- 1State Key Laboratory of Mariculture Breeding, College of Ocean and Earth Sciences, Xiamen University, Xiamen, China
- 2Fujian Key Laboratory of Genetics and Breeding of Marine Organisms, College of Ocean and Earth Sciences, Xiamen University, Xiamen, China
- 3Key Laboratory of Cultivation and High-Value Utilization of Marine Organisms, Fisheries Research Institute of Fujian, Xiamen, China
- 4Biotechnology Institute, Fujian Academy of Agricultural Sciences, Fuzhou, Fujian, China
Despite the high cost of genotyping and the limited economic return per individual that restricts the use of genome selection technology in aquatic species breeding, integrating multiple traits in a selection index can reduce costs and present a promising direction for fish breeding. The aim of this study was to evaluate the feasibility and performance of breeding for dual resistance traits against C. irritans and V. alginolyticus in large yellow croaker, using a selection index approach. A candidate stock population from the C. irritans resistance strain (GS3) of the F2 generation (n = 560) was genotyped using a 55 K SNP array. A total of 50 broodstock with the highest genomic estimated breeding value (GEBV) ranking for the C. irritans resistance trait were selected to produce the F3 generation, while 50 broodstock with the highest selection index were selected to breed the double trait strain (GS9). In parallel, two populations without selective breeding population (C1and C2) were used as the control group to produce the offspring. The genetic improvement effect of all the breeding lines (GS3, GS9, C1, and C2) was assessed through artificial challenge experiments with C. irritans and V. alginolyticus, respectively. The GWAS result indicated that resistance to V. alginolyticus was a polygenic trait and the heritability of resistance to V. alginolyticus was estimated to be approximately 0.1. The survival rates of GS9 and GS3 were 43.74 and 86.81%, respectively, after 120 h of challenge with C. irritans infection, whereas those of C1 and C2 remained at 0%. Furthermore, the survival rates of GS9 and GS3 were 27.03 and 15.48%, respectively, after 48 h of V. alginolyticus infection, while the survival rates of C1 and C2 were 12.67 and 14.17%, respectively. These results indicate that genetic improvement of multiple traits in large yellow croaker using genomic selection technology is indeed feasible. Moreover, this study provides a valuable reference for advancing multi-trait breeding applications in aquatic species.
1 Background
The selection index has been widely used in animal breeding since it was introduced by Hazel (1). Compared to single-trait selection, the selection index allows each trait to be weighted according to its economic value and then allows simultaneous genetic improvement of each trait to maximize the genetic effect (1). According to national conditions, a total of 13 vital traits such as milk yield, protein yield, fertility, yield and health are included in the comprehensive index for genetic improvement of dairy cattle varieties in many countries (2). Over several decades, the average milk yield has nearly doubled from 6,619 to 12,662 kg in US Holstein dairy cattle breeding, and genetic gains for milk, fat, and protein yields were 109, 6.0 and 4.1 kg per year after the implementation of genomic selection (3). Furthermore, an analysis based on the genetic progress made by dairy cattle on the selection index over the last decade to predict genetic progress over the next 50 years showed that milk yield, fat and protein could also increase by 34, 53, and 41%, respectively (2). In aquaculture, O’Flynn et al. conducted two generations of genetic improvement on Atlantic salmon, focusing on growth, seedling survival rate, and harvest weight using the selection index. The results showed an average weight gain of 0.88 kg, an increase in body length by an average of 4.48 cm, and a 9% increase in survival rate (4). Similarly, Friars et al. also demonstrated that higher genetic gains can be achieved on Atlantic salmon based on selection index compared to mass selection (5). In the blue tilapia (Oreochromis aureus) selective breeding program, the results indicate that after four successive generations of index selection, the selection response for growth traits in each generation reached 14%. Additionally, the survival rate of tilapia in soil pond and cold-water tolerance experiments increased by 8.4 and 1.2%, respectively (6).
As the most productive marine fish in China, the large yellow croaker (Larimichthys crocea) achieved a production level of 280,997 tonnes in 2024 (7). Despite this success, persistent diseases are still the primary obstacle in healthy culture of the large yellow croaker. Among these, parasitic diseases like Cryptocarya irritans (C. irritans) have significant implications on the survival rate of large yellow croaker. Currently, the application of genomic selection for resistance against C. irritans in large yellow croaker has shown promising results. The infection challenge experiment results indicated that the survival rate of selective lines has been significantly enhanced compared to the control group (8, 9). However, the single trait genetic improvement of resistance against C. irritans in large yellow croaker does not sufficiently address the complex environmental challenges in the culture region, such as bacterial diseases.
Vibrio alginolyticus (V. alginolyticus), a member of the Vibrionaceae family and the genus Vibrio, is classified as a halophilic thermotolerant, facultatively anaerobic vibrio (10). Infections of V. alginolyticus in large yellow croakers and the stimulation of C. irritans parasitism often occur concurrently during the summer. The global distribution of V. alginolyticus includes seawater and estuaries, with marine vibrio being its dominant species (11).
A 2013 study utilized data on the ulcerative condition of large yellow croaker from sea nets in Xiangshan County, Zhejiang Province, to perform a diagnostic tracing analysis of pathogenous vibrios. The findings revealed that the infection rate of V. alginolyticus was significantly higher than that of V. harveyi and V. parahaemolyticus (12). V. alginolyticus infects fish through attachment to compromised fish skin and oral mucosa. Infected tissues often exhibit symptoms such as congestion, inflammation, and skin erosion. Currently, there are reports of vibrio-associated diseases in the fish species such as Larimichthys crocea (13), Epinephelus coioides (14) and Litopenaeus Vannamei (15). Another study analyzing 116 randomly sampled seafood products from the aquatic product market in Beijing found a positive detection rate of V. alginolyticus at a staggering 82% (16). At present, research on controlling the disease caused by V. alginolyticus has led to the development of inactivated vaccines for key farmed species such as whiteleg shrimp, halibut, and pompano. However, the industry’s wide variety of cultivated species makes it challenging and costly to administer and inject thesevaccines. Therefore, starting with genetic breeding based on the selection index and integrating resistance to V. alginolyticus and C. irritans into genetic improvement target traits presents another effective approach to mitigate losses in large yellow croaker.
Prior to the establishment of a breeding line with resistant toward both C. irritans and V. alginolyticus, it is essential to devise a challenge standard for V. alginolyticus, assess the genetic correlation between those two disease traits and establish a reference population. The objective of this study was to explore the feasibility and practical improvement effect of using a selection index to enhance the resistant to C. irritans and V. alginolyticus traits in large yellow croaker.
2 Methods
2.1 Ethics statement
This study was approved by the Animal Care and Use committee at College of Ocean and Earth Sciences, Xiamen University. The methods were carried out in accordance with approved guidelines.
2.2 Technical route of breeding application
The candidate stock population (n = 560) for genetic improvement of the dual resistance traits based on the selection index originated from the F2 generation of the breeding line designated as GS3 of the large yellow croaker, which was previously subjected to selective breeding for resistance to C. irritans in F1 generation (GS3) in the laboratory. The reference population (n = 474) for resistance to C. irritans in large yellow croaker was constructed based on the F1 population (GS3) in the laboratory earlier. This close genetic relationship between the reference and candidate stock populations enabled a higher accuracy in calculating the GEBV. Another reference population (n = 496) for resistance to V. alginolyticus was established in this experiment, which derived from the unselected breeding population that had no genetic connection to GS3 line. The selection index was calculated based on the genomic estimated breeding value (GEBV) of the two disease traits, where each GEBV was standardized and given a weighting of 0.5. Candidates with the superior comprehensive index ranking were selected from the GS3 line in F2 generation stocks population (n = 560), resulting in the creation of the disease-resistant breeding line titled GS9. In addition, the GS3 breeding line underwent continued genomic selection for resistance to C. irritans and produced the F3 progeny. Simultaneously, two populations (C1 and C2) that had not undergone selective breeding were used to produce offspring as the control group. Finally, the genetic improvement effect of all breeding lines (GS3, GS9, C1, C2) was assessed through artificially challenge experiments with C. irritans and V. alginolyticus by using their offpsring, respectively. For further details on the application of genomic selective breeding program, please refer to Figure 1.
2.3 Built up the standard artificial challenge for Vibrio alginolyticus
Before to conducted the genomic selection breeding of resistance to double traits by using the selection index, three preliminary experiments on large yellow croaker resistance to V. alginolyticus were carried out to determine the artificial infection method, V. alginolyticus strain and concentration, respectively. V. alginolyticus was provided by Fujian Academy of Agricultural Sciences (Deposit No.: H2017041302).
