
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Vet. Sci., 13 February 2025
Sec. Veterinary Clinical, Anatomical, and Comparative Pathology
Volume 12 - 2025 | https://doi.org/10.3389/fvets.2025.1521705
This article is part of the Research TopicEmerging Diseases and Diagnostics in Poultry ProductionView all 7 articles
 Jialong Chen1†
Jialong Chen1† Wenchang Xue1†
Wenchang Xue1† Zhanxin Yao1
Zhanxin Yao1 Chao Wang1
Chao Wang1 Wanjun Zhu2
Wanjun Zhu2 He Wang1,3
He Wang1,3 Jipei Zhang1
Jipei Zhang1 Yi Tang4
Yi Tang4 Rongchang Liu5
Rongchang Liu5 Jidang Chen1,6*
Jidang Chen1,6*Goose circovirus (GoCV) is a recently identified pathogen in geese that is known to cause slow growth, feather disorder syndrome, and immunosuppression. Infection with GoCV may increase the risk of coinfections with multiple pathogens, leading to significant economic losses in the goose industry. However, due to a lack of serological detection methods, analysis of viral nucleic acids has been widely used in GoCV epidemiological surveys, which has limited accurate monitoring of the prevalence of GoCV. In this study, we developed and optimized an indirect ELISA method based on the prokaryotic-expressed recombinant GoCV capsid protein (△Cap-iELISA). The △Cap-iELISA was then used to test 349 goose serum samples collected from Guangdong, Shandong, and Fujian provinces during 2023 and 2024. The results showed that the positive rate of GoCV antibodies in the sampled geese was 71.06%. Further analysis indicated that the positive rate of GoCV antibodies increased with the age of the geese. In conclusion, we have developed a novel iELISA method that is well-suited for large-scale clinical detection and early diagnosis of GoCV infection. Notably, a significant correlation between age and the positive rate of GoCV antibodies among geese was observed based on this newly established method.
Goose circovirus (GoCV) is a non-enveloped virus with a circular single-stranded DNA genome 1.8 kb in size (1), making it the smallest known animal virus. GoCV belongs to the Circovirus the genus Circovirus with in the family Circoviridae (2). Porcine circovirus (PCV), duck circovirus (DuCV), and GoCV share a similar genome structure that features three open reading frames (ORFs), ORFV1, ORFC1, and ORFC2, on both complementary DNA strands. The transcription of circoviruses is bidirectional. The sense strand encodes the replicase protein (Rep/Rep0) and the virus Capsid by ORFV1 and ORFC1, respectively. In addition, the apoptosis-related protein is encoded by ORFC2 located on the complementary strand (3, 4).
Cap protein is the main structural protein of GoCV (5). There are five major B-cell antigenic epitopes on Cap protein, which are located at 28aa–47aa, 129aa–148aa, 105aa–124aa, 156aa–175aa, and 231aa–250aa; followed by two glycosylation sites at 49aa and 141aa. proteins play an essential role in neutralizing antibodies against GoCV. In our previous sequence analysis we found that the nucleotide similarity of GoCV Cap proteins ranged from 86.3 to 98.5%, and the amino acid sequence similarity ranged from 92.4 to 100%, with the highest upper limits of amino acid sequence similarity between PCV2 Cap, BFDV Cap, and GoCV Cap being 37.09 and 31.13%, respectively (6). In addition we found that the Cap protein contains 17 arginine residues at the N-terminus, which correlates with the nuclear localization of the protein. It has been shown that a high percentage of arginine residues can greatly reduce the expression level of recombinant proteins in E. coli (7). This may be an important reason for the non-expression or low expression of the Cap protein of circovirus in E. coli (8, 9).
GoCV was firstly discovered in a flock of geese in Germany in 1999 by Soike et al. (10). Subsequently, it was reported in Hungary (11), the United Kingdom (12), Poland (13) and other countries. In China, GoCV was first detected in a flock of geese in Taiwan Province in 2003 (14). Subsequently, infected geese were also found in Zhejiang (15), Fujian, Guangxi and Guangdong (16). Based on early observations of GoCV infection and associated symptoms, the main clinical signs in geese include growth retardation, feather stunting, shedding, and feather follicular necrosis (17). These symptoms can severely affect the appearance of a goose’s carcass, which can have a significant impact on yield. However, GoCV has a low direct lethality in geese and clinical signs and lesions are not readily detectable (18). In addition, current nucleic acid-based GoCV detection methods require more time, more procedural steps, and are costly, leading to limitations in monitoring and preventing GoCV (19). Therefore, there is an urgent need for sensitive and efficient serologic methods to detect GoCV to diagnose the disease.
In this study, we report the establishment and optimization of an indirect ELISA (△Cap-iELISA) detection method that is based on coating a truncated Capsid protein (△Cap) to evaluate the seroprevalence of GoCV. A total of 349 serum samples from geese were tested in the three major goose breeding areas of Guangdong, Shandong, and Fujian provinces. The ELISA developed in this study, is suitable for large-scale clinical detection and thus provides a new method for the early diagnosis of GoCV infection. Furthermore, a notable correlation was observed between age and the positive rate of GoCV antibodies among geese, indicating an escalating trend with advancing age. The utilization of △Cap-iELISA has enriched the seroepidemiological survey data of GoCV in goose flocks, thus holding significant value for determining the current status of the GoCV epidemic and devising targeted prevention and control strategies.
BL21 (DE3) competent cells (TaKaRa, China) and the expression vector pET-30a(+) (Unibio, China) were stored at −20°C in the laboratory. The reference positive and negative sera for GoCV, validated by Western blots of the GoCV △Cap protein, were previously stored in the lab. Additionally, reference goose serum samples for specificity analysis against H5 and H7 subtypes of Avian influenza virus (AIV), Newcastle disease virus (NDV), Goose paramyxovirus (GPMV), Avian reovirus (ARV), Tembusu virus (TMUV) Goose parvovirus (GPV). Sera samples for filed study, were randomly collected from geese breeding-intensive areas in Guangdong, Fujian, and Shandong Provinces between 2023 and 2024. A total of 349 samples were obtained from goslings, meat geese, and breeding geese. There were 239 serum samples from Jiangmen, Zhaoqing, Enping, and Yangjiang in Guangdong, 60 serum samples from Hua’an City in Fujian, and 50 serum samples from Wulong City in Shandong. Serum samples mentioned above were stored at −80°C.
