
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Vet. Sci., 07 March 2025
Sec. Animal Nutrition and Metabolism
Volume 12 - 2025 | https://doi.org/10.3389/fvets.2025.1558856
This article is part of the Research TopicAdvancements in Synthetic Microbiomes for Enhancing Animal HealthView all 9 articles
Heat stress significantly impacts dairy cow productivity, health, and welfare. This study evaluated a self-developed herbal formula as a dietary intervention to mitigate heat stress. A total of 198 lactating cows were divided into two groups: a Control group receiving standard total mixed rations and a Herbs group supplemented with herbal formula for 60 days. Various parameters were assessed, including milk yield and composition, antioxidant capacity, immune responses, stress-related gene expression, and rumen microbial composition. Compared to the Control group, cows in the Herbs group showed improved feed intake, milk yield and quality, rumination frequency, and enhanced antioxidant activity and immune response. Rumen microbiome analysis revealed a reduced relative abundance of Proteobacteria and Ochrobactrum in the Herbs group, along with an enrichment of beneficial genera such as Lachnospira. Functional predictions indicated that the Herbs group exhibited enhanced glycolysis/gluconeogenesis, pyruvate metabolism, and starch and sucrose metabolism, reflecting improved fermentation efficiency and energy utilization. In conclusion, the herbal formula improved physiological and biochemical attributes, boosted antioxidant and immune responses, and modulated the rumen microbiome, contributing to the alleviation of heat stress in dairy cows. These findings highlight its potential as a natural dietary strategy to support dairy cow health and productivity under heat stress conditions.
Heat stress is a critical challenge faced by the dairy industry worldwide, significantly compromising milk production, reproductive efficiency, and overall animal health (1–3). Dairy cows are particularly vulnerable to rising environmental temperatures due to their intensive metabolic heat production and small surface:volume ratio (4). Heat stress not only disrupts the physiological and hormonal balance of cows but also alters their feeding behavior, leading to reduced feed intake and nutrient absorption (2, 5, 6). In recent years, dietary strategies have emerged as an effective approach to mitigate the negative impacts of various types of stress (7–9). Among these, herbal supplements have gained increasing attention due to their potential to enhance antioxidant capacity, modulate immune responses, and regulate stress-related metabolic pathways (10–13). For example, Turmeric has shown significant potential in poultry nutrition, with its active compound, Curcumin, improving digestive enzyme activity, leading to better nutrient absorption and utilization. Additionally, Curcumin exhibits immunomodulatory and antioxidant properties, enhancing immune responses, reducing infections, and mitigating oxidative stress (14). Furthermore, Astragalus and ginseng polysaccharide supplementation have demonstrated significant benefits in improving growth performance, liver function, and intestinal villus morphology in weaned piglets. These plant polysaccharides improve nutrient absorption by promoting healthier intestinal structures and regulate immune function through activation of the TLR4-mediated MyD88-dependent signaling pathway (15).
The gut microbiome has recently gained significant attention for its vital role in regulating host health, immunity, and metabolism (16–20). In animals, the gut microbiota plays a key role in nutrient absorption, disease resistance, and overall productivity, drawing considerable interest from researchers aiming to enhance health and performance by modulating its composition and functions (21–23). Among ruminants, a large portion of the gut microbiome is housed in the rumen, a specialized digestive compartment. The rumen microbiome, distinct from the broader gut microbiome, is essential for breaking down plant fibers and fermenting nutrients into metabolites critical for energy production and growth (24).
Heat stress, however, can disrupt the microbial balance, impairing fermentation efficiency and overall digestive function. Investigating how dietary interventions influence rumen microbial composition and functions under heat stress is essential for developing targeted strategies to support microbial stability and enhance resilience in dairy cows. This study investigated a self-developed herbal formula as a dietary intervention to mitigate heat stress. By assessing its effects on physiological and biochemical attributes, antioxidant and immune responses, stress-related gene expression, and rumen microbial composition, this research evaluated whether the herbal formula could alleviate heat stress and support sustainable dairy farming practices.
All animal procedures used in this experiment were reviewed and approved by the Institutional Animal Care and Use Committee of Hebei Agricultural University (Baoding, China).
Herbal formula composition: Astragalus Root (Huangqi) 175 parts, Codonopsis Root (Dangshen) 60 parts, Angelica Sinensis (Danggui) 60 parts, Prepared Rehmannia Root (Shudi) 60 parts, Dried Tangerine Peel (Chenpi) 60 parts, Oriental Arborvitae Leaves (Cebaiye) 80 parts, Atractylodes Rhizome (Baizhu) 45 parts, Licorice Root (Gancao) 30 parts, Sichuan Lovage Rhizome (Chuanxiong) 30 parts, Ophiopogon Root (Maidong) 40 parts, White Peony Root (Baishao) 40 parts, Roasted Hawthorn Fruit (Zhishanzha) 100 parts, Roasted Radish Seeds (Laifuzi) 60 parts, Wine-processed Rhubarb Root (Jiu-Zhi Dahuang) 10 parts, Oriental Wormwood (Yincheng) 60 parts, Roasted Pig Hoof Shells (Zhi Zhutijia) 60 parts. All the components were purchased from Anguo Jufu Herbal Medicine Co., Ltd. (Hebei, China) and prepared following the guidelines of the classical Chinese Pharmacopoeia (State Pharmacopoeia Commission of the PRC, 2005).
This study was conducted on a commercial dairy farm in Hebei Province, China. A total of 198 lactating cows, with a parity distribution of 1 to 3 and an average milk production of 41.22 ± 5.05 kg/day, were selected for a 60-day experiment. The cows were divided into two groups: the Control group (n = 100), which received the farm standard total mixed ration (TMR), and the Herbs group (n = 98), which was fed the same TMR supplemented with the prepared herbal formula. The preliminary research has determined the optimal dosage of this self-developed traditional Chinese medicine formula for alleviating heat stress in dairy cows (25). Based on the findings, this study administered 20 g per head per day. The herbal formulation was evenly divided into three portions and thoroughly mixed into the TMR before each feeding, with each cow fed three times daily. Details of the TMR ingredients and nutritional composition are provided in Supplementary Table S1. Cows were housed in an open shed equipped with electric fans and provided with free access to water.
Ambient temperature (AT) and relative humidity (RH) in the cowshed were measured using KTH-350-I temperature and humidity data-logger (Kimo Industry Co., French) at 30-min intervals over 24 h for a 60-day experimental period to calculate the temperature-humidity index (THI) (26):
Cows were fed three times daily at approximately 5:30 am, 1:30 pm, and 9:30 pm. Feed intake for each group was recorded on days 1–3, 14–16, 29–31, 44–46, and 58–60 of the experiment. On the same days, milk samples (100 mL) were collected from each group before feeding. The samples were then pooled in a 4:3:3 ratio (morning, midday, and evening milkings), stored at 4°C, and sent to the Dairy Herd Improvement (DHI) center (Shijiazhuang, China) within 12 h for composition analysis, including milk fat percentage (FP), protein percentage (PP), lactose percentage (LP), total solids (TS), somatic cell count (SCC), and blood urea nitrogen (BUN).
At the end of the experiment, blood samples were collected from 10 randomly selected cows in each group before the morning feeding. For each animal, two 5 mL blood samples were collected via the jugular vein: one into a vacuum tube without anticoagulant and the other into a tube containing heparin sodium. The sample in the anticoagulant-free tube was centrifuged at 3,000 g for 20 min at 4°C to obtain serum, which was stored at −20°C for subsequent analyses. Alanine aminotransferase (ALT), aspartate aminotransferase (AST), alkaline phosphatase (ALP), lactate dehydrogenase (LDH), creatine kinase (CK), blood urea nitrogen (BUN), total antioxidant capacity (T-AOC), superoxide dismutase (SOD), glutathione peroxidase (GSH-Px), and malondialdehyde (MDA) in serum were measured using a fully automatic biochemical analyzer (Gaomi Rainbow Analytical Instrument Co., Ltd., Shandong, China). Catalase (CAT) levels were determined using ELISA kit (Nanjing Jiancheng Biotechnology Research Institute Co., Ltd), following the manufacturer’s instructions. Interleukins (IL-1, IL-2, IL-6, IL-12), tumor necrosis factor-α (TNF-α), gamma-interferon (IFN-γ), and immunoglobulins (IgG, IgM, IgA) were analyzed using ELISA kit according to the manufacturer’s protocols (Dakemei Technology Co., Ltd., Beijing, China).
Blood samples collected in heparinized tubes were used to isolate peripheral blood lymphocytes (PBLCs) for the measurement of heat stress-related gene expression. Total RNA was extracted from PBLCs using a magnetic tissue/cell/blood total RNA kit (Tiangen Biotech, Beijing, China). Purified total RNA (1 μg) was reverse transcribed into cDNA using the PrimeScript™ RT Reagent Kit with gDNA Eraser (Takara Biotechnology, Dalian, China). RT-qPCR amplification was performed using SYBR® Premix Ex Taq™ II (Tli RNase H Plus) (Takara Biotechnology, Dalian, China), according to the manufacturer’s protocol. Each gene was analyzed in three technical replicates. The relative changes in target gene expression were calculated using the method. The RT-qPCR primers for each gene were designed using Primer Premier (version 5.0, Premier, Canada) and are listed in the Supplementary Table S2.
