- Department of Avian Diseases, Faculty of Veterinary Medicine, University of Tehran, Tehran, Iran
Introduction: Poultry products are considered an important source of Salmonella infections. Transmission of non-typhoidal Salmonella enterica serovars to humans has been a great concern worldwide. Occurrence of multi-drug resistance, adding to the presence of various virulence factors, which facilitate the pathogenesis of Salmonella, would cause tremendous risk for both human and animals’ health.
Methods and results: During 2023, out of a total number of 1,274 samples from broilers in Iran, 114 isolates of Salmonella spp. (8.94%) were detected from which 97 isolates were confirmed as Salmonella Enteritidis (SE). Eight virulence genes including invA, sefA, sopE, spvC, hilA, agfA, sivH and lpfA, were detected among SE isolates and it was found that all isolates harbored these genes at the rate of 100% except for spvC, which was present in 96.90% of the SE isolates. In phenotypic evaluation of resistance against 16 antimicrobial agents, high resistance rates were observed against nalidixic acid, ampicillin, amoxicillin–clavulanate and ciprofloxacin. While resistance to tetracycline, streptomycin and chloramphenicol was found to be moderate, it was very low to azithromycin, sulfamethoxazole-trimethoprim, amikacin, gentamicin, ceftriaxone and cefotaxime. However, all isolates were sensitive to meropenem, ceftazidime and aztreonam. The mean of MAR index values was 0.26 and 72.15% of the isolates were found to be highly resistant. In detection of 14 resistance genes among SE isolates, five genes including blaTEM, tetA, tetB, sul1 and strA/B were found with prevalence rates of 63.92, 36.08, 61.85, 10.30 and 14.43%, respectively.
Discussion: The high prevalence rates of MDR in SE, along with the overwhelming presence of major virulence factors raise public health concerns. These data highlight the great potential risks of the presence and transmission of highly pathogenic MDR Salmonella to humans from chicken meat sources, as well as the need for more effective surveillance for antimicrobial use in the poultry industry. Reducing/optimizing the use of antimicrobials, improving poultry management procedures, using probiotics and biosecurity or vaccines are essential to deal with this issue.
1 Introduction
Salmonella, a Gram negative, motile bacterium from the Enterobacteriaceae family, is a primary pathogen which can infect a wide range of animals. As a zoonotic disease, salmonellosis has been one of the most common causes of gastroenteritis and food poisoning in humans in recent years affecting most parts of the world including developed countries (1). Salmonellosis in humans has been reported from almost all countries and the rates of occurrence vary, but usually it has not been decreased significantly in the last few years, even in well-developed countries (2). For example, in 2010, in the United States the incidence of salmonellosis was higher than any other food-borne pathogens (17.6 infections /100,000 population) (3). In that year, the World Health Organization (WHO) reported 153 million cases of Salmonella gastroenteric infections worldwide. Additionally, in 2019, WHO estimated 26 million cases of Salmonella gastroenteritis and 118,000 deaths globally (4).
Poultry products are a major reservoir of Salmonella, posing risks to human health, poultry production and food products (5, 6). Salmonella spreads vertically through eggs and horizontally via direct or indirect contact, persisting on farms for extended periods (7). Poultry salmonellosis causes high mortality, reduced flock performance, and increased susceptibility to other diseases leading to economic losses (8). Surveillance programs worldwide aim to control Salmonella and reduce its entry into the food chain (9). In the U.S., foodborne salmonellosis costs an estimated $4–11 billion annually in medical care, lost production, and premature deaths (10).
In poultry production, using antimicrobial agents is very common for different purposes such as growth promotion, prophylactic and control of infections (11). However, antimicrobial usage contributes to development of drug resistance; and posing risks to public health, the poultry industry, and the environment (12). It was estimated that in 2019, around five million people around the world died because of antimicrobial resistance (13). In addition to the pathogenic potential of Salmonella, these bacteria can develop resistance to several antimicrobials, which may make medical treatment of the infections even more challenging (12, 14). Antimicrobial resistant bacteria can transmit resistance either vertically to their progeny or horizontally to other bacterial populations through mobile genetic elements; thereby, facilitating the dissemination of resistance (13–15).
As a primary pathogen, Salmonella is equipped with many virulence properties. Every virulence property may play a distinct role in the complex pathogenicity, the ability to survive, and/or transmission of the bacteria. The genes which encode these virulence properties are integrated into the plasmid or chromosomal genome and their expressions and interactions are yet to be well-understood (16, 17). Chromosomal virulence-associated genes helping Salmonella with its attack and invasion capabilities include invA, hilA and sivH which are essential for the intrusion of epithelial cells (18). Salmonella effector protein attached by sopE gene help Salmonella in the disorganizing host cell membrane (19). The aggregative fimbria, agf operon, takes part in an essential interaction of Salmonella with the digestive tract cells of the host and facilitates microbial self-aggregation for higher rates of survival (20). The Salmonella-encoded fimbria (sef operon), encodes the major subunit of the fimbrial protein SEF14 which supports interaction between the organisms and the macrophages of the host (21). The plasmid-mediated spvC gene counts liable for vertical transmission of Salmonella through eggs (22). Long polar fimbria (lpf operon), is a plasmid-mediated virulence factor which encodes an important part of a fimbria and is associated with the fascination of the organism for Peyer’s patches and its attachment to intestinal M cells (20).
Among more than 2,500 recognized Salmonella serovars, about 10% are found in poultry, with Salmonella Enteritidis (SE) and S. Typhimurium (ST) being the most prevalent worldwide (23–25). Salmonella Enteritidis infections in humans are often linked to the consumption of contaminated poultry products; especially eggs, while S. Typhimurium infections are mostly associated with the consumption of pork, poultry, beef and even seafood (26–28). Given the public health and economic impact of salmonellosis, along with its complex pathogenesis, studying virulence genes and antimicrobial resistance in poultry-origin Salmonella is very critical. This study was conducted to provide data and updates on poultry-origin Salmonella from Iran, in order to achieve a better understanding for control and treatment of salmonellosis.
2 Materials and methods
2.1 Sample collection and bacteriological procedures
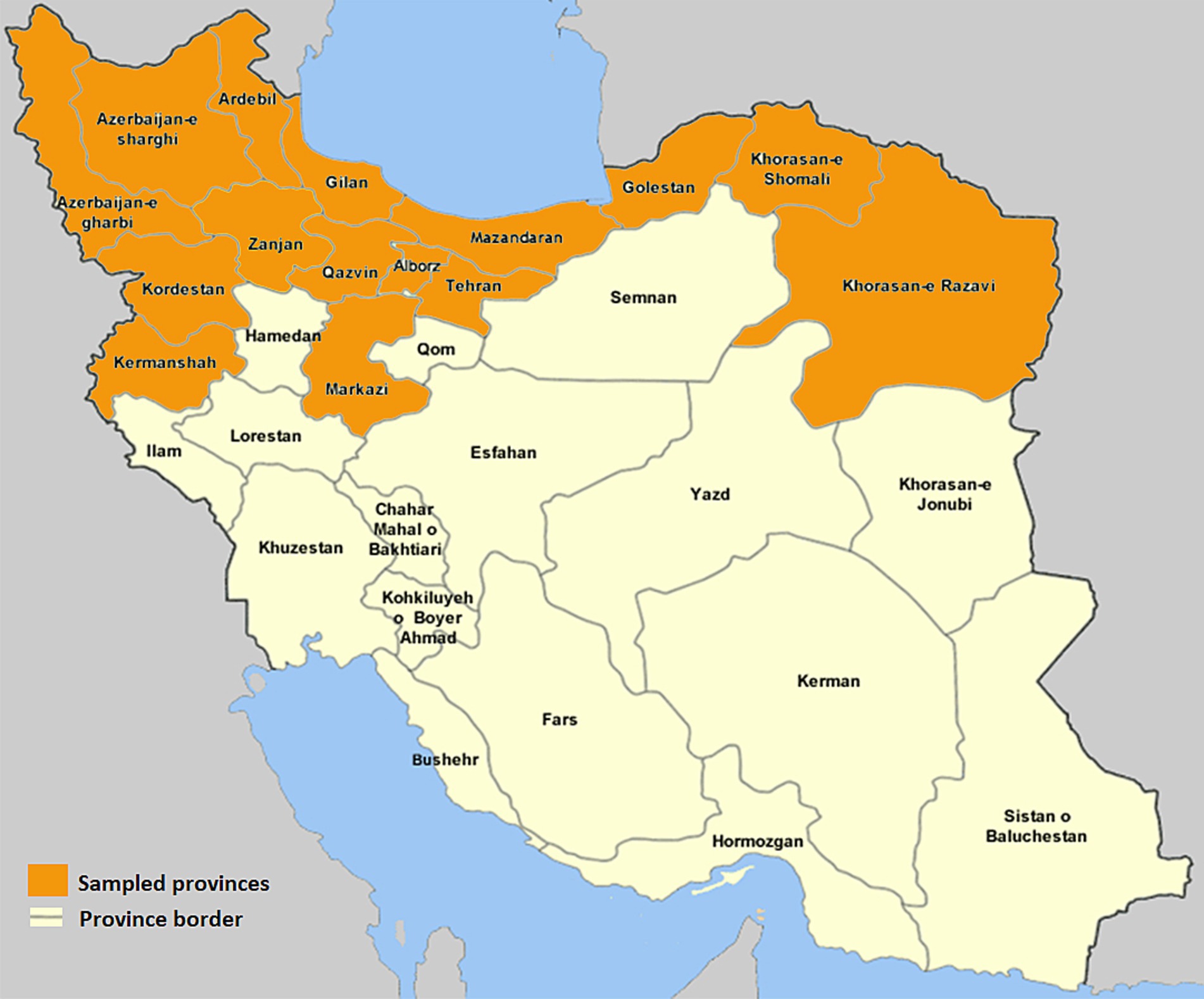
This cross-sectional study was completed in 2023. The geographical regions from which the samples were received are shown in Figure 1. The sampled provinces have a high density of poultry farms. Our laboratory and our collaborative laboratories in Tehran city often receives samples from those flocks for Salmonella isolation. The population of broiler chickens in the sampling areas was estimated to be more than 1,000,000,000 birds in 2023. Sterile cotton swabs contained in 10 mL of Selenite F as enrichment medium were used for swabbing from every submitted sample from the broiler chicken flocks. Samples were collected aseptically and brought to the microbiology laboratory in an insulated icebox. Salmonella isolation and identification were carried out according to standard procedures previously described (29). Briefly, samples were inoculated onto selenite F enrichment broth at 41°C for 24 h, followed by sub-cultivation on Salmonella-Shigella (SS) and MacConkey agar at 37°C for 24 h. Typical black-centered colonies on SS and colorless colonies on MacConkey plates were picked and subsequently cultured onto nutrient agar plate (NA; Oxoid, UK). The biochemical confirmation was done by using triple sugar iron (TSI), motility indole urea (MIU), catalase and oxidase tests.
