
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Vet. Sci., 21 March 2025
Sec. Veterinary Epidemiology and Economics
Volume 12 - 2025 | https://doi.org/10.3389/fvets.2025.1481822
This article is part of the Research TopicAntimicrobial Resistance in Veterinary Medicine: Epidemiology, Economic Impact, and Mitigation StrategiesView all 4 articles
 Shaqiu Zhang1,2,3,4*†
Shaqiu Zhang1,2,3,4*† Jing Yang1†
Jing Yang1† Qian Yang1†
Qian Yang1† Qianlong Li1
Qianlong Li1 Zhijun Zhong3
Zhijun Zhong3 Mingshu Wang1,2,3,4
Mingshu Wang1,2,3,4 Renyong Jia1,2,3,4
Renyong Jia1,2,3,4 Shun Chen1,2,3,4
Shun Chen1,2,3,4 Mafeng Liu1,2,3,4
Mafeng Liu1,2,3,4 Dekang Zhu1,2,3,4
Dekang Zhu1,2,3,4 Xinxin Zhao1,2,3,4
Xinxin Zhao1,2,3,4 Ying Wu1,2,3,4
Ying Wu1,2,3,4 Qiao Yang1,2,3,4
Qiao Yang1,2,3,4 Juan Huang1,2,3,4
Juan Huang1,2,3,4 Xumin Ou1,2,3,4
Xumin Ou1,2,3,4 Di Sun1,2,3,4
Di Sun1,2,3,4 Bin Tian1,2,3,4
Bin Tian1,2,3,4 Zhen Wu1,2,3,4
Zhen Wu1,2,3,4 Yu He1,2,3,4
Yu He1,2,3,4 Anchun Cheng1,2,3,4*
Anchun Cheng1,2,3,4*Fosfomycin (FOS) is a critical antibiotic for treating multi-drug resistant (MDR) Enterobacteriaceae infections, but its effectiveness is jeopardized by the dissemination of plasmids encoding enzymes that modify FOS. Despite the prohibition on its use in animal breeding in China, 100 strains of Escherichia coli (E. coli) exhibiting high resistance to FOS (MIC≥512 mg/L) were isolated from samples of waterfowl origin collected in Hainan, Sichuan, and Anhui. These strains commonly carried the fosA3 (88/100, 88.0%). In addition, 21 other antimicrobial resistance genes (ARGs) were detected in these strains, with high positivity rates for tetA, aphA1, sul2, folR, qnrS, and blaCTX-M. It is noteworthy that there was a significant positive correlation between the fosA3 and blaCTX-M (OR = 15.162, 95% CI: 1.875–122.635). The results of pulsed-field gel electrophoresis (PFGE) demonstrated the existence of multiple dispersed clonal clusters. Multilocus sequence typing (MLST) analysis identified 45 ST types, with ST48 and ST10 representing the most dominant clones. In the conjugation experiments, 53 fosA-like genes positive transconjugants were obtained with measurable conjugation frequency, which strongly demonstrated that these fosA3 may mainly locate on different types of plasmids possessing an efficient transmission ability. Whole genome sequencing (WGS) analysis further showed that the fosA3 was co-localized with the blaCTX-M on plasmids that showed a high degree of similarity in genetic structure. Of particular interest is the observation that the fosA3 is frequently accompanied by IS26 on either side of the gene. This structure may play a pivotal role in the horizontal transfer of the fosA3. The study revealed the alarming prevalence of FOS resistance in E. coli of waterfowl origin and delved deeply into the genetic characteristics and transmission mechanisms of the fosA3. The discovery of plasmid-mediated, transmissible FOS resistance in waterfowl E. coli poses a threat to “One Health”. There’s an urgent need for thorough monitoring and control measures against FOS resistance.
Antimicrobial resistance (AMR) represents a significant challenge to global public health (1). It is projected that by 2050, AMR will be responsible for the deaths of approximately 10 million individuals (2). In contrast to the rapid spread and development of AMR, the development of novel antimicrobial has been relatively sluggish (3). Consequently, the scientific community has initiated a re-evaluation of the potential benefits of older antimicrobial. Fosfomycin (FOS), an antimicrobial agent first discovered in the 1960s, has garnered renewed interest in recent years (4). FOS is bactericidal against a variety of multi-drug resistant bacteria (5). For example, carbapenem-resistant Enterobacteriaceae (CRE), extended-spectrum β-lactamase producing Enterobacteriaceae (ESBL-PE), methicillin-resistant Staphylococcus aureus, and vancomycin-resistant Enterococcus faecalis, all of which are classified as priority pathogens by the World Health Organization (6). However, with the increasing use of FOS, there has also been a rise in instances of FOS-resistant pathogens, particularly those harboring plasmid-encoded FOS resistance modifier genes (7).
Previous studies have demonstrated that two primary mechanisms mediate resistance to FOS in Enterobacteriaceae: (i) Chromosomal mutations in the FOS target gene murA or in FOS uptake genes (e.g., uhpT, uhpA, glpT, cyaA and ptsI); and (ii) Plasmid encoded inactivating enzymes (including FosA variants, FosB, FosC, FosX, FosK, FosD, FosE, FosI, FosL, FomA and FomB). Although the use of FOS in animals has long been prohibited in China, there has been a notable increase in the incidence of FOS resistance among bacteria of animal origin (8). In addition, recent studies indicate that plasmid-mediated fosA3 is the predominant mechanism conferring resistance to FOS within Enterobacteriaceae in China (9). The presence of bacteria harboring the fosA3 poses a significant risk to both animal and human health under the “One Health” concept. Notably, The fosA3 has been identified in Enterobacteriaceae isolated from a diverse array of animal sources in China, including pigeons, pigs, chickens, companion animals, cattle, and rodents (10, 11). It has been observed that livestock and poultry populations within China have become reservoirs for FOS-resistant bacteria. Waterfowl excrement serves as a reservoir for novel ARGs, with various ARGs having been identified in the feces of waterfowl (12–16). The rapid dissemination of fosA3 is evidently associated with IncF plasmid and IS26 (9, 17). Plasmids carrying the fos gene are frequently found to co-occur with other ARGs, thereby increasing the likelihood that FOS resistance will be selected concurrently alongside these additional resistance mechanisms. Previous studies have shown that coexistence between fosA3 and other ARGs is mediated by IS26-type complex transposons (18).
Our research team has a long-standing interest in the study of bacterial resistance in waterfowl, and has produced a series of findings, particularly in the area of MDR (15, 16, 19). Among our findings, we have made significant strides in understanding the global distribution of E. coli harboring the mcr-1, and have conducted a comprehensive genomic analysis to elucidate the spread of mcr-1 and its broader public health implications (13, 20). In the field of tigecycline resistance, our initial research elucidated the molecular propagation of tet(X4) in the avian environment of Sichuan Province (15, 21). In the area of carbapenem resistance, we have identified for the first time E. coli carrying the blaNDM-1 in waterfowl-origin bacteria from a tropical island in China, a finding that underscores the urgency of tightening control of the spread of blaNDM-1-producing E. coli in the region (22).
The prevalence of drug resistance in waterfowl is increasing on a global scale. The spread of AMR microorganisms and associated genes is not geographically restricted and can easily cross different regions and hosts. It is therefore imperative that global cooperation should be established to address this challenge, through the development of policies, the implementation of preventive measures, and the involvement of all relevant parties in the entire chain.
Despite some advances in research on FOS resistance, our knowledge of FOS resistance in E. coli in waterfowl remains limited. FOS, a long-established antimicrobial agent, has recently been the subject of renewed interest from the research community due to its demonstrated bactericidal activity against a wide range of antibiotic-resistant bacteria (ARB). However, with the increased utilization of FOS, the prevalence of ARB harboring plasmid-encoded FOS resistance related genes has also increased. Accordingly, the objective of this study was to conduct a comprehensive investigation into the resistance characteristics of E. coli derived from waterfowl to FOS, and to analyze the genetic characteristics of the fosA3 and its transmission risk. This is crucial for enhancing our comprehension of and ability to control ARB in waterfowl.
Our research subjects are strains of E. coli isolated and preserved in the laboratory from 2019 to 2023, which all originate from representative waterfowl species in China, including Sichuan mallard ducks, Hainan Jiaji ducks, and Anhui white geese (16, 23, 24). The sample collection sites cover the main breeding areas of these waterfowl, ensuring the geographical representativeness of the samples. Specifically, the sources of the samples include duck intestinal contents or pericardial effusion, cloacal swabs and fresh feces, all of which were collected onsite at breeding farms. During the collection process, we had detailed exchanges with local veterinarians and learned that these breeding farms have a history of using antibiotics.
The authenticated E. coli strains were meticulously stored in 20% glycerol at −80°C. Subsequently, these isolates were cultured on MacConkey agar plates supplemented with 128 mg/L of FOS and 25 mg/L of glucose-6-phosphate (G-6-P), with the aim of identifying FOS-resistant E. coli strains among our previously collected strains.