The first experiment was to explore the effects of immersion infection and intraperitoneal injection on resistance to V. alginolyticus in large yellow croaker. For the immersion experiment, 406 fish (body weight (g): 5.2 ± 1.7, body length (cm): 7.10 ± 0.82) were randomly sampled from the marine aquaculture cages and transported to the 8 tons cement pond. After a week of acclimation, fish were divided into three groups and placed into respective tanks, where a seawater level of 200 L maintained during the infection, and infected by immersion for 2 h in a bacterial suspension (1.0 × 105 CFU/mL). The immersion infection experiment lasted for 6 days, and the observation and recordings were made every 2 h. For the intraperitoneal injection experiment, 397 fish (body weight (g): 6.26 ± 1.84, body length (cm): 7.13 ± 0.79) were divided into three groups, anesthetized with tricaine methanesulfonate at a concentration of 80 ppm (MS222; Sigma, St. Louis, MO, USA), and artificially injected with 0.1 mL of V. alginolyticus MTH strain at a concentration of 1.0 × 109 CFU/mL. In additional, 30 fish from control group were injected with 0.1 mL of normal saline solution. The experiment was continuously observed, and the mortality was recorded for 48 h. Survival time and survival status were used as indicators of the antibacterial resistance of large yellow croaker during artificial infection challenges. For the survival status trait, surviving fish were recorded as 1, while dead fish were recorded as 0. If a fish loses its balance after infection and does not recover within 10 s, the corresponding time is recorded as the survival time, and the survival status is indicated as 0 If a fish remains alive and in good condition 48 h after infection, it is considered to have strong disease resistance, and the survival status is recorded as 1. This standard was adopted in subsequent experiments to assess the disease resistance of large yellow croaker.
The second experiment was to identify the strains suitable for large yellow croaker to infect V. alginolyticus artificially. Four strain named CZH, DWH, MTH and SXJH were randomly selected for this experiment. The concentrations of the four strains ranged from 5.3 to 6.2 × 109 CFU/mL, and each strain infected 37 to 42 fish (body weight (g): 9.68 ± 2.86), with an injection dose of 0.1 mL for each fish. The mortality of experiment was continuously observed and recorded for 48 h.
After the infection strain of V. alginolyticus was confirmed, the third experiment was conducted to ascertain the appropriate concentration level of V. alginolyticus infection in large yellow croaker. The initial strain concentration (6.78 × 109 CFU/mL) was diluted 2, 5, 25 and 125 times using normal saline solution (corresponding to 3.4 × 109, 1.3 × 109, 2.7 × 108, 5.4 × 107 CFU/mL, respectively). An average of 22 fish were infected with each diluted concentration (using 0.1 mL injection), and the experiment was continuously observed for 48 h.
2.4 Construction of reference population
After determining the V. alginolyticus strain, challenge method and concentration, 708 fish (body weight (g): 10.75 ± 3.70, body length (cm): 9.02 ± 1.05) were randomly selected from an unselected breeding population and divided into four cylindrical tanks (1 m3). The fish were anesthetized with MS222 and intraperitoneally injected with MTH V. alginolyticus strain (3.16 × 108, 0.1 mL). The experimental water temperature was maintained at 30°C, and mortality was continuously observed and recorded for 48 h that used as the reference population phenotype data of resistance against V. alginolyticus. The fish lost their balance and could not recover within 10 s, which meant that the fish had a weak resistance to V. alginolyticus. On the contrary, 48 h after infection, the fish were still alive and showed good vitality, which indicated that the fish had strong antibacterial properties. Therefore, both survival time and survival state were used as the indicators of resistance performance. Samples (n = 496) were randomly selected from the phenotype reference population for genotyped.
2.5 Genotyping and quality control
Samples (n = 496) were randomly selected from the resistance against V. alginolyticus reference population and 560 (body weight (g): 664.20 ± 159.47, body length (cm): 32.94 ± 3.04) healthy broodstock fish were randomly selected from F2 generation (GS3) strain which was undergoing two rounds of selective breeding for resistance to C. irritans. Meanwhile, 415 broodstock fish (body weight (g): 812.20 ± 122.20, body length (cm):32.94 ± 3.04) were randomly selected from the unselected breeding population as the control group (C2). Fin samples of these reference and broodstock fish were collected and genotyped by using “NingXin-III” 55 K genotype array (17) in MolBreeding Biotechnology Co., LTD (Shijiazhuang, HeBei). The raw genotype data of reference and candidate population were obtained and the quality control of the genotypes was conducted using Plink (version 1.9) software (18) with the criteria of individual call rate > 0.9, minor allele frequency > 0.05, SNP call rate > 0.9. The quality genotype data was used for next analysis. The CMplot (version 4.5.1) package (19) of R software was used to map the probability density distribution of SNP loci in the genome. The genome referenced for this analysis is available at https://www.ncbi.nlm.nih.gov/datasets/genome/GCA_003845795.1/.
2.6 Genetics analysis of resistance to Vibrio alginolyticus
The univariate mixed linear model of GEMMA 0.94.1 software (20) was used to conduct genome-wide association analysis (GWAS) on the survival time and survival status traits of large yellow croaker resistant to V. alginolyticus, respectively. The tank effect was added to the model as a fixed effect in the univariate mixed linear model. The p-value threshold of 0.0001 was taken as the significance association threshold. The 50 kb upstream and downstream regions of significant SNPs were identified as candidate regions, and R software was used to screen related genes in these candidate regions. The results of genome-wide association analysis were plotted by qqman package (21) of R software. The heritability of survival time trait of large yellow croaker for resistance to V. alginolyticus was calculated by using HIBLUP software (22). In additional, the genetic correlation between resistance to V. alginolyticus and C. irritans trait in large yellow croaker was also calculated based on the GEBV of two reference population.
2.7 Construction the resistance strain and control group
The reference population (n = 474) with the phenotype of resistance to C. irritans in large yellow croaker was constructed in the laboratory earlier, and genotype by 55 K SNP array. Another reference population (n = 496) that resistance to V. alginolyticus was built up in this experiment. Based on the two references genotype and phenotype data, the GEBV of two candidate populations of large yellow croaker (GS3-F2 broodstock (n = 560) and unselected broodstock fish named C2 (n = 415)) was calculated by using the BayesB model in the BGLR package in R software (23). The final selection index was calculated based on the standardized GEBV of each disease trait and with a weighting of 0.5, respectively.
Based on the GEBV ranking of resistance to C. irritans in GS3 breeding line F2 broodstock fish, 50 individuals with the top ranking were selected as the candidate parents for produced the F3 offspring (GS3-F3). Additionally, the top 50 ranking individuals were selected from the GS3 breeding line F2 broodstock fish based on the selection index and produced the F1 generation of double trait breeding line (GS9-F1). If the screened GS9 candidate was the same as the GS3 candidate, the GS3 candidate was given priority. Control groups were derived from two different unselected breeding candidate populations, with one being genotyped for genetic improvement of swimming performance traits. The GEBV of the selected candidate parents for two disease resistance traits was also calculated. All candidate group were being randomly mating to produce offspring in January 2022. According to the culture standard of large yellow croaker (NY/T 5061–2002), the fish in each group were fed for 2 months in a cement pond with 100 tons of water, with the water temperature kept at 24°C, oxygen concentration maintained at 5.0 mg/L, and water changed daily to remove waste.
2.8 Offspring challenge test
The fish were randomly selected from each group for the C. irritans infection experiment. The number of challenged fish in each group was as follows: GS9 (body weight (g): 2.97 ± 1.05, body length (cm): 5.32 ± 0.66), GS3 (body weight (g): 4.28 ± 1.28, body length (cm): 6.15 ± 0.62), C1 (body weight (g): 2.55 ± 0.79, body length (cm): 5.21 ± 0.63) and C2 (body weight (g):2.97 ± 1.02, body length (cm):5.39 ± 0.76), with 487, 508, 510, and 483 fish, respectively. The fish were then transported to the pond and acclimated for a week before the experiment. During this period, the individuals were fed normally and feeding was stopped 3 days before the start of the experiment. After stabilization, a challenge test for resistance to C. irritans was conducted in 1-ton tanks, with each group having 3 replicates. Based on the results of the preliminary challenge test, the infection concentration of C. irritans was set at 500 larvae per liter of water in the formal test, and the experimental water temperature was maintained at 25–26°C. Observations were recorded every 2 h, and the experiment lasted for 5 days.
In the offspring resistance to V. alginolyticus challenge experiment, a total of 1,566 7-month-old offspring were randomly selected from the double trait bred strain (GS9, body weight (g): 22.67 ± 6.54, body length (cm): 11.38 ± 1.21), single trait bred strain (GS3, body weight (g): 16.03 ± 3.90, body length (cm): 10.21 ± 0.84), C1 control group (body weight (g): 16.70 ± 5.55, body length (cm): 10.22 ± 1.30), and C2 control group (body weight (g): 13.82 ± 4.37, body length (cm): 9.71 ± 1.09), and then transported to the pond and acclimated for a week before the experiment. After stabilization, a challenge test for resistance to V. alginolyticus was conducted in 1-ton tanks, with each group having 3 replicates. Based on the results of the preliminary challenge test and the size of the fish in this experiment, 0.2 mL bacterial solution with a concentration of 3.32 × 107 CFU/mL was injected into the abdominal cavity of large yellow croaker for the main experiment. The experimental water temperature was maintained at 28–29°C and data were continuously observed and recorded every 2 h for 2 days.