2.2 Antimicrobial susceptibility test
The susceptibility of the SE isolates to a panel of 16 antimicrobial agents was determined by the agar disk diffusion method and the interpretation of results was carried out according to the National Committee for Clinical Laboratory Standards guidelines (30). The antimicrobial agents were selected because of their importance in human and veterinary medicine. The evaluated antimicrobials and their concentrations were: amikacin, AN (30 μg), gentamicin, GM (10 μg), streptomycin, S (10 μg), meropenem, MEN (10 μg), ceftriaxone, CRO (30 μg), cefotaxime, CTX (10 μg), ceftazidime, CAZ (30 μg), aztreonam, AZT (30 μg), amoxicillin–clavulanate, AMC (30 μg), ampicillin, AM (10 μg), azithromycin, AZM (15 μg), ciprofloxacin, CP (5 μg), nalidixic acid, NA (30 μg), sulfamethoxazole-trimethoprim, SXT (1.25–23.75 μg), tetracycline, TE (10 μg), chloramphenicol, C (30 μg). All antibacterial disks were provided from Padtan Teb Co (Tehran, Iran). The ATCC reference strains Escherichia coli ATCC 25922, Pseudomonas aeruginosa, ATCC 27853, and E. coli ATCC 35218 were used for quality control purposes. In this study, the SE isolates with intermediate susceptibility classification were considered to be resistant to that drug and multi-resistance was defined as resistance to 3 or more classes of antibacterials.
2.3 Multiple antibiotic resistance indexing
Multiple antibiotic resistance (MAR) indexing has been considered a suitable and valid method for tracking the source of bacteria. To calculate the MAR index, the number of resistant antibiotics for an organism would be divided by the total number of antibiotics to which the organism has been exposed. MAR index values larger than 0.2 indicate high resistance of the organism, where antibiotics are often used. The multiple antibiotic resistance (MAR) index of all isolates was calculated and the results were interpreted using a proven method as described previously (31).
2.4 Confirmation of Salmonella genus and Salmonella Enteritidis by PCR
To extract bacterial DNA, 1 mL pure overnight culture of each SE isolate grown overnight at 37°C for 16 h was transferred to a clean 1.5 mL microtube and centrifuged for 5 min at 10,000×g. The supernatants were carefully removed and discarded. The pellet was re-suspended in 300 μL sterile double distilled water by vortexing, incubated for 15 min at 100°C, chilled on ice immediately, and centrifuged again for 5 min at 14,000×g at 4°C. The supernatant was removed and used as template DNA. The concentration of DNA was determined by Biophotometer (Eppendorff, Germany) and adjusted to approximately 200 ng for each PCR reaction. The supernatant was stored at −20°C for further use.
In this study, invA gene specific primers were used to confirm the Salmonella genus (Table 1). Also, in order to identify serovar Enteritidis one pair of specific primers for amplification of sdf-ι gene were used (Table 1). All primers were synthesized by Bioneer (South Korea). Amplification reactions for Salmonella genus and serovar Enteritidis confirmation were carried out in a 25 μL reaction volume containing 12.5 μL of 2x mastermix (Taq 2x Red Master Mix, Ampliqon, Denmark), 0.5 μL each of forward and reverse primers (10 pmol/μL), 2 μL of DNA template, and 9.5 μL nuclease-free water. Salmonella Enteritidis PT21 strain (32) and dH2O (instead of template DNA) were used as positive and negative controls, respectively, in all PCR reaction sets. Amplifications were programmed in a thermocycler (SensoQuest, Germany) as described below. For Salmonella genus, 95°C for 1 min followed by 38 cycles of 95°C for 30 s, 64°C for 30 s, 72°C for 30 s, and a final extension at 72°C for 4 min was used (33). For serovar Enteritidis, program was as follows: 95°C for 2 min followed by 30 cycles of 95°C for 60 s, 57°C for 60 s, 72°C for 2 min, and a final extension at 72°C for 5 min (34). The amplified products were detected by gel electrophoresis in 1.5% agarose gel containing Safe Stain® (SinaClon) at 100 V for 30 min in 1x TAE buffer and visualized under UV illumination. A 100 bp DNA ladder (Yekta Tajhiz Azma, Iran) was used as a molecular weight marker for the PCR products in gel electrophoresis.

Table 1. Targeted genes, primer sequences and expected amplicon sizes for identification of Salmonella genus and Salmonella Enteritidis.
2.5 Detection of virulence genes
All isolates were examined for the presence of seven important virulence genes namely: hilA, agfA, lpfA, sivH, sefA, sopE and spvC. Each of seven virulence genes was amplified by using specific primer pairs and according to the PCR protocols described in Table 2. The preparation of reaction mixtures and gel electrophoresis were done as described above. The positive control (SE PT21 strain) and negative control (dH2O instead of template DNA) were used in all PCR reaction sets for validation. All primers of virulence genes were synthetized by SinaClon (Tehran, Iran).
2.6 Detection of antimicrobial resistance genes
All isolates were screened by using PCR to investigate the presence of 14 antimicrobial resistance genes including seven β-lactamase genes (blaTEM, blaSHV, blaOXA, blaCTX-M-1, blaCTX-M-2, blaCTX-M-9 and blaCTX-Mg8/25), three tetracycline resistant genes (tetA, tetB and tetC), three sulfonamide resistant genes (sul1, sul2 and sul3) and one streptomycin resistant gene (strA/B). For the detection of β-lactam genes, two cycles of multiplex PCR were carried out as described previously (35). Multiplex PCR were also performed to detect the resistance genes for sulfonamides (sul1, sul2 and sul3), tetracycline (tetA, tetB and tetC), and a single PCR for streptomycin (strA/B) according to the methods described previously (36). The PCR reaction mixture preparations and gel electrophoresis were done as described above. The specific primers used to detect antimicrobial resistance genes were synthetized by SinaClon (Table 3). More details about the PCR procedures are provided in Supplementary Table S1.
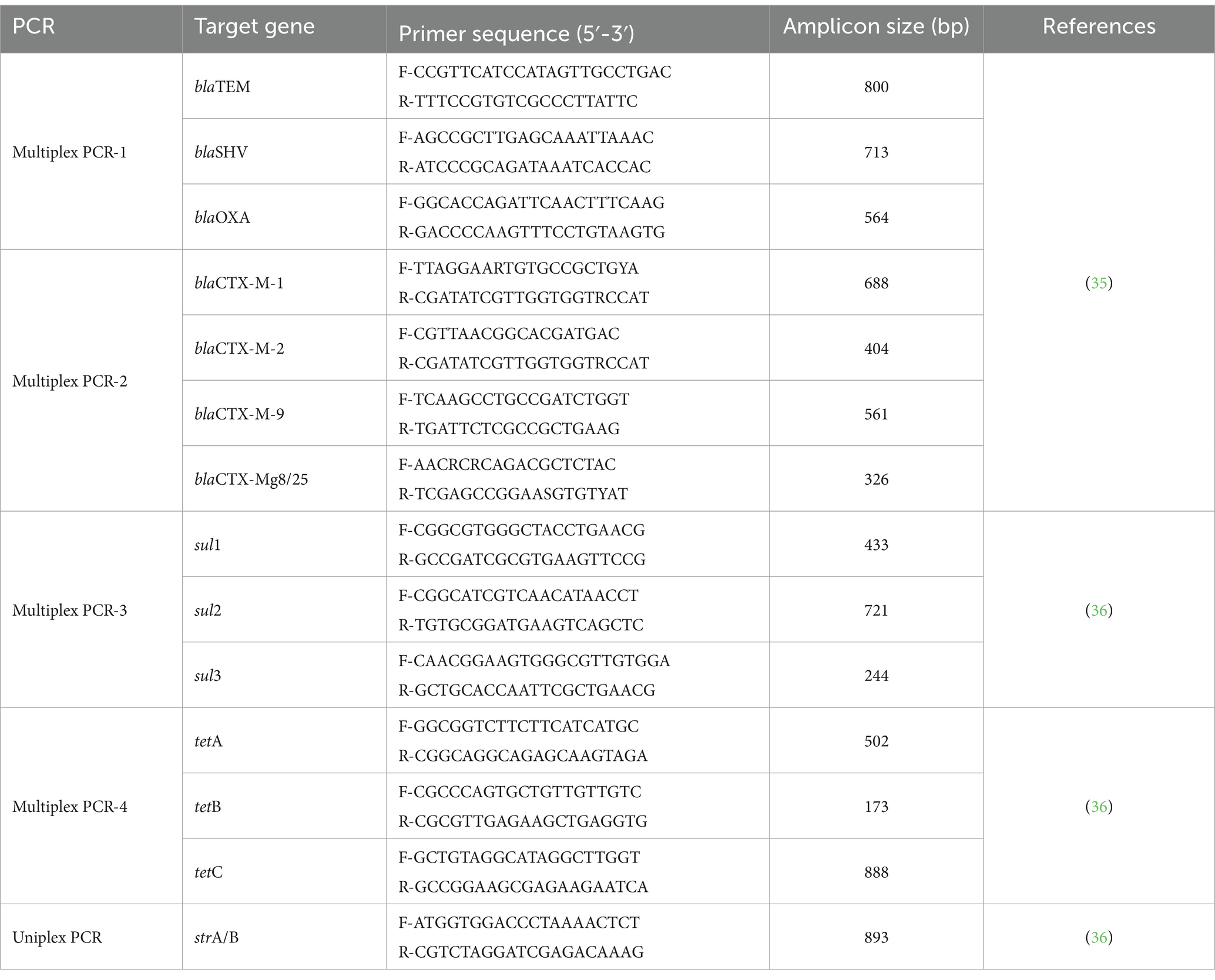
Table 3. Targeted genes, primer sequences and expected amplicon sizes for detection of antimicrobial resistance genes.
2.7 Statistical analysis
The attained data on antimicrobial susceptibility was presented in Excel worksheets (MS-2017). The prevalence was calculated using descriptive analysis.