The △Cap gene (37aa-250aa) of strain GoCV/369/GD/2020 (GenBank Accession No. MT831925, referred to as GoCV/369) with the nuclear localization sequence (NLS) removed was synthesized by Genscript (Nanjing, China) using codon optimization software (GenSmart™ Codon Optimization) (the modified sequence is included in the Supplementary material). Two primers with restriction enzyme cutting sites were designed to amplify the synthetic △Cap gene: Cap-NdeI-F (GCTACATATGCACCACCACCACCACCACTCAAAGTACACT-3′) and Cap-HindIII-R (5′-AGCTAAGCTTTCATTACGGCGCCAGGCCGGT-3′). The amplified gene was subsequently cloned into the plasmid pET-30a (+) with a His tag for further expression. The resulting plasmid, termed pET30a-△Cap, underwent verification through PCR, double enzyme digestion using NdeI and HindIII enzymes from New England Biolabs (United States), and sequencing conducted by Genewiz (China).
For protein expression, the pET30a-△Cap plasmid was initially transformed into E. coli BL21 (DE3) competent cells and then cultured on LB solid medium plates with 50 μg/mL of Kanamycin sulfate at 37°C. Five colonies were selected and cultured in LB medium with 50 μg/mL of Kanamycin sulfate and incubated at 37°C until the optical density at 600 nm (OD600 nm) reached 0.6 to 0.7. Following this, 0.5 mM IPTG (isopropyl-β-D-thiogalactopyranoside, Beyotime Biotechnology) was added to induce protein expression. The ultrasonic-disrupted bacterial lysates were then analyzed by 12% SDS-PAGE (sodium dodecyl sulfate-polyacrylamide gel electrophoresis). The optimal concentration of IPTG (0.25, 0.50, 0.75, or 1.00 mM) and induction time (1, 2, 3, 4, 5, 6, 7, and 8 h) were determined through a gradient test based on equal loading quantities of total protein in each group.
The △Cap protein was expressed and identified by Western blotting followed purification using a Ni+-agarose His-tagged protein purification kit (Beyotime, China). The concentration of the purified protein was determined using the BCA method and analyzed by SDS-PAGE. To quantify the expression level of the △Cap protein, Western blotting was performed using a mouse anti-His tag monoclonal antibody (Beyotime, China) as the primary antibody (dilution 1:1000), sheep anti-mouse IgG labeled with HRP (Proteintech, United States) as the secondary antibody (dilution = 1:10000), and BeyoECL Plus (Beyotime, China) as the chromogenic agent.
Two 10-week-old female New Zealand rabbits were selected for the study and divided into an immunization group and a blank control group, and negative serum samples were collected using marginal ear vein blood collection prior to immunization. The experimental rabbits were immunized with purified recombinant protein via subcutaneous multipoint injection at a dose of 1 mg per rabbit. Freund’s complete adjuvant was used for the first immunization, followed by emulsification with Freund’s incomplete adjuvant for the second and third immunizations. Serum samples were collected on the seventh day after each immunization, with subsequent booster shots administered every 10 days. After three rounds of immunization, blood was collected from the heart to obtain a large amount of serum, which was then frozen at −80°C. The prepared rabbit polyclonal antibody served as the primary antibody, while sheep anti-rabbit IgG was used as the secondary antibody for Western blot identification.
A checkerboard titration method was utilized to determine the optimal concentration of the coated antigen and the proper serum dilution (20), Both antigen and serum were added to 96-well plates with 100 μL/well, respectively. Briefly, the purified △Cap protein was diluted with a coating solution to 0.5, 1, 2, 3, and 4 μg /mL and then coated onto a 96-well plate. GoCV-positive and negative serum samples were employed to establish the optimal working conditions at dilutions of 1:50, 1:100, 1:200, and 1:400. The HRP labeled goat anti-Duck IgG (H + L) (KPL, USA) antibody was used at different dilutions (1:1000, 1:2000, 1:5000, and 1:8000). Following color development with TMB (Solarbio, China), the reaction was stopped with ELISA stop solution (Solarbio, China). The absorbance of each well at OD450 nm was measured using a microplate reader. The dilution that exhibited the greatest difference in absorbance of P/N (i.e., positive and negative sera) at OD450 nm was determined as the optimal working dilution for testing experimental serum samples. Thirteen GoCV-negative serum samples that had been prevalidated by Western blotting and PCR were tested using the optimized iELISA reaction conditions to determine the cut-off value. The cut-off value was calculated based on the following formula: cut-off value = mean OD450 nm value +3 standard deviations (21).
Various anti-sera, including GoCV, AIV, NDV, GMPV, ARV, GPV, and TMUV, were employed to evaluate the antigenic cross-reactivity of the △Cap-iELISA. Six serum samples were randomly selected for intra-batch and between-batch replicates, and the mean, standard deviation, and coefficient of variation of each sample were calculated.
A total of 118 serum samples were collected from geese that were suspected of being infected with GoCV (i.e., exhibiting feather growth disorder and dysplasia). The serum samples underwent △Cap-ELISA and Real-time Quantitative PCR assays simultaneously for comparative evaluation of the △Cap-ELISA test. Each sample was tested in triplicate for technical accuracy. The primers used for the Real-time Quantitative PCR detection were qGoCV-F: 5′-GGTCTGCCGATAACTGA-3′ and qGoCV-R: 5′-GGCCGACCAATCAGAACGA-3′.
The information of sera samples for field study was mentioned previously at the begining of this section. The △Cap-iELISA method established in this study was utilized to conduct serological surveys of goose circovirus capsid protein antibodies in the aforementioned regions.
Successful insertion of the optimized ΔCap gene sequence into the prokaryotic expression vector pET-30a (+) was confirmed by double digestion (Figure 1). The recombinant Cap protein was expressed as an inclusion body formation with a partially soluble protein with a molecular weight of approximately 27 kDa, as demonstrated by SDS-PAGE (Figure 2A). After optimizing the induction conditions, the highest yield of the △Cap protein was achieved with 0.5 mM IPTG at 37°C for 5 h (Figures 2B,C). The inclusion body protein was purified using a Ni-Agarose His tag protein purification kit, and this method yielded a purer protein sample compared to gel purification (Figures 3A,B). The immunoreactivity of the △Cap protein was subsequently assessed via Western blotting (Figure 4A). Further Western blot assays using rabbit polyclonal antibodies prepared with the recombinant protein also confirmed the immunogenicity and reactogenicity of the △Cap protein (Figure 4B).