Total genomic DNA was extracted from bacterial cultures using the TIANamp Bacteria DNA Kit (TianGen Biotech, Beijing, China, #DP302) according to the manufacturer’s instructions, and the DNA quality was measured by UV absorbance (ND1000, Nanodrop, Thermo Fisher Scientific). The DNA samples were stored at −20°C.
In order to determine which types of fosA-like genes are carried by FOS -resistant E. coli, 10 previously reported fosA-like genes (fosA to fosA10) were identified through PCR. Furthermore, these strains were tested for the presence of other ARGs using primers that had previously been employed for this purpose. All primers are shown in Supplementary Table S1. PCR experiments were performed in a 20 μL volume containing 2 μL of template DNA, 10 μL of 2 × Hieff® Robust PCRMaster Mix (With Dye) (Yeasen Biotechnology, Shanghai, China), 6 μL of ddH2O, and 1 μL of each primer. The PCR amplification program was set according to the instructions of the amplification enzyme. PCR products were separated by gel electrophoresis in a 1.0% agarose gel stained with GoldView™ (Sangon Biotech, Shanghai, China), visualized under UV light and photographed with a gel documentation system (Bio Rad, Hercules, United States). The positive PCR products were sent to Tsingke Biotechnology Co., Ltd. for DNA sequencing. Finally, the obtained sequences were analyzed by using BLAST tools of online gene database.1
FOS-resistant strains were subjected to susceptibility testing for 15 antibacterial agents according to Clinical & Laboratory Standards Institute (CLSI) standards, employing micro broth dilution and agar dilution methodologies. The following antibacterial agents were included in the study: imipenem (IPM, #S24020), tigecycline (TGC, #MB1246), polymyxin B (PB, #MB1188), enrofloxacin (ENR, #MB1359-1), ofloxacin (OFX, #MB1659-1) and fosfomycin (FOS, #MB1110) were procured from Dalian Meilun Biotechnology (Dalian, China). Ampicillin (AMP, #A830931), cefotaxime (CTX, #C804340), florfenicol (FLM, #F809685), nitrofurantoin (NIT, #N814882), amikacin (AMK, #A837351) were obtained from Shanghai Macklin Biochemical Technology (Shanghai, China). Additionally, tetracycline (TET, #B25579), sulfamethoxazole (SMZ, #B24157), gentamicin (GEN, #V30125) and ceftazidime (CAZ, #S17139) were also purchased from Shanghai yuanye Bio-Technology (Shanghai, China). In order to idenitfy the MIC values of FOS, the agar dilution assay must be performed on Mueller-Hinton agar, which has been supplemented with 25 mg/L G-6-P. The MIC breakpoints were determined according to CLSI criteria (25). For TGC, breakpoints were referenced against the European Committee on Antimicrobial Susceptibility Testing (EUCAST) criteria, while those for ENR were based on a previous study (26).
XbaI-PFGE was conducted using XbaI endonuclease in accordance with the previously described methodology (22). In summary, Genomic DNA of E. coli was prepared using the restriction enzyme XbaI (TaKaRa, Japan) and electrophoresed in SeaKem Gold Agarose (Lonza, Swiss). DNA fragments were separated by PFGE on a CHEF-Mapper XA PFGE system (Bio-Rad, United States) with a pulse angle of 120° (pulse times 6.76–35.38 s) for 18 h at 6 V/cm and 14°C. After electrophoresis, the gel was stained with SuperRed/GelRed nucleic acid stain (Biosharp, China) and imaged using a gel imaging system. Subsequent to this, phylogenetic analyses of the PFGE patterns were performed utilizing Gel Jv 2.0 software, and clustering analyses were conducted using the unweighted group average method (UPGMA).
A classification of E. coli into six types, A, B1, B2, C, D, and E, as well as two branches, clade I and clade II, was established by employing a quadruple PCR and two individual PCRs, in accordance with the method initially proposed (27). In accordance with the guidelines set forth by the MLST database2 as proposed (28), 7 housekeeping genes (adk, fumC, gyrB, icdF, mdh, purE, recA) were subjected to PCR amplification for MLST analysis. All primers are shown in Supplementary Table S1.
Conjugation experiments were conducted using the liquid ligation method with E. coli J53 as the recipient bacterium. Specifically, E. coli J53 was co-cultured with fosA-like gene-positive E. coli, after which the mixture was centrifuged and processed. The resulting solution was then spread on agar plates containing specific components, including 256 mg/L FOS, 150 mg/L NaN3, and 25 mg/L G-6-P. Subsequently, the colonies were picked from the plates and subjected to PCR identification to detect the presence of the fosA-like gene. The positive strain was retained as a transconjugant. In addition, the transconjugants were tested for their MIC against antimicrobial agents, and the ARGs carried by these transconjugants were analyzed. Furthermore, the conjugation frequency was determined in accordance with the previous study (14). Briefly, donor and recipient bacteria were activated and diluted to OD600 = 0.5, mixed 1:1 and incubated at 37°C for 24 h, and then diluted with saline to appropriate concentrations. Hundred microliter of the mixture was spread on LB agar containing 256 mg/L FOS and 150 mg/L NaN3, and 100 μL of E. coli J53 was spread on LB agar containing 150 mg/L NaN3, both incubated at 37°C for 24 h. Individual colonies on the FOS + NaN3 LB agar were the transconjugant (note as T), and those on the NaN3 LB agar were the receptor (note as T). On NaN3 LB agar were the acceptors (note as R), and the frequency of conjugation was calculated as T/R. The experiment was repeated three times.
The PCR-based replicon typing (PBRT) technique was employed for the detection of plasmid incompatibility groups in transconjugants as well as their donor bacteria, as previously outlined (19). All primers are shown in Supplementary Table S1.
In this study, 3 strains were selected for genome sequencing: HN68, carrying the fosA3 + blaCTX-M-55; LA3, carrying the fosA3 + blaNDM-5; and BHD6, carrying the fosA3 + mcr-1.1. We commissioned Paysono Bioscience and Technology Co., Ltd. to perform the precise sequencing work using Illumina NovaSeq and Oxford Nanopore ONT sequencing platforms. The process of annotating bacterial genomes utilizing the NCBI prokaryotic genome annotation pipeline (29). Additionally, an in-depth analysis of ARGs, MGEs, and serotypes was conducted using the Centre for Genomic Epidemiology.3 In order to visualize the genomic structure and differences between plasmids, CGview4 was employed to map the plasmid comparative genomic circles. Finally, a linear comparative genome map was plotted using the Easyfig soft.
We have submitted the genomic data of three fosA3 positive strains to the NCBI database. The strains are HN68, LA3, and BHD6, and the corresponding BioProject IDs are PRJNA1057772, PRJNA1057764, and PRJNA1057743 respectively, which also including plasmids information.
Based on the isolation results of the FOS-resistant bacteria, the data of the antimicrobial sensitivity test and the detection results of the ARGs, the associations between antimicrobial resistance and the ARGs, and within the ARGs between different regions were statistically analyzed by using the χ2-test in the SPSS (Version 27.0) software. These differences were considered statistically significant when p < 0.05. To further quantify this association, it was calculated by odds ratio (OR) and 95% confidence interval (95% CI). In this case, an OR value of less than 1 indicates a negative correlation, whereas an OR value of greater than 1 indicates a positive correlation. To analyze the results of plasmid conjugation transfer frequency, one-way ANOVA was performed using GraphPad Prism 8 software. These results were considered statistically significant when p < 0.05.
A total of 100 FOS-resistant E. coli isolates were obtained from waterfowl isolates obtained from Hainan, Sichuan, and Anhui. The results showed that the isolation rate of resistant strains in Sichuan was significantly higher than that in Hainan and Anhui (p = 0.047), while no significant difference was observed between the latter two regions (Table 1).
In this study, we identified 10 types of fosA-like genes (fosA-fosA10) in 100 strains of FOS-resistant bacteria. The results revealed that the prevalence of fosA3 was the highest at 88.0%, followed by fosA (11.0%), fosA7 (3.0%), fosA10 (3.0%), and fosA6 (2.0%). It is noteworthy that all resistant strains carried the fosA-like genes, while the presence of fosA2, fosA4, fosA5, fosA8, and fosA9 was not observed in this study (Supplementary Table S2).
