
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Vet. Sci., 27 January 2025
Sec. Parasitology
Volume 12 - 2025 | https://doi.org/10.3389/fvets.2025.1521055
 Yun Zhang1,2†
Yun Zhang1,2† Guangxu Ren1,3†
Guangxu Ren1,3† Qingqing Lu1†
Qingqing Lu1† Jiaqi Li1,4†
Jiaqi Li1,4† Yu Qiang1
Yu Qiang1 Youyou Li1
Youyou Li1 Xiuyi Lai1
Xiuyi Lai1 Yuan Wang1
Yuan Wang1 Xingyue Yu1
Xingyue Yu1 Sheng Lei1
Sheng Lei1 Yu Li4
Yu Li4 Yunxing Chang5
Yunxing Chang5 Xianrong Liu5
Xianrong Liu5 Xuning Qi6
Xuning Qi6 Zhi Xie6
Zhi Xie6 Tingting Li1
Tingting Li1 Jiang Du1
Jiang Du1 Rui Duan1
Rui Duan1 Xinyu Chang1
Xinyu Chang1 Hesheng Wang5,7*
Hesheng Wang5,7* Gang Lu1,2*
Gang Lu1,2*Introduction: Enterocytozoon bieneusi is one of the most frequent microsporidia species causing digestive disorder mainly diarrhea in humans and animals. Eld’s deer (Rucervus eldii) is the class I national key protected wildlife and only distributed on Hainan Island in China. No report on the prevalence and molecular characterization of E. bieneusi in wild Eld’s deer worldwide.
Methods: 217 fecal samples were collected from Eld’s deer in two isolated habitats of a nature reserve in Hainan, and examined by nested Polymerase Chain Reaction (PCR) targeting the internal transcribed spacer (ITS) region.
Results and discussion: The overall prevalence of E. bieneusi in Eld’s deer was 17.5% (38/217), with 13.5% (12/89) and 20.3% (26/128) in habitats 1 and 2, respectively. Seven ITS genotypes were identified, including five known genotypes: D (n = 19), Peru11 (n = 10), EbpC (n = 5), Peru8 (n = 1) and Type IV (n = 1), and two novel genotypes: HNED-I and HNED-II (one each). Genotypes Peru8 and Peru11 were firstly identified in cervids. Phylogenetic analysis showed that all the detected genotypes belonged to zoonotic Group 1. The results implied that the further research on threaten of E. bieneusi to endangered Eld’s deer and potential risks for public health is necessary.
Microsporidia are widely spread obligate intracellular pathogens that infect a broad range of hosts, including both vertebrates, such as humans, and invertebrates (1, 2). There are about 220 genera and 1,700 species of microsporidia, which are classified based on their ultrastructural features, developmental cycle, host–parasite relationship, and molecular analysis (3). Of the 17 microsporidian species known to infect humans, Enterocytozoon bieneusi is by far the most frequent species in the clinical setting and generally presents as chronic diarrhea and wasting syndrome, particularly in immunocompromised individuals such as those with AIDS or transplant recipients, as well as travelers, children, and the elderly (4–6). It was transmitted by fecal-oral route, mainly by ingestion of contaminated food and water with spores (7–9). Due to the difficulty of microscopic identification for small size, E. bieneusi is mainly detected and genotyped by the method of nested polymerase chain reaction (PCR) targeted internal transcribed spacer (ITS) region and sequence analysis (10). To date, around 900 different genotypes of E. bieneusi have been identified and classed into 13 phylogenetic groups (group 1–13) (11). The first two clusters (Groups 1 and 2) accounted for a significant proportion (94%) of the total genotypes, encompassing the majority of known human-pathogenic genotypes and zoonotic genotypes (12). Group 3–13 were host adaptation groups and might be present in specific hosts and wastewater (5, 12).
Eld’s deer (Rucervus eldii) is a rare and globally endangered tropical deer species, belonging to Artiodactyla, Family Cervidae and Subfamily Cervinae. It is distributed across Southeast Asia, Southern China and the northeastern part of India. Because of illegal poaching and severe habitat encroachment, the global population of Eld’s deer has sharply declined (13). It has been listed in Appendix I of the Convention on International Trade in Endangered Species of Wild Fauna and Flora (CITES) and classified as endangered on the Red List of Threatened Species by the International Union for Conservation of Nature (IUCN) and the class I national key protected wildlife in China (14–17). In China, Eld’s deer is only distribute in Hainan Island. Due to the rapid destruction of habitats and intense hunting by humans, only 26 individuals was remained in Hainan at end of 1970s (18). Despite fact that the Eld’s deer population has recovered and grown after over 40 years of development and preservation, it continues to be extremely vulnerable to extinction because of inbreeding, poor genetic diversity, the diminishing evolutionary capacity of tiny populations, high population density, and infectious diseases (19). At present, no information about E. bieneusi in endangered wild Eld’s deer was reported. The aims of this study were to investigate the prevalence and molecular characterization of E. bieneusi in wild Eld’s deer in Hainan, and provide valuable information for development and preservation of this endangered wildlife.
The collection of fecal samples from Eld’s deer have been permitted by Hainan Bangxi Provincial Nature Reserve without human disturbance to the animals. The non-invasive sampling strategy did not involve hunting or otherwise manipulating the experimental animals.