3 Results
3.1 Bacteriology
Out of 1,274 samples that were cultured, 114 (8.94%) Salmonella isolates were recovered and from which 97 (85.09%) isolates were identified to be Salmonella Enteritidis. The remaining 17 (14.91%) Salmonella isolates were excluded from this study.
3.2 Virulence factors
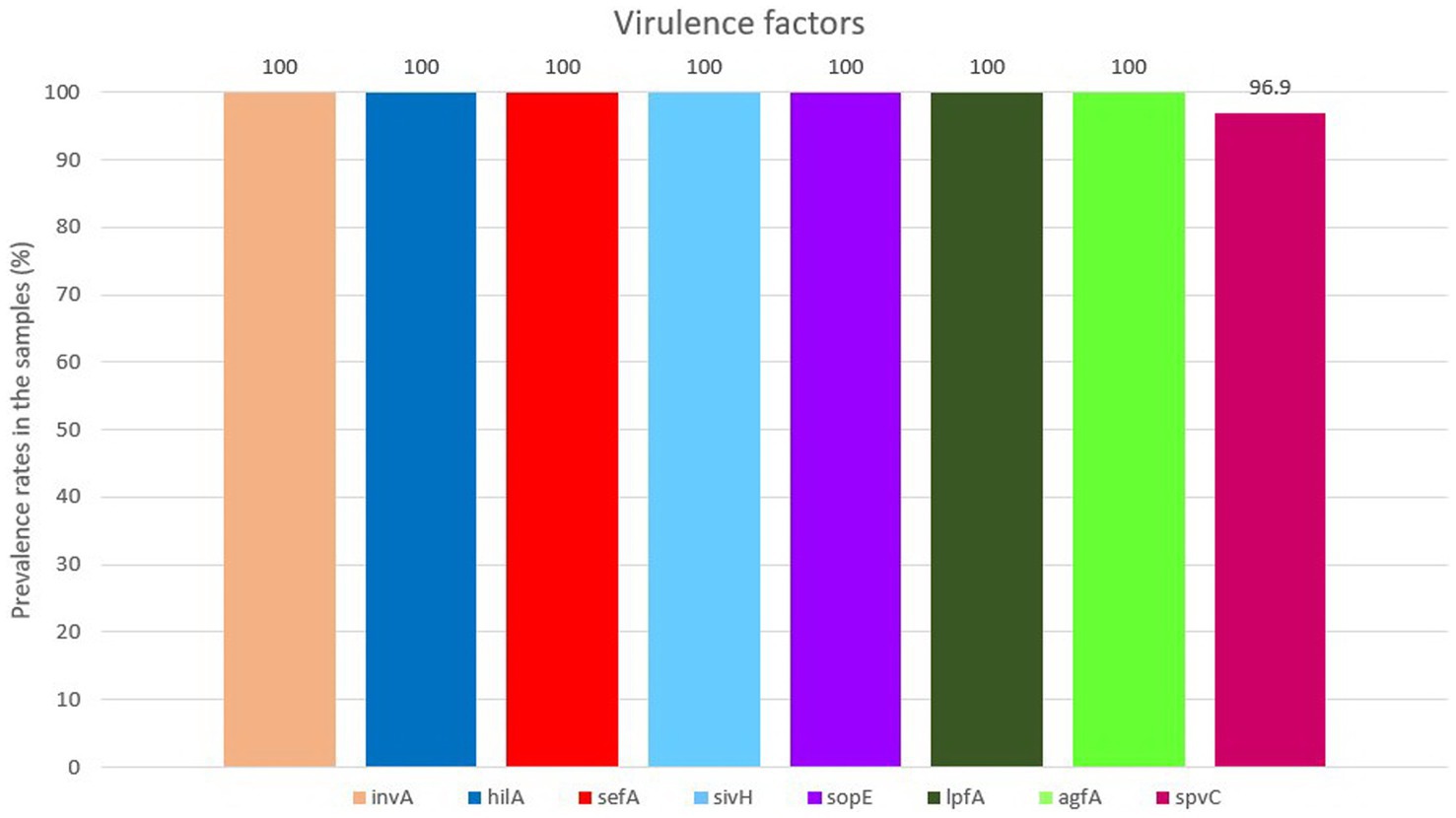
All isolates were positive for the presence of all eight virulence genes, except for spvC gene which was found in 94 (96.90%) out of 97 isolates (Figure 2). For detailed information, refer to Supplementary Table S2.
3.3 MAR index and phenotypic resistance profiles
The antimicrobial susceptibility evaluation of 16 antimicrobial agents belonging to 10 different classes of antimicrobials revealed that 100% of isolates showed resistance to nalidixic acid, followed by 83.5 and 80.41% resistance observed to ampicillin and amoxicillin–clavulanate, respectively. In addition, 79.38% of isolates showed resistance to ciprofloxacin. Moderate to low resistance rates were found against tetracycline (38.14%), streptomycin (12.37%) and chloramphenicol (11.3%). All isolates were 100% sensitive to meropenem, ceftazidime and aztreonam and a high sensitivity was observed to azithromycin (95.88%), sulfamethoxazole-trimethoprim (96.91%), amikacin (97.94%), gentamicin (97.94%), ceftriaxone (97.94%) and cefotaxime (98.97%). Details are given in Supplementary Table S3.
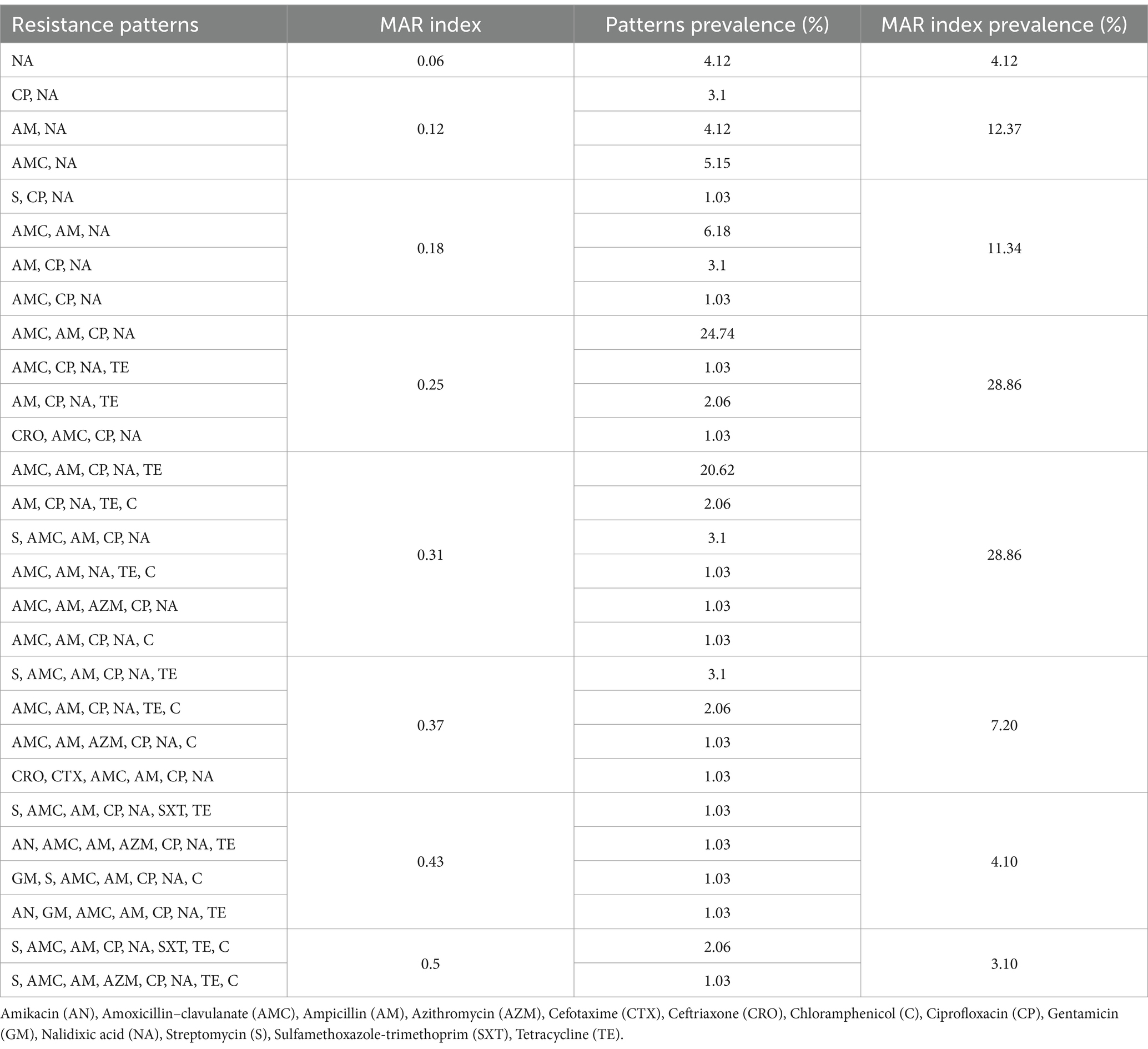
The calculated mean of MAR index for all isolates was 0.26, the lowest MAR index was 0.06 and the highest MAR index was 0.5. Considering MAR indexes above 0.2 are highly resistant, the prevalence of these isolates was 72.15%. The most prevalent antimicrobial resistance pattern (24.74%) was AMC, AM, CP, NA (MAR index = 0.25). Resistance patterns and MAR indexes are shown in Table 4.
3.4 Genotypic resistance patterns
Among 14 investigated resistance genes, only five genes including blaTEM, tetA, tetB, sul1 and strA/B were variably detected among isolates. No resistance genes were found in 11 isolates, 10 of which had a MAR index below 0.2. In four isolates which had MAR indexes above 0.4, all five genes of blaTEM, tetA, tetB, sul1 and strA/B were simultaneously present. The most frequent detected resistance gene was blaTEM (63.92%), followed by tetB (61.85%), tetA (36.08%), strA/B (14.43%) and sul1 (10.30%). The related details are given in Supplementary Table S3.
4 Discussion
Different serovars of Salmonella have been detected among various poultry sources with a large variation in prevalence. For instance, only in poultry egg samples, studies around the world have reported Salmonella prevalence of 0% in Egypt (37), 0.3% in Bangladesh (38), 2.9% in Eastern Ethiopia (39), 3% in Belgium (40), 3.3% in North India (41), 5.4% in China (42), 7.7% in South India (43), 24.17% in Nigeria (44) and 34% in Spain (45). In Iran, a Salmonella contamination rate of 3.8% (46) and 6.3% (47) in poultry eggs have been indicated. Additionally, Bahramianfard and co-workers (47) found a SE prevalence of 1.3 and 2.3% in egg and poultry samples, respectively. These variations in Salmonella prevalence could be related to differences in geographical regions, periods of sampling, hygienic measures and management programs.
