
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Vet. Sci., 05 February 2025
Sec. Veterinary Epidemiology and Economics
Volume 12 - 2025 | https://doi.org/10.3389/fvets.2025.1493928
This article is part of the Research TopicParasites in One Health InterfaceView all 18 articles
 Shuo Liu1,2†
Shuo Liu1,2† Jing-Hao Li3†
Jing-Hao Li3† Si-Yuan Qin3
Si-Yuan Qin3 Jing Jiang1*
Jing Jiang1* Zhen-Jun Wang4
Zhen-Jun Wang4 Tao Ma4
Tao Ma4 Jun-Hui Zhu4
Jun-Hui Zhu4 Hong-Li Geng2
Hong-Li Geng2 Wei-Lan Yan2
Wei-Lan Yan2 Nian-Yu Xue5
Nian-Yu Xue5 Yan Tang6
Yan Tang6 He-Ting Sun3*
He-Ting Sun3*Introduction: Pentatrichomonas hominis is a conditional pathogen that parasitizes the intestines of vertebrates and has been detected in various wild animals. However, its infection rate in Tibetan antelopes has not been previously studied.
Methods: In this study, 503 fecal samples from Tibetan antelopes were analyzed to determine the prevalence and molecular characteristics of P. hominis.
Results: Results showed that 1.19% (6/503) of the samples tested positive, and although the prevalence was low, this finding underscores the importance of monitoring wild animals population as hosts of zoonotic pathogens. Additionally, the highest prevalence in Nima County (6.25%, 4/64), followed by Shenza County (2.44%, 2/82). No P. hominis was detected in samples from Shuanghu, Ruoqiang, Qiemo, and Qumarlêb Counties. Seasonally, the highest prevalence was recorded in autumn (1.42%, 6/423). Interestingly, P. hominis was only detected in 2020 (2%, 6/300), with no infections found in 2023 (0/50) or 2024 (0/153). Additionally, the phylogenetic analysis indicated that most islolates belonged to the CC1 genotype, with one representing a potential novel genotype.
Discussion: This is the first s to report the presence of P. hominis in Tibetan antelopes, revealing that Tibetan antelopes may be a potential transmitter of zoontic P. hominis. These findings offer new insights into its epidemiology and contribute valuable data for Tibetan antelope conservation efforts.
Pentatrichomonas hominis is an anaerobic, unicellular protozoan characterized by five anterior flagella and a single recurrent flagellum. It belongs to the genus Trichomonadidae within the phylum Parabasalia (1). P. hominis demonstrates broad host adaptability, commonly parasitizing the intestines of vertebrates, including dogs, cats, and pigs (2–4). Pathological reports have also confirmed its presence in the urinary tract of bulls (5). While P. hominis is typically associated with gastrointestinal disturbances in non-human primates, it is increasingly recognized as a potential pathogen in human gastrointestinal diseases (6, 7). Moreover, P. hominis has been linked to respiratory diseases, suggesting its involvement in a broader range of health conditions beyond gastrointestinal issues (8). These observations imply that P. hominis could act as a zoonotic pathogen, rather than merely existing as a commensal organism. In recent years, extensive epidemiological research has focused on P. hominis in a wide range of animals, such as owls, boa constrictors, Siberian tigers, cattle, sheep, and macaques (9–13). Notably, there is a related report in huamans in northern China. However, the prevalence of P. hominis in Tibetan antelopes remains unexplored.
The Tibetan antelope (Pantholops hodgsonii) is the only species within the genus Pantholops of the subfamily Antilopinae, and it is classified as a Class I protected animal in China (14). In 2016, the International Union for Conservation of Nature (IUCN) listed the Tibetan antelope as Near Threatened (15). Tibetan antelopes are known to harbor various pathogens, such as Brucella, Blastocystis, and Cryptosporidium. They may serve as an intermediate host, facilitating the transmission of these pathogens from livestock to humans (16–18). Therefore, this study hypothesized that Tibetan antelope could serve as a host for P. hominis to promote the transmission of zoonotic diseases.
This study aimed to assess the prevalence of P. hominis, phylogenetic analysis, and potential risk factors in Tibetan antelopes. Nested PCR was employed to detect P. hominis in fecal samples collected from Tibetan antelopes across Xinjiang, Tibet, and Qinghai. This study is the first to focus on the infection and prevalence of P. hominis in Tibetan antelopes, and provides new evidence of P. hominis infection in wild animals.