The results revealed that all strains exhibited high levels of resistance to FOS (MIC≥512 mg/L). Among the remaining 14 antimicrobial agents, AMP, FLM, CTX, and TET demonstrated significantly higher resistance rates of up to 98.0, 94.0, 90.0, and 89.0%, respectively. SMZ, ENR, OFX, and NIT also displayed relatively high resistance rates of 79.0, 60.0, 59.0, and 41.0%, respectively. Additionally, CAZ, GEN, TGC, PB, and AMK showed resistance rates of 33.0, 29.0, 18.0, 16.0, and 15.0% respectively; whereas IMP exhibited the lowest resistance rate at 10.0% (Figure 1A). Strains positive for the fosA demonstrated variable resistance to different types of antimicrobials with a staggering 99.0% exhibiting multi-drug resistance. The antimicrobial-resistant-patterns (ARP) ranged from 3 to 9 indicating the breadth and severity of the resistance (Figure 1B). The only exception was a strain that was solely resistant to SMZ and FOS. Among the multi-resistant bacteria, a large proportion were resistant to antimicrobial drugs belonging to classes 5 through 8, especially classes 6 and 7 reaching 36.0 and 22.0%, respectively. These fosA positive strains displayed various phenotypic patterns with a total of 69 resistance profiles detected. Of these, the most common pattern was the 6-ARP, which encompassed 36 strains in total (Figure 1B).
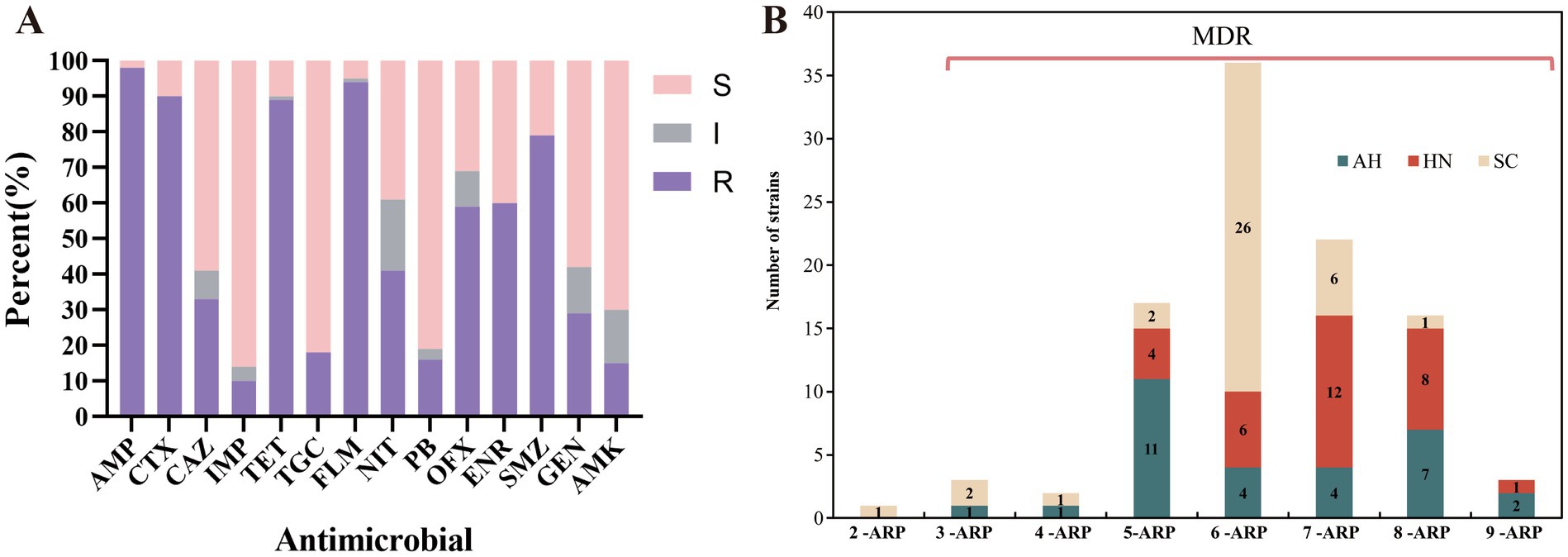
Figure 1. Antimicrobial resistance rate and MDR classification of FOS resistant strains. In panel (A), the histogram with different colors represents the percentages of sensitivity (s), intermediation (I) and resistance (R), respectively. In panel (B), the histogram shows the number of strains with different ARP.
Highly diverse ARGs were detected in the genomes of the isolated strains, suggesting that waterfowl have the potential to serve as a reservoir of ARGs. Of the 26 ARGs, 21 were detected (Figure 2), with the major genotypes including: tetracyclines: tetA (79.0%); phenicols: floR (62.0%); quinolones: qnrS (58.0%); sulfonamides: sul2 (65.0%); aminoglycosides: aphA1 (69.0%); and β-lactams: blaTEM (56.0%). Other ARGs were distributed in the following proportions: tetracyclines (tetB, 16.0%; tetC, 1.0%), phenicols (cmlA, 34.0%), quinolones (qnrA, 14.0%; qnrB, 1.0%), sulfonamides (sul1, 26.0%; sul3, 1.0%), aminoglycosides (aadA1, 36.0%; aac(6′)-ib-cr, 25.0%; rmtB, 18.0%; aac(3′)-III, 8.0%), β-lactams (blaCTX-M, 52.0%; blaSHV, 2.0%; blaNDM, 1.0%), peptides (mcr-1. 1.0%).
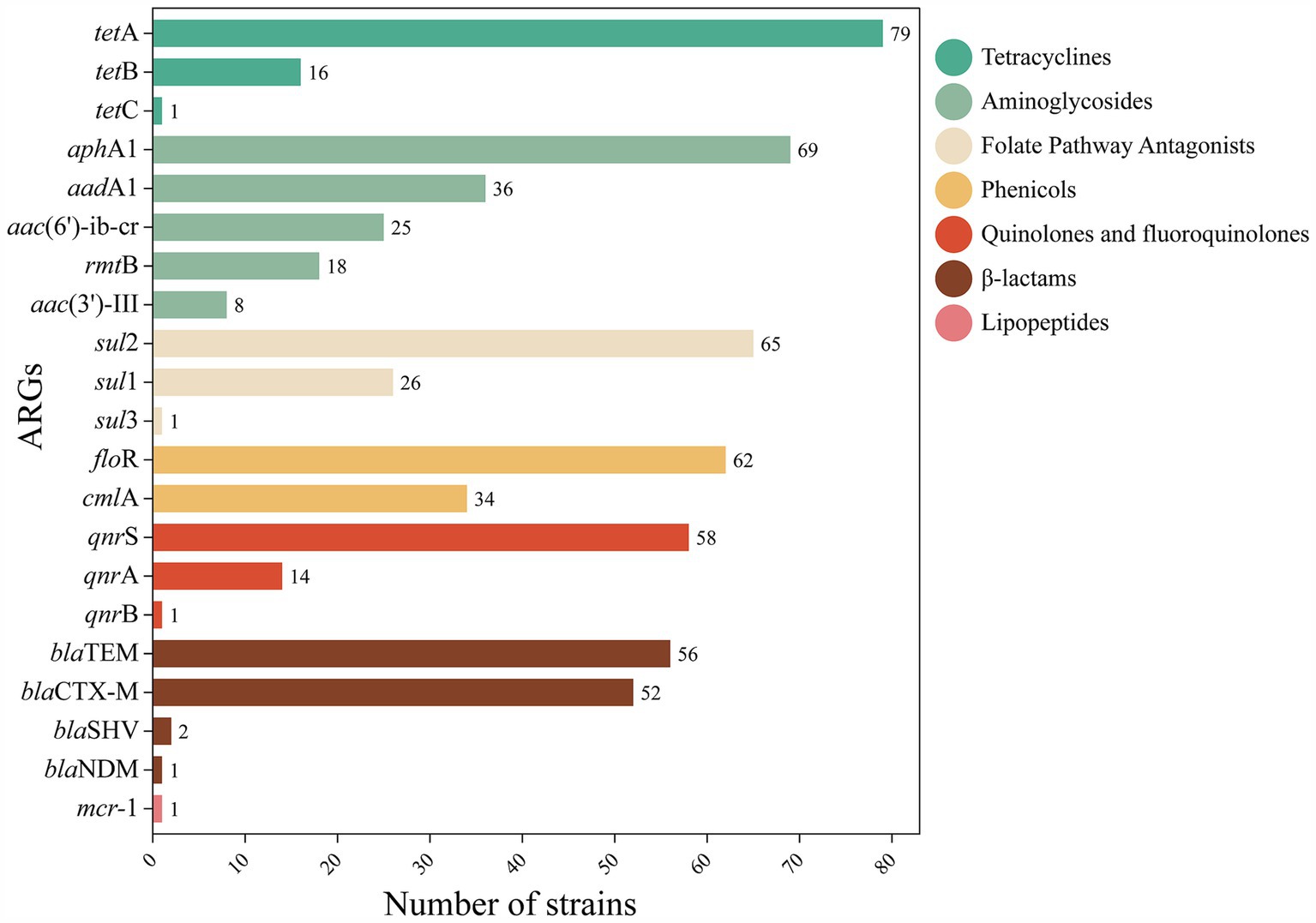
Figure 2. Distribution of number of ARGs. The distribution and quantity of different ARGs in FOS resistant strains were described. Color codes represent different classes of antimicrobial. Each bar represents a specific gene, and its length represents the number of genes detected in strains.