The importance of Salmonella Enteritidis among non-typhoid serovars is undoubtable and unquestionable. In 2005, Velge et al. ranked Salmonella Enteritidis as the most prevalent serovar in poultry and since then, it has been confirmed by many studies performed in different regions of the world (48). Results from 37 countries revealed prevalence rates from 19.2% (in Cameroon) to 49% (in Tunisia) in Africa, and 5 to 93.7% in Europe and Asia (3, 47, 49).
Many researchers in Iran have conducted studies on Salmonella prevalence in Iranian poultry flocks. Zahraei Salehi et al. found 30 (15.6%) Salmonella isolates in 192 samples taken from liver and intestine of broiler chickens (50). The prevalence of Salmonella in broiler chicken carcasses in abattoir was reported to be 8.3% (51). In 2011–2012, 25.8% of the fecal samples originated from poultry slaughterhouses were positive for Salmonella, from which 40.4% of the isolates were identified as SE (52). In another study, Afshari et al. found 14 (14%) Salmonella isolates among 100 samples taken from poultry, in which six (43%) isolates were confirmed as serovar Enteritidis (53). Recent studies on Salmonella contamination of poultry meat samples provided from retail stores in Iran have reported the prevalence of Salmonella to be 8.75% in 80 samples (54) or 2.7% in 150 samples (55). Even though the geographical regions of the latter studies were not included in our study, our results are in harmony with these findings. This high prevalence is correlated with the complexity of the transmission of Salmonella. This complexity could be explained by the ability to survive in different environments, the vast spectrum of hosts and carriers, being equipped with different virulence factors and resistance against antimicrobial agents (56–60). These results confirm the point that new hygienic measures and protocols should be implemented to control this zoonosis pathogen.
We found all eight detected virulence genes at very high rates in SE isolates including genes of Salmonella pathogenicity islands 1 and 2 (SPI-1 and SPI-2), genes related to cell adhesion and genes which play important roles in pathologic mechanisms. It could be interpreted from previous studies that among non-typhoid serovars, the presence of investigated virulence factors in SE was higher compared to other common serovars such as ST. For instance, in another study with the same framework on poultry-origin Salmonella from wet markets in Bangladesh, Siddiky et al. detected the presence of eight virulence genes including invA, agfA, lpfA, hilA, sivH, sefA, sopE and spvC with the rate of 100% presence in SE isolates (20). Our results are compatible with the results of Siddiky et al. except for spvC, a plasmid-mediated gene. We found spvC in 96.90% of SE isolates compared with that of in 100% (20, 61), 92% (62) and 88.6% (63) of SE isolates in previous studies (20, 61). These variations in the results could be related to the geographical distribution of the strains and also to the developments and optimizations made over the years in molecular methods and materials.
The sefA gene is associated with the production of an SE fimbrial protein with a molecular mass of 17 kDa (SEF 17), which inhibits the binding of the extracellular matrix protein named fibronectin to SE (64). The 100% prevalence of sefA gene among SE isolates of this study confirmed the previous findings and as some researchers have suggested, the sefA gene can be considered a proper candidate for identification of serovar Enteritidis (20, 61–63). Aside from the invA gene role in cell invasion, the sefA gene plays an important role in diagnosis of Salmonella species (65, 66). Additionally, for invasion into the host’s cells, expression of the hilA gene also increases the virulence of Salmonella, and this gene is usually 100% present in SE isolates (67, 68). Both invA and hilA genes could be considered as symbols of the invasive nature of Salmonella (20, 61, 62). The 100% presence of lpfA, agfA, and sopE genes among SE isolates of the present study is consistent with the findings of previous investigations (20, 64, 69). The agfA gene takes part in the development of biofilm, which is crucial for the survival of the organism and, due to its relation to coding a fimbrial protein, it is liable for cell adhesion (70). Previous findings also confirm our data (20, 61, 70), except the results of Borges and co-workers in Brazil, which found agfA gene only in 96% of SE isolates from chickens (62). In addition, the 100% prevalence of sopE gene –encoded in SPI-1- is accordant to previous studies (20, 62, 71, 72). The high rate of virulence genes detected in this study, reinforces the necessity of practical hygienic measures to control and reduce Salmonella infections, considering the vigorous invasive nature of this pathogen. The synergism between virulence genes and antimicrobial resistance could escalate the risk of infection and its consequences, and facilitate the spread of resistant pathogenic Salmonella in human and animals (71, 73, 74).
In this study, we evaluated the recent situation of antimicrobial resistance among Salmonella Enteritidis isolated from broiler chickens from both phenotypic and genetic aspects. Phenotypically, SE isolates were found to have high resistance rates against nalidixic acid (100%), ampicillin (83.5%), amoxicillin–clavulanate (80.41%), and ciprofloxacin (79.38%); moderate resistance to tetracycline (38.14%), and low resistance to streptomycin (12.37%), and chloramphenicol (11.34%). In comparing these results to other studies in Iran, Bahramianfard et al. (47) observed resistance to nalidixic acid (87.3%), kanamycin (25.4%), colistin sulphate (23.8%) and trimethoprim-sulfamethoxazole (20.6%) among SE isolates from poultry meat and egg samples. In another study, Khademi et al. also reported high rates of antimicrobial resistance to Salmonella serovars recovered from clinical samples in Iran (1983–2019) including resistance to tetracycline (54.3%), ceftizoxime (50.6%), streptomycin (50.2%), and nalidixic acid (48.1%) (75). In a study by Vaez et al., Salmonella isolates from animals were mostly resistant to nalidixic acid (67%), tetracycline (66.9%), streptomycin (49.6%), and trimethoprim-sulfamethoxazole (41.6%) (76). Moreover, Besharati et al. showed higher resistance rates against antimicrobial agents among Salmonella serovars originated from poultry processed meat compared to those of obtained from human stool samples (77). The maximum resistance rates among Salmonella isolates from poultry processed meat were as follows tetracycline (59%), trimethoprim-sulfamethoxazole (43%), azithromycin (42%), chloramphenicol (27%); while the resistance rates were significantly lower in human stool samples indicating tetracycline (13.6%), trimethoprim-sulfamethoxazole (9.1%), azithromycin (9.1%), and chloramphenicol (0%) (77). Nemati and Ahmadi reported the antimicrobial resistance rates among Salmonella isolates from western regions of Iran as such ampicillin (100%), nalidixic acid (73.13%), trimethoprim-sulfamethoxazole (58.20%), streptomycin (47.76%), and tetracycline (43.28%) (78).
In other countries, in China, Salmonella isolates recovered from abattoirs was shown to be resistant mostly against nalidixic acid (99.5%), ampicillin (87.8%), tetracycline (51.9%), ciprofloxacin (48.7%), and trimethoprim-sulfamethoxazole (48.1%) (79). In Bangladesh, Parvin et al. found a high resistance among Salmonella isolates originated from frozen chicken meat samples against oxytetracycline (100%), trimethoprim-sulfamethoxazole (89.2%), tetracycline (86.5%), nalidixic acid (83.8%), amoxicillin (74.3%), and pefloxacin (74.3%) (80). In a Chinese study, it was demonstrated that Salmonella isolates recovered from hatcheries were highly resistant to ciprofloxacin (77%), sulfisoxazole (73%), and ampicillin (55.6%) (81). Furthermore, in another study from Bangladesh, Siddiky et al. detected high resistance levels against streptomycin (100%), ciprofloxacin, tetracycline and gentamicin (80%); moderate resistance to amikacin, amoxicillin–clavulanate, azithromycin and sulphamethoxazole-trimethoprim (20%) in SE isolated from broilers (20). In comparison, Siddiky and coworkers detected 100% resistance against ciprofloxacin and streptomycin, 86.66% resistance to tetracycline, nalidixic acid and gentamicin, 66.66% resistance to ampicillin and 40% resistance against amoxicillin–clavulanate in ST isolates from broiler chickens (20). Our other findings from antimicrobial susceptibility test results indicated low resistance levels against azithromycin (4.12%), sulfamethoxazole-trimethoprim (3.09%), ceftriaxone, gentamicin and amikacin (2.06%) and cefotaxime (1.03%) and full sensitivity against meropenem, ceftazidime and aztreonam that are comparable with those of from other studies (20).
A high MAR index (> 0.2) indicates a frequent use of antibiotics suggesting poultry products as a high-risk source for multi-drug resistant (MDR) Salmonella strains. In addition, Mishra et al. indicated that MAR index values >0.2 were associated with a high-risk source of contamination and MAR > 0.4 indicated a fecal source of contamination (82). In this study, isolates had a mean MAR index of 0.259 with the highest MAR of 0.5. Seven out of 97 (7.21%) isolates had MAR index values of >0.4 and 70 (72.16%) isolates had a MAR index value of >0.2. Moreover, we detected 28 different resistance patterns and the most prevalent pattern (20.62%) was resistance to amoxicillin–clavulanate, ampicillin, ciprofloxacin, nalidixic acid, and tetracycline. Moreover, one finding that drew our attention was that resistance to sulfamethoxazole-trimethoprim that was observed only in isolates showing MAR index values of >0.43. Strict surveillances should be applied in the regions where isolates with high MAR index values were detected.
