- 1Research Institute for Biological Safety Problems, Gvardeisky, Kazakhstan
- 2Institute of Ionosphere, Almaty, Kazakhstan
- 3Kazakh National Research Technical University named after K.I. Satbayev, Almaty, Kazakhstan
- 4National Centre for Foreign Animal Disease, Canadian Food Inspection Agency, Winnipeg, MB, Canada
- 5Department of Immunology, University of Manitoba, Winnipeg, MB, Canada
Introduction: Bluetongue virus (BTV) is a significant vector-borne pathogen affecting ruminants, leading to substantial economic losses, and adversely impacting livestock production worldwide. Recently, Bluetongue (BT) has emerged as a growing concern for European and Asian countries, including Kazakhstan. This study examines the prevalence and distribution of BTV in Kazakhstan during 2023-2024, providing up-to-date information on its occurrence in livestock and Culicoides species. The findings aim to contribute to better understanding and management of BT in the region.
Methods: A total of 972 whole blood and 972 serum samples were collected from cattle, sheep, and goats in the southern and northern regions of Kazakhstan, alongside 11,859 Culicoides midges in the autumn of 2023 and Spring of 2024. The serum samples were tested for BT virus (BTV)-specific antibodies using ELISA, while the whole blood and Culicoides specimens were analyzed for BTV RNA by Real-time RT-PCR (rRT-PCR). Morphological and molecular identification of Culicoides species was also conducted.
Result: The overall seroprevalence of BTV in Southern Kazakhstan increased across all animal species in 2024 compared to 2023, with goats showing the most notable rise (from 3.8% to 29.5%). In the northern regions, seroprevalence remained zero in 2023 but reached 10.0% in cattle by 2024. rRT-PCR results confirmed active virus circulation, with rRT-PCR-positive samples significantly higher in 2024, especially among goats (from 4.2% in 2023 to 62.0% in 2024) and cattle (from 9.2% to 34.4%). Based on morphology, nine species of Culicoides midges were identified, including C. obsoletus a known BTV vector in European countries. Four of them were genetically confirmed, and BTV RNA was detected in all four species (C. miutissimus, C. sphagnumensis, C. newsteadi, and C. pectipennis), suggesting their potential vectorial role in BTV transmission.
Discussion: This study provides new insights into the epidemiology of BT in Kazakhstan and serves as a valuable resource for veterinary professionals. The findings emphasize the need for continued surveillance and vector control strategies to mitigate the spread of BTV in the region.
Introduction
Culicoides biting midges are the smallest blood-sucking two-winged insects (1–4 mm) in the order Diptera. They belong to the family Ceratopogonidae and have mottled wings with dark patterns that can be used to identify the species (1).
Culicoides are important vectors for arboviruses in both animals and humans. The Culicoides midges transmit bluetongue (BT), an infectious, non-contagious, vector-borne hemorrhagic viral disease of wild and domestic ruminants. BT is characterized by fever, inflammatory-necrotic lesions in the oral cavity, tongue, the digestive tract, and the epithelium of the coronary band, along with degenerative changes in skeletal and cardiac muscles, subintimal hemorrhage in the pulmonary artery, oedema of the lungs and pericardial, pleural, and abdominal effusions (2, 3). The causative agent, BT virus (BTV) is a double-stranded RNA virus that belongs to the genus Orbivirus in the family Reoviridae. The genome of BTV is composed of 10 segments of dsRNA. This segmented structure allows the virus to undergo reassortment, leading to the emergence of variants with novel biological characteristics (4). Currently, 29 serotypes of BTV are known to exist (5).
The distribution of BTV has traditionally been observed within the geographical belt between latitudes 40° N and 35° S (6), corresponding to the habitat of Culicoides vectors (7). The mild, warm climates of Africa, southern Europe, and Southeast Asia favor the year-round activity of Culicoides, facilitating the overwintering of the BTV. Over the past 20 years, the distribution of BTV in Europe has changed significantly. Global climate change has influenced the emergence of different serotypes in geographic areas above 50° and identification of new Culicoides vector species (8–10). Currently, the disease has been reported on all continents, with a noticeable trend of expanding to more northern regions (8, 9, 11).
The unprecedented spread of BTV in Europe (6 serotypes across 12 EU countries since 1998), is believed to be driven by the global warming. Reports of BTV detection in countries neighboring Kazakhstan (12–14), has prompted a renewed focus on the epidemiology of BT, and the Culicoides vectors in the region.
Historically Kazakhstan was designated as a BT free country. However, serological studies using samples collected during 1996–1998 showed 23.2% BT seroprevalence in livestock (15, 16). Recently Zhigailov et al., reported the detection of BTV antibodies in livestock and BTV genetic material in sheep and Culicoides midges in the southeastern region of Kazakhstan. Through molecular genetic analysis and partial sequencing, they determined that the BTV belonged to the “western” topotype of the BTV-9 strain (17). In another study, using mathematical modeling showed that the transmission of BTV in Kazakhstan is not possible between October and March. The winter in Kazakhstan prevents the transmission of BT during this time. Assuming there are endemic BTV competent Culicoides midges in Kazakhstan, the modeling predicted risk of BT appearing in the south of Kazakhstan in April and spread north reaching maximum levels in northern Kazakhstan in July, decline in September and disappear by October (18).
Currently, systematic studies of the range and density of the primary vectors of BTV infection in Kazakhstan are either not conducted or are limited to serological monitoring studies. In this study we report, based on the samples collected during 2023–2024, seroprevalence of BT in livestock, the Culicoides species associated and their potential role as BTV vectors in Northern and Southern region of Kazakhstan.
Materials and methods
Collection and transport of animal biological samples
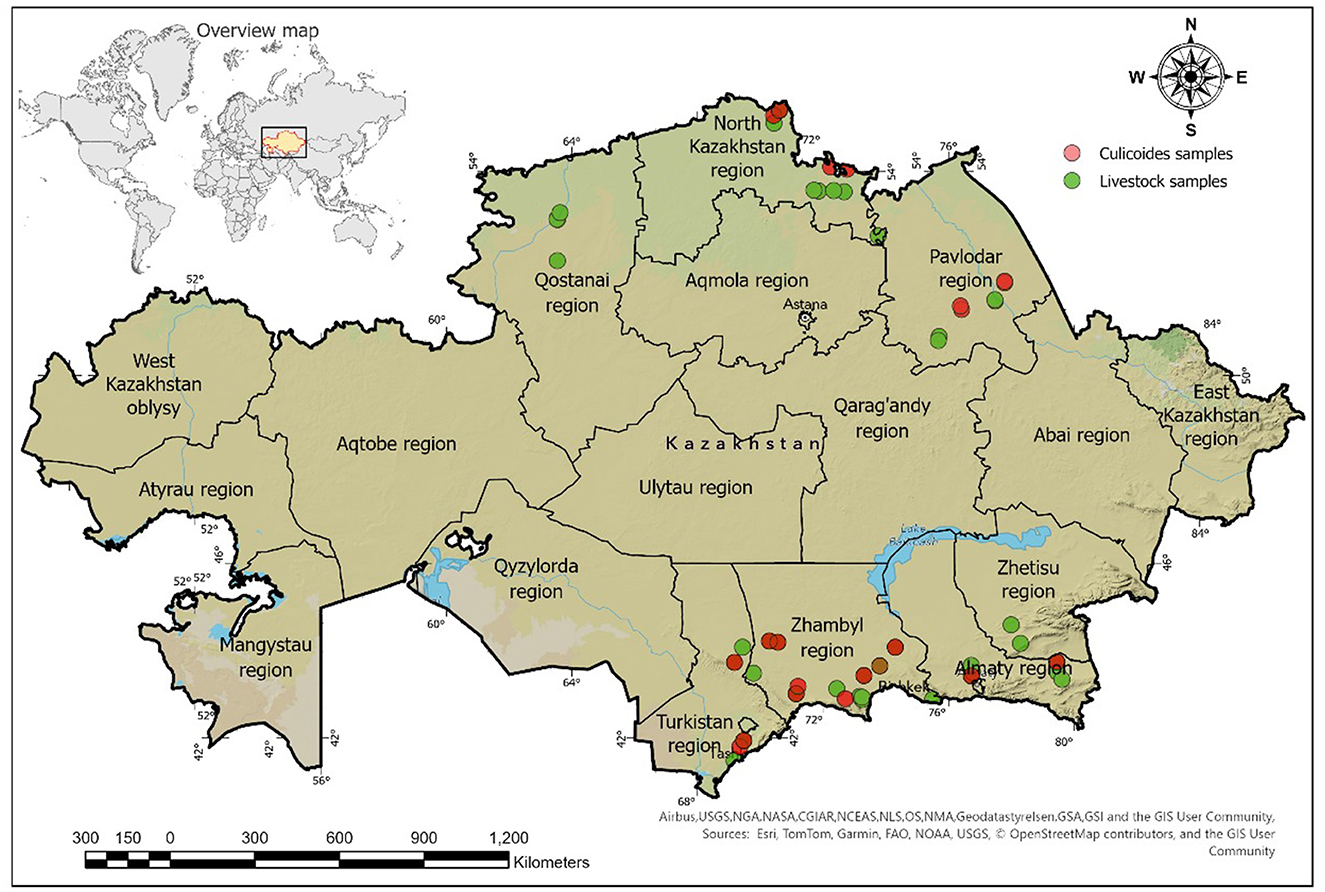
A total of 972 whole blood and serum samples were randomly collected from clinically healthy sheep, goats, and cattle from the southern regions of Kazakhstan, namely Zhetysu, Almaty, Zhambyl, and Turkestan and the northern regions of Kazakhstan, namely Kostanay, Pavlodar, and North Kazakhstan during August–September 2023 and May–June 2024 (Figure 1). The sample sizes and volumes were determined according to OIE guidelines (19), with modifications based on prevalence levels (Supplementary Table 1) (20).