In September 2020 and from July 2023 to May 2024, a total of 503 fecal samples were collected from Tibetan antelopes in the Tibet Autonomous Region, Xinjiang Uygur Autonomous Region, and Qinghai Province, China (Figure 1). In the field, fresh fecal samples were randomly collected using disposable sterile gloves, immediately placed in sterile sample tubes, and stored in ice boxes. They were then transported to the laboratory under cold chain conditions and stored at −80°C.
For each sample, 200 mg of feces was placed in 2 mL centrifuge tubes containing 200 mg of glass beads, and 0.9% saline solution was added. The mixture was homogenized using a vortex mixer (JOANLAB, Zhejiang, China). Genomic DNA was extracted following the manufacturer’s protocol using the E.Z.N.A.® Stool DNA Kit (Omega Biotek, Inc., Norcross, GA, USA). The extracted DNA was stored at −20°C until PCR analysis. This study followed the guidelines and recommendations of the Animal Welfare Committee of Qingdao Agricultural University, China. The positivity rate of P. hominis was determined using nested PCR (19). In the first round of amplification, small subunit ribosomal RNA was amplified using forward primer FF (5’-GCGCCTGAGAGATAGCGACTA-3′) and reverse primer RR (5’-GGACCTGTTATTGCTACCCTCTTC-3′) to detect Trichomonas spp. A second round of amplification was performed using 2 μL of the first-round product, with forward primer HF (5’-TGTAAACGATGCCGACAGAG-3′) and reverse primer HR (5’-CAACACTGAAGCCAATGCGAGG-3′) to specifically detect P. hominis (339 bp). Both negative and positive controls were included in each PCR. For the second-round product, 5 μL was analyzed via electrophoresis on a 1.5% agarose gel and visualized under ultraviolet light.
PCR products positive for P. hominis were sent to Anhui General Biotech Co., Ltd. (Anhui, China) for sequencing. The resulting contigs from bidirectional sequencing were aligned with reference sequences in GenBank using BLAST.1 Phylogenetic trees were constructed using the neighbor-joining (NJ) method (20), with genetic distances calculated using the Kimura 2-parameter model in MEGA11 (21). The reliability of the phylogenetic trees was assessed with 1,000 bootstrap replicates. Representative nucleotide sequences were submitted to GenBank under accession numbers PQ276129-PQ276131.
The effects of sampling region (x1), sampling season (x2), and sampling year (x3) on the prevalence of P. hominis in Tibetan antelopes were analyzed using chi-square tests in the Statistical Analysis System (SAS, v9.0), with the best model selected using Fisher scoring. Logistic regression analysis was performed using Statistical Product and Service Solutions (SPSS, IBM Corp., Armonk, NY, USA), with 95% confidence intervals (95% CI) calculated. A p-value of less than 0.05 was considered statistically significant.
In this study, 6 out of 503 fecal samples were identified as positive for P. hominis, with an overall prevalence of 1.19% (6/503, 95% CI: 0.44–2.56). Nima County had the highest infection rate at 6.25% (4/64, 95% CI: 1.38–13.76), followed by Shenza County at 2.44% (2/82, 95% CI: 0.40–7.21). No P. hominis was detected in the remaining four counties. Seasonally, no infections were detected in spring or summer, with all positive cases found in autumn (1.42%, 6/423, 95% CI: 0.47–2.10). The prevalence of P. hominis was higher in 2020 (2%, 6/300, 95% CI: 0.67–3.95) compared to 2023 (0/50) and 2024 (0/153) (Table 1 and Figure 2).

Table 1. Prevalence of Pentatrichomonas hominis infection in Tibetan antelope (Pantholops hodgsonii) by various factors.