The χ2-test was utilized to investigate the association between ARGs and AMR. The findings indicated that the blaCTX-M was significantly correlated with β-lactams resistance (p = 0.014), the tetA had a significant correlation with tetracyclines resistance (p = 0.001), the floR exhibited a significant correlation with phenicols resistance (p = 0.028), and the sul2 displayed a significant correlation with sulphonamides resistance. Moreover, Supplementary Table S3 summarizes the prevalence of ARGs and their associations in waterfowl-derived FOS-resistant E. coli. Statistical analysis revealed multiple noteworthy correlations (p < 0.05) among these ARGs, with positive correlations being more prevalent, totaling 16 pairs of positive correlations and 11 pairs of negative correlations, resulting in a total of 27 pairs. of particular note is the strongest positive correlation observed between blaCTX-M and fosA3 (OR, 15.162; 95% CI, 1.875–122.635).
In order to investigate whether the spread of FOS resistance is closely related to clonal transmission, 100 FOS-resistant strains were subjected to PFGE typing in this study. The results demonstrated that 96 strains were successfully typed, including 39 strains from Sichuan, 28 strains from Anhui, and 29 strains from Hainan (Figure 3). 4 strains were unable to be typed. The evolutionary tree of PFGE profiles, constructed using Gel Jv 2.0 software, was employed to identify strains with greater than 90% similarity of PFGE profiles, which were then designated as belonging to the same clonal group. In the Anhui region, 5 clonal groups (A–E) were identified among 28 strains, all of which exhibited greater than 90% similarity. Among these, group D comprised 6 clonal strains. 3 clonal groups (F, G, and H) were identified among the 29 strains in the Hainan region, with similarities exceeding 90.0%. The remaining strains exhibited a PFGE spectral similarity that was primarily distributed between 70.0 and 85.0%. In Sichuan, 8 clonal groups (I–P) were identified among 39 strains, with the PFGE spectral similarity of the remaining strains concentrated in the range of 80.0–90.0%. The PFGE analysis revealed that the transmission of FOS resistance was clonally transmitted in the 3 regions, although this was not the main mode of transmission. The MLST results showed that the ST types of 83 strains were identified, involving a total of 45 ST types. The most prevalent types were ST48 (n = 10), ST10 (n = 5), ST206 (n = 4), and ST115 (n = 4). The minimum spanning tree constructed using Phyloviz 2.0 software demonstrated that these strains primarily formed 2 major clusters, centered on ST10 and ST1727, respectively. The genetic distances between the strains in Sichuan were found to be more closely related than those in Anhui and Hainan (Figure 4).
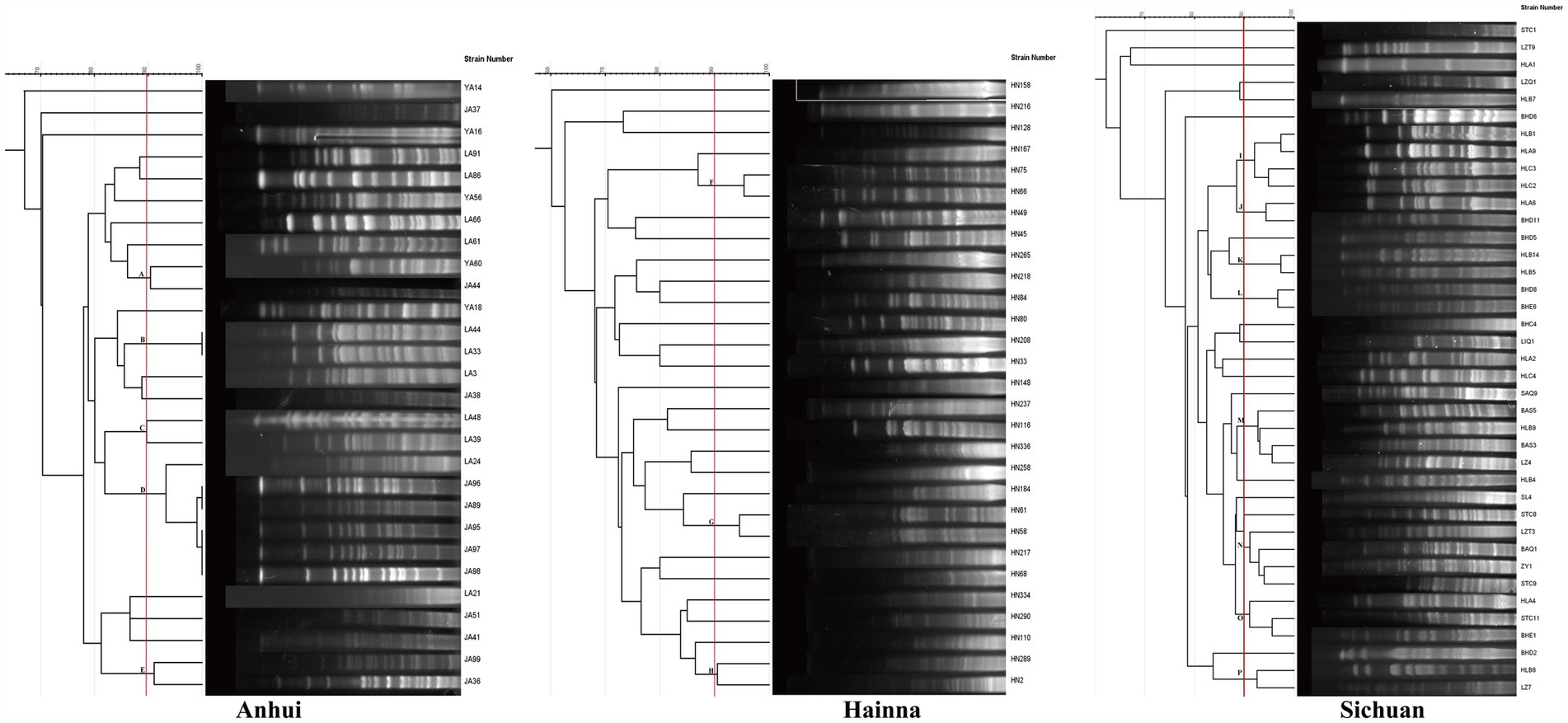
Figure 3. Strain typing based on PFGE. The figure shows the molecular typing of strains from Anhui, Hainan and Sichuan by PFGE. The strains in each region were clustered according to similarity, and their genetic relationships were displayed through a tree view. The bright colored bands in the figure represent DNA fragments, and the pattern of bands can reflect the genetic similarity between strains.
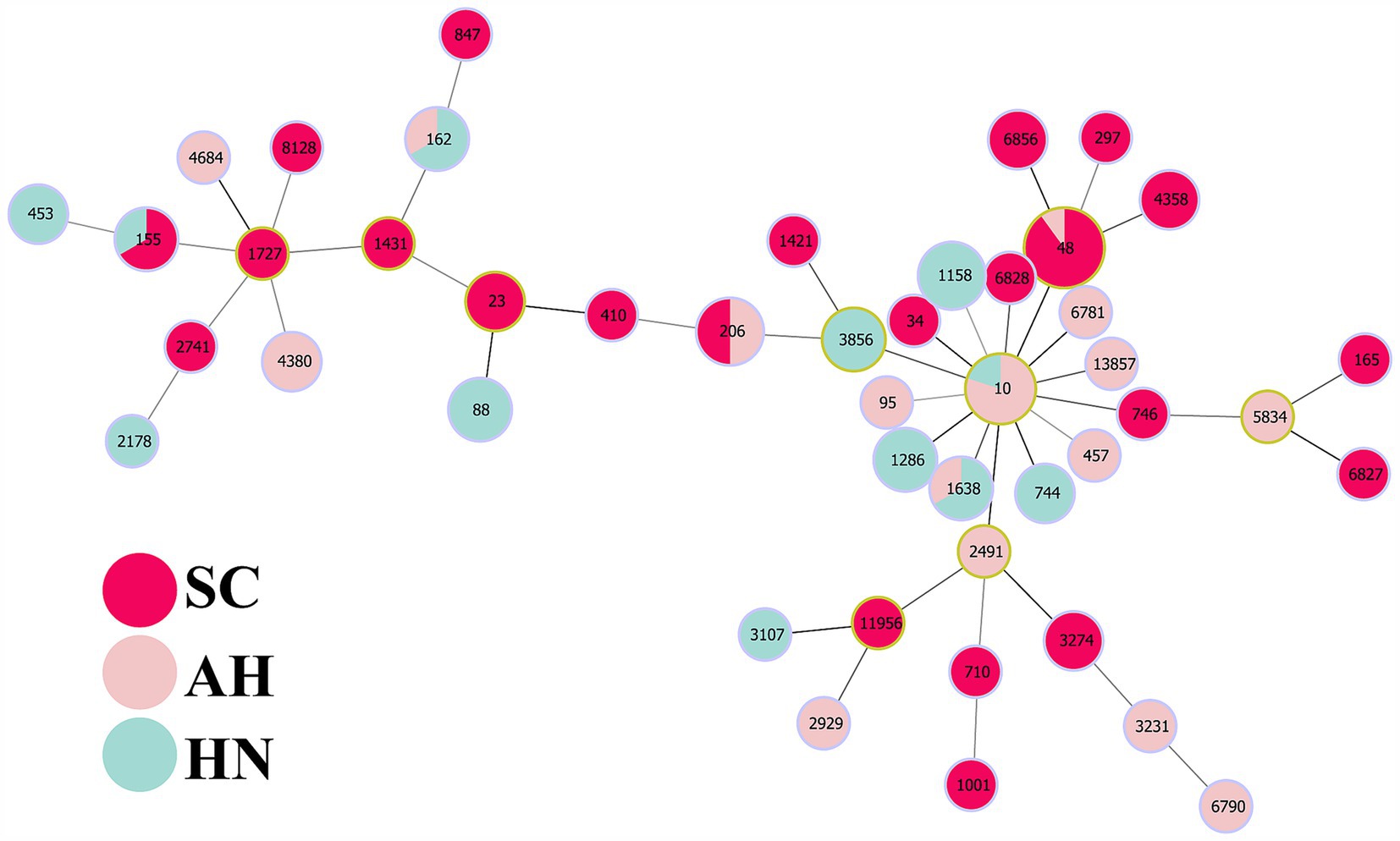
Figure 4. MLST minimum spanning tree of FOS resistant strains. This figure shows the minimum spanning tree of MLST of MDR FOS resistant strains. Nodes represent different sequence types, the size of nodes represents the number of strains of the sequence type, and the line between nodes represents the genetic distance between sequence types. Nodes with different colors represent different regions: red (SC), pink (AH), lightcyan (HN).