From March to August 2021, a total of 217 fresh fecal samples were collected from wild Eld’s deer in two completely isolated areas of Hainan Bangxi Provincial Nature Reserve: Habitat 1 (n = 89) and Habitat 2 (n = 128) (Figure 1). Fresh specimens (approximately 20 g) were immediately collected in sterilized 5-mL tubes with the assistance of experienced staff of the nature reserve, after observing the leaving of Eld’s deer. Each collected fecal sample should be kept more than 3 m apart to ensure that they were not from the same deer, and temporarily stored in a refrigerated insulated tank. All the samples were taken back to the laboratory for storage at −80°C until analysis.
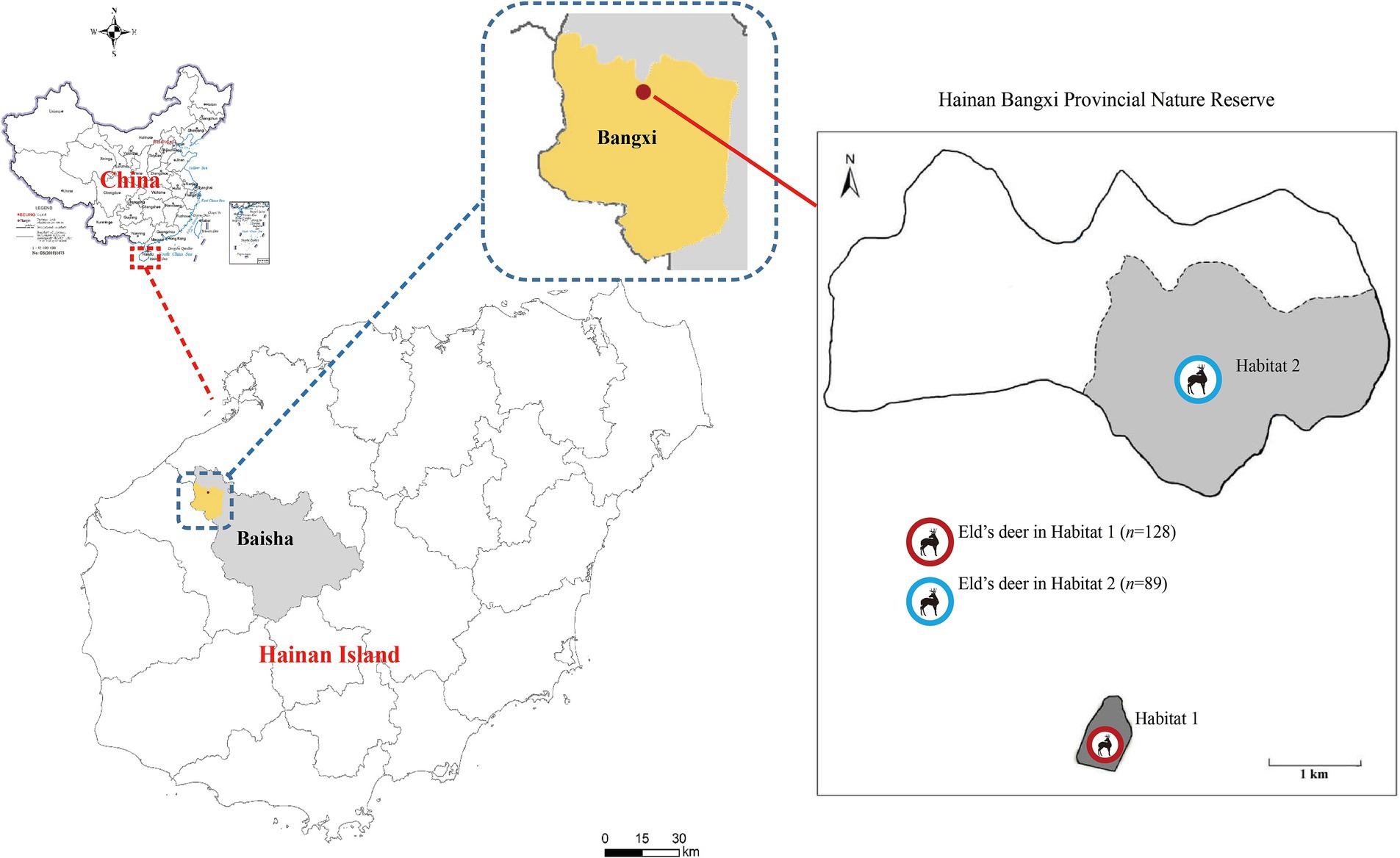
Figure 1. Distribution of sampling sites of Eld’s deer in the Hainan Bangxi Provincial Nature Reserve in the present study.
Fecal samples were washed with distilled water and centrifuged at 1500×g for 10 min. This process was repeated three times. Genomic DNA was extracted directly from 200 mg of each processed fecal specimen using the QIAamp DNA stool mini kit (Qiagen, Hilden, Germany). The extraction procedure adhered to the manufacturer’s recommended protocol, with an elevated lysis temperature of 95°C to guarantee a high DNA yield. The extracted DNA was stored at −20°C until PCR analysis.
To assess the prevalence and genotypes of E. bieneusi, nested PCR assays were used to amplify a 390 bp fragment encompassing the ITS region as described in primers previously reported (20). Each PCR run included a positive control with DNA of the E. bieneusi BEB6 genotype from goat and a negative control (reagent-grade water without DNA). All the secondary PCR products were run on a 1.5% agarose gel and visualized by staining the gel with Goldenview.