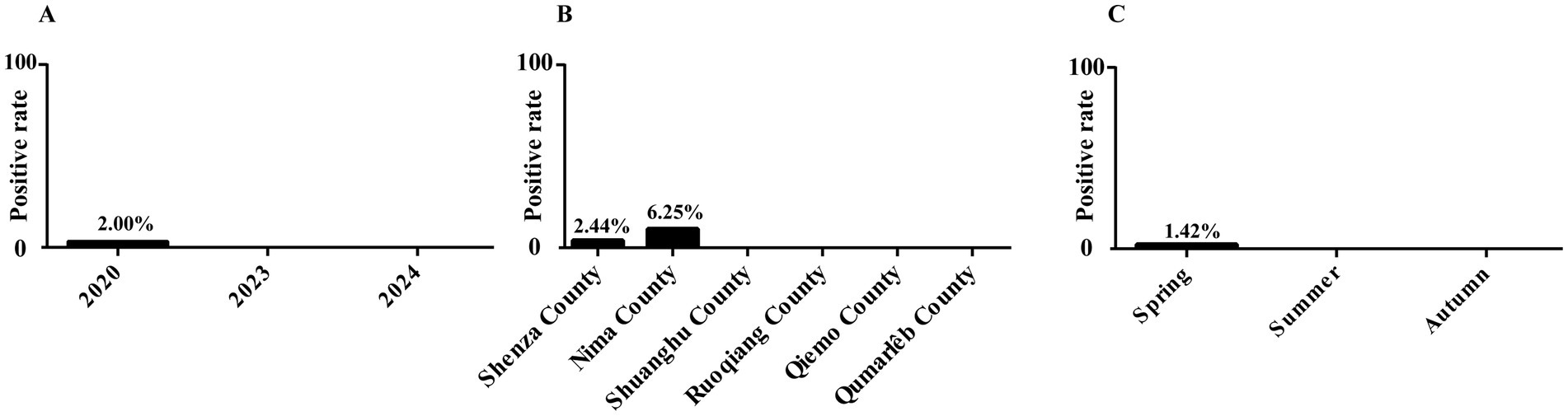
Figure 2. Infection rate of P. homins in Tibetan antelope under various factors. (A) Infection rate of P. homins in Tibetan antelope in different years. (B) Infection rate of P. homins in Tibetan antelope in different regions. (C) Infection rate of P. homins in Tibetan antelope in different seasons.
Logistic regression analysis using the Fisher scoring method was performed to assess the influence of sampling region, season, and year on P. hominis prevalence. The final model did not retain any of these factors, suggesting that sampling region, season, and year had no statistically significant impact on the infection rate of P. hominis.
Confirmed that the three representative sequences obtained in this study were all classified as P. hominis. Among them, sequences PQ276129 and PQ276130 showed 100% homology with the CC1 genotype (KJ408929). However, PQ276131 formed a distinct branch on the phylogenetic tree, indicating that this sequence may represent a novel genetic variant (Figure 3).
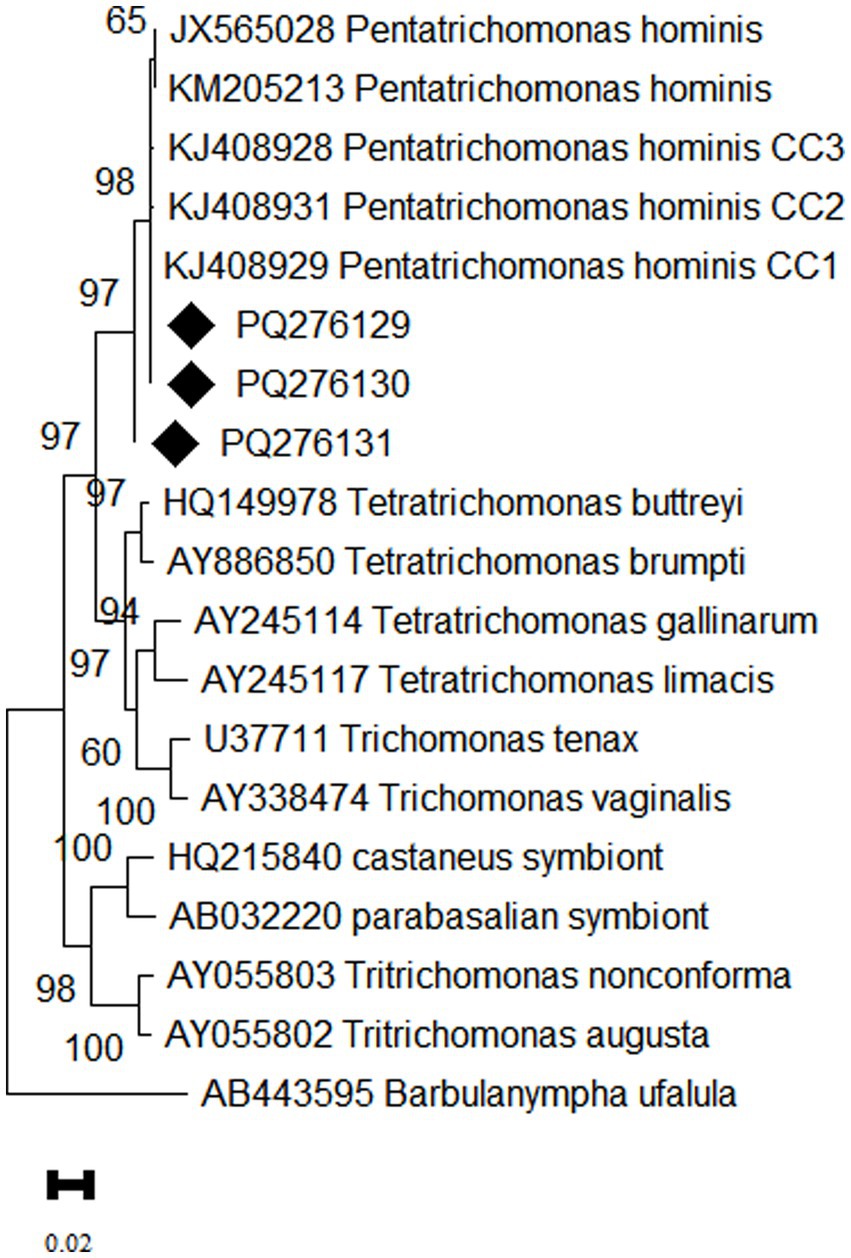
Figure 3. Phylogenetic tree based on the 18S rRNA gene of P. hominis. The phylogenetic relationship between P. hominis obtained in this study and other known trichomonads were inferred using the maximum likelihood analysis based on the genetic distance calculated by the Kimura 2-parameter model. Bootstrap values more than 50% are shown. The diamond shape denotes isolates from the present study.
Multiple case reports have demonstrated that P. hominis is often isolated from patients with diarrhea, suggesting it is a likely cause of gastrointestinal symptoms (6). Furthermore, research indicates that up to 41.54% of gastrointestinal cancer patients are infected with P. hominis, which may imply a potential role in the development of gastrointestinal cancer in some cases (22, 23). These findings underscore the importance of not underestimating the potential dangers of P. hominis. There is a critical need for further research into its hosts, transmission mechanisms, pathogenicity, and specific role in gastrointestinal diseases. Such studies could lead to the development of effective prevention and treatment strategies for P. hominis.
