
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Vet. Sci., 26 March 2025
Sec. Veterinary Infectious Diseases
Volume 12 - 2025 | https://doi.org/10.3389/fvets.2025.1561533
 Xinru Zhao1
Xinru Zhao1 Min Li1
Min Li1 Yingying Yang1
Yingying Yang1 Yidan Wang1
Yidan Wang1 Xiaoru Zheng1
Xiaoru Zheng1 Dehua Yin2
Dehua Yin2 Haihui Gao3
Haihui Gao3 Huatao Li1
Huatao Li1 Kaiqiang Fu1
Kaiqiang Fu1 Zhi Cao1*
Zhi Cao1*Introduction: Bovine rotavirus (BRV), bovine coronavirus (BCoV), Escherichia coli K99+ (E. coli K99+), and Cryptosporidium parvum (C. parvum) are the most common pathogens involved in calf production. These pathogens can cause calf diarrhea, leading to significant economic losses in the cattle farming industry. These four pathogens have similar clinical symptoms, making them difficult to distinguish. Therefore, we established a one-step quadruple TaqMan fluorescence quantitative PCR method capable of simultaneously and rapidly detecting BRV, BCoV, E. coli K99+, and C. parvum.
Methods: Specific primers and TaqMan probes were designed for the BRV VP-6 gene, BCoV N gene, E. coli K99+ K99 gene, and C. parvum 18S rRNA gene. Standard positive plasmids were constructed, and the reaction conditions of the method were optimized. The sensitivity, specificity, and repeatability of the method were validated, and clinical samples were tested.
Results: The minimum detection limits of this method for BRV, BCoV, E. coli K99+, and C. parvum were 5.8 × 101, 2.3 × 101, 4.5 × 102, and 2.6 × 101 copies/μL, respectively. The intra- and intergroup coefficients of variation were all less than 1.2%. This method has the advantages of strong specificity, reproducibility, low cost, and no cross-reaction with other bovine pathogens. Compared with the commercial reagent kit method were used to analyze clinical samples, and both the diagnostic sensitivity (DSe) and diagnostic specificity (DSp) were above 90%, with kappa values greater than 0.9.
Discussion: The one-step multiplex RT-qPCR method developed in this study for detecting BRV, BCoV, E. coli K99+, and C. parvum is expected to be an effective tool for the rapid and economical diagnosis and monitoring of diarrhoeal diseases in calves.
Neonatal calf diarrhea (NCD) is one of the most serious diseases of calves worldwide due to high mortality and stunted growth of affected animals, resulting in productivity and economic losses for cattle producers worldwide (1–3). The causes of bovine diarrhea include both infectious and noninfectious factors. Infectious factors include bacteria, viruses, parasites and other pathogens. Bovine rotavirus, Bovine coronavirus, E. coli K99+ and Cryptosporidium are the most common pathogenic factors of calf diarrhea. BRV is frequently involved in mixed infections (4, 5).
Bovine rotavirus, a member of the genus Rotavirus within the Reoviridae family and is the main cause of calf diarrhea. It causes symptoms such as diarrhea, loss of appetite, dehydration and depression. Often leading to secondary bacterial infection due to compromised immune resistance. In addition, there are potential zoonotic and economic impacts (6–8).
Bovine coronavirus, belonging to the genus Coronavirus within the Coronaviridae family, is an enveloped, linear single-stranded RNA virus. Most calves develop yellow watery diarrhea accompanied by mucus and blood clots, dehydration, hypothermia, depression, reduced food intake and electrolyte disturbance, which can lead to metabolic acidosis and hypoglycaemia (9, 10).
Bovine colibacillosis, an economically significant disease in newborn calves, is caused by enterotoxigenic Escherichia coli (ETEC). The most common strain of ETEC expresses F5 (K99) fimbriae (11), which allows the bacteria to attach to intestinal cells. It can cause severe diarrhea and rapid dehydration of newborn calves, and hinder the growth and development of calves (12).
The main Cryptosporidium species infecting cattle include C. parvum, C. bovis, C. andersoni, and C. ryanae. C. parvum mainly infects young calves, and it is the most important zoonotic disease (13–15). Clinical symptoms include malabsorption and secretory diarrhea, which usually resolve on their own within 2 weeks. Diarrhea severity can range from mild to severe, with yellowish watery or mucoid feces. Affected calves may exhibit dehydration, depression, and anorexia, depressed, and anorexic (16).
Studies on calf diarrhea in Ningxia (2021–2022) found detection rates of 50.46% for Cryptosporidium, 23.18% for BRV, 20% for E. coli K99, and 11.82% for BCoV (17); A systematic review and meta-analysis estimated the prevalence of BRV in 25 provinces of China from 1984 to 2021, with a prevalence rate of 35.7% (8,176/17,292) (18). Another study collected 1,646 cow fecal samples, with a detection rate of 34.02% for BCoV (19). A study investigated diarrheal samples from calves treated at the University of Etchiers, screening for BCoV, Cryptosporidium, E. coli K99+, and BRV. The results showed that Cryptosporidium was the most common pathogen (61.5%), followed by rotavirus (56.4%) (20). A global meta-analysis (2000–2021) on bovine C. parvum in newborn calves reported that 33.6% (1,637/6,077) of diarrheic calves were infected (21).
These pathogens cause bovine diarrhea in clinical practice, and the symptoms are similar; moreover, they are difficult to distinguish, and there are many mixed infections, so it is necessary to carry out pathogen detection for differential diagnosis to improve targeted prevention and treatment. The common detection methods for these four pathogens include PCR, ELISA and LAMP, etc. (22–27). PCR detection may require a longer time to obtain results, with complex operating steps and the need to run agarose gel electrophoresis to visualize the results. ELISA has relatively lower detection precision and sensitivity compared to molecular biology methods, and involves more experimental steps. LAMP has several disadvantages, such as complex primer design, complicated product structure, difficulty in quantification, and higher risk of false positives. In comparison, real-time quantitative PCR (qPCR) is a well-established method for the detection, quantification, and typing of different microbial agents in the areas of clinical and veterinary diagnostics and food safety (28). At present, different methods have been established to detect the above four pathogens (29–32); however, there is no rapid simultaneous detection method to distinguish among the four pathogens at the same time. Therefore, the aim of this study was to establish a one-step multiplex TaqMan fluorescent quantitative PCR method for BCoV, BRV, E. coli K99+, and C. parvum to make the laboratory diagnosis of bovine more convenient and rapid and minimize the impact of bovine diarrhoeal diseases.
The VP6 gene of representative BRV strains (JN790188.1, AF317127.1, EU873011.1, etc.), the N gene of BCoV strains (EF424617.1, OP037373.1, LC494170.1, etc.), the K99 gene of E. coli K99 + strains (JX987524.1, MF467447.1, MF467448.1, etc.), and the 18S rRNA gene of C. parvum strains (MZ892386.1, AB513858.1, MZ377024.1, etc.) were analyzed using Primer Express 5.0 software to identify conserved regions (Table 1). Primers and probes were designed based on these conserved sequences and synthesized by Sangon Biotech (Shanghai) Co., Ltd.