The 100 strains were classified into 7 phylogenetic clusters, indicating that FOS-resistant bacteria have diverse phylogenetic backgrounds. In particular, 36 strains were assigned to cluster A, 18 strains to cluster B1, 7 strains to cluster B2, 17 strains to cluster C, 9 strains to cluster D, and 10 strains to cluster E. Additionally, 1 strain was classified as belonging to cluster F, 1 strain was unassigned in Anhui, and 1 strain was assigned to branch I/II in Sichuan. These findings provide an important basis for understanding the mechanisms of FOS resistance transmission and for the development of prevention and control strategies.
The plasmid conjugation transfer assay yielded 53 fosA-like gene positive transconjugants, representing a success rate of 53.0%. The conjugation transfer frequencies of the 53 transconjugants under investigation ranged from 1.3 × 10−3 to 1.1 × 10−1 (Figure 5). Among the various locations, the conjugation transfer frequencies of the transconjugants in Anhui were found to be significantly higher than those in Sichuan and Hainan. MICs of FOS for all transconjugants were ≥ 512 mg/L, and transconjugants exhibited MDR. In addition, other types of ARGs were found to be co-transferred in the transconjugants, including aadA1, cmlA, floR, blaCTX-M, blaTEM, qnrS, sul1, sul2, and tetA (Table 2).
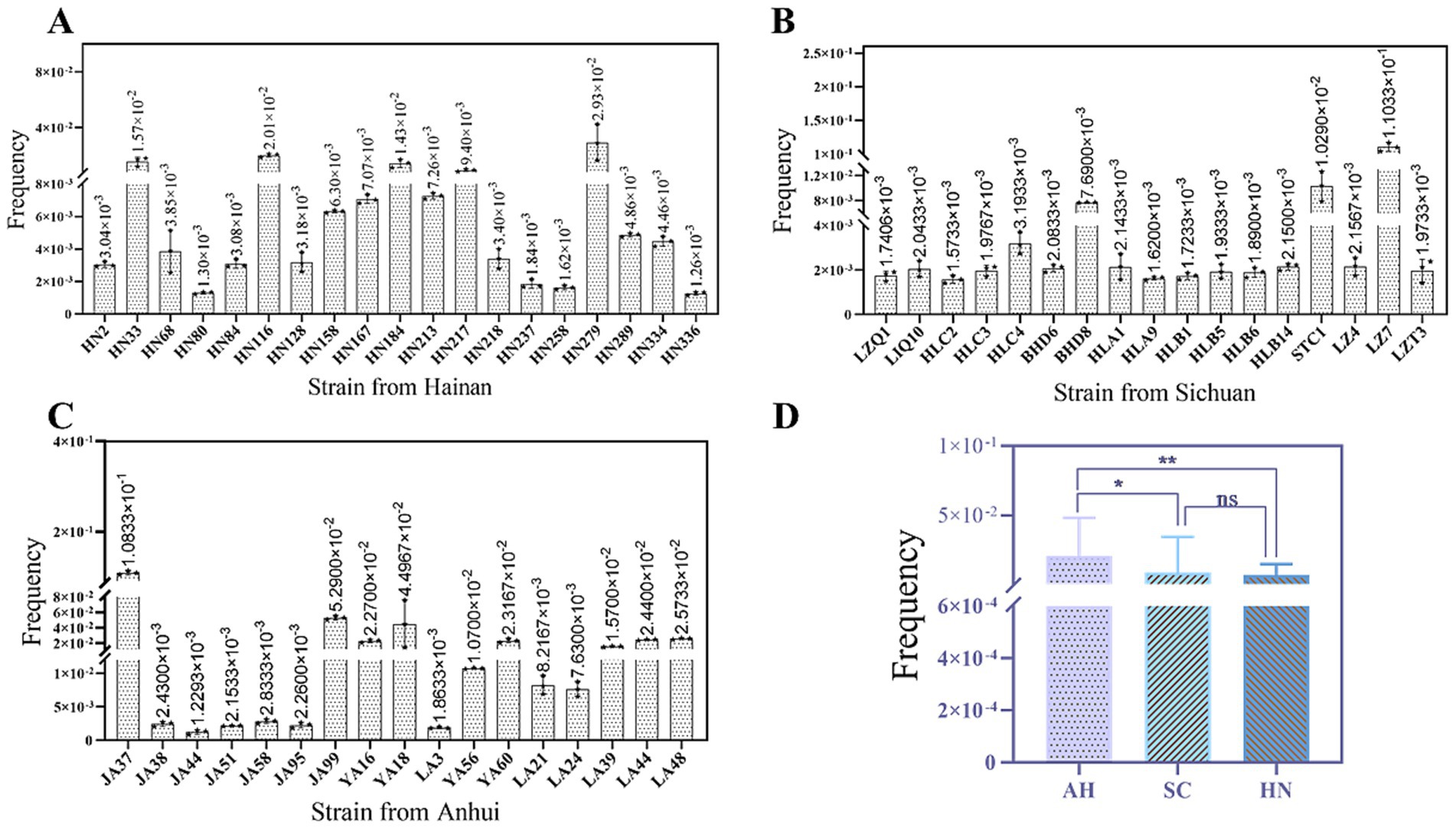
Figure 5. Conjugation transfer frequency of transconjugants in different regions. This figure shows the conjugation transfer frequency of 53 transconjugants. (A) The transfer frequency of transconjugants in Hainan; (B) The transfer frequency of transconjugants in Sichuan; (C) The transfer frequency of transconjugants in Anhui; (D) Comparison of transfer frequency in 3 regions. In panels (A-C), the histogram in the figure shows the transfer frequency of each transconjugants, and the error line represents the standard error. In panel (D), asterisk indicates statistical significance: *indicates P < 0.05, **indicates P < 0.01, and NS indicates no significant difference.
The plasmid carriage of the 53 transconjugants and their corresponding donor bacteria was investigated. The findings revealed that a total of 16 distinct plasmid replicon types were carried by the 53 donor bacteria, with the majority plasmid replicon types falling into the FIB and frepB (Table 3).
In strain LA3, the fosA3 was localized in the IncX1-type plasmid pfosA3-LA3 (47,481 bp, CP141843.1), which lacks the type IV secretion system but contains genes related to stabilizing, replicating and relaxase (Figure 6). This plasmid is very similar to the Enterobacteriaceae IncX1 plasmid, but only partially carries the fosA3, the transfer of which may be related to the IS26 insertion sequence. In HN68 and BHD6, the fosA3 was located within the IncFII (F16:A-:B) type plasmids, pfosA3-HN68 (77,578 bp, CP141846.1) and pfosA3-BHD6 (76,461 bp, CP141794.1), respectively, with a GC content of 52.6%. Both contained a MDR region (12.3 Kbp) with 4 ARGs and multiple insertion sequences, and shared a 40.4 Kbp plasmid backbone region containing replication, conjugation-transfer and relaxase-related genes. The 2 were highly similar (99.0% coverage, 100.0% similarity) but differed in ST type and serotype, suggesting that they were from different sources (Figure 7).