Statistical analysis was performed using an independent samples t-test in SPSS 18.0 software (SPSS, Chicago, IL, United States). The bar charts in this study display mean values with standard deviations, with significant differences between groups indicated by an asterisk (*) for p < 0.05.
Six cows from each group, previously used for blood collection, were selected for rumen fluid sampling via oral lavage. Approximately 10 mL of rumen content was collected from each cow and stored at −80°C for microbial sequencing. Genomic DNA from rumen content samples was extracted using the DNeasy PowerSoil Kit (Qiagen, Valencia, CA, United States) following the manufacturer’s protocol. DNA concentration and quality were assessed using a NanoDrop One spectrophotometer (Thermo Fisher Scientific, Madison, WI, United States) and 1% agarose gel electrophoresis. The DNA was subsequently diluted to the appropriate concentration for PCR.
The 16S rRNA gene regions of bacteria and archaea were amplified using specific primers. For the bacterial 16S V3–V4 region, the forward primer (ACTCCTACGGGAGGCAGCAG) and reverse primer (GGACTACHVGGGTWTCTAAT) were used, while for the archaeal 16S V3–V4 region, the forward primer (ACGGGGYGCAGCAGGCGCGA) and reverse primer (GGACTACVSGGGTATCTAAT) were employed. PCR conditions for bacteria included an initial denaturation at 94°C for 5 min, followed by 28 cycles of denaturation at 94°C for 30 s, annealing at 55°C for 30 s, and extension at 72°C for 60 s, with a final extension at 72°C for 7 min. For archaea, PCR conditions included an initial denaturation at 94°C for 5 min, followed by 35 cycles of denaturation at 94°C for 30 s, annealing at 63°C for 30 s, and extension at 72°C for 60 s, with a final extension at 72°C for 7 min. All reactions were held at 4°C after completion.
The PCR products were electrophoresed on a 1% agarose gel to confirm the expected size of the amplicons. Purification of the PCR products was performed using Agencourt AMPure XP beads, and the purified products were used to create libraries, which were sequenced on the Illumina MiseqPE300 platform (AllweGene Technology Company, Beijing, China).
The 16S rRNA gene sequences for bacteria and archaea were analyzed using QIIME 2 (2019.7 release) (27). Paired-end reads were imported into QIIME 2, merged to combine forward and reverse reads, and filtered to exclude low-quality sequences based on quality scores. The Deblur plugin was used to denoise sequences, remove chimeric and low-abundance reads, and generate amplicon sequence variants (ASVs) with 100% sequence identity, providing high-resolution insights into microbial communities. The ASVs, along with their read counts per sample, were used to construct an ASV table, which was subsequently rarefied to standardize sequencing depth across all samples for diversity analyses. Representative sequences derived from the ASVs were used for taxonomic classification (28). Taxonomic classification was achieved using a naive Bayes classifier trained on the Greengenes database (version 13_8) tailored to the primer region used in this study (29).
Alpha and beta diversity metrics were calculated using rarefied data to standardize sequencing depth across all samples, reducing biases from differences in sequencing effort. In this study, alpha diversity metrics, including Shannon, Observed_ASVs, and Chao1 indices, and beta diversity metrics, including Bray–Curtis and Jaccard distances, were assessed. Statistical significance of beta diversity differences between groups was evaluated using ANOSIM. Functional prediction was performed using PICRUSt2, which generated KEGG pathway-based outputs to infer the functional potential of the microbial communities (30, 31).
Differences in alpha diversity between the two groups were assessed using the Kruskal–Wallis test in R to evaluate statistical significance. Principal coordinate analysis (PCoA) plots were used to visualize Bray–Curtis and Jaccard distances. Stacked bar plots of microbial composition at the phylum, family, and genus levels were created in R using the ggplot2 package (32). Differentially abundant taxa between the two groups were identified using the LEfSe algorithm on the Galaxy platform1 with default settings (LDA score >2). The importance of ASVs in differentiating the control and Herbs groups was evaluated using the randomForest package in R. Random Forest classification models were built with 1,000 trees to calculate the importance of each ASV based on the mean decrease in accuracy. The varImpPlot function was used to visualize the top 20 ASVs with the highest importance values, and the pheatmap package was employed to display the relative abundance of these ASVs across the two groups (33). Functional predictions based on PICRUSt2 results were visualized using STAMP (Statistical Analysis of Metagenomic Profiles) software (34).
The diurnal temperature in the experimental barn averaged 26.51°C, with an average relative humidity of 71.65% and an average temperature-humidity index of 75.71 throughout the experimental period, indicating potential heat stress in the animals (Supplementary Figure S1). The Herbs group showed significant improvements in feed intake, rumination frequency, and milk production compared to the Control group on days 29–31, 44–46, and 58–60 (p < 0.05; Figures 1A–C), with no significant differences observed on days 1–3 and 14–16 (p > 0.05). Similarly, the effects of the herbal formula on milk composition were primarily evident at these later time points (29–31 days, 44–46 days, and 58–60 days; Table 1). Compared to the Control group, the Herbs group showed significantly higher FP, PP, LP, and TS at these time points (p < 0.05), except for TS at 29–31 days and PP at 44–46 days, where no significant differences were observed (p > 0.05). Additionally, SCC and BUN levels were consistently reduced in the Herbs group at these three later time points (p < 0.05). Together, these findings demonstrate that the effects of herbal formula become more pronounced over time, resulting in significant improvements in milk yield, milk quality, and health indicators in heat-stressed dairy cows.
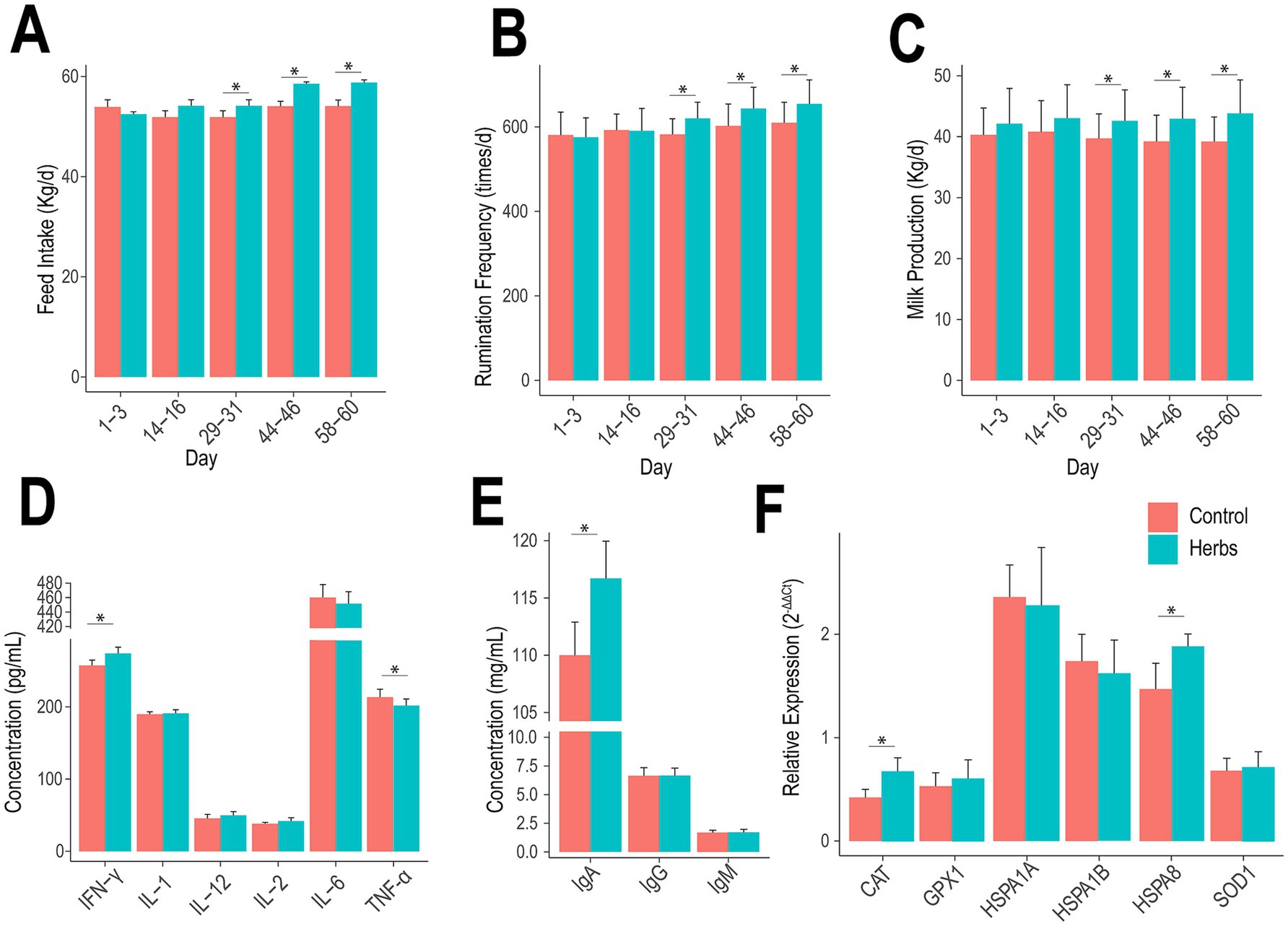
Figure 1. Effects of herbal formula supplementation on feed intake (A), rumination frequency (B), milk production (C), cytokine levels (D), immunoglobulin levels (E), and relative expression of heat stress-related genes (F).