Four hundred clinical diarrhea samples were obtained from intensive farms in Shandong Province. The nucleic acid samples of BRV, BCoV, E. coli K99+, C. parvum, Bovine Parvovirus (BPV), Infectious Bovine Rhinotracheitis Virus (IBRV), and Bovine Nebovirus (BNeV) were obtained from field isolates and preserved at the College of Veterinary Medicine, Qingdao Agricultural University (QAU). In addition, the samples of Salmonella typhimurium (S. typhimurium) (ATCC13311) and Pasteurella multocida (P. multocida) (ATCC12948) were obtained from the College of Veterinary Medicine, Qingdao Agricultural University (QAU).
The collected feces were placed in physiological saline, and pathogenic DNA and RNA were manually extracted from clinical samples according to the instructions of the VAMNE Magnetic Pathogen DNA/RNA Kit (Vazyme, China). The extracted DNA/RNA samples were used as templates for qPCR detection.
Genomic RNA from BRV and BCoV was extracted and reverse-transcribed into cDNA using the FastKing RT Kit (TIANGEN Biotech, China). Genomic DNA from E. coli K99+ and C. parvum was also extracted. Specific primers were used to amplify the target genes using cDNA/DNA as templates, the target products were purified, the target genes were transferred into DH5α competent cells (Thermo Fisher, China) for enhanced culture, and the cells were cultured in a rocking incubator at 37°C overnight. Positive clones were selected through blue-white screening and identified as positive by PCR. Subsequently, the samples were sent to Sangon Biotech Co., Ltd. in Shanghai for sequencing. Use the nanodrop 2000 spectrophotometer (Thermo Fisher, China) to measure the concentration and calculate the copy number.
Using BRV, BCoV, E. coli K99+, and C. parvum positive plasmids as templates, 5 μL of each template was added to different reaction systems to screen for optimal primer and probe concentrations. The amplification conditions were as follows: 37°C for 2 min; 52°C for 5 min; 95°C for 1 min; 95°C for 3 s; and 60°C for 30 s. Using an orthogonal design for optimization, qPCR amplification was performed for 40 cycles with the TOROIVD® Probe 1-step RT-qPCR 5G Kit 3.0 (TOROIVD, China) on a Gentier 96R instrument (TIANLONG, China) to determine the optimal concentrations of primers and probes in the reaction system.
The four recombinant standard plasmids were serially diluted to 108–102copies/μL in a 10-fold gradient and used as templates for qPCR amplification, with ddH2O as a negative control. Amplification was performed using the CFX Opus Real-Time PCR System (BIO-RAD, China). A standard curve was generated by plotting the template concentration on the x-axis and the corresponding Ct values on the y-axis to evaluate the linear range and amplification efficiency of the assay.
The specificity of the optimized TaqMan qPCR method was validated using E. coli K99+, C. parvum, BPV, S. typhimurium, and P. multocida genomic DNA, BRV, BCoV, IBRV, and BNeV cDNA as templates, with ddH2O as a negative control.
The prepared positive plasmids were serially diluted in a 10-fold manner. Singleplex RT-qPCR amplifications were performed using positive plasmids with concentrations ranging from 104 to 101 copies/μL as templates, with ddH₂O serving as the negative control. Additionally, multiplex RT-qPCR amplifications were conducted using a mixture of positive plasmids for BRV, BCoV, E. coli K99+, and C. parvum, with copy numbers ranging from 104 to 101 as templates, and ddH₂O as the negative control.
The four recombinant plasmids were diluted, and three different concentrations 104–102 copies/μL were selected as positive templates, with nuclease-free water as the negative control. Multiplex fluorescent quantitative PCR amplification was performed within the same batch and across different batches. The coefficient of variation calculation formula CV = (standard deviation s/mean x) × 100% was used to calculate the coefficient of variation in the intrabatch and interbatch repeatability tests, and the repeatability of the method was evaluated.
A total of 400 samples of calf diarrhea collected from large-scale cattle farms in Shandong Province from December 2023 to March 2024 were detected using the proposed method to investigate the prevalence of the four pathogens in this area and the results were compared with TOROIVD® Gene Test Tube (TOROIVD®, China). DSe and DSp evaluations were conducted via the proposed method. Kappa was further used to test the consistency of the two detection methods.
The formula for calculating diagnostic specificity (DSe) and diagnostic sensitivity (DSp) is DSe = TP/(TP + FN) and DSp = TN/(TN + FP), where a represents a true positive, c represents a false-negative, b represents a false-positive, and d represents a true negative. Kappa = [Pr(a)-Pr(e)]/[1-Pr(e)], where Pr(a) represents the actual observed consistency rate and Pr(e) represents the chance consistency rate.
After the recombinant plasmid was constructed, amplified, and purified, BLAST comparison was carried out, and if the results were consistent with the target sequence, the recombinant plasmid was successfully constructed. The concentration of the recombinant plasmid was calculated, and the concentrations of BRV, BCoV, E. coli K99+ and C. parvum were adjusted to the following: 5.8 × 1010, 2.3 × 1010, 4.5 × 1010 and 2.6 × 1010 copies/μL, respectively.
The optimal primers (50 μmol/L) for BRV, BCoV, E. coli K99+ and C. parvum were 0.18 μL, 0.22 μL, 0.18 μL and 0.20 μL, and the optimal probe combinations were 0.6 μL, 0.8 μL, 0.6 μL and 0.4 μL. The reaction system consisted of 12.5 μL of 2 × 5G qPCR Buffer BB, 1.3 μL of RT–qPCR Enzyme Mix, 5 μL of template and ddH2O to bring the final volume to 25 μL.
Copy numbers 108–102 copies/μL were selected as the reaction template for qPCR amplification, and the standard curve was established with positive plasmid copy numbers as the horizontal coordinate and corresponding Ct values as the vertical coordinate (Figure 1). The BRV linear regression equation was y = −3.5389x + 40.365, and the correlation coefficient R2 = 0.9992. The linear regression equation of BCoV was y = −3.4014x + 39.549, and the correlation coefficient R2 = 0.9977. The linear regression equation of E. coli K99+ was y = −3.5225x + 40.355, and the correlation coefficient R2 = 0.9993. The linear regression equation of C. parvum was y = −3.6379x + 40.486, and the correlation coefficient R2 = 0.9978.