In this study, P. hominis was detected in Tibetan antelopes with an infection rate of 1.19% (6/503). This rate is lower compared to the infection rates of Blastocystis (4.8%, 30/627) and Cryptosporidium (3.0%, 19/627) in Tibetan antelopes (17, 18). The difference may be attributed to the varying survival and transmission capabilities of these parasites in extreme environments. Although Tibetan antelopes are ruminants, their P. hominis infection rate is lower than that of dairy cattle (6.8%, 36/526) and beef cattle (4.6%, 15/32), indicating variability in susceptibility among species (13). Notably, the infection rates of P. hominis in Siberian tigers (31.3%, 41/131) and silver foxes (43.33%, 26/60) were significantly higher than that in Tibetan antelopes, suggesting that different dietary habits shape distinct gut microbiota, resulting in varying parasite susceptibility (24).
In this study, P. hominis was detected in Nima County (6.25%, 4/64) and Shenza County (2.44%, 2/82), while it was not found in the other four counties. Considering the small difference in altitude among the sampling sites, the influence of altitude on the parasite infection rate can be excluded. The differences in infection rates may be related to local ecological and sanitary conditions affecting parasite infection rates. Seasonally, autumn on the Qinghai-Tibet Plateau appears more conducive to P. hominis transmission, with an infection rate of 1.42% (6/423) observed in autumn, compared to no infections detected in summer and spring. This may relate to the migratory behavior of Tibetan antelopes, which migrate from April to June and return between August and September (25). Studies suggest that migration not only helps Tibetan antelopes evade predators but may also be a strategy to avoid parasites, thus reducing infection risk in offspring and potentially aiding population growth (26). In conclusion, although no significant effects of seasonal and regional variations on infection rates were observed in this study, the observed trends warrant further investigation with larger sample sizes.