The administration of herbal formula significantly affected serum biochemical parameters in heat-stressed dairy cows (Table 2). Serum BUN levels were significantly reduced in the herbs group (p < 0.05), consistent with the previously observed decrease in milk BUN levels. Additionally, the herbs group exhibited significantly lower MDA levels and higher T-AOC, SOD, GSH-Px, and CAT activities (p < 0.05), indicating enhanced antioxidant defenses and reduced oxidative stress. No significant differences were observed in ALT, AST, ALP, LDH, or CK levels between the two groups (p > 0.05).
The pro-inflammatory cytokine analysis showed that the Herbs group had significantly higher levels of IFN-γ and lower levels of TNF-α compared to the Control group (p < 0.05), with no significant differences observed for IL-1, IL-2, IL-6, and IL-12 (p > 0.05; Figure 1D). Immunoglobulin analysis revealed significantly higher IgA concentrations in the Herbs group (p < 0.05), while IgG and IgM concentrations remained similar between the two groups (p > 0.05; Figure 1E). For heat stress-related gene expression, the Herbs group displayed significantly higher expression of CAT and HSPA8 (p < 0.05), whereas no significant differences were detected for GPX1, HSPA1A, HSPA1B, and SOD1 (p > 0.05; Figure 1F).
Alpha diversity was assessed using the Shannon index, observed ASVs, and Chao1 index for both bacterial (Supplementary Figure S2A) and archaeal (Supplementary Figure S2B) communities. In the bacterial community, the Herbs group exhibited slightly lower Shannon index and observed ASVs compared to the Control group, while the Chao1 index was slightly higher. For the archaeal community, the Herbs group showed slightly lower values for all three indices compared to the Control group. However, statistical analysis revealed that none of these differences were significant (p > 0.05).
The PCoA plots illustrate differences in community structure between the Control and Herbs groups, supported by ANOSIM results. For bacteria (Figure 2A), Bray–Curtis distance showed significant differences (R = 0.22, p = 0.04), while Jaccard distance did not show significant separation (R = 0.09, p = 0.19). For archaea (Figure 2B), both Bray–Curtis (R = 0.22, p = 0.04) and Jaccard distances (R = 0.49, p = 0.004) indicated significant differences.
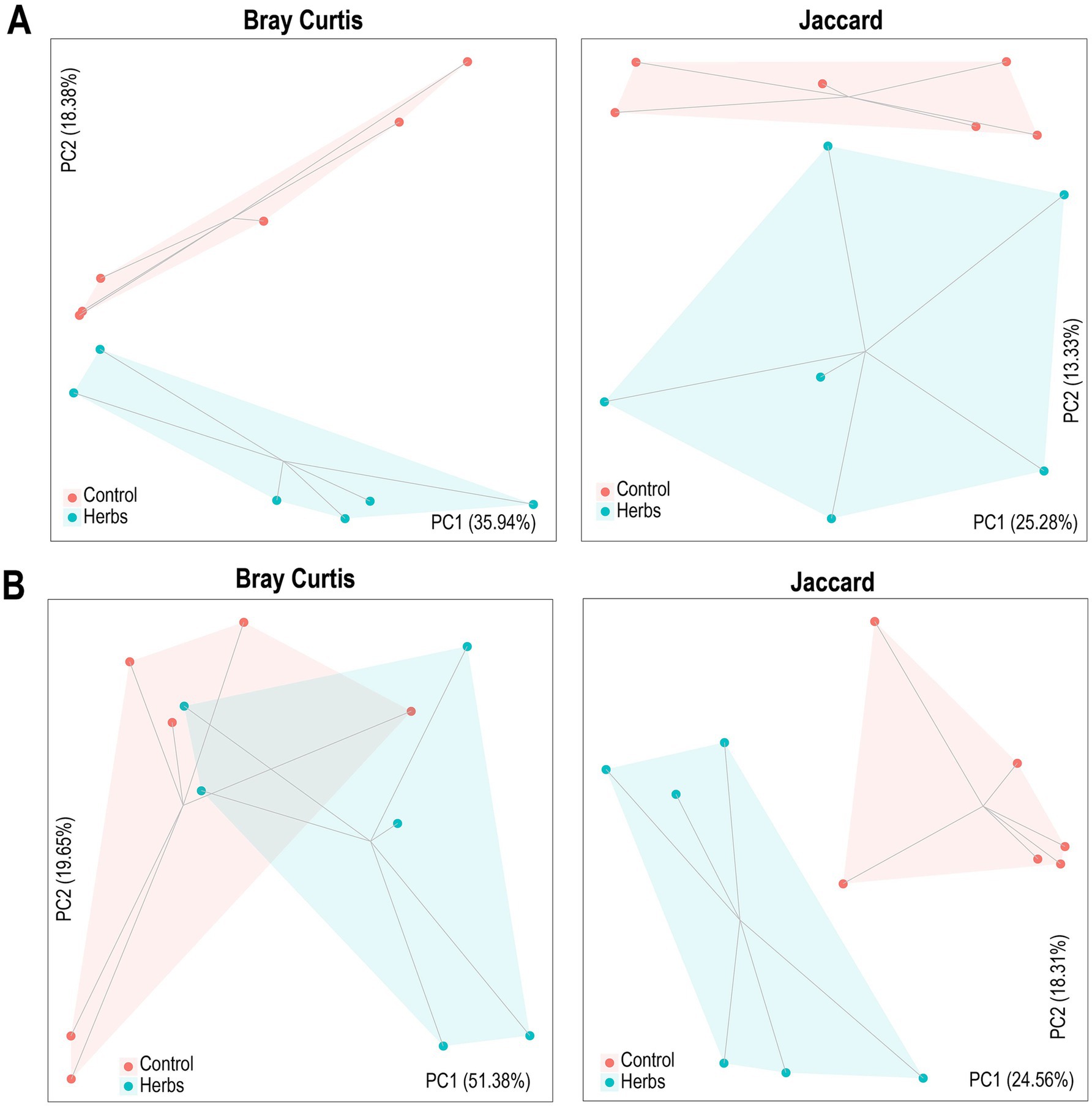
Figure 2. Principal coordinate analysis (PCoA) of Bray–Curtis and Jaccard distances for bacterial (A) and archaeal (B) communities. Each point represents a unique sample, with shaded areas indicating group clustering. Lines connect samples within each group, and statistical differences were evaluated using ANOSIM.
For the rumen bacteria, Firmicutes and Proteobacteria were the dominant phyla across all samples, accounting for 73.0 and 10.1% of the sequences in the Herbs group, compared to 61.7 and 19.1% in the Control group, respectively (Figure 3A). The relative abundance of Bacteroidetes slightly increased in the Herbs group (10.2%) compared to the Control group (7.7%), while Actinobacteria decreased (Control: 6.1%, Herbs: 3.4%). At the family level (Figure 3B), Lachnospiraceae and Ruminococcaceae were consistently dominant, with a slight decrease in Lachnospiraceae from 17.0% (Control) to 16.2% (Herbs) and stable levels of Ruminococcaceae around 13%. Staphylococcaceae, however, exhibited an increase in the Herbs group (Control: 2.6%, Herbs: 19.4%). At the genus level (Figure 3C), Staphylococcus exhibited the most pronounced change, increasing significantly in the Herbs group (19.1%) compared to the Control group (2.5%) (Figure 3C). Other genera showed more variable and inconsistent patterns between groups, with no clear trends emerging.