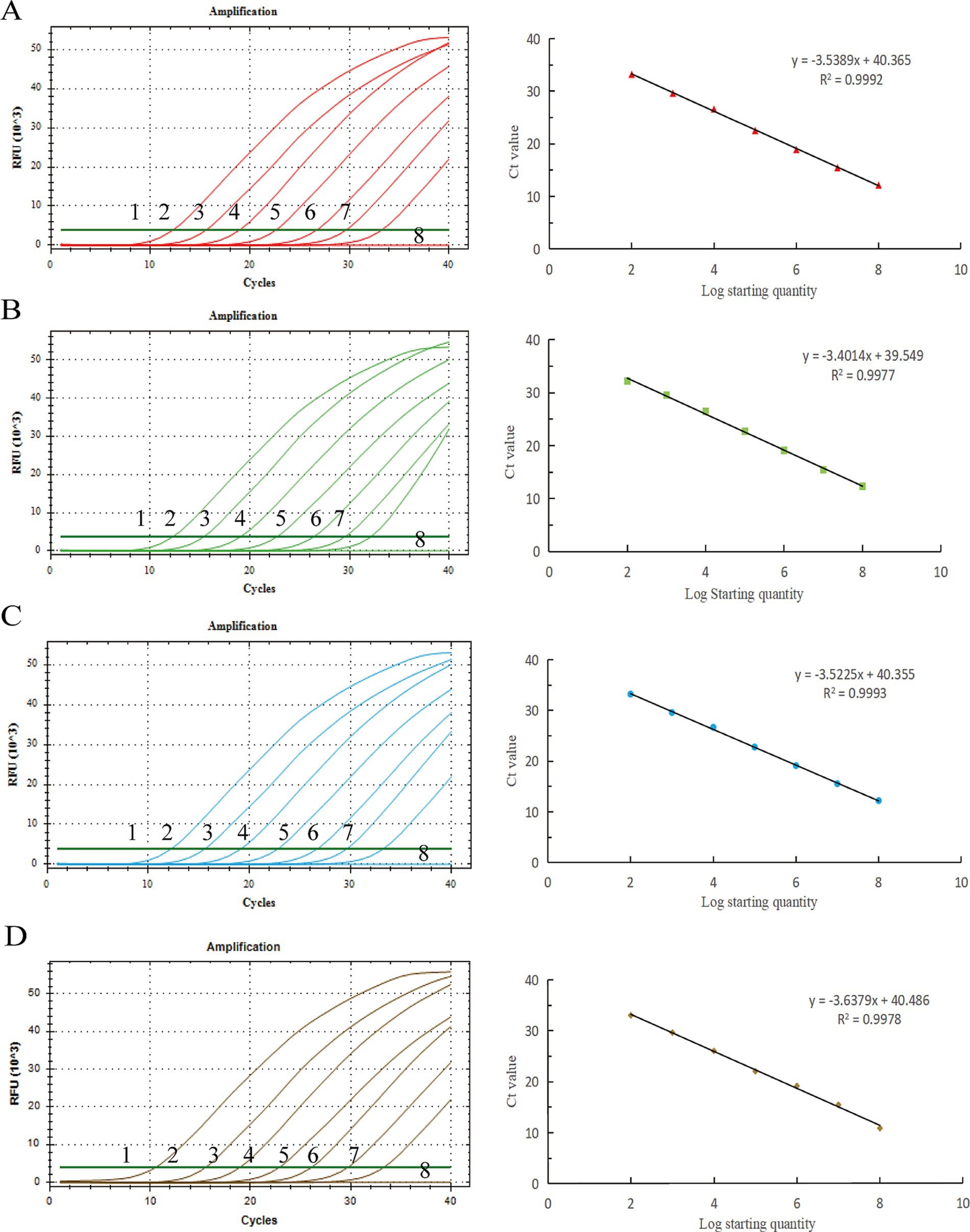
Figure 1. Standard curves of the developed TaqMan real-time PCR system for detection. X-axis: copy number; Y-axis: cycle number. (A) BRV, (B) BCoV, (C) E. coli K99+, (D) C. parvum. y: the curve from the equation, R2: correlation coefficient.
BRV, BCoV, E. coli K99+, and C. parvum all presented Ct values and corresponding amplification curves, whereas no amplification curves were found for the negative reference products, indicating that the experiment was successful (Figure 2).
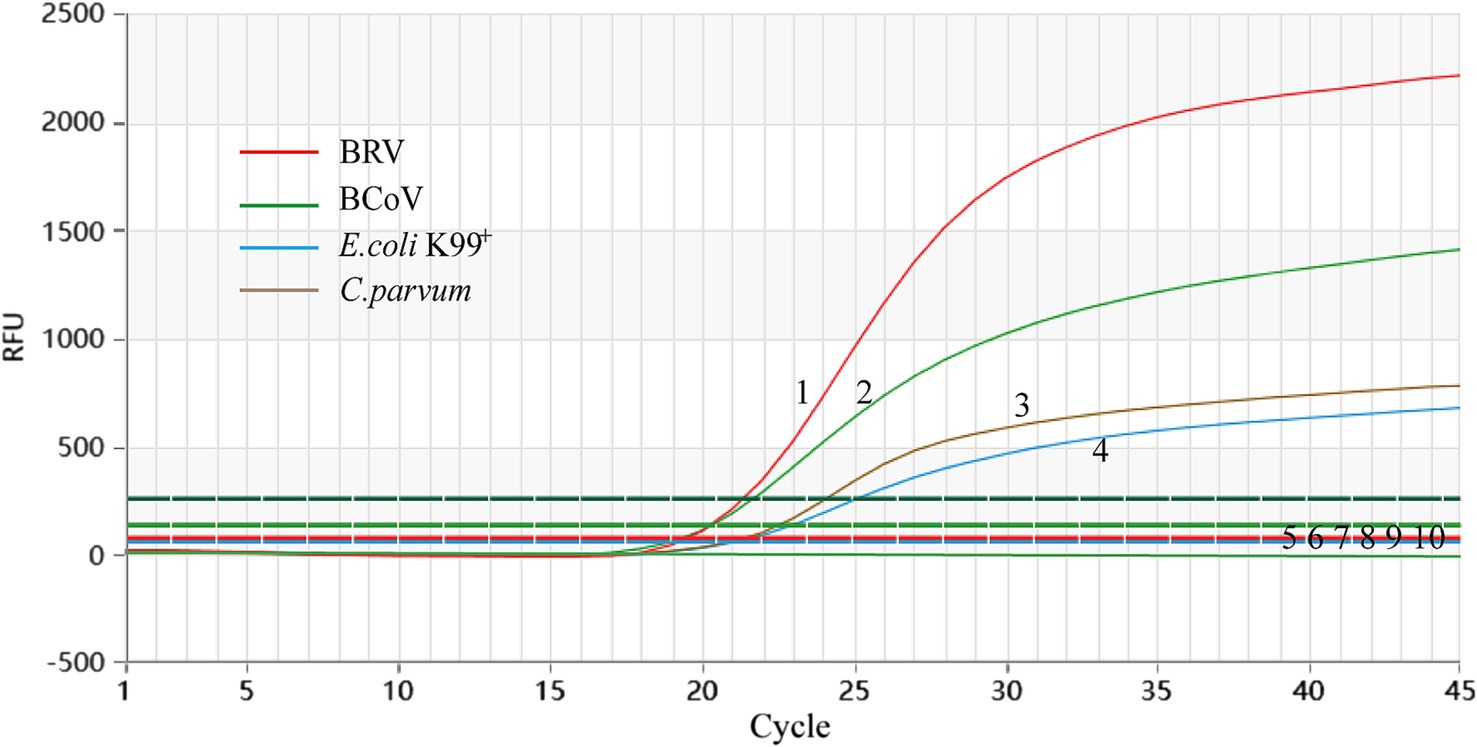
Figure 2. Analytical specificity of the primer/probe sets used in the one-step real-time RT-PCR assay. 1–4: BRV, BCoV, C. parvum, and E. coli K99+. 5–10: S. typhimurium, P. multocida, IBRV, BNeV, BPV, ddH2O.