Secondary PCR products of positive samples were sequenced in both directions using Big Dye Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems, USA) and an ABI PRISM 3730 XL DNA Analyzer (Thermo Fisher Scientific, Waltham, MA, USA). Sequence accuracy was verified through bidirectional sequencing. The obtained nucleotide sequences were aligned with each other and compared to the reference sequences downloaded from GenBank using the Basic Local Alignment Search Tool (BLAST)1 and ClustalX 1.832 in order to determine the genotypes. According to the established nomenclature system, the nucleotide sequences of the ITS region identical to known genotypes were given the first published name; the nucleotide sequences with single nucleotide substitutions, deletions, or insertions as compared to the known ITS genotypes were considered novel genotypes (21). Meanwhile, the novel genotypes were confirmed by sequencing another two separate PCR products of the same preparations.
A phylogenetic analysis was performed using the Neighbor-joining (NJ) method as implemented in MEGA 7,3 which was calculated by the Kimura 2-parameter model with 1,000 bootstrap replicates. The nucleotide sequences representative of the present study have been deposited in the GenBank database, with the corresponding accession numbers of OL603973 and OL603974 for E. bieneusi.
Statistical analysis were performed using Statistical Package for the Social Sciences (SPSS) version 22.0 (SPSS Inc., Chicago, IL, USA). Chi-square analysis was performed to compare the prevalence of E. bieneusi among different areas. The difference was considered statistically significant when the p < 0.05.
The overall prevalence of E. bieneusi in Eld’s deer was 17.5% (38/217) in this study. Specifically, the infection rates were 13.5% (12/89) in Habitat 1, and 20.3% (26/128) in Habitat 2 (Table 1). There was no significant differences in infection rates between the two completely independent areas under investigation (p > 0.05).
Seven genotypes were obtained from ITS sequencing of 38 E. bieneusi isolates, including five known genotypes: genotype D (n = 19), Peru 11 (n = 10), EbpC (n = 5), Peru 8 (n = 1) and Type IV (n = 1), and two novel genotypes: HNED-I (n = 1) and HNED-II (n = 1). Notably, the detected genotypes were different between two completely isolated habitats of Eld’s deer. The genotypes Peru 11, HNED-I and HNED-II were all detected in samples from Habitat 1, but the genotypes D, EbpC, Peru 8 and Type IV were all detected in samples from Habitat 2 (Table 1). The phylogenetic analysis of the ITS region of E. bieneusi divided the genotypes, which were identified in Eld’s deer in this study, all into Group 1 (Figure 2).
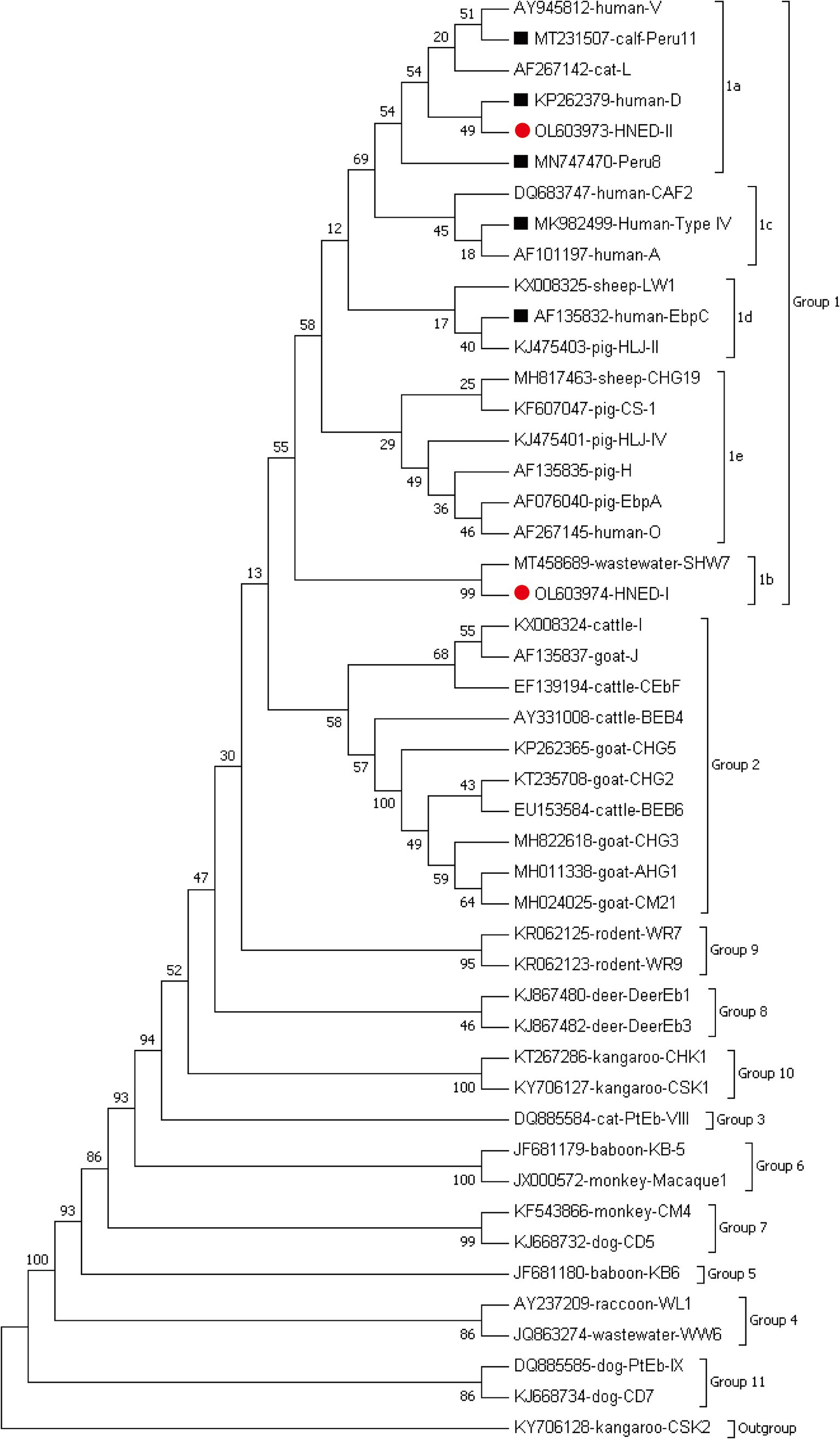
Figure 2. Phylogenetic relationships of representative sequences for the ITS genotypes of E. bieneusi identified from Eld’s deer in present study with reference sequences using maximum likelihood analysis. The known and novel genotypes identified in this study were indicated by black squares (■) and red circles(●), respectively. Genotype CSK2 from white kangaroo (KY706128) is used as the outgroup.