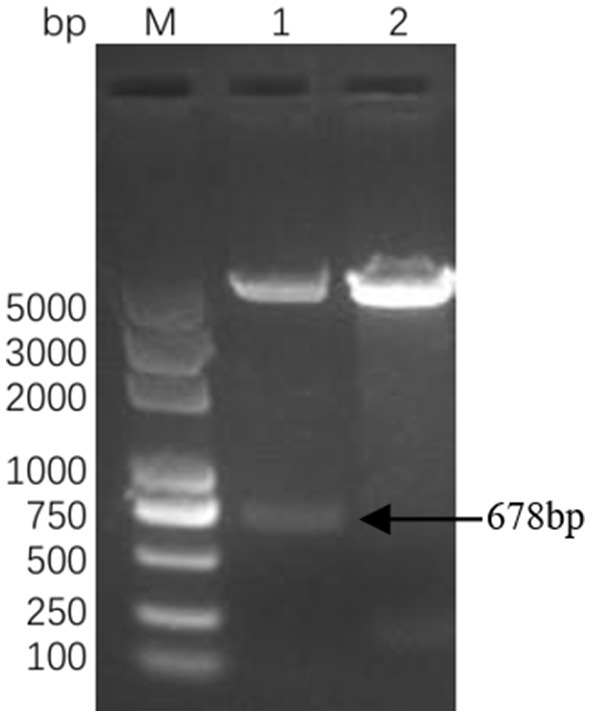
Figure 1. Identification of the recombinant plasmid pET-30a-△Cap. Lane M DL2000 Puls DNA marker; Lane 1 pET-30a-△Cap enzyme double digestion, Lane 2 pET-30a-(+) (NdeI + HindIII).
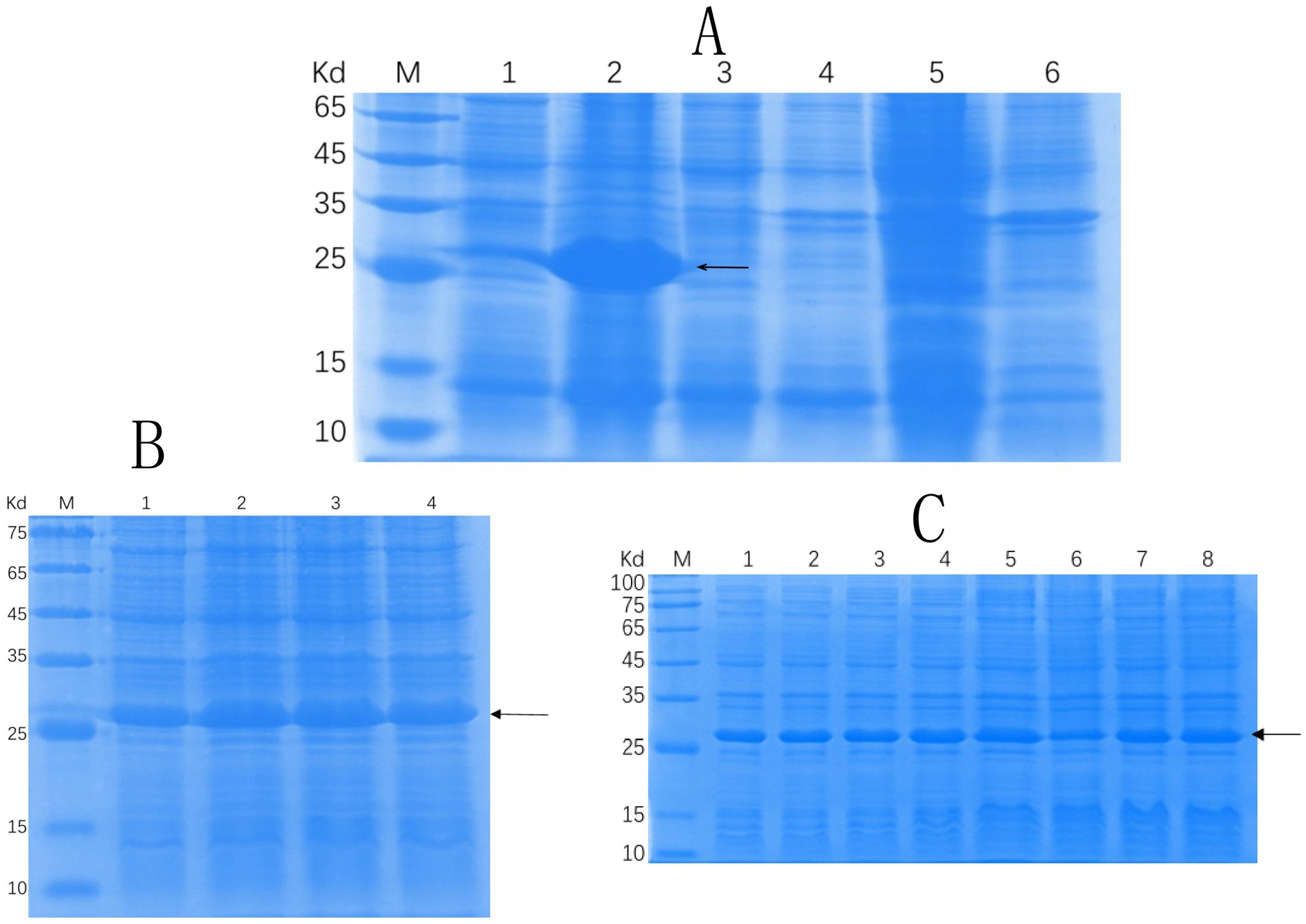
Figure 2. SDS-PAGE analysis of solubility and the optimization of expression conditions of the recombinant protein Cap. (A) SDS-PAGE analysis of △Cap protein solubility. Lane M, protein molecular quality standard. Lanes 1–2, the supernatant and precipitate after crushing and centrifugation of pET30a-△Cap-BL21 (DE3), respectively. Lanes 3–4, the supernatant and precipitate after crushing and centrifugation of pET30a--BL21 (DE3), respectively; Lanes 5–6: the supernatant and precipitate after crushing and centrifugation of BL21 (DE3), respectively. (B) SDS-PAGE analysis of the △Cap protein at different IPTG-induced concentrations. Lane M, protein molecular quality standard. Lanes 1–4, the samples with IPTG concentrations of 0.25, 0.5, 0.75, and 1 mM. (C) SDS-PAGE analysis of the △Cap protein at different induction times. Lane M, protein molecular quality standard. Lanes 1–8: Samples induced for IPTG for 1, 2, 3, 4, 5, 6, 7, and 8 h. The arrows pointed band are the expected protein.