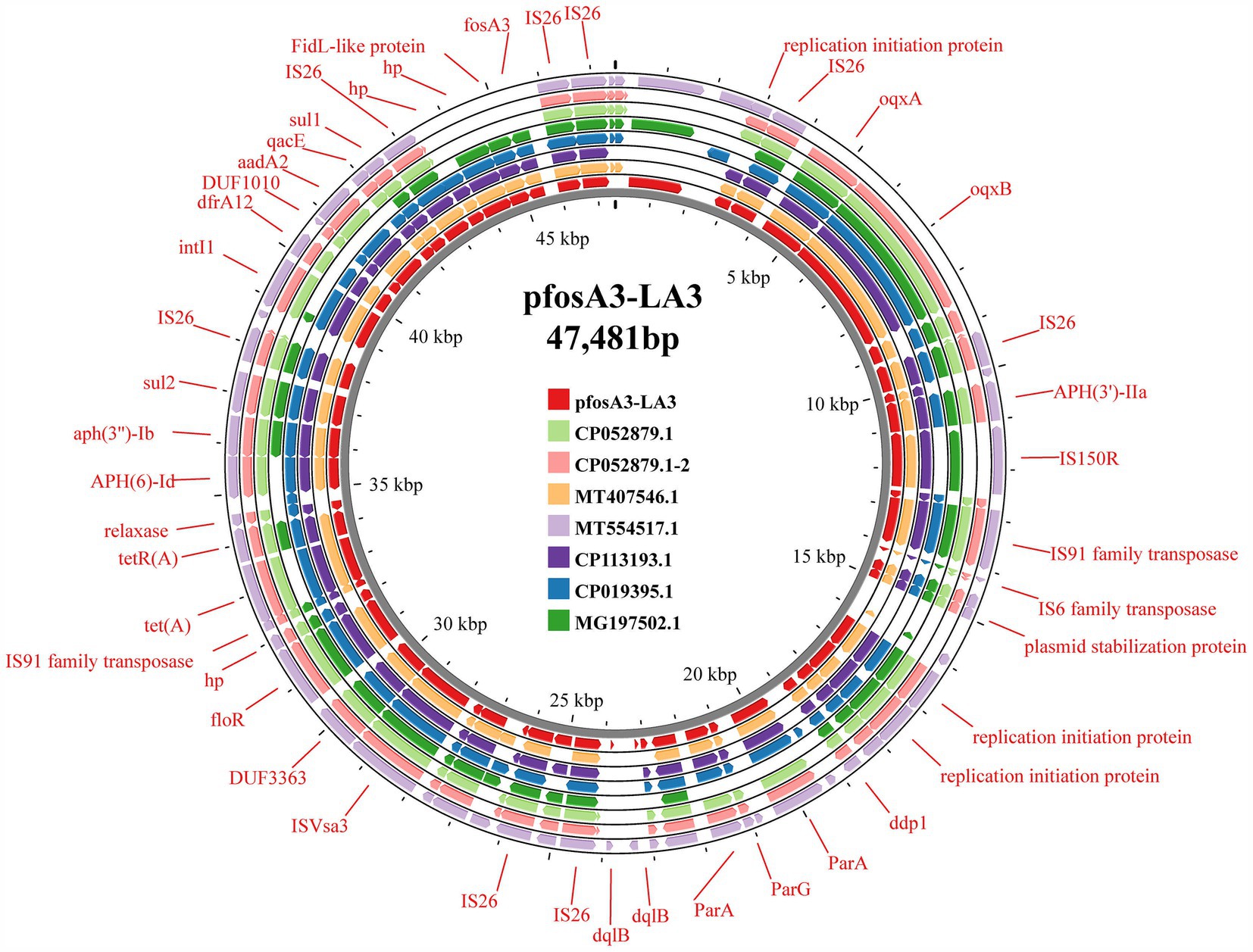
Figure 6. Comparison diagram of pfosA3-LA3 plasmid and other similar plasmids. The figure shows the comparison between pfosA3-LA3 plasmid and other similar plasmids. The ring chart in the figure shows the genome structure of each plasmid, and different colors represent different plasmids. The illustration highlights key genes and functional areas, including replication initiation proteins, transposases, and ARGs.
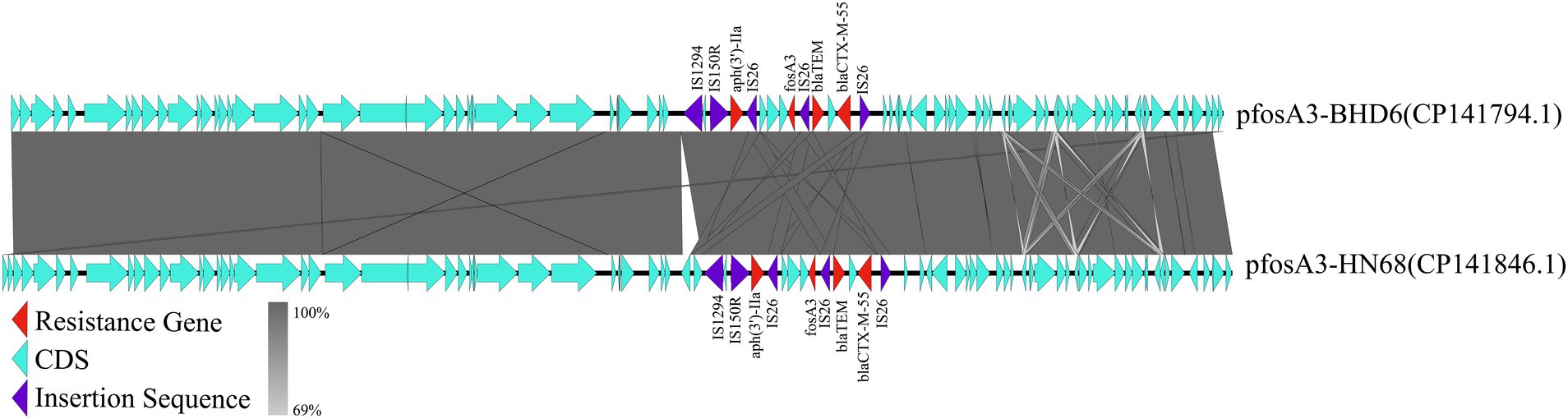
Figure 7. Co-linearity analysis of plasmid pfosA3-BHD6 with plasmid pfosA3-HN68. In the figure, different colors and shapes of markers are used to represent different genes and sequence characteristics: the red triangle represents the ARGs, the light blue arrow represents the coding DNA sequence (CDS), and the purple diamond represents the insertion sequence. The gray shaded area indicates the degree of collinearity between the 2 plasmids, and the line connection indicates the homology between genes. This figure reveals the similarities and differences between the 2 plasmids in genome structure.
Comparison with the NCBI database revealed that pfosA3-HN68 and pfosA3-BHD6 were highly similar (>85.0% coverage, >90.0% similarity) to several E. coli plasmids in which the fosA3 coexisted with elements such as IS26 and blaTEM on a genomic island, allowing further study of the distribution of the fosA3 in E. coli plasmids from different sources and the propagation of the fosA3 in different sources of E. coli plasmids (Figure 8).