3 Results
3.1 Standard for artificial challenge of Vibrio alginolyticus infection in large yellow croaker
In the first preliminary experiment to determine the artificial challenge method for infecting large yellow croaker with V. alginolyticus, the results showed that the mortality rate was less than 2% within 48 h following the immersion infection challenge. In contrast, the fish subjected to intraperitoneal injection of V. alginolyticus showed a peak death rate at 6 h post-challenge, which continued until 24 h and then stabilized. Meanwhile, the control group injected with normal saline solution had no deaths within 48 h. These results indicated that intraperitoneal injection was the most effective method for challenging large yellow croaker with V. alginolyticus to test resistance (Figure 2A).
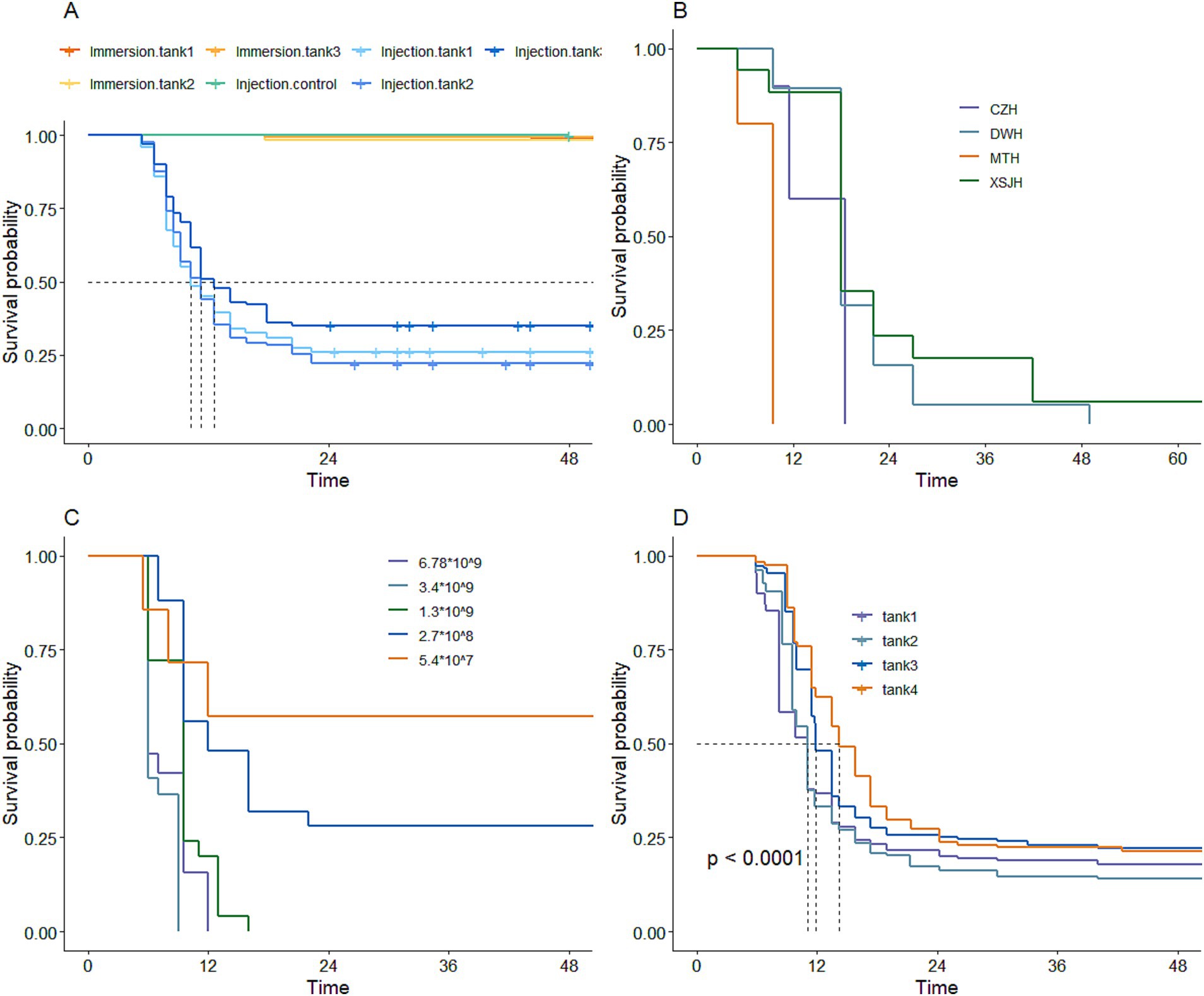
Figure 2. The standard of artificial infection of resistance against V. alginolyticus in large yellow croaker. (A) Challenge method. (B) Different strains. (C) Different concentrations. (D) Reference population constructed.
The results of the experiment using involving diverse strains and the intraperitoneal infection method on large yellow croaker demonstrated that the virulence of various V. alginolyticus strains varied. Among these strains, the MTH strain exhibited the strongest virulence, whereas the XSJH strain exhibited the weakest (Figure 2B). The experiment investigated the impact of different concentrations of MTH strain on large yellow croaker mortality rates. The results showed that the higher infection concentrations led to more rapid and substantial mortality rates, with the optimal concentration of MTH strains for the infection challenge in large yellow croaker identified as 107–108 CFU/mL (Figure 2C).
3.2 Reference and candidate population resource
To construct a phenotypic reference population of large yellow croaker resistant to V. alginolyticus, 708 individuals were artificially intraperitoneally injected with V. alginolyticu strain (MTH strain, 0.1 mL) at a concentration of 3.16 × 108 CFU/mL, based on the pre-experimental results (Figure 2D). Among them, 496 individuals were randomly selected and genotyped by using 55 K SNP array, and 45,133 SNPs were obtained after quality control. The average distance between SNPs was 12.4 kb (Supplementary Figure S1). Additionally, 44,449 SNPs were obtained from the 930 candidate genotypes (523 GS3 F2 candidate parents and 407 unbred control candidate parents) after quality control.
3.3 Genome wide association of resistance to Vibrio alginolyticus
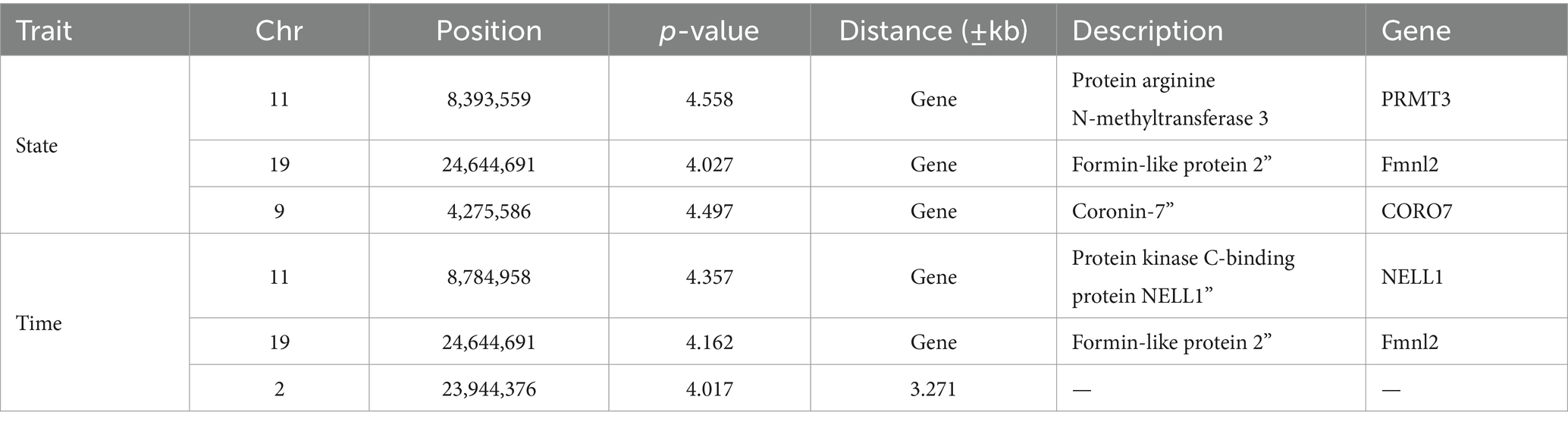
GWAS was performed by using the reference population (sample = 496) for the survival time and survival status of resistance to V. alginolyticus in large yellow croaker. The results showed that significant SNPs were identified in chromosome 2, 9, 11 and 19 (Figure 3). Except for the SNPs in chromosome 2, the rest of SNPs were located in the Prmt3, Fmnl2, Coro7, and Nell1 genes, respectively. The heritability of resistance to V. alginolyticus was approximately 0.1 (Standard error 0.008), and the genetic correlation between resistance to V. alginolyticus and resistance to C. irritans was 0.05 (Figure 4).