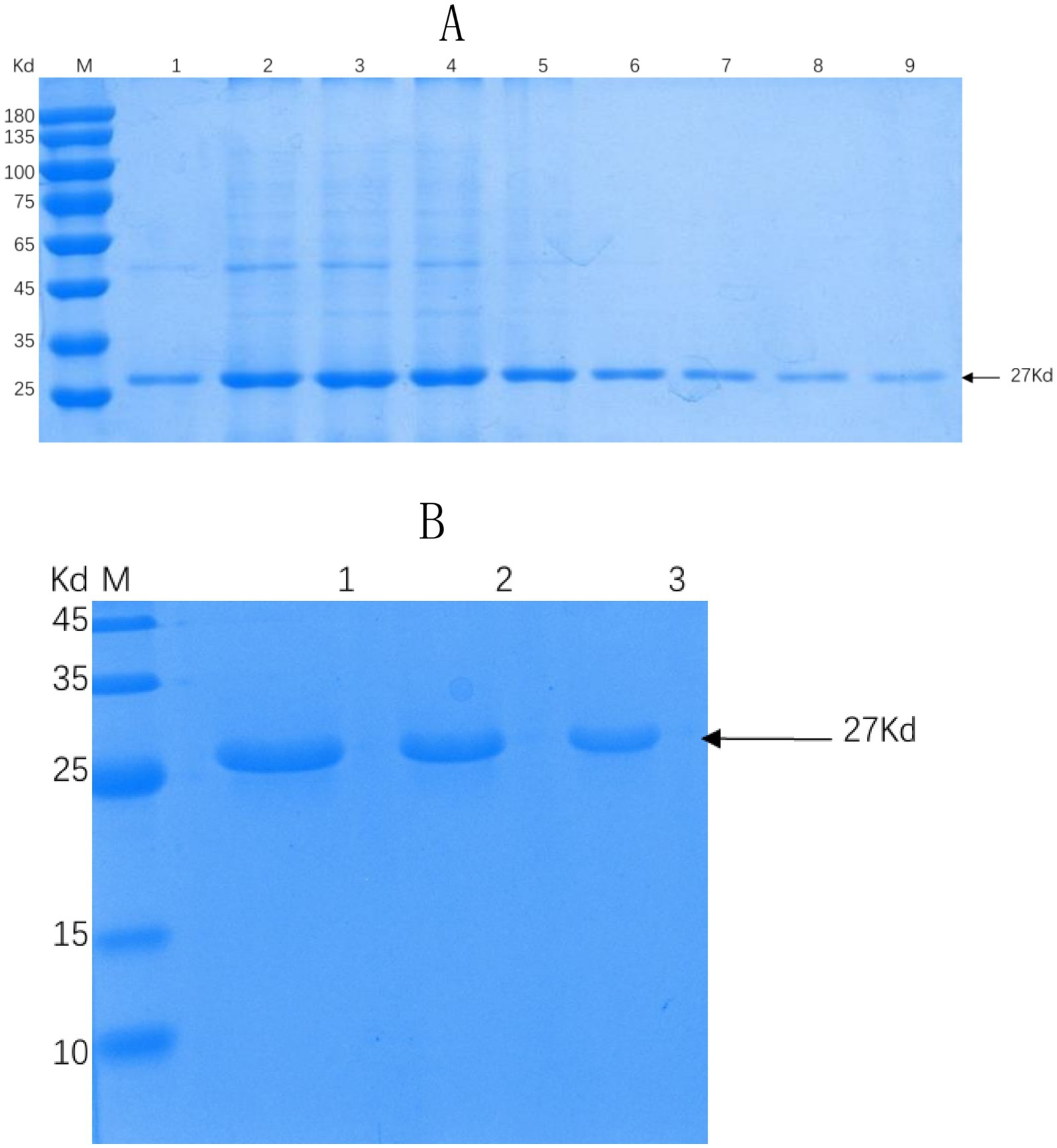
Figure 3. SDS-PAGE analysis of recombinant proteins under different purification conditions. (A) SDS-PAGE analysis of △Cap protein purified by nickel column affinity chromatography. Lane M, protein molecular quality standard. Lanes 1–9, the collected △Cap proteins at different elution concentrations. (B) SDS-PAGE analysis of the △Cap protein purified by gel purification protein. Lane M, protein molecular quality standard. Lanes 1–3, △Cap protein samples of different concentrations were purified by cutting rubber. The arrows pointed band are the expected protein.
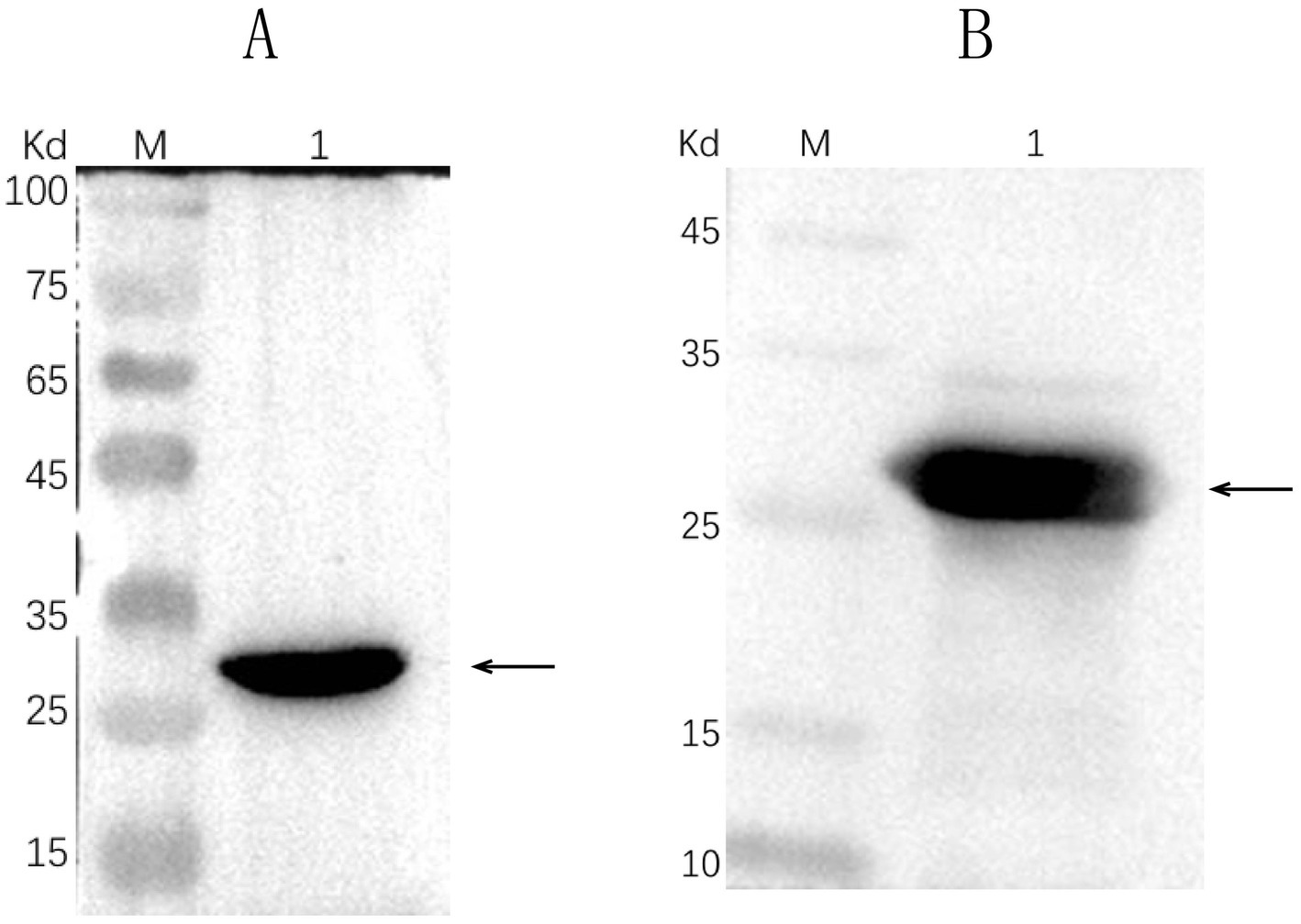
Figure 4. Western blot analysis of the △Cap fusion protein. M, protein molecular weight marker. (A) Lane 1, purified fusion protein blot incubated with mouse anti-His tag monoclonal antibody. (B) Lane 1, purified fusion protein blot incubated with prepared rabbit polyclonal antibody. The arrows pointed band are the expected protein.
