- College of Veterinary Medicine, Inner Mongolia Agricultural University, Hohhot, China
Introduction: In lambs, the function of the rumen is incompletely developed at weaning, and the inclusion of yeast cultures in the diet can profoundly influence the morphological and functional development of the rumen.
Methods: In this study, the effects of Saccharomyces cerevisiae and Kluyveromyces marxianus (NM) yeast co-cultures on ruminal histomorphology were assessed, and corresponding transcriptomic changes within the rumen epithelium were identified. In total, 24 lambs were grouped into four groups of six lambs including a control (C) group fed a basal diet, and N, M, and NM groups in which lambs were fed the basal diet, respectively, supplemented with Saccharomyces cerevisiae yeast cultures (30 g/d per head), Kluyveromyces marxianus yeast cultures (30 g/d per head), and co-cultures of both yeasts (30 g/d per head), the experiment lasted for 42 d.
Results: In morphological analyses, lambs from the NM group presented with significant increases in papilla length, papilla width, and epithelial thickness in the rumen relative to lambs in the C group (p < 0.05). Transcriptomic analyses revealed 202 genes that were differentially expressed between samples from the C and NM groups, with the largest proportion of these genes being associated with the oxidative phosphorylation pathway. In a weighted gene coexpression network analysis, a positive correlation was observed between the MEgreen and MEpurple modules and rumen morphology. Of these modules, the MEgreen module was found to be more closely linked to fatty acid metabolism and oxidative phosphorylation, whereas the MEpurple module was linked to oxidative phosphorylation and fatty acid degradation. Ultimately, these results suggest that dietary supplementation with NM has driven the degradation of fatty acids, the induction of oxidative phosphorylation, the acceleration of lipid metabolism, the production of ATP to sustain ruminal growth, and the maintenance of intracellular NADH/NAD+ homeostasis on weaned lambs and is superior to single yeast fermentation.
Discussion: These results thus offer a theoretical foundation for further studies examining the mechanisms through which NM cultures can influence ruminal development in lambs.
1 Introduction
The weaning process is a critical stage in lamb development, and the rumen plays a central role in the coordination of the uptake and use of nutrients in weaned lambs (1), with an appropriately developed rumen being vital to metabolic activity, immune function, and overall health (2). The rumen epithelium is composed of lobular papillae that facilitate absorption while also presenting an epithelial barrier that can provide protection against the entry of toxins or microbes from the rumen (3). Rumen epithelial morphology is central to the ways in which ruminants respond to changes in dietary composition (4). The development of this epithelial layer, in turn, is influenced by a wide variety of factors that include dietary physico-mechanical stimulation (5) as well as chemical stimulation mediated by the products of feed fermentation (6). Yeast cultures have been reported to influence the composition of the rumen microflora in lambs, stimulating their overall growth and enhancing immunological activity (7, 8). As they can be stored easily and or of high levels of economic value, these cultures are often used for the preparation of feed (9). These cultures can alter the morphology of the rumen epithelium in weaned lambs as a result of shifts in the expression of epithelial genes (10). The impacts of different strains of yeast culture on the rumen epithelial barrier, however, differs markedly (11). Research shows that higher levels of overall rumen epithelial thickness have been observed for goats fed S. cerevisiae yeast cultures, with changes in the expression of the cyclin D1 gene having been linked to increases in teat growth (12, 13). Adding Bacillus subtilis and S. cerevisiae co-cultures has been shown to increase ruminal gene expression related to proliferative activity while reducing expression levels linked to the process of VFA uptake (14). The addition of different yeast cultures may have specific effects on the molecular processes that shape rumen functional and morphological characteristics in ruminants after weaning.
RNA sequencing (RNA-seq) is a high-throughput transcriptomic analytical technique that can offer abundant quantitative and qualitative insights into activities in eukaryotic or prokaryotic cells and associated conditions (15, 16). Such approaches have been used extensively when analyzing rumen development (17). Zhuang et al. (18) for instance, demonstrated that β-hydroxybutyric acid supplementation resulted in increases in rumen papillae length and width, and they further identified hub genes enriched in the nicotinamide adenine dinucleotide (NADH) dehydrogenase (ubiquinone) activity Gene Ontology (GO) pathway. Baldwin et al. (19) also performed transcriptomic studies of rumen samples from Holstein cows prior to and at 14, 42, 56, and 70 days after weaning, revealing a shift in the reliance of the rumen epithelium from glucose toward VFA as a source of energy. Sun et al. (20) demonstrated that introducing fermentation concentrates led to the enhancement of fatty acid and amino acid metabolism within the rumen in pre-weaned lambs. Introducing hay together with ferment concentrate preparations resulted in the activation of rumen epithelial growth and development-related gene pathways in lambs (21). Additionally, the altered thickness of the rumen epithelium has been found to be associated with a range of cellular functions including proliferative activity (22) and the differentiation and proliferation of the epithelium (23). Transcriptionally, rumen epithelial morphology has been found to be correlated with particular gene-targeted functions including cellular development (24), the proliferation of the epithelium (25), papilla size and surface area, and tight junctions (26). Many recent studies have shown that co-culture can provide benefits to animals, such as supplementation with Kluyveromyces marxianus and Pichia kudriavzevii co-cultures that improve feed intake and milk quality in cows, Co-culture with Lactobacillus rhamnosus and Saccharomyces cerevisiae increases the content of short-chain fatty acids in mammal guts (27, 28). S. cerevisiae and K. marxianus cultures are both rich sources of metabolites (29), but there has been little research exploring how these two yeast species can modulate the development of the rumen epithelium.
As such, in this study, differentially expressed genes were identified in the rumen epithelium of lambs fed NM yeast cultures through a transcriptomics approach, ultimately enabling the identification of pathways associated with particular rumen epithelial features. Overall, these analyses provide insight into how dietary NM supplementation can influence the morphology of the rumen epithelium and the associated molecular mechanisms.
2 Materials and methods
2.1 Yeast culture preparation
The natural fermented mare’s milk was filtered and diluted in the laboratory, and then separated and purified in the yeast extract peptone glucose medium (YPD). S. cerevisiae and K. marxianus strains with good fermentation characteristics were identified, pre-grown at 30°C on YPD for 24 h, and inoculated into the fermentation medium, and pilot production efforts were conducted at Kehong Feed Co., Ltd. (Inner Mongolia, China). The basal diet for lambs in this study was composed of 12% bran, 12% spraying corn bran, 10% corn, 10% rice bran, 10% cottonseed meal, 8% wheat shorts, 28% corn germ meal, and 10% soybean meal. S. cerevisiae (3 × 108 CFU/g), K. marxianus (3 × 108 CFU/g), or a 1: 1 mixture of the two (3 × 108 CFU/g) were used to inoculate the diet at 8% per 1,000 kg wet mixed matrix through the addition of sterile water with constant stirring for a final system moisture content level of 40%. Aerobic fermentation at 30°C was then performed for 72 h. The nutrient compositions of the yeast cultures are presented in Supplementary Table S1 (30).
2.2 Experimental design and diet
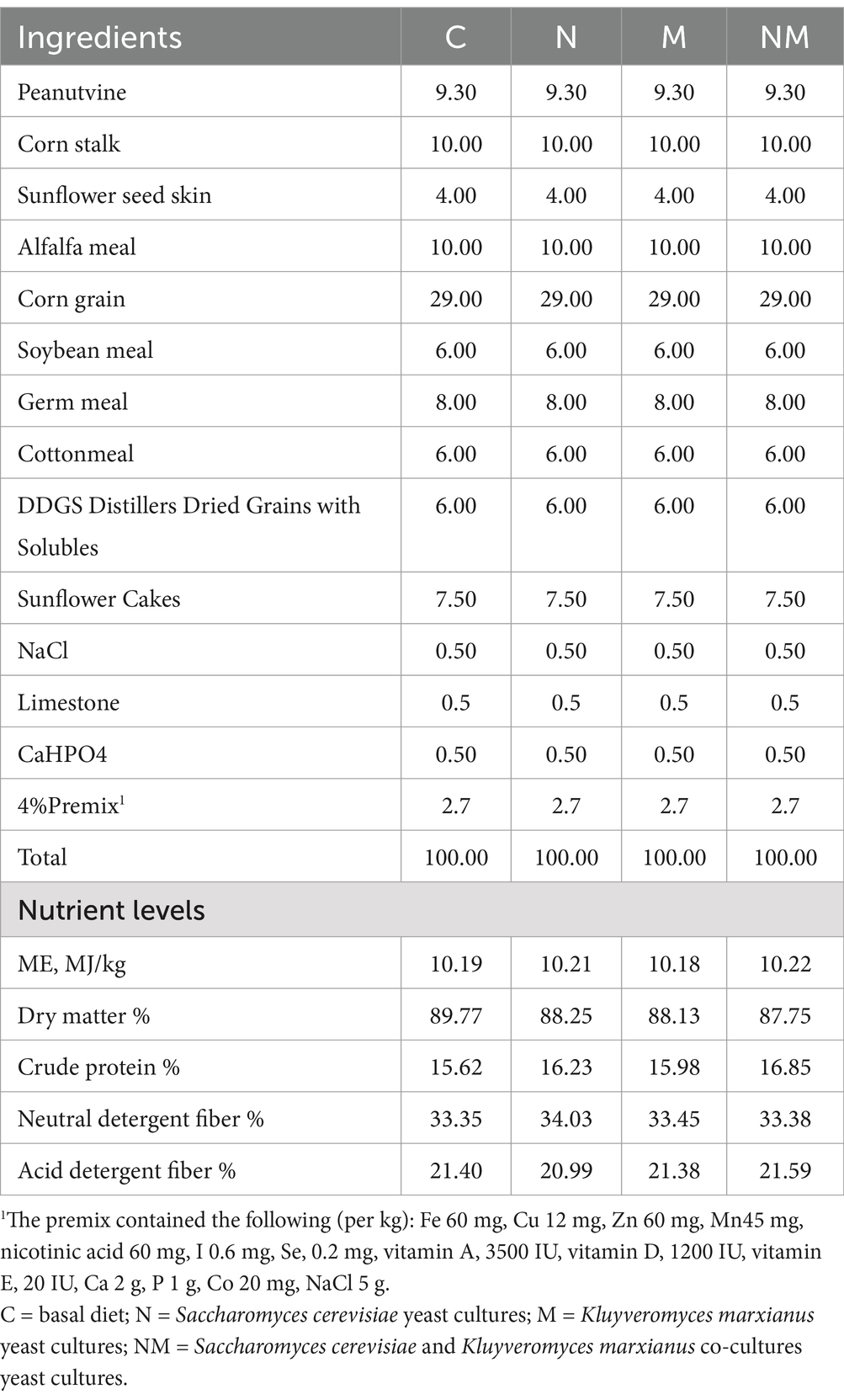
The experiments in this study were performed at Fuchuan Farm in Bayannaoer, Inner Mongolia, China. In total, 24 healthy (23.5 ± 2.85 kg) 2-month-old weaning (Duber × thin-tail sheep) lambs were randomly assigned to four groups (n = 6/group) while maintaining comparable average body weight levels in each of these groups. The animals in the control (C) group were fed a basal diet, while those in the N, M, and NM groups were, respectively, fed diets supplemented with fermentation cultures of S. cerevisiae, K. marxianus, or a combination of the two (30 g/head/day). These lambs were fed at 08: 00 and 19: 00 daily in equal amounts. The study was performed over the course of 42 days, with the first 7 being the pre-test period. Animals had free food and water access during this time. The composition and nutrient content in the basal diet are presented in Table 1.
