
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Vet. Sci., 06 February 2025
Sec. Veterinary Pharmacology and Toxicology
Volume 12 - 2025 | https://doi.org/10.3389/fvets.2025.1534498
This article is part of the Research TopicInsights in Veterinary Pharmacology and Toxicology: 2024View all 4 articles
 Qiuru Chen1†
Qiuru Chen1† Yulei Liang1†
Yulei Liang1† Yanbin Dong1
Yanbin Dong1 Junling Cui1
Junling Cui1 Kun He1
Kun He1 Xiaoyuan Ma1
Xiaoyuan Ma1 Jinfeng Zhao1,2
Jinfeng Zhao1,2 Yajun Zhai1,2*
Yajun Zhai1,2* Li Yuan1,2,3,4*
Li Yuan1,2,3,4*H-NS is a histone-like nucleoid-structuring protein that regulates gene expressions, particularly acquired foreign genes, however, little is known about whether H-NS can modulate bacterial susceptibility by regulating its intrinsic genes. The hns-deleted mutant EΔhns, the hns-complemented strain EΔhns/phns and the hns-overexpressed strain E/phns were derivatives of Escherichia coli ATCC 25922, the susceptibility of which were assessed by the broth microdilution method and time-kill curves assays. We found that the MICs for strain EΔhns against gentamicin and amikacin were significantly decreased by 8–16 folds in contrast to E. coli ATCC 25922. Further studies displayed that the absence of hns caused damage to the bacterial outer membrane and increased the expression levels of porin-related genes, such as ompC, ompF, ompG, and ompN, thus obviously enhancing aminoglycosides uptake of strain EΔhns. Meanwhile, hns deletion also led to remarkable inhibition of the efflux pumps activity and decreased expressions of efflux-related genes clbM, acrA, acrB, acrD, and emrE, which reduced the efflux of aminoglycosides. In addition, the activation of glycolysis and electron transport chain, as well as the reduction of Δψ dissipation, could lead to a remarkable increase in proton motive force (PMF), thus further inducing more aminoglycosides uptake by strain EΔhns. Our findings reveal that H-NS regulates the resistance of E. coli to aminoglycosides by influencing its uptake and efflux, which will enrich our understanding of the mechanism by which H-NS modulates bacterial resistance.
The histone-like nucleoid structuring (H-NS) protein is 137 amino acids in length in Escherichia coli and closely related bacteria and is present at very high levels, with approximately 20,000 to 60,000 molecules per cell (1, 2). It is generally believed that the H-NS protein, as a global regulator, preferentially binds to AT-rich DNA by preventing RNA polymerase from accessing or escaping promoters (3), thereby silencing the expression of corresponding genes, notably acquired foreign genes, such as resistance genes acquired by horizontal transfer (4, 5). To date, H-NS has received considerable attention in regulating acquired foreign genes, and it has been proved that H-NS protects bacteria and drives their evolution via regulating the expression of foreign DNA (6–8). Meanwhile, some literatures have clarified that H-NS can also modulate genes associated with virulence, stress response, quorum sensing, biosynthesis pathways and cell adhesion by recognizing intrinsically curved DNA (9–11).
Up to now, there are a few reports on H-NS regulating multidrug resistance by controlling the inherent genes in the bacterial host. Nishino et al. proposed that H-NS contributed to multidrug resistance E. coli by regulating the expression of multidrug exporter genes such as acrEF and mdtEF (12, 13). In 2018, Deveson Lucas team found that H-NS inactivation resulted in an increased resistance to colistin in a clinical isolate of Acinetobacter baumannii (14). Rodgers et al. proved that H-NS modulated antibiotic resistance in Acinetobacter baumannii by governing genes codifying for biofilms and efflux pumps (15). Antibiotic resistance is achieved through a variety of mechanisms, such as alteration or bypass of the drug target, production of antibiotic-modifying enzymes, decreased drug uptake, increased drug efflux, and biofilm formation (16). However, little is known about whether H-NS can influence multidrug resistance by interfering drug uptakes.
Aminoglycosides have been one of the important antibiotics for preventing Gram-negative bacteria infection since 1940. Nevertheless, the widespread presence of resistance bacteria has led to a sharp decline in its efficacy. Recently, we occasionally found that the deletion of hns increased the susceptibility of E. coli to many antibiotics, especially aminoglycosides, with their minimal inhibitory concentrations (MICs) decreased by 8–16 folds. The work described below elucidates that H-NS regulates the resistance of E. coli ATCC 25922 to aminoglycosides by influencing its uptake and efflux, which will enrich our understanding of the regulatory mechanism of H-NS on bacterial resistance, and also contribute to the development of new drugs to curb bacterial resistance.
Bacteria and plasmids used in this study are listed in Table 1. Escherichia coli ATCC 25922 was obtained from the China Institute of Veterinary Drug Control. Strain EΔhns is a derivative of E. coli ATCC 25922 via the one-step inactivation of chromosomal gene hns (17). The complementary strain EΔhns/phns was constructed as follows: Firstly, the complete open reading frame of hns was amplified by PCR from the genomic DNA of E. coli ATCC 25922 using primers XhoI-hns–F/HindIII-hns-R (Table 2). Thereafter, the expression plasmid pBAD::hns was generated by inserting the target fragment to the vector pBAD (Karsbad, CA, the United States) and then was introduced to EΔhns by electroporation. The overexpressed hns strain E/phns is a derivative of E. coli ATCC 25922 that was introduced of pBAD::hns by electroporation. All strains were cultured in fresh Luria-Bertani (LB) broth (Beijing Land Bridge Technology Co., Ltd.) at 37°C, where strains EΔhns, EΔhns/phns and E/phns were induced by 0.2% L-arabinose (18).
A single colony of strains, such as E. coli ATCC 25922, EΔhns, EΔhns/phns and E/phns were cultured in LB medium at 37°C. After growth overnight, the cultures were diluted 1:100 in fresh medium and grown to an OD600 of 0.5. Following this, the total bacterial RNA was extracted with a TaKaRa MiniBEST universal RNA extraction kit (TaKaRa Bio, Inc., Shiga, Japan). The quantity of extracted RNA was determined by A260 measurements and purity was evaluated by the A260/A280 ratio using a NanoDrop 1000 Spectrophotometer (Thermo Scientific, Hvidovre, Denmark). The cDNA samples were synthesized using the cDNA reverse transcription kit with gDNA Eraser (TaKaRa Bio, Inc.) and then the RT-qPCR was performed with an annealing temperature of 60°C according to our previous study (18). The relative expression levels were calculated by the 2−ΔΔCt method and compared with the levels of E. coli ATCC 25922. The 16S rRNA gene was chosen as a housekeeping gene, and the primers of RT-qPCR are listed in Table 1. Three independent biological replicates were performed.
Growth curves for E. coli ATCC 25922, EΔhns, EΔhns/phns and E/phns were plotted. Overnight cultures were inoculated in fresh LB medium and grown to an OD600 of 0.5. Then the cultures were diluted (1:1,000) in preheated fresh LB broth and inoculated with shaking at 37°C. The OD600 was measured periodically at 1-h intervals using an ultraviolet spectrophotometer and shown as means ± standard deviations. Experiments were performed with at least three biological replicates.
Antimicrobial susceptibility testing was tested using the broth microdilution method according to the guidelines of Clinical and Laboratory Standards Institute (CLSI) guidelines (19). The antimicrobial agents are cefotaxime, gentamicin, amikacin, doxycycline, tigecycline, florfenicol, and enrofloxacin. Experiments were performed with at least three biological replicates.
Overnight cultures of E. coli ATCC 25922, EΔhns and EΔhns/phns were diluted (1:1,000) in fresh LB broth and grown to an OD600 of 0.5. Bacteria were cultured in the LB broth with different concentrations of gentamicin (0.25, 0.5, 1, and 2 μg/mL) for 24 h. At regular intervals, the culture broths were serially diluted with 0.9% saline and plated onto LB agar plates. Colony forming units (CFU) were counted after 18 h of incubation at 37°C, and the Log10 CFU values were calculated according to previous study (20). Take the Log10 CFU values as the vertical axis, and the incubation time (h) as the horizontal axis to establish the time sterilization curve. Three independent biological replicates were performed.