The purified △Cap protein was utilized as a diagnostic antigen for the development of an indirect ELISA method. The optimal coating antigen concentration determined by checkerboard titration was 4 μg/mL, with the optimal serum dilution being 1:100 (Table 1). Other optimized reaction conditions included coating the antigen at 37°C for 2 h and using a blocking solution of 2% skim milk powder at 37°C for 1 h. The HRP labeled goat anti-duck IgG (H + L) was diluted to 1:1000. The reaction times for serum, HRP labeled goat anti-duck IgG (H + L), and TMB were set at 60, 30, and 10 min, respectively according to the results of optimal tests (Supplementary Tables S1–S3). The mean OD value at 450 nm in 13 negative goose serum samples was 0.052, with a standard deviation of 0.032. Thus, the cut-off value (mean + 3 SD) for determining the state of serum samples in ELISA was 0.147.
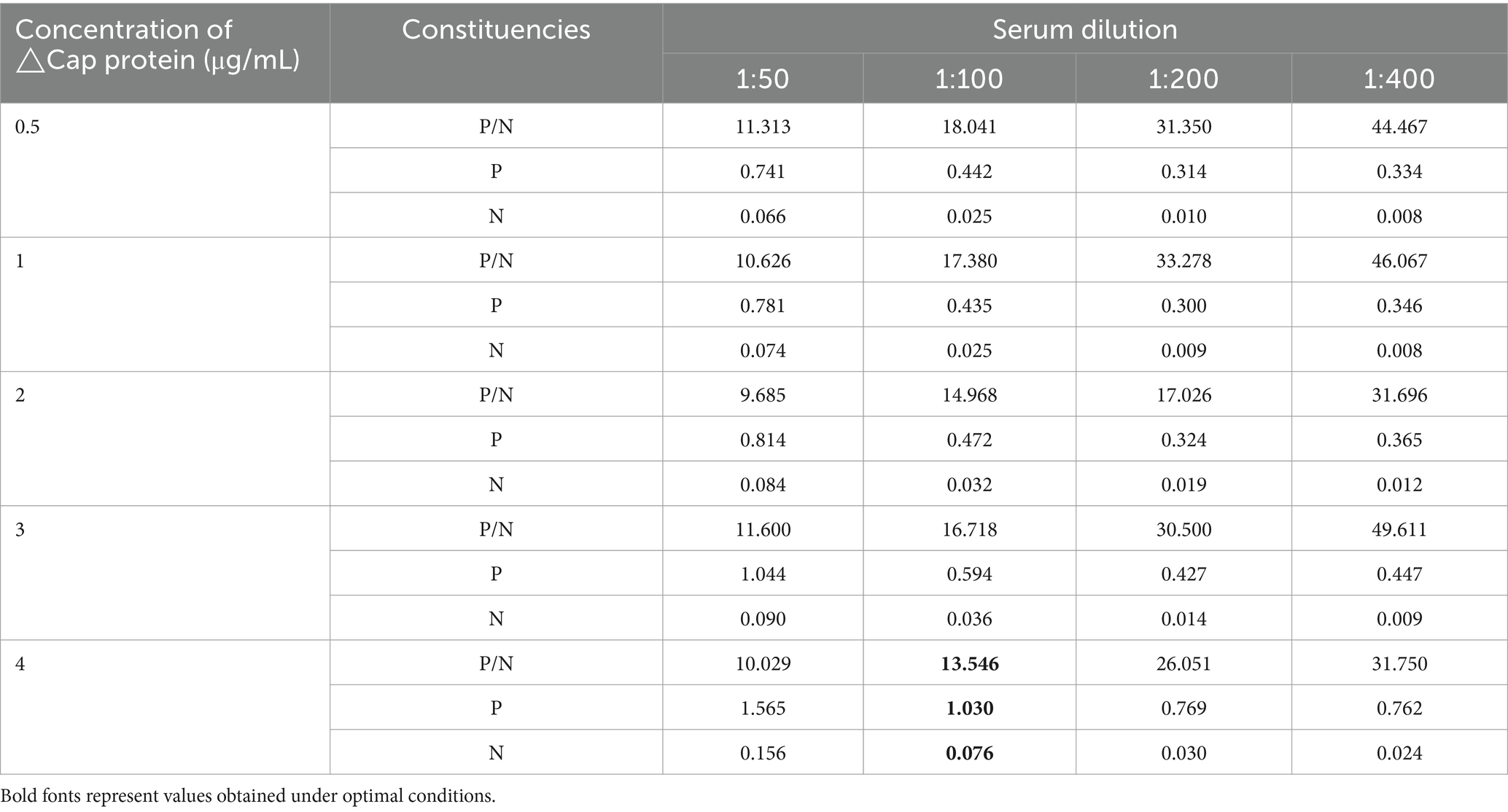
Table 1. The determination of optimal coating conditions and secondary antibody dilution results of △Cap-iELISA.