2.3 Analyses of the ruminal epithelium
After the 42-day feeding period, animals were harvested before the morning feeding at 7: 00. The rumen was then harvested and separated from the omasum as in prior reports (31). Previous methods were then used to conduct a detailed characterization of the histomorphology of the rumen papillae (32). Briefly, the ventral rumen sac was chosen as it has been found to exhibit the highest mucous capillary blood flow per unit weight of any portion of the rumen (33). Sterile surgical scissors were used to collect 800 mg of rumen tissue, which was then washed with cold PBS (pH 7.2) 20 times. Then, ~2 cm2 samples of rumen epithelial tissue including rumen atria, ventral rumen sac, and ventral caecum were collected, fixed with 4% paraformaldehyde overnight, dehydrated, paraffin-embedded, cut into 8 μm sections, and stained with a conventional hematoxylin and eosin (H&E) approach. Six slices were produced for each lamb rumen and repeated three times. Papillae of similar size and shape from paraffin sections were selected for histomorphological analyses, analyzing the morphological features of these papillae from four sections with the optimal papillary orientation being in the median sagittal plane. Analyses were conducted with Image-Pro Plus 6.0 (Media Cybernetics Inc., Bethesda, MD, USA). Rumen papilla width and length were measured as reported previously (32). Rumen papillae were also separated from the basal epithelial layer, snap-frozen in liquid nitrogen, and stored at −80°C in the lab for subsequent transcriptomic analyses.
2.4 Transcriptomic sequencing
TRIzol (Tiangen, Beijing, China) was used for the isolation of RNA, followed by the use of a NanoDrop spectrophotometer (Agilent, Cary 60 UV–Vis, CA, USA) and 1% agarose gel electrophoresis assay to measure RNA quantity and integrity. Then, 3 μg of total RNA was selected as input for sequencing library preparation. Initially, magnetic beads were used to purify the mRNA, followed by its fragmentation at high temperatures using divalent cations in fragmentation buffer from Illumina. SuperScript II and random oligonucleotides were then employed for first-strand DNA preparation, with DNA polymerase I and RNase h being used for second-strand cDNA preparation. Nucleic acid cleavage enzymes and polymerase activity were then used to convert any overhangs into blunt ends, followed by the removal of these enzymes. Next, 3′ adenylation was performed, followed by ligation of the Illumina PE adapter oligonucleotides in preparation for hybridization. Fragments of cDNA that were 400–500 bp long were selected through the use of the AMPure XP system (Beckman Coulter, CA, USA) for purification. Selective enrichment of DNA fragments for junction molecules attached at both ends was achieved through a 15-cycle PCR reaction with an Illumina PCR primer cocktail. After using the AMPure XP system to purify the resultant products, they were subjected to high-sensitivity DNA analysis-based quantification with a Bioanalyzer 2100 system (Agilent). A NovaSeq 6000 platform (Illumina) was used for sequencing, which was performed by Shanghai personalbio biotechnology Co. Ltd. (Shanghai, China). An in-house Perl script was used for the removal of low-quality reads. Then, HISAT2 (34) was used to align the remaining clean reads with the Ovis aries reference genome (Oar v3.1). StringTie (version 1.3.4d) was used for assembly (35) and the fragments per kilobase per million mapped fragments (FPKM) values were calculated to evaluate the expression of gene transcripts. The Personalbio Cloud Platform1 was used to compile volcano plots, and Gene Ontology (GO) functional classification and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analyses were performed. In addition, KOBAS (version 2.0) was utilized to evaluate the significant enrichment of differentially expressed genes (DEGs) in KEGG pathways (36).
2.5 qRT-PCR verified the expression of genes associated with transcriptomic data
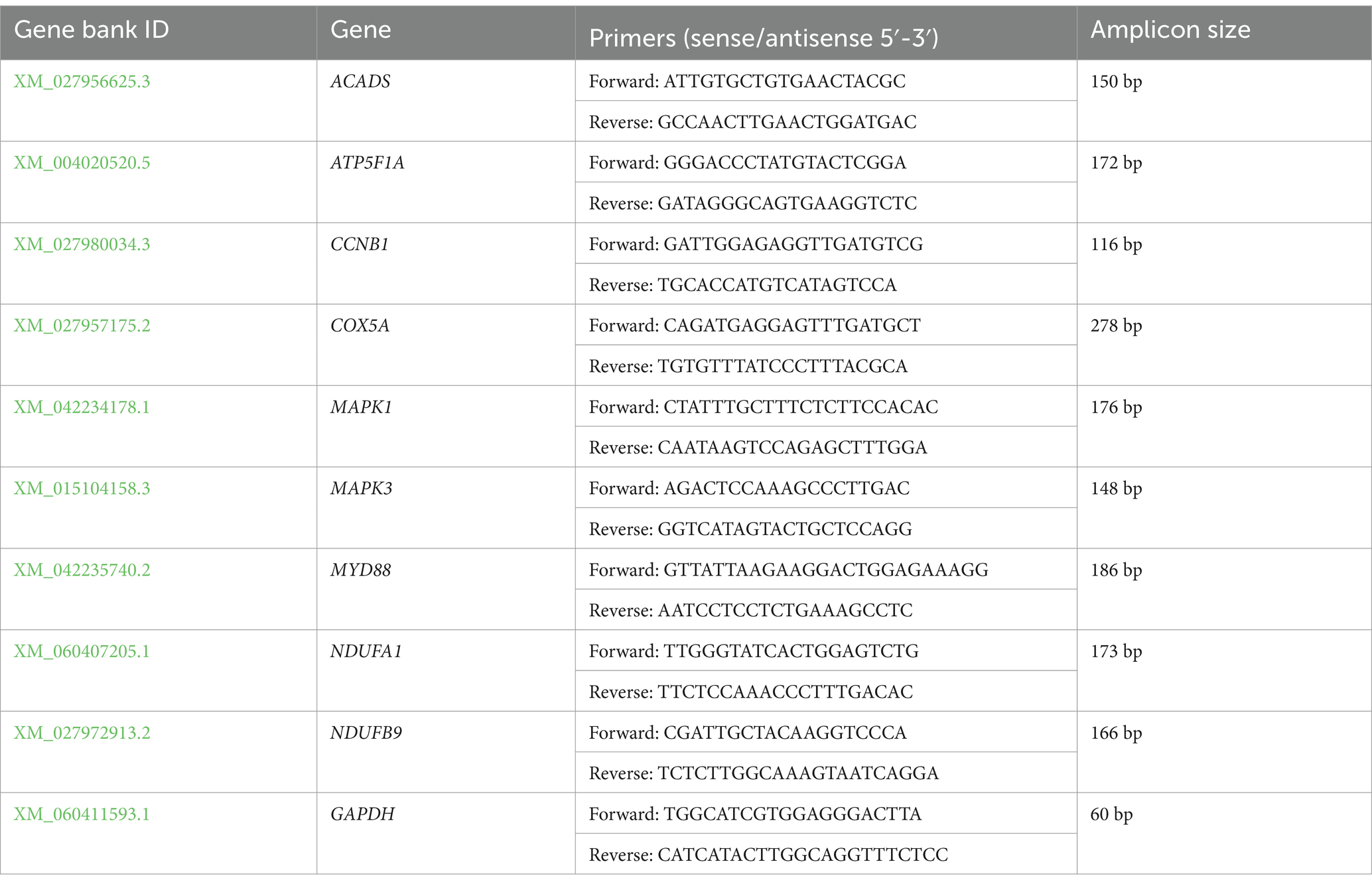
The results of transcriptomic analyses were validated by selecting 9 DEGs to analyze via qPCR. TRIzol (Tiangen) was used for the extraction of RNA from samples of ruminal epithelium. Primer designs were generated in Primer Premier 5 (Table 2), with GADPH as a reference control. Each qPCR reaction (20 μL) included 2 × ChamQ Universal SYBR qPCR Master Mix (10 μL), cDNA template, and forward and reverse primers (each 0.4 μL at 10 μM), and thermocycler settings were: 95°C for 30 s, and cycles of 95°C for 5 s, 60°C for 20 s, and GAPDH served as a normalization control. Amplicon specificity was confirmed through a melt curve analysis, and relative expression was assessed via the 2−ΔΔCt method (37).
2.6 Statistical analyses
Differences in parameters pertaining to the ruminal epithelium were compared between groups via one-way ANOVAs in SPSS 25.0 (SPSS Inc., IL, USA), with p < 0.05 defining significance. The weighted gene co-expression network analysis (WGCNA) algorithm in the R package was used to access the modules of genes related to the characterization of the rumen epithelium development and different groups. The qRT-PCR data were analyzed using one-way ANOVA for expression levels in each group and compared using Tukey’s honestly significant difference test (Tukey’s HSD). p < 0.05 was considered to be statistically significant. For the analysis of gene expression data, RNA-Seq data were initially subjected to log2 transformation to approximate a normal distribution better and stabilize the variance. The False Discovery Rate (FDR) was calculated using the Benjamini-Hochberg method to control the rate of false positives in the context of multiple comparisons. Mann–Whitney tests were then employed to compare log2-transformed FPKM values between groups. The fold-change cutoff for identifying differentially expressed genes (DEGs) was set at >1.2, which, in the context of log2-transformed data, this choice was made to balance sensitivity and specificity in detecting biologically meaningful changes while controlling for multiple comparisons. DEGs identified when comparing the C vs. N, C vs. M, C vs. NM, N vs. NM and M vs. NM groups were detected based on this fold-change >1.2 in the log2-transformed data and an FDR < 0.05.
3 Results
3.1 Morphology of the rumen epithelium
Rapid, healthy ruminal development is essential for weaned lamb growth and development. Accordingly, the development of the rumen in weaned lambs from the NM group was characterized, revealing several changes in morphological features in the epithelium attributable to yeast supplementation (Figure 1 and Table 3). The rumen papilla lengths were significantly greater in the NM group compared to the control (C) group (p = 0.01). N, M, and NM supplementation also led to significant increases in the width of these papillae (p = 0.002). Significant increases in the thickness of the rumen epithelium were evident in the N, M, and NM groups relative to the C group (p < 0.001), and significantly greater thickness was evident in the NM group relative to either of the individual yeast supplementation groups. Lamina propria thickness was unchanged across groups (p = 0.05). These data thus highlighted the ability of NM supplementation to promote rumen papillary development.
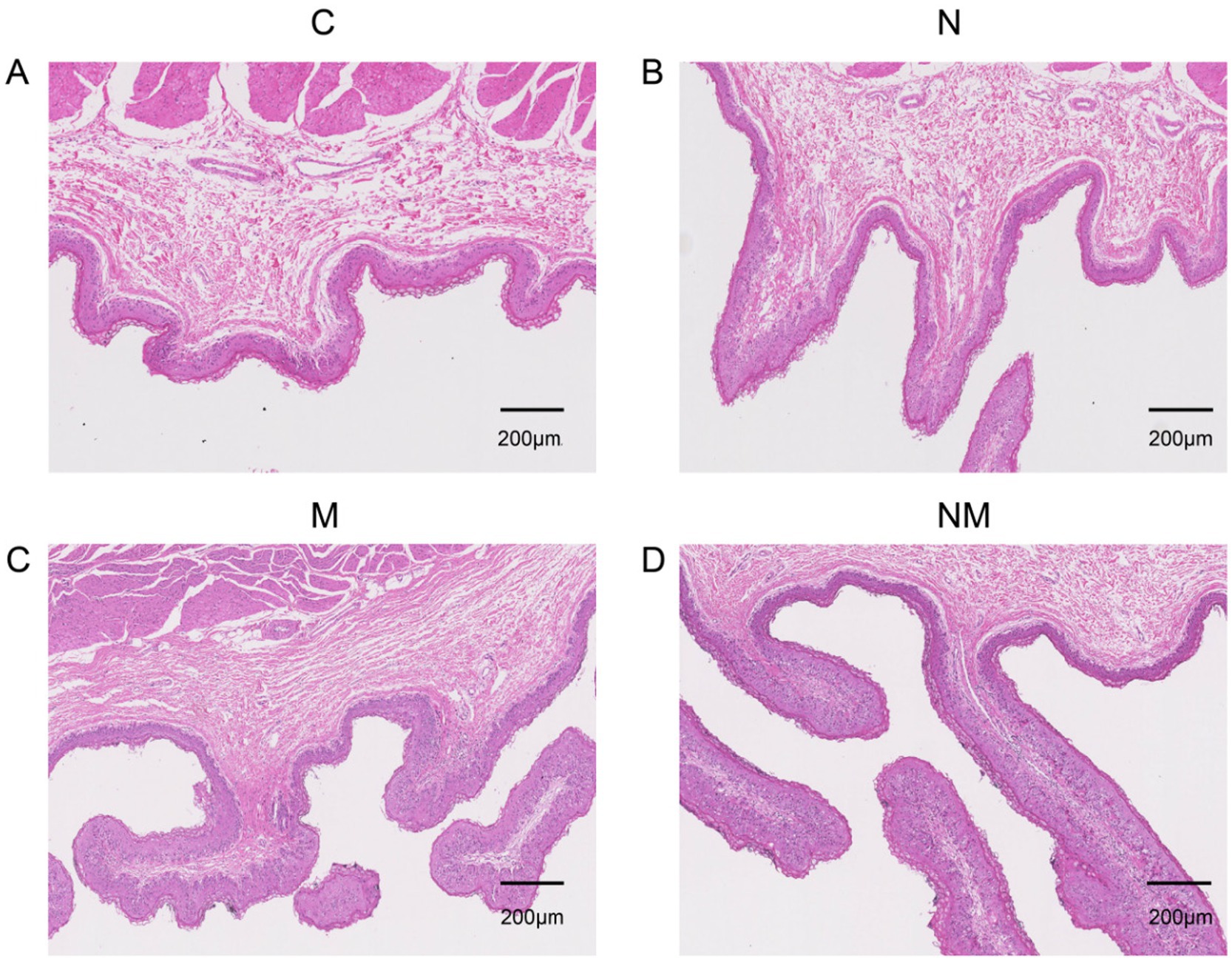
Figure 1. The impact of dietary N, M, and NM on ruminal epithelial characteristics in lambs. Representative rumen epithelium images are shown for comparisons for groups (A) Basal diet (B) Saccharomyces cerevisiae yeast cultures (C) Kluyveromyces marxianus yeast cultures (D) NM = Saccharomyces cerevisiae and Kluyveromyces marxianus co-cultures yeast cultures.