Using a gradient dilution of standard plasmids ranging from 104 to 101 copies/μL as templates, with ddH2O as a negative control, both singleplex and multiplex TaqMan fluorescent quantitative PCR amplifications were performed. The detection limits of the established method for BRV, BCoV, E. coli K99+, and C. parvum were determined to be 5.8 × 101, 2.3 × 101, 4.5 × 102, and 2.6 × 101 copies/μL, respectively (Figure 3). The established multiplex TaqMan quantitative PCR detection method still shows good amplification curves at this plasmids concentration (Figure 4).
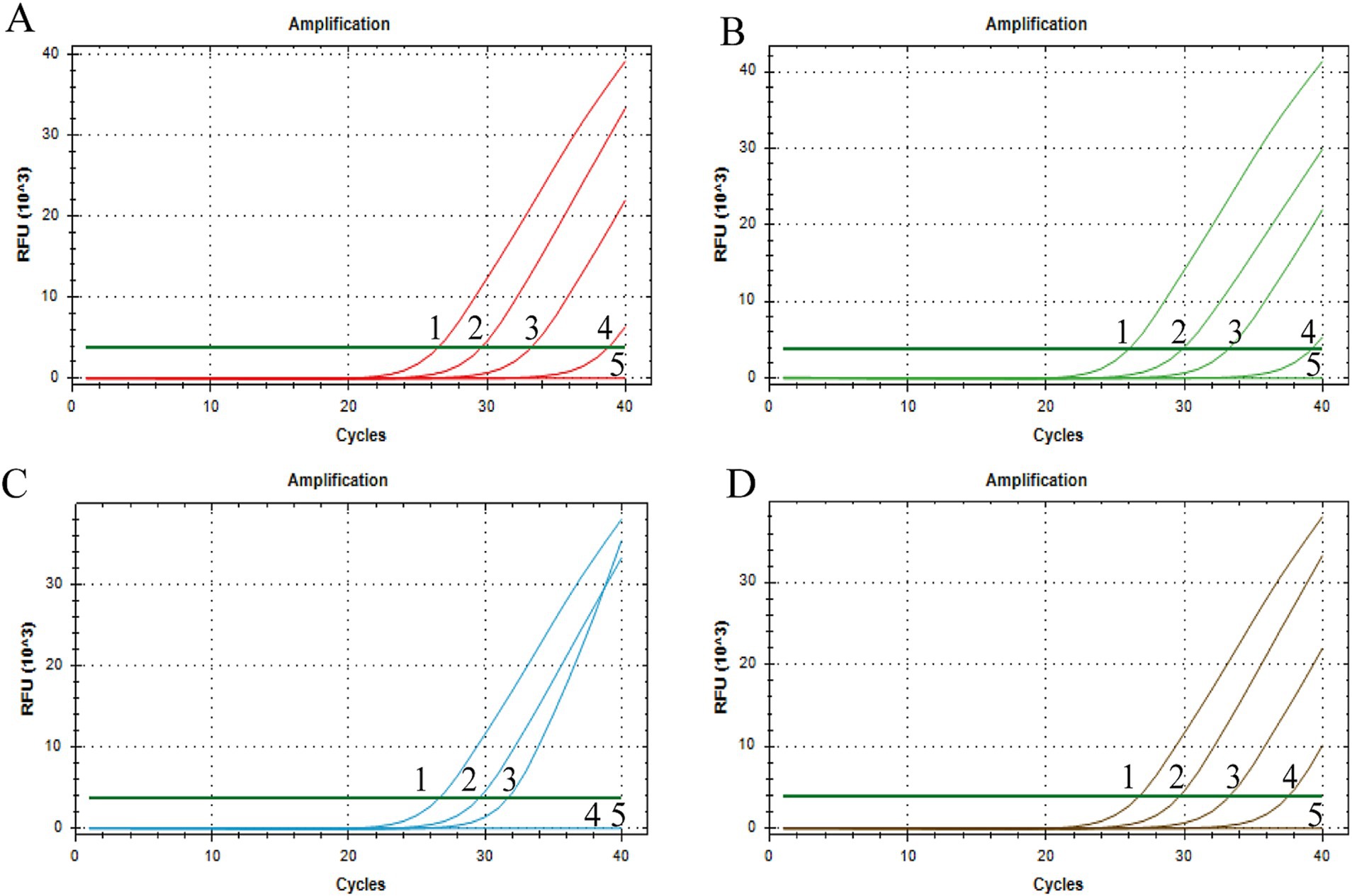
Figure 3. Sensitivity test of singleplex TaqMan Real-Time PCR Assay. X-axis: number of cycles; Y-axis: RFU; (A) BRV, (B) BCoV, (C) E. coli K99+, (D) C. parvum; 1–4: fluorescence quantitative PCR amplification curve of each standard positive recombinant plasmid from 104 to 101 copies/μL of plasmid content; 5: ddH2O.
