronavirus in the infection of differentiated porcine airway epithelial cells. Virulence. (2021) 12:1111–21. doi: 10.1080/21505594.2021.1911148
32. Van Reeth, K, and Pensaert, MB. Porcine respiratory coronavirus-mediated interference against influenza virus replication in the respiratory tract of feeder pigs. Am J Vet Res. (1994) 55:1275–81. doi: 10.2460/ajvr.1994.55.09.1275
33. Van Reeth, K, Nauwynck, H, and Pensaert, M. Dual infections of feeder pigs with porcine reproductive and respiratory syndrome virus followed by porcine respiratory coronavirus or swine influenza virus: a clinical and virological study. Vet Microbiol. (1996) 48:325–35. doi: 10.1016/0378-1135(95)00145-x
34. Niazi, AM, ZiHeng, Z, Fuke, N, Toyama, K, Habibi, WA, Kawaguchi, N, et al. Detection of swine influenza a and porcine reproductive and respiratory syndrome viruses in nasopharynx-associated lymphoid tissue. J Comp Pathol. (2022) 197:23–34. doi: 10.1016/j.jcpa.2022.06.006
35. Kaden, V, Lange, E, Hänel, A, Hlinak, A, Mewes, L, Hergarten, G, et al. Retrospective serological survey on selected viral pathogens in wild boar populations in Germany. Eur J Wildl Res. (2009) 55:153. doi: 10.1007/s10344-008-0229-0
36. Watzinger, F, Ebner, K, and Lion, T. Detection and monitoring of virus infections by real-time PCR. Mol Asp Med. (2006) 27:254–98. doi: 10.1016/j.mam.2005.12.001
37. Gutiérrez-Aguirre, I, Rački, N, Dreo, T, and Ravnikar, M. Droplet digital PCR for absolute quantification of pathogens. Methods Mol Biol. (2015) 1302:331–47. doi: 10.1007/978-1-4939-2620-6_24
38. Kojabad, AA, Farzanehpour, M, Galeh, HEG, Dorostkar, R, Jafarpour, A, Bolandian, M, et al. Droplet digital PCR of viral DNA/RNA, current progress, challenges, and future perspectives. J Med Virol. (2021) 93:4182–97. doi: 10.1002/jmv.26846
39. Hudecova, I. Digital PCR analysis of circulating nucleic acids. Clin Biochem. (2015) 48:948–56. doi: 10.1016/j.clinbiochem.2015.03.015
40. Belmonte, FR, Martin, JL, Frescura, K, Damas, J, Pereira, F, Tarnopolsky, MA, et al. Digital PCR methods improve detection sensitivity and measurement precision of low abundance mtDNA deletions. Sci Rep. (2016) 6:25186. doi: 10.1038/srep25186
41. Hindson, BJ, Ness, KD, Masquelier, DA, Belgrader, P, Heredia, NJ, Makarewicz, AJ, et al. High-throughput droplet digital PCR system for absolute quantitation of DNA copy number. Anal Chem. (2011) 83:8604–10. doi: 10.1021/ac202028g
42. Tan, LL, Loganathan, N, Agarwalla, S, Yang, C, Yuan, W, Zeng, J, et al. Current commercial dPCR platforms: technology and market review. Crit Rev Biotechnol. (2023) 43:433–64. doi: 10.1080/07388551.2022.2037503
43. Madic, J, Zocevic, A, Senlis, V, Fradet, E, Andre, B, Muller, S, et al. Three-color crystal digital PCR. Biomol Detect Quantif. (2016) 10:34–46. doi: 10.1016/j.bdq.2016.10.002
44. Li, H, Bai, R, Zhao, Z, Tao, L, Ma, M, Ji, Z, et al. Application of droplet digital PCR to detect the pathogens of infectious diseases. Biosci Rep. (2018) 38:BSR20181170. doi: 10.1042/BSR20181170
45. Yang, Q, Xi, J, Chen, X, Hu, S, Chen, N, Qiao, S, et al. The development of a sensitive droplet digital PCR for quantitative detection of porcine reproductive and respiratory syndrome virus. Int J Biol Macromol. (2017) 104:1223–8. doi: 10.1016/j.ijbiomac.2017.06.115