Mapping sampling sites using ArcGIS pro
The GPS coordinates of sampling locations were imported and mapped using the WGS 84 projection (EPSG: 4326), and ArcGIS Pro (21) was used to compile and generate the maps. Briefly, a layer of sampling points was created and overlaid on satellite imagery (22). The results were exported as a map with a legend and geographic grid for inclusion in the publication.
Culicoides collection method
Culicoides were captured utilizing specialized CDC light traps containing ultraviolet lamp (Light trap, LI-MR-512, BioQuip, USA). The trapping was conducted in the evening at sunset, under calm and windless conditions, in wet, marshy regions adjacent to pastures and livestock facilities. The collected specimens were then transferred to 70% ethanol solution and stored at −20°C for subsequent sorting and morphological identification.
ELISA
BTV antibodies were detected using the ID Screen® BT Competition kit (Ref: BTC-5P, ID-Vet, France), a competitive ELISA with 96.5% sensitivity and 99.3% specificity, designed to identify anti-VP7 antibodies in serum or plasma from cattle, sheep, and goats (23). Testing and result interpretation followed the manufacturer's instructions.
Optical density (OD) was measured at 450 nm using a Chromate®-4300 reader (Awareness Technology, USA). Samples were classified as positive if their OD was <40% of the mean negative control value (S/N) and negative if OD was ≥40% of the mean negative control value (S/N).
RNA extraction
Viral RNA was extracted from whole blood using Qiagen RNeasy Mini Kit (Cat number, Qiagen, Hilden, Germany), in accordance with the manufacturer's guidelines. Briefly, 560 μl of AVL buffer and 5.6 μl of carrier RNA were added to 140 μl of the whole blood sample, followed by thorough mixing at room temperature for 10 min. Next, 560 μl of 96% ethanol was added, and mixed thoroughly. The resulting solution was then transferred to RNA binding columns and centrifuged at 8,000 rpm for 30 s. The columns were then washed with 700 μl of AW1 buffer each, followed by 500 μl of AW2 buffer involving by centrifugation at 8,000 rpm for 30 s. To remove any residual buffer, the columns were centrifuged at 12,000 rpm for additional 1 min, and viral RNA was eluted by adding 40 μl of AVE buffer, followed by centrifugation at 8,000 rpm for 1 min. The purity and concentration of the isolated RNA were assessed spectrophotometrically at 260/280 nm, with samples yielding an A260/A280 ratio between 1.8 and 2.0 considered pure and suitable for subsequent rRT-PCR use.
rRT-PCR setup
ID Gene™ Bluetongue Duplex rRT-PCR (Cat number: IDBTV-50, Innovative Diagnostics, Grabels, France) was conducted using a Rotor-Gene Q thermal cycler (Qiagen), and the results were analyzed using Rotor-Gene Q software version 1.8.187.5. rRT-PCR setup was performed in accordance with the ID Gene BT Duplex kit instructions and the following thermal cycling conditions were followed: reverse transcription at 45°C for 10 min, initial denaturation at 95°C for 10 min followed by 40 amplification cycles (denaturation at 95°C for 15 s, annealing and extension at 60°C for 30 s). Detection of the fluorescent signal occurred during the annealing phase in the “Green” channel. Upon completion of the reaction, the amplification curve was analyzed, and a final report was generated. The threshold line was established at a maximum level of 0.05. The reliability of the obtained data was verified through appropriate values of negative and positive controls.
Morphological and genetic identification of midges
Morphological identification of midges was performed based on wing patterns and size (24), under a stereomicroscope (Optika®, Model SZO-T, S/N 651480-482, Ponteranica, Italy). Identification keys (25, 26) and the interactive specialized software Culicoides Xper2 Version 2.0 (www.iikculicoides.net) were utilized for precise species determination.
For molecular characterization of the Culicoides species, DNA from individual midge specimen was extracted using the DNeasy Blood & Tissue Kit (Qiagen, Ref: 69506, Hilden, Germany). Briefly, each midge specimen was added to ATL buffer, supplemented with Proteinase K, and mixed with 200 μl of Buffer AL. The mixture was vortexed for 15 s and incubated at 56°C for 10 min to ensure complete lysis. Subsequently, 200 μl of 96%−100% ethanol was added, and the mixture was vortexed for another 15 s. The lysate was transferred to a DNeasy Mini Spin Column and centrifuged at 6,000 × g (8,000 rpm) for 1 min. The column was then washed with 500 μl of Buffer AW1, centrifuged at 6,000 × g for 1 min, followed by a second wash with 500 μl of Buffer AW2, and centrifuged at 20,000 × g (14,000 rpm) for 3 min. DNA was eluted with 200 μl of nuclease-free water, incubated for 1 min, and centrifuged at 6,000 × g for 1 min. The eluted DNA was stored at −20°C until further analysis. The 28S rDNA is a molecular marker that can be used to differentiate closely related Culicoides species. The primers (Forward 28S rDNA 28S_C'1 5′-ACCCGCTGAATTTAAGCAT-3′, and Reverse 28S_D2 5′-TCCGTGTTTCAAGACGGG-3′) were used to amplify 28S rRNA gene from each specimen (27–29). The cycling conditions used were; initial denaturation at 98°C for 30 s, succeeded by 35 cycles comprising denaturation at 98°C for 10 s, annealing at 55–58°C for 90 s, and extension at 72°C for 45 s. This process concluded with a final extension at 72°C for 5 min. The PCR products, ~700 bp in length, were subsequently assessed using gel electrophoresis on a 1.5% agarose gel and subjected to Sanger sequencing.