Overnight cultures of E. coli ATCC 25922, EΔhns and EΔhns/phns were diluted 1:100 in fresh LB medium and incubated at 37°C. Samples equivalent to an OD600 of 0.5 were removed and suspended with 5 mmol·L−1 N-2-hydroxyethylpiperazine-N'-2- ethanesulfonic acid (HEPES, pH 7.0, plus 5 mmol·L−1glucose). Thereafter, the OD600 of the above bacteria suspension was standardized to 0.5 using the same buffer. Fluorescent probe NPN (10 μM) was used to evaluate the OM integrity of strains (21). Fluorescence intensity was measured using a Sparpk 10M Microplate reader (Tecan) with the excitation wavelength at 350 nm and the emission wavelength at 420 nm.
The culture suspension of E. coli ATCC 25922 and EΔhns were prepared according to the method described in 2.6. Thereafter, the bacterial suspensions were centrifuged, washed softly with phosphate-buffered saline (PBS) buffer for three times, and mixed with 2.5% (w/v) glutaraldehyde in 0.1 M PBS buffer at 25°C for about 2 h away from light. Subsequently, the samples were dehydrated in sequential with a graded ethanol series (30%, 50%, 70%, 90%, and 100%), transferred to a culture dish, dried overnight, and observed using SEM (Hitachi, Japan).
Bacteria were cultured and standardized to an OD600 of 0.5 using similar protocols described in 2.6. DiSC3 (5) (0.5 μM) (Aladdin, Shanghai, China) was added to achieve a final concentration of 2 μM in the mixture to determine the transmembrane electric potential (Δψ). The dissipation for Δψ in E. coli ATCC 25922, EΔhns and EΔhns/phns was measured with the excitation wavelength of 622 nm and emission wavelength of 670 nm according to the method described in the previous study (22).
The accumulation of ethidium bromide (Beyotime, Shanghai, China) in the cells was monitored as previously described with some modifications (21). Strains of E. coli ATCC 25922, EΔhns and EΔhns/phns were grown to an OD600 of 0.5, then centrifuged and suspended with 5 mmol·L−1 HEPES (pH 7.0, plus 5 mmol·L−1glucose). Subsequently, the OD600 of the bacterial suspension was standardized to 0.3 using the same buffer. 5 μM of ethidium bromide was added to determine the efflux pumps activity. Fluorescence intensity was determined with a Spark multifunctional microplate reader with excitation wavelength 530 nm and emission wavelength 600 nm. Each experiment was conducted in triplicate at least three times.
Total RNA of strains E. coli ATCC 25922 and EΔhns were extracted by the RNAprep Pure Cell/Bacteria Kit (TIANGEN Biotech (Beijing) Co., Ltd) and sequenced using the Illumina NovaSeq 2000 system by Novogene Bioinformatics Technology Co., Ltd. Differentially expressed genes (DEGs) were identified by using the fragments per kilobase of transcript per million mapped reads method with Padj < 0.05 and fold change (FC) of | log2FC | > 1. RNA-sequencing reads were aligned to E. coli ATCC 25922 (Ref-Seq accession no. CP025268).
Statistical analysis and the generation of plots were performed using GraphPad Prism 8. Unpaired t-test (normally distributed data) between two groups was used to calculate P-values. Differences with P < 0.05 were considered as significant difference. Significance levels are indicated by numbers of asterisks (ns, No significant difference, * P < 0.05, ** P < 0.01, *** P < 0.001, **** P < 0.0001).
The mRNA expression levels of hns gene in E. coli ATCC 25922, hns deletion strain EΔhns, complemented strain EΔhns/phns and overexpressed strain E/phns were measured by RT-qPCR (Figure 1). Compared with strain EΔhns, the hns expressions of strain EΔhns/phns increased by 44-fold, from 0.006 ± 0.001 increasing to 0.264 ± 0.015, although they were still sharply lower than that of E. coli ATCC 25922 (P < 0.0001), demonstrating that the complement of hns could only partially restore the function of H-NS in the deletion strain EΔhns, which may be closely linked to the location of H-NS, such as on chromosome or plasmids (1, 2, 23). Meanwhile, we observed that the hns expressions of strain E/phns (52.232 ± 4.182) were 52.23-fold increments in comparison with the reference strain E. coli ATCC 25922, proving that strain E/phns overexpressed hns.
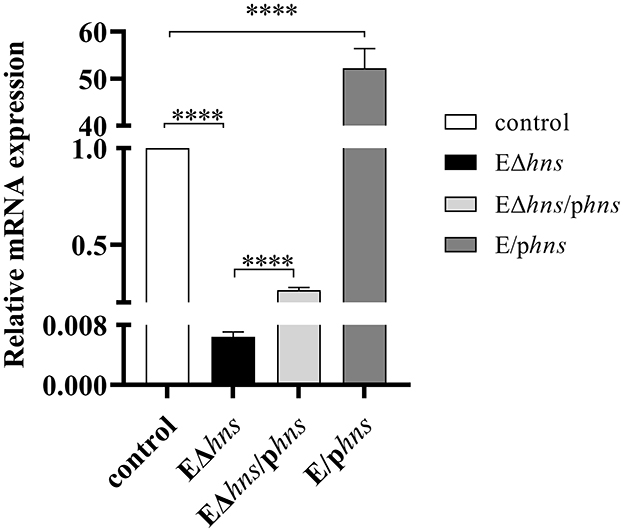
Figure 1. The mRNA expression levels of hns in the strains E. coli ATCC 25922, EΔhns, EΔhns/phns and E/phns. E. coli ATCC 25922 is the control. The strain EΔhns is the derivative of E. coli ATCC 25922 that lacks hns, the strain EΔhns/phns is the derivative of EΔhns that carries the recombined plasmid pBAD::hns, while the strain E/phns is the derivative of E. coli ATCC 25922 that carries pBAD::hns. Strains EΔhns, EΔhns/phns and E/phns are induced by 0.2% L-arabinose. Data are expressed as the mean ± standard deviation (SD) (**** P < 0.0001).
Growth kinetics of strains E. coli ATCC 25922, EΔhns, EΔhns/phns and E/phns were established respectively based on optical density at 600 nm values (Figure 2). No difference in growth was observed between E. coli ATCC 25922 and E/phns, which inferred that hns overexpressions had no effect on the growth of E. coli. It's worth noting that the OD600 values of strains EΔhns and EΔhns/phns were significantly lower than those of E. coli ATCC 25922 at 3–6 h incubation time, although the OD600 values rose to the control level at the later incubation, in line with previous studies (24). The aforementioned results reflect that hns deletion will lead to reduced adaptability of E. coli, although it does not affect the bacterial survival.
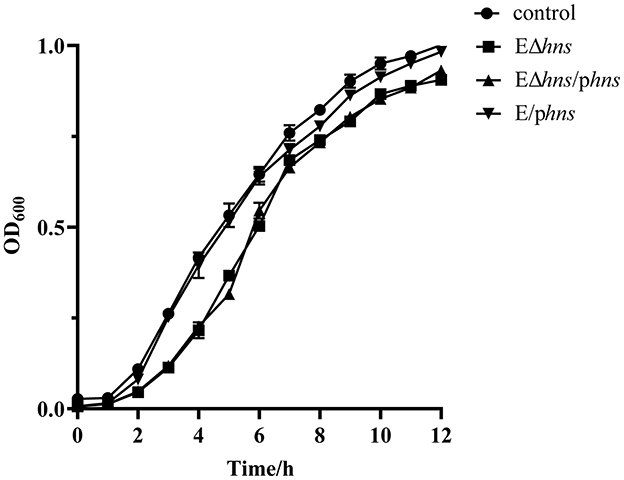
Figure 2. Growth curves of the strains E. coli ATCC 25922, EΔhns, EΔhns/phns and E/phns. E. coli ATCC 25922 is the control. The strain EΔhns is the derivative of E. coli ATCC 25922 that lacks hns, the strain EΔhns/phns is the derivative of EΔhns that carries the recombined plasmid pBAD::hns, while the strain E/phns is the derivative of E. coli ATCC 25922 that carries pBAD::hns. Curves indicate the means of three independent experiments. Strains EΔhns, EΔhns/phns, and E/phns are induced by 0.2% L-arabinose.