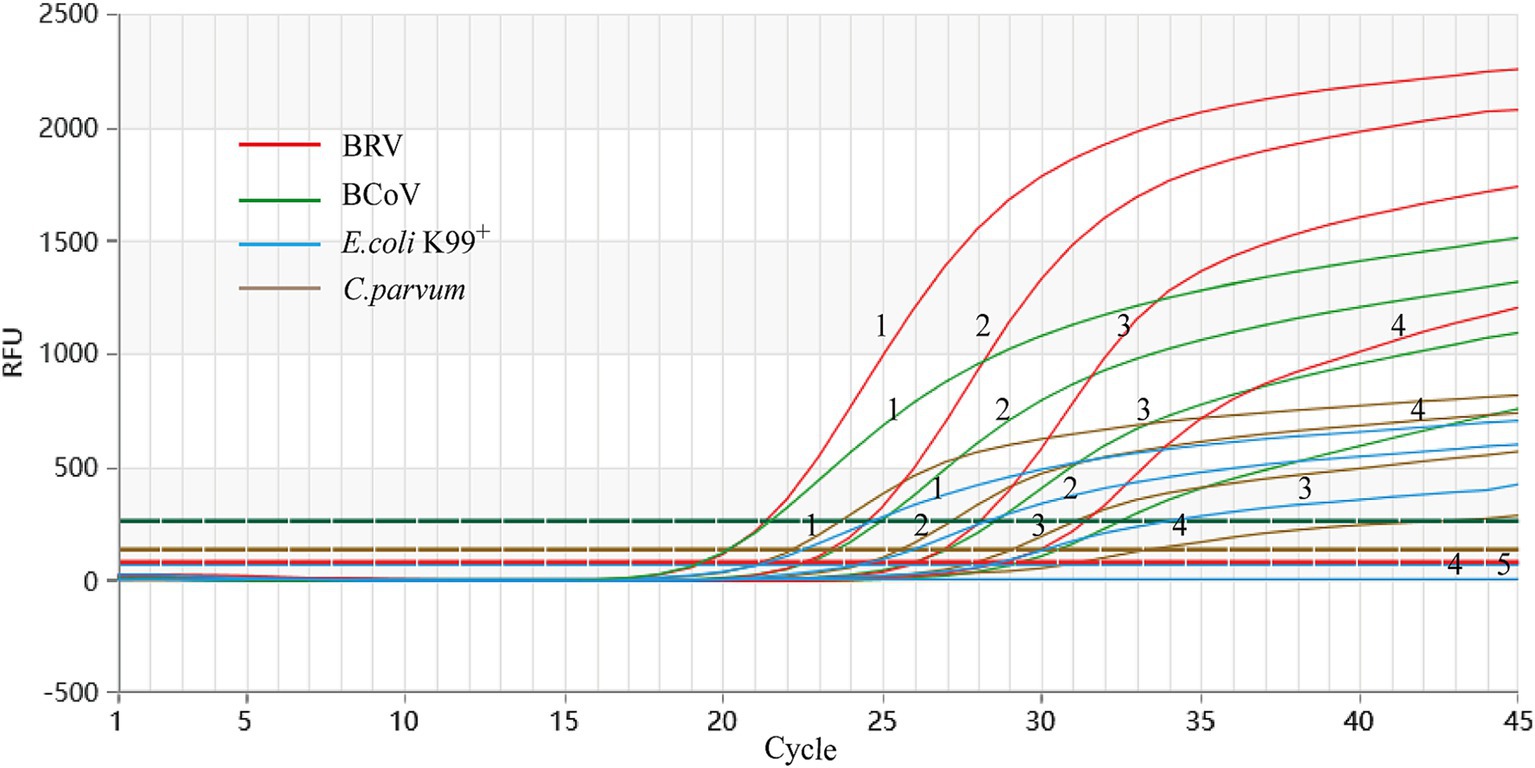
Figure 4. Sensitivity test of singleplex TaqMan Real-Time PCR Assay. X-axis: number of cycles; Y-axis: RFU; 1–4: fluorescence quantitative PCR amplification curve of each standard positive recombinant plasmid from 104 to 101 copies/μL of plasmid content; 5: ddH2O.
The highest variation coefficient Cv value of the quadruple qPCR detection group was 1.08%, and the highest variation coefficient Cv value of the intergroup was 1.2%, indicating that the quadruple TaqMan fluorescence quantitative PCR detection method had good detection repeatability and high accuracy (Table 2).
The qPCR method established in this study and the commercial qPCR kit method were used to analyze 400 clinical samples collected from intensive farms in Shandong Province. The results of quadruple fluorescence quantitative PCR revealed that 72 of the 400 samples were BRV positive, with a positive detection rate of 18%; 43 were BCoV positive, with a positive detection rate of 10.75%; 82 were E. coli K99 + positive, with a positive detection rate of 20.5%; and 19 were C. parvum positive, with a detection rate of 4.75%. The detection rates of BRV + BCoV, BRV + E. coli K99+, BRV + C. parvum, BCoV+E. coli K99+, BCoV+C. parvum, and E. coli K99++C. parvum were 6, 7, 1, 4.5, 0.5, and 1%, respectively. The detection rates of BRV + BCoV+E. coli K99+, BRV + BCoV+C. parvum, BRV + E. coli K99++C. parvum, and BCoV+E. coli K99++C. parvum were 1.75, 0, 0.25, and 0%, respectively. The detection rate of BRV + BCoV+E. coli K99++C. parvum was 0.5% (Figure 5).
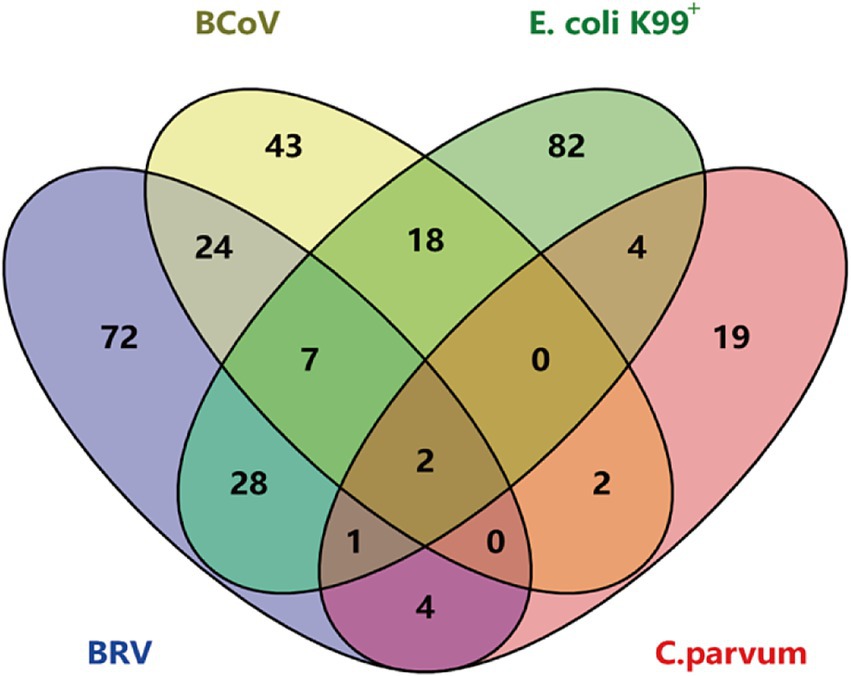
Figure 5. Results of multiple TaqMan fluorescence quantitative PCR for 400 clinical samples. The diagram illustrates the distribution of samples infected with individual pathogens and co-infected samples. Overlapping regions represent samples infected with multiple pathogens simultaneously.