We investigated the presence of seven ESBL genes among our isolates. Only one, the blaTEM gene, was present in 63.92% of isolates, and it was the most prevalent gene. Other β-lactamase resistance genes were not detected in any of isolates from this study. In this regard, Sales et al. detected blaTEM gene in 34.61%, and blaSHV in 11.53% of ST isolates from children with diarrhea, and a total rate of 57.69% of their isolates were positive for ESBL (83). According to Lai et al., 89.9% of 129 Salmonella isolated from fecal samples of pigs, goats, cattle, rabbits, chickens and ducks between September 2016 and May 2019 in China possessed β-lactamase resistance genes and the blaTEM gene was detected in 82.9% of those isolates. Other β-lactamase genes including blaOXA (20.2%), blaCTX-M (6.2%), and blaCMY (2.3%) were also detected (84). Furthermore, Das et al. found a high prevalence of blaTEM (95.4%) in Salmonella isolated from broiler flocks of Bangladesh (85). In Bangladesh, Siddiky et al. also detected blaTEM gene in 73.3% of Salmonella isolated from broiler chickens (86) and 62.06 to 69.62% of Salmonella isolated from different parts of wet markets (20). In Iraq, Hassan et al. detected blaTEM in 52.6% of Salmonella enterica isolates from 28 broiler chicken farms, with no trace of blaSHV, blaCTX-M, and blaOXA (87). In contrast, Ramatla et al. detected blaTEM in 7% of ST and 28% of SE isolates from chickens and rats in layer farms of South Africa. Among SE isolates, they also detected other ESBL genes including blaCTX-M (39%), blaCTX-M1 (44%), and blaCTX-M9 (33%). The total number of ESBL encoding genes was higher in ST compared to SE isolates (88). Moreover, Hardiati et al. identified a 100% presence of blaTEM in Salmonella isolates from chicken farms of Java, Indonesia (89).
In evaluating the prevalence of antimicrobial resistance genes related to tetracyclines, we detected tetA gene in 36.08% and tetB in 61.85% of the isolates. No tetC positive isolate was detected in any isolates. Moreover, the strA/B gene, which is related to resistance against streptomycin was identified in 14.43% of SE isolates. Similarly, a lower prevalence of 10.30% for sulphonamide-related resistance gene sul1 was found in SE isolates. No isolates harbored sul2 and sul3 resistance genes. Four (4.12%) isolates possessed all of these five detected genes with the genotypic pattern of “blaTEM, tetA, tetB, sul1, strA/B,” while in 11 (11.34%) isolates no resistance gene was detected at all. Based on our findings, it was interesting that the mean of MAR index values for those five isolates with the mentioned genotypic resistance pattern was 0.465, while the mean of MAR index values for the 11 isolates with no detected resistance gene was 0.126. This significant difference in the latter results can confirm the correlation and harmony between the phenotypic and genotypic resistance results. Keeping in mind that modulation of gene expression could depend on several different factors, it is a nonnegligible point, and it can justify the higher prevalence of some resistance genes, compared to phenotypic resistance rates against related classes of antimicrobial drugs (90).
The given data in the previous paragraph are comparable to those of Siddiky et al. study (86). They found prevalence values of 100% for tetA and 20% for sul1 and strA/B in Salmonella isolates from chicken samples provided from wet markets in Bangladesh (86). Moreover, Das et al. detected the tetA, tetB, and tetC genes in 81.4, 19.8, and 10.47% of Salmonella isolates in commercial broiler farms of Bangladesh, respectively. In addition, 37.2% of their isolates harbored the sul1 gene (85). Consequently, Hardiati et al. identified tetA in 33.3% of the Salmonella isolates from chicken farms in Indonesia (89). Wang et al. detected tetA, blaTEM, sul1, and sul2 genes with prevalence rates of 81.3, 62.5, 25, and 100%, respectively, among Salmonella isolated from retail meat samples in China (91). Recently, Nazari Moghadam et al. reported the presence of tetracycline-resistant (tetA, tetB, tetC, tetG) and sulphonamide-resistant (sul1, sul2, and sul3) genes in 100, 23, 27, 39% for tetA-tetG and 84, 50, and 17% for sul1-sul3, respectively, among Salmonella isolated from poultry meat (54). Our genotypic resistance patterns were also consistent with earlier findings in Iran and other countries.
It is important to note the fact that this study performed on samples received from only 15 out of 31 Iranian provinces and; moreover, due to different regional antimicrobial use practices, the interpretation and generalizability of our findings may have some limitations. Working on additional samples from other provinces may help for a better understanding of SE distribution among broiler chicken population in Iran and of the relevant genetic data. The variability in regional antimicrobial use practices, including varying levels of antibiotic stewardship, accessibility, and usage policies, creates discrepancies in the selection pressure exerted on bacterial populations (92). These variations can result in significant regional differences in MAR index values, making it challenging to extrapolate our findings to broader populations or global settings. To address these challenges, we emphasize the importance of adopting standardized sampling protocols and incorporating regional antibiotic usage data to provide a more comprehensive understanding of AMR trends. Despite these limitations, our study contributes valuable insights into the resistance patterns of Salmonella and underscores the need for harmonized methodologies to enable better cross-regional comparisons and the development of targeted mitigation strategies.
The observed resistance patterns in Salmonella isolates may result from factors beyond direct antibiotic use. Horizontal gene transfer in shared environments, such as farms or processing facilities, could facilitate the spread of resistance genes among bacterial populations. Environmental contamination with antibiotics or residues may create sub-lethal selection pressures, favoring resistant strains (93). Additionally, historical antibiotic use and regional differences in host immune responses or ecological factors may contribute to resistance dynamics (94). Methodological variations, including sampling protocols and resistance classification criteria, could also influence the results. To better understand these patterns, future studies should integrate environmental, genetic, and methodological data alongside comprehensive antimicrobial use surveillance, enabling more targeted and effective mitigation strategies.
To control AMR in Salmonella, vaccination and stricter biosecurity measures are essential. Vaccination reduces Salmonella prevalence and minimizes the need for antibiotics by enhancing immunity. Multivalent and live-attenuated vaccines should be integrated into routine poultry health programs, with regular monitoring to assess their effectiveness. Also, There have been several trials to develop a vaccine against SE (95). Stricter biosecurity measures, including enhanced hygiene, restricted farm access, pest control, and clean feed and water management, prevent Salmonella introduction and spread. Implementing all-in/all-out flock systems further reduces cross-contamination (96). Combining these strategies with robust antimicrobial stewardship and surveillance can effectively reduce Salmonella infections and mitigate the spread of AMR in poultry systems.
To effectively mitigate AMR, we propose implementing several targeted policies. First, non-therapeutic antibiotic use should be banned, with therapeutic applications allowed only under strict veterinary supervision. Vaccination programs targeting key Salmonella serovars should be mandated to lower infection rates and reduce reliance on antibiotics, supported by financial incentives to encourage adoption. Enhanced biosecurity measures, including improved hygiene, pest control, and controlled access to farms, should be enforced through regular inspections. Additionally, a centralized surveillance system to monitor antibiotic usage and resistance trends in poultry farming is crucial for guiding evidence-based interventions. Training programs for farmers and veterinarians on AMR, sustainable practices, and biosecurity protocols should be prioritized to build compliance and awareness. Finally, farms that adopt sustainable, antibiotic-free practices should receive financial incentives, while penalties for non-compliance will ensure accountability. These combined measures can significantly reduce Salmonella-related AMR risks and promote sustainable poultry farming practices.
Whole genome sequencing (WGS) surpasses classical PCR in detecting virulence and antimicrobial resistance (AMR) genes by providing comprehensive genomic analysis. Unlike PCR, which targets specific genes, WGS identifies both known and novel genes, their genetic context (e.g., plasmids or mobile elements), and mutations linked to resistance. It enables full characterization of gene clusters and regulatory elements, offering deeper insights into pathogen biology and gene transfer mechanisms. Additionally, WGS is scalable for epidemiological studies and outbreak tracking, making it a more powerful and versatile tool than PCR for studying virulence and AMR (97).
5 Conclusion
Data obtained from this study highlights the great potential risks of the presence and transmission of highly pathogenic MDR Salmonella to humans from chicken meat sources as well as the need for more effective surveillance program for antibiotic use in the poultry industry. Strict hygiene and sanitation standards in poultry product chains should be reprogrammed to reduce transmission. High associations among the virulence genes, phenotypic resistance and genotypic resistance were evident in the results. The high prevalence rates of MDR in Salmonella Enteritidis isolates, along with the overwhelming presence of major virulence factors raise public health concerns. Therapeutic, preventive and imprudent uses of antimicrobial agents have resulted in the exposure of pathogens to these drugs and increasing the risk of developing resistance. Reducing/optimizing the use of antimicrobials, improving poultry management procedures, strict clean and disinfection measures, proper use of probiotics and biosecurity—especially considering the possibility of using vaccines—are essential to deal with this issue.
To address antimicrobial resistance in Salmonella, we recommend studying molecular mechanisms like plasmids and efflux pumps, tracking resistance trends through longitudinal studies, and exploring the role of horizontal gene transfer and environmental reservoirs. Omics-based research can identify resistance determinants and therapeutic targets, while investigations into host-microbiota interactions and non-antibiotic interventions, such as vaccines and probiotics, can support sustainable mitigation strategies. These efforts are essential for effective AMR control.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary material, further inquiries can be directed to the corresponding author.
Ethics statement
The animal studies were approved by the ethical committee of the Faculty of Veterinary Medicine of the University of Tehran. The studies were conducted in accordance with the local legislation and institutional requirements. Written informed consent was not obtained from the owners for the participation of their animals in this study because the samples were sent by poultry farm owners to Diagnostic Veterinary Laboratory of University of Tehran for Salmonella isolation.
Author contributions
MP: Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Validation, Writing – original draft, Writing – review & editing. SP: Conceptualization, Formal analysis, Funding acquisition, Methodology, Project administration, Resources, Supervision, Validation, Visualization, Writing – original draft, Writing – review & editing. JR: Conceptualization, Formal analysis, Methodology, Supervision, Visualization, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This research was funded by the grant no. 7508007.6.48 from the Research Council of the University of Tehran.
Acknowledgments
We would like to thank Dr. A. Barin for his help in providing samples, Ms. A. Yazdani and Ms. F. Naderinezhad for their assistance in laboratory works.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The authors declare that no Gen AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fvets.2025.1542313/full#supplementary-material
SUPPLEMENTARY TABLE S1 | Thermal profiles of the studied genes.
SUPPLEMENTARY TABLE S2 | Laboratory codes, isolation dates and details of virulence factors results.
SUPPLEMENTARY TABLE S3 | Origin of the isolates, phenotypic results, patterns of drug resistance, MAR index and genotypic results of resistance genes.
References
1. Hawker, J, Begg, N, Reintjes, R, Ekdahl, K, Edeghere, O, and Van Steenbergen, JE. Communicable disease control and health protection handbook. 4th ed. Hoboken, NJ: John Wiley & Sons (2019).