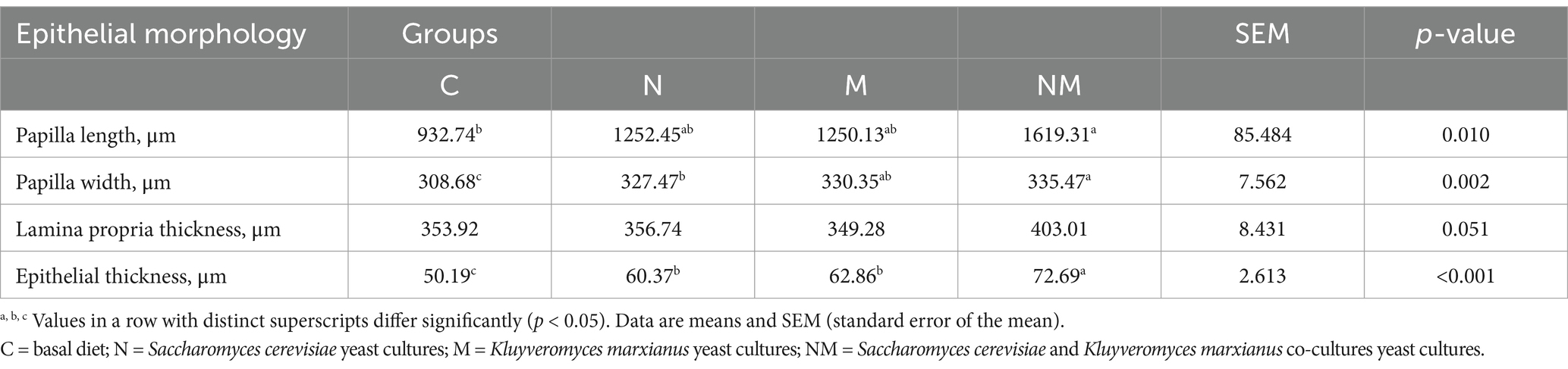
Table 3. The impact of dietary N, M, and NM supplementation on ruminal epithelial characteristics in lambs.
3.2 RNA-seq analysis and DEG identification
Next, efforts were made to explore the effects of NM supplementation on gene expression in the ruminal epithelium. These analyses yielded 79.00 GB of clean data, with an average of 6.58 GB per sample. The expression levels for 17.032 genes were measured across samples. Relative to the C group, the N group exhibited 413 DEGs (231 upregulated, 182 downregulated) (Figure 2A), the M group exhibited 716 DEGs (199 upregulated, 517 downregulated) in group M (Figure 2B), and the NM group exhibited 393 DEGs (166 upregulated, 227 downregulated) (Figure 2C). Relative to the NM group, the N group exhibited 490 DEGs (198 upregulated, 292 downregulated), and the M group exhibited 759 DEGs (538 upregulated, 221 downregulated) (Supplementary Figures S1A,B). Venn diagrams also revealed 7 DEGs shared between these comparisons, whereas there were 123, 191, 78, 163 and 220 unique DEGs, respectively, associated with the C vs. N, C vs. M, C vs. NM, N vs. NM, and M vs. NM comparisons (Figure 2D).
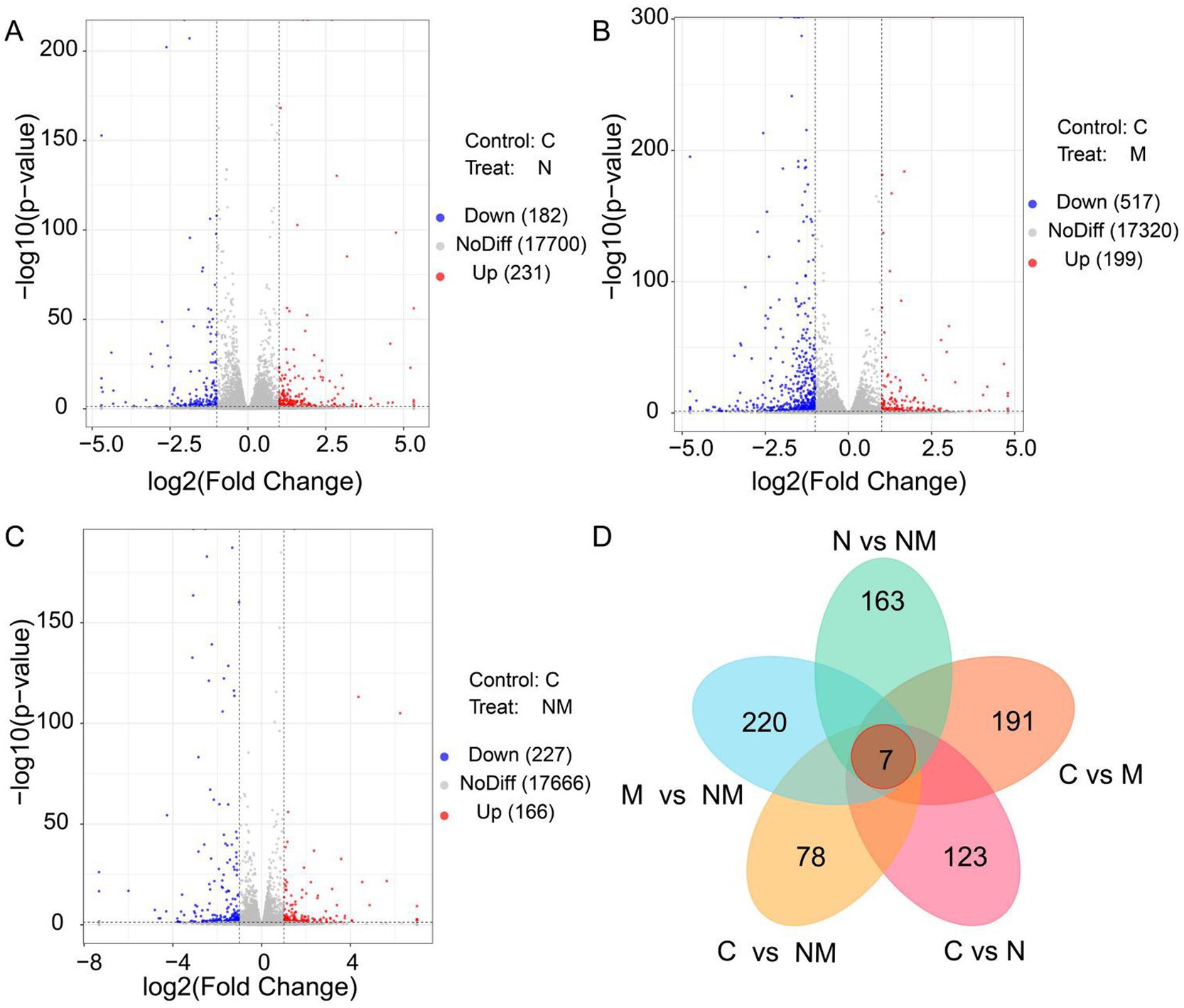
Figure 2. Differentially expressed gene identification in the ruminal epithelium for the three different group comparisons. (A) C vs. N, (B) C vs. M, and (C) C vs. NM. In the generated volcano plots, the X-and Y-axis, respectively, represent log2 fold change and log10 p-values, with blue, red, and gray dots indicating downregulated DEGs, upregulated DEGs, and non-DEGs, respectively. (D) A Venn diagram of the overlap in DEGs for the indicated comparisons. C = basal diet; N = Saccharomyces cerevisiae yeast cultures; M = Kluyveromyces marxianus yeast cultures; NM = Saccharomyces cerevisiae and Kluyveromyces marxianus co-cultures yeast cultures.
3.3 Functional enrichment analyses
To gain additional insight into the shifts in biological activity that may related to the DEGs from the NM group, GO and KEGG enrichment analyses were next performed for the DEGs identified for the C vs. N, C vs. M, C vs. NM, N vs. NM, and M vs. NM comparisons.
For GO analyses (Figure 3), the C vs. N DEGs were associated with biological process (BP) terms including “negative regulation of endopeptidase activity” and “keratinization,” cellular component (CC) terms including “extracellular space” and “apical plasma membrane,” and molecular function (MF) terms including “serine-type endopeptidase inhibitor activity” and “endopeptidase inhibitor activity” (Figure 3A). For the C vs. M comparison, DEGs were enriched in BP terms including “defense response to virus” and “negative regulation of viral genome replication,” CC terms including “sarcolemma” and “Z disc,” and MF terms including “calcium ion binding” and “chemokine activity” (Figure 3B). For the C vs. NM comparison, the DEGs were enriched in BP terms including “defense response to virus” and “innate immune response,” CC terms including “extracellular region” and “extracellular space,” and MF terms including “2′-5′-oligoadenylate synthetase activity” and “calcium ion binding” (Figure 3C). For the N vs. NM comparison, the DEGs were enriched in BP terms including “negative regulation of viral genome replication” and “defense response to virus,” CC terms including “extracellular space,” and MF terms including “protein-arginine deiminase activity.” For the M vs. NM comparison, the DEGs were enriched in BP terms including “chemical synaptic transmission” and “myofibril assembly,” CC terms including “Z disc,” and MF terms including “transmembrane transporter binding”(Supplementary Figures S2A,B).