46. Long, F, Chen, Y, Shi, K, Yin, Y, Feng, S, and Si, H. Development of a multiplex crystal digital RT-PCR for differential detection of classical, highly pathogenic, and NADC30-like porcine reproductive and respiratory syndrome virus. Animals. (2023) 13:594. doi: 10.3390/ani13040594
47. Shi, K, Chen, Y, Yin, Y, Long, F, Feng, S, Liu, H, et al. A multiplex crystal digital PCR for detection of African swine fever virus, classical swine fever virus, and porcine reproductive and respiratory syndrome virus. Front Vet Sci. (2022) 9:926881. doi: 10.3389/fvets.2022.926881
48. Shi, K, Qian, X, Shi, Y, Wei, H, Pan, Y, Long, F, et al. A triplex crystal digital PCR for the detection of genotypes I and II African swine fever virus. Front Vet Sci. (2024) 11:1351596. doi: 10.3389/fvets.2024.1351596
49. Shi, K, Hu, X, Yin, Y, Shi, Y, Pan, Y, Long, F, et al. Development of a triplex crystal digital RT-PCR for the detection of PHEV, PRV, and CSFV. Front Vet Sci. (2024) 11:1462880. doi: 10.3389/fvets.2024.1462880
50. Fang, K, Liu, S, Li, X, Chen, H, and Qian, P. Epidemiological and genetic characteristics of porcine reproductive and respiratory syndrome virus in South China between 2017 and 2021. Front Vet Sci. (2022) 9:853044. doi: 10.3389/fvets.2022.853044
51. Huang, B, Xu, T, Luo, Z, Deng, L, Jian, Z, Lai, S, et al. Prevalence and genetic diversity of PRRSV in Sichuan province of China from 2021 to 2023: evidence of an ongoing epidemic transition. Virology. (2024) 600:110213. doi: 10.1016/j.virol.2024.110213
52. Li, C, Fan, A, Liu, Z, Wang, G, Zhou, L, Zhang, H, et al. Prevalence, time of infection, and diversity of porcine reproductive and respiratory syndrome virus in China. Viruses. (2024) 16:774. doi: 10.3390/v16050774
53. Guo, Z, Chen, XX, Li, R, Qiao, S, and Zhang, G. The prevalent status and genetic diversity of porcine reproductive and respiratory syndrome virus in China: a molecular epidemiological perspective. Virol J. (2018) 15:2. doi: 10.1186/s12985-017-0910-6
54. Zhao, Y, Han, L, Chen, T, Sang, H, Ding, G, Li, Y, et al. Serological surveillance of the H1N1 and H3N2 swine influenza A virus in Chinese swine between 2016 and 2021. Biomed Res Int. (2022) 2022:5833769. doi: 10.1155/2022/5833769
55. Li, H, Leng, H, Tang, S, Su, C, Xu, Y, Wang, Y, et al. Prevalence, genetics and evolutionary properties of Eurasian avian-like H1N1 swine influenza viruses in Liaoning. Viruses. (2022) 14:643. doi: 10.3390/v14030643
56. Deng, YM, Wong, FYK, Spirason, N, Kaye, M, Beazley, R, Grau, MLL, et al. Locally acquired human infection with swine-origin influenza A (H3N2) variant virus, Australia, 2018. Emerg Infect Dis. (2020) 26:143–7. doi: 10.3201/eid2601.191144
Keywords: porcine respiratory coronavirus, porcine reproductive and respiratory syndrome virus, swine influenza virus, triplex crystal digital PCR, co-infection
Citation: Shi Y, Shi K, Ma Y, Yin Y, Long F, Feng S, Mo M, He J and Wei Z (2025) Development of a triplex crystal digital PCR for the detection of PRCoV, PRRSV, and SIV. Front. Vet. Sci. 12:1562444. doi: 10.3389/fvets.2025.1562444
Received: 17 January 2025; Accepted: 13 March 2025;
Published: 31 March 2025.
Edited by:
Simon Paul Graham, The Pirbright Institute, United KingdomReviewed by:
Jean-Pierre Frossard, Animal and Plant Health Agency (United Kingdom), United KingdomCopyright © 2025 Shi, Shi, Ma, Yin, Long, Feng, Mo, He and Wei. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jiakang He, amtoZUBneHUuZWR1LmNu; Zuzhang Wei, enV6aGFuZ3dlaUBneHUuZWR1LmNu
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.