To determine whether hns deletion alters the susceptibility of E. coli to a variety of drugs, such as gentamicin, amikacin, cefotaxime, doxycycline, tigecycline, florfenicol, and enrofloxacin, the MIC values were determined for E. coli ATCC 25922, EΔhns, EΔhns/phns and E/phns (Table 1). In contrast to E. coli ATCC 25922 (MICs = 0.008 ~ 2 μg/mL), the MICs for strain EΔhns were significantly decreased, with gentamicin decreased by 16 folds, followed by amikacin (8-fold) and enrofloxacin (4-fold), while cefotaxime, doxycycline, tigecycline and florfenicol displaying 2-fold decreases. As expected, the MICs for strain EΔhns/phns partially recovered, while for strain E/phns was the same as that of the control. Collectively, our data illustrate that protein H-NS negatively regulates the multidrug resistance of E. coli.
To further investigate the impact of hns deletion on the susceptibility of strains, we conducted in vitro time-dependent killing assay of gentamicin against E. coli ATCC 25922, EΔhns and EΔhns/phns. As shown in Figure 3, strains E. coli ATCC 25922, EΔhns and EΔhns/phns were inhibited in a concentration-dependent manner after incubation with different concentrations of gentamicin from 0 h to 24 h. In addition, the bactericidal effect of gentamicin against EΔhns displayed markedly stronger than that against E. coli ATCC 25922. For strain EΔhns, the number of viable bacteria began to decrease obviously after incubation with 0.25 μg/mL of gentamicin for 3 h (Figure 3A), and all died after incubation with 2 μg/mL for 12 h (Figures 3C, D), whereas for E. coli ATCC 25922, the bactericidal effect was observed only after incubation with 2 μg/mL of gentamicin for 4 h (Figure 3D). In conclusion, hns deletion leads to increased susceptibility of E. coli ATCC 25922 to gentamicin.
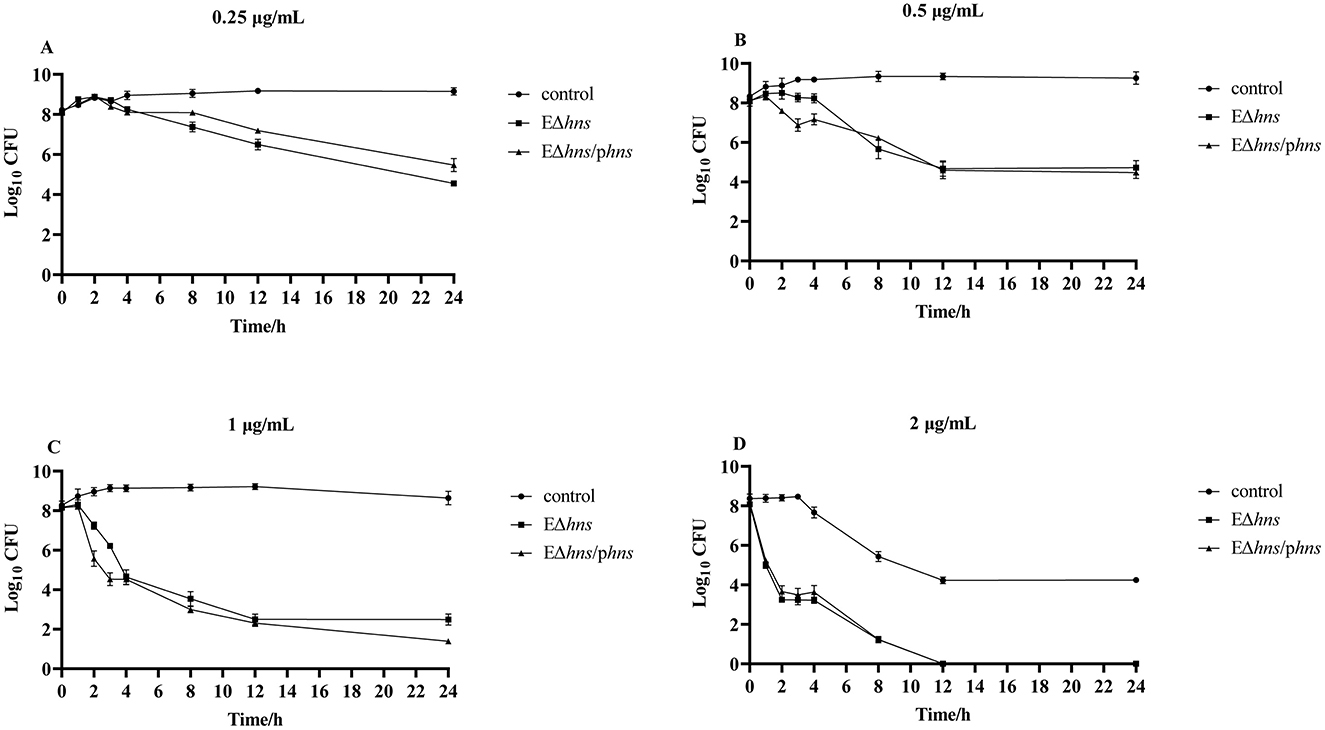
Figure 3. Time-kill curves for exponential phase strains E. coli ATCC 25922, EΔhns and EΔhns/phns, in the presence of gentamicin. The values of Log10 CFU are plotted as a function of time for different concentrations of gentamicin: (A) 0.25, (B) 0.5, (C) 1, and (D) 2 μg/mL). E. coli ATCC 25922 is the control. The strain EΔhns is the derivative of E. coli ATCC 25922 that lacks hns and the strain EΔhns/phns is the derivative of EΔhns that carries the recombined plasmid pBAD:hns. Strains EΔhns and EΔhns/phns are induced by 0.2% L-arabinose.
Notably, there was no significant difference in the bactericidal effect of gentamicin against strains EΔhns and EΔhns/phns, suggesting that (1) the expressions of hns located on pBAD could not completely complement the deletion of hns on chromosome, which had been also confirmed by the expression of hns in recombinant bacteria (Figure 1) and the results of in vitro antimicrobial susceptibility testing (Table 1); (2) the deletion of hns gene might lead to bacterial damages that were difficult to reverse even after the gene hns was complemented, such as membrane damages, although further studies were needed.
To understand the molecular mechanism by which H-NS regulates the susceptibility of E. coli, we determined the OM permeability changes in E. coli ATCC 25922, EΔhns and EΔhns/phns using 1-N-phenylnaphthylamine (NPN) as a hydrophobic fluorescent probe (Figure 4A). The results demonstrated that strain EΔhns exhibited dramatically stronger NPN uptakes than that of E. coli ATCC 25922, implying that the absence of hns led to the elevated OM permeability of E. coli. In parallel, compared with strain EΔhns, the fluorescence values of complementary strain EΔhns/phns decreased significantly, although it still showed higher than E. coli ATCC 25922, which was in agreement with the expression levels of hns gene in the complementary strain. Soon afterwards, we further determined the permeability of inner membrane (IM) of the aforementioned bacteria using propidium iodide (PI) as a fluorescent probe (21) and no evident changes were observed (Figure 4B). In summary, hns deletion increases OM permeability of E. coli, however has little effect on IM.