2. Jain, P, Chowdhury, G, Samajpati, S, Basak, S, Ganai, A, Samanta, S, et al. Characterization of non-typhoidal Salmonella isolates from children with acute gastroenteritis, Kolkata, India, during 2000–2016. Brazilian J Microbiol. (2020) 51:613–27. doi: 10.1007/s42770-019-00213-z
3. Hendriksen, RS, Vieira, AR, Karlsmose, S, Lo, D, Jensen, AB, Wegener, HC, et al. Global monitoring of Salmonella serovar distribution from the World Health Organization global foodborne infections network country data bank: results of quality assured aboratories from 2001 to 2007. Foodborne Pathog Dis. (2011) 8:887–900. doi: 10.1089/fpd.2010.0787
4. World Health Organization. WHO releases the 2019 aware classification antibiotics. New York, NY, USA: World Health Organization (2019).
5. Sornplang, P, Aieamsaard, J, Saksangawong, C, and Suayroop, N. Risk factors associated with Salmonella prevalence, its antibiotic resistance, and egg antibiotic residues in the layer farming environment. Vet World. (2022) 15:543–50. doi: 10.14202/vetworld.2022.543-550
6. Boiko, O, Garkavenko, T, Musiiets, I, Nedosekov, V, and Kozytska, T. Salmonellosis in Ukraine: an analysis of food products contamination, salmonella transmission, and serovar diversity during 2012–2023. Ger J Vet Res. (2024) 4:65–74. doi: 10.51585/gjvr.2024.2.0085
7. Wales, A, and Davies, R. Review of hatchery transmission of bacteria with focus on Salmonella, chick pathogens and antimicrobial resistance. Worlds Poult Sci J. (2020) 76:517–36. doi: 10.1080/00439339.2020.1789533
8. Kaonga, N, Hang’ombe, B, Lupindu, A, and Hoza, A. Detection of CTX-M-type extended-spectrum beta-lactamase producing Salmonella Typhimurium in commercial poultry farms in Copperbelt province, Zambia. Ger J Vet Res. (2021) 1:27–34. doi: 10.51585/gjvr.2021.2.0011
9. Gast, R.K., and Porter, JR R.E. Salmonella infections, in: D.E. Swayne, M. Boulianne, C. M. Logue, L. R. McDougald, V. Nair, and D. L. Suarez, et al., Eds., Diseases of poultry, 14th edition. Hoboken, NJ: John Wiley & Sons, Inc. (2020) 719–753
10. Scharff, RL. Economic burden from health losses due to foodborne illness in the United States. J Food Prot. (2012) 75:123–31. doi: 10.4315/0362-028X.JFP-11-058
11. Agyare, C, Boamah, VE, Zumbi, CN, and Osei, FB. Antibiotic use in poultry production and its effects on bacterial resistance In: Y Kumar, editor. Antimicrobial resistance—A global threat. London: IntechOpen Limited (2018). 33–51.
12. Castro-Vargas, RE, Herrera-Sánchez, MP, Rodríguez-Hernández, R, and Rondón-Barragán, IS. Antibiotic resistance in Salmonella spp. isolated from poultry: a global overview. Vet World. (2020) 13:2070–84. doi: 10.14202/vetworld.2020.2070-2084
13. Zhou, K, Sun, L, Zhang, X, Xu, X, Mi, K, Ma, W, et al. Salmonella antimicrobials inherited and the non-inherited resistance: mechanisms and alternative therapeutic strategies. Front Microbiol. (2023) 14:1176317. doi: 10.3389/fmicb.2023.1176317
14. Igbinosa, IH, Amolo, CN, Beshiru, A, Akinnibosun, O, Ogofure, AG, El-Ashker, M, et al. Identification and characterization of MDR virulent Salmonella spp. isolated from smallholder poultry production environment in Edo and Delta states, Nigeria. PLoS One. (2023) 18:e0281329. doi: 10.1371/journal.pone.0281329
15. Igbinosa, EO, Beshiru, A, Igbinosa, IH, and Okoh, AI. Antimicrobial resistance and genetic characterisation of Salmonella enterica from retail poultry meats in Benin city, Nigeria. LWT. (2022) 169:114049. doi: 10.1016/j.lwt.2022.114049
16. Johnson, R, Mylona, E, and Frankel, G. Typhoidal Salmonella: distinctive virulence factors and pathogenesis. Cell Microbiol. (2018) 20:e12939. doi: 10.1111/cmi.12939
17. Lou, L, Zhang, P, Piao, R, and Wang, Y. Salmonella pathogenicity island 1 (SPI-1) and its complex regulatory network. Front Cell Infect Microbiol. (2019) 9:270. doi: 10.3389/fcimb.2019.00270
18. Han, J, Aljahdali, N, Zhao, S, Tang, H, Harbottle, H, Hoffmann, M, et al. Infection biology of Salmonella enterica. EcoSal Plus. (2024) 12:eesp00012023. doi: 10.1128/ecosalplus.esp-0001-2023
19. Azimi, T, Zamirnasta, M, Sani, MA, Soltan Dallal, MM, and Nasser, A. Molecular mechanisms of Salmonella effector proteins: a comprehensive review. Infect Drug Resist. (2020) 13:11–26. doi: 10.2147/IDR.S230604
20. Siddiky, NA, Sarker, S, Khan, SR, Rahman, T, Kafi, A, and Samad, MA. Virulence and antimicrobial resistance profile of non-typhoidal Salmonella enterica serovars recovered from poultry processing environments at wet markets in Dhaka, Bangladesh. PLoS One. (2022) 17:e0254465. doi: 10.1371/journal.pone.0254465
21. Wellawa, RMDH. Characterizing the role of putative virulence genes associated with infection, colonization and persistence of Salmonella Enteritidis in chicken using a bioluminescent reporter: University of Saskatchewan. (2022).
22. Silva, C, Puente, JL, and Calva, E. Salmonella virulence plasmid: pathogenesis and ecology. Pathog Dis. (2017) 75:ftx070. doi: 10.1093/femspd/ftx070
23. Wagenaar, J, Hendriksen, RS, and Carrique-Mas, J. Practical considerations of surveillance of Salmonella serovars other than Enteritidis and Typhimurium. Rev Sci Tech. (2013) 32:509–19. doi: 10.20506/rst.32.2.2244
24. Shivaning Karabasanavar, N, Benakabhat Madhavaprasad, C, Agalagandi Gopalakrishna, S, Hiremath, J, Shivanagowda Patil, G, and Barbuddhe, SB. Prevalence of Salmonella serotypes S. Enteritidis and S. Typhimurium in poultry and poultry products. J Food Saf. (2020) 40:e12852. doi: 10.1111/jfs.12852
25. Patra, SD, Mohakud, NK, Panda, RK, Sahu, BR, and Suar, M. Prevalence and multidrug resistance in Salmonella enterica Typhimurium: an overview in south East Asia. World J Microbiol Biotechnol. (2021) 37:185–17. doi: 10.1007/s11274-021-03146-8
26. Putturu, R, Eevuri, T, Ch, B, and Nelapati, K. Salmonella Enteritidis-foodborne pathogen-a review. Int J Pharm Biol Sci. (2015) 5:86–95.
27. Antunes, P, Mourão, J, Campos, J, and Peixe, L. Salmonellosis: the role of poultry meat. Clin Microbiol Infect. (2016) 22:110–21. doi: 10.1016/j.cmi.2015.12.004
28. Beshiru, A, Igbinosa, IH, and Igbinosa, EO. Prevalence of antimicrobial resistance and virulence gene elements of Salmonella serovars from ready-to-eat (rte) shrimps. Front Microbiol. (2019) 10:1613. doi: 10.3389/fmicb.2019.01613
29. Swayne, DE, Glisson, JR, Jackwood, MW, Pearson, JE, and Reed, WM. A laboratory manual for the isolation and identification of avian pathogens, 4th edition. American Association of Avian Pathologists; (1998)
30. Yoon, J. Focused commentary; about revision of CLSI antimicrobial breakpoints, 2018-2021. J Bacteriol Virol. (2022) 52:41–53. doi: 10.4167/jbv.2022.52.2.041
31. Chitanand, M, Kadam, T, Gyananath, G, Totewad, N, and Balhal, D. Multiple antibiotic resistance indexing of coliforms to identify high risk contamination sites in aquatic environment. Indian J Microbiol. (2010) 50:216–20. doi: 10.1007/s12088-010-0042-9
32. Morshed, R, and Peighambari, SM. Drug resistance, plasmid profile and random amplified polymorphic DNA analysis of Iranian isolates of Salmonella Enteritidis. New Microbiol. (2010) 33:47.
33. Malorny, B, Tassios, PT, Rådström, P, Cook, N, Wagner, M, and Hoorfar, J. Standardization of diagnostic PCR for the detection of foodborne pathogens. Int J Food Microbiol. (2003) 83:39–48. doi: 10.1016/s0168-1605(02)00322-7
34. Agron, PG, Walker, RL, Kinde, H, Sawyer, SJ, Hayes, DC, Wollard, J, et al. Identification by subtractive hybridization of sequences specific for Salmonella enterica serovar Enteritidis. Appl Environ Microbiol. (2001) 67:4984–91. doi: 10.1128/AEM.67.11.4984-4991.2001
35. Dallenne, C, Da Costa, A, Decré, D, Favier, C, and Arlet, G. Development of a set of multiplex PCR assays for the detection of genes encoding important β-lactamases in Enterobacteriaceae. J Antimicrob Chemother. (2010) 65:490–5. doi: 10.1093/jac/dkp498
36. Kozak, GK, Boerlin, P, Janecko, N, Reid-Smith, RJ, and Jardine, C. Antimicrobial resistance in Escherichia Coli isolates from swine and wild small mammals in the proximity of swine farms and in natural environments in Ontario, Canada. Appl Environ Microbiol. (2009) 75:559–66. doi: 10.1128/AEM.01821-08
37. Abdel-Maksoud, M, Abdel-Khalek, R, El-Gendy, A, House, BL, Gamal, RF, and Abdelhady, HM. Genetic characterisation of multidrug-resistant Salmonella enterica serotypes isolated from poultry in Cairo, Egypt. Afr J Lab Med. (2015) 4:1–7. doi: 10.4102/ajlm.v4i1.158
38. Begum, K, Reza, TA, Haque, M, Hossain, A, Hassan, FK, Hasan, SN, et al. Isolation, identification and antibiotic resistance pattern of Salmonella spp. from chicken eggs, intestines and environmental samples. Bangladesh Pharm J. (2010) 13:23–7.