Sanger sequencing
Sanger sequencing method, which employs terminating dideoxynucleotides on a 3,500 × l Genetic Analyzer (Applied Biosystems, Thermo Fisher Scientific, Japan. Serial number: 35395-010) was used to sequence the 28S rDNA amplicons. This process utilizes the BigDye™ Terminator v3.1 Cycle Sequencing Kit (Thermo Fisher Scientific), following the manufacturer's protocols. Capillary electrophoresis is facilitated using POP-7 (Thermo Fisher Scientific) as the polymer. DNA termination products are produced through cyclic sequencing. To purify the DNA from unbound dyes, gel filtration is carried out using Centri-Sep columns or with CleanSeq Reagent, in accordance with the kit instructions. The sequencing operations on the sequencer conform to the guidelines provided with the instrument. The resulting chromatograms were analyzed using the Sequencher software package (Gene Codes Corporation, Ann Arbor, MI), and the sequences were analyzed against the GenBank (NCBI) database with MEGA11 software to identify midge species based on percent identity (30).
Statistical analysis
The statistical analysis was performed using GraphPad Prism version 9 (GraphPad Software, Inc., La Jolla, CA, U.S.A.). In a univariable analysis, the chi-square test was performed to examine the relationships between risk factors and BTV seroprevalence. The chi-square test was used to assess the strength of association between independent variables, effectively evaluating differences in BTV prevalence across different regions and animal species in our study.
Using multivariable logistic regression, potential risk factors with p-values < 0.05 were further evaluated. Logistic regression analysis was employed to assess the relationship between the dependent variable (presence of BTV infection) and independent variables (e.g., age, sex, geographic location). This method allowed for the simultaneous consideration of multiple variables. For the outcome variable, a multivariable model was constructed using logistic regression analysis. Confidence intervals (95% CI) and odds ratios (OR) were calculated.
Results
Prevalence of BTV in animals
Thus far, animals showing BT clinical signs have never been reported in the northern regions. However, in the southern regions, sheep with clinical manifestations of BT have been reported. In this study, all the samples (a total of 972 samples of whole blood and sera) were collected from clinically healthy animals in the southern and northern Kazakhstan in the autumn of 2023 and Spring of 2024. This included 642 samples from sheep, 87 from goats, and 243 from cattle. All sera samples were evaluated by ELISA for BTV-specific antibodies (Table 1). The overall serology results revealed a higher number of positive samples in the southern regions compared to the northern regions (p < 0.0001) (Supplementary Table 2). When the data was analyzed based on animal species, the overall seroprevalence was 4.2% (95% CI: 2.9–6.0) among sheep, 21.8% (95% CI: 14.4–31.6) among goats, and 14.4% (95% CI: 10.5–19.4) among cattle (Supplementary Table 3). When compared the year of collection, seroprevalence in the southern regions for 2023 Autumn was 5.4% (95% CI: 2.9–9.9) among sheep, 4.2% (95% CI: 0.7–20.2) among goats, and 10.5% (95% CI: 5.4–19.4) among cattle. In 2024 Spring, the seroprevalence was 6.8% (95% CI: 4.3–10.5) for sheep, 36.0% (95% CI: 24.1–49.9) for goats, and 22.9% (95% CI: 15.6–32.3) for cattle (Supplementary Table 4).
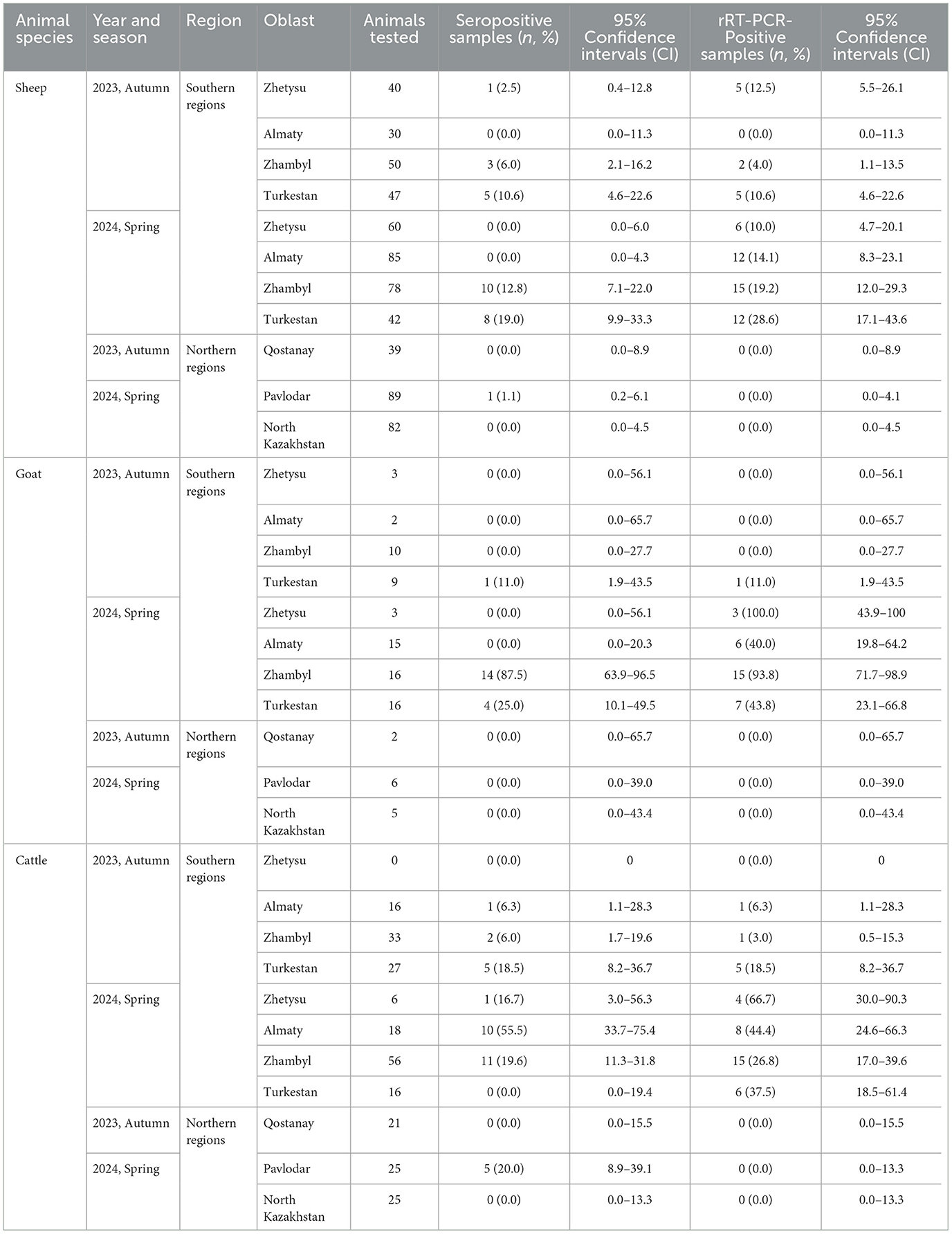
Table 1. Detection of seropositive and rRT-PCR-positive animal species in the southern and northern regions of Kazakhstan for the years 2023–2024.
In northern regions, overall seroprevalence was zero in samples collected in Autumn 2023. In 2024, the overall seroprevalence was 0.6% (95% CI: 0.1–3.2) for sheep and 10.0% (95% CI: 4.3–21.4) for cattle, with no seroprevalence in goats (Supplementary Table 4). Detailed serological prevalence by oblasts is presented in Table 1.
rRT-PCR testing revealed that the number of positive samples for bluetongue virus (BTV) RNA was significantly higher in southern regions compared to northern regions (p < 0.0001). Additionally, the overall number of rRT-PCR-positive samples was higher in 2024 than in 2023 (Supplementary Table 2). BTV RNA was detected in the different animal species as follows: sheep-−8.9% (95% CI: 6.9–11.3; p = 0.57), goats-−36.8% (95% CI: 27.4–47.3), and cattle-−16.5% (95% CI: 12.3–21.6). Notably, the number of rRT-PCR-positive samples exceeded those identified as positive by ELISA (Supplementary Table 3).