Figure 4. Effects of hns deleted from E. coli ATCC 25922 on bacterial membrane permeability, membrane potential (Δψ), efflux pumps activity and cellular respiration. (A) The permeabilities of outer membrane are evaluated by the fluorescence intensity with 1-N-phenylnaphthalamine. (B) The permeabilities of inner membrane are evaluated by the fluorescence intensity with propidium iodide. (C) The dissipations for Δψ are analyzed by the fluorescence intensity with 3,3′-Dipropylthiadicarbocyanine Iodide. (D) Membrane proton gradient is assessed by fluorescence dye BCECF-AM. (E) Efflux pumps activity is assessed by fluorescence dye ethidium bromide. (F) The changes of NAD+/NADH ratio are measured using an NAD+/NADH Assay Kit. E. coli ATCC 25922 is the control. The strain EΔhns is the derivative of E. coli ATCC 25922 that lacks hns and the strain EΔhns/phns is the derivative of EΔhns that carries the recombined plasmid pBAD::hns. Strains EΔhns and EΔhns/phns are induced by 0.2% L-arabinose. All experiments are performed with biological replicates and present as the mean ± SD. The significances are determined by unpaired t-test between two groups. ns, No significant difference by Student's t-test; *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001.
To clarify the impact of hns deletion on the morphology of E. coli, morphological changes of strains E. coli ATCC 25922 and EΔhns were examined by SEM. In comparison with E. coli ATCC 25922, the cell surface of strain EΔhns exhibited different degree of deformations and devastation (Figure 5). As shown in Figure 5, long rod-shaped bacteria increased significantly in strain EΔhns, speculating that it might be related to the slow growth caused by the deletion of the hns gene (Figures 5C, D). In addition, some cells of strain EΔhns displayed observable membrane damage, which may be one of the reasons for the increased uptake of aminoglycosides by strain EΔhns. Recently, Lv et al. proved that hypoionic shock-induced cell membrane damage could dramatically increase the bacterial uptake of aminoglycosides, which enhanced bactericidal action of the antibiotics (25).
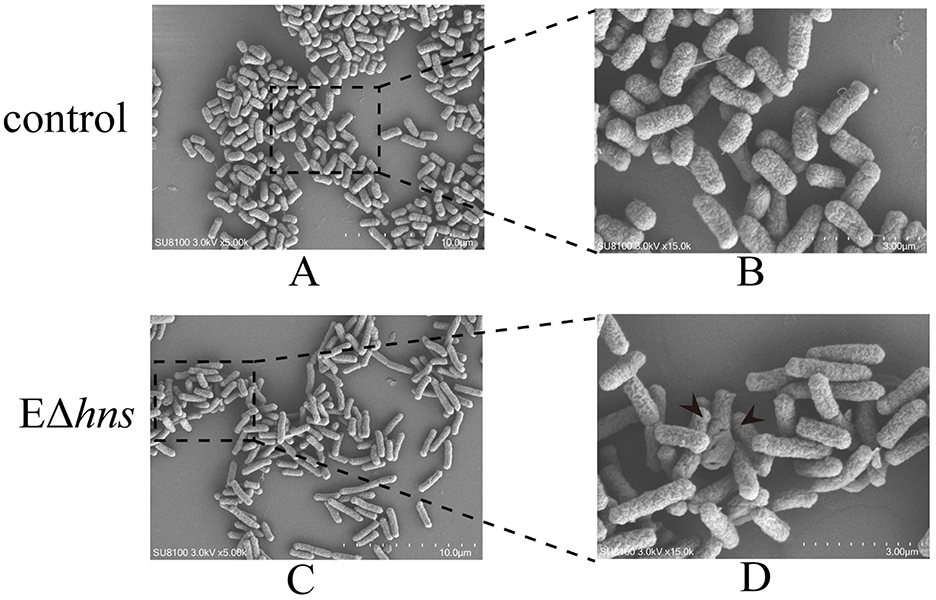
Figure 5. Morphological changes of the strains E. coli ATCC 25922 and EΔhns. (A, B) Bacterial morphology of E. coli ATCC 25922; (C, D) bacterial morphology of EΔhns. E. coli ATCC 25922 is the control. The strain EΔhns is the derivative of E. coli ATCC 25922 that lacks hns and is induced by 0.2% L-arabinose.
Earlier studies have documented that the SOS response is an extremely important molecular instrument of bacteria which allows it to deal with diverse DNA damages (26), but at the same time, activation of the SOS response accelerates the synthesis of the SulA protein, which can arrest cell division (27). In the present study, strain EΔhns not only grew slowly, but also showed cell membrane damage. Therefore, we speculate that the deletion hns gene in E. coli ATCC 25922 leads to bacterial membrane damage, thus activating the SOS response, which in turn hinders bacterial growth.
It is well-known that aminoglycosides do not require energy to cross the OM of bacteria, where OM damage and increased expression of porin-related genes can accelerate absorption, while PMF is required to provide energy when they pass through the IM (28–30). Bacterial PMF, an energetic pathway located on the bacterial membrane, consists of Δψ and the transmembrane proton gradient (ΔpH).
To verify whether PMF altered among strains E. coli ATCC 25922, EΔhns and EΔhns/phns, we first assessed the dissipation of Δψ by the fluorescent probe 3,3′-Dipropylthiadicarbocyanine Iodide [DiSC3 (5)]. When Δψ dissipates, the fluorescent probe DiSC3 (5) is released into the buffer solution, resulting in a significant increase in fluorescence intensity. Our findings showed that compared with E. coli ATCC 25922, the fluorescence intensity of strain EΔhns was decreased dramatically (P < 0.0001) (Figure 4C), indicating that the dissipation of Δψ in strain EΔhns was sharply repressed. Further, the fluorescence intensity of complemented strain EΔhns/phns reverted to the levels of E. coli ATCC 25922. Similarly, ΔpH was also determined by the fluorescent probe BCECF-AM (20 μM, Beyotime, Shanghai, China) and the results demonstrated that no observable changes were found in the tested bacteria (Figure 4D). These results imply that the elevated PMF of strain EΔhns is mainly due to the inhibition of Δψ dissipations, which enhances the uptake of aminoglycosides.
Given that the intracellular accumulation of antibiotics is determined by both influx and efflux (31), we also determined efflux pumps activity changes in E. coli ATCC 25922, EΔhns and EΔhns/phns using ethidium bromide as a hydrophobic fluorescent probe (Figure 4E). Compared with E. coli ATCC 25922 and complementary strain EΔhns/phns, the fluorescence intensity of strain EΔhns exhibited higher (P < 0.0001), suggesting that its efflux activity was significantly inhibited.
To gain a deeper understanding of H-NS regulation mechanism on the susceptibility of E. coli, we further performed the transcriptomic analysis. In comparison with the control E. coli ATCC 25922, Gene Ontology (GO) annotation analysis (Figure 6A) showed that DGEs of strain EΔhns were mainly correlated with adhesion, stress response and chemotaxis of biological processes, lyase activity of molecular function, and pili of cellular component. Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis (Figure 6B) revealed that DEGs in strain EΔhns were enriched significantly in energy metabolisms.