39. Tessema, K, Bedu, H, Ejo, M, and Hiko, A. Prevalence and antibiotic resistance of Salmonella species isolated from chicken eggs by standard bacteriological method. J Vet Sci Technol. (2017) 8:2. doi: 10.4172/2157-7579.1000421
40. De Vylder, J, Dewulf, J, Van Hoorebeke, S, Pasmans, F, Haesebrouck, F, Ducatelle, R, et al. Horizontal transmission of Salmonella Enteritidis in groups of experimentally infected laying hens housed in different housing systems. Poult Sci. (2011) 90:1391–6. doi: 10.3382/ps.2010-00944
41. Singh, R, Yadav, A, Tripathi, V, and Singh, R. Antimicrobial resistance profile of Salmonella present in poultry and poultry environment in North India. Food Control. (2013) 33:545–8. doi: 10.1016/j.foodcont.2013.03.041
42. Xie, T, Wu, G, He, X, Lai, Z, Zhang, H, and Zhao, J. Antimicrobial resistance and genetic diversity of Salmonella enterica from eggs. Food Sci Nutr. (2019) 7:2847–53. doi: 10.1002/fsn3.1126
43. Suresh, T, Hatha, A, Sreenivasan, D, Sangeetha, N, and Lashmanaperumalsamy, P. Prevalence and antimicrobial resistance of Salmonella Enteritidis and other salmonellas in the eggs and egg-storing trays from retails markets of Coimbatore, South India. Food Microbiol. (2006) 23:294–9. doi: 10.1016/j.fm.2005.04.001
44. Ekundayo, E, and Ezeoke, J. Prevalence and antibiotic sensitivity profile of Salmonella species in eggs from poultry farms in Umudike, Abia state. J Anim Vet Adv. (2011) 10:206–9. doi: 10.3923/javaa.2011.206.209
45. García, C, Soriano, J, Benítez, V, and Catalá-Gregori, P. Assessment of Salmonella spp. in feces, cloacal swabs, and eggs (eggshell and content separately) from a laying hen farm. Poult Sci. (2011) 90:1581–5. doi: 10.3382/ps.2010-01104
46. Askari Badouei, M, Mohammadian Ghalejooghi, B, and Madadgar, O. Study on Salmonella contamination of traditionally produced edible poultry eggs. Comp Clin Path. (2012) 21:1093–7. doi: 10.1007/s00580-011-1238-z
47. Bahramianfard, H, Derakhshandeh, A, Naziri, Z, and Khaltabadi, FR. Prevalence, virulence factor and antimicrobial resistance analysis of Salmonella Enteritidis from poultry and egg samples in Iran. BMC Vet Res. (2021) 17:196. doi: 10.1186/s12917-021-02900-2
48. Velge, P, Cloeckaert, A, and Barrow, P. Emergence of Salmonella epidemics: the problems related to Salmonella enterica serotype Enteritidis and multiple antibiotic resistance in other major serotypes. Vet Res. (2005) 36:267–88. doi: 10.1051/vetres:2005005
49. Gritli, A, Daboussi, T, Moussa, MB, and Abassi, M. Prevalence and characterizaton of Salmonella in chicken consumed in military cantines. J New Sci. (2015) 12:908–14.
50. Salehi, TZ, Mahzounieh, M, and Saeedzadeh, A. The isolation of antibiotic-resistant Salmonella from intestine and liver of poultry in shiraz province of Iran. Int J Poult Sci. (2005) 4:320–2. doi: 10.3923/ijps.2005.320.322
51. Jamshidi, AE, Basami, M, and Afshari, NS. Identification of Salmonella spp and Salmonella Typhimurium by a multiplex PCR-based assay from poultry carcasses in Mashhad-Iran. Iran J Vet Med. (2009) 3:43–8. doi: 10.22059/ijvm.2009.19608
52. Ghaderi, R, Moradi Bidhendi, S, and Khaki, P. Occurrence of multidrug-resistant Salmonella enterica serovar Enteritidis isolates from poultry in Iran. Arch Razi Inst. (2016) 71:43–9. doi: 10.22034/ari.2016.105997
53. Afshari, A, Baratpour, A, Khanzade, S, and Jamshidi, A. Salmonella Enteritidis and Salmonella Typhimorium identification in poultry carcasses. Iranian J Microbiol. (2018) 10:45–50.
54. Nazari Moghadam, M, Rahimi, E, Shakerian, A, and Momtaz, H. Prevalence of Salmonella Typhimurium and Salmonella Enteritidis isolated from poultry meat: virulence and antimicrobial-resistant genes. BMC Microbiol. (2023) 23:168. doi: 10.1186/s12866-023-02908-8
55. Mir, R, Salari, S, Najimi, M, and Rashki, A. Determination of frequency, multiple antibiotic resistance index and resistotype of Salmonella spp. in chicken meat collected from southeast of Iran. Vet Med Sci. (2022) 8:229–36. doi: 10.1002/vms3.647
56. Alali, WQ, Thakur, S, Berghaus, RD, Martin, MP, and Gebreyes, WA. Prevalence and distribution of Salmonella in organic and conventional broiler poultry farms. Foodborne Pathog Dis. (2010) 7:1363–71. doi: 10.1089/fpd.2010.0566
57. Dewaele, I, Rasschaert, G, Wildemauwe, C, Van Meirhaeghe, H, Vanrobaeys, M, De Graef, E, et al. Polyphasic characterization of Salmonella Enteritidis isolates on persistently contaminated layer farms during the implementation of a national control program with obligatory vaccination: a longitudinal study. Poult Sci. (2012) 91:2727–35. doi: 10.3382/ps.2012-02218
58. Roche, A, Cox, N, Richardson, L, Buhr, R, Cason, J, Fairchild, B, et al. Transmission of Salmonella to broilers by contaminated larval and adult lesser mealworms, Alphitobius Diaperinus (Coleoptera: Tenebrionidae). Poult Sci. (2009) 88:44–8. doi: 10.3382/ps.2008-00235
59. Shirota, K, Umali, DV, Suzuki, T, and Katoh, H. Epizootiologic role of feeds in the epidemiology of Salmonella Senftenberg contamination in commercial layer farms in eastern Japan. Avian Dis. (2012) 56:516–20. doi: 10.1637/9964-101611-Reg.1
60. Wales, A, Carrique-Mas, J, Rankin, M, Bell, B, Thind, B, and Davies, R. Review of the carriage of zoonotic bacteria by arthropods, with special reference to Salmonella in mites, flies and litter beetles. Zoonoses Public Health. (2010) 57:299–314. doi: 10.1111/j.1863-2378.2008.01222.x
61. Crăciunaş, C, Keul, A-L, Flonta, M, and Cristea, M. DNA-based diagnostic tests for Salmonella strains targeting hilA, agfA, spvC and sef genes. J Environ Manag. (2012) 95:S15–8. doi: 10.1016/j.jenvman.2010.07.027
62. Borges, KA, Furian, TQ, Borsoi, A, Moraes, HL, Salle, CT, and Nascimento, VP. Detection of virulence-associated genes in Salmonella Enteritidis isolates from chicken in south of Brazil. Pesq Vet Bras. (2013) 33:1416–22. doi: 10.1590/S0100-736X2013001200004
63. Amini, K, Salehi, TZ, Nikbakht, G, Ranjbar, R, Amini, J, and Ashrafganjooei, SB. Molecular detection of invA and spv virulence genes in Salmonella Enteritidis isolated from human and animals in Iran. Afr J Microbiol Res. (2010) 4:2202–10.
64. Collinson, S, Doig, P, Doran, J, Clouthier, S, Trust, T, and Kay, W. Thin, aggregative fimbriae mediate binding of Salmonella Enteritidis to fibronectin. J Bacteriol. (1993) 175:12–8. doi: 10.1128/jb.175.1.12-18.1993
65. Nayak, R, Stewart, T, Wang, R-F, Lin, J, Cerniglia, C, and Kenney, P. Genetic diversity and virulence gene determinants of antibiotic-resistant Salmonella isolated from preharvest Turkey production sources. Int J Food Microbiol. (2004) 91:51–62. doi: 10.1016/S0168-1605(03)00330-1
66. Elkenany, R, Elsayed, MM, Zakaria, AI, El-sayed, SA-E-S, and Rizk, MA. Antimicrobial resistance profiles and virulence genotyping of Salmonella enterica serovars recovered from broiler chickens and chicken carcasses in Egypt. BMC Vet Res. (2019) 15:124–9. doi: 10.1186/s12917-019-1867-z
67. Ramatla, T, Ngoma, L, Adetunji, M, and Mwanza, M. Evaluation of antibiotic residues in raw meat using different analytical methods. Antibiotics. (2017) 6:34. doi: 10.3390/antibiotics6040034
68. Cardona-Castro, N, Restrepo-Pineda, E, and Correa-Ochoa, M. Detection of hilA gene sequences in serovars of Salmonella enterica subspecies enterica. Mem Inst Oswaldo Cruz. (2002) 97:1153–6. doi: 10.1590/s0074-02762002000800016
69. Borsoi, A, Santin, E, Santos, L, Salle, C, Moraes, H, and Nascimento, V. Inoculation of newly hatched broiler chicks with two Brazilian isolates of Salmonella Heidelberg strains with different virulence gene profiles, antimicrobial resistance, and pulsed field gel electrophoresis patterns to intestinal changes evaluation. Poult Sci. (2009) 88:750–8. doi: 10.3382/ps.2008-00466
70. Yoo, AY, Yu, JE, Yoo, H, Lee, TH, Lee, WH, Oh, J-I, et al. Role of sigma factor E in regulation of Salmonella agf expression. Biochem Biophy Res Commun. (2013) 430:131–6. doi: 10.1016/j.bbrc.2012.11.025
71. Huehn, S, la, R, Anjum, M, Saunders, M, Woodward, MJ, Bunge, C, et al. Virulotyping and antimicrobial resistance typing of Salmonella enterica serovars relevant to human health in Europe. Foodborne Pathog Dis. (2010) 7:523–35. doi: 10.1089/fpd.2009.0447
72. Batchelor, M, Hopkins, K, Threlfall, E, Clifton-Hadley, F, Stallwood, A, Davies, R, et al. BlaCTX-M genes in clinical Salmonella isolates recovered from humans in England and Wales from 1992 to 2003. Antimicrob Agents Chemother. (2005) 49:1319–22. doi: 10.1128/AAC.49.4.1319-1322.2005
73. Salam, M, Al-Amin, M, Salam, M, Pawar, J, Akhter, N, Rabaan, A, et al. Antimicrobial resistance: a growing serious threat for global public health. Healthcare. (2023) 11:1946. doi: 10.3390/healthcare11131946
74. Aslam, M, Checkley, S, Avery, B, Chalmers, G, Bohaychuk, V, Gensler, G, et al. Phenotypic and genetic characterization of antimicrobial resistance in Salmonella serovars isolated from retail meats in Alberta, Canada. Food Microbiol. (2012) 32:110–7. doi: 10.1016/j.fm.2012.04.017
75. Khademi, F, Vaez, H, Ghanbari, F, Arzanlou, M, Mohammadshahi, J, and Sahebkar, A. Prevalence of fluoroquinolone-resistant Salmonella serotypes in Iran: a meta-analysis. Pathog Glob Health. (2020) 114:16–29. doi: 10.1080/20477724.2020.1719701
76. Vaez, H, Ghanbari, F, Sahebkar, A, and Khademi, F. Antibiotic resistance profiles of Salmonella serotypes isolated from animals in Iran: a meta-analysis. Iranian J Vet Res. (2020) 21:188–97. doi: 10.22099/ijvr.2020.36252.5296
77. Besharati, S, Sadeghi, A, Ahmadi, F, Tajeddin, E, Mohammad, R, Fani, F, et al. Serogroups, and drug resistance of non-typhoidal Salmonella in symptomatic patients with community-acquired diarrhea and chicken meat samples in Tehran. Iranian J Vet Res. (2020) 21:269–78. doi: 10.22099/ijvr.2020.36912.5387
78. Nemati, F, and Ahmadi, E. Class1-3 integrons and antimicrobial resistance profile in Salmonella spp. isolated from broiler chicken in western Iran. J Hellenic Vet Med Soc. (2020) 71:2471–82. doi: 10.12681/jhvms.25922
79. Zhu, Y, Lai, H, Zou, L, Yin, S, Wang, C, Han, X, et al. Antimicrobial resistance and resistance genes in Salmonella strains isolated from broiler chickens along the slaughtering process in China. Int J Food Microbiol. (2017) 259:43–51. doi: 10.1016/j.ijfoodmicro.2017.07.023
80. Parvin, MS, Hasan, MM, Ali, MY, Chowdhury, EH, Rahman, MT, and Islam, MT. Prevalence and multidrug resistance pattern of Salmonella carrying extended-spectrum β-lactamase in frozen chicken meat in Bangladesh. J Food Prot. (2020) 83:2107–21. doi: 10.4315/JFP-20-172
81. Xu, X, Biswas, S, Gu, G, Elbediwi, M, Li, Y, and Yue, M. Characterization of multidrug resistance patterns of emerging Salmonella enterica serovar Rissen along the food chain in China. Antibiotics. (2020) 9:660. doi: 10.3390/antibiotics9100660
82. Mishra, M, Patel, AK, and Behera, N. Prevalence of multidrug resistant E. coli in the river Mahanadi of Sambalpur. Curr Res Microbiol Biotechnol. (2013) 1:239–44.