Among the 38 recognized sequences, two were novel and labeled as genotypes HNED-I (GenBank accession no: OL603974) and HNED-II (GenBank accession no: OL603973). Genotype HNED-I exhibited 97.53% similarity with genotype SHW7 (MT458689) from urban wastewater in China, and has four nucleotide substitutions at positions 128 (T → C), 198 (T → G), 218 (A → G) and 232 (C → G). Compared to genotype D (MN704918) from donkeys in China, genotype HNED-II exhibited 99.18% similarity and has two nucleotide substitutions at positions 3 (A → G) and positions 217 (G → A) (Table 2).
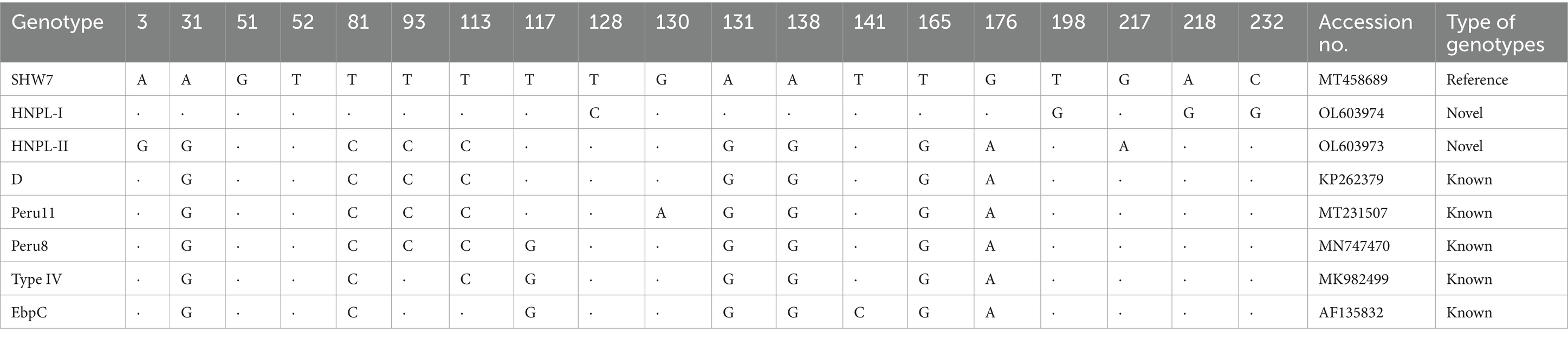
Table 2. Positions of nucleotide changes of known and novel genotypes of E. bieneusi isolates in present study.
To date, there have been near 20 reports on the molecular epidemiological research of E. bieneusi involving 13 cervid species worldwide, and the infection rates varied from 0 to 75.0% (Table 3). In present study, the overall prevalence of E. bieneusi in wild Eld’s deer in Hainan was 17.5%, which was higher than infection rate of captive Eld’s deer (14.3%) (22), sika deer (5.7–16.0%) (9, 22–24), red deer (6.8–8.3%), Siberian roe deer (11.1%) (25) and free-ranging Chinese water deer (7.5%) in China (23), wild red deer (1.5%) in Spain (26), Sambar deer (4.8%) in Australia (27) and white-tailed deer (12.2%) in the USA (28). However, it was considerably lower than the prevalence in captive hog deer (75.0%) (29), fallow deer (27.3%) (23), sika deer (28.6–44.1%), and red deer (20.0–37.5%) (29–31), free-ranging and wild Père David’s deer (24.5–35.2%) (23, 32–34) in China, wild Korean water deer (53.6%) in Korea (35), and white-tailed deer (32.5%) in the USA (36). Notably, the infection rate of E. bieneusi in wild Eld’s deer in this study not only was similar to those in wild reindeers (16.8%) (34) and captive sika deer (17.8%) in China (9), captive red deer (19.4%) in Spain (26), but also in the average rate of cervid species in China (19.3%) (37) and around the world (19.7%) (95% CI: 0.021–0.310, I2 = 97.651%, p = 0.001, Table 3). The different infection rates of E. bieneusi in cervids not only were significantly associated with deer species (23), but also were influenced by various living conditions, biogeographic distributions, age, susceptibilities and health status of individuals (9, 20, 29).