Interestingly, P. hominis prevalence was 2% (6/300) in 2020, with no infections detected in 2023 (0/50) and 2024 (0/153). This fluctuation may be linked to global warming, which can alter precipitation patterns and humidity, affecting parasite survival (27). Additionally, changes in vegetation may lead Tibetan antelopes to modify their migration routes and habitats, influencing P. hominis infection rates (28). However, the geographical scope of this study is limited, and the sample size is small in some years. Future studies can combine annual sampling with wider geographical coverage and larger sample size to verify this conjecture.
Epidemiological studies have shown that the CC1 genotype was first found in dogs from Changchun, later wildly distribute in P. hominis infection in dogs across eastern China. It has also been detected in Siberian tigers, foxes in Henan Province, and primates, including humans in north China, highlighting the strong cross-host transmission ability and potential zoonotic nature of this genotype (1, 7, 10). In this study, two representative P. hominis sequences from Tibetan antelopes belonged to the CC1 genotype, indicating that this genotype is the predominant one in Tibetan antelopes, further suggesting that Tibetan antelopes may serve as a potential source of infection of P. hominis in humans. Notably, PQ276131 exhibited genetic differences from the CC1, CC2, and CC3 genotypes, indicating that it may represent a novel genotype of P. hominis. The newly discovered genotype in Tibetan antelopes may represent a unique adaptation to high-altitude environments, warranting further genetic studies to gain deeper insights into its adaptation mechanisms.
Tibetan antelopes infected with P. hominis can easily disperse trophozoites into the environment (29). Given the lifestyle of pastoralists in the Tibetan region, there is a heightened risk of contact with contaminated water or food, increasing the risk of infection. This not only threatens the local ecosystem but also poses potential health risks to residents. Effective measures are needed to monitor and reduce environmental contamination by P. hominis and its impact on human health.
This study is the first to report P. hominis infection in Tibetan antelopes. Despite the low prevalence, which may be influenced by sampling and detection methods, it indicates that Tibetan antelopes could be potential vectors of CC1 genotype. This finding provides important data for understanding the epidemiology of P. hominis and suggests that Tibetan antelopes should be considered in studies and monitoring of zoonoses in Tibetan areas. Addtionally, this study recommends including other wild animals and livestock in the Tibetan region in the scope of investigation and emphasizes the importance of incorporating P. hominis into public health surveillance programs in areas with high human-wildlife interaction.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/supplementary material.
The animal study was approved by the Research Ethics Committee for the Care and Use of Laboratory Animals in Qingdao Agricultural University, China. The study was conducted in accordance with the local legislation and institutional requirements.
SL: Methodology, Software, Writing – original draft. J-HL: Conceptualization, Resources, Writing – review & editing. S-YQ: Resources, Writing – review & editing. JJ: Conceptualization, Writing – review & editing. Z-JW: Resources, Writing – review & editing. TM: Resources, Writing – review & editing. J-HZ: Resources, Writing – review & editing. H-LG: Methodology, Writing – review & editing. W-LY: Methodology, Writing – review & editing. N-YX: Data curation, Writing – review & editing. YT: Data curation, Writing – review & editing. H-TS: Conceptualization, Funding acquisition, Resources, Writing – review & editing.
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This work was supported by National Key Research and Development Program of China (2021YFC2300903).
We would like to extend our deepest gratitude to Chair Professor Hany Elsheikha, from University of Nottingham, for his invaluable assistance in revising and improving our manuscript. His insightful feedback, expert advice, and meticulous attention to detail have significantly enhanced the quality of our work.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
1. Song, P, Guo, Y, Zuo, S, Li, L, Liu, F, Zhang, T, et al. Prevalence of Pentatrichomonas hominis in foxes and raccoon dogs and changes in the gut microbiota of infected female foxes in the Hebei and Henan provinces in China. Parasitol Res. (2023) 123:74. doi: 10.1007/s00436-023-08099-5
2. Li, WC, Wang, K, Zhang, W, Wu, J, Gu, YF, and Zhang, XC. Prevalence and molecular characterization of intestinal Trichomonads in pet dogs in East China. Korean J Parasitol. (2016) 54:703–10. doi: 10.3347/kjp.2016.54.6.703
3. Itoh, N, Iijima, Y, Ogura, I, Yonekura, N, Kameshima, S, and Kimura, Y. Molecular prevalence of Trichomonad species from pet shop puppies and kittens in Japan. Rev Bras Parasitol Vet. (2020) 29:e014820. doi: 10.1590/S1984-29612020098
4. Li, WC, Wang, K, Li, Y, Zhao, LP, Xiao, Y, and Gu, YF. Survey and molecular characterization of Trichomonads in pigs in Anhui Province, East China, 2014. Iran J Parasitol. (2018) 4:602–10.