Figure 3. Genome-wide association analysis of V. alginolyticus resistance traits in large yellow croaker.
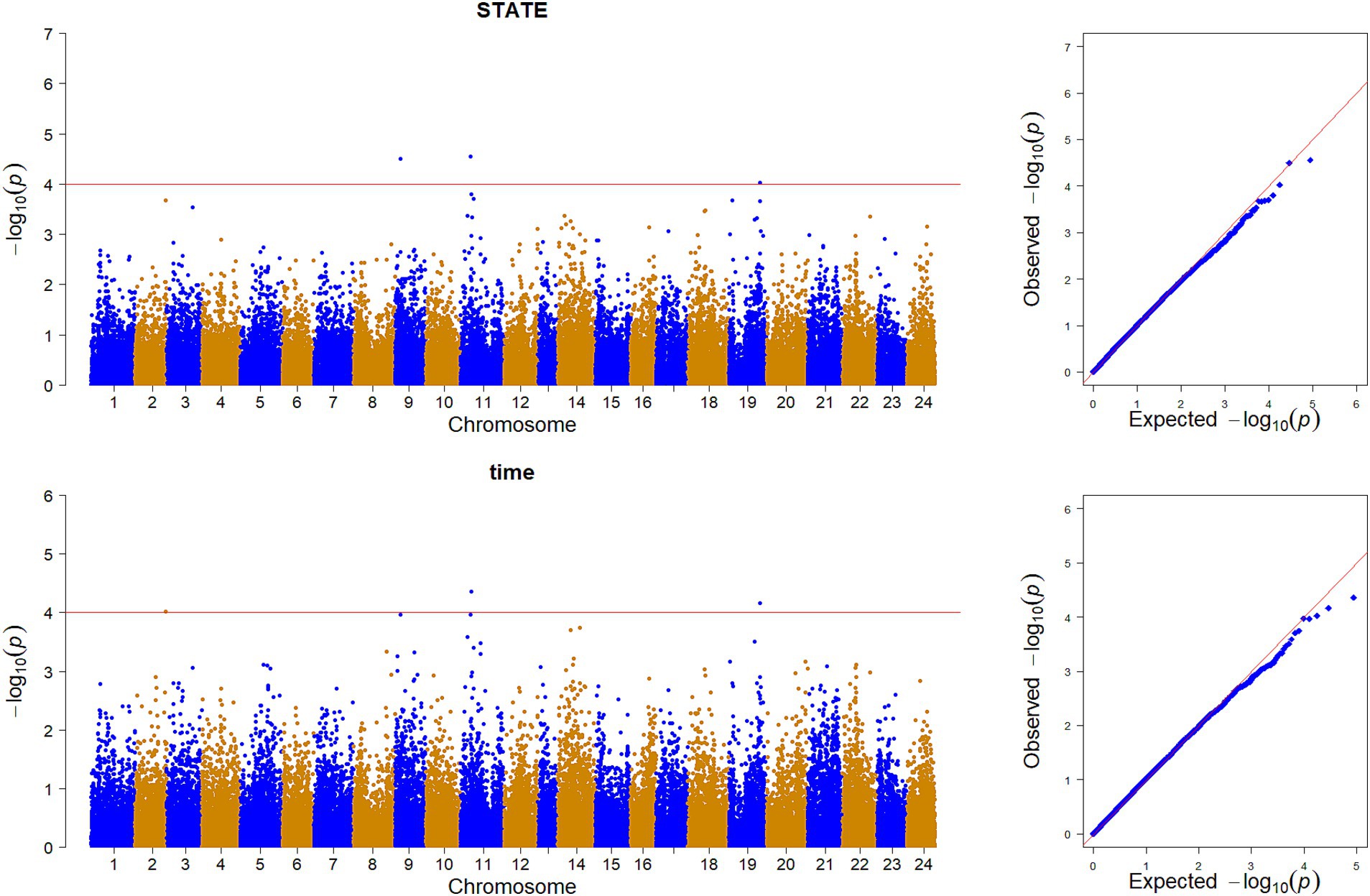
Figure 4. Genetic correlation between resistance to V. alginolyticus and C. irritans in large yellow croaker.
3.4 Offspring produced
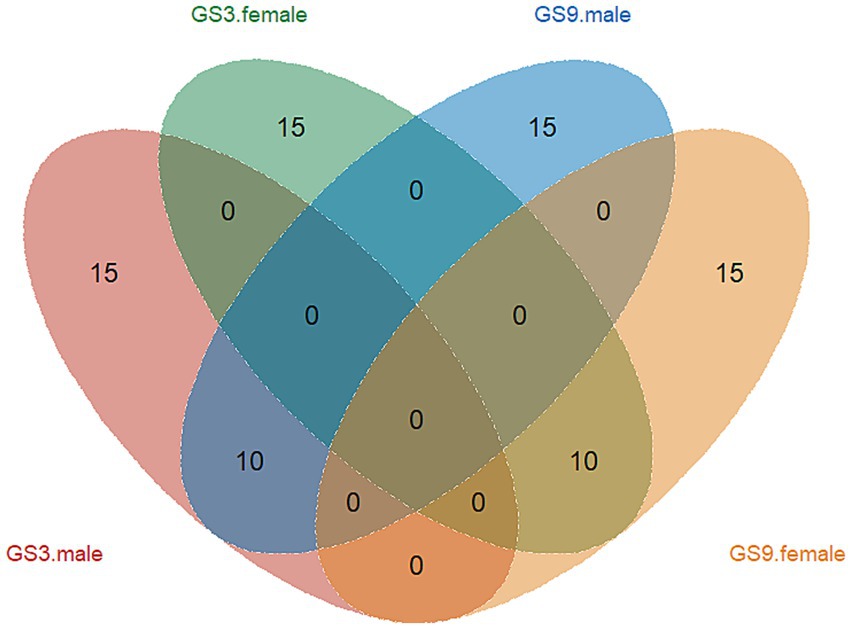
The GEBV and selection index were calculated by using the reference population of resistance to V. alginolyticus (sample = 496) and C. irritans (sample = 474) in large yellow croaker. Finally, 50 broodstock candidates (33 females, 17 males) with top GEBV rankings and 50 broodstock candidates (33 females, 17 male) with top selection index were selected from the GS3-F2 population. To evaluate the coincidence between the selection index and the GEBV, the top 25 male and female candidates ranked in GEBV and selection index were selected for further analysis. The results showed that 10 individuals of each sexoverlapped (Figure 5). The average GEBV of resistance to C. irritans traits of GS3, GS9 and C2 candidates were 0.83, 0.77 and 0.58, respectively. For resistance to V. alginolyticus trait, the average GEBV of GS3, GS9 and C2 broodstock were 20.53, 22.35 and 20.60, respectively (Figure 6). The GS3, GS9, and C2 broodstock produced 2.7 kg, 3.1 kg, and 4.0 kg of floating eggs, respectively. After 2 months of standard culture feeding in a 100 m3 pond, approximately 500 thousand, 576 thousand and 480 thousand juvenile fish of the GS3, GS9, and C2 strains, were released into marine cages, respectively (Table 1).