The ELISA detection method was utilized for the detection of NDV, GPV, GPMV, ARV, AIV(H5), AIV(H7), and TMUV positive serum, Controls for GoCV positive and negative serum were also established. Except for the GoCV-positive serum, the OD450 nm values of the remaining sera below the cut-off value (Figure 5), indicating that the ELISA detection method established in this study did not cross-react with antibodies induced by common pathogens in geese. The established △Cap-iELISA was applied to six serum samples, with a coefficient of variation of the OD450 nm value in the batch ranging from 4.72 to 7.90%, and a coefficient of variation of the inter-batch replicate value ranging from 1.98 to 8.06% (Table 2). All coefficients of variation were below 10%, indicating that the ELISA detection method established in this study did not cross-react with antibodies induced by common pathogens in geese.
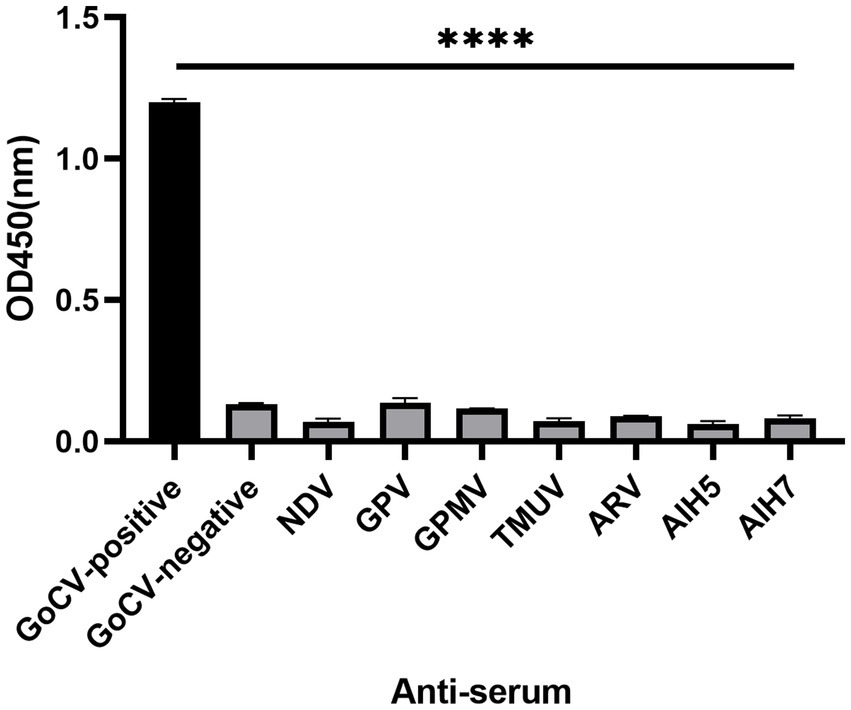
Figure 5. The specificity analysis of △Cap-iELISA against the common pathgens in geese. The reference anti-serum samples included GoCV positive anti-serum (GoCV-positive), GoCV negative anti-serum (GoCV-negative), Newcastle disease virus positive anti-serum (NDV), Goose parvovirus positive anti-serum (GPV), Goose paramyxovirus positive anti-serum (GPMV), Tembusu virus positive anti-serum (TMUV), Avian reovirus positive anti-serum (ARV) and H5 and H7 subtypes of Avian influenza virus positive anti-serum (AIH5, AIH7). **** for p < 0.0001.
One hundred and eighteen serum samples randomly selected from total samples were analyzed using △Cap-iELISA and confirmed by Real-time Quantitative PCR (RT-qPCR). Of these, 78 samples tested positive with △Cap-iELISA, while 48 tested positive by RT-qPCR. The Kappa value of 0.48835 for the two diagnostic methods indicated moderate consistency between the △Cap-iELISA assay and the RT-qPCR assay (Table 3).
A total of 349 clinical serum samples were tested using the △Cap-iELISA method (Table 4). The results revealed that 248 out of 349 samples tested positive for GoCV antibodies, resulting in a high positive rate of 71.06%. Among these, sera from 163 breeding geese were examined, with 150 testing positive for the antibodies, yielding a positive rate of 92.02% (150/163). Additionally, 88 meat goose, day age between 40d and 100d, serum samples were analyzed, and 71 were positive for the antibodies, resulting in a high positive rate of up to 80.68% (71/88). Of the 98 gosling serum samples tested, only 27 were positive for the antibodies. Moreover, the positive rates of GoCV antibodies in breeding geese, meat geese, and gosling in Guangdong province were 96.24% (128/133), 86.20% (50/58), and 33.33% (16/48) respectively. These rates were significantly higher than those observed in Fujian and Shandong provinces at 73.33% (22/30), 70.00% (21/30), and 22.00% (11/50), respectively. The overall positive rate of GoCV antibodies in goose serum samples from Guangdong was higher than in samples from Fujian, and both were higher than in samples from Shandong (81.17, 76.67, and 22%, respectively).
Studying the pathogenesis of GoCV has been challenging due to the lack of virus isolation and culture technology. Currently, research on GoCV primarily focuses on epidemiological investigation. The main methods for detecting GoCV are nucleic acid techniques, including routine PCR (14), nested broad-spectrum PCR (22), TaqMan real-time PCR, competitive PCR, dual PCR, Loop-mediated isothermal amplification (LAMP) (23), dot blot hybridization tests (DBH) (11), in situ hybridization (ISH) (24), and indirect fluorescent assays (IFA) (12). Serological assays, in contrast, are relatively rare. Therefore, it is necessary to establish an effective serological test to monitor GoCV and control the spread of the virus.
As the primary structural protein of GoCV, the cap protein contains five main B-cell antigenic epitopes (16) that may inducing an immune response and eliciting antibodies against the Cap protein in the body. Serological tests based on a recombinant capsid protein could serve as a valuable tool for epidemiological investigation. Studies have demonstrated that the entire Cap gene could not be expressed in E. coli due to the presence of NLS (25). Therefore, similar to beak feather disease virus (BFDV) (26), pigeon circovirus (PiCV) (27), and duck circovirus (DuCV) (28), We prepared truncated GoCV coat protein prokaryotic expression vectors, and subsequently, rabbit polyclonal antibodies were generated from the gel-purified proteins and their reactivity was assessed by Western blot analysis. The results showed that the purified target proteins maintained acceptably good immunogenicity and reactivity.