At present, a total of 100 ITS genotypes of E. bieneusi with high genotypic heterogeneity and phenotypic diversity have been identified in cervid species, including 61 genotypes in Group 1, 38 genotypes in Group 2 and one in Group 3 (Table 3). Genotypes HLJD-V and BEB6 were the most popular genotypes in deer from China, and many other genotypes also have been detected in deer from Australia, Korea, Spain and the USA, such as D, MWC_d1, J, Korea-WL-, WL-, CHN- and JLD- associated genotypes. Many genotypes in Groups 1 and 2 have been previously discovered both in humans and animals, which implied that E. bieneusi might be spread from deer to humans (Table 3). In our research, 7 distinct genotypes were identified, including five known (D, EbpC, Peru11, Peru8 and Type IV) and two novel genotypes (HNED-I and HNED-II) (Table 1). All genotypes of were categorized into Group 1 (Figure 1). This result indicates a possible risk of zoonotic transmission, where these genotypes could potentially pass from Eld’s deer to humans. Genotype D was the most prevalent genotypes in Eld’s deer with the rate of 50.0% (19/38), which was similar to the results of previous studies on wild Korean water deer (35). Genotype D also were identified in wild Sambar deer in Australia (27), free-ranging Père David’s deer (Elaphurus davidianus) (34) and captive Sika deer (31) in China. Genotype D was known as the most prevalent zoonotic genotype and not only distributed in humans but also in livestock (sheep, goat, cattle, and pig), companion animals (cat and dog), wild animals (wild boar, wild deer, non-human primates, and tiger), and water sources worldwide (12). Genotypes Peru11, EbpC, Peru8 and Type IV have been frequently observed in humans and various animal hosts, including nonhuman primates, domesticated animals, and avian species (11, 38). To our knowledge, genotypes Peru11 and Peru8 have not been documented in deer previously. This work represented the initial detection of these two genotypes in cervid species, broadening their recognized range of hosts. Genotype EbpC has been detected in wild Père David’s deer (32) and captive Sika deer in China (9, 24, 31). Remarkably, genotypes Peru8 and EbpC have been reported in diarrheic livestocks, and genotype EbpC was the main genotype and demonstrating higher genetic diversity than others in diarrheic pigs in China (39–42), which implied that these 2 genotypes might be associated with intestinal disease in artiodactyl animals, including deer. Genotype Type IV was dominant genotype in wild Père David’s deer in Henan, China (32), which also was identified in wild Sambar deer in Australia (27) and Red deer in Spain (26). In our study, the novel genotype HNED-I showed the highest match (97.53% identity) with E. bieneusi genotype SHW7, obtained from urban wastewater in China in 2020 (43). Genotype SHW7 also has been found in civets and bamboo rats in Hainan (44, 45), and wild rats in Zhejiang, China (46). The novel genotype HNED-II exhibited 99.18% similarity with genotype D, obtained from donkeys in China in 2020 (47).
Despite no significant difference between infection rates of E. bieneusi in Eld’s deer from two completely isolated habitats, the ITS genotypes carried by Eld’s deer in perfectly independent habitats were rather different. Genotypes Peru11, HNED-I and HNED-II were detected in samples from Habitat 1, but genotypes D, EbpC, Peru8 and Type IV were identified from Habitat 2 in the nature reserve. Moreover, the genotype HNED-III was identified in captive Eld’s deer in Hainan Tropical Wildlife Park in our previous research (22). The similar results were found in research on E. bieneusi in Père David’s deer from Henan, Hubei and Beijing (23, 32, 33), and in giant pandas from Sichuan and Shaanxi in China (30, 48). These data suggest that the difference among genotypes of E. bieneusi in the same animal species may be related to living status, habitant environment and sources of infection. Currently, there were no reports on direct evidence of deer’s diarrhea caused by E. bieneusi, but it was crucial to persistently observe and comprehend the epidemiology of E. bieneusi in endangered Eld’s deer to acquire a more profound comprehension of its transmission patterns and prospective consequences on health and survival of Eld’s deer.
In summary, E. bieneusi infection was detected in wild globally endangered Eld’s deer for the first time. Seven ITS genotypes were identified and all belonging to zoonotic Group 1. The discovery of novel genotypes HNED-I and HNED-II offered more genetic diversity of E. bieneusi. Genotypes Peru11 and Peru8 were first identified in cervids in this study. The future studies should systematically focus on revealing the biological characteristics of E. bieneusi and assessing its potential threats to public health, veterinary, and Eld’s deer conservation.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/supplementary material.
The animal study was approved by Ethics Committee of the Hainan Medical University (approval no. HMUEC20180059). The study was conducted in accordance with the local legislation and institutional requirements.
YZ: Conceptualization, Data curation, Formal analysis, Methodology, Project administration, Visualization, Writing – original draft, Writing – review & editing, Funding acquisition. GR: Data curation, Formal analysis, Investigation, Methodology, Visualization, Writing – original draft. QL: Investigation, Methodology, Writing – original draft. JL: Formal analysis, Methodology, Writing – original draft. YQ: Investigation, Methodology, Writing – original draft. YoL: Formal analysis, Validation, Writing – original draft. XLa: Investigation, Methodology, Writing – original draft. YW: Investigation, Writing – original draft. XY: Investigation, Writing – original draft. SL: Investigation, Methodology, Writing – original draft. YuL: Formal analysis, Writing – original draft. YC: Investigation, Resources, Writing – original draft. XLi: Investigation, Resources, Writing – original draft. XQ: Investigation, Resources, Writing – original draft. ZX: Investigation, Resources, Writing – original draft. TL: Formal analysis, Project administration, Writing – original draft. JD: Data curation, Formal analysis, Writing – review & editing. RD: Investigation, Writing – original draft. XC: Investigation, Writing – original draft. HW: Investigation, Resources, Writing – review & editing. GL: Funding acquisition, Project administration, Writing – review & editing.