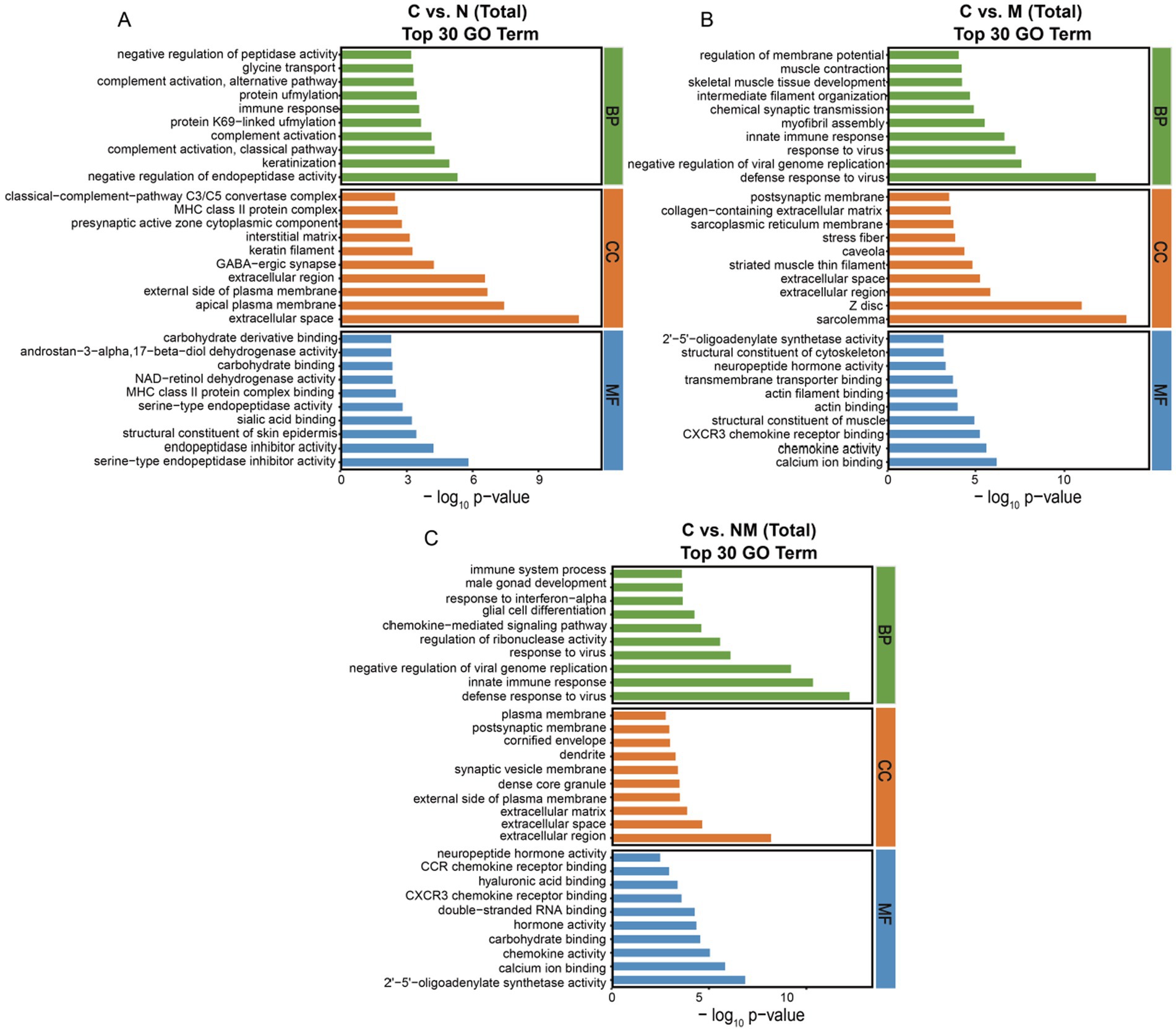
Figure 3. Analyses of DEG enrichment in specific GO terms. The top 30 biological process, cellular component, and molecular function terms are shown (p < 0.05; the DEGs number of GO terms was >2). (A) C vs. N. (B) C vs. M. (C) C vs. NM. C = basal diet; N = Saccharomyces cerevisiae yeast cultures; M = Kluyveromyces marxianus yeast cultures; NM = Saccharomyces cerevisiae and Kluyveromyces marxianus co-cultures yeast cultures.
For the C vs. N comparison, the oxidative phosphorylation, cell adhesion molecules, and protein digestion and absorption KEGG pathways exhibited significant enrichment (p < 0.05) (Figure 4A). For the C vs. M comparison these enriched pathways included the oxidative phosphorylation, neuroactive ligand-receptor interaction, and chemokine signaling pathways (p < 0.05) (Figure 4B). For the C vs. NM comparison they included the oxidative phosphorylation, viral protein interaction with cytokine and cytokine receptors, and chemokine signaling pathways (p < 0.05) (Figure 4C). For the N vs. NM comparison, the complement and coagulation cascades, protein digestion and absorption, and oxidative phosphorylation KEGG pathways exhibited significant enrichment (p < 0.05) (Supplementary Figure S3A). Lastly, for the M vs. NM comparison, the calcium signaling pathway, cell adhesion molecules and dopaminergic synapse KEGG pathways exhibited significant enrichment (p < 0.05) (Supplementary Figure S3B). Given its consistent enrichment, the oxidative phosphorylation pathway was selected for further analysis.
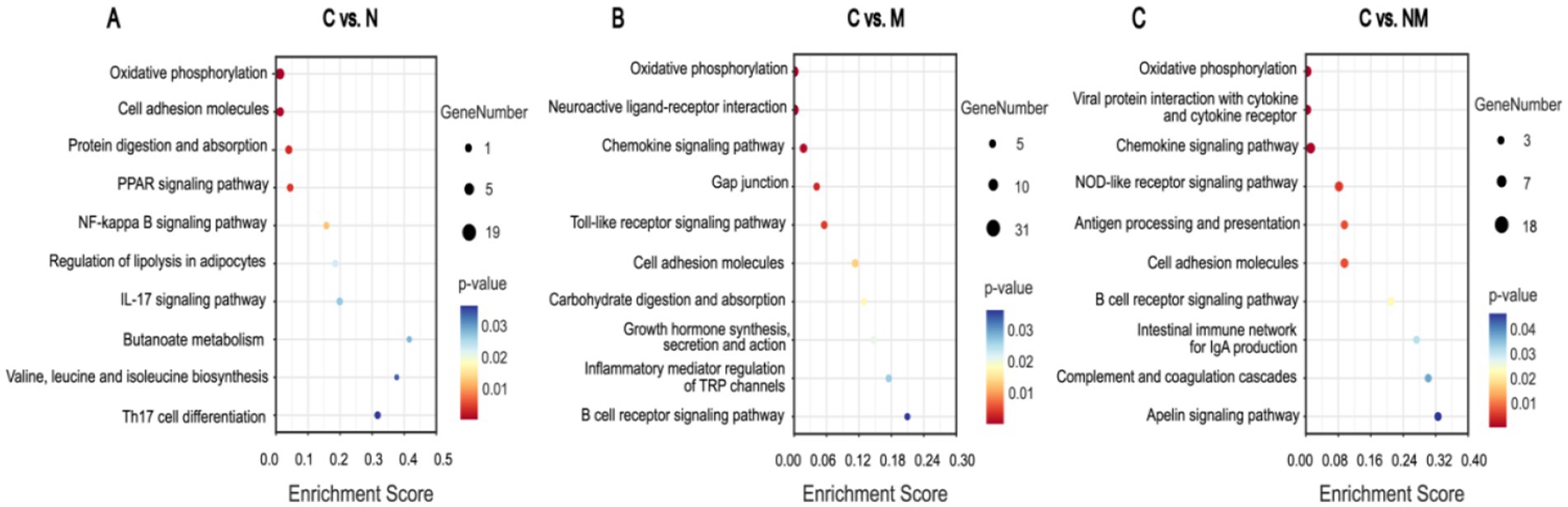
Figure 4. Analysis of DEG enrichment in KEGG pathways. Pathways and enrichment scores are, respectively, shown on the Y-and X-axes. Bubble sizes and colors correspond to significance levels and number of genes in each pathway, respectively (p < 0.05). (A) C vs. N. (B) C vs. M. (C) C vs. NM. C = basal diet; N = Saccharomyces cerevisiae yeast cultures; M = Kluyveromyces marxianus yeast cultures; NM = Saccharomyces cerevisiae and Kluyveromyces marxianus co-cultures yeast cultures.
3.4 DEGs associated with the oxidative phosphorylation pathway
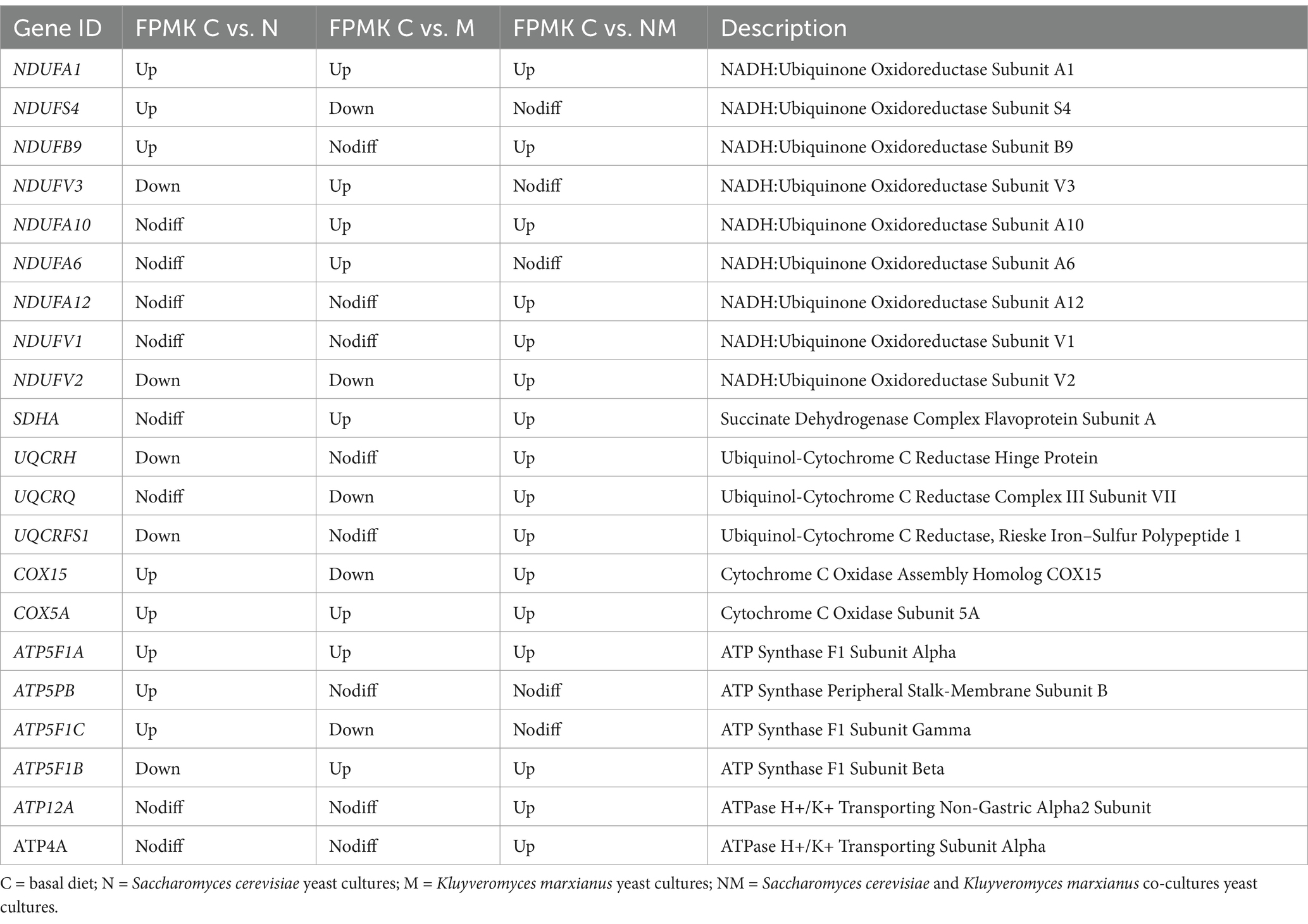
For the C vs. N comparison, 8 upregulated DEGs in the oxidative phosphorylation pathway were evident (NDUFA1, NDUFS4, NDUFB9, COX15, COX5A, ATP5F1A, ATP5PB and ATP5F1C) while 5 were downregulated (NDUFV3, NDUFV2, UQCRH, UQCRFS1, and ATP5F1B). Similarly, for the C vs. M comparison, there were 8 upregulated (NDUFA1, NDUFV3, NDUFA10, NDUFA6, SDHA, COX5A, ATP5F1A, and ATP5F1B) and 5 downregulated (NDUFS4, NDUFV2, UQCRQ, COX15, and ATP5F1C) DEGs in this pathway. Lastly, for the C vs. NM comparison, 16 upregulated DEGs (NDUFA1, NDUFB9, NDUFA10, NDUFA12, NDUFV1, NDUFV2, SDHA, UQCRH, UQCRQ, UQCRFS1, COX15, COX5A, ATP5F1A, ATP5F1B, ATP12A, ATP4A) that were associated with this pathway were identified (Table 4).