The ELISA detection method offers several advantages, including high sensitivity, strong specificity, good repeatability, and fast and convenient operation (29). Importantly, only a small volume of goose serum is required for detection. Our established ELISA method can complete antibody detection within 2 h when using the coated antigen, allowing for rapid results. Additionally, GoCV is often found alongside multiple pathogens such as TMUV and GPV (6, 18, 29); therefore, an assessment of cross-reactivity is necessary. The OD450 nm values of all groups except for GoCV antibodies were below the cut-off value, indicating the relatively good specificity of this △Cap-iELISA for GoCV antiserum (Figure 5). In comparison to the qPCR assay, the kappa value was 0.48835, indicating moderate consistency (Table 3). Repeatability tests also demonstrated that this method has good repeatability and is suitable for large-scale clinical trials.
The positive rate of GoCV antibodies in breeding and meat geese in Guangdong Province was significantly higher than that in Fujian Province. The positive rate of GoCV antibodies in goslings was also higher in Guangdong Province compared to Shandong Province. Overall, the positive rate of GoCV antibodies in goose serum samples from Guangdong Province is higher than that in Fujian Province, which is further higher than that in Shandong Province. Admittedly, due to the variation in the difficulty of sampling among different regions, the number of serum samples collected from geese at each age stage was not the same. However, even when the number of serum samples collected from goslings in Guangdong and Shandong was equal, the positive rate of GoCV antibodies remained higher in gosling serum from Guangdong compared to Shandong. This suggests a higher probability of goose flocks being infected with GoCV in Guangdong compared to other provinces. A similar trend of infection was observed among different goose breeds and age groups across Guangdong, Fujian, and Shandong provinces. The positive rate of GoCV serum antibodies increased gradually with the age of the geese. This finding aligns with research conducted by Scott et al. indicating that susceptibility to infection varies among geese of various breeds and age groups. In addition, our test detected a seroprevalence rate exceeding 80% (194/239) for geese from Guangdong, while our previous studies reported nucleic acid positivity rates exceeding 50% for goose flocks within this region (6). This indicates that GoCV infection is widely prevalent in Guangdong Province, China and that it is more severe compared to other provinces, even becoming an endemic disease among the geese raised in Guangdong. The above results suggest that △Cap-ELISA can be applied for large-scale clinical serological diagnosis of GoCV. Furthermore, China is the world’s largest goose producer. Given the high prevalence of GoCV infection in some parts of China, combined with the economic losses from growth retardation and feather dysplasia caused by GoCV infection in geese as well as the difficulties in disease prevention and control caused by co-infection and immunosuppression, it is necessary to develop effective diagnostic tests.
In conclusion, the △Cap-iELISA method has been successfully employed for the detection of antibodies to GoCV in geese. The method demonstrated high sensitivity, specificity, and reproducibility. The serological data of GoCV in goose flocks from Guangdong, Fujian, and Shandong were obtained through the detection of clinical samples. This study lays the groundwork for seroepidemiological investigations in these areas and provides valuable reference data for establishing standardized control measures.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary material.
The animal study was approved by Foshan University Committee for Animal Experiments. The study was conducted in accordance with the local legislation and institutional requirements.
JiaC: Data curation, Investigation, Methodology, Writing – original draft. WX: Methodology, Writing – review & editing, Investigation, Project administration. ZY: Methodology, Writing – review & editing, Data curation, Formal analysis. CW: Writing – review & editing. WZ: Data curation, Investigation, Methodology, Writing – review & editing. HW: Data curation, Investigation, Methodology, Writing – review & editing. JZ: Conceptualization, Supervision, Writing – review & editing. YT: Investigation, Writing – review & editing. RL: Investigation, Writing – review & editing. JidC: Conceptualization, Funding acquisition, Supervision, Writing – original draft, Writing – review & editing.
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This study was supported by the Young Scientists Fund of National Natural Science Foundation of China (NSFC) (31800141); the Natural Science Foundation of Guangdong Province, China (2017A030310573); the Rural Revitalization Strategy Special Foundation of Guangdong Province (Rural Science and Technology Commissioner Program) (163-2019-XMZC-0009-02-0075); the Science and Technology Innovation Program of Gaoming District, Foshan City (2017C01) and the Collaboration Project with WANMUZHOU Biotechnology Limited Company (GDKTP2021002400, kh23279). The authors declare that this study received funding from WANMUZHOU Biotechnology Limited Company. The funder was not involved in the study design, collection, analysis, interpretation of data, the writing of this article, or the decision to submit it for publication.
WZ was employed by WANMUZHOU Biotechnology Limited Company. HW was employed by Shenzhen Kingkey Smart Agriculture Times Co., Ltd.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The handling editor CW declared a past co-authorship with the author RL.