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This work was supported by the National Natural Science Foundation of China (No. 82060375), High-level talents project of Hainan Natural Science Foundation (Nos. 822RC695 and 824RC516), Hainan Medical University Talent Development Project (No. XRC2021002), Hainan Province Science and Technology Special Fund (No. ZDYF2023SHFZ146), College Student Innovation and Entrepreneurship Training Program Project (Nos. X202311810139 and S202411810053).
We would like to extend our gratitude to all the institution and individuals who participate and provided their kind assistance, especially generous permission and collaboration in the sample collection process from the Hainan Bangxi Provincial Nature Reserve Administration and Hainan Tropical Infectious Diseases Biobank.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The authors declare that no Gen AI was used in the creation of this manuscript.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
1. Moratal, S, Magnet, A, Izquierdo, F, del Águila, C, López-Ramon, J, and Dea-Ayuela, MA. Microsporidia in commercially harvested marine fish: a potential health risk for consumers. Animals. (2023) 13:2673. doi: 10.3390/ani13162673
2. Nourrisson, C, Lavergne, R-A, Moniot, M, Morio, F, and Poirier, P. Enterocytozoon bieneusi, a human pathogen. Emerg Microbes Infect. (2024) 13:2406276. doi: 10.1080/22221751.2024.2406276
3. Han, B, Pan, G, and Weiss, LM. Microsporidiosis in humans. Clin Microbiol Rev. (2021) 34:e0001020. doi: 10.1128/CMR.00010-20
4. Naguib, D, Roellig, DM, Arafat, N, and Xiao, L. Prevalence and genetic characterization of Enterocytozoon bieneusi in children in Northeast Egypt. Parasitol Res. (2022) 121:2087–92. doi: 10.1007/s00436-022-07546-z
5. Li, W, Feng, Y, and Santin, M. Host specificity of Enterocytozoon bieneusi and public health implications. Trends Parasitol. (2019) 35:436–51. doi: 10.1016/j.pt.2019.04.004
6. Abdoli, A, Olfatifar, M, Zaki, L, Asghari, A, Hatam-Nahavandi, K, Nowak, O, et al. The global prevalence of microsporidia infection in rabbits as a neglected public health concern: a systematic review and meta-analysis. Prev Vet Med. (2025) 234:106380. doi: 10.1016/j.prevetmed.2024.106380
7. Li, W, and Xiao, L. Ecological and public health significance of Enterocytozoon bieneusi. One Health. (2021) 12:100209. doi: 10.1016/j.onehlt.2020.100209
8. Zajaczkowska, Z, Akutko, K, Kvac, M, Sak, B, Szydlowicz, M, Hendrich, AB, et al. Enterocytozoon bieneusi infects children with inflammatory bowel disease undergoing immunosuppressive treatment. Front Med (Lausanne). (2021) 8:741751. doi: 10.3389/fmed.2021.741751
9. Tao, WF, Ni, HB, Du, HF, Jiang, J, Li, J, Qiu, HY, et al. Molecular detection of Cryptosporidium and Enterocytozoon bieneusi in dairy calves and sika deer in four provinces in northern China. Parasitol Res. (2020) 119:105–14. doi: 10.1007/s00436-019-06498-1
10. Santín, M, and Fayer, R. Microsporidiosis: Enterocytozoon bieneusi in domesticated and wild animals. Res Vet Sci. (2011) 90:363–71. doi: 10.1016/j.rvsc.2010.07.014
11. Koehler, AV, Zhang, Y, and Gasser, RB. A perspective on the molecular identification, classification, and epidemiology of Enterocytozoon bieneusi of animals. Exp Suppl. (2022) 114:389–415. doi: 10.1007/978-3-030-93306-7_14
12. Zhao, W, Zhou, H, Yang, L, Ma, T, Zhou, J, Liu, H, et al. Prevalence, genetic diversity and implications for public health of Enterocytozoon bieneusi in various rodents from Hainan Province, China. Parasit Vectors. (2020) 13:438. doi: 10.1186/s13071-020-04314-9
13. Pumpitakkul, V, Roytrakul, S, Phaonakrop, N, Thongphakdee, A, Sanannu, S, Nipanunt, T, et al. Analysis of serum proteomic profiles of endangered Siamese and Burmese Eld's deer infected with subclinical Babesia bovis in Thailand. Acta Trop. (2024) 257:107294. doi: 10.1016/j.actatropica.2024.107294
14. CITES. Appendix I of the convention on international trade in endangered species of wild Fauna and Flora. United Nations Environment Programme. Federal Register/FIND. (2023):81. https://www.cites.org/eng/app/appendices.php
15. Ghazi, MG, Sharma, SP, Tuboi, C, Angom, S, Gurumayum, T, Nigam, P, et al. Population genetics and evolutionary history of the endangered Eld's deer (Rucervus eldii) with implications for planning species recovery. Sci Rep. (2021) 11:2564. doi: 10.1038/s41598-021-82183-7
16. Gray, T, Brook, S, McShea, W, Mahood, S, Ranjitsingh, M, Miyunt, A, et al. Rucervus eldii. The IUCN red list of threatened species. e: T4265A22166803. (2015). Available at: https://iucn.org/content/verge-extinction-a-look-endangered-species-indo-burma-hotspot (Accessed June 15, 2015).