In the southern regions in 2023, the proportions of rRT-PCR-positive results were: sheep-−7.2% (95% CI: 4.2–12.1), goats-−4.2% (95% CI: 0.7–20.2), and cattle-−9.2% (95% CI: 4.5–17.8). In 2024, the percentage of rRT-PCR-positive samples among sheep increased to 17.0% (95% CI: 12.9–22.0), among goats to 62.0% (95% CI: 48.1–74.1), and among cattle to 34.4% (95% CI: 25.6–44.3; Supplementary Table 4). Conversely, in the northern regions, no rRT-PCR-positive animals for BTV were detected in either 2023 or 2024 (Supplementary Table 4). The distribution of BTV RNA detection across oblasts is presented in Table 1.
Distribution of Culicoides spp. in the southern and northern regions of Kazakhstan
A total of 11,859 Culicoides spp. were collected in the southern and northern regions of Kazakhstan. Specifically, 209 specimens (1.8%) were from the Zhetysu, 564 (4.8%) from the Almaty oblast, 2,201 (18.6%) from the Turkestan, 6,350 (53.5%) from the Zhambyl, 1,287 (10.9%) from the North Kazakhstan oblast, and 1,248 (10.5%) from the Pavlodar oblast. Detailed quantitative and percentage metrics of captured Culicoides in each oblast are presented in Figure 2 and Table 2. The observed variations in the capture rates of Culicoides across different oblasts are attributed to changes in weather conditions.
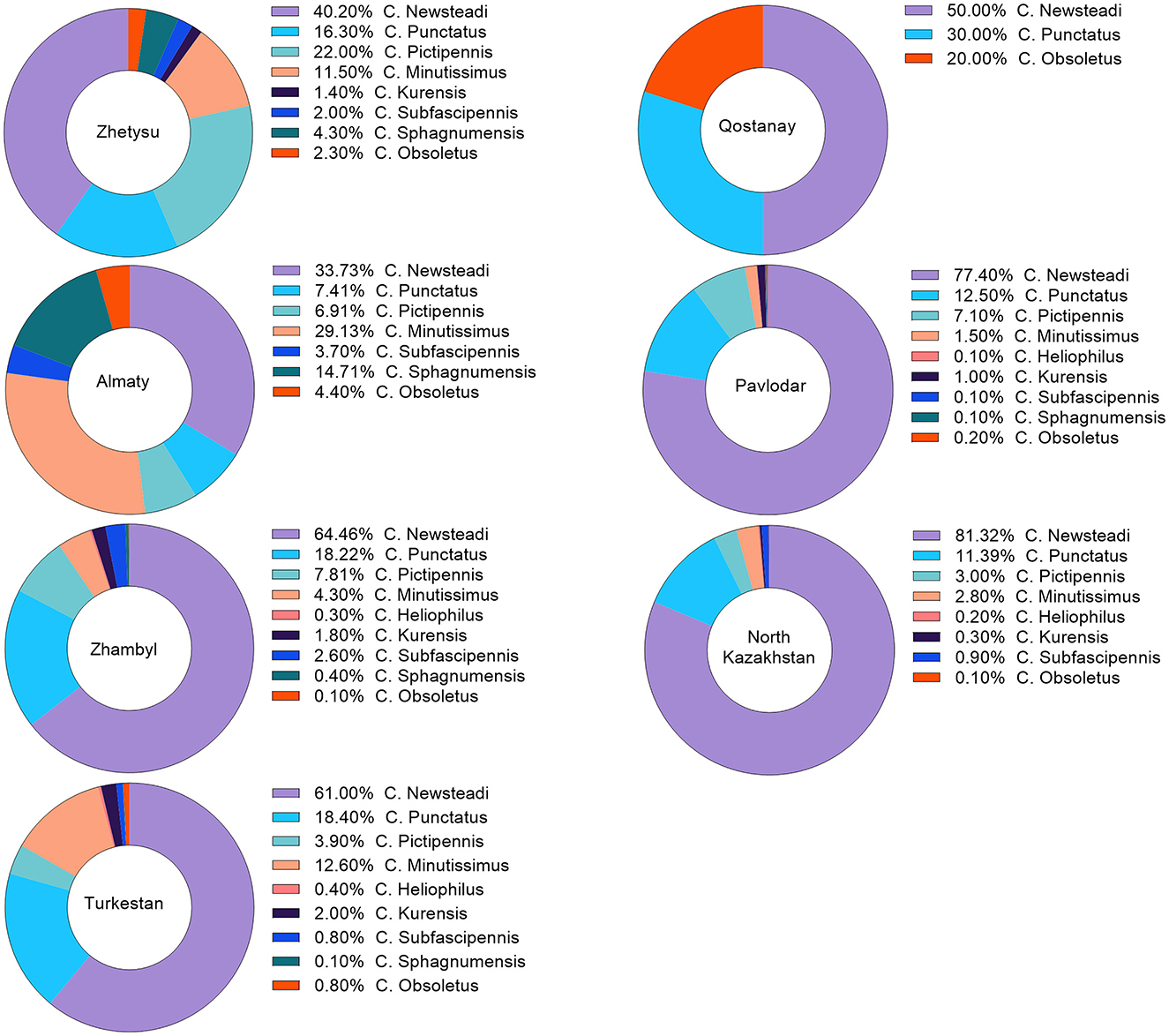
Figure 2. Percentage distribution of Culicoides species occurrence by region. The percentage distribution of Culicoides species occurrence was calculated based on the captured species. This percentage may vary according to the differing weather conditions in each region.
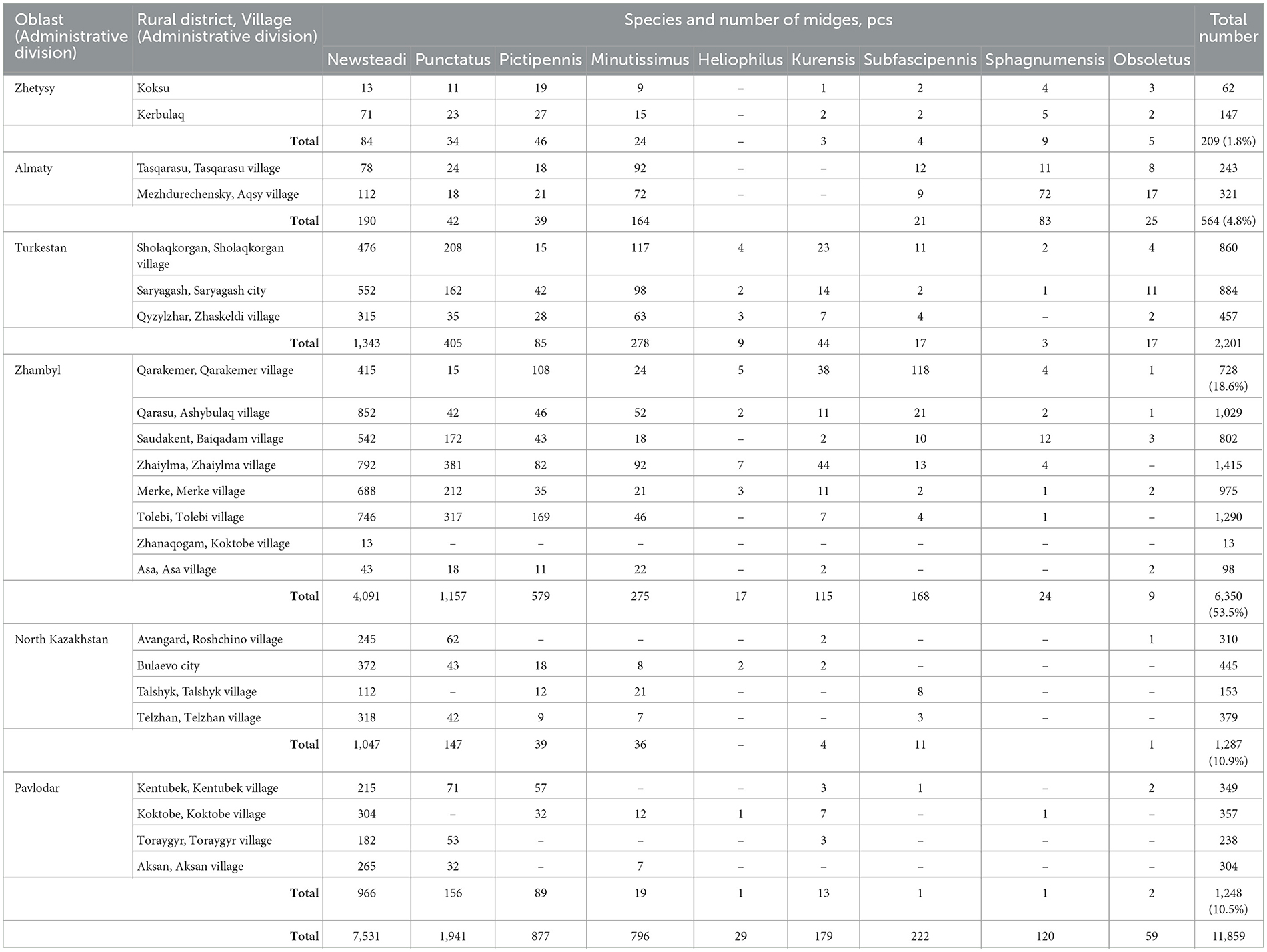
Table 2. Quantitative metrics of Culicoides capture in the southern and northern regions of Kazakhstan for 2024.