Diarrhea is one of the most serious diseases of newborn calves, causing significant economic losses due to mortality, poor growth, and treatment costs (33). During diarrhea outbreaks in herds, it is important to identify the infectious cause so that further appropriate control measures can be targeted (34). Real-time PCR is easily adapted to detect nucleic acid targets specific to each given pathogen, with high specificity and sensitivity; making it a vital tool for infection identification and detection (35). Therefore, we developed a multiple TaqMan qPCR protocol to simultaneously detect single or mixed infections of the four main pathogens that cause diarrhea in calves. The nucleic acid extraction procedure can simultaneously extract DNA and RNA from viruses, bacteria, and protozoa, saving time and cost. In this study, primers and probes were designed for the BRV VP6 gene, BCoV N gene, E. coli K99+ K99 gene and C. parvum 18S rRNA gene. After the selection of probes and primers and the optimization of the concentration ratio, a fluorescence quantitative PCR detection method based on a TaqMan probe was successfully constructed. Standard plasmids showed a good linear relationship in the range of 108 ~ 102copies/μL, with R2 values greater than 0.997. The method has good specificity and does not exhibit cross-reactions with other pathogens. The experimental results are stable within and between groups, and the coefficient of variation is less than 1.2%, indicating that the method has good repeatability. The methods used to detect E. coli K99+ and C. parvum are mostly common PCR (36, 37). Wang et al. established a triple qPCR method for BCoV, BRV and E. coli K99+, with a minimum detection limit of 10 copies/μL (27). Compared with Wang’s method, this method did not achieve higher sensitivity, likely due to the inherent limitations of multiplex fluorescence quantification. Establishing quadruple qPCR is more challenging. In this study, we screened various primer-probe combinations. Since the four signals have different wavelengths, their fluorescence signals can be detected simultaneously in the same reaction tube. High-specificity multiplex real-time PCR requires optimizing the concentration ratios of primers and probes to prevent mutual interference. Pedroso et al. established a fivefold PCR detection method for BRV, BCoV, E. coli K99+, Pasteurella and Cryptosporidium (26). In contrast, the introduction of the TaqMan probe in this method enhances the specificity of the target gene. This method relies on the collection of fluorescence signals and can determine the presence or absence of pathogenic infection on the basis of Ct values. The experimental steps are simple, do not easily contaminate the environment, and can effectively shorten the detection time (28). Demirci et al. established conventional molecular kits for rotavirus Group A, E. coli K99+, and C. parvum species based on LAMP method, which can be used under field conditions without a laboratory environment and provides results in a short time. However, compared with the PCR method, this method was less sensitive, which can result in false-positive or false-negative results (38).
In this study, 400 clinical diarrhea samples from Shandong Province were used to evaluate the performance of the developed multiplex qPCR method. Compared to commercial single pathogen-detection kits, the method showed diagnostic sensitivity and specificity greater than 90%, with a kappa value exceeding 0.9. Despite some false negatives and false positives, these results indicate a high level of agreement between the two methods (Table 3). The occurrence of false-positive and false-negative samples may be attributed to various factors. False positives could arise from sample or reagent contamination, non-specific amplification, or cross-reactivity of primers/probes with non-target sequences. Conversely, false negatives might be caused by inhibitors present in the sample, such as heme or polysaccharides in feces, or by incomplete nucleic acid extraction. We did not perform further tests to investigate the false positive and false negative samples, and a key limitation of this study is the lack of internal controls, which could have improved the reliability of the results. In this study, we used the established method to test 400 diarrhoeal fecal samples from Shandong. The detection rates for BRV, BCoV, E. coli K99+, and C. parvum were 18% (72/400), 10.75% (43/400), 20.5% (82/400), and 4.75% (19/400), respectively. Among these pathogens, E. coli K99+ had the highest detection rate. These results demonstrate that the four pathogens play a significant role in the occurrence of calf diarrhea.
One limitation of this study is the lack of cross-reactivity testing to assess potential interactions between the primers, probes and non-target organisms. Although in analysis indicated high specificity, experimental validation would strengthen the evidence. This limitation may impact result interpretation, especially in cases of co-infections or closely related organisms. Future studies will include comprehensive cross-reactivity testing to further validate the assay’s specificity and reliability.
In future studies, we propose a new farm detection scheme that includes a qPCR device and a set of reagent kits. The reagent kit contains lysis buffer, primers and probes for BRV, BCoV, E. coli K99+ and C. parvum, reverse transcriptase, and sampling swabs. This scheme does not require a centrifuge, professional operation, or complex nucleic acid extraction steps. The swabs are used to collect samples and then the swabs are placed in lysis buffer for 3 min. A pipette is used to transfer 5 μL of the sample to the reaction tube, and then qPCR detection is performed. The results can be obtained in approximately 1 h (Figure 6). This method eliminates many complex steps, reducing manual operations in sample processing and pipetting. In scenarios requiring simultaneous detection of multiple pathogens, quadruplex qPCR can improve detection efficiency. In the future, it has the potential to achieve automation and high-throughput detection.
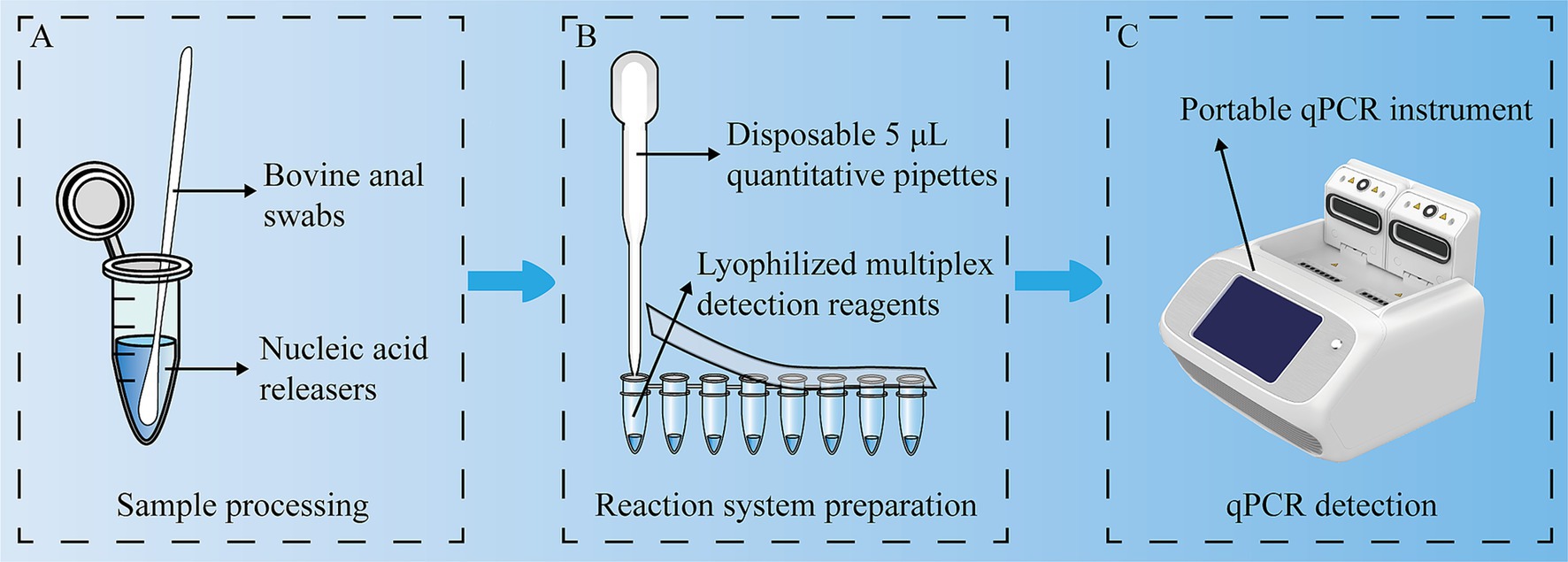
Figure 6. Experimental flow chart. A multiplex, one-step RT–qPCR assay was used to detect BRV, BCoV, E. coli K99+ and C. parvum simultaneously. (A) Sample processing; (B) Reaction system preparation; (C) qPCR detection.