17. Groves, C. Systematics of the Artiodactyla of China in the 21(st) century. Zool Res. (2016) 8:441–5. doi: 10.1128/EC.00302-08
18. Pan, D, Song, YL, Zeng, ZG, and Bravery, BD. Habitat selection by Eld's deer following relocation to a patchy landscape. PLoS One. (2014) 9:e91158. doi: 10.1371/journal.pone.0091158
19. Wong, MHG, Mo, Y, and Chan, BPL. Past, present and future of the globally endangered Eld’s deer (Rucervus eldii) on Hainan Island, China. Glob Ecol Conserv. (2021) 26:e01505. doi: 10.1016/J.GECCO.2021.E01505
20. Zhang, Q, Zhang, Z, Ai, S, Wang, X, Zhang, R, and Duan, Z. Cryptosporidium spp., Enterocytozoon bieneusi, and Giardia duodenalis from animal sources in the Qinghai-Tibetan plateau area (QTPA) in China. Comp Immunol Microbiol Infect Dis. (2019) 67:101346. doi: 10.1016/j.cimid.2019.101346
21. Santin, M, and Fayer, R. Enterocytozoon bieneusi genotype nomenclature based on the internal transcribed spacer sequence: a consensus. J Eukaryot Microbiol. (2009) 56:34–8. doi: 10.1111/j.1550-7408.2008.00380.x
22. Ren, G, Li, J, Xiong, J, Lai, X, Wang, Y, Lei, S, et al. Molecular detection and public health risk assessment of Cryptosporidium spp., Giardia duodenalis, Enterocytozoon bieneusi, and Blastocystis sp. of animals in a tropical wildlife park of Hainan Island, China. One Health Bull. (2023) 3:1–10. doi: 10.4103/2773-0344.383636
23. Zhang, Q, Zhong, Z, Xia, Z, Meng, Q, Shan, Y, Guo, Q, et al. Molecular epidemiology and genetic diversity of Enterocytozoon bieneusi in Cervids from Milu Park in Beijing, China. Animals (Basel). (2022) 12:1539. doi: 10.3390/ani12121539
24. Zhang, XX, Cong, W, Liu, GH, Ni, XT, Ma, JG, Zheng, WB, et al. Prevalence and genotypes of Enterocytozoon bieneusi in sika deer in Jilin province, Northeastern China. Acta Parasitol. (2016) 61:382–8. doi: 10.1515/ap-2016-0050
25. Zhao, W, Wang, J, Yang, Z, and Liu, A. Dominance of the Enterocytozoon bieneusi genotype BEB6 in red deer (Cervus elaphus) and Siberian roe deer (Capreolus pygargus) in China and a brief literature review. Parasite. (2017) 24:54. doi: 10.1051/parasite/2017056
26. Dashti, A, Santín, M, Köster, PC, Bailo, B, Ortega, S, Imaña, E, et al. Zoonotic Enterocytozoon bieneusi genotypes in free-ranging and farmed wild ungulates in Spain. Med Mycol. (2022) 60:myac070. doi: 10.1093/mmy/myac070
27. Zhang, Y, Koehler, AV, Wang, T, Haydon, SR, and Gasser, RB. First detection and genetic characterisation of Enterocytozoon bieneusi in wild deer in Melbourne's water catchments in Australia. Parasite Vector. (2018) 11:2. doi: 10.1186/s13071-017-2577-7
28. Guo, Y, Alderisio, KA, Yang, W, Cama, V, Feng, Y, and Xiao, L. Host specificity and source of Enterocytozoon bieneusi genotypes in a drinking source watershed. Appl Environ Microb. (2014) 80:218–25. doi: 10.1128/AEM.02997-13
29. Li, W, Deng, L, Yu, X, Zhong, Z, Wang, Q, Liu, X, et al. Multilocus genotypes and broad host-range of Enterocytozoon bieneusi in captive wildlife at zoological gardens in China. Parasite Vector. (2016) 9:395. doi: 10.1186/s13071-016-1668-1
30. Zhao, W, Zhang, W, Wang, R, Liu, W, Liu, A, Yang, D, et al. Enterocytozoon bieneusi in sika deer (Cervus nippon) and red deer (Cervus elaphus): deer specificity and zoonotic potential of ITS genotypes. Parasitol Res. (2014) 113:4243–50. doi: 10.1007/s00436-014-4100-9
31. Huang, J, Zhang, Z, Yang, Y, Wang, R, Zhao, J, Jian, F, et al. New genotypes of Enterocytozoon bieneusi isolated from sika deer and Red Deer in China. Front Microbiol. (2017) 8:879. doi: 10.3389/fmicb.2017.00879
32. Zhang, Z, Huang, J, Karim, MR, Zhao, J, Dong, H, Ai, W, et al. Zoonotic Enterocytozoon bieneusi genotypes in Père David's deer (Elaphurus davidianus) in Henan, China. Exp Parasitol. (2015) 155:46–8. doi: 10.1016/j.exppara.2015.05.008
33. Xie, F, Zhang, Z, Zhao, A, Jing, B, Qi, M, and Wang, R. Molecular characterization of Cryptosporidium and Enterocytozoon bieneusi in Pere David's deer (Elaphurus davidianus) from Shishou, China. Int J Parasitol Parasites Wildl. (2019) 10:184–7. doi: 10.1016/j.ijppaw.2019.09.001
34. Zhang, P, Zhang, Q, Han, S, Yuan, G, Bai, J, and He, H. Occurrence and genetic diversity of the zoonotic enteric protozoans and Enterocytozoon bieneusi in pere David's deer (Elaphurus davidianus) from Beijing, China. Pathogens. (2022) 11:1223. doi: 10.3390/pathogens11111223