Morphological and molecular identification of Culicoides spp.
According to the morphological identification based on wing patterns, the following nine species of Culicoides were identified (Figure 3A): C. Newsteadi, C. Punctatus, C. Pictipennis, C. Minutissimus, C. Heliophilus, C. Kurensis, C. Subfascipennis, C. Sphagnumensis, C. Obsoletus. Among them, the most dominant species were C. Newsteadi, C. Punctatus, C. Pictipennis, C. Minutissimus (Figure 2).

Figure 3. Morphological and Genetic Identification of Culicoides spp. (A)—Wings of Culicoides species captured in Kazakhstan. Photographs of the wings were taken using a camera mounted on a stereomicroscope (Ken-A-Vesion, USA) and processed with specialized software, PowerPoint. Scale bar: 0.5 mm. (B)—Phylogenetic tree of Culicoides species captured in the southern and northern regions of Kazakhstan. Evolutionary relationships of the taxa were analyzed using the Neighbor-Joining method (30). The optimal phylogenetic tree displays the relationships among 43 nucleotide sequences. The reliability of taxa clustering was assessed using a bootstrap test (1,000 iterations), with corresponding percentages indicated next to the branches (31). The branch lengths on the tree are proportional to evolutionary distances, which were calculated using the Tamura-Nei method (32) and expressed as the number of nucleotide substitutions per site. Compositional bias differences among sequences were taken into account in evolutionary comparisons (32).
During the molecular identification, it was confirmed that the species C. newsteadi, C. punctatus, C. pictipennis, and C. minutissimus aligned with their morphological identification (Figure 3B). Five other species (C. heliophilus, C. kurensis, C. subfascipennis, C. sphagnumensis, and C. obsoletus) could not be confirmed molecularly due to insufficient quality and or quantity of data obtained.
Prevalence of BTV in Culicoides species
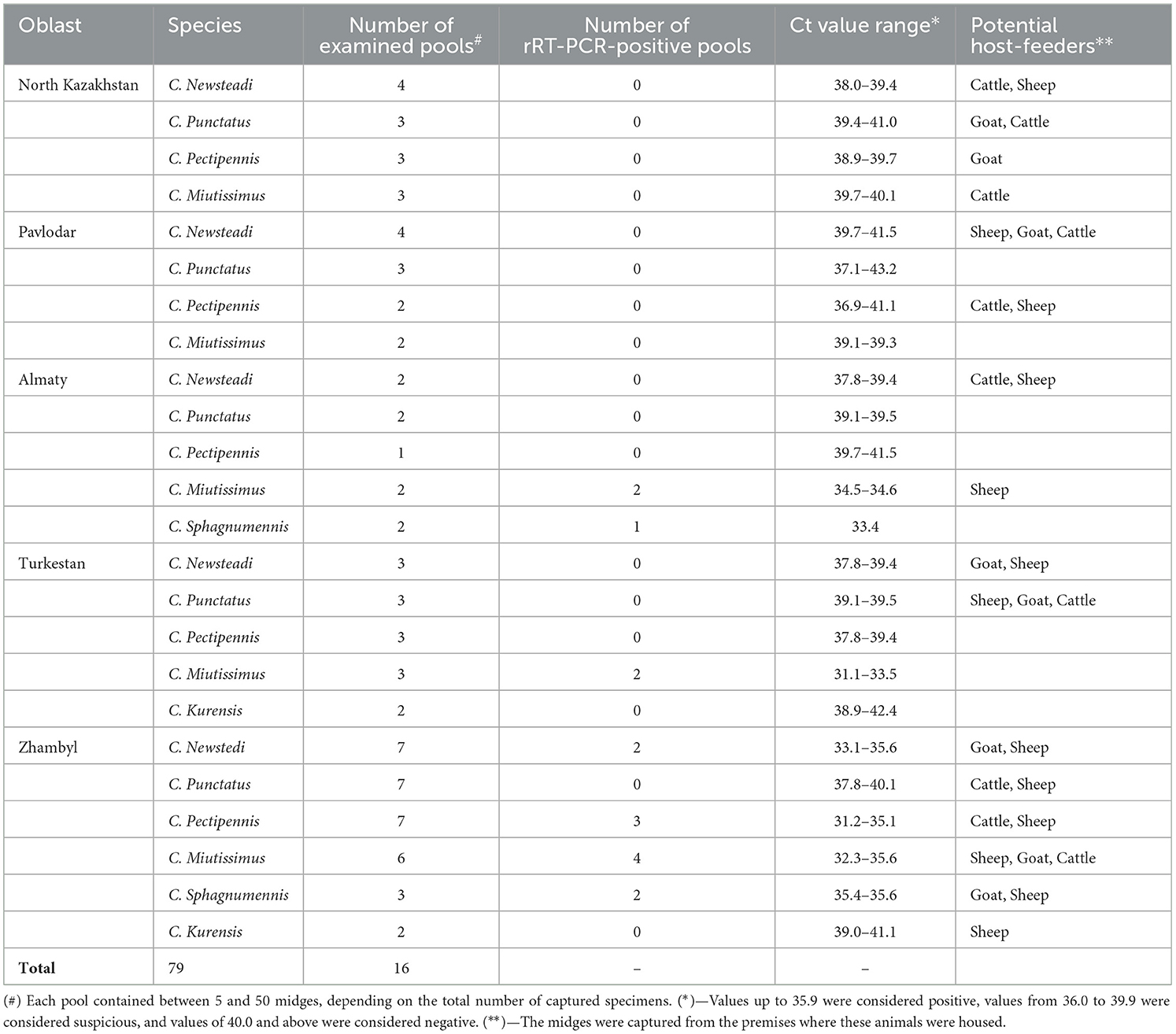
Morphological and genetic identification of Culicoides species facilitated the creation of 79 pooled samples categorized by species and collection sites. The number of pooled samples collected in each region is presented in Table 3.
During the study, no BTV RNA was detected in Culicoides specimens collected from the North Kazakhstan and Pavlodar regions. In the Almaty region, RNA was identified in C. miutissimus (Ct 34.5–34.6) and C. sphagnumennis (Ct 33.4). In contrast, in the Turkestan region, RNA was detected only in C. miutissimus (Ct 31.1–33.5).
In the Zhambyl region, BTV RNA was identified in the following midge species: C. newsteadi (Ct 33.1–35.6), C. pectipennis (Ct 31.2–35.1), C. miutissimus (Ct 32.3–35.6), and C. sphagnumennis (Ct 35.4–35.6; see Table 3).