3.5 Weighted gene co-expression network analyses of the correlative relationship between host transcriptional features and indicators of ruminal epithelial development
To explore the relationships between rumen epithelial indices and the effects of NM supplementation on gene expression, WGCNA screening was next performed, leading to the identification of 12 modules of co-expressed genes named after colors (Figures 5A,B). Of these, the MEgreen and MEpurple modules were the most strongly correlated with rumen epithelial measures (papilla length, papilla width, lamina propria thickness, and epithelial thickness). Both the MEgreen (451 genes, 2.6% of total genes) and MEpurple (144 genes, 0.8% of total genes) modules exhibited positive correlations with the NM group whereas they were negatively correlated with the C and M groups.
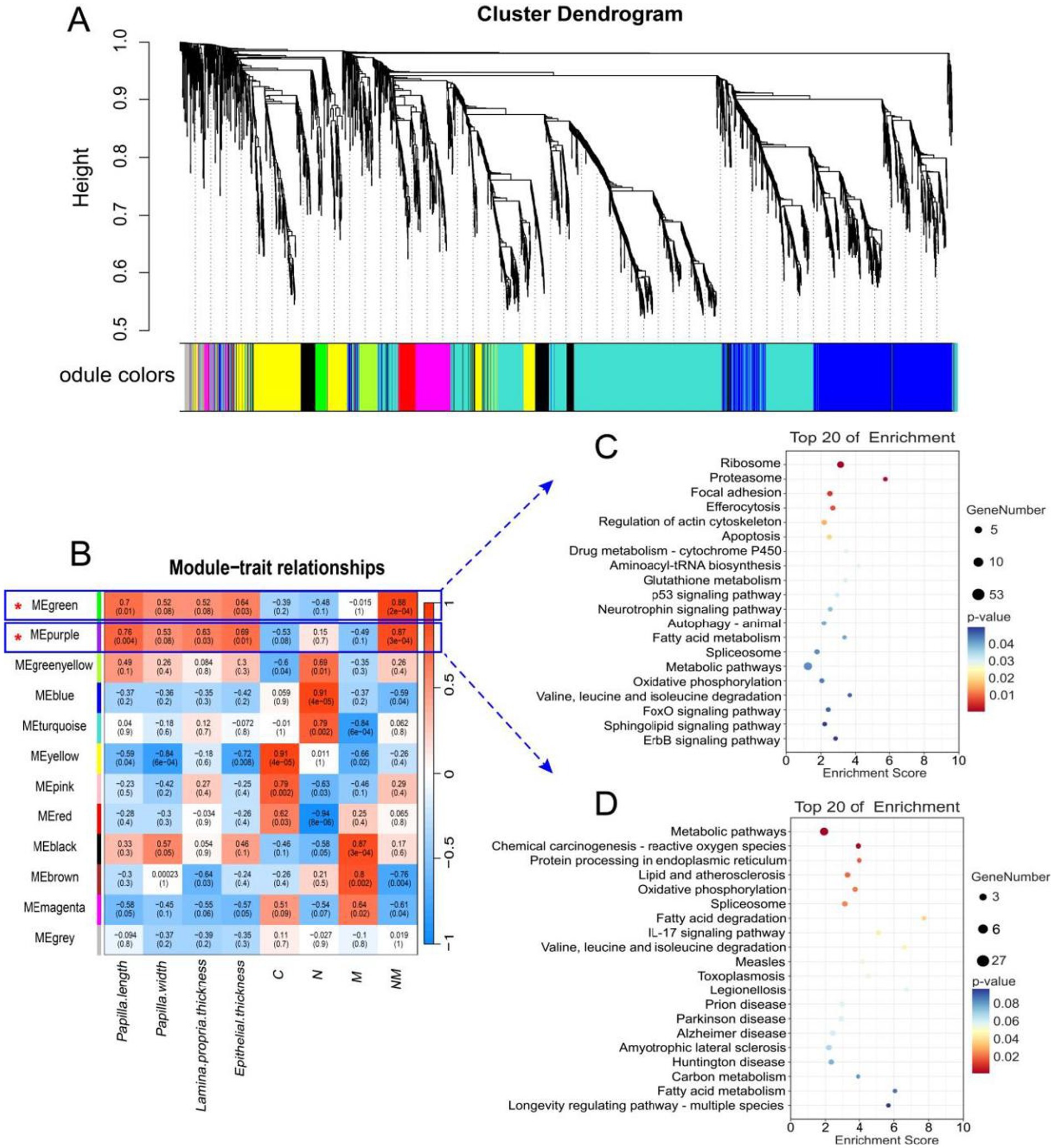
Figure 5. WGCNA identification of rumen tissue gene modules correlated with the rumen wall morphological and development indexes, and KEGG analysis of significant module. (A) The dendrogram derived from the gene co-expression network of samples in the four groups (C, N, M, and NM). (B) WGCNA of the correlation of host transcriptome with the rumen wall morphological and development indexes. (C) Top KEGG pathway of genes significantly in the MEgreen module. (D) Top KEGG pathway of genes significantly in the MEpurple module. The significance of identified KEGG pathways was determined by p < 0.05. KEGG, Kyoto Encyclopedia of Genes and Genomes; WGCNA, weighted gene co-expression network analysis. C = basal diet; N = Saccharomyces cerevisiae yeast cultures; M = Kluyveromyces marxianus yeast cultures; NM = Saccharomyces cerevisiae and Kluyveromyces marxianus co-cultures yeast cultures.
Additionally, the induction of certain GO terms associated with these modules was evaluated (Table 5). The primary annotated functions attributed to the MEgreen module included translation, mitochondrial electron transport, NADH to ubiquinone, cytoplasmic translation, and glutathione metabolic processes, while the corresponding enriched KEGG pathways included the fatty acid metabolism, oxidative phosphorylation, and FoxO signaling pathways (Figure 5C). DEGs associated with these pathways included NDUFB9, NDUFB8, NDUFA1, MAPK1, MAPK3, CCNB1, CCND2, and ATP5F1D. The functions of the MEpurple module were primarily associated with protein refolding, cellular response to unfolded protein, chaperone-mediated protein folding requiring cofactor and positive regulation of IκB/NF-κB signaling (Table 5), while corresponding enriched KEGG pathways included the oxidative phosphorylation and fatty acid degradation pathways (Figure 5D). DEGs related to these pathways included NDUFS3, ACADS, and ACAA1.
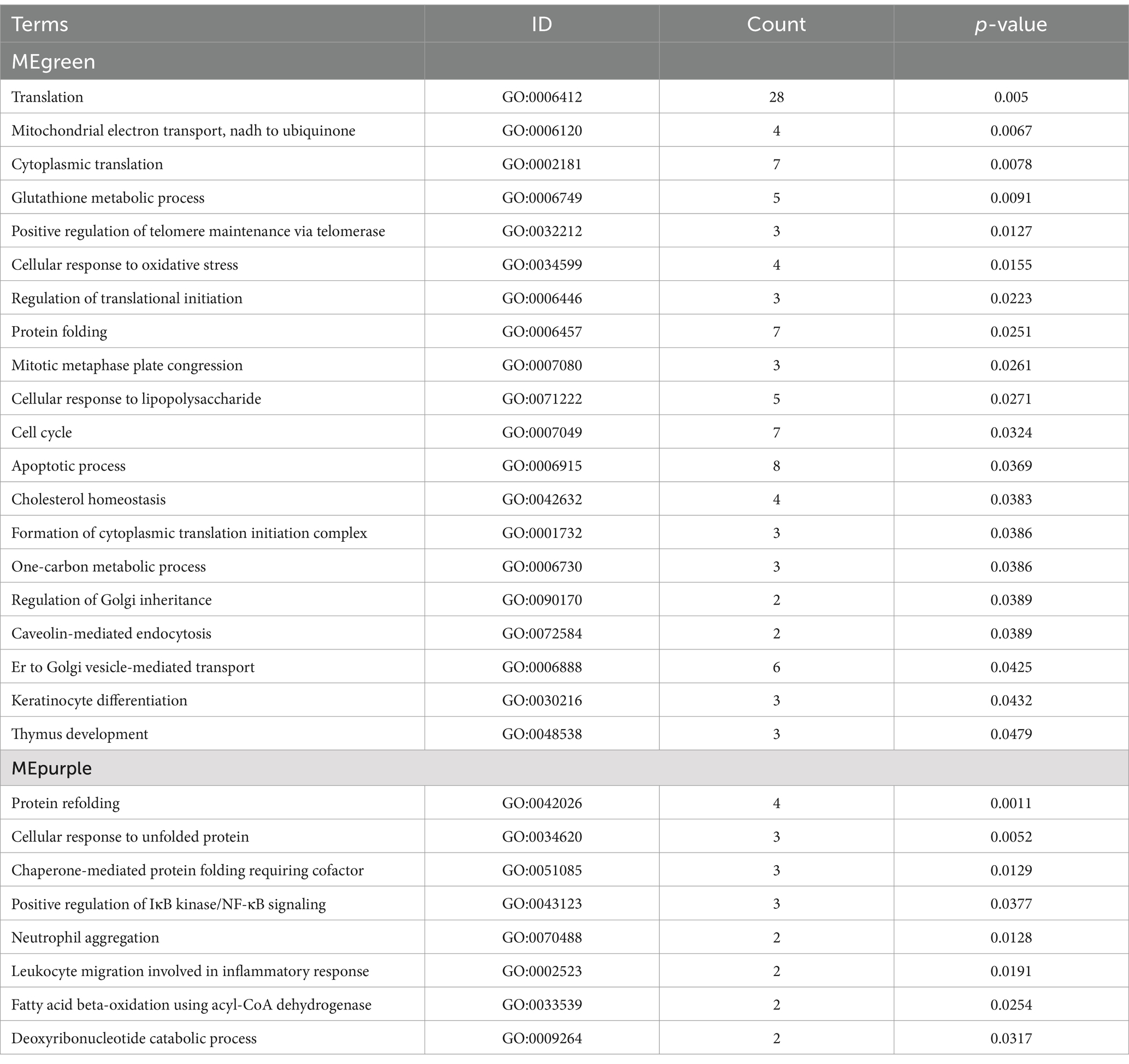
Table 5. GO enrichment analyses of key hub genes in the MEgreen and MEpurple transcriptomic modules.
3.6 qPCR-based result verification
Specific DEGs associated with rumen morphology and the oxidative phosphorylation pathway identified in the WGCNA analyses above were selected for qPCR-based validation, including NDUFA1, NDUFB9, ATP5F1A, COX5A, CCND2, SDHA, CCNB1, ACADS, and ATP12A, the results showed that the expression pattern was consistent with that of RNA-seq (Figure 6). These analyses revealed that relative to the C group, rumen epithelial tissues from lambs in the N group exhibited increased ACADS, ATP5F1A, COX5A, NDUFA1 and NDUFB9 expression (p < 0.05), while group M exhibited ACADS, ATP5F1A, COX5A, SDHA, NDUFA1 and NDUFB9 upregulation (p < 0.05), and group NM exhibited the upregulation of ATP5F1A, CCNB1, COX5A, NDUFA1, CCND2, SDHA, ATP12A, NDUFB9 and ACADS (p < 0.05).