83. Sales, AJ, Naebi, S, Nasiri, R, and Bannazadeh-Baghi, H. The antibiotic resistance pattern and prevalence of blaTEM, blaSHV, blaCTX-M, blaPSE-1, sipB/C, and cmlA/tetR genes in Salmonella Typhimurium isolated from children with diarrhea in Tabriz, Iran. Int J Health Life Sci. (2021) 7:e118523. doi: 10.5812/ijhls.118523
84. Lai, J, Mu, H, Zhou, B, He, J, Cheng, X, Gan, Y, et al. BlaTEM-positive Salmonella enterica serovars Agona and Derby are prevalent among food-producing animals in Chongqing, China. Front Microbiol. (2023) 14:1011719. doi: 10.3389/fmicb.2023.1011719
85. das, T, Rana, EA, Dutta, A, Bostami, MB, Rahman, M, Deb, P, et al. Antimicrobial resistance profiling and burden of resistance genes in zoonotic Salmonella isolated from broiler chicken. Vet Med Sci. (2022) 8:237–44. doi: 10.1002/vms3.648
86. Siddiky, NA, Sarker, MS, Khan, MSR, Begum, R, Kabir, ME, Karim, MR, et al. Virulence and antimicrobial resistance profiles of Salmonella enterica serovars isolated from chicken at wet markets in Dhaka, Bangladesh. Microorganisms. (2021) 9:952. doi: 10.3390/microorganisms9050952
87. Hassan, ER, Alhatami, AO, Abdulwahab, HM, and Schneider, BS. Characterization of plasmid-mediated quinolone resistance genes and extended-spectrum beta-lactamases in non-typhoidal Salmonella enterica isolated from broiler chickens. Vet World. (2022) 15:1515–22. doi: 10.14202/vetworld.2022.1515-1522
88. Ramatla, T, Mileng, K, Ndou, R, Mphuti, N, Syakalima, M, Lekota, KE, et al. Molecular detection of integrons, colistin and β-lactamase resistant genes in Salmonella enterica serovars Enteritidis and Typhimurium isolated from chickens and rats inhabiting poultry farms. Microorganisms. (2022) 10:313. doi: 10.3390/microorganisms10020313
89. Hardiati, A, Safika, S, Pasaribu, FH, and Wibawan, IWT. Multidrug-resistant Salmonella sp. isolated from several chicken farms in West Java, Indonesia. Indonesian J Vet Sci. (2022) 16:6–11. doi: 10.21157/j.ked.hewan.v16i1.18944
90. Depardieu, F, Podglajen, I, Leclercq, R, Collatz, E, and Courvalin, P. Modes and modulations of antibiotic resistance gene expression. Clin Microbiol Rev. (2007) 20:79–114. doi: 10.1128/CMR.00015-06
91. Wang, W, Chen, J, Shao, X, Huang, P, Zha, J, and Ye, Y. Occurrence and antimicrobial resistance of Salmonella isolated from retail meats in Anhui, China. Food Sci Nutr. (2021) 9:4701–10. doi: 10.1002/fsn3.2266
92. Zanichelli, V, Monnier, AA, Gyssens, IC, Adriaenssens, N, Versporten, A, Pulcini, C, et al. Variation in antibiotic use among and within different settings: a systematic review. J Antimicrob Chemother. (2018) 73:vi17–29. doi: 10.1093/jac/dky115
93. Ifedinezi, OV, Nnaji, ND, Anumudu, CK, Ekwueme, CT, Uhegwu, CC, Ihenetu, FC, et al. Environmental antimicrobial resistance: implications for food safety and public health. Antibiotics. (2024) 13:1087. doi: 10.3390/antibiotics13111087
94. Uddin, TM, Chakraborty, AJ, Khusro, A, Zidan, BRM, Mitra, S, Emran, TB, et al. Antibiotic resistance in microbes: history, mechanisms, therapeutic strategies and future prospects. J Infect Public Health. (2021) 14:1750–66. doi: 10.1016/j.jiph.2021.10.020
95. Shehata, AA, Tarabees, R, Elsayed, M, Wareth, G, and Basiouni, S. Development of Salmonella Enteritidis vaccine candidate based on streptomycin independent suppressor and metabolic drift rifampicin resistance-attenuating markers. Heliyon. (2020) 6:e04810. doi: 10.1016/j.heliyon.2020.e04810
96. Denagamage, TN. Epidemiology, risk factors, and control of Salmonella Enteritidis contamination of commercial layer flocks and shell eggs. (2016)
97. Akinyemi, KO, Fakorede, CO, Linde, J, Methner, U, Wareth, G, Tomaso, H, et al. Whole genome sequencing of Salmonella enterica serovars isolated from humans, animals, and the environment in Lagos, Nigeria. BMC Microbiol. (2023) 23:164. doi: 10.1186/s12866-023-02901-1
98. Cesco, MAO, Zimermann, FC, Giotto, DB, Guayba, J, Borsoi, A, Rocha, SL, et al. Pesquisa de genes de virulência em Salmonella Hadar em amostras provenientesde material avícola. Vet foco. (2009) 6:159–64. doi: 10.13140/2.1.2720.6082
99. Bäumler, AJ, and Heffron, F. Identification and sequence analysis of lpfABCDE, a putative fimbrial operon of Salmonella Typhimurium. J Bacteriol. (1995) 177:2087–97. doi: 10.1128/jb.177.8.2087-2097.1995
100. Guo, X, Chen, J, Beuchat, LR, and Brackett, RE. PCR detection of Salmonella enterica serotype Montevideo in and on raw tomatoes using primers derived from hilA. Appl Environ Microbiol. (2000) 66:5248–52. doi: 10.1128/AEM.66.12.5248-5252.2000
101. Kingsley, RA, Humphries, AD, Weening, EH, de, M, Winter, S, Papaconstantinopoulou, A, et al. Molecular and phenotypic analysis of the CS54 island of Salmonella enterica serotype Typhimurium: identification of intestinal colonization and persistence determinants. Infect Immun. (2003) 71:629–40. doi: 10.1128/IAI.71.2.629-640.2003
102. Oliveira, S, Santos, L, Schuch, D, Silva, A, Salle, C, and Canal, C. Detection and identification of salmonellas from poultry-related samples by PCR. Vet Microbiol. (2002) 87:25–35. doi: 10.1016/s0378-1135(02)00028-7
103. Prager, R, Rabsch, W, Streckel, W, Voigt, W, Tietze, E, and Tschäpe, H. Molecular properties of Salmonella enterica serotype Paratyphi B distinguish between its systemic and its enteric pathovars. J Clin Microbiol. (2003) 41:4270–8. doi: 10.1128/JCM.41.9.4270-4278.2003
Keywords: antimicrobial resistance, broiler chicken, poultry, Salmonella Enteritidis, virulence factors
Citation: Piryaei MR, Peighambari SM and Razmyar J (2025) Drug resistance and genotyping studies of Salmonella Enteritidis isolated from broiler chickens in Iran. Front. Vet. Sci. 12:1542313. doi: 10.3389/fvets.2025.1542313
Edited by:
Faham Khamesipour, Ministry of Health and Medical Education, IranReviewed by:
Gamal Wareth, Friedrich Loeffler Institut, GermanyAbeni Beshiru, Federal Institute for Risk Assessment (BfR), Germany
Yesim Soyer, Middle East Technical University, Türkiye
Copyright © 2025 Piryaei, Peighambari and Razmyar. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Seyed Mostafa Peighambari, bXBlaWdoYW1AdXQuYWMuaXI=
 Mohammad Reza Piryaei
Mohammad Reza Piryaei Seyed Mostafa Peighambari
Seyed Mostafa Peighambari Jamshid Razmyar
Jamshid Razmyar