35. Amer, S, Kim, S, Han, JI, and Na, KJ. Prevalence and genotypes of Enterocytozoon bieneusi in wildlife in Korea: a public health concern. Parasite Vector. (2019) 12:160. doi: 10.1186/s13071-019-3427-6
36. Santin, M, and Fayer, R. Enterocytozoon bieneusi, Giardia, and Cryptosporidium infecting white-tailed deer. J Eukaryot Microbiol. (2015) 62:34–43. doi: 10.1111/jeu.12155
37. Qiu, L, Xia, W, Li, W, Ping, J, Ding, S, and Liu, H. The prevalence of microsporidia in China: a systematic review and meta-analysis. Sci Rep. (2019) 9:3174. doi: 10.1038/s41598-019-39290-3
38. Ruan, Y, Xu, X, He, Q, Li, L, Guo, J, Bao, J, et al. The largest meta-analysis on the global prevalence of microsporidia in mammals, avian and water provides insights into the epidemic features of these ubiquitous pathogens. Parasites Vectors. (2021) 14:186. doi: 10.1186/s13071-021-04700-x
39. Ghebremichael, ST, Meng, X, Wei, J, Yang, Y, Huang, Q, Luo, L, et al. Prevalence and genotyping distribution of Enterocytozoon bieneusi in diarrheic pigs in Chongqing and Sichuan provinces, China. Front Microbiol. (2022) 13:1025613. doi: 10.3389/fmicb.2022.1025613
40. Ghebremichael, ST, Meng, X, Yang, Y, Andegiorgish, AK, Wu, Z, Chen, J, et al. First identification and coinfection detection of Enterocytozoon bieneusi, Encephalitozoon spp., Cryptosporidium spp. and Giardia duodenalis in diarrheic pigs in Southwest China. BMC Microbiol. (2023) 23:334. doi: 10.1186/s12866-023-03070-x
41. Li, S, Zou, Y, Wang, P, Han, RY, Wang, CB, Song, DP, et al. A high genetic diversity of Enterocytozoon bieneusi in diarrheic pigs in southern China. Transbound Emerg Dis. (2022) 69:3562–70. doi: 10.1111/tbed.14719
42. Luo, R, Xiang, L, Liu, H, Zhong, Z, Liu, L, Deng, L, et al. First report and multilocus genotyping of Enterocytozoon bieneusi from Tibetan pigs in southwestern China. Parasite. (2019) 26:24. doi: 10.1051/parasite/2019021
43. Jiang, W, Roellig, DM, Li, N, Wang, L, Guo, Y, Feng, Y, et al. Contribution of hospitals to the occurrence of enteric protists in urban wastewater. Parasitol Res. (2020) 119:3033–40. doi: 10.1007/s00436-020-06834-w
44. Zhao, W, Ren, GX, Qiang, Y, Li, J, Pu, J, Zhang, Y, et al. Molecular-based detection of Enterocytozoon bieneusi in farmed masked palm civets (Paguma larvata) in Hainan, China: a high-prevalence, specificity, and zoonotic potential of ITS genotypes. Front Vet Sci. (2021) 8:714249. doi: 10.3389/fvets.2021.714249
45. Zhao, W, Wang, T, Ren, G, Li, J, Tan, F, Li, W, et al. Molecular detection of Enterocytozoon bieneusi in farmed Asiatic brush-tailed porcupines (Atherurus macrourus) and bamboo rats (Rhizomys pruinosus) from Hainan Province, China: common occurrence, wide genetic variation and high zoonotic potential. Acta Trop. (2023) 242:106915. doi: 10.1016/j.actatropica.2023.106915
46. Zhang, T, Yu, K, Xu, J, Cao, W, Wang, Y, Wang, J, et al. Enterocytozoon bieneusi in wild rats and shrews from Zhejiang Province, China: occurrence, genetic characterization, and potential for zoonotic transmission. Microorganisms. (2024) 12:811. doi: 10.3390/microorganisms12040811
47. Li, F, Wang, R, Guo, Y, Li, N, Feng, Y, and Xiao, L. Zoonotic potential of Enterocytozoon bieneusi and Giardia duodenalis in horses and donkeys in northern China. Parasitol Res. (2020) 119:1101–8. doi: 10.1007/s00436-020-06612-8
48. Tian, GR, Zhao, GH, Du, SZ, Hu, XF, Wang, HB, Zhang, LX, et al. First report of Enterocytozoon bieneusi from giant pandas (Ailuropoda melanoleuca) and red pandas (Ailurus fulgens) in China. Infect Genet Evol. (2015) 34:32–5. doi: 10.1016/j.meegid.2015.06.015
Keywords: Eld’s deer, Enterocytozoon bieneusi , prevalence, genotype, Hainan Island
Citation: Zhang Y, Ren G, Lu Q, Li J, Qiang Y, Li Y, Lai X, Wang Y, Yu X, Lei S, Li Y, Chang Y, Liu X, Qi X, Xie Z, Li T, Du J, Duan R, Chang X, Wang H and Lu G (2025) Prevalence and molecular characterization of Enterocytozoon bieneusi in endangered Eld’s deer (Rucervus eldii) in Hainan, China. Front. Vet. Sci. 12:1521055. doi: 10.3389/fvets.2025.1521055
Received: 01 November 2024; Accepted: 13 January 2025;
Published: 27 January 2025.
Edited by:
Calin Mircea Gherman, University of Agricultural Sciences and Veterinary Medicine of Cluj-Napoca, RomaniaCopyright © 2025 Zhang, Ren, Lu, Li, Qiang, Li, Lai, Wang, Yu, Lei, Li, Chang, Liu, Qi, Xie, Li, Du, Duan, Chang, Wang and Lu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hesheng Wang, aGVzaGVuZ3dAMTI2LmNvbQ==; Gang Lu, bHVnYW5naG5AMTYzLmNvbQ==
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.