The authors declare that no Gen AI was used in the creation of this manuscript.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fvets.2025.1521705/full#supplementary-material
1. Todd, D, Weston, JH, Soike, D, and Smyth, JA. Genome sequence determinations and analyses of novel circoviruses from goose and pigeon. Virology. (2001) 286:354–62. doi: 10.1006/viro.2001.0985
2. Todd, D, Weston, J, Ball, NW, Borghmans, BJ, Smyth, JA, Gelmini, L, et al. Nucleotide sequence-based identification of a novel circovirus of canaries. Avian Pathol. (2001) 30:321–5. doi: 10.1080/03079450120066322
3. Breitbart, M, Delwart, E, Rosario, K, Segalés, J, Varsani, A, and Ictv, RC. ICTV virus taxonomy profile: Circoviridae. J Gen Virol. (2017) 98:1997–8. doi: 10.1099/jgv.0.000871
4. Rosario, K, Breitbart, M, Harrach, B, Segalés, J, Delwart, E, Biagini, P, et al. Revisiting the taxonomy of the family Circoviridae: establishment of the genus Cyclovirus and removal of the genus Gyrovirus. Arch Virol. (2017) 162:1447–63. doi: 10.1007/s00705-017-3247-y
5. Mankertz, A, and Hillenbrand, B. Replication of porcine circovirus type 1 requires two proteins encoded by the viral rep gene. Virology. (2001) 279:429–38. doi: 10.1006/viro.2000.0730
6. Chen, J, Wang, H, Pei, H, Wang, J, Wu, H, Zhong, J, et al. The prevalence, coinfection, and evolutionary and molecular characteristics of prevalent goose circovirus in Guangdong. China Avian Dis. (2021) 65:559–71. doi: 10.1637/aviandiseases-D-21-00045
7. Zahn, K. Overexpression of an mRNA dependent on rare codons inhibits protein synthesis and cell growth. J Bacteriol. (1996) 178:2926–33. doi: 10.1128/jb.178.10.2926-2933.1996
8. Yang, C, Xu, Y, Jia, R, Li, P, Zhang, L, Wang, M, et al. Prokaryotic expression of a codon-optimized capsid gene from duck circovirus and its application to an indirect ELISA. J Virol Methods. (2017) 247:1–5. doi: 10.1016/j.jviromet.2017.05.003
9. Zhou, JY, Shang, SB, Gong, H, Chen, QX, Wu, JX, Shen, HG, et al. In vitro expression, monoclonal antibody and bioactivity for capsid protein of porcine circovirus type II without nuclear localization signal. J Biotechnol. (2005) 118:201–11. doi: 10.1016/j.jbiotec.2005.02.017
10. Soike, D, Kohler, B, and Albrecht, K. A circovirus-like infection in geese related to a runting syndrome. Avian Pathol. (1999) 28:199–202. doi: 10.1080/03079459994939
11. Ball, NW, Smyth, JA, Weston, JH, Borghmans, BJ, Palya, V, Glávits, R, et al. Diagnosis of goose circovirus infection in Hungarian geese samples using polymerase chain reaction and dot blot hybridization tests. Avian Pathol. (2004) 33:51–8. doi: 10.1080/03079450310001610613
12. Scott, AN, Beckett, A, Smyth, JA, Ball, NW, Palya, V, and Todd, D. Serological diagnosis of goose circovirus infections. Avian Pathol. (2006) 35:495–9. doi: 10.1080/03079450601087841
13. Kozdruń, W, Woźniakowski, G, Samorek-Salamonowicz, E, and Czekaj, H. Viral infections in goose flocks in Poland. Pol J Vet Sci. (2012) 15:525–30. doi: 10.2478/v10181-012-0080-9
14. Chen, CL, Chang, PC, Lee, MS, Shien, JH, Ou, SJ, and Shieh, HK. Nucleotide sequences of goose circovirus isolated in Taiwan. Avian Pathol. (2003) 32:165–71. doi: 10.1080/0307945021000071614
15. Yu, XP, Zheng, XT, He, SC, Zhu, JL, and Shen, QF. Cloning and analysis of the complete genome of a goose circovirus from Yongkang Zhejiang. Acta Microbiol Sin. (2005) 45:860–4. doi: 10.13343/j.cnki.wsxb.2005.06.009
16. Chen, J, Chen, G, Niu, S, Zhu, W, Zhang, Y, Liu, M, et al. Epidemiological investigation and genome analysis of goose circovirus in Guangdong of China. China Poult. (2020) 42:97–102. doi: 10.16372/j.issn.1004-6364.2020.04.018
17. Ting, CH, Lin, CY, Huang, YC, Liu, SS, Peng, SY, Wang, CW, et al. Correlation between goose circovirus and goose parvovirus with gosling feather loss disease and goose broke feather disease in southern Taiwan. J Vet Sci. (2021) 22:e1. doi: 10.4142/jvs.2021.22.e1
18. Yu, X, Zhu, C, Zheng, X, He, S, and Liu, X. Genome analysis and epidemiological investigation of goose circovirus detected in eastern China. Virus Genes. (2007) 35:605–9. doi: 10.1007/s11262-007-0112-1
19. Yang, KK, Yin, DD, Xu, L, Liang, YQ, Tu, J, Song, XJ, et al. A TaqMan-based quantitative real-time PCR assay for identification of the goose circovirus. Mol Cell Probes. (2020) 52:101564. doi: 10.1016/j.mcp.2020.101564
22. Halami, MY, Nieper, H, Müller, H, and Johne, R. Detection of a novel circovirus in mute swans (Cygnus olor) by using nested broad-spectrum PCR. Virus Res. (2008) 132:208–12. doi: 10.1016/j.virusres.2007.11.001
23. Woźniakowski, G, Kozdruń, W, and Samorek-Salamonowicz, E. Loop-mediated isothermal amplification for the detection of goose circovirus. Virol J. (2012) 9:110. doi: 10.1186/1743-422x-9-110
24. Smyth, J, Soike, D, Moffett, D, Weston, JH, and Todd, D. Circovirus-infected geese studied by in situ hybridization. Avian Pathol. (2005) 34:227–32. doi: 10.1080/03079450500112419
25. Xu-ping, Y, Dong-jian, H, Xin-tian, Z, Chun, Z, Xiao-ning, L, Hong-tao, Y, et al. Expression of the recombinant capsid protein of goose circovirus in E.Coli system. Chinese J Prev Vet Med. (2008) 16:534–6. doi: 10.3724/SP.J.1011.2008.00534
26. Johne, R, Raue, R, Grund, C, Kaleta, EF, and Müller, H. Recombinant expression of a truncated capsid protein of beak and feather disease virus and its application in serological tests. Avian Pathol. (2004) 33:328–36. doi: 10.1080/0307945042000220589
27. Daum, I, Finsterbusch, T, Härtle, S, Göbel, TW, Mankertz, A, Korbel, R, et al. Cloning and expression of a truncated pigeon circovirus capsid protein suitable for antibody detection in infected pigeons. Avian Pathol. (2009) 38:135–41. doi: 10.1080/03079450902737797
28. Liu, SN, Zhang, XX, Zou, JF, Xie, ZJ, Zhu, YL, Zhao, Q, et al. Development of an indirect ELISA for the detection of duck circovirus infection in duck flocks. Vet Microbiol. (2010) 145:41–6. doi: 10.1016/j.vetmic.2010.03.010
Keywords: goose circovirus, capsid protein, serological detection methods, prokaryotic expression, indirect ELISA
Citation: Chen J, Xue W, Yao Z, Wang C, Zhu W, Wang H, Zhang J, Tang Y, Liu R and Chen J (2025) Epidemiological investigation of goose circovirus based on a newly developed indirect ELISA method. Front. Vet. Sci. 12:1521705. doi: 10.3389/fvets.2025.1521705
Received: 02 November 2024; Accepted: 24 January 2025;
Published: 13 February 2025.
Edited by:
Chunhe Wan, Fujian Academy of Agricultural Sciences, ChinaReviewed by:
Yang Zhan, Hunan Agricultural University, ChinaCopyright © 2025 Chen, Xue, Yao, Wang, Zhu, Wang, Zhang, Tang, Liu and Chen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jidang Chen, amlkYW5nY2hlbkBmb3N1LmVkdS5jbg==
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.