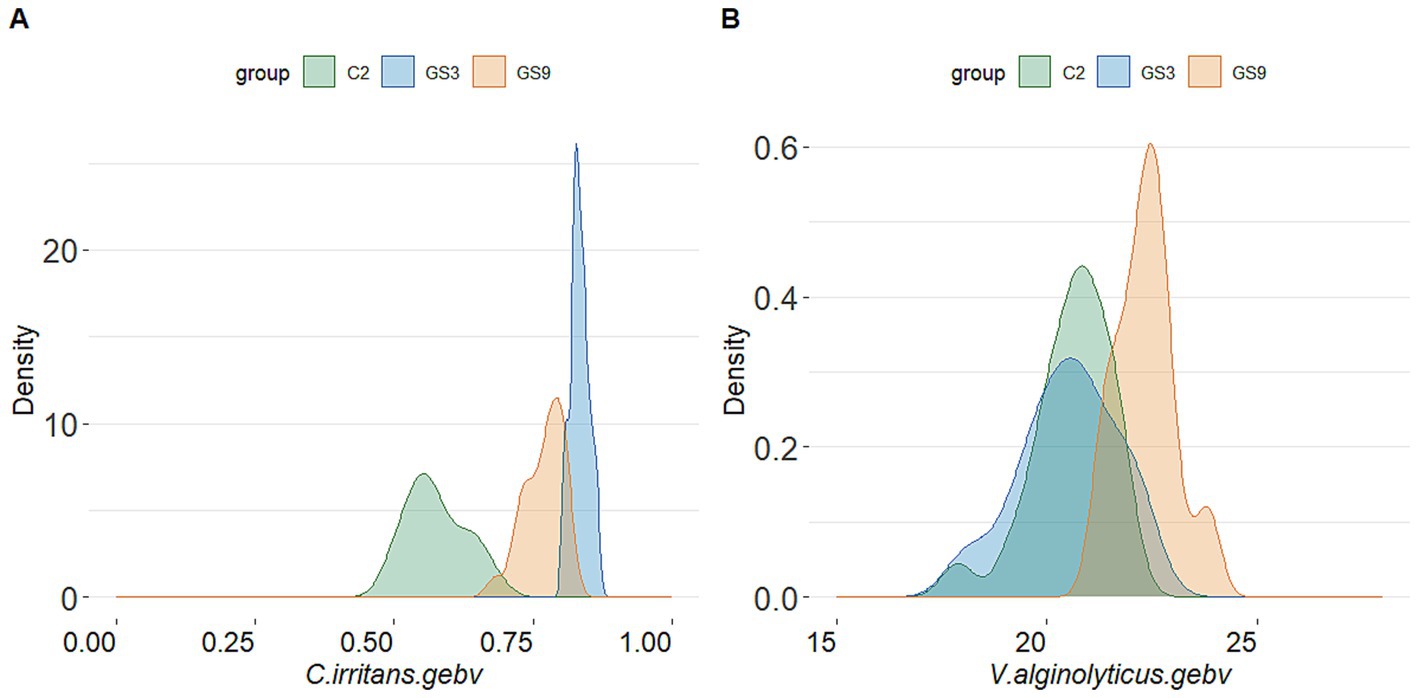
Figure 6. GEBV probability density of candidate fish of each breeding line of large yellow croaker. (A) C. irritans. (B) V. alginolyticus.
3.5 Offspring challenge test
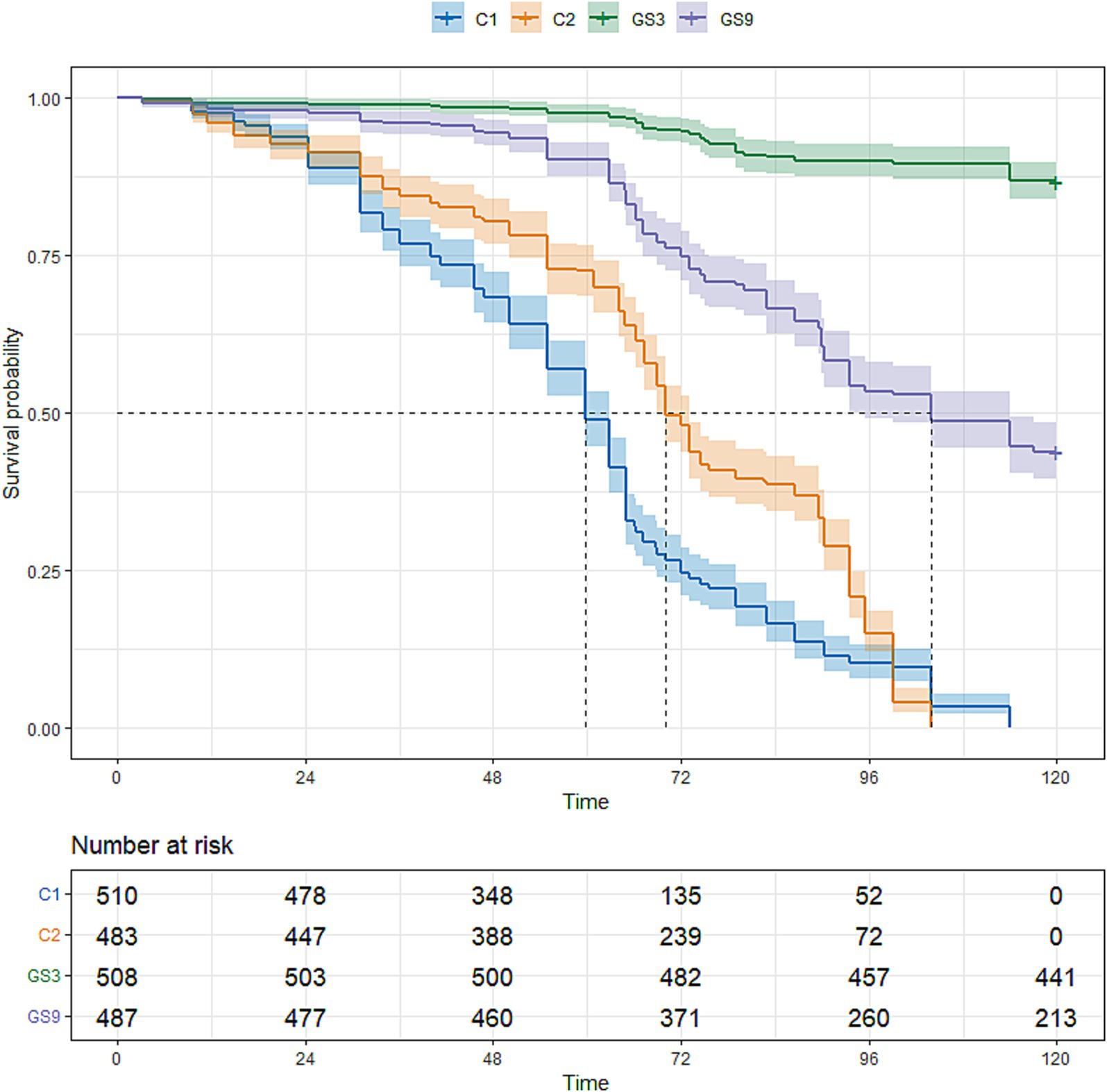
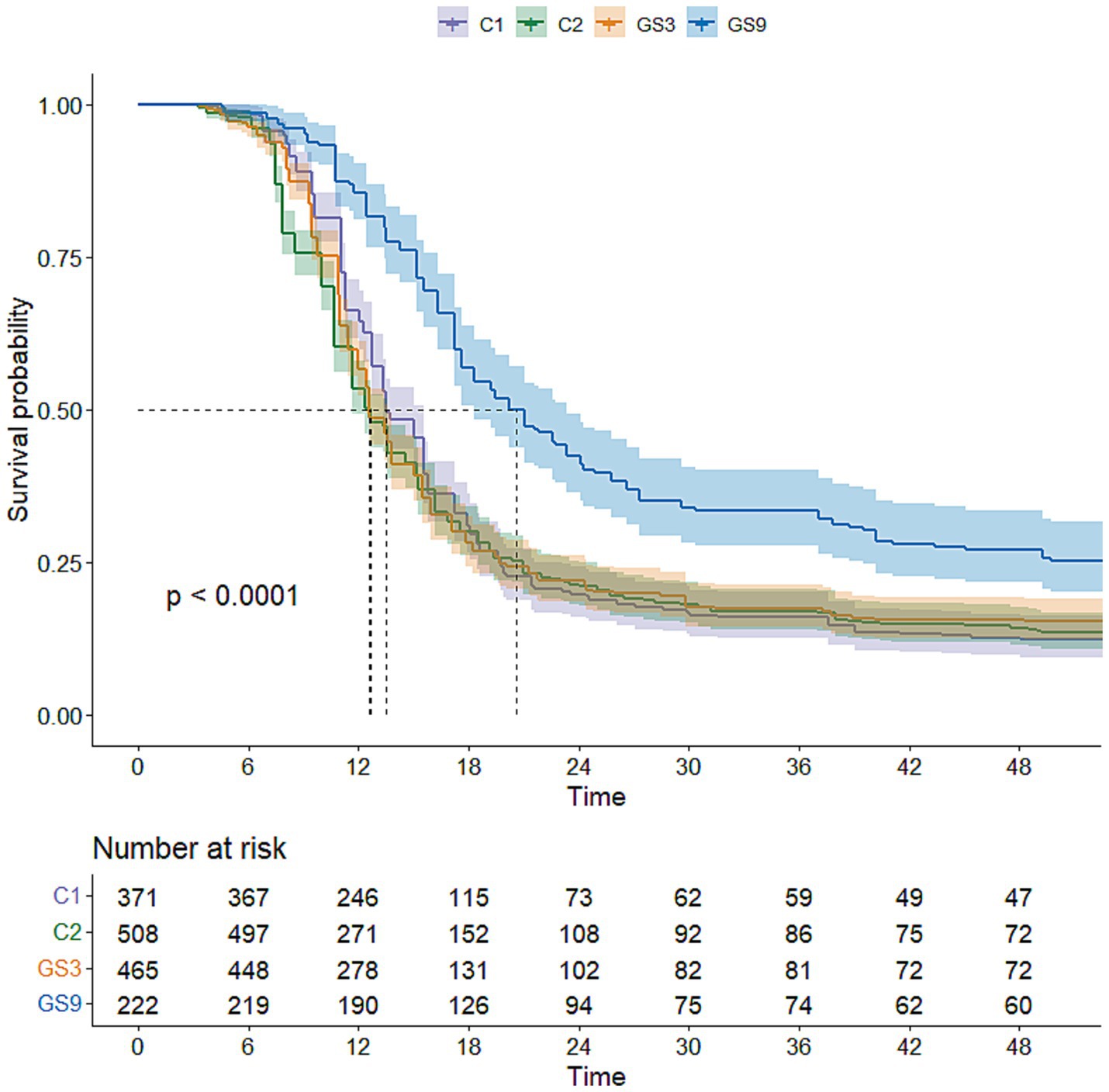
To evaluate the genetic improvement of resistance to C. irritans in the double-trait bred strain and single-trait bred strain, 487, 503, 510 and 483 5-month-old fish selected from GS9, GS3, C1 and C2, respectively, were tested using an artificial infection challenge. The survival rates of GS9 and GS3 were 43.74 and 86.81%, respectively, after 120 h of challenge, whereas those of C1 and C2 remained at 0 % (Figure 7). Additionally, 222, 465, 371 and 508 8-month-old fish from GS9, GS3, C1 and C2 were tested in a V. alginolyticus infection experiment. The survival rates of GS9 and GS3 were 27.03 and 15.48%, respectively, while the survival rates of C1 and C2 were 12.67 and 14.17% (Figure 8).