5. Silva, ORE, Ribeiro, L, Jesus, VLT, McIntosh, D, Silenciato, LN, Ferreira, JE, et al. Identification of Pentatrichomonas hominis in preputial washes of bulls in Brazil. Rev Bras Parasitol Vet. (2022) 31:e005322. doi: 10.1590/S1984-29612022034
6. Abdo Mohamed, S, Ibrahim Ghallab, MM, Elhawary Mohammed, N, and Elhadad, H. Pentatrichomonas hominis and other intestinal parasites in school-aged children: coproscopic survey. J Parasit Dis. (2022) 46:896–900. doi: 10.1007/s12639-022-01506-1
7. Li, WC, Ying, M, Gong, PT, Li, JH, Yang, J, Li, H, et al. Pentatrichomonas hominis: prevalence and molecular characterization in humans, dogs, and monkeys in northern China. Parasitol Res. (2016) 115:569–74. doi: 10.1007/s00436-015-4773-8
8. Mantini, C, Souppart, L, Noel, C, Duong, TH, Mornet, M, Carroger, G, et al. Molecular characterization of a new Tetratrichomonas species in a patient with empyema. J Clin Microbiol. (2009) 47:2336–9. doi: 10.1128/JCM.00353-09
9. Dimasuay, KG, and Rivera, WL. Molecular characterization of Trichomonads isolated from animal hosts in the Philippines. Vet Parasitol. (2013) 196:289–95. doi: 10.1016/j.vetpar.2013.03.019
10. Zhang, H, Zhang, N, Gong, P, Cheng, S, Wang, X, Li, X, et al. Prevalence and molecular characterization of Pentatrichomonas hominis in Siberian tigers (Panthera tigris altaica) in Northeast China. Integr Zool. (2022) 17:543–9. doi: 10.1111/1749-4877.12629
11. Dib, LV, Barbosa, ADS, Correa, LL, Torres, BDS, Pissinatti, A, Moreira, SB, et al. Morphological and molecular characterization of parabasilids isolated from ex-situ nonhuman primates and their keepers at different institutions in Brazil. Int J Parasitol Parasites Wildl. (2024) 24:100946. doi: 10.1016/j.ijppaw.2024.100946
12. Li, WC, Wang, K, and Gu, Y. Occurrence of Blastocystis sp. and Pentatrichomonas hominis in sheep and goats in China. Parasit Vectors. (2018) 11:93. doi: 10.1186/s13071-018-2671-5
13. Li, WC, Huang, JM, Fang, Z, Ren, Q, Tang, L, Kan, ZZ, et al. Prevalence of Tetratrichomonas buttreyi and Pentatrichomonas hominis in yellow cattle, dairy cattle, and water buffalo in China. Parasitol Res. (2020) 119:637–47. doi: 10.1007/s00436-019-06550-0
14. Song, X, Shen, W, Wan, H, Hou, P, and Lin, G. Dynamic monitoring of Tibetan antelope habitat suitability in the Hoh Xil nature reserve using remote sensing images. Res Sci. (2016) 38:17–23. doi: 10.18402/resci.2016.08.03
15. International Union for Conservation of Nature and Natural Resources. The IUCN red list of threatened species. Gland, Switzerland: International Union for Conservation of Nature and Natural Resources (2016).