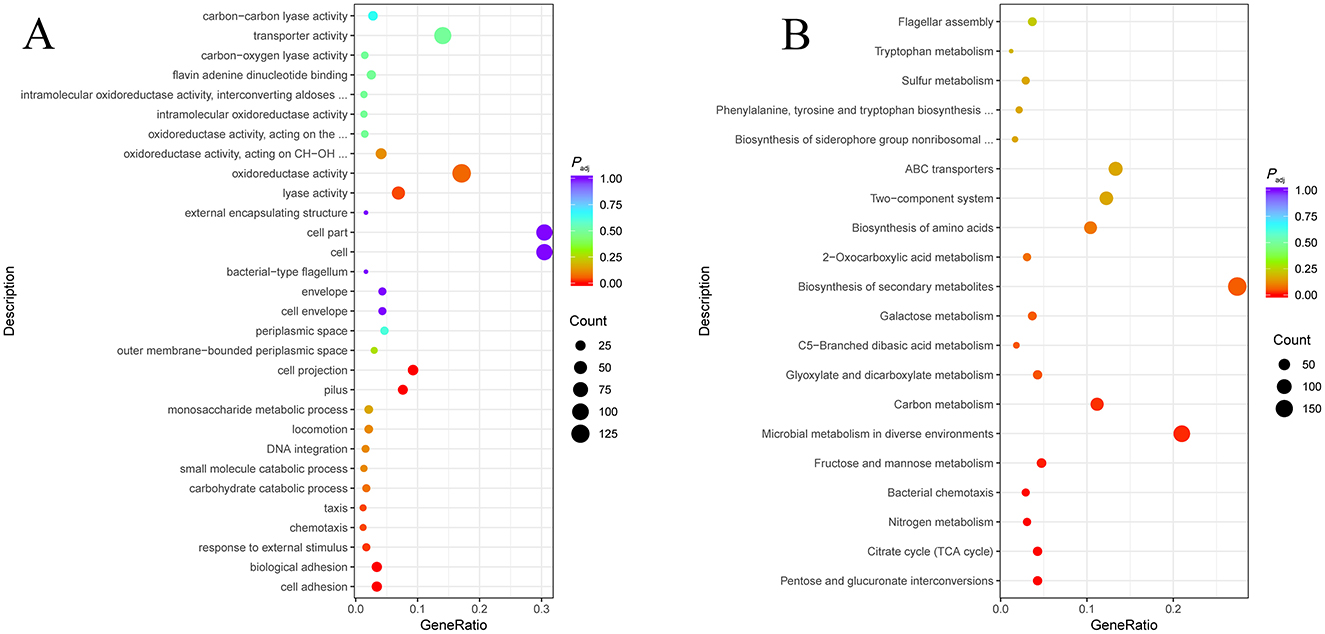
Figure 6. Transcriptomic analysis of strain EΔhns against the reference genome of E. coli ATCC 25922. (A) Gene Ontology (GO) function classification of differentially expressed genes (DEGs). (B) Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis of DEGs. Padj < 0.05 is statistically significant. The x axis and y axis indicate the name of the pathway and the Gene ratio, respectively. The Gene ratio is the ratio of the number of differentially expressed genes enriched into a particular pathway or functional class to the number of all annotated genes in that pathway or functional class. The size of the dot indicates the number of differentially expressed genes in this pathway. The colors of the points correspond to different Padj ranges. The strain EΔhns is the derivative of E. coli ATCC 25922 that lacks hns and is induced by 0.2% L-arabinose.
Specifically, the genes with significantly increased expression were involved in glycolysis and electron transport chain of respiratory process (Figure 6) in strain EΔhns. As shown in Figure 7A, the expression levels of glycolytic-related genes presented distinctly elevated. In parallel, the genes involved in respiratory chain, such as NADH-Q oxidoreductase-related gene ndh and cytochrome oxidase-related genes appA, appB, appC, cydA, and cydB, were all obviously up-regulated (Figure 7B). Previous studies have verified that an increase in glycolytic metabolites can facilitate the conversion of redox energy to PMF, thereby promoting aminoglycosides uptake (28, 32, 33). Accordingly, to identify whether the respiratory chain is activated, we determined the intracellular NAD+/NADH ratio in strains EΔhns, EΔhns/phns and E. coli ATCC 25922 using an NAD+/NADH Assay Kit (WST-8, Beyotime, Shanghai, China). As expected, the NAD+/NADH ratio of strain EΔhns was remarkably higher than that of E. coli ATCC 25922 (P < 0.0001) (Figure 4F), suggesting that the deletion hns gene in E. coli ATCC 25922 can stimulate glycolysis and promote the conversion of redox energy into PMF, thus facilitating the uptake of aminoglycosides.
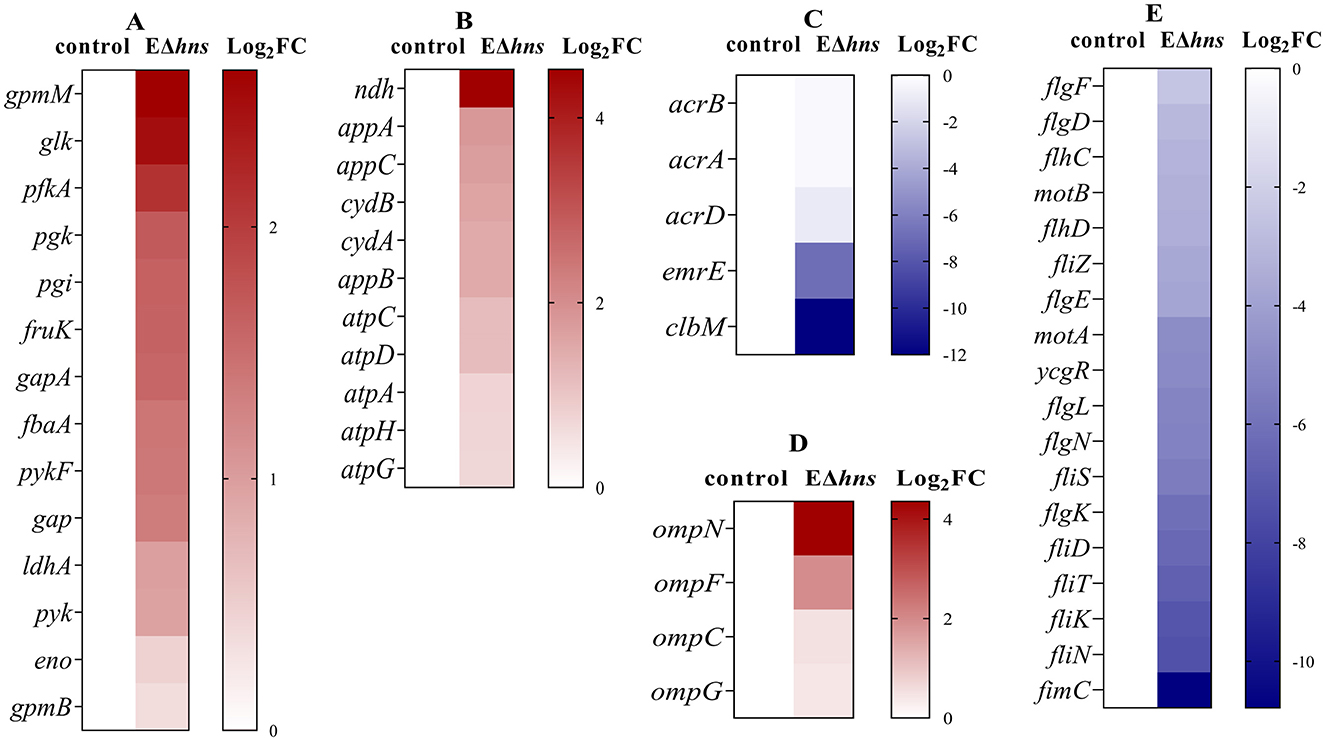
Figure 7. Differentially expressed genes (DEGs) obtained from E. coli ATCC 25922 and EΔhns. E. coli ATCC 25922 is the control. The strain EΔhns is the derivative of E. coli ATCC 25922 that lacks hns. (A) DEGs involved in the glycolysis. (B) DEGs involved in the electron transport chain. (C) DEGs involved in efflux systems. (D) DEGs involved in porins. (E) DEGs involved in flagellar systems. Log2FC means log2 (Fold Change), blue for decreased expression, brown for increased expression. The strain EΔhns is the derivative of E. coli ATCC 25922 that lacks hns and is induced by 0.2% L-arabinose.
Further analysis found that many efflux-related genes (Figure 7C) presented down-regulated, such as clbM (34), emrE (35, 36), acrD (37), and acrAB (38), which were helpful to reduce drugs efflux. To confirm whether the expression levels of above genes were altered in strain EΔhns, we examined their mRNA relative expression levels using RT-qPCR (Figure 8A). The results demonstrated that the expression levels in strain EΔhns were evidently lower than those in E. coli ATCC 25922, approximately decreased by 99.0% (clbM), 68.3% (emrE), 49.7% (acrB), and 39.2% (acrA), respectively. Thus, reduced expression levels of genes associated with the efflux system can inhibit efflux activity, thereby helping to slow drugs efflux.