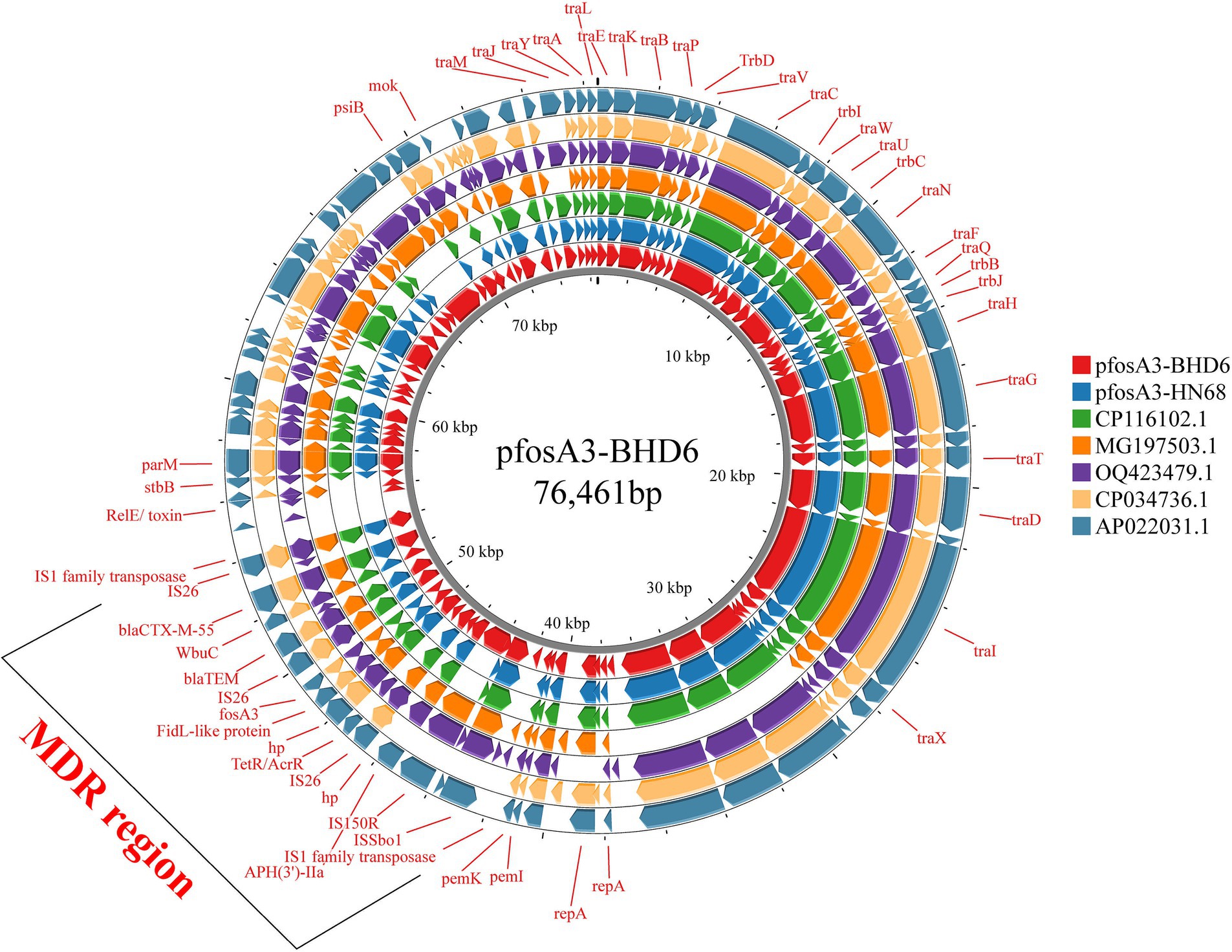
Figure 8. Comparison of plasmids pfosA3-BHD6 and pfosA3-HN68 with other similar plasmids. It illustrates the genomic similarities and differences among these plasmids. The different colors in the figure represent distinct plasmids, with key genes and the MDR region highlighted. The MDR region contains multiple ARGs.
The 3 fosA3-positive plasmids share a common fosA3 core genetic environment surrounding the fosA3. The gene is flanked by IS26 both upstream and downstream. Notably, the IS26 sequences upstream of fosA3 are identical at 385 bp in length, whereas the downstream IS26 encompasses 3 open reading frames (ORFs) situated between its 3′ end and the IS26 element. These ORFs encode transcriptional regulators belonging to the TetR/AcrR family, hypothetical proteins, and FidL-like proteins, respectively. Collectively, the 2 IS26 elements may constitute a composite transposon that promotes the horizontalTransconjugantssfer of the fosA3 (Figure 9).
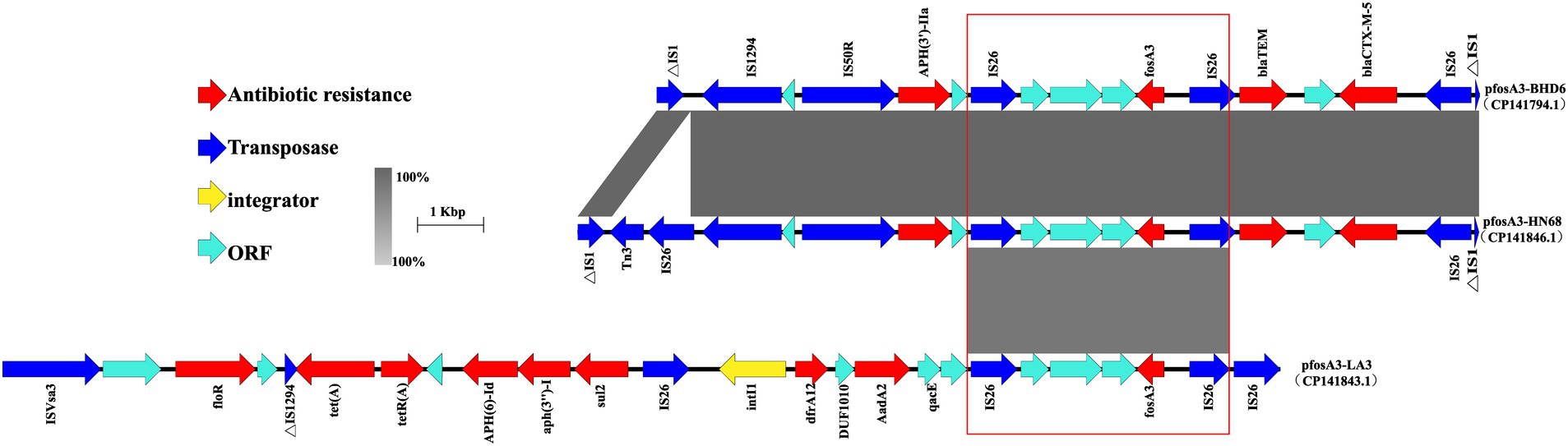
Figure 9. Genetic structure of the fosA3 environment. In the figure, different colors and shapes of markers are used to represent different genes and sequence characteristics: the red arrow represents the ARG, the blue arrow represents the transposase gene, the yellow diamond represents the integron, and the light blue arrow represents the ORF. The gray shaded area indicates the high similarity area, and the part marked by the red box shows the conservative structure near the fosA3 in different plasmids.
In plasmids pfosA3-BHD and pfosA3-HN68, there is a MDR region near the fosA3, accompanied by the ESBL genes blaTEM and blaCTX-M-55 upstream and the aminoglycoside resistance gene aph (3′)-IIa downstream, surrounded by the IS50R, ISSbo1 and the truncated IS1 family insertion element, forming a stable genetic structure. In plasmid pfosA3-HN68, transposons Tn3 and IS26 were inserted between ΔIS1 and IS1294.
In plasmid pfosA3-LA3 there is a type I integron structure downstream of the fosA3, containing gene cassettes such as the folate pathway inhibitor resistance gene dfrA12, the aminoglycoside resistance gene aadA2 and the structural domain protein DUF1010. The 3′ end of the integron consists of the quaternary ammonium efflux transporter protein qacE and the truncated folate pathway inhibitor sul1, surrounded by IS26. Downstream of the integrase gene intI1 is the multidrug resistance region, consisting of IS26, ISVsA3 and a number of ARGs such as floR, tet(A), aph (6)-Id, aph(3″)-Ib, sul2, etc., which together form a complex drug resistance network.
Despite the prohibition of FOS in veterinary medicine in China since 2005, a survey of 833 E. coli strains of waterfowl origin from Hainan, Sichuan and Anhui revealed that the resistance rate to FOS remained alarmingly high at 12.0%. Among the 100 resistant strains, fosA3 exhibited the highest positivity rate (88.0%). This finding aligns with previous studies, indicating that fosA3 is the most prevalent plasmid-mediated resistance gene for FOS (30). The fosA3 positive E. coli were detected across various sample types; Notably, a detection rate of 21.9% was observed in E. coli sourced from ducks in Shandong (31), while a positive rate of 27.4% was reported for E. coli isolated from poultry farms (32). The tested strains demonstrated significantly high levels of resistance to FOS (MIC≥512 mg/L) and varying degrees of resistance to an additional 14 antimicrobial agents. Notably, high levels of resistance were observed against AMP, FLM, CTX and TET indeed, 99.0% of these strains were classified as MDR bacteria. MDR means resistance to 3 or more unique antimicrobial drug classes (33, 34). This elevated level of AMR may be closely associated with environmental contamination stemming from residual tetracyclines, phenicols and sulfonamides antimicrobials (35).
It has been demonstrated that waterfowl farm waste contains a significant amount of ARGs, which pose potential threat to public health and the environment (12). The importance of ARGs, which confer bacterial resistance to antimicrobials, cannot be overstated. They represent a considerable challenge for antimicrobial therapy at present (36). In this study, 26 ARGs were subjected to comprehensive testing and analysis. The study revealed that the prevalence of tetA, aphA1, sul2, folR, qnrS, and blaTEM in E. coli derived from waterfowl was significantly elevated. It was demonstrated that high levels of veterinary tetracyclines and sulfonamides discharged into the environment substantially contributed to the widespread presence of tet and sul ARGs (35). Further analyses revealed that although there were differences between bacterial resistance phenotypes and ARGs, some genes exhibited significant associations with specific resistance phenotypes. In particular, blaCTX-M was found to be significantly associated with β-lactams resistance, tetA with tetracyclines resistance, floR with phenicols resistance, and sul2 with sulfonamides resistance (p < 0.05). These studies have attributed this phenomenon to abnormal expression or low expression levels of ARGs (37). The present study demonstrated a significant positive correlation between the fosA3 and tetA, qnrS, and blaCTX-M, as well as fosA3 frequently coexisted with these ARGs (7, 38). However, the fosA3 exhibited a negative correlation (OR < 1) with rmtB (39). Given the extensive use of tetracyclines, β-lactams, aminoglycosides, and chloramphenicols in Chinese animal husbandry, it is plausible that fosA3 may coexist alongside multiple ARGs, such as tetA, qnrS, blaCTX-M, and folR, within the same strain. This may, in turn, facilitate the dissemination of fosA3 (40). Despite the explicit prohibition on FOS as a veterinary drug in China, the co-existence of plasmid-borne fosA3 alongside other ARGs may have contributed to the observed increase of FOS resistance in bacteria of animal origin and the widespread dispersal of the fosA3 under co-selection pressure (41).