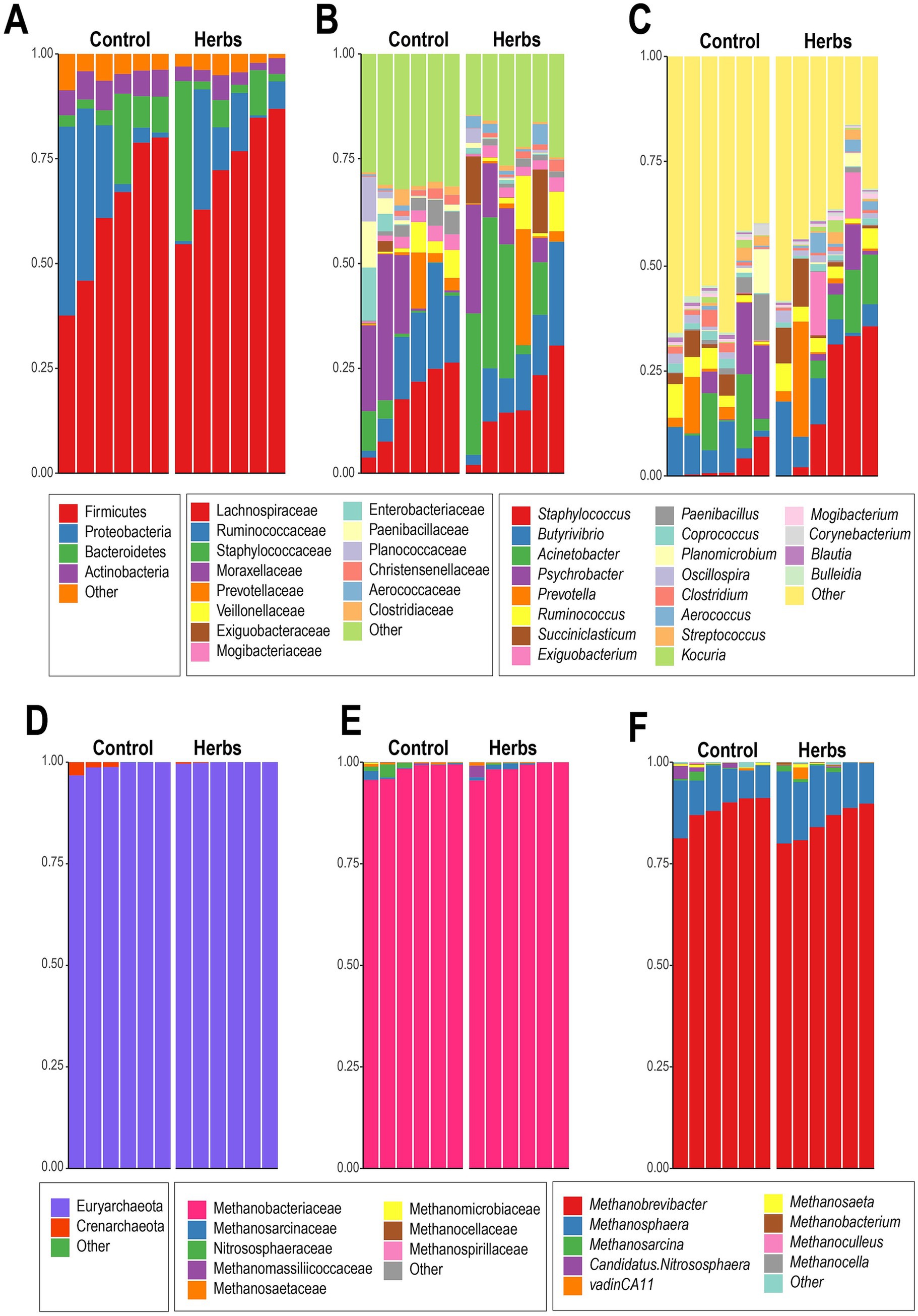
Figure 3. Taxonomic composition of bacterial and archaeal communities in the Control and Herbs groups. Bacterial composition at the phylum level (A), family level (B), and genus level (C). Archaeal composition at the phylum level (D), family level (E), and genus level (F).
The archaeal community was dominated by Euryarchaeota, accounting for around 99% of the sequences in both the Control and Herbs groups (Figure 3D). At the family level, Methanobacteriaceae was the predominant family, representing approximately 98% of the sequences in both groups (Figure 3E). At the genus level, the community was primarily composed of Methanobrevibacter, which accounted for 88.1% in the Control group and 85.0% in the Herbs group, and Methanosphaera, contributing 9.5 and 13.2% in the Control and Herbs groups, respectively (Figure 3F). These results indicated that the archaeal community was dominated by similar taxa in both groups, although beta diversity analysis revealed differences in community structure.
LEfSe analysis revealed distinct taxa enriched in the Control and Herbs groups (Figure 4A). The Herbs group showed significant enrichment of Aerococcus, Lachnospira, and Kocuria, along with taxa associated with Chloroplast and Streptophyta (p < 0.05). In contrast, the Control group had higher abundances of Ochrobactrum, Sanguibacteraceae, and Proteobacteria (p < 0.05). The relative abundances of Aerococcus, Lachnospira, Ochrobactrum, and Proteobacteria were further displayed in boxplots, highlighting the greater abundance of Aerococcus and Lachnospira in the Herbs group and Ochrobactrum and Proteobacteria in the Control group (Figure 4B).
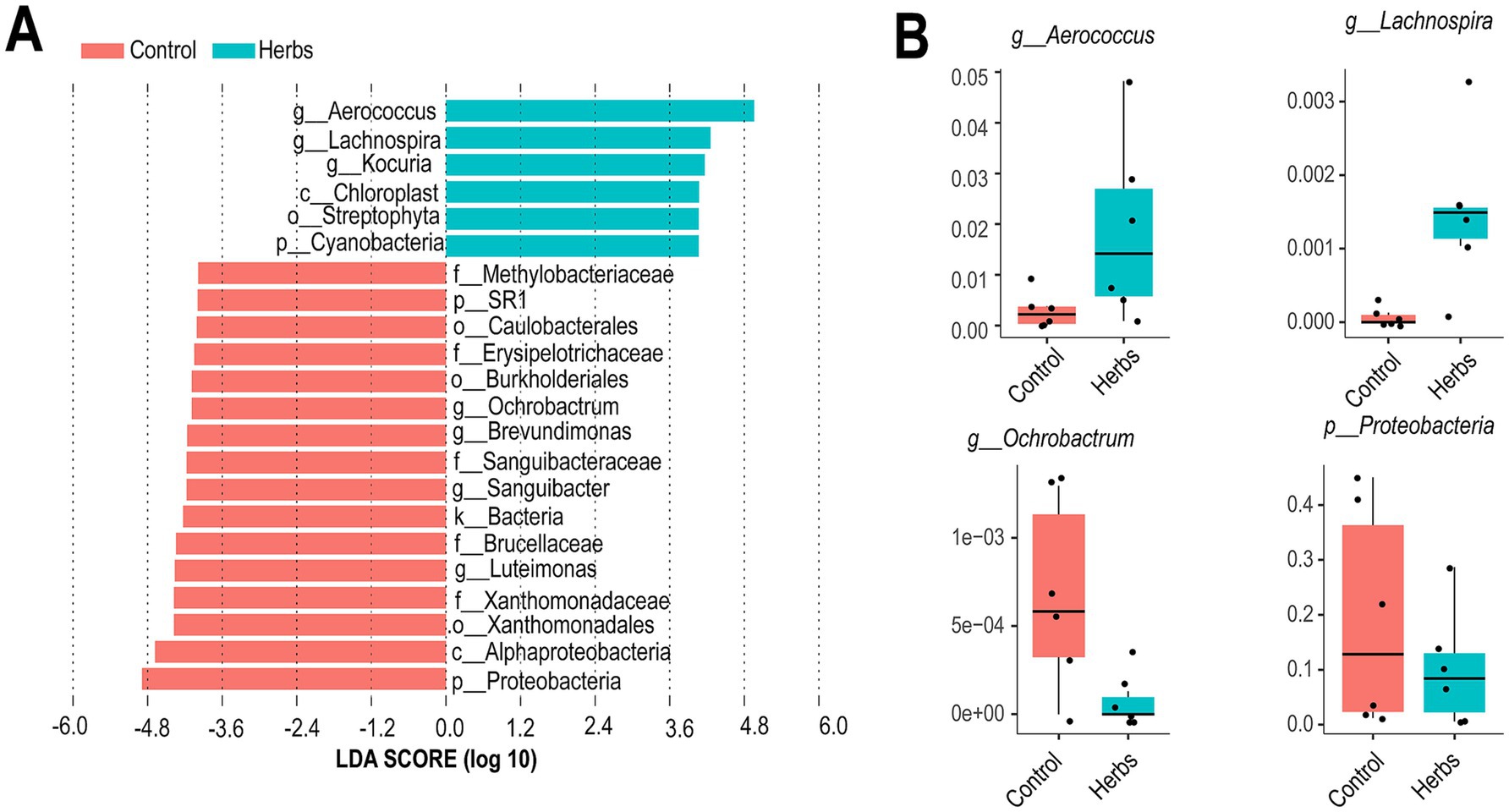
Figure 4. Differentially abundant taxa between the Control and Herbs groups identified by LEfSe analysis. Linear discriminant analysis (LDA) scores show the taxa enriched in the Control (red) and Herbs (blue) groups (A). Relative abundances of selected taxa in the two groups (B).
The random forest analysis and heatmap visualization identified key ASVs that best differentiate the Control and Herbs groups. Among the top 20 ASVs identified by random forest analysis, bacterial taxa included Butyrivibrio, Aerococcus, and Staphylococcus (Figure 5A), while archaeal taxa included Methanosphaera, Methanobrevibacter, and Nitrososphaera (Figure 5B).