4 Discussion
4.1 Standard for artificial challenge of Vibrio alginolyticu infection in large yellow croaker
The objective of this study was to explore the effect of genetic improvement of double traits in resistance to V. alginolyticus and C. irritans. Before implementing genomic selection for both disease traits, it is imperative to establish standardized artificial challenges of V. alginolyticus resistance in large yellow croaker and build up a reference population. In studies on artificial challenge infection of aquatic species with bacterial diseases globally, the majority have used bacterial solution injection as the method for infection challenge. For instance, Yan et al. injected V. alginolyticus into the abdominal cavity of large yellow croakers (body weight: 300–500 g) to investigate its immune efficacy. They found the number of red blood cells, white blood cells, and lymphocytes in the infected group was significantly higher compared with the control group, while the number of granulocytes in peripheral blood was significantly lower than control group 1 day after the injection. Antibody titer began to increase on day 7 and reached its peak on day 15 (24). Fevolden et al. evaluated the resistance performance of individuals with high and low stress levels by injecting vibrio into rainbow trout (Oncorhynchus mykiss) (25). Leeds et al. injected Flavobacterium psychrophilum into rainbow trout to explore the improvement of rainbow trout resistance to bacterial cold water disease (BCWD) (26). The mortalities record data at day 21 showed that the survival rate of select-line increased by 24.6% after one generation of selection and by 44.7% after two generations of selection. Fraslin et al. found that immersion and injection affected the genetic mechanism of resistance to Flavobacterium psychrophilum in rainbow trout, which might be caused by epistatic interactions between resistance QTL (27). The result showed that the overall survival rate was 77% on day 48 for immersion infection and 55% survival rate on day 34 for injection challenge. Additionally, regardless of the route of infection, fish body weight at the time of infection did not have a significant effect on either time to death or survival rate. Degremont et al. injected Vibrio aestuarianus into Pacific oysters (Crassostrea gigas) offspring to evaluate the resistance performance of selectively bred strains (28). The effect of two infection methods (immersion and intraperitoneal injection) was compared in large yellow croaker in this study, and results showed a significant difference in challenge test between two infection methods, revealing that intraperitoneal injection is more suitable for resistance to V. alginolyticus in large yellow croaker. Unlike the C. irritans infection expreiment in large yellow croaker, intraperitoneal injection of V. alginolyticus is an acute infection experiment with short experiment period and fast death rate in our study, which is consistent with the research showing that non-specific immune factors in large yellow croakers played an important role on the first day after injection of V. alginolyticus (24). Furthermore, a study showed that intraperitoneal injection of V. alginolyticus in Pacific white shrimp (Litopenaeus vannamei) significantly increased its non-specific immune antibacterial proteins within 12 h (29, 30). The multiple injections experimental results in large yellow croaker showed that the injection concentration could significantly affect the survival rate of large yellow croaker infected with V. alginolyticus. When the injection concentration was 109 CFU/mL, all fish died within 12 h. At an injection concentration of 108 CFU/mL, the 24-h survival rate of experimental fish was 25% when the injection concentration was 107 CFU/mL, the 24-h survival rate of experimental fish exceeded 50%. Therefore, for the specifications of large yellow croakers in this experiment, the optimal concentration of MTH strains for the infection challenge in large yellow croaker identified as 107-c108CFU/mL. Consequently, prior to formal V. alginolyticus infection experiments, preliminary trials are necessary to determine the appropriate dose and concentration.
4.2 Genetic analysis of resistance to Vibrio alginolyticus infection in large yellow croaker
As is well known, traits with high heritability often exhibit substantial genetic variation, indicating a significant potential for trait improvement. However, most disease resistance traits in aquaculture exhibit low to medium heritability. For example, the heritability of resistance to V. alginolyticus ranged from 0.133 to 0.257 (31) in Pacific Oyster (Crassostrea giga). The heritability of resistance to Streptococcus agalactis of tilapia ranged from 0.23 to 0.25 (32), and the heritability of resistance to BCWD of rainbow trout ranged from 0.22 to 0.35 (26). In this study, the heritability of survival time trait related to V. alginolyticus resistance in large yellow croaker was 0.1, indicating it as a low heritability trait. This means that it is difficult to screen out resistant individuals through pedigree selection for genetic breeding of large yellow croaker against V. alginolyticus, making genomic selection technology more suitable for breeding applications targeting this disease trait (33). Moreover, this may require multiple generations of continuous genetic improvement to achieve significant genetic progress. Based on the phenotypic data and genotype data of the reference population of V. alginolyticus resistant established by random population, genome-wide association analysis was carried out and the results showed that resistance to V. alginolyticus in large yellow croaker is a polygenic trait. Only a common SNP was identified on both survival status and survival time traits. This SNP is located in the Fmnl2 gene in chromosome 19, which encodes a formin protein involved in cytoplasmic division and cell polarity. Compared with random population, the genetic structure of family population may powerful for the genome-wide association analysis to identified the candidate gene related with the important trait. For example, Sukhavachana et al. conducted genome-wide association analysis on Streptococcus agalactis traits based on tilapia family population data, and the SNP loci located on chromosomes 11 and 16 could explain 10% genetic variation of this trait (34). Vallejo et al. conducted genome-wide association analysis of BCWD trait in rainbow trout based on family population data, and identified 9 significant QTL regions, which could explain 88% genetic variation (35). Yang et al. identified 3 QTL association with V. alginolyticus resistance trait in Pacific oysters based on family population data (36).
Prior to applying the genetic improvement of multi-trait, it is essential to evaluated the genetic correlation between each trait. Positive correlations among the traits show that the changes of two traits are in the same direction, while negative correlations indicate two traits cannot be improved by breeding at the same time, and strategies like index selection or multi-trait selection models can help identify optimal compromises. However, for most aquatic species, the complex underwater environment greatly increases the difficulty of acquiring disease traits and it is impossible for the same individual to be artificially infected with both diseases at the same time, which means that the phenotypic correlation between two disease traits is difficult to accurately calculate. In this case, most studies have calculated genetic correlations based on family and half-sib populations to reflect the relationship between two disease traits. For example, the genetic correlation between BCWD and columnaris disease in rainbow trout was 0.39 ~ 0.40 based on family population data (37). Similar results were also been found in Evenhuis et al. study that the genetic correlation and phenotypic correlation was 0.35 and 0.2, respectively (38). The genetic correlation between the resistance of rainbow trout to Piscirickettsia salmonis and sea louse resistance (Caligus rogercresseyi) is 0.39, which is possible to improvement of these two disease resistance traits simultaneously (39). In Atlantic salmon, the genetic correlation between Aeromonas salmonicida infection resistance and anemia traits was 0.15 in a family population (40). Gjedrem et al. found the genetic correlation between the ability to survive the diseases for the furunculosis, bacterial kidney disease and BCWD in Atlantic salmon were all positive (41). In this study, considering the high cost of constructing families of large yellow croaker, the genetic correlation of two disease traits was calculated based on the genotypic and phenotypic data from two reference populations. The results showed that there was a weak positive correlation between V. alginolyticus resistance and C. irritans resistance of large yellow croaker, which supports the application of a selection index for these two disease traits.