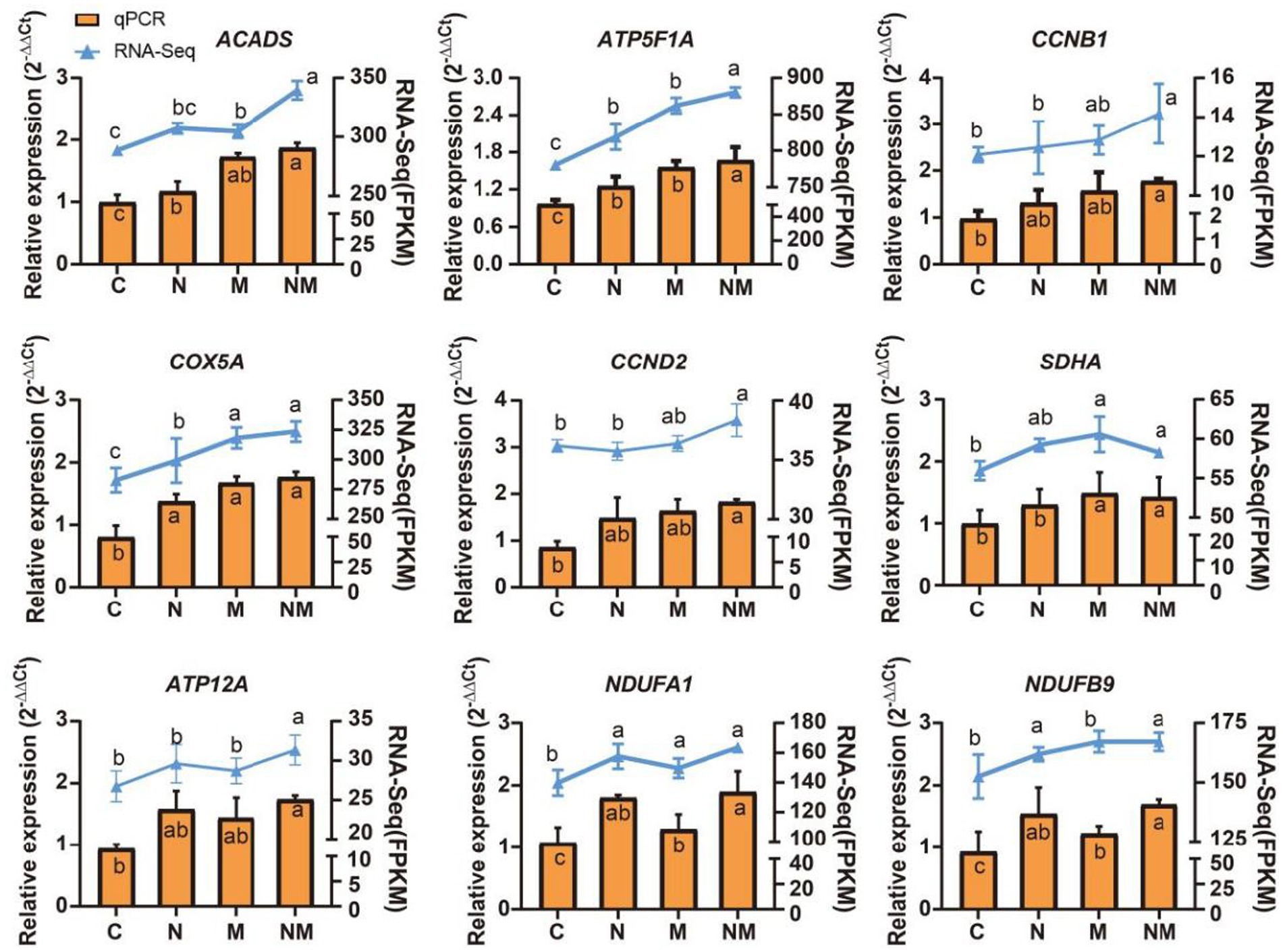
Figure 6. qPCR-based analyses of ACADS, ATP5F1A, CCNB1, COX5A, MAPK1, MAPK3, MYD88, NDUFA1, and NDUFB9 expression were used to validate RNA-seq results. a-cindicates the difference between different superindexes within the same index column (p < 0.05). C = basal diet; N = Saccharomyces cerevisiae yeast cultures; M = Kluyveromyces marxianus yeast cultures; NM = Saccharomyces cerevisiae and Kluyveromyces marxianus co-cultures yeast cultures.
4 Discussion
The development of the rumen is a vital physiologic process in ruminants, as it is responsible for transforming substances that subsequently pass into the intestines, liver, and periphery (38). The application of yeast cultures as a means of enhancing ruminal development has been a recent focus of growing interest (39). Previous studies have shown that dietary supplementation of S. cerevisiae and K. marxianus yeast co-culture can increase the average daily gain in lambs, and alter the composition and metabolite profiles of the ruminal microbiome (30), but no prior studies have performed transcriptomic analyses exploring the effects of S. cerevisiae and K. marxianus yeast co-cultures yeast cultures on the morphology and functionality of the rumen wall. The papillae of the rumen contribute to an increase in the surface area with which the rumen wall can interact with the chyme, leading to better nutrient absorption (40). Rumen development is most commonly evaluated based on the width and length of these papillae (41). In the present analyses, the yeast co-cultures contributed to significant beneficial effects on epithelial thickness and papillary width/length in the rumen of lambs in the NM group. Prior reports have noted that the introduction of yeast cultures can lead to improved epithelial thickness (42). This may be attributable to large quantities of nutritionally active substances in the yeast cell walls of NM yeast cultures such as β-glucan, mannan, and organic acids which can provide the fuel for the development of the rumen (43).
A series of transcriptomic analyses were conducted to assess changes in the patterns of gene expression in weaned lambs fed an NM-supplemented diet. Through GO enrichment analyses, the DEGs identified in group N were found to be related to the negative regulation of endopeptidase activity, keratinization, and complement activation, classical pathway, in partial agreement with prior results (44, 45). The DEGs in the M and NM groups were associated with defense responses to virus and negative regulation of viral genome replication. These underlying molecular associations and mechanisms require further investigation in the future. KEGG analyses additionally revealed that DEGs associated with the oxidative phosphorylation pathway were significantly enriched for all three analyzed comparisons (C vs. N, C vs. M, C vs. NM, and N vs. NM,). The oxidative phosphorylation process is catalyzed by complexes I, II, III, IV, and V, oxidizing nutrients and producing chemical energy (46), which is used to produce ATP through the phosphorylation of ADP by ATP synthase (47). Mitochondrial oxidative phosphorylation is a hallmark of most aerobic eukaryotes, providing the majority of the energy required by cells (48). Complex I, also referred to as NADH dehydrogenase, is responsible for the conversion of NADH to NAD+. Complex I is the largest enzymatic complex within the electron transport chain (49, 50), consisting of 46 subunits in mammals (51), with these subunits being encoded by both nuclear and mitochondrial genes. Complex I allows for the binding of NADH to the distal portion of its hydrophilic arm, with two electrons being transferred to coenzyme Q after it has bound (52). The reduction of coenzyme Q results in a change in membrane arm conformation, with four proteins being transferred through the membrane (53). Complex I is required for normal cellular function such that, when impaired, defects will arise including conditions such as growth retardation and metabolic disorders (54). Here, the dietary supplementation in group N was associated with NDUFA1, NDUFS4, and NDUFB9 upregulation, while group M exhibited NDUFA1, NDUFV3, NDUFA10, and NDUFA6, and group NM exhibited the upregulation of NDUFA1, NDUFA12 NDUFA10, NDUFV1, NDUFV2, and NDUFB9, demonstrating the close link between dietary yeast intake, shifts in the expression of genes related to the first step in the electron transport chain, and the growth and development of the rumen epithelium.
Complex II, also referred to as succinate dehydrogenase, is composed of the SDHA, SDHB, SDHC, and SDHD subunits (55). It is the only enzymatic complex involved in both the oxidative phosphorylation-related electron transport chain and the mitochondrial TCA cycle through the oxidation of succinate to fumarate (56). In the present study, SDHA upregulation was observed in the rumen epithelium from the M and NM groups, indicating that these dietary supplementation practices impacted complex II. Complex III (ubiquinol-cytochrome c reductase) is an essential component of the mitochondrial respiratory chain responsible for transferring electrons to cytochrome C from coenzyme Q (57). This complex is composed of the mitochondrially-encoded MTCYTB and 10 other subunits that are encoded by genes in the nucleus (58). In the current study, upregulation of UQCRH, UQCRQ, and UQCRFS1 in the ruminal epithelium upon NM group that the assembly of complex III was affected, and its structure and function were possibly changed. Complex IV (cytochrome C oxidase) received electrons from cytochrome c molecules and transfers electrons to dioxygen molecules to produce H2O (59), while also driving the flow of protons across the membrane, leading to an increase in transmembrane electrochemical potential (60). Here, COX15 and COX5A upregulation was noted in the N, M, and NM groups, suggesting that these diets modulated complex IV structural and functional characteristics. Complex V (ATP synthase) can take advantage of this proton gradient to produce ATP using substrates consisting of ADP and Pi (61). Here, the upregulation of ATP5F1A, ATP5PB, and ATP5F1C was noted in group N, while ATP5F1A and ATPF1B were upregulated in group M, and group NM exhibited ATP12A, ATP4A, ATP5F1A, and ATP5F1B upregulation, suggesting that yeast co-cultures can alter this pathway in the context of rumen development by controlling ATP production. Given its efficiency as a means of producing ATP, oxidative phosphorylation plays an essential role in various biological processes (62). The dietary supplementation in group N impacted complexes I, IV, and V, while that in group M affected complexes I, II, IV, and V, and that in group NM impacted complexes I, II, III, IV, and V. These analyses highlighted the ability of co-culture yeast fermentation to promote oxidative phosphorylation within the rumen epithelium in lambs, leading to enhanced ATP production and a more robust intracellular supply of energy that can impact rumen epithelial growth and homeostasis (Figure 7).
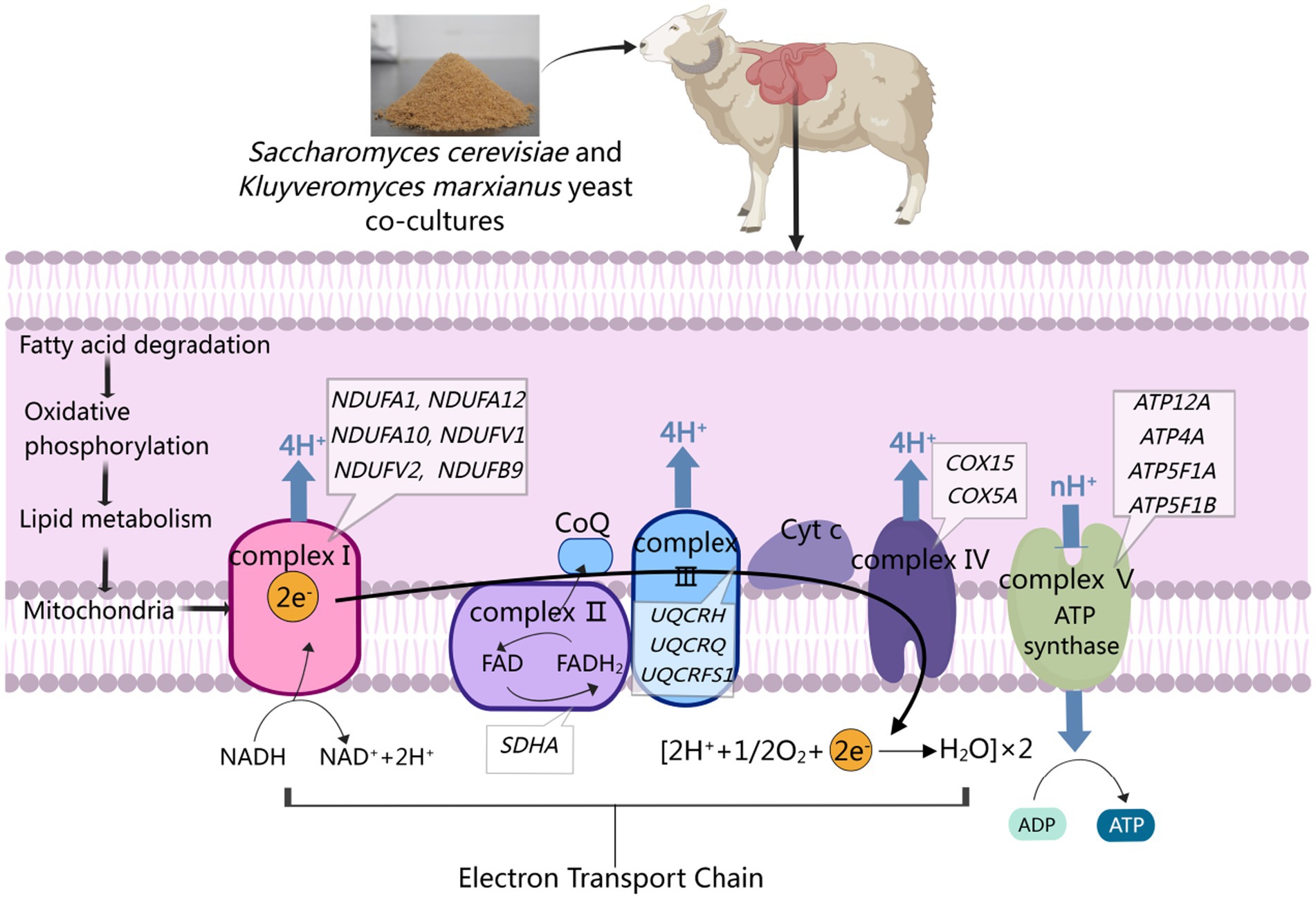
Figure 7. Adding co-cultured yeast to weaned lamb diets can enhance oxidative phosphorylation through the activation of fatty acid degradation, leading to accelerated lipid metabolism, the production of the ATP necessary for rumen growth, and the maintenance of NADH/NAD+ homeostasis within cells.