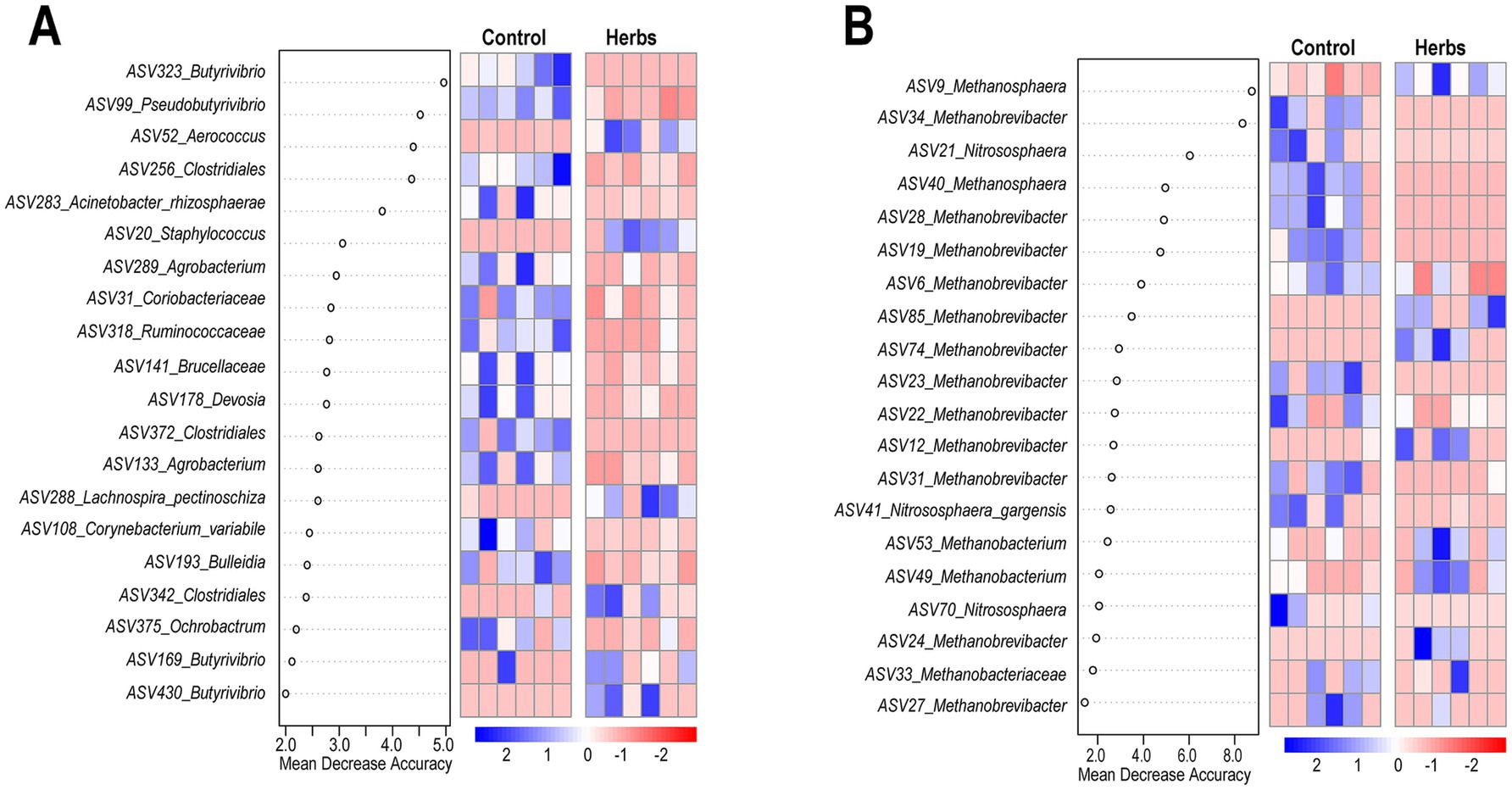
Figure 5. Gut microbiota signature of the Control and Herbs groups determined by Random Forest. Top 20 ASVs from the bacterial community that best differentiate the Herbs group from the Control group (A). Top 20 ASVs from the archaeal community (B). Heatmaps display the relative abundance (log10 transformed) of ASVs selected by Random Forest, with blue and red indicating higher and lower abundance, respectively.
The PICRUSt analysis revealed significant differences (p < 0.05) in predicted KEGG pathways between the Control and Herbs groups in both bacterial (Figure 6A) and archaeal (Figure 6B) communities. In the bacterial community (Figure 6A), genes related to nicotinate and nicotinamide metabolism were predicted to be more abundant in the Control group, while genes involved in D-arginine and D-ornithine metabolism and glycine, serine, and threonine metabolism were enriched in the Herbs group. In the archaeal community (Figure 6B), 10 pathways were identified as significantly different between groups (p < 0.05), with nine pathways, including those involved in starch and sucrose metabolism, pyruvate metabolism and glycolysis/gluconeogenesis, predicted to be enriched in the Herbs group. Only one pathway, bacterial secretion system, was predicted to be more abundant in the Control group. These results suggested that the herbal formula influenced key metabolic pathways in both bacterial and archaeal communities.
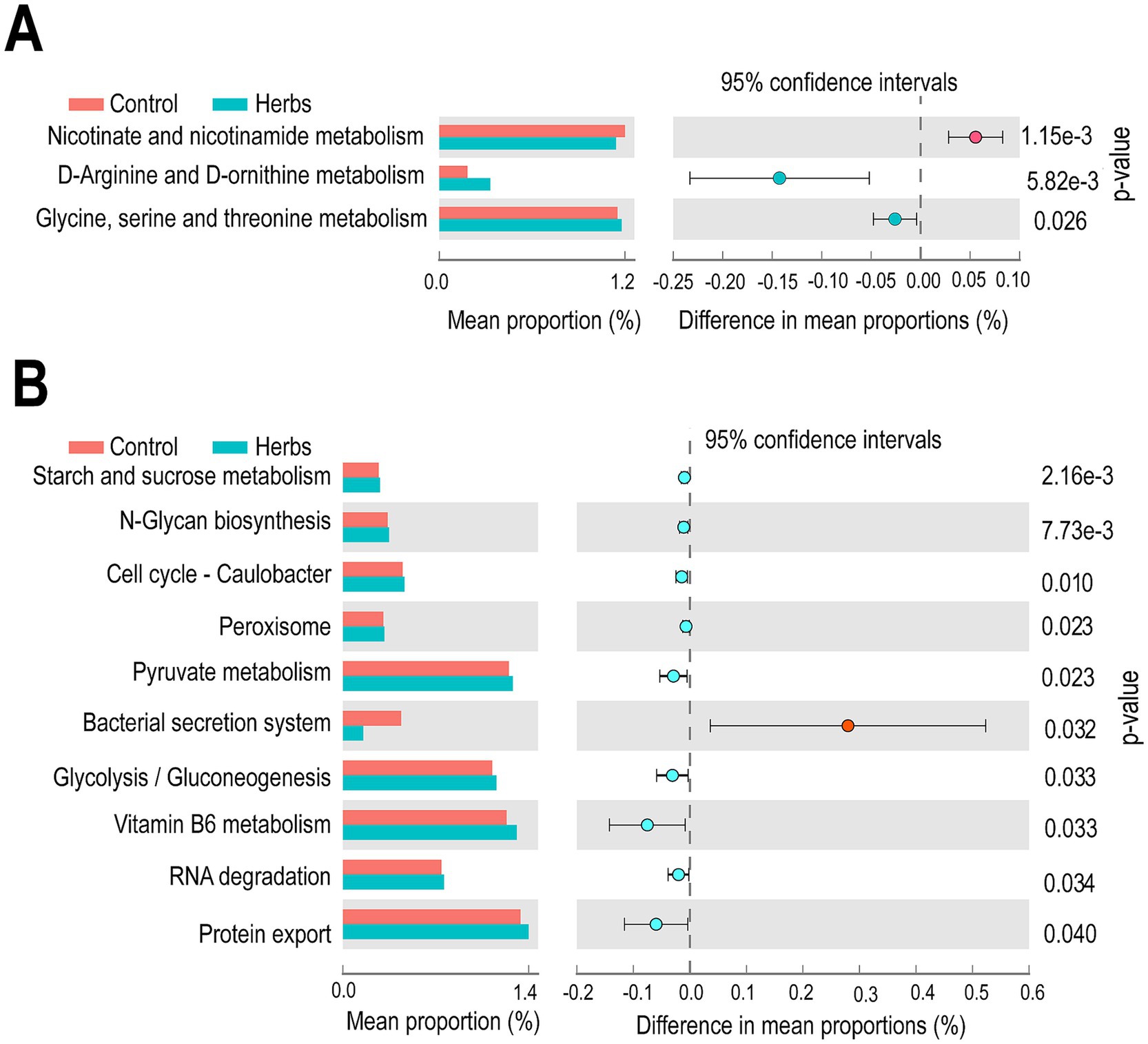
Figure 6. Predicted KEGG functional pathways of bacterial (A) and archaeal (B) communities in the Control (red) and Herbs (blue) groups. The third-level KEGG pathways are shown, with mean proportions and 95% confidence intervals. Significant differences between the groups were determined using a t-test (p < 0.05).
Heat stress is a significant challenge in the livestock industry, particularly for dairy cattle, due to its impact on both animal welfare and economic productivity. As global temperatures rise and greenhouse gas emissions increase, the impact of heat stress is expected to become more severe, posing ongoing challenges for dairy farming (1). Dairy cattle are especially vulnerable to heat stress because of their high metabolic heat production associated with milk synthesis, making them less able to cope with elevated temperatures compared to other livestock species (35). This susceptibility not only reduces milk production but also impacts overall health and fertility, leading to substantial economic losses (2).
Heat stress is known to induce oxidative stress by generating excessive reactive oxygen species (ROS), which can damage tissues and cells (36). Antioxidant enzymes, such as CAT, SOD, GSH-Px, and T-AOC, play a critical role in scavenging ROS, enhancing the body antioxidant defenses, and maintaining overall health (37, 38). In this study, supplementation with herbal formula significantly enhanced the activities of these enzymes, which likely contributed to the observed reduction in serum MDA levels, a key marker of oxidative damage (39). In addition, heat stress is well-documented to elevate BUN levels, indicative of disrupted protein metabolism (40, 41). Notably, herbal formula supplementation significantly reduced BUN levels in both serum and milk under heat-stress conditions (p < 0.05), suggesting an improvement in protein metabolism.