Discussion
This study provides critical data on the latest seroprevalence of BT and identification of potential BTV vectors in Kazakhstan. In this study, 972 blood and serum samples were collected from sheep, goat and cattle from southern and northern Kazakhstan in Autumn 2023 and Spring 2024. The testing results indicated that seroprevalence and virus distribution were higher in the southern regions compared to the northern regions. It is important to note that sample collection was based on an estimated seroprevalence of ~5%. According to previous studies (15–18), the seroprevalence in the southern regions of Kazakhstan was at least 20%. However, the results of this study confirmed the proposed hypothesis, demonstrating a seroprevalence rate of 5% or higher. The overall seroprevalence of BTV across all animal species was as follows: 4.2% among sheep, 21.8% among goats, and 14.4% among cattle. It is important to note that, despite the trend of higher seroprevalence in goats and cattle, no statistically significant differences were observed among the animal species (p > 0.05). Compared to previous years, the seroprevalence in the southern regions increased across all animal species in 2024 compared to 2023. This increase was particularly notable among goats (from 3.8% in 2023 to 29.5% in 2024) and cattle (from 7.8% to 18.5%). In the northern regions, seroprevalence was zero for all animal species in 2023. However, in 2024, seroprevalence of 10.0% was observed in cattle, indicating the onset of virus circulation in these regions.
To confirm the serological data obtained, rRT-PCR testing was conducted. The rRT-PCR results corroborated the findings from ELISA, demonstrating a significantly higher number of positive BTV RNA samples in the southern regions compared to the northern regions (p < 0.0001), highlighting active virus circulation in these areas. It is noteworthy that the number of rRT-PCR-positive samples was considerably higher in 2024 than in 2023. For instance, the proportion of rRT-PCR-positive samples in sheep increased from 7.2% in 2023 to 17.0% in 2024, while in cattle, the increase was from 9.2% to 34.4%. A particularly sharp rise in rRT-PCR-positive samples was observed among goats, where the percentage rose from 4.2% in 2023 to 62.0% in 2024. This growth may be associated with more intense virus circulation among this species or changes in risk factors. Additionally, in some cases, the number of PCR-positive samples exceeded the number of ELISA seropositive samples. This could indicate recent infections, where animals had not yet developed antibodies, or reflect the higher sensitivity of rRT-PCR in detecting active infections.
In this study, sequencing and virus isolation from BTV rRT-PCR-positive samples were not performed due to financial and resource limitations. However, given the importance, detailed characterization of circulating strains will be focused in future studies. The information on the genetic diversity of BTV in Kazakhstan would help to trace the origin and spread of BTV within different ecological and climatic zones within the country.
The presence and the abundance of Culicoides midges, influence iBTV transmission, and understanding their regional diversity is essential for assessing the risk of BTV spread and designing effective control measures.
An ~1,400 species of Culicoides midges have been identified globally, with 130 species reported in the former USSR. Among these, C. obsoletus, C. dewulfi, C. scoticus, C. pulicaris, C. stigma, C. nubeculosus, C. chiopterus, and C. punctatus are widely distributed in Russia, with their range extending up to 72° latitude (26). In Kazakhstan, members of Culicoides subgenera Avaritia, Culicoides, Hofmania, Monoculicoides, Oaecata, Wirthomyia, Silvaticulicoides (Beltranmyia have been reported (26, 33)). According to the literature, the most abundant species in southern Kazakhstan are C. punctatus, C. newsteadi, and C. obsoletus (34, 35). In central Kazakhstan, C. obsoletus, C. dewulfi, and C. montanus dominate (36); in northern Kazakhstan, C. obsoletus, C. punctatus, and C. chiopterus are prevalent (33, 36); while in eastern Kazakhstan, C. grisescens and C. obsoletus are the most common (33, 36).
According to the literature, C. imicola is the primary vector of BTV in Africa, the Middle East, and southern Europe, C. sonorensis in North America, and C. brevitarsis in Australia. It is worth noting that C. imicola is absent from the listed species in Kazakhstan; however, C. obsoletus a known BTV vector in European countries, is present in Kazakhstan. In Europe, the main BTV vectors are midges from the obsoletus and pulicaris complexes (6). C. pulicaris was not detected in any region during this study.
Adult females Culicoides require blood during their gonotrophic cycle. Therefore, they feed on warm-blooded animals every 3–4 days, staying close to the ground. Different species exhibit distinct host preferences: some are anthropophilic, feeding on humans, others are zoophilic, preferring livestock, or ornithophilic, feeding on birds. Their activity patterns also vary, with some being active predominantly at twilight and nighttime, while others feed during the day, either in open spaces or livestock enclosures. Based on the samples collected in this study, host preferences of the Culicoides were; Sheep-−42%, Cattle-−34%, and Goats-−24%.
In this study, nine Culicoides spp. were morphologically identified and the identity of four of the species were confirmed by sequencing 28S rDNA. The 28S marker, is commonly employed in examining the evolutionary relationships among insects. Nevertheless, the effectiveness of the primer employed for amplifying this marker was observed to be diminished in relation to some species. This issue may be attributable to the genetic traits of the taxa influencing the primer binding process, the presence of polymorphisms in the amplification regions specific to these species, or the degradation or low quality of the DNA in the initial samples.
For further research, it is planned to revise the approach to primer selection, including the use of alternative sequences that are more specific to the species in question, or to modify the PCR conditions to improve amplification efficiency.
Out of the nine Culicoides species four (C. Miutissimus, C. Newstedi, C. Pectipennis and C. Sphagnumennis) were identified as potential BTV vectors in Kazakhstan. In southern regions of Kazakhstan, BTV RNA was primarily detected in C. miutissimus and C. sphagnumennis. These findings align with previous studies indicating active virus circulation in these warm and humid regions (34, 35). These species play a critical role in the epidemiological situation in the southern regions, as the environmental conditions there are favorable for their reproduction and activity (33).
In northern Kazakhstan, specifically in regions such as North Kazakhstan and Pavlodar, viral RNA was not detected in any Culicoides spp., likely due to less favorable climatic conditions for the vectors in these areas (18). Climatic factors, such as cold winters and relatively dry summer months, limit their activity and ability to transmit the virus.
As for C. newstedi and C. pectipennis, they exhibited a high level of BTV infection in the Zhambyl region, strongly suggesting their potential role as vectors (34). Previously, these species have also been reported in Russia and Central Asia as important BTV carriers (1).
Additionally, the literature indicates that C. imicola and C. obsoletus, though not widely present in Kazakhstan, are primary vectors of BTV in Europe and Africa (6). These species possess significant epidemiological potential in the global spread of the virus, and if they were to be detected in Kazakhstan, their role in the epidemiological situation could become crucial.
Notably, despite the detection of seropositive animals for BT in the northern regions of Kazakhstan, no PCR-positive animals or vectors were found among susceptible livestock (sheep, goats, and cattle). The presence of seropositive animals in could possibly be explained through animal movement of seropositive animals from the southern regions to the northern regions, or lower level of BT infections. In contrast, the BT situation in southern regions was the presence of both seropositive and PCR-positive animals, as well as PCR-positive midges, were identified. In all southern regions, the RNA of BTV was predominantly detected in C. miutissimus among PCR-positive Culicoides species. However, the prevalence of C. miutissimus in the southern regions varied significantly, ranging from 4.3% to 29.1% (see Figure 2).
These findings have significant and multifaceted implications for the epidemiology of the diseases they transmit (37). Analyzing the obtained results, we aimed to assess the current epidemiological situation of BT in the northern and southern regions of Kazakhstan. However, this study has highlighted the need for a more comprehensive analysis taking into account the climatic heterogeneity of the country. For a more accurate assessment of the risk of virus transmission, it is necessary to consider multiple factors, including climate zones, temperature, the presence and density of susceptible animals, age, sex, vector populations, insect biological activity, prevailing wind directions, and other epidemiologically significant parameters (18, 33, 38).