4.3 Disease resistance performance of double trait strain
In this study, the double-trait strain was constructed based on the selection index in F2 generation of C. irritans resistance strain (GS3) in large yellow croaker. Meanwhile, the F3 generation of C. irritans resistance strain in large yellow croaker was also produced based on the GEBV ranking of F2 candidate population. The average GEBV of C. irritans resistance trait of broodstock fish was ranked as follows: GS3 (0.83) > GS9 (0.77) > C2 (0.58), indicating that the F3 offspring of GS3 strain exhibited greater resistance to C. irritans than those of other breeding strains. The survival rate of all breeding strain offspring in the artificial challenge infection supported this conclusion. The GS3 breeding strain had the strongest resistance to C. irritans, followed by GS9, while C1 and C2 exhibited weaker resistance. The survival rate of GS9 and GS3 was 43.74 and 86.81%, respectively, and that of C1 and C2 was 0% in resistance to C. irritans.
In terms of resistance to V. alginolyticus, the average GEBV of GS9 broodstock fish was the highest (22.35), while the average GEBV of GS3 broodstock fish and C2 broodstock fish were similar (20.53 and 20.60, respectively). The survival rate of GS9 offspring infected with V. alginolyticus was 11.55 percentage points higher than that of GS3, 14.36 and 12.86 percentage points higher than that of C1 and C2 controls, respectively. These findings was consistent with the average GEBV ranking of all candidate fish strains. All experimental groups showed good repeatability in the artificial infection challenge of two diseases (Supplementary Figures S2, S3). Similar results have been found in the genomic selection breeding of tilapia against Streptococcus agalactiae. The average GEBV of the candidate fish of the breeding strain is higher than that of the control group, and the survival rate of the offspring of the breeding strain is 21% higher than that the control group (32).
The performance of artificial infection experiment of the breeding strain offspring was consistent with the expectations. However, the disease risk and challenge of large yellow croaker cultured in sea area are much higher than the experimental conditions of land-based artificial infection experiment. Only the survival rate and growth performance and other indicators observed in the cultured sea area can accurately reflect the genetic improvement effect of all breeding strains. Although tracking this trait takes more time, energy and cost, it has important breeding significance and value for the genetic improvement application of large yellow croaker.
5 Conclusion
This study establishes the technical standard for artificial infection of V. alginolyticus in large yellow croaker, and selectively breeding for the double trait of resistance to V. alginolyticus and C. irritans by using the selection index based on the C. irritans resistance strain in large yellow croaker. The experiment on artificial infection challenge demonstrated a significant and simultaneous improvement in the survival rate of the double trait strain in resistance to both C. irritans and V. alginolyticus. In contrast, the single-trait selection strain only demonstrated a significant improvement in resistance to C. irritans. These results indicate that genetic improvement of the double trait in large yellow croaker using genome selection technology is a viable approach. This provides a reference for the application of double trait breeding in aquatic species.
Data availability statement
The datasets presented in this article are not readily available because the data (genotypes and phenotypes) are part of a reference population used for genomic selection and have commercial value. Requests to access the datasets should be directed to corresponding author.
Ethics statement
The animal study was approved by the Animal Care and Use Committee at College of Ocean and Earth Sciences, Xiamen University. The study was conducted in accordance with the local legislation and institutional requirements.
Author contributions
JW: Writing – review & editing, Data curation, Formal analysis, Investigation, Writing – original draft. JZh: Data curation, Writing – review & editing. BL: Data curation, Writing – review & editing. HC: Data curation, Writing – review & editing. PJ: Data curation, Writing – review & editing. JX: Data curation, Writing – review & editing. ZJ: Data curation, Writing – review & editing. LM: Data curation, Writing – review & editing. QH: Data curation, Writing – review & editing. JZe: Data curation, Writing – review & editing. QK: Data curation, Writing – review & editing. TZ: Data curation, Writing – review & editing. PX: Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This study was funded by the Seed Industry Innovation and Industrialization Project of Fujian Province (2021FJSCZY01), the National Science Fund for Distinguished Young Scholars (32225049), and National Natural Science Foundation of China (NSFC) (U21A20264).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The authors declare that no Gen AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fvets.2025.1524914/full#supplementary-material
SUPPLEMENTARY FIGURE S1 | SNP density distribution of large yellow croaker reference population.
SUPPLEMENTARY FIGURE S2 | Result of resistance to C. irritans in each replicate of large yellow croaker.
SUPPLEMENTARY FIGURE S3 | Result of resistance to V. alginolyticus in each replicate of large yellow croaker.
SUPPLEMENTARY TABLE S1 | The situation of offspring of different breeding line of large yellow croaker.
References
1. Hazel, LN. The genetic basis for constructing selection indexes. Genetics. (1943) 28:476–90. doi: 10.1093/genetics/28.6.476
2. Cole, JB, and Van Raden, PM. Symposium review: possibilities in an age of genomics: the future of selection indices. J Dairy Sci. (2018) 101:3686–701. doi: 10.3168/jds.2017-13335
3. García-Ruiz, A, Cole, JB, Van Raden, PM, Wiggans, GR, Ruiz-López, FJ, and Van Tassell, CP. Changes in genetic selection differentials and generation intervals in US Holstein dairy cattle as a result of genomic selection. Proc Natl Acad Sci USA. (2016) 113:E3995–4004. doi: 10.1073/pnas.1519061113
4. O'Flynn, FM, Bailey, JK, and Friars, GW. Responses to two generations of index selection in Atlantic salmon (Salmo salar). Aquaculture. (1999) 173:143–7. doi: 10.1016/S0044-8486(98)00482-7
5. Friars, GW, Bailey, JK, and Oflynn, FM. Applications of selection for multiple traits in cage-reared Atlantic salmon (Salmo salar). Aquaculture. (1995) 137:213–7. doi: 10.1016/0044-8486(95)01101-3
6. Thodesen, J, Rye, M, Wang, YX, Li, SJ, Bentsen, HB, and Gjedrem, T. Genetic improvement of tilapias in China: genetic parameters and selection responses in growth, pond survival and cold-water tolerance of blue tilapia (Oreochromis aureus) after four generations of multi-trait selection. Aquaculture. (2013) 396-399:32–42. doi: 10.1016/j.aquaculture.2013.02.010
7. Bureau of Fishery Administration of the Ministry of Agriculture and Rural Affairs, N.F.T.E.C. China fishery statistical yearbook 2024. China Society of Fisheries. Beijing: China Agriculture Press, pp 21–22. (2024).
8. Wang, J, Zhao, J, Tong, B, Ke, Q, Bai, Y, Gong, J, et al. Effects of artificial mating on genomic selection of resistance against Cryptocaryon irritans in large yellow croaker. Aquaculture. (2022) 561:738617. doi: 10.1016/j.aquaculture.2022.738617
9. Zhao, J, Bai, H, Ke, Q, Li, B, Zhou, Z, Wang, H, et al. Genomic selection for parasitic ciliate Cryptocaryon irritans resistance in large yellow croaker. Aquaculture. (2021) 531:735786. doi: 10.1016/j.aquaculture.2020.735786
10. Hörmansdorfer, S, Wentges, H, Neugebaur-Büchler, K, and Bauer, J. Isolation of Vibrio alginolyticus from seawater aquaria. Int J Hyg Environ Health. (2000) 203:169–75. doi: 10.1078/S1438-4639(04)70024-3
11. Ripabelli, G, Sammarco, ML, Grasso, GM, Fanelli, I, Caprioli, A, and Luzzi, I. Occurrence of Vibrio and other pathogenic bacteria in Mytilus galloprovincialis (mussels) harvested from Adriatic Sea, Italy. Int J Food Microbiol. (1999) 49:43–8. doi: 10.1016/S0168-1605(99)00056-2
12. Ge, M, Zheng, X, and Wang, G. Detection of pathogenic vibrios infection in Larimichthys crocea and its forecast and warning of disease. J Fish China. (2014) 38:2068–74.
13. Yan, Q, Wang, J, Su, Y, and Zhang, J. Studies on vibriosis in caged-cultured Pseudosciaena crocea (Richardson). J Jimei Univ. (2001) 6:191–6. doi: 10.19715/j.jmuzr.2001.03.001
14. Chen, X, Hu, C, Chen, C, Zhang, L, and Ren, C. Characteristics of pathogenic bacteria of vibriosis in caged malabra grouper, Epinephelus malabaricus. Mar Sci. (2002) 27:68–72.
15. Cao, J, Jian, J, and Wu, Z. Effects of Vibrio alginolyticus vaccine on white legged shrimp (Litopenaeus vannamei) immunity. Fish Sci. (2006) 25:330–333. doi: 10.16378/j.cnki
16. Zhao, X, Wang, Q, Wang, Z, Liu, L, Han, X, Cao, J, et al. Comparison of the identification methods of VITEKRT-PCR and rpoB sequencing on Vibrio alginolyticus isolated from seafood. Chinese J Food Hygiene. (2017) 29:671–5.