Herbal formula supplementation significantly upregulated the expression of the HSPA8 gene (p < 0.05), a key member of the heat shock protein family that is critical for cellular stress responses. Heat shock proteins like HSPA8 stabilize proteins as intra-cellular chaperones, prevent aggregation, and assist in protein refolding, which are essential functions under heat stress conditions (42–44). The increased expression of HSPA8 observed in the herbs group suggested that our self-developed herbal formula enhanced the cell ability to cope with heat-induced protein damage. This upregulation likely strengthened cellular resilience, mitigating the impact of heat stress on cellular function and viability. In addition, several studies have demonstrated the pivotal role of HSPA8 in milk protein synthesis in cows (45, 46). In this study, PP showed significantly increased in Herbs group (p < 0.05), it may be related to the significant increase in HSPA8 expression level.
In this study, the Herbs group showed significantly higher levels of IgA (p < 0.05), an important serum immunoglobulin that mediates various protective functions through interactions with specific receptors and immune mediators (47). Elevated IgA levels may further contribute to maintaining overall health and mitigating the adverse effects of heat stress by strengthening the immune system and providing additional protection against stress-related vulnerabilities. The Herbs group also exhibited significantly reduced levels of TNF-α (p < 0.05), a pro-inflammatory cytokine. Lower TNF-α levels suggested that the herbal formula may help alleviate systemic inflammation and support immune regulation under heat stress. Together, these findings demonstrated the effectiveness of herbal formula in mitigating heat stress by enhancing antioxidant defenses, improving immune responses, reducing oxidative damage, supporting metabolic function, and reducing systemic inflammation.
The beneficial effects of herbal formula on heat-stressed cows may be attributed to its key components, such as Astragalus (Huangqi), which has been widely recognized for its antioxidant, immune-enhancing, and anti-inflammatory properties (48). Its bioactive components, such as polysaccharides and saponins, have been shown to scavenge free radicals, enhance the activities of antioxidant enzymes like CAT and SOD, and reduce oxidative stress in livestock (48, 49). Astragalus polysaccharides are also known for their immune-modulatory properties, which may have contributed to the elevated IgA levels observed in the Herbs group. Furthermore, Astragalus calycosin exhibits significant anti-inflammatory activity by modulating inflammatory pathways, such as the NF-κB signaling pathway, and reducing the production of pro-inflammatory cytokines, including TNF-α (48, 50).
Beyond its benefits in mitigating oxidative damage and supporting immune regulation, herbal formula significantly impacted the rumen microbiome, a key player in nutrient digestion and absorption (51). Notably, this self-developed herbal formula reduced the relative abundance of Proteobacteria, a phylum increasingly recognized as a microbial marker of disease. Increased relative abundance of Proteobacteria has been associated with inflammation-driven diseases such as metabolic disorders, inflammatory bowel disease, and respiratory illnesses (52). In our study, the reduction of Proteobacteria in the Herbs group suggested that herbal formula may contribute to a healthier rumen microbial environment, potentially reducing the risk of inflammation. At the genus level, the rumen microbiome composition in the Control group showed considerable variability between samples, whereas the Herbs group exhibited a relatively more stable composition, with Staphylococcus consistently emerging as the dominant genus. Although Staphylococcus is generally not a major or abundant member of the rumen microbiome (53–56), herbal formula appeared to create favorable conditions for its proliferation within the rumen environment. While some members of Staphylococcus, such as S. aureus, are associated with diseases like mastitis in cattle (57, 58), others play beneficial roles. For example, S. xylosus contributes to lactic acid fermentation (59), S. carnosus has nitrate-reducing and antimicrobial properties (60), and S. chromogenes may inhibit pathogenic colonization through microbial competition (61). Although the specific Staphylococcus species in the Herbs group could not be identified, their increased abundance likely contributed to a more balanced and efficient microbial ecosystem, coinciding with the improved performance and health observed in Herbs group.
Other genera with increased abundance in the Herbs group included Aerococcus and Lachnospira. Aerococcus is a genus of Gram-positive bacteria primarily studied in human, where certain species are recognized as opportunistic pathogens (62–64). However, its role within the rumen microbiome remains largely unexplored. Its increased presence in Herbs group may reflect a microbial response to herbal formula, though its specific role in rumen environment warrants further investigation. In contrast, Lachnospira is well-known for its ability to ferment dietary fibers into short-chain fatty acids (SCFAs) such as acetate, propionate, and butyrate (65). These SCFAs are critical for maintaining gut health, regulating immune responses, promoting anti-inflammation, and supporting metabolic health (66, 67). The higher abundance of Lachnospira in the Herbs group could suggest enhanced fiber degradation, potentially improving energy utilization. Additionally, Ochrobactrum, a genus that includes opportunistic pathogens (68), was significantly reduced in the Herbs group. This reduction, along with changes in other microbial populations, highlighted herbal formula potential to improve microbial balance and support overall health.
The PICRUSt analysis revealed distinct differences in the predicted functional potential of metabolic pathways between the Herbs and Control groups, with a notably greater number of pathways altered in archaea compared to bacteria. However, it should be noted that PICRUSt-based functional predictions derive potential metabolic pathways from 16S rRNA gene data rather than whole metagenomes. One of the most notable changes among the archaeal pathways was the significant reduction in the bacterial secretion system in Herbs group. This system is essential for transporting proteins and mediating interactions between microbes and their environment or host. In bacterial pathogens, secretion systems are essential for virulence, facilitating diverse functions such as enhancing bacterial adhesion, scavenging environmental resources, and directly disrupting host cell functions (69). However, their role in archaea remains unclear, and the observed reduction may reflect broader shifts in microbial interactions or metabolic adaptation rather than pathogenic mechanisms. Additionally, pathways related to glycolysis/gluconeogenesis and pyruvate metabolism in archaea were significantly enriched in the Herbs group. These pathways are fundamental to energy production and carbon cycling in the rumen, suggesting that herbal formula enhanced microbial energy utilization and fermentation efficiency. This was further supported by the observed enrichment in the starch and sucrose metabolism pathway, reflecting an increased capacity for carbohydrate degradation.
This study showed that our self-developed herbal formula enhanced antioxidant capacity, strengthened immune responses, and improved cellular resilience in dairy cows under heat stress. Additionally, they helped mitigate heat stress by regulating the composition, function, and metabolites of the rumen microbiota, ultimately improving overall health. These findings highlighted their potential as a natural dietary strategy for alleviating heat stress in dairy cows.
Raw data were submitted to the National Center for Biotechnology Information (NCBI) Short Read Archive database and are available with BioProject accession number PRJNA1225285 (https://www.ncbi.nlm.nih.gov/bioproject/PRJNA1225285/).
The animal study was reviewed and approved by the Institutional Animal Care and Use Committee of Hebei Agricultural University (Baoding, China). The study was conducted in accordance with the local legislation and institutional requirements.
XW: Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Writing – original draft, Writing – review & editing. YW: Data curation, Formal analysis, Methodology, Writing – original draft, Writing – review & editing. MF: Methodology, Software, Writing – review & editing. JL: Data curation, Formal analysis, Writing – review & editing. ZL: Methodology, Validation, Writing – review & editing. LF: Methodology, Software, Writing – review & editing. NZ: Data curation, Formal analysis, Writing – review & editing. HZ: Methodology, Software, Writing – review & editing. JQ: Conceptualization, Investigation, Supervision, Writing – review & editing.
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This work was supported by Hebei Cows/Beef Cattle Innovation Team Comprehensive Experiment and Promotion Station of Modern Agro-Industry Technology Research System (HBCT2024230406) and High-Level Talent Support Program of Hebei Province (B20231013).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The authors declare that no Gen AI was used in the creation of this manuscript.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fvets.2025.1558856/full#supplementary-material
SUPPLEMENTARY FIGURE S1 | Dynamics of the thermal environment in dairy barns. The temperature unit is °C and the relative humidity unit is %. Cows suffer from heat stress almost 24 h a day.
SUPPLEMENTARY FIGURE S2 | Comparison of alpha diversity metrics between the Control and Herbs groups for bacterial (A) and archaeal (B) communities. Diversity indices include the Shannon index, observed ASVs, and Chao1 index, illustrating microbial diversity and richness. No statistically significant differences were detected between the two groups.