According to previous mathematical modeling studies (18), it has been scientifically established that the spread of Bluetongue virus is most likely in the southern and southeastern regions of Kazakhstan. The results obtained in our study confirm this prediction, demonstrating a high level of seroprevalence in these areas. This pattern may be associated with favorable climatic conditions that facilitate virus circulation and vector activity. A study conducted in Southern Italy (Campania region) demonstrated that moderately mild winters and high seasonal Culicoides activity contribute to sustained virus circulation (39). Similarly, a meta-analysis of BTV seroprevalence in Africa (40) emphasized that the year-round warm climate and high Culicoides population density ensure continuous virus transmission among various animal species, including cattle, sheep, goats, and camels. However, unlike these regions, BTV circulation in Kazakhstan is restricted exclusively to the warm season, necessitating the development of region-specific control and prevention measures.
Furthermore, the transboundary spread of BTV remains a significant risk factor. However, data on the epidemiological situation in neighboring countries bordering our study regions, such as Russia, Kyrgyzstan, and Uzbekistan, are extremely limited. Therefore, future research should incorporate an assessment of the epizootic situation in border areas and the influence of climatic factors on virus circulation.
Nevertheless, the data obtained not only confirm the current distribution of vectors but also allow for the prediction of BTV dynamics in the northern and southern regions of Kazakhstan. This, in turn, may contribute to the improvement of preventive and anti-epizootic measures in the future.
Conclusion
The results of this study highlight the importance of systematic monitoring of Culicoides spp. in Kazakhstan. The identification of species such as C. miutissimus, C. sphagnumennis, C. newstedi, and C. pectipennis as potential BTV vectors calls for further investigation to develop effective measures to prevent the spread of BTV in the region. The abundance of Culicoides species can vary during the vector season. This study was conducted in the Spring and the Autumn. Therefore, in future studies samples collected in summer months might provide information on additional species of Culicoides that could serve as BTV vectors. To confirm the vector competence of Culicoides species, it is essential to demonstrate not only their ability to acquire the virus from a host but also their capacity to retain and transmit it to a new, uninfected susceptible host. Therefore, detecting the virus in the body of a blood-feeding insect alone is insufficient evidence of its vector competence. Hence, a more precise evaluation requires separate analysis of the insect's head, salivary glands, and body. Additionally, it is crucial to consider climatic and ecological factors that influence vector activity and their ability to transmit the virus. These factors must be integrated into future studies to provide a comprehensive understanding of vector behavior and disease transmission dynamics.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary material, further inquiries can be directed to the corresponding author/s.
Ethics statement
Ethical approval was not required for the study involving animals in accordance with the local legislation and institutional requirements because According to the Law of the Republic of Kazakhstan No. 97-VII ZRK “On Responsible Treatment of Animals” dated December 30, 2021, Ethical Committee approval is not required when sampling animals for the purpose of infectious disease monitoring.
Author contributions
KZ: Conceptualization, Data curation, Formal analysis, Funding acquisition, Investigation, Methodology, Project administration, Resources, Software, Supervision, Validation, Visualization, Writing – original draft, Writing – review & editing. DM: Formal analysis, Methodology, Visualization, Writing – original draft. NS: Formal analysis, Methodology, Writing – original draft. MM: Formal analysis, Methodology, Writing – original draft. SK: Formal analysis, Methodology, Writing – original draft. MA: Formal analysis, Methodology, Writing – original draft. MK: Methodology, Writing – original draft. ST: Methodology, Writing – original draft. MA: Formal analysis, Methodology, Writing – original draft. AM: Formal analysis, Methodology, Writing – original draft. NR: Formal analysis, Methodology, Writing – original draft. KS: Data curation, Methodology, Writing – original draft. SB: Methodology, Validation, Visualization, Writing – review & editing, Conceptualization, Data curation, Formal analysis. AA: Conceptualization, Data curation, Formal analysis, Methodology, Visualization, Writing – review & editing. AK: Investigation, Resources, Supervision, Writing – original draft.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This research was funded by the Science Committee of the Ministry of Science and Higher Education of the Republic of Kazakhstan (Grant Number: AP19676490).
Acknowledgments
The authors express their gratitude to the Science Committee of the Ministry of Science and Higher Education of the Republic of Kazakhstan for funding this research.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as potential conflicts of interest.
Generative AI statement
The author(s) declare that Gen AI was used in the creation of this manuscript. Artificial intelligence was utilized during the preparation of the article to ensure clarity when translating sentences from Kazakh to English.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fvets.2025.1559636/full#supplementary-material
References
1. Sprygin AV, Fedorova OA, Babin YY, Kononov AV, Karaulov AK. Midges of the genus Culicoides (Diptera: Ceratopogonidae) and their role in the spread of bluetongue and Schmallenberg disease in Russia. Agric Biol. (2015) 2:183–197. doi: 10.15389/agrobiology.2015.2.183rus
2. Saminathan M, Singh KP, Khorajiya JH, Dinesh M, Vineetha S, Maity M, et al. An updated review on bluetongue virus: epidemiology, pathobiology, and advances in diagnosis and control with special reference to India. Vet Q. (2020) 40:258–321. doi: 10.1080/01652176.2020.1831708
3. Sperlova A, Zendulkova D. Bluetongue: a review. Vet Med. (2011) 56:430–52. doi: 10.17221/3206-VETMED
4. Maan NS, Maan S, Guimera M, Nomikou K, Morecroft E, Pullinger G, et al. The genome sequence of a reassortant bluetongue virus serotype 3 from India. J Virol. (2012) 86:6375–6. doi: 10.1128/JVI.00671-12
5. Joardar SN, Sanyal A, Abd El Wahed A, Ray S. Posterior positivity distribution analysis of subclinical bluetongue in the eastern and north-eastern states of India: a wakeup call for outbreak preparedness. Viruses. (2025) 17:18. doi: 10.3390/v17010018
6. Purse BV, Mellor PS, Rogers DJ, Samuel AR, Mertens PP, Baylis M. Climate change and the recent emergence of bluetongue in Europe. Nat Rev Microbiol. (2005) 3:171–81. doi: 10.1038/nrmicro1090
7. Guichard S, Guis H, Tran A, Garros C, Balenghien T, Kriticos DJ. Worldwide niche and future potential distribution of Culicoides imicola, a major vector of bluetongue and African horse sickness viruses. PLoS ONE. (2014) 9:e112491. doi: 10.1371/journal.pone.0112491
8. Gibbs EP, Greiner EC. The epidemiology of bluetongue. Comp Immunol Microbiol Infect Dis. (1994) 17:207–20. doi: 10.1016/0147-9571(94)90044-2
9. Maclachlan NJ. Bluetongue: history, global epidemiology, and pathogenesis. Prev Vet Med. (2011) 102:107–11. doi: 10.1016/j.prevetmed.2011.04.005
11. Mayo C, McDermott E, Kopanke J, Stenglein M, Lee J, Mathiason C, et al. Ecological dynamics impacting bluetongue virus transmission in North America. Front Vet Sci. (2020) 7:186. doi: 10.3389/fvets.2020.00186
12. Ma J, Gao X, Liu B, Xiao J, Chen H, Wang H. Spatial patterns and risk factors of bluetongue virus infection in inner Mongolia, China. Vector Borne Zoonotic Dis. (2019) 19:525–32. doi: 10.1089/vbz.2018.2361
13. Li J, Li K, Shahzad M, Han Z, Nabi F, Gao J, et al. Seroprevalence of bluetongue virus in domestic yaks (Bos grunniens) in Tibetan regions of China based on circulating antibodies. Trop Anim Health Prod. (2015) 47:1221–3. doi: 10.1007/s11250-015-0853-0
14. Avci O. Detection of antibodies against Blue tongue virus in yaks (Bos grunniens) in Issyk kul, first report. J Anim Plant Sci. (2014) 24:1220–3.