In WGCNA analyses probing the link between NM co-culture supplementation and shifts in the morphology of the rumen wall, the MEgreen and MEpurple modules were identified as being most important. GO terms associated with the genes in the MEgreen module related to the development of the rumen epithelium included translation, mitochondrial electron transport, NADH to ubiquinone, and cytoplasmic translation are highlighted as features of rumen epithelial development. Shared DEG analyses also revealed significant changes in the fatty acid metabolism, oxidative phosphorylation, and FoxO signaling pathways associated with the development of the rumen wall. Previously, mitosis has been shown to play an essential role in rumen epithelial growth and development, particularly in phases G1 and G2 (63). Strikingly, the CCB1 and CCND2 genes, which are closely linked to the cell cycle, were upregulated in the rumen epithelium of lambs from the NM group. The stability of CCNB1 and CCND2 is dependent on the capacity for ATP biogenesis (64). CCNB1 and CCND2 stabilization during the G1-S transition and G2 phase may thus be linked to NM supplementation. Genes in the MEpurple module were primarily associated with the oxidative phosphorylation and fatty acid degradation pathways. Based on these findings, the yeast cultures in the NM group were speculated to activate fatty acid degradation pathways, promoting oxidative phosphorylation and greater lipid metabolism so as to produce higher levels of ATP necessary to fuel the growth of the rumen epithelium.
5 Conclusion
In conclusion, supplementing the diets of weaned lambs with S. cerevisiae and K. marxianus yeast co-cultures can drive improvements in rumen epithelial development and morphology. These transcriptomic analyses demonstrated that the utilized yeast co-cultures were able to promote enhanced oxidative phosphorylation through the activation of the fatty acid degradation pathway, resulting in enhanced lipid metabolism such that higher levels of ATP necessary for rumen development were generated while sustaining appropriate NADH/NAD+ homeostasis within cells. These results emphasize the benefits that yeast co-cultures can have on the development of the rumen, offering new insight into the gene expression change that control this developmental process.
Data availability statement
The data presented in this study are openly available in Sequence Read Archive at https://www.ncbi.nlm.nih.gov/sra (accessed on 10 December 2023), reference number PRJNA1031991.
Ethics statement
The animal study was approved by the animal welfare and research ethics committee of Inner Mongolia Agricultural University. The study was conducted in accordance with the local legislation and institutional requirements.
Author contributions
ZX: Writing – original draft. LY: Writing – review & editing. HC: Writing – review & editing. PB: Writing – review & editing. XL: Writing – review & editing. DL: Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by the Inter-Governmental International Science and Technology Innovation Cooperation Project (2023YFE0100400); National Natural Science Foundation of China (32060774); Science and Technology Project of Inner Mongolia Autonomous Region (2020GG0036); Basic Scientific Research Business Project of Universities directly under the Inner Mongolia Autonomous Region (BR22-11-17); and Higher Education Reform and Development Program – Graduate Student Research and Innovation Funding Program (B20231071Z).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The authors declare that no Gen AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fvets.2025.1510689/full#supplementary-material
Footnotes
References
1. Yu, S, Shi, W, Yang, B, Gao, G, Chen, H, Cao, L, et al. Effects of repeated oral inoculation of artificially fed lambs with lyophilized rumen fluid on growth performance, rumen fermentation, microbial population and organ development. Anim Feed Sci Technol. (2020) 264:114465. doi: 10.1016/j.anifeedsci.2020.114465
2. Sha, Y, Liu, X, He, Y, Zhao, S, Hu, J, Wang, J, et al. Multi-omics revealed rumen microbiota metabolism and host immune regulation in Tibetan sheep of different ages. Front Microbiol. (2024) 15:1339889. doi: 10.3389/fmicb.2024.1339889
3. Steele, MA, Penner, GB, Chaucheyras-Durand, F, and Guan, LL. Development and physiology of the rumen and the lower gut: targets for improving gut health. J Dairy Sci. (2016) 99:4955–66. doi: 10.3168/jds.2015-10351
4. Tian, C, Wu, J, Jiao, J, Zhou, C, and Tan, Z. Short communication: a high-grain diet entails alteration in nutrient chemosensing of the rumen epithelium in goats. Anim Feed Sci Technol. (2020) 262:114410. doi: 10.1016/j.anifeedsci.2020.114410
5. Zhuang, Y, Lv, X, Cui, K, Chai, J, and Zhang, N. Early solid diet supplementation influences the proteomics of rumen epithelium in goat kids. Biology. (2023) 12:684. doi: 10.3390/biology12050684
6. Na, SW, and Guan, LL. Understanding the role of rumen epithelial host-microbe interactions in cattle feed efficiency. Anim Nutr. (2022) 10:41–53. doi: 10.1016/j.aninu.2022.04.002
7. Ansari, F, Alian Samakkhah, S, Bahadori, A, Jafari, SM, Ziaee, M, Khodayari, MT, et al. Health-promoting properties of Saccharomyces cerevisiae var. boulardii as a probiotic; characteristics, isolation, and applications in dairy products. Crit Rev Food Sci Nutr. (2021) 63:457–85. doi: 10.1080/10408398.2021.1949577
8. Amin, AB, and Mao, S. Influence of yeast on rumen fermentation, growth performance and quality of products in ruminants: a review. Anim Nutr. (2021) 7:31–41. doi: 10.1016/j.aninu.2020.10.005
9. Russell, B, Miro-Quesada, G, Limin, Q, and Ahuja, S. Characterizing the preparation of a concentrated nutrient feed solution for a large-scale cell culture process. Biochem Eng J. (2018) 134:120–8. doi: 10.1016/j.bej.2018.03.009
10. Sun, DM, Mao, SY, Zhu, WY, and Liu, JH. Effect of starter diet supplementation on rumen epithelial morphology and expression of genes involved in cell proliferation and metabolism in pre-weaned lambs. Animal. (2018) 12:2274–83. doi: 10.1017/s1751731118000290
11. Pang, Y, Zhang, H, Wen, H, Wan, H, Wu, H, Chen, Y, et al. Yeast probiotic and yeast products in enhancing livestock feeds utilization and performance: an overview. J Fungi. (2022) 8:1191. doi: 10.3390/jof8111191
12. Wang, J, Zhao, G, Zhuang, Y, Chai, J, and Zhang, N. Yeast (Saccharomyces cerevisiae) culture promotes the performance of fattening sheep by enhancing nutrients digestibility and rumen development. Fermentation. (2022) 8:719. doi: 10.3390/fermentation8120719
13. Gui, H, and Shen, Z. Concentrate diet modulation of ruminal genes involved in cell proliferation and apoptosis is related to combined effects of short-chain fatty acid and pH in rumen of goats. J Dairy Sci. (2016) 99:6627–38. doi: 10.3168/jds.2015-10446
14. Jia, P, Cui, K, Ma, T, Wan, F, Wang, W, Yang, D, et al. Influence of dietary supplementation with bacillus licheniformis and Saccharomyces cerevisiae as alternatives to monensin on growth performance, antioxidant, immunity, ruminal fermentation and microbial diversity of fattening lambs. Sci Rep. (2018) 8:16712. doi: 10.1038/s41598-018-35081-4
15. Hall, NAL, Carlyle, BC, Haerty, W, and Tunbridge, EM. Roadblock: improved annotations do not necessarily translate into new functional insights. Genome Biol. (2021) 22:320. doi: 10.1186/s13059-021-02542-5
16. Hrdlickova, R, Toloue, M, and Tian, B. RNA-Seq methods for transcriptome analysis. Wiley Interdiscip Rev RNA. (2016) 8:10. doi: 10.1002/wrna.1364
17. Elekwachi, CO, Wang, Z, Wu, X, Rabee, A, and Forster, RJ. Total rRNA-Seq analysis gives insight into bacterial, fungal, protozoal and archaeal communities in the rumen using an optimized RNA isolation method. Front Microbiol. (2017) 8:1814. doi: 10.3389/fmicb.2017.01814
18. Zhuang, Y, Chai, J, Abdelsattar, MM, Fu, Y, and Zhang, N. Transcriptomic and metabolomic insights into the roles of exogenous β-hydroxybutyrate acid for the development of rumen epithelium in young goats. Anim Nutr. (2023) 15:10–21. doi: 10.1016/j.aninu.2023.02.012
19. Baldwin Vi, RL, Liu, M, Connor, EE, Ramsay, TG, Liu, GE, and Li, C-J. Transcriptional reprogramming in rumen epithelium during the developmental transition of pre-ruminant to the ruminant in cattle. Animals. (2021) 11:2870. doi: 10.3390/ani11102870
20. Sun, D, Yin, Y, Guo, C, Liu, L, Mao, S, Zhu, W, et al. Transcriptomic analysis reveals the molecular mechanisms of rumen wall morphological and functional development induced by different solid diet introduction in a lamb model. J Anim Sci Biotechnol. (2021) 12:33. doi: 10.1186/s40104-021-00556-4
21. Wang, J, Zhao, K, Li, M, Fan, H, Wang, M, Xia, S, et al. A preliminary study of the potential molecular mechanisms of individual growth and rumen development in calves with different feeding patterns. Microorganisms. (2023) 11:2423. doi: 10.3390/microorganisms11102423
22. Chai, J, Lv, X, Zhuang, Y, Diao, Q, Cui, K, Deng, F, et al. Dataset of the rumen microbiota and epithelial transcriptomics and proteomics in goat affected by solid diets. Sci Data. (2024) 11:749. doi: 10.1038/s41597-024-03584-7
23. Connor, EE, Baldwin, RL, Li, CJ, Li, RW, and Chung, H. Gene expression in bovine rumen epithelium during weaning identifies molecular regulators of rumen development and growth. Funct Integr Genomics. (2013) 13:133–42. doi: 10.1007/s10142-012-0308-x
24. Liu, Y, Ma, L, Riqing, D, Qu, J, Chen, J, Zhandu, D, et al. Microbial metagenomes and host transcriptomes reveal the dynamic changes of rumen gene expression, microbial colonization and co-regulation of mineral element metabolism in yaks from birth to adulthood. Animals. (2024) 14:1365. doi: 10.3390/ani14091365