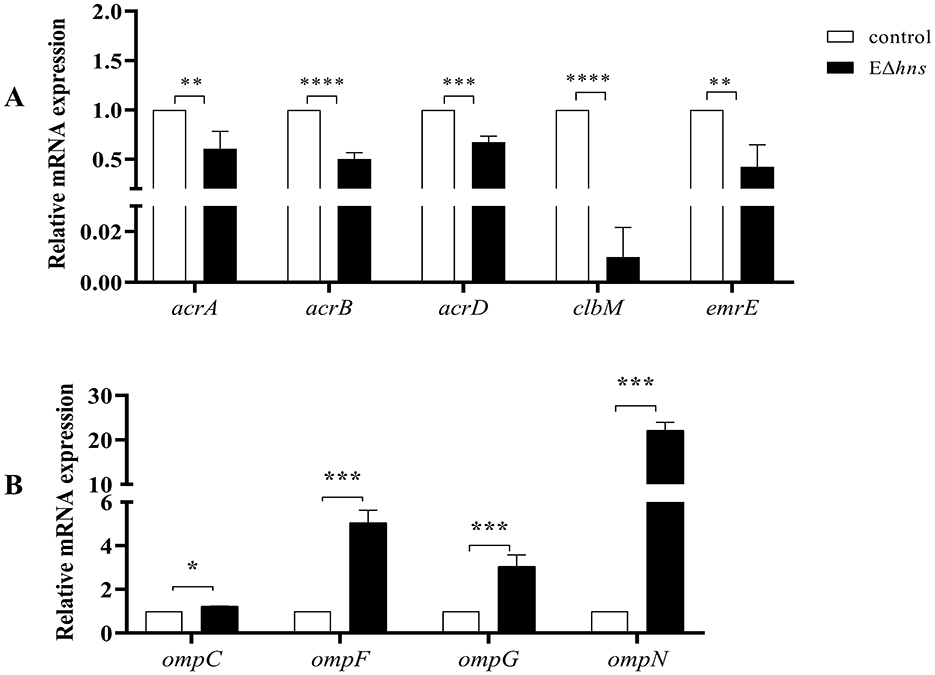
Figure 8. The mRNA expression levels of strains E. coli ATCC 25922, EΔhns and EΔhns/phns. (A) Efflux-related genes. (B) Porin-related genes. E. coli ATCC 25922 is the control. The strain EΔhns is the derivative of E. coli ATCC 25922 that lacks hns and is induced by 0.2 % L-arabinose. ns, No significant difference by Student's t-test. *; P < 0.05, **; P < 0.01; ***, P < 0.001; ****, P < 0.0001.
Due to porin-related genes ompN, ompF, ompC, and ompG (Figure 7D) showed obvious upregulations, we also examined their mRNA expression levels in strains E. coli ATCC 25922 and EΔhns using RT-qPCR. We found that the expression levels of these genes were markedly higher in strain EΔhns than that in E. coli ATCC 25922 (Figure 8B), with the highest levels of ompN (i.e., 22.21-fold higher), followed by ompF (5.06-fold higher) and ompG (3.06-fold higher), whereas the levels of ompC exhibited only a marginal increase. Porins OmpC and OmpF passively diffuse small hydrophilic molecules (≤ 500 Da) into the periplasm (39), and their expressions are mutually exclusive, i.e., when ompF is on, ompC is off, and vice versa (40). OmpG, which belongs to the subclass of porins, harbors a larger channel than OmpF and OmpC, and also allows for translocation (41). OmpN is one of the minor porins, although its translocation function has not been reported (42). Therefore, it is conceivable that the elevated expression levels of porin-related genes will further destabilize the OM barrier and contribute to the increase of OM permeability of strain EΔhns.
Intriguingly, there were many down-regulated DEGs in the flagellar system in strain EΔhns (Figure 7E), compared to the reference strain E. coli ATCC 25922. Further, it can also be seen from Figure 5 that the flagella of E. coli ATCC 25922 are clearly visible, while that of strain EΔhns is almost invisible. Many studies have revealed that flagellum is a locomotive organelle and can affect bacterial adhesion, invasion and biofilm formation (37, 43). To further confirm whether the motility of strain EΔhns has changed, we fulfilled the swimming motility assay of strains E. coli ATCC 25922, EΔhns, EΔhns/phns, and E/phns using LB plates with 0.3% agar according to the method of previous studies (44, 45). The results demonstrated that strain EΔhns had the shortest swimming distance (≈ 9.3 mm), followed by EΔhns/phns (≈ 12.8 mm) and E. coli ATCC 25922 (≈ 13.8 mm), while E/phns had the longest distance (≈ 14.7 mm), suggesting that H-NS modulates the motility of E. coli by positively governing the expression of flagellate-related genes.
Taken together, our findings highlight the deletion of hns in E. coli can strengthen antibacterial activity of aminoglycosides by increasing intracellular drug concentrations (Figure 9). On the one hand, the inhibition of efflux pump activity in strain EΔhns can reduce the efflux of aminoglycosides. On the other hand, in addition to elevated OM permeability can promote drugs uptake, the increase of PMF induced by glycolysis and Δψ can further accelerate the uptake of aminoglycosides.
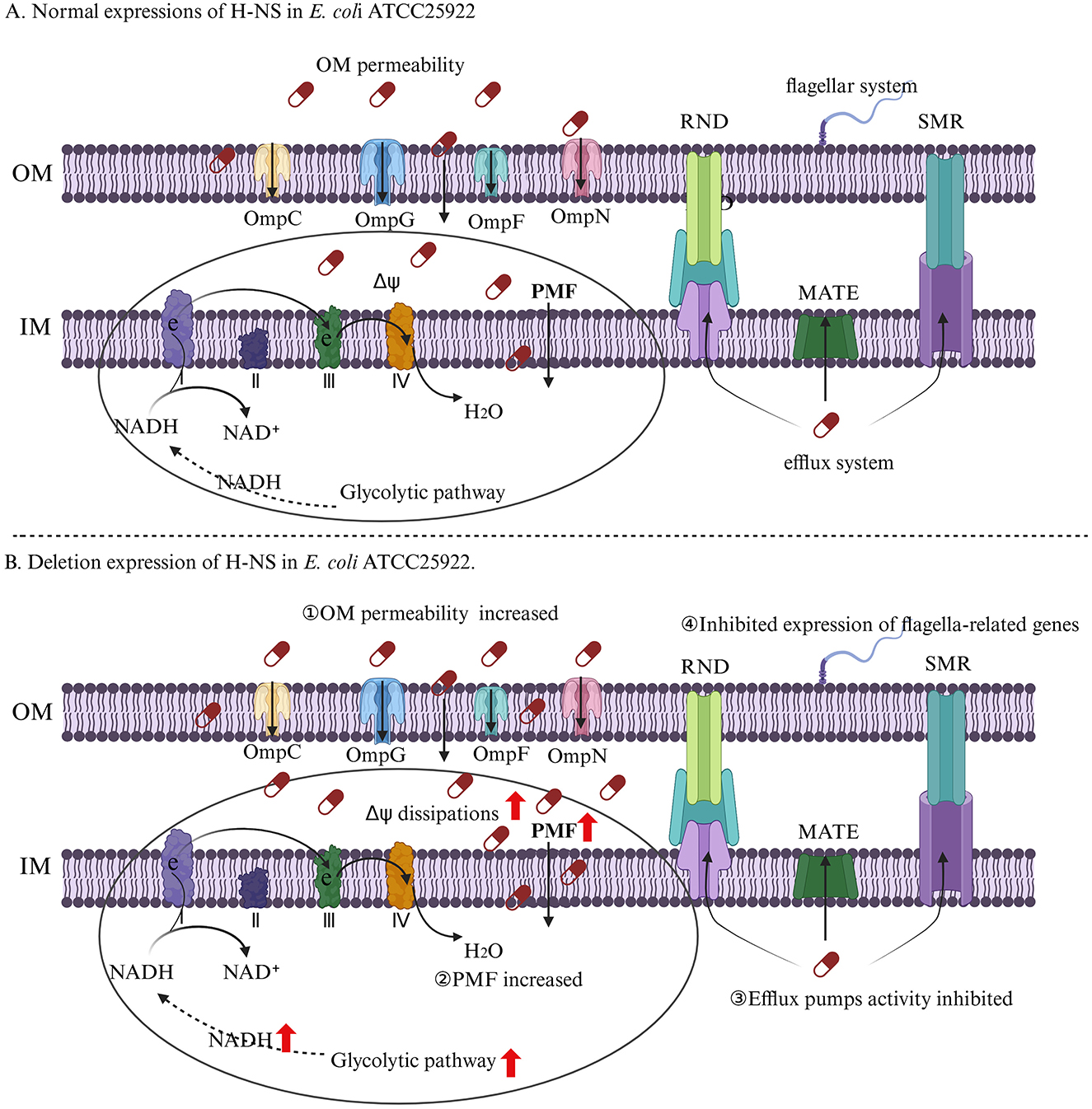
Figure 9. The regulation mechanism of H-NS on aminoglycosides resistance of E. coli. (A) E. coli ATCC 25922. (B) E. coli ATCC 25922 that lacks hns. I, II, III, and IV represent complexes I, complexes II, complexes III, and complexes IV, which are respiratory chain complex and carries out electron transport. The OM permeability of strain EΔhns is improved by membrane damages and increased expression levels of porin-related genes, thus increasing the uptake of aminoglycosides. The activation of glycolysis and electron transport chain and the inhibition of Δψ dissipations in strain EΔhns result in an increase of PMF, which can further facilitate the uptake of aminoglycosides. The efflux pumps activity of strain EΔhns is inhibited that can reduce drugs efflux. The flagella of strain EΔhns is apparently reduced, which can inhibit the formation of bacterial biofilms, thereby enhancing bacterial susceptibility to aminoglycosides.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/supplementary material.