15. Lundervold M, Milner-Gulland EJ, O'Callaghan CJ, Hamblin C. First evidence of bluetongue virus in Kazakhstan. Vet Microbiol. (2003) 92:281–7. doi: 10.1016/S0378-1135(02)00365-6
16. Lundervold M, Milner-Gulland EJ, O'Callaghan CJ, Hamblin C, Corteyn A, Macmillan AP. A serological survey of ruminant livestock in Kazakhstan during post-Soviet transitions in farming and disease control. Acta Vet Scand. (2004) 45:211–24. doi: 10.1186/1751-0147-45-211
17. Zhigailov AV, Perfilyeva YV, Maltseva ER, Ostapchuk YO, Cherusheva AS, Naizabayeva DA, et al. Identification and characterization of bluetongue virus in Culicoides spp. and clinically healthy livestock in southeastern Kazakhstan. Comp Immunol Microbiol Infect Dis. (2022) 90–91:101895. doi: 10.1016/j.cimid.2022.101895
18. Abdrakhmanov SK, Beisembayev KK, Sultanov AA, Mukhanbetkaliyev YY, Kadyrov AS, Ussenbayev AY, et al. Modelling bluetongue risk in Kazakhstan. Parasit Vectors. (2021) 14:491. doi: 10.1186/s13071-021-04945-6
19. “Chapter 1.1.2. – Collection, submission and storage of diagnostic specimens,” in Manual of Diagnostic Tests Vaccines for Terrestrial Animals (OIE) (2018). Available online at: https://www.woah.org/fileadmin/Home/eng/Health_standards/tahm/1.01.02_COLLECTION_DIAG_SPECIMENS.pdf (accessed December 04, 2024).
20. Gulenkin VM, Karaulov AK, Varkentin AV. Representative and randomized assessment of the required number of diagnostic samples to prove the freedom of domestic poultry populations from infection at low disease prevalence. Proc Fed Cent Anim Health. (2020) 17:207–21.
22. Smith M, Jones R. Using GIS to analyze spatial data in ecology. J Environ GIS. (2020) 15:245–60.
23. Niedbalski W. Evaluation of commercial ELISA kits for the detection of antibodies against bluetongue virus. Pol J Vet Sci. (2011) 14:615–9. doi: 10.2478/v10181-011-0091-y
24. Rawlings P. A key, based on wing patterns of biting midges (genus Culicoides Latreille-Diptera: Ceratopogonidae) in the Iberian Peninsula, for use in epidemiological studies. Graellsia. (1996) 52:57–71. doi: 10.3989/graellsia.1996.v52.i0.376
25. Mirzaeva AG. Blood Sucking Diptera: Ceratopodonidae in Siberia and the Far East. Novosibirsk: Nauka (Science) (1989).
26. Glukhova VM. “Blood-sucking midges of the genera Culicoides and Forcipomyia (Ceratopogonidae),” In:Cêtre-Sossah C, Baldet T, Delécolle JC, Mathieu B, Perrin A, Grillet C, Albina E, , editors. Fauna of the USSR. Dipteran Insects, Vol. 3. Leningrad: Nauka (1989), p. 408.
27. Lehmann K, Werner D, Hoffmann B, Kampen H. PCR identification of culicoid biting midges (Diptera, Ceratopogonidae) of the Obsoletus complex including putative vectors of bluetongue and Schmallenberg viruses. Parasites Vectors. (2012) 5:213. doi: 10.1186/1756-3305-5-213
28. Cêtre-Sossah C, Baldet T, Delécolle JC, Mathieu B, Perrin A, Grillet C, et al. Molecular detection of Culicoides spp. and Culicoides imicola, the principal vector of bluetongue (BT) and African horse sickness (AHS) in Africa and Europe. Vet Res. (2004) 35:325–37. doi: 10.1051/vetres:2004015
29. Lassen SB, Nielsen SA, Skovgård H, Kristensen M. Molecular identification of bloodmeals from biting midges (Diptera: Ceratopogonidae: Culicoides Latreille) in Denmark. Parasitol Res. (2011) 108:823–9. doi: 10.1007/s00436-010-2123-4
30. Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. (1987) 4:406–25.
31. Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. (1985) 39:783–91. doi: 10.1111/j.1558-5646.1985.tb00420.x
32. Tamura K, Nei M. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol Biol Evol. (1993) 10:512–26.
33. Tamura K., Stecher G., Kumar S. MEGA 11: molecular evolutionary genetics analysis Version 11. Mol Biol Evol. (2021) 38:3022–7. doi: 10.1093/molbev/msab120
34. Zhigaylov AV, Ostapchuk EO, Perfilieva YV, Abdolla N, Maltseva ER, Naizabayeva DA, et al. Risk analysis of bluetongue virus spread in Kazakhstan. Bull Med Geogr Ser. (2022) 106:71–81. doi: 10.31489/2022BMG1/71-81
35. Bulatov Y, Zhugunissov K, Taranov D, Amanova Z, Koshemetov G, Nakhanova G, et al. Prevalence of bluetongue virus among goats, sheep, and cattle in the southern regions of Kazakhstan. In: 99th Conference of Research Workers in Animal Diseases. Chicago, IL: Chicago Marriott, Downtown Magnificent Mile (2018).
36. Amanova Z, Bulatov Y, Zhugunissov K. The species composition and numerical ratio of bluetongue vectors (Culicoides midges) in Southern Kazakhstan. In:Zakarya K, , editor. Proceedings of the International Scientific and Practical Conference Biotechnology and Biological Safety: Achievements and Development Prospects. Almaty: RIBSP (2023). p. 172–3.
37. Aubakirova AM. Fauna and ecology of midges on high-altitude pastures in the southwestern Altai. Bull Altai State Univ. (2017) 10:92–5.
38. Sick F, Beer M, Kampen H, Wernike K. Culicoides biting midges-underestimated vectors for arboviruses of public health and veterinary importance. Viruses. (2019) 11:376. doi: 10.3390/v11040376
39. Selim A, Alsubki RA, Albohairy FM, Attia KA, Kimiko I. A survey of bluetongue infection in one-humped camels (Camelus dromedarius); seroprevalence and risk factors analysis. BMC Vet Res. (2022) 18:322. doi: 10.1186/s12917-022-03421-2
Keywords: bluetongue, Kazakhstan, Culicoides, serology, RNA detection
Citation: Zhugunissov K, Muzarap D, Sarsenkulova N, Mambetaliyev M, Kilibayev S, Azanbekova M, Kenzhebayeva M, Tabys S, Abayeva M, Melisbek A, Rametov N, Sultankulova K, Babiuk S, Ambagala A and Kerimbayev A (2025) Prevalence of Bluetongue and the distribution of Culicoides species in northern and southern regions of Kazakhstan in 2023–2024. Front. Vet. Sci. 12:1559636. doi: 10.3389/fvets.2025.1559636
Received: 13 January 2025; Accepted: 13 February 2025;
Published: 06 March 2025.
Edited by:
Francesco Mira, Istituto Zooprofilattico Sperimentale della Sicilia “A. Mirri”, ItalyReviewed by:
Siddhartha Narayan Joardar, West Bengal University of Animal and Fishery Sciences, IndiaGianmarco Ferrara, University of Messina, Italy
Gamil S. G. Zeedan, National Research Centre, Egypt
Ouafaa Fassi Fihri, Agronomic and Veterinary Institute Hassan II, Morocco
Copyright © 2025 Zhugunissov, Muzarap, Sarsenkulova, Mambetaliyev, Kilibayev, Azanbekova, Kenzhebayeva, Tabys, Abayeva, Melisbek, Rametov, Sultankulova, Babiuk, Ambagala and Kerimbayev. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Kuandyk Zhugunissov, ay56aHVndW5pc292QGJpb3NhZmV0eS5reg==; Aruna Ambagala, YXJ1bmEuYW1iYWdhbGFAaW5zcGVjdGlvbi5nYy5jYQ==
 Kuandyk Zhugunissov
Kuandyk Zhugunissov Dias Muzarap1
Dias Muzarap1 Nurkuisa Rametov
Nurkuisa Rametov Shawn Babiuk
Shawn Babiuk Aruna Ambagala
Aruna Ambagala