1. Cartwright, SL, Schmied, J, Karrow, N, and Mallard, BA. Impact of heat stress on dairy cattle and selection strategies for thermotolerance: a review. Front Vet Sci. (2023) 10:1198697. doi: 10.3389/fvets.2023.1198697
2. Wolfenson, D, and Roth, Z. Impact of heat stress on cow reproduction and fertility. Anim Front. (2019) 9:32–8. doi: 10.1093/af/vfy027
3. Das, R, Sailo, L, Verma, N, Bharti, P, Saikia, J, Imtiwati,, et al. Impact of heat stress on health and performance of dairy animals: a review. Vet World. (2016) 9:260–8. doi: 10.14202/vetworld.2016.260-268
4. Koch, F, Thom, U, Albrecht, E, Weikard, R, Nolte, W, Kuhla, B, et al. Heat stress directly impairs gut integrity and recruits distinct immune cell populations into the bovine intestine. Proc Natl Acad Sci USA. (2019) 116:10333–8. doi: 10.1073/pnas.1820130116
5. Collier, RJ, Dahl, GE, and VanBaale, MJ. Major advances associated with environmental effects on dairy cattle. J Dairy Sci. (2006) 89:1244–53. doi: 10.3168/jds.S0022-0302(06)72193-2
6. Becker, CA, Collier, RJ, and Stone, AE. Invited review: physiological and behavioral effects of heat stress in dairy cows. J Dairy Sci. (2020) 103:6751–70. doi: 10.3168/jds.2019-17929
7. Wei, X, Tsai, T, Howe, S, and Zhao, J. Weaning induced gut dysfunction and nutritional interventions in nursery pigs: a partial review. Animals. (2021) 11:1279. doi: 10.3390/ani11051279
8. Wei, X, Bottoms, KA, Stein, HH, Blavi, L, Bradley, CL, Bergstrom, J, et al. Dietary organic acids modulate gut microbiota and improve growth performance of nursery pigs. Microorganisms. (2021) 9:110. doi: 10.3390/microorganisms9010110
9. Yang, Y, Yan, G, Meng, X, Wang, X, Zhao, Z, Zhou, S, et al. Effects of Lactobacillus plantarum and Pediococcus acidilactici co-fermented feed on growth performance and gut microbiota of nursery pigs. Front Vet Sci. (2022) 9:1076906. doi: 10.3389/fvets.2022.1076906
10. Di Sotto, A, Vitalone, A, and Di Giacomo, S. Plant-derived nutraceuticals and immune system modulation: an evidence-based overview. Vaccines. (2020) 8:468. doi: 10.3390/vaccines8030468
11. Zheng, Y, Ren, W, Zhang, L, Zhang, Y, Liu, D, and Liu, Y. A review of the pharmacological action of Astragalus polysaccharide. Front Pharmacol. (2020) 11:349. doi: 10.3389/fphar.2020.00349
12. Saleh, AA, Soliman, MM, Yousef, MF, Eweedah, NM, El-Sawy, HB, Shukry, M, et al. Effects of herbal supplements on milk production quality and specific blood parameters in heat-stressed early lactating cows. Front Vet Sci. (2023) 10:1180539. doi: 10.3389/fvets.2023.1180539
13. Rahman, MA, Redoy, MRA, Shuvo, AAS, Chowdhury, R, Hossain, E, Sayem, SM, et al. Influence of herbal supplementation on nutrient digestibility, blood biomarkers, milk yield, and quality in tropical crossbred cows. PLoS One. (2024) 19:e0313419. doi: 10.1371/journal.pone.0313419
14. Aderemi, FA, and Alabi, OM. Turmeric (Curcuma longa): an alternative to antibiotics in poultry nutrition. Transl Anim Sci. (2023) 7:txad133. doi: 10.1093/tas/txad133
15. Yang, CM, Han, QJ, Wang, KL, Xu, YL, Lan, JH, and Cao, GT. Astragalus and ginseng polysaccharides improve developmental, intestinal morphological, and immune functional characters of weaned piglets. Front Physiol. (2019) 10:418. doi: 10.3389/fphys.2019.00418
16. Deng, F, Li, Y, Peng, Y, Wei, X, Wang, X, Howe, S, et al. The diversity, composition, and metabolic pathways of archaea in pigs. Animals. (2021) 11:2139. doi: 10.3390/ani11072139
17. Wang, X, Tsai, T, Deng, F, Wei, X, Chai, J, Knapp, J, et al. Longitudinal investigation of the swine gut microbiome from birth to market reveals stage and growth performance associated bacteria. Microbiome. (2019) 7:109. doi: 10.1186/s40168-019-0721-7
18. Yang, J, Fan, Y, Jin, R, Peng, Y, Chai, J, Wei, X, et al. Exploring the intestinal microbial community of Lantang Pigs through metagenome-assembled genomes and carbohydrate degradation genes. Fermentation. (2024) 10:207. doi: 10.3390/fermentation10040207
19. Shreiner, AB, Kao, JY, and Young, VB. The gut microbiome in health and in disease. Curr Opin Gastroenterol. (2015) 31:69–75. doi: 10.1097/MOG.0000000000000139
20. de Vos, WM, Tilg, H, Van Hul, M, and Cani, PD. Gut microbiome and health: mechanistic insights. Gut. (2022) 71:1020–32. doi: 10.1136/gutjnl-2021-326789
21. Deng, F, Han, Y, Huang, Y, Li, D, Chai, J, Deng, L, et al. A comprehensive analysis of antibiotic resistance genes in the giant panda gut. iMeta. (2024) 3:e171. doi: 10.1002/imt2.171
22. Deng, F, Wang, C, Li, D, Peng, Y, Deng, L, Zhao, Y, et al. The unique gut microbiome of giant pandas involved in protein metabolism contributes to the host’s dietary adaption to bamboo. Microbiome. (2023) 11:180. doi: 10.1186/s40168-023-01603-0
23. Wei, X, Tsai, T, Knapp, J, Bottoms, K, Deng, F, Story, R, et al. ZnO modulates swine gut microbiota and improves growth performance of nursery pigs when combined with peptide cocktail. Microorganisms. (2020) 8:146. doi: 10.3390/microorganisms8020146
24. Qi, W, Xue, MY, Jia, MH, Zhang, S, Yan, Q, and Sun, HZ. Invited review—understanding the functionality of the rumen microbiota: searching for better opportunities for rumen microbial manipulation. Anim Biosci. (2024) 37:370–84. doi: 10.5713/ab.23.0308
25. Wang, X, Wang, Y, Li, J, Fu, L, Zhang, N, and Qin, J. Screening and application research of traditional Chinese medicine formulas for anti-heat stress in dairy cows. China Anim Husb Vet Med. (2024) 51:5064–73.
26. Naderi, N, Ghorbani, GR, Sadeghi-Sefidmazgi, A, Nasrollahi, SM, and Beauchemin, KA. Shredded beet pulp substituted for corn silage in diets fed to dairy cows under ambient heat stress: feed intake, total-tract digestibility, plasma metabolites, and milk production. J Dairy Sci. (2016) 99:8847–57. doi: 10.3168/jds.2016-11029
27. Bolyen, E, Rideout, JR, Dillon, MR, Bokulich, NA, Abnet, CC, Al-Ghalith, GA, et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat Biotechnol. (2019) 37:852–7. doi: 10.1038/s41587-019-0209-9
28. Amir, A, McDonald, D, Navas-Molina, JA, Kopylova, E, Morton, JT, Zech, XZ, et al. Deblur rapidly resolves single-nucleotide community sequence patterns. mSystems. (2017) 2:e00191. doi: 10.1128/mSystems.00191-16
29. DeSantis, TZ, Hugenholtz, P, Larsen, N, Rojas, M, Brodie, EL, Keller, K, et al. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microbiol. (2006) 72:5069–72. doi: 10.1128/AEM.03006-05
30. Douglas, GM, Maffei, VJ, Zaneveld, JR, Yurgel, SN, Brown, JR, Taylor, CM, et al. PICRUSt2 for prediction of metagenome functions. Nat Biotechnol. (2020) 38:685–8. doi: 10.1038/s41587-020-0548-6
31. Kanehisa, M, Furumichi, M, Sato, Y, Matsuura, Y, and Ishiguro-Watanabe, M. KEGG: biological systems database as a model of the real world. Nucleic Acids Res. (2024) 53:D672–7. doi: 10.1093/nar/gkae909
32. Wickham, H. ggplot2: elegant graphics for data analysis In: Use R. Cham: Springer (2009). 1–212.
33. Kolde, R. (2018). Package ‘pheatmap’. Available online at: https://cranr-projectorg/web/packages/pheatmap/pheatmappdf (Accessed October 14, 2022).