In conclusion, this study successfully established a four-way detection method for BRV, BCoV, E. coli K99+ and C. parvum. This method has good specificity, high sensitivity and good repeatability and can quickly detect infections and mixed infections caused by bovine diarrhea pathogens via laboratory detection, providing convenience for the timely clinical treatment of bovine diarrhea. Consequently, the adverse effect of diarrhoeal pathogens on calves can be reduced.
The original contributions presented in the study are included in the article/supplementary material, further inquiries can be directed to the corresponding author.
The animal studies were approved by Research Ethics Committee of Qingdao Agricultural University. The studies were conducted in accordance with the local legislation and institutional requirements. Written informed consent was obtained from the owners for the participation of their animals in this study.
XZha: Methodology, Validation, Writing – original draft, Writing – review & editing, Investigation. ML: Validation, Writing – review & editing, Investigation. YY: Validation, Writing – review & editing, Investigation. YW: Writing – review & editing, Formal analysis, Visualization. XZhe: Writing – review & editing, Formal analysis, Visualization. DY: Data curation, Writing – review & editing, Resources. HG: Data curation, Writing – review & editing, Resources. HL: Resources, Writing – review & editing, Data curation. KF: Resources, Writing – review & editing, Data curation. ZC: Methodology, Writing – review & editing, Conceptualization, Supervision.
The author(s) declare that financial support was received for the research and/or publication of this article. This work was supported by the Natural Science Foundation of Shandong Province (grant numbers ZR2023MC027 & ZR2020QC200), A Youth Innovation Team Project of Shandong Province Higher Educational Science and Technology Program (2021KJ102), and Ningxia Hui Autonomous Region Agricultural Science and Technology Independent Innovation Project (NKYG-24-19).
DY was employed by Innovus Solarex Biotech Co., Ltd.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The authors declare that no Gen AI was used in the creation of this manuscript.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
1. Maier, GU, Breitenbuecher, J, Gomez, JP, Samah, F, Fausak, E, and Van Noord, M. Vaccination for the prevention of neonatal calf diarrhea in cow-calf operations: a scoping review. Vet Anim Sci. (2022) 15:100238. doi: 10.1016/j.vas.2022.100238
2. Cho, Y, and Yoon, K. An overview of calf diarrhea - infectious etiology, diagnosis, and intervention. J Vet Sci. (2014) 15:1–17. doi: 10.4142/jvs.2014.15.1.1
3. Uhde, FL, Kaufmann, T, Sager, H, Albini, S, Zanoni, R, Schelling, E, et al. Prevalence of four enteropathogens in the faeces of young diarrhoeic dairy calves in Switzerland. Vet Rec. (2008) 163:362–6. doi: 10.1136/vr.163.12.362
4. Geletu, US, Usmael, MA, and Bari, FD. Rotavirus in calves and its zoonotic importance. Vet Med Int. (2021) 2021:1–18. doi: 10.1155/2021/6639701
5. Brunauer, M, Roch, F, and Conrady, B. Prevalence of worldwide neonatal calf diarrhoea caused by bovine rotavirus in combination with bovine coronavirus, escherichia coli k99 and cryptosporidium spp.: a meta-analysis. Animals. (2021) 11:1014. doi: 10.3390/ani11041014
6. Seid, U, Dawo, F, Tesfaye, A, and Ahmednur, M. Isolation and characterization of coronavirus and rotavirus associated with calves in central part of Oromia, Ethiopia. Vet Med Int. (2020) 2020:1–10. doi: 10.1155/2020/8869970
7. Wang, M, Yan, Y, Wang, R, Wang, L, Zhou, H, Li, Y, et al. Simultaneous detection of bovine rotavirus, bovine parvovirus, and bovine viral diarrhea virus using a gold nanoparticle-assisted pcr assay with a dual-priming oligonucleotide system. Front Microbiol. (2019) 10:10. doi: 10.3389/fmicb.2019.02884
8. Ha, S, Kang, S, Park, K, Ku, J, Choi, K, and Park, J. Comparison of blood electrolyte and biochemical parameters between single infections of rotavirus andcryptosporidium parvumin diarrheic hanwoo calves. J Vet Sci. (2022) 23:e85. doi: 10.4142/jvs.22196
9. Zhu, Q, Li, B, and Sun, D. Advances in bovine coronavirus epidemiology. Viruses. (2022) 14:1109. doi: 10.3390/v14051109
10. Hodnik, JJ, Ježek, J, and Starič, J. Coronaviruses in cattle. Trop Anim Health Prod. (2020) 52:2809–16. doi: 10.1007/s11250-020-02354-y
11. Jay, CM, Bhaskaran, S, Rathore, KS, and Waghela, SD. Enterotoxigenic K99+ escherichia coli attachment to host cell receptors inhibited by recombinant pili protein. Vet Microbiol. (2004) 101:153–60. doi: 10.1016/j.vetmic.2004.03.019
12. Sahagun-Ruiz, A, Velazquez, LV, Bhaskaran, S, Jay, CM, Morales-Salinas, E, Rathore, K, et al. Reduction of enterotoxin induced fluid accumulation in ileal loops of neonatal calves with anti-f5 fimbriae recombinant antibody. Vet Res Commun. (2015) 39:229–36. doi: 10.1007/s11259-015-9646-1
13. Li, N, Wang, R, Cai, M, Jiang, W, Feng, Y, and Xiao, L. Outbreak of cryptosporidiosis due to cryptosporidium parvum subtype iida19g1 in neonatal calves on a dairy farm in China. Int J Parasitol. (2019) 49:569–77. doi: 10.1016/j.ijpara.2019.02.006
14. Ryan, U, Zahedi, A, Feng, Y, and Xiao, L. An update on zoonotic cryptosporidium species and genotypes in humans. Animals. (2021) 11:3307. doi: 10.3390/ani11113307
15. Zhang, Z, Su, D, Meng, X, Liang, R, Wang, W, Li, N, et al. Cryptosporidiosis outbreak caused bycryptosporidium parvum subtype iida20g1 in neonatal calves. Transbound Emerg Dis. (2022) 69:278–85. doi: 10.1111/tbed.13976