All experiments were cared for strictly in accordance with the guidelines set by the State Council of the People's Republic of China and the Henan Science and Technology Department (protocol number SCXK 2019-0002). Also, informed consent was obtained from all of the owners. The study was conducted in accordance with the local legislation and institutional requirements.
QC: Data curation, Investigation, Validation, Writing – original draft, Writing – review & editing. YL: Conceptualization, Data curation, Methodology, Writing – original draft, Writing – review & editing. YD: Formal analysis, Validation, Writing – original draft. JC: Methodology, Software, Writing – original draft. KH: Formal analysis, Software, Writing – original draft. XM: Methodology, Supervision, Writing – review & editing. JZ: Methodology, Supervision, Writing – review & editing. YZ: Methodology, Supervision, Writing – review & editing. LY: Funding acquisition, Project administration, Writing – review & editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by grants from the National Natural Science Foundation of China (grant number 32373074), the Program for Innovative Research Team (in Science and Technology) in the University of Henan Province (grant number 23IRSTHN021), and the Key Projects of the Natural Science Foundation of Henan Province (grant number 232300421111).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declare that no Gen AI was used in the creation of this manuscript.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
1. Dorman CJ. H-NS: a universal regulator for a dynamic genome. Nat Rev Microbiol. (2004) 2:391–400. doi: 10.1038/nrmicro883
2. Grainger DC. Structure and function of bacterial H-NS protein. Biochem Soc Trans. (2016) 44:1561–9. doi: 10.1042/BST20160190
3. Forrest D, Warman EA, Erkelens AM, Dame RT, Grainger DC. Xenogeneic silencing strategies in bacteria are dictated by RNA polymerase promiscuity. Nat Commun. (2022) 13:1149. doi: 10.1038/s41467-022-28747-1
4. Picker MA, Karney MMA, Gerson TM, Karabachev AD, Duhart JC, McKenna JA, et al. Localized modulation of DNA supercoiling, triggered by the Shigella anti-silencer VirB, is sufficient to relieve H-NS-mediated silencing. Nucleic Acids Res. (2023) 51:3679–95. doi: 10.1093/nar/gkad088
5. Choi J, Schmukler M, Groisman EA. Degradation of gene silencer is essential for expression of foreign genes and bacterial colonization of the mammalian gut. Proc Natl Acad Sci U S A. (2022) 119:e2210239119. doi: 10.1073/pnas.2210239119
6. Hüttener M, Paytubi S, Juárez A. Success in incorporating horizontally transferred genes: the H-NS protein. Trends Microbiol. (2015) 23:67–9. doi: 10.1016/j.tim.2014.12.009
7. Lamberte LE, Baniulyte G, Singh SS, Stringer AM, Bonocora RP, Stracy M, et al. Horizontally acquired AT-rich genes in Escherichia coli cause toxicity by sequestering RNA polymerase. Nat Microbiol. (2017) 2:16249. doi: 10.1038/nmicrobiol.2016.249
8. Cooper C, Legood S, Wheat RL, Forrest D, Sharma P, Haycocks JRJ, et al. H-NS is a bacterial transposon capture protein. Nat Commun. (2024) 15:7137. doi: 10.1038/s41467-024-51407-5
9. Shahul Hameed UF, Liao C, Radhakrishnan AK, Huser F, Aljedani SS, Zhao X, et al. H-NS uses an autoinhibitory conformational switch for environment-controlled gene silencing. Nucleic Acids Res. (2019) 47:2666–80. doi: 10.1093/nar/gky1299
10. Figueroa-Bossi N, Sánchez-Romero MA, Kerboriou P, Naquin D, Mendes C, Bouloc P, et al. Pervasive transcription enhances the accessibility of H-NS-silenced promoters and generates bistability in Salmonella virulence gene expression. Proc Natl Acad Sci U S A. (2022) 119:e2203011119. doi: 10.1073/pnas.2203011119
11. Pratama A, Ishii E, Kodama T, Iida T, Matsuda S. The xenogeneic silencer histone-like nucleoid-structuring protein mediates the temperature and salinity- dependent regulation of the type III secretion system 2 in Vibrio parahaemolyticus. J Bacteriol. (2023) 205:e0026622. doi: 10.1128/jb.00266-22
12. Cho H, Misra R. Mutational activation of antibiotic-resistant mechanisms in the absence of major drug efflux systems of Escherichia coli. J Bacteriol. (2021) 203:e0010921. doi: 10.1128/JB.00109-21
13. Nishino K, Hayashi-Nishino M, Yamaguchi A. H-NS modulates multidrug resistance of Salmonella enterica serovar Typhimurium by repressing multidrug efflux genes acrEF. Antimicrob Agents Chemother. (2009) 53:3541–3. doi: 10.1128/AAC.00371-09
14. Deveson Lucas D, Crane B, Wright A, Han M-L, Moffatt J, Bulach D, et al. Emergence of high level colistin resistance in an Acinetobacter baumannii clinical isolate mediated by inactivation of the global regulator H-NS. Antimicrob Agents Chemother. (2018) 62:e02442. doi: 10.1128/AAC.02442-17
15. Rodgers D, Le C, Pimentel C, Tuttobene MR, Subils T, Escalante J, et al. Histone-like nucleoid-structuring protein (H-NS) regulatory role in antibiotic resistance in Acinetobacter baumannii. Sci Rep. (2021) 11:18414. doi: 10.1038/s41598-021-98101-w
16. Darby EM, Trampari E, Siasat P, Gaya MS, Alav I, Webber MA, et al. Molecular mechanisms of antibiotic resistance revisited. Nat Rev Microbiol. (2023) 21:280–95. doi: 10.1038/s41579-022-00820-y
17. Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A. (2000) 97:6640–5. doi: 10.1073/pnas.120163297
18. Jia YT, Hu HH, Zhai YJ, Zhao B, Sun HR, Hu GZ, et al. CpxR negatively regulates IncFII-replicon plasmid pEC011 conjugation by directly binding to multi-promoter regions. Res Vet Sci. (2022) 150:98–106. doi: 10.1016/j.rvsc.2022.05.016
19. Clinical and Laboratory Standards Institute (CLSI). Performance Standards for Antimicrobial Susceptibility Testing 30th ed. Wayne, PA, USA: CLSI supplement M100. (2021).