16. Wu, JY, Li, JJ, Wang, DF, Wei, YR, Meng, XX, Tuerxun, G, et al. Seroprevalence of five zoonotic pathogens in wild ruminants in Xinjiang, Northwest China. Vector Borne Zoonotic Dis. (2020) 20:882–7. doi: 10.1089/vbz.2020.2630
17. Qin, SY, Sun, HT, Lyu, C, Zhu, JH, Wang, ZJ, Ma, T, et al. Prevalence and characterization of Cryptosporidium species in Tibetan Antelope (Pantholops hodgsonii). Front Cell Infect Microbiol. (2021) 11:713873. doi: 10.3389/fcimb.2021.713873
18. Geng, HL, Sun, YZ, Jiang, J, Sun, HT, Li, YG, Qin, SY, et al. The presence of Blastocystis in Tibetan Antelope (Pantholops hodgsonii). Front Cell Infect Microbiol. (2021) 11:747952. doi: 10.3389/fcimb.2021.747952
19. Wang, ZR, Fan, QX, Wang, JL, Zhang, S, Wang, YX, Zhang, ZD, et al. Molecular identification and survey of Trichomonad species in pigs in Shanxi Province, North China. Vet Sci. (2024) 11:203. doi: 10.3390/vetsci11050203
20. Saitou, NNM. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. (1987) 4:406–25.
21. Tamura, K, Stecher, G, and Kumar, S. MEGA11: molecular evolutionary genetics analysis version 11. Mol Biol Evol. (2021) 38:3022–7. doi: 10.1093/molbev/msab120
22. Zhang, N, Zhang, H, Yu, Y, Gong, P, Li, J, Li, Z, et al. High prevalence of Pentatrichomonas hominis infection in gastrointestinal cancer patients. Parasit Vectors. (2019) 12:423. doi: 10.1186/s13071-019-3684-4
23. Zhang, H, Yu, Y, Li, J, Gong, P, Wang, X, Li, X, et al. Changes of gut microbiota in colorectal cancer patients with Pentatrichomonas hominis infection. Front Cell Infect Microbiol. (2022) 12:961974. doi: 10.3389/fcimb.2022.961974
24. Li, X, Li, J, Zhang, X, Yang, Z, Yang, J, and Gong, P. Prevalence of Pentatrichomonas hominis infections in six farmed wildlife species in Jilin, China. Vet Parasitol. (2017) 244:160–3. doi: 10.1016/j.vetpar.2017.07.032
25. Buho, H, Jiang, Z, Liu, C, Yoshida, T, Mahamut, H, Kaneko, M, et al. Preliminary study on migration pattern of the Tibetan Antelope (Pantholops hodgsonii) based on satellite tracking. Adv Space Res. (2011) 48:43–8. doi: 10.1016/j.asr.2011.02.015
26. Cao, Y, Foggin, M, and Zhao, X. Tibetan Antelope migration during mass calving as parasite avoidance strategy. Innovations. (2022) 3:100326. doi: 10.1016/j.xinn.2022.100326
27. Wood, CL, Welicky, RL, Preisser, WC, Leslie, KL, Mastick, N, Greene, C, et al. A reconstruction of parasite burden reveals one century of climate-associated parasite decline. Proc Natl Acad Sci USA. (2023) 120:e2211903120. doi: 10.1073/pnas.2211903120
28. Shi, F, Liu, S, An, Y, Sun, Y, Zhao, S, Liu, Y, et al. Climatic factors and human disturbance influence ungulate species distribution on the Qinghai-Tibet plateau. Sci Total Environ. (2023) 869:161681. doi: 10.1016/j.scitotenv.2023.161681
Keywords: Pentatrichomonas hominis , prevalence, Tibetan antelope (Pantholops hodgsonii), risk factors, China
Citation: Liu S, Li J-H, Qin S-Y, Jiang J, Wang Z-J, Ma T, Zhu J-H, Geng H-L, Yan W-L, Xue N-Y, Tang Y and Sun H-T (2025) Existence of Pentatrichomonas hominis in Tibetan Antelope (Pantholops hodgsonii). Front. Vet. Sci. 12:1493928. doi: 10.3389/fvets.2025.1493928
Received: 12 September 2024; Accepted: 24 January 2025;
Published: 05 February 2025.
Edited by:
Mughees Aizaz Alvi, University of Agriculture, Faisalabad, PakistanReviewed by:
Hongchao Sun, Zhejiang Academy of Agricultural Sciences, ChinaCopyright © 2025 Liu, Li, Qin, Jiang, Wang, Ma, Zhu, Geng, Yan, Xue, Tang and Sun. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jing Jiang, amlhbmdqaW5neGlhb3lhb0AxNjMuY29t; He-Ting Sun, eGlhb2ZlbmdzaHRAMTYzLmNvbQ==
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.