16. Weldemariam, DT, Awoke, MG, and Aklilu, MA. Review on bovine cryptosporidiosis, its associated risk factors and diagnostics methods. Sm Trop Med J. (2024) 6:1–08. doi: 10.36876/2573-363X.smtmj.886873
17. Wang, D, Gao, H, Zhao, L, Lv, C, Dou, W, Zhang, X, et al. Detection of the dominant pathogens in diarrheal calves of Ningxia, China in 2021–2022. Front Vet Sci. (2023) 10:1155061. doi: 10.3389/fvets.2023.1155061
18. Qin, Y, Gong, Q, Zhang, M, Sun, Z, Wang, W, Wei, X, et al. Prevalence of bovine rotavirus among bovidae in China during 1984–2021: a systematic review and meta-analysis. Microb Pathog. (2022) 169:105661. doi: 10.1016/j.micpath.2022.105661
19. Li, S, Huang, J, Cai, X, Mao, L, Xie, L, Wang, F, et al. Prevalence and evolutionary characteristics of bovine coronavirus in China. Vet Sci. (2024) 11:230. doi: 10.3390/vetsci11060230
20. Berber, E, Çanakoğlu, N, Sözdutmaz, O, Simsek, E, Sursal, N, Ekinci, G, et al. Seasonal and age-associated pathogen distribution in newborn calves with diarrhea admitted to icu. Vet Sci. (2021) 8:128. doi: 10.3390/vetsci8070128
21. Chen, Y, Huang, J, Qin, H, Wang, L, Li, J, and Zhang, L. Cryptosporidium parvum and gp60 genotype prevalence in dairy calves worldwide: a systematic review and meta-analysis. Acta Trop. (2023) 240:106843. doi: 10.1016/j.actatropica.2023.106843
22. Cheng, X, Wu, W, Teng, F, Yan, Y, Li, G, Wang, L, et al. Isolation and characterization of bovine rva from Northeast China, 2017–2020. Life. (2021) 11:1389. doi: 10.3390/life11121389
23. Dhama, K, Chauhan, RS, Mahendran, M, and Malik, SVS. Rotavirus diarrhea in bovines and other domestic animals. Vet Res Commun. (2009) 33:1–23. doi: 10.1007/s11259-008-9070-x
24. Liu, Q, Niu, X, Jiang, L, Zhang, G, Wang, P, Zhang, S, et al. Establishment of an indirect elisa method for detecting bovine coronavirus antibodies based on n protein. Front Vet Sci. (2025) 12:12. doi: 10.3389/fvets.2025.1530870
25. Zhao, L, Wang, D, Jiang, H, Gu, Q, Gao, H, Zhang, L, et al. Isolation and characterization of bovine coronavirus variants with mutations in the hemagglutinin-esterase gene in dairy calves in China. BMC Vet Res. (2025) 21:92. doi: 10.1186/s12917-025-04538-w
26. Pedroso, NH, Silva Júnior, JVJ, Becker, AS, Weiblen, R, and Flores, EF. An end-point multiplex pcr/reverse transcription-pcr for detection of five agents of bovine neonatal diarrhea. J Microbiol Methods. (2023) 209:106738. doi: 10.1016/j.mimet.2023.106738
27. Wang, C, Wang, F, Chang, J, Jiang, Z, Han, Y, Wang, M, et al. Development and application of one-step multiplex real-time pcr for detection of three main pathogens associated with bovine neonatal diarrhea. Front Cell Infect Microbiol. (2024) 14:14. doi: 10.3389/fcimb.2024.1367385
28. Kralik, P, and Ricchi, M. A basic guide to real time pcr in microbial diagnostics: definitions, parameters, and everything. Front Microbiol. (2017) 8:8. doi: 10.3389/fmicb.2017.00108
29. van den Hurk, S, Regmi, G, Naikare, HK, and Velayudhan, BT. Advances in laboratory diagnosis of coronavirus infections in cattle. Pathogens. (2024) 13:524. doi: 10.3390/pathogens13070524
30. Decaro, N, Elia, G, Campolo, M, Desario, C, Mari, V, Radogna, A, et al. Detection of bovine coronavirus using a taqman-based real-time rt-pcr assay. J Virol Methods. (2008) 151:167–71. doi: 10.1016/j.jviromet.2008.05.016
31. Jothikumar, N, Da Silva, AJ, Moura, I, Qvarnstrom, Y, and Hill, VR. Detection and differentiation of cryptosporidium hominis and cryptosporidium parvum by dual taqman assays. J Med Microbiol. (2008) 57:1099–105. doi: 10.1099/jmm.0.2008/001461-0
32. Jesser, KJ, and Levy, K. Updates on defining and detecting diarrheagenic escherichia coli pathotypes. Curr Opin Infect Dis. (2020) 33:372–80. doi: 10.1097/QCO.0000000000000665
33. Blanchard, PC. Diagnostics of dairy and beef cattle diarrhea. Vet Clin N Am Food Anim Pract. (2012) 28:443–64. doi: 10.1016/j.cvfa.2012.07.002
34. Yimer, M, Gezhagne, M, Biruk, T, and Dinaol, B. A review on major bacterial causes of calf diarrhea and its diagnostic method. J Vet Med Anim Health. (2015) 7:173–85. doi: 10.5897/JVMAH2014.0351
35. Engstrom-Melnyk, J, Rodriguez, PL, Peraud, O, and Hein, RC. Clinical applications of quantitative real-time pcr in virology. Methods Microbiol. (2015) 42:161–97. doi: 10.1016/bs.mim.2015.04.005
36. Bouzid, M, Steverding, D, and Tyler, KM. Detection and surveillance of waterborne protozoan parasites. Curr Opin Biotechnol. (2008) 19:302–6. doi: 10.1016/j.copbio.2008.05.002
37. Rekha, HKM, Puttalakshmamma, GC, and PE, DS. Comparison of different diagnostic techniques for the detection of cryptosporidiosis in bovines. Vet World. (2016) 9:211–5. doi: 10.14202/vetworld.2016.211-215
Keywords: BRV, BCoV, E. coli K99+, C. parvum , multiplex TaqMan real-time PCR
Citation: Zhao X, Li M, Yang Y, Wang Y, Zheng X, Yin D, Gao H, Li H, Fu K and Cao Z (2025) Multiplex one-step RT–qPCR assays for simultaneous detection of BRV, BCoV, Escherichia coli K99+ and Cryptosporidium parvum. Front. Vet. Sci. 12:1561533. doi: 10.3389/fvets.2025.1561533
Received: 16 January 2025; Accepted: 10 March 2025;
Published: 26 March 2025.
Edited by:
Jerry William Simecka, University of North Texas Health Science Center, United StatesReviewed by:
Gang Wang, Shandong Agricultural University, ChinaCopyright © 2025 Zhao, Li, Yang, Wang, Zheng, Yin, Gao, Li, Fu and Cao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhi Cao, MjAxOTAxMjUyQHFhdS5lZHUuY24=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.