17. Wang, J, Miao, L, Chen, B, Zhao, J, Ke, Q, Pu, F, et al. Development and evaluation of liquid SNP array for large yellow croaker (Larimichthys crocea). Aquaculture. (2023) 563:739021. doi: 10.1016/j.aquaculture.2022.739021
18. Purcell, S, Neale, B, Todd-Brown, K, Thomas, L, Ferreira, MA, Bender, D, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. (2007) 81:559–75. doi: 10.1086/519795
19. Yin, L, Zhang, H, Tang, Z, Xu, J, Yin, D, Zhang, Z, et al. rMVP: a memory-efficient, visualization-enhanced, and parallel-accelerated tool for genome-wide association study. Genomics Proteomics Bioinformatics. (2021) 19:619–668. doi: 10.1016/j.gpb.2020.10.007
20. Zhou, X, and Stephens, M. Genome-wide efficient mixed-model analysis for association studies. Nat Genet. (2012) 44:821–4. doi: 10.1038/ng.2310
21. Turner, S. qqman: an R package for visualizing GWAS results using Q-Q and Manhattan plots. J Open Source Softw. (2018) 3:731. doi: 10.21105/joss.00731
22. Yin, L, Zhang, H, Tang, Z, Yin, D, Fu, Y, Yuan, X, et al. HIBLUP: an integration of statistical models on the BLUP framework for efficient genetic evaluation using big genomic data. Nucleic Acids Res. (2023) 51:3501–12. doi: 10.1093/nar/gkad074
23. Perez, P, and de los Campos, G. Genome-wide regression and prediction with the BGLR statistical package. Genetics. (2014) 198:483–95. doi: 10.1534/genetics.114.164442
24. Yan, Q, Zhang, J, Zou, W, Chen, Q, Zhuang, Z, and Wang, X. Effect of experimental infection with Vibrio alginolyticus on immune parameters of Pseudosciaena crocea. J Fish China. (2007) 31:250–6.
25. Fevolden, SE, Refstie, T, and Røed, KH. Disease resistance in rainbow trout (Oncorhynchus mykiss) selected for stress response. Aquaculture. (1992) 104:19–29. doi: 10.1016/0044-8486(92)90134-7
26. Leeds, TD, Silverstein, JT, Weber, GM, Vallejo, RL, Palti, Y, Rexroad, CE III, et al. Response to selection for bacterial cold water disease resistance in rainbow trout. J Anim Sci. (2010) 88:1936–46. doi: 10.2527/jas.2009-2538
27. Fraslin, C, Dechamp, N, Bernard, M, Krieg, F, Hervet, C, Guyomard, R, et al. Quantitative trait loci for resistance to Flavobacterium psychrophilum in rainbow trout: effect of the mode of infection and evidence of epistatic interactions. Genet Sel Evol. (2018) 50:60. doi: 10.1186/s12711-018-0431-9
28. Degremont, L, Azema, P, Maurouard, E, and Travers, MA. Enhancing resistance to Vibrio aestuarianus in Crassostrea gigas by selection. Aquaculture. (2020) 526:735429. doi: 10.1016/j.aquaculture.2020.735429
29. Han-Ching Wang, KC, Tseng, C-W, Lin, H-Y, Chen, IT, Chen, Y-H, Chen, Y-M, et al. RNAi knock-down of the Litopenaeus vannamei toll gene (LvToll) significantly increases mortality and reduces bacterial clearance after challenge with Vibrio harveyi. Dev Comparat Immunol. (2010) 34:49–58. doi: 10.1016/j.dci.2009.08.003
30. Luo, C, Huang, X, Li, S, Zhao, L, Wei, L, Chen, C, et al. Expressions of toll receptor, IMD and lysozyme mRNA in the gills of the white shrimp (Litopenaeus vannamei) after acute challenge with Vibrio alginolyticus. J Fishery Sci China. (2014) 21:189–96.
31. Zhai, SY, Yang, B, Zhang, FQ, Li, Q, and Liu, SK. Estimation of genetic parameters for resistance to Vibrio alginolyticus infection in the Pacific oyster (Crassostrea gigas). Aquaculture. (2021) 538:736545. doi: 10.1016/j.aquaculture.2021.736545
32. Joshi, R, Skaaurd, A, and Tola Alvarez, A. Experimental validation of genetic selection for resistance against Streptococcus agalactiae via different routes of infection in the commercial Nile tilapia breeding programme. J Anim Breed Genet. (2021) 138:338–48. doi: 10.1111/jbg.12516
33. Crossa, J, Pérez-Rodríguez, P, Cuevas, J, Montesinos-López, O, Jarquín, D, de los Campos, G, et al. Genomic selection in plant breeding: methods, models, and perspectives. Trends Plant Sci. (2017) 22:961–75. doi: 10.1016/j.tplants.2017.08.011
34. Sukhavachana, S, Tongyoo, P, Massault, C, McMillan, N, Leungnaruemitchai, A, and Poompuang, S. Genome-wide association study and genomic prediction for resistance against Streptococcus agalactiae in hybrid red tilapia (Oreochromis spp.). Aquaculture. (2020) 525:735297. doi: 10.1016/j.aquaculture.2020.735297
35. Vallejo, RL, Evenhuis, JP, Cheng, H, Fragomeni, BO, Gao, GT, Liu, SX, et al. Genome-wide mapping of quantitative trait loci that can be used in marker-assisted selection for resistance to bacterial cold water disease in two commercial rainbow trout breeding populations. Aquaculture. (2022) 560:738574. doi: 10.1016/j.aquaculture.2022.738574
36. Yang, B, Zhai, SY, Zhang, FQ, Wang, HB, Ren, LT, Li, YJ, et al. Genome-wide association study toward efficient selection breeding of resistance to Vibrio alginolyticus in Pacific oyster, Crassostrea gigas. Aquaculture. (2022) 548:737592. doi: 10.1016/j.aquaculture.2021.737592
37. Silva, RMO, Evenhuis, JP, Vallejo, RL, Tsuruta, S, Wiens, GD, Martin, KE, et al. Variance and covariance estimates for resistance to bacterial cold water disease and columnaris disease in two rainbow trout breeding populations. J Anim Sci. (2019) 97:1124–32. doi: 10.1093/jas/sky478
38. Evenhuis, JP, Leeds, TD, Marancik, DP, LaPatra, SE, and Wiens, GD. Rainbow trout (Oncorhynchus mykiss) resistance to columnaris disease is heritable and favorably correlated with bacterial cold water disease resistance. J Anim Sci. (2015) 93:1546–54. doi: 10.2527/jas.2014-8566
39. Bassini, LN, Lhorente, JP, Oyarzun, M, Bangera, R, Yanez, JM, and Neira, R. Genetic parameters for Piscirickettsia salmonis resistance, sea lice (Caligus rogercresseyi) susceptibility and harvest weight in rainbow trout (Oncorhynchus mykiss). Aquaculture. (2019) 510:276–82. doi: 10.1016/j.aquaculture.2019.05.008
40. Odegard, J, Olesen, I, Gjerde, B, and Klemetsdal, G. Positive genetic correlation between resistance to bacterial (furunculosis) and viral (infectious salmon anaemia) diseases in farmed Atlantic salmon (Salmo salar). Aquaculture. (2007) 271:173–7. doi: 10.1016/j.aquaculture.2007.06.006
Keywords: selection index, disease resistance trait, genomic selection, large yellow croaker, aquaculture
Citation: Wang J, Zhao J, Liu B, Chi H, Jiang P, Xiao J, Jiang Z, Miao L, He Q, Zeng J, Ke Q, Zhou T and Xu P (2025) Application of selection Index for enhancing resistance to Cryptocarya irritans and Vibrio alginolyticus in large yellow croaker. Front. Vet. Sci. 12:1524914. doi: 10.3389/fvets.2025.1524914
Edited by:
Yidi Wu, Chinese Academy of Fishery Sciences (CAFS), ChinaReviewed by:
Jian Xu, Chinese Academy of Fisheries Sciences, ChinaYubang Shen, Shanghai Ocean University, China
Copyright © 2025 Wang, Zhao, Liu, Chi, Jiang, Xiao, Jiang, Miao, He, Zeng, Ke, Zhou and Xu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Peng Xu, eHVwZW5nNzdAeG11LmVkdS5jbg==
†These authors have contributed equally to this work
 Jiaying Wang
Jiaying Wang Ji Zhao1,2†
Ji Zhao1,2† Bo Liu
Bo Liu Pengxing Jiang
Pengxing Jiang Zhou Jiang
Zhou Jiang Lingwei Miao
Lingwei Miao Junjia Zeng
Junjia Zeng Tao Zhou
Tao Zhou Peng Xu
Peng Xu