20. Zhang MK, Zhang MY, Liu SB, Yang YY, Zhai YJ, He DD, et al. Double deletion of cpxR and tolC significantly increases the susceptibility of Salmonella enterica serovar Typhimurium to colistin. J Antimicrob Chemother. (2021) 76:3168–74. doi: 10.1093/jac/dkab332
21. Liu Y, Jia Y, Yang K, Tong Z, Shi J, Li R, et al. Melatonin overcomes MCR-mediated colistin resistance in Gram-negative pathogens. Theranostics. (2020) 10:10697–711. doi: 10.7150/thno.45951
22. Hamamoto H, Urai M, Ishii K, Yasukawa J, Paudel A, Murai M, et al. Lysocin E is a new antibiotic that targets menaquinone in the bacterial membrane. Nat Chem Biol. (2015) 11:127–33. doi: 10.1038/nchembio.1710
23. Jia YT, Hu HH, Zhai YJ, Zhao B, He K, Pan YS, et al. Molecular mechanism of regulation by H-NS on IncFII plasmid transmission of multi-drug resistant chicken Escherichia coli. Sci Agric Sinica. (2022) 55:3675–84. doi: 10.3864/j.issn.0578-1752.2022.18.016
24. Huang F, Fitchett N, Razo-Gutierrez C, Le C, Martinez J, Ra G, et al. The H-NS regulator plays a role in the stress induced by carbapenemase expression in Acinetobacter baumannii. mSphere. (2020) 5:e00793. doi: 10.1128/msphere.00793-20
25. Lv B, Zeng Y, Zhang H, Li Z, Xu Z, Wang Y, et al. Mechanosensitive channels mediate hypoionic shock-induced aminoglycoside potentiation against bacterial persisters by enhancing antibiotic uptake. Antimicrob Agents Chemother. (2022) 66:e0112521. doi: 10.1128/aac.01125-21
26. Baharoglu Z, Mazel D. SOS, the formidable strategy of bacteria against aggressions. FEMS Microbiol Rev. (2014) 38:1126–45. doi: 10.1111/1574-6976.12077
27. Vedyaykin A, Rumyantseva N, Khodorkovskii M, Vishnyakov I. SulA is able to block cell division in Escherichia coli by a mechanism different from sequestration. Biochem Biophys Res Commun. (2020) 525:948–53. doi: 10.1016/j.bbrc.2020.03.012
28. Lang M, Carvalho A, Baharoglu Z, Mazel D. Aminoglycoside uptake, stress, and potentiation in Gram-negative bacteria: new therapies with old molecules. Microbiol Mol Biol Rev. (2023) 87:e0003622. doi: 10.1128/mmbr.00036-22
29. Yang B, Tong Z, Shi J, Wang Z, Liu Y. Bacterial proton motive force as an unprecedented target to control antimicrobial resistance. Med Res Rev. (2023) 43:1068–90. doi: 10.1002/med.21946
30. Lv B, Huang X, Lijia C, Ma Y, Bian M, Li Z, et al. Heat shock potentiates aminoglycosides against gram-negative bacteria by enhancing antibiotic uptake, protein aggregation, and ROS. Proc Natl Acad Sci U S A. (2023) 120:e2217254120. doi: 10.1073/pnas.2217254120
31. Wan Y, Wang M, Chan EWC, Chen S. Membrane transporters of the major facilitator superfamily are essential for long-term maintenance of phenotypic tolerance to multiple antibiotics in E. coli Microbiol Spectr. (2021) 9:e0184621. doi: 10.1128/Spectrum.01846-21
32. Allison KR, Brynildsen MP, Collins JJ. Metabolite-enabled eradication of bacterial persisters by aminoglycosides. Nature. (2011) 473:216–20. doi: 10.1038/nature10069
33. Papa S, Capitanio G, Papa F. The mechanism of coupling between oxido-reduction and proton translocation in respiratory chain enzymes. Biol Rev Camb Philos Soc. (2018) 93:322–49. doi: 10.1111/brv.12347
34. Mousa JJ, Newsome RC, Yang Y, Jobin C, Bruner SD. ClbM is a versatile, cation-promiscuous MATE transporter found in the colibactin biosynthetic gene cluster. Biochem Biophys Res Commun. (2017) 482:1233–9. doi: 10.1016/j.bbrc.2016.12.018
35. Robinson AE, Thomas NE, Morrison EA, Balthazor BM, Henzler-Wildman KA. New free-exchange model of EmrE transport. Proc Natl Acad Sci U S A. (2017) 114:E10083–91. doi: 10.1073/pnas.1708671114
36. Shcherbakov AA, Spreacker PJ, Dregni AJ, Henzler-Wildman KA, Hong M. High-pH structure of EmrE reveals the mechanism of proton-coupled substrate transport. Nat Commun. (2022) 13:991. doi: 10.1038/s41467-022-28556-6
37. Prajapati JD, Kleinekathöfer U, Winterhalter M. How to enter a bacterium: bacterial porins and the permeation of antibiotics. Chem Rev. (2021) 121:5158–92. doi: 10.1021/acs.chemrev.0c01213
38. Jang S. AcrAB-TolC, a major efflux pump in Gram negative bacteria: toward understanding its operation mechanism. BMB Rep. (2023) 56:326–34. doi: 10.5483/BMBRep.2023-0070
39. Vergalli J, Bodrenko IV, Masi M, Moynié L, Acosta-Gutiérrez S, Naismith JH, et al. Porins and small-molecule translocation across the outer membrane of Gram-negative bacteria. Nat Rev Microbiol. (2020) 18:164–76. doi: 10.1038/s41579-019-0294-2
40. Sleator RD, Hill C. Bacterial osmoadaptation: the role of osmolytes in bacterial stress and virulence. FEMS Microbiol Ecol. (2002) 26:49–71. doi: 10.1111/j.1574-6976.2002.tb00598.x
41. Schmitt C, Bafna JA, Schmid B, Klingl S, Baier S, Hemmis B, et al. Manipulation of charge distribution in the arginine and glutamate clusters of the OmpG pore alters sugar specificity and ion selectivity. Biochim Biophys Acta Biomembr. (2019) 1861:183021. doi: 10.1016/j.bbamem.2019.07.009
42. Zhang S, Shao Y, Zhao X, Li C, Guo M, Lv Z, et al. Indole contributes to tetracycline resistance via the outer membrane protein OmpN in Vibrio splendidus. World J Microbiol Biotechnol. (2020) 36:36. doi: 10.1007/s11274-020-02813-6
43. Duan Q, Zhou M, Zhu L, Zhu G. Flagella and bacterial pathogenicity. J Basic Microbiol. (2013) 53:1–8. doi: 10.1002/jobm.201100335
44. He K, Lin J, Liang Y, Cui J, Chen Q, Dong Y, et al. Coexistence of a nonresistance-conferring IncI1 plasmid favors persistence of the blaCTX − M-bearing IncFII plasmid in Escherichia coli. Microbiol Spectr. (2024) 12:e0424023. doi: 10.1128/spectrum.04240-23
Keywords: H-NS, aminoglycosides, uptake, efflux, glycolysis, proton motive force
Citation: Chen Q, Liang Y, Dong Y, Cui J, He K, Ma X, Zhao J, Zhai Y and Yuan L (2025) H-NS controls the susceptibility of Escherichia coli to aminoglycosides by interfering its uptake and efflux. Front. Vet. Sci. 12:1534498. doi: 10.3389/fvets.2025.1534498
Received: 26 November 2024; Accepted: 16 January 2025;
Published: 06 February 2025.
Edited by:
Arturo Anadón, Complutense University of Madrid, SpainReviewed by:
Weiwei Zhang, Ningbo University, ChinaCopyright © 2025 Chen, Liang, Dong, Cui, He, Ma, Zhao, Zhai and Yuan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Li Yuan, bGl5dWFuaG4wM0BoZW5hdS5lZHUuY24=; Yajun Zhai, enlqOTA1MThAMTI2LmNvbQ==
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.