25. Xiang, R, McNally, J, Rowe, S, Jonker, A, Pinares-Patino, CS, Oddy, VH, et al. Gene network analysis identifies rumen epithelial cell proliferation, differentiation and metabolic pathways perturbed by diet and correlated with methane production. Sci Rep. (2016) 6:39022. doi: 10.1038/srep39022
26. Sha, Y, He, Y, Liu, X, Zhao, S, Hu, J, Wang, J, et al. Rumen epithelial development-and metabolism-related genes regulate their micromorphology and VFAs mediating plateau adaptability at different ages in Tibetan sheep. Int J Mol Sci. (2022) 23:16078. doi: 10.3390/ijms232416078
27. Nenciarini, S, Reis-Costa, A, Pallecchi, M, Renzi, S, D’Alessandro, A, Gori, A, et al. Investigating yeast–lactobacilli interactions through co-culture growth and metabolite analysis. Fermentation. (2023) 9:933. doi: 10.3390/fermentation9110933
28. Intanoo, M, Kongkeitkajorn, MB, Suriyasathaporn, W, Phasuk, Y, Bernard, JK, and Pattarajinda, V. Effect of supplemental Kluyveromyces marxianus and Pichia kudriavzevii on aflatoxin M1 excretion in Milk of lactating dairy cows. Animals. (2020) 10:709. doi: 10.3390/ani10040709
29. Motey, GA, Johansen, PG, Owusu-Kwarteng, J, Ofori, LA, Obiri-Danso, K, Siegumfeldt, H, et al. Probiotic potential of Saccharomyces cerevisiae and Kluyveromyces marxianus isolated from West African spontaneously fermented cereal and milk products. Yeast. (2020) 37:403–12. doi: 10.1002/yea.3513
30. Xu, Z, Yang, L, Chen, H, Liu, S, Li, X, Li, S, et al. Saccharomyces cerevisiae and Kluyveromyces marxianus yeast co-cultures modulate the ruminal microbiome and metabolite availability to enhance rumen barrier function and growth performance in weaned lambs. Anim Nutr. (2024) 19:139–52. doi: 10.1016/j.aninu.2024.06.005
31. Krueger, WK, Gutierrez-Bañuelos, H, Carstens, GE, Min, BR, Pinchak, WE, Gomez, RR, et al. Effects of dietary tannin source on performance, feed efficiency, ruminal fermentation, and carcass and non-carcass traits in steers fed a high-grain diet. Anim Feed Sci Technol. (2010) 159:1–9. doi: 10.1016/j.anifeedsci.2010.05.003
32. Malhi, M, Gui, H, Yao, L, Aschenbach, JR, Gäbel, G, and Shen, Z. Increased papillae growth and enhanced short-chain fatty acid absorption in the rumen of goats are associated with transient increases in cyclin D1 expression after ruminal butyrate infusion. J Dairy Sci. (2013) 96:7603–16. doi: 10.3168/jds.2013-6700
33. Wang, B, Wang, D, Wu, X, Cai, J, Liu, M, Huang, X, et al. Effects of dietary physical or nutritional factors on morphology of rumen papillae and transcriptome changes in lactating dairy cows based on three different forage-based diets. BMC Genomics. (2017) 18:353. doi: 10.1186/s12864-017-3726-2
34. Kim, D, Langmead, B, and Salzberg, SL. HISAT: a fast spliced aligner with low memory requirements. Nat Methods. (2015) 12:357–60. doi: 10.1038/nmeth.3317
35. Pertea, M, Kim, D, Pertea, GM, Leek, JT, and Salzberg, SL. Transcript-level expression analysis of RNA-seq experiments with HISAT. StringTie and Ballgown Nat Protoc. (2016) 11:1650–67. doi: 10.1038/nprot.2016.095
36. Xie, C, Mao, X, Huang, J, Ding, Y, Wu, J, Dong, S, et al. KOBAS 2.0: a web server for annotation and identification of enriched pathways and diseases. Nucleic Acids Res. (2011) 39:W316–22. doi: 10.1093/nar/gkr483
37. Livak, KJ, and Schmittgen, TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods. (2001) 25:402–8. doi: 10.1006/meth.2001.1262
38. Pan, X, Li, Z, Li, B, Zhao, C, Wang, Y, Chen, Y, et al. Dynamics of rumen gene expression, microbiome colonization, and their interplay in goats. BMC Genomics. (2021) 22:288–15. doi: 10.1186/s12864-021-07595-1
39. Baker, LM, Kraft, J, Karnezos, TP, and Greenwood, SL. Review: the effects of dietary yeast and yeast-derived extracts on rumen microbiota and their function. Anim Feed Sci Technol. (2022) 294:115476. doi: 10.1016/j.anifeedsci.2022.115476
40. Jing, XP, Peng, QH, Hu, R, Zou, HW, Wang, HZ, Yu, XQ, et al. Dietary supplements during the cold season increase rumen microbial abundance and improve rumen epithelium development in Tibetan sheep. J Anim Sci. (2018) 96:293–305. doi: 10.1093/jas/skx032
41. Nishihara, K, Kato, D, Suzuki, Y, Kim, D, Nakano, M, Yajima, Y, et al. Comparative transcriptome analysis of rumen papillae in suckling and weaned Japanese black calves using RNA sequencing. J Anim Sci. (2018) 96:2226–37. doi: 10.1093/jas/skx016
42. Wang, H, Su, M, Wang, C, Li, D, Li, Q, Liu, Z, et al. Yeast culture repairs rumen epithelial injury by regulating microbial communities and metabolites in sheep. Front Microbiol. (2023) 14:1305772. doi: 10.3389/fmicb.2023.1305772
43. Liu, Y, Wu, Q, Wu, X, Algharib, SA, Gong, F, Hu, J, et al. Structure, preparation, modification, and bioactivities of β-glucan and mannan from yeast cell wall: a review. Int J Biol Macromol. (2021) 173:445–56. doi: 10.1016/j.ijbiomac.2021.01.125
44. Wang, S, Zhu, S, Zhang, J, Li, H, Yang, D, Huang, S, et al. Supplementation with yeast culture improves the integrity of intestinal tight junction proteins via NOD1/NF-κB P65 pathway in weaned piglets and H2O2-challenged IPEC-J2 cells. J Funct Foods. (2020) 72:104058. doi: 10.1016/j.jff.2020.104058
45. Jia, W, Zhang, R, Zhu, Z, and Shi, L. LC-Q-Orbitrap HRMS-based proteomics reveals potential nutritional function of goat whey fraction. J Funct Foods. (2021) 82:104502. doi: 10.1016/j.jff.2021.104502
46. Rui, L. New antidiabetes agent targeting both mitochondrial uncoupling and pyruvate catabolism: two birds with one stone. Diabetes. (2019) 68:2195–6. doi: 10.2337/dbi19-0024
47. Srivastava, AP, Luo, M, Zhou, W, Symersky, J, Bai, D, Chambers, MG, et al. High-resolution cryo-EM analysis of the yeast ATP synthase in a lipid membrane. Science. (2018) 360:eaas9699. doi: 10.1126/science.aas9699
48. Ji, W, Tang, X, Du, W, Lu, Y, Wang, N, Wu, Q, et al. Optical/electrochemical methods for detecting mitochondrial energy metabolism. Chem Soc Rev. (2022) 51:71–127. doi: 10.1039/d0cs01610a
49. Chinopoulos, C. Acute sources of mitochondrial NAD+ during respiratory chain dysfunction. Exp Neurol. (2020) 327:113218. doi: 10.1016/j.expneurol.2020.113218
50. Chaban, Y, Boekema, EJ, and Dudkina, NV. Structures of mitochondrial oxidative phosphorylation supercomplexes and mechanisms for their stabilisation. Biochim Biophys Acta. (2014) 1837:418–26. doi: 10.1016/j.bbabio.2013.10.004
51. Lenaz, G, Fato, R, Genova, ML, Bergamini, C, Bianchi, C, and Biondi, A. Mitochondrial complex I: structural and functional aspects. Biochim Biophys Acta. (2006) 1757:1406–20. doi: 10.1016/j.bbabio.2006.05.007
52. Efremov, RG, Baradaran, R, and Sazanov, LA. The architecture of respiratory complex I. Nature. (2010) 465:441–5. doi: 10.1038/nature09066
53. Baradaran, R, Berrisford, JM, Minhas, GS, and Sazanov, LA. Crystal structure of the entire respiratory complex I. Nature. (2013) 494:443–8. doi: 10.1038/nature11871
54. Mallik, B, and Frank, CA. Roles for mitochondrial complex I subunits in regulating synaptic transmission and growth. Front Neurosci. (2022) 16:846425. doi: 10.3389/fnins.2022.846425
55. Courage, C, Jackson, CB, Hahn, D, Euro, L, Nuoffer, JM, Gallati, S, et al. SDHA mutation with dominant transmission results in complex II deficiency with ocular, cardiac, and neurologic involvement. Am J Med Genet A. (2016) 173:225–30. doi: 10.1002/ajmg.a.37986
56. Goetzman, E, Gong, Z, Zhang, B, and Muzumdar, R. Complex II biology in aging, health, and disease. Antioxidants. (2023) 12:1477. doi: 10.3390/antiox12071477
57. Hargreaves, IP. Coenzyme Q10 in mitochondrial and lysosomal disorders. J Clin Med. (2021) 10:1970. doi: 10.3390/jcm10091970
58. Sloan, DB, Warren, JM, Williams, AM, Wu, Z, Abdel-Ghany, SE, Chicco, AJ, et al. Cytonuclear integration and co-evolution. Nat Rev Genet. (2018) 19:635–48. doi: 10.1038/s41576-018-0035-9
59. Schmidt-Rohr, K. Oxygen is the high-energy molecule powering complex multicellular life: fundamental corrections to traditional bioenergetics. ACS Omega. (2020) 5:2221–33. doi: 10.1021/acsomega.9b03352
60. Yoshikawa, S, Muramoto, K, Shinzawa-Itoh, K, Aoyama, H, Tsukihara, T, Shimokata, K, et al. Proton pumping mechanism of bovine heart cytochrome c oxidase. Biochim Biophys Acta. (2006) 1757:1110–6. doi: 10.1016/j.bbabio.2006.06.004
61. Senkler, J, Rugen, N, Eubel, H, Hegermann, J, and Braun, H-P. Absence of complex I implicates rearrangement of the respiratory chain in European mistletoe. Curr Biol. (2018) 28:1606–13.e4. doi: 10.1016/j.cub.2018.03.050
62. Singh, A, Kukreti, R, Saso, L, and Kukreti, S. Oxidative stress: a key modulator in neurodegenerative diseases. Molecules. (2019) 24:1583. doi: 10.3390/molecules24081583
63. Yang, T, Zhan, K, Ning, L, Jiang, M, and Zhao, G. Short-chain fatty acids inhibit bovine rumen epithelial cells proliferation via upregulation of cyclin-dependent kinase inhibitors 1A, but not mediated by G protein-coupled receptor 41. J Anim Physiol Anim Nutr. (2019) 104:409–17. doi: 10.1111/jpn.13266
Keywords: weaned lamb, transcriptome, co-cultures, oxidative phosphorylation, rumen
Citation: Xu Z, Yang L, Chen H, Bai P, Li X and Liu D (2025) Transcriptomic characterization of the functional and morphological development of the rumen wall in weaned lambs fed a diet containing yeast co-cultures of Saccharomyces cerevisiae and Kluyveromyces marxianus. Front. Vet. Sci. 12:1510689. doi: 10.3389/fvets.2025.1510689
Edited by:
Yanfeng Xue, Anhui Agricultural University, ChinaReviewed by:
Haji Akbar, Langston University, United StatesYi Ma, Jiangsu University, China
Zhuo Zhao, Anhui Agricultural University, China
Copyright © 2025 Xu, Yang, Chen, Bai, Li and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Dacheng Liu, bm1nbGRjQDE2My5jb20=
 Zixuan Xu
Zixuan Xu Lan Yang
Lan Yang