34. Parks, DH, Tyson, GW, Hugenholtz, P, and Beiko, RG. STAMP: statistical analysis of taxonomic and functional profiles. Bioinformatics. (2014) 30:3123–4. doi: 10.1093/bioinformatics/btu494
35. Polsky, L, and von Keyserlingk, MAG. Invited review: effects of heat stress on dairy cattle welfare. J Dairy Sci. (2017) 100:8645–57. doi: 10.3168/jds.2017-12651
36. Slimen, IB, Najar, T, Ghram, A, Dabbebi, H, Ben Mrad, M, and Abdrabbah, M. Reactive oxygen species, heat stress and oxidative-induced mitochondrial damage. A review. Int J Hyperther. (2014) 30:513–23. doi: 10.3109/02656736.2014.971446
37. Jena, AB, Samal, RR, Bhol, NK, and Duttaroy, AK. Cellular Red-Ox system in health and disease: the latest update. Biomed Pharmacother. (2023) 162:114606. doi: 10.1016/j.biopha.2023.114606
38. Matés, JM, Pérez-Gómez, C, and de Castro, N I. Antioxidant enzymes and human diseases. Clin Biochem. (1999) 32:595–603. doi: 10.1016/S0009-9120(99)00075-2
39. de Paz, NM, García-González, M, Gómez-Bernal, F, Quevedo-Abeledo, JC, de Vera-González, A, López-Mejias, R, et al. Relationship between malondialdehyde serum levels and disease features in a full characterized series of 284 patients with systemic lupus erythematosus. Antioxidants. (2023) 12:1535. doi: 10.3390/antiox12081535
40. Kim, WS, Ghassemi Nejad, J, Peng, DQ, Jo, YH, Kim, J, and Lee, HG. Effects of different protein levels on growth performance and stress parameters in beef calves under heat stress. Sci Rep. (2022) 12:8113. doi: 10.1038/s41598-022-09982-4
41. Gao, ST, Guo, J, Quan, SY, Nan, XM, Fernandez, MVS, Baumgard, LH, et al. The effects of heat stress on protein metabolism in lactating Holstein cows. J Dairy Sci. (2017) 100:5040–9. doi: 10.3168/jds.2016-11913
42. Murshid, A, Prince, TL, Lang, B, and Calderwood, SK. Role of heat shock factors in stress-induced transcription. Methods Mol Biol. (2018) 1709:23–34. doi: 10.1007/978-1-4939-7477-1_2
43. Lindquist, S. The heat-shock response. Annu Rev Biochem. (1986) 55:1151–91. doi: 10.1146/annurev.bi.55.070186.005443
44. Joy, A, Dunshea, FR, Leury, BJ, Clarke, IJ, DiGiacomo, K, and Chauhan, SS. Resilience of small ruminants to climate change and increased environmental temperature: a review. Animals. (2020) 10:867. doi: 10.3390/ani10050867
45. Jiangfeng, F, Yuzhu, L, Sijiu, Y, Yan, C, Gengquan, X, Libin, W, et al. Transcriptional profiling of two different physiological states of the yak mammary gland using RNA sequencing. PLoS One. (2018) 13:e0201628. doi: 10.1371/journal.pone.0201628
46. Paten, AM, Duncan, EJ, Pain, SJ, Peterson, SW, Kenyon, PR, Blair, HT, et al. Functional development of the adult ovine mammary gland—insights from gene expression profiling. BMC Genomics. (2015) 16:748. doi: 10.1186/s12864-015-1947-9
47. Woof, JM, and Kerr, MA. The function of immunoglobulin A in immunity. J Pathol. (2006) 208:270–82. doi: 10.1002/path.1877
48. Yao, J, Peng, T, Shao, C, Liu, Y, Lin, H, and Liu, Y. The antioxidant action of Astragali radix: its active components and molecular basis. Molecules. (2024) 29:1691. doi: 10.3390/molecules29081691
49. Samuel, AO, Huang, BT, Chen, Y, Guo, FX, Yang, DD, and Jin, JQ. Antioxidant and antibacterial insights into the leaves, leaf tea and medicinal roots from Astragalus membranaceus (Fisch.) Bge. Sci Rep. (2021) 11:19625. doi: 10.1038/s41598-021-97109-6
50. Ma, R, Yuan, F, Wang, SX, Liu, YP, Fan, TT, and Wang, FL. Calycosin alleviates cerulein-induced acute pancreatitis by inhibiting the inflammatory response and oxidative stress via the p38 MAPK and NF-κB signal pathways in mice. Biomed Pharmacother. (2018) 105:599–605. doi: 10.1016/j.biopha.2018.05.080
51. Matthews, C, Crispie, F, Lewis, E, Reid, M, O’Toole, PW, and Cotter, PD. The rumen microbiome: a crucial consideration when optimising milk and meat production and nitrogen utilisation efficiency. Gut Microbes. (2019) 10:115–32. doi: 10.1080/19490976.2018.1505176
52. Rizzatti, G, Lopetuso, LR, Gibiino, G, Binda, C, and Gasbarrini, A. Proteobacteria: a common factor in human diseases. Biomed Res Int. (2017) 2017:9351507. doi: 10.1155/2017/9351507
53. da Cunha, LL, Monteiro, HF, Figueiredo, CC, Canisso, IF, Bicalho, RC, Cardoso, FC, et al. Characterization of rumen microbiome and metabolome from oro-esophageal tubing and rumen cannula in Holstein dairy cows. Sci Rep. (2023) 13:5854. doi: 10.1038/s41598-023-33067-5
54. Wen, Z, Chen, Y, Wu, L, Tian, H, Zhu, N, Guo, Y, et al. Effects of Broussonetia papyrifera silage on rumen fermentation parameters and microbes of Holstein heifers. AMB Express. (2022) 12:62. doi: 10.1186/s13568-022-01405-x
55. Torres Manno, MA, Gizzi, FO, Martin, M, Espariz, M, Magni, C, and Blancato, VS. Metagenomic approach to infer rumen microbiome derived traits of cattle. World J Microbiol Biotechnol. (2023) 39:250. doi: 10.1007/s11274-023-03694-1
56. Lima, J, Martinez-Alvaro, M, Mattock, J, Auffret, MD, Duthie, CA, Cleveland, MA, et al. Temporal stability of the rumen microbiome and its longitudinal associations with performance traits in beef cattle. Sci Rep. (2024) 14:20772. doi: 10.1038/s41598-024-70770-3
57. Monistero, V, Graber, HU, Pollera, C, Cremonesi, P, Castiglioni, B, Bottini, E, et al. Staphylococcus aureus isolates from bovine mastitis in eight countries: genotypes, detection of genes encoding different toxins and other virulence genes. Toxins. (2018) 10:247. doi: 10.3390/toxins10060247
58. Keinprecht, H, Irimaso, E, Rosel, AC, Stessl, B, Ntakirutimana, C, Marek, L, et al. Diversity of Staphylococcus aureus associated with mastitis from dairy cows in Rwanda. J Glob Antimicrob Resist. (2024) 36:326–35. doi: 10.1016/j.jgar.2024.01.017
59. Vermassen, A, Dordet-Frisoni, E, de La Foye, A, Micheau, P, Laroute, V, Leroy, S, et al. Adaptation of Staphylococcus xylosus to nutrients and osmotic stress in a salted meat model. Front Microbiol. (2016) 7:87. doi: 10.3389/fmicb.2016.00087
60. Neubauer, H, and Gotz, F. Physiology and interaction of nitrate and nitrite reduction in Staphylococcus carnosus. J Bacteriol. (1996) 178:2005–9. doi: 10.1128/jb.178.7.2005-2009.1996
61. Chin, D, Goncheva, MI, Flannagan, RS, Deecker, SR, Guariglia-Oropeza, V, Ensminger, AW, et al. Coagulase-negative staphylococci release a purine analog that inhibits Staphylococcus aureus virulence. Nat Commun. (2021) 12:1887. doi: 10.1038/s41467-021-22175-3
62. Sihvonen, R, Turunen, M, Lehtola, L, Pakarinen, L, Grönroos, JO, Rantakokko-Jalava, K, et al. Clinical and microbiological characterization of bacteraemias at Helsinki metropolitan area, Finland. Eur J Clin Microbiol. (2022) 41:751–60. doi: 10.1007/s10096-022-04415-6
63. Senneby, E, Eriksson, B, Fagerholm, E, and Rasmussen, M. Bacteremia with Aerococcus sanguinicola: case series with characterization of virulence properties. Open Forum Infect Dis. (2014) 1:ofu025. doi: 10.1093/ofid/ofu025
64. Varshini, K, Ganesan, V, and Charles, J. Bacteremia: a rare case report from India. Indian J Crit Care M. (2022) 26:127–8. doi: 10.5005/jp-journals-10071-24072
65. Vinelli, V, Biscotti, P, Martini, D, Del Bo, C, Marino, M, Merono, T, et al. Effects of dietary fibers on short-chain fatty acids and gut microbiota composition in healthy adults: a systematic review. Nutrients. (2022) 14:2559. doi: 10.3390/nu14132559
66. Furusawa, Y, Obata, Y, Fukuda, S, Endo, TA, Nakato, G, Takahashi, D, et al. Commensal microbe-derived butyrate induces the differentiation of colonic regulatory T cells. Nature. (2013) 504:446–50. doi: 10.1038/nature12721
67. Louis, P, and Flint, HJ. Diversity, metabolism and microbial ecology of butyrate-producing bacteria from the human large intestine. FEMS Microbiol Lett. (2009) 294:1–8. doi: 10.1111/j.1574-6968.2009.01514.x
68. Ryan, MP, and Pembroke, JT. The genus as major opportunistic pathogens. Microorganisms. (2020) 8:1797. doi: 10.3390/microorganisms8111797
Keywords: heat stress, dairy cow, herbal formula, antioxidant capacity, immune response, rumen microbiome
Citation: Wang X, Wang Y, Feng M, Li J, Liu Z, Fu L, Zhang N, Zhang H and Qin J (2025) Herbal formula alleviates heat stress by improving physiological and biochemical attributes and modulating the rumen microbiome in dairy cows. Front. Vet. Sci. 12:1558856. doi: 10.3389/fvets.2025.1558856
Received: 11 January 2025; Accepted: 17 February 2025;
Published: 07 March 2025.
Edited by:
Jianmin Chai, Foshan University, ChinaReviewed by:
Bin Zuo, South China Agricultural University, ChinaCopyright © 2025 Wang, Wang, Feng, Li, Liu, Fu, Zhang, Zhang and Qin. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jianhua Qin, cWpocXFxQDEyNi5jb20=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.