This study subjected 100 FOS-resistant strains to PFGE typing. The resulting data demonstrated that Anhui, Hainan, and Sichuan were distinctly different. While the majority of strains exhibited unique PFGE profiles, these findings nonetheless underscore a significant clonal spread characteristic of FOS-resistant strains. Notably, this phenomenon of clonal spread was particularly conspicuous in Sichuan. Several studies have highlighted the potential role of migratory birds and poultry exposure to wildlife in facilitating cross-regional transmission of AMR (42). One study has shown that even in the absence of ARGs contamination at hatcheries, the introduction of ARGs into the flock can occur rapidly through vectors such as dogs, flies, wild birds, and others (43). Our previous research has shown that waterfowl farming in open environments can also lead to the development of antibiotic resistance among carrying bacteria (23). Thus, inadequate environmental control and biosecurity measures on farms may facilitate the dissemination of FOS-resistant bacteria and fosA3 among waterfowl.
In epidemiological investigations, MLST and PFGE are commonly employed methods for the molecular typing of E. coli (44). MLST analysis effectively classified 83 E. coli strains into 45 known sequence types (STs), with ST48 (n = 10) and ST10 (n = 5) being the most prevalent types. Notably, all 10 ST48 strains were found to be positive for the fosA3 and originated from Sichuan Province. PFGE analysis revealed that some strains belonged to the same clonal group, suggesting a clonal spread within Sichuan; however, this was not indicative of a broader trend. The geographical distribution of ST types s across three regions was diverse, with strains sharing the same ST type potentially belonging different PFGE clonal groups. Reports on fosA3-positive E. coli strains associated with ST48 and ST10 are rare in existing literature. A previous study indicated that the most prevalent type of fosA3-positive E. coli sourced from waterfowl in Shandong was ST69 (31). Furthermore, ST156 and ST115 were the most prevalent types among fosA3-carrying E. coli of animal origin (10). Typically, E. coli ST48 isolates are associated with the spread of blaNDM in poultry farms and retail meat (45). In contrast, E. coli ST10 is a low virulence commensal that is widely present in the animal and human intestinal tract (46). Furthermore, E. coli ST10 is regarded as a significant reservoir for the mcr-1 (47). Among three identified fosA7-positive strains, three distinct STs were observed, such as ST847, ST1727, and ST2741; these have not been previously reported in the reference. Notably, although ST847 was previously reported in a blaCTX-M-positive plant-derived E. coli strain, it does not carry fosA-like genes (48). Moreover, out of 100 analyzed FOS-resistant strains categorized into seven phylogenetic groups, with groups A and B1 representing the majority, while groups B2 and D were the least numerous. The findings of the current study align with those of previous research conducted in Pakistan, which similarly identified groups B1 and A as the most prevalent strains of E. coli in both human and animal populations (49). Notably, the identification of an ST410 type D group fosA3-positive strain, as it is associated with ExPEC pathogens and represents a high-risk clonal group (50, 51).
Plasmids serve as a molecular cornerstone in the dissemination of AMR in the era of “One Health” (52). The first report of plasmid-mediated horizontal transfer of the fosA3 was published by Japanese researchers in 2010 (53). Subsequent studies have confirmed that the fosA3 is typically located on plasmids and spreads among various bacteria through horizontal transfer (7, 13). Despite the fact that FOS is not approved for use in veterinary clinics, the prevalence of the fosA3 remains significant across Asia (7). In this study, we found that the transconjugants from fosA3-positive strains transferred genes such as blaCTX-M, floR, and aadA1 simultaneously during the plasmid conjugation transfer. This co-transfer enabled these strains to acquire resistance to cephalosporins, chloramphenicols, sulfonamides, and aminoglycosides, which are commonly used in veterinary clinics. CTX-Ms is at the pinnacle of the pyramid of ESBLs, which are distributed in different ecosystems and exhibit resistance to a broad spectrum of antibiotics (54). Due to the slow development of antibacterial agents, researchers are compelled to revisit the utilization of established antibiotics like FOS to combat MDR bacteria, particularly those within the Enterobacteriaceae family known for their ability to produce ESBLs and carbapenemases (55). However, prior research indicates that the fosA3 is generally co-located on plasmids with blaCTX-M, and they tend to co-transfer and disseminate together (56). FosA3-positive E. coli were identified in pets that had not been treated with FOS. In these instances, it was found that the fosA3, flanked by IS26 resided on an F33:A-:B-type plasmid also carrying rmtB. This suggests that transmission of the fosA3 is predominantly driven by horizontal transfer involving F33:A-:B- plasmid closely associated with rmtB, rather than through clonal transmission (57). Furthermore, co-transfer of the floR with the fosA3 was observed in chicken-derived fosA3-positive E. coli (40). The concerning co-occurrence of the fosA with key ARGs on plasmids presents an extra hurdle in treating Enterobacteriaceae infections (55). In this study, transconjugants of fosA and fosA7 positive strains could not be successfully obtained. The findings indicate that the fosA is frequently located on the chromosomes of various Gram-negative bacteria, including Klebsiella pneumoniae, Streptococcus mucinosus, and Pseudomonas aeruginosa (58). In contrast, the fosA7 is predominantly located on the chromosome of Salmonella (10). Furthermore, it was demonstrated that the fosA7 on the chromosome of Salmonella conferred a low level of resistance to FOS. However, the transfer of this gene into a plasmid resulted in a significant increase in resistance and fitness (59).
One of the key factors contributing to the spread of AMR is the high frequency of conjugation transfer. In this study, we observed that the conjugation transfer frequency of 53 transconjugants ranged from 1.3 × 10−3 to 1.1 × 10−1. This suggests that these plasmids possess a potential for dissemination in natural environments, particularly when their frequency approaches or exceeds values between 10−1 and 10−2 (60). This study identified multiple plasmid replicon types in the spliceosome, including FIB, FIA, frpB, HI1, HI2, and others. These findings indicate that the fosA3 can adapt to diverse plasmid environments encompassing both single-replicon and multi-replicon plasmids (9). Previous studies have demonstrated the prevalence of the fosA3 in a diverse range of plasmids, including the IncC-IncN-type plasmids (lacking horizontal transfer genes) of multidrug-resistant Salmonella (61). Additionally, the IncN-type and IncHI2-type plasmids of vegetable-derived E. coli were identified (48), as well as mucin-resistant E. coli harboring IncFII/FIA/FIB-type plasmids (62). Finally, coexisting with blaCTX-M in chicken-derived E. coli were identified either IncHI2-type or IncI1-type plasmids (40). Furthermore, it was noted the fosA3 was disseminated in retail vegetable-derived ESBL-producing E. coli through various plasmid vectors, including IncHI2/ST3, IncN1-F33:A-:B-, F33:A-:B-, IncFIIK, and F24:A-:B1 (63). In conclusion, the rapid spread of FOS resistance in pathogenic bacteria of originating from animals is closely associated with the transfer of multiple plasmid-mediated resistance.
Sequencing results revealed that the blaNDM-5, mcr-1, and blaCTX-M are all located on plasmids. However, in strains BHD6 and LA3, the mcr-1 and blaNDM-5 do not coexist with fosA3 on the same plasmid. In contrast, in strains BHD6 and HN68, the fosA3 is located on the same plasmid as blaCTX-M. Notably, these two plasmids show high similarity and also carry other ARGs. The emergence of plasmid-mediated mcr-1 and blaNDM poses a serious threat to antibiotic treatment efficacy, presenting unprecedented challenges to public health and animal husbandry (64, 65). Although FOS retains some bactericidal activity against CRE, ESBL-EC, and colistin-resistant bacteria, its effectiveness can be severely compromised when coexisting with ARGs such as blaNDM-5, mcr-1, and blaCTX-M. This may lead to treatment failure. The presence of multiple resistance plasmids in a single strain frequently leads to resistance against nearly all available antibiotics while facilitating the dissemination of ARGs. Some studies have reported instances where fosA3, mcr-1, and blaNDM coexist on a single plasmid, this phenomenon contributes to broad-spectrum resistance to multiple antibiotics in related strains (66–68).
In the 3 sequenced strains, the fosA3 and blaCTX-M are located on typical IncFII plasmids, except for strain LA3. Previous studies have indicated that IncFII plasmids represent the primary type of plasmid harboring the fosA3 (9). Multiple studies have confirmed a strong association between the co-dissemination of fosA3 and blaCTX-M on IncFII plasmids (69, 70), which is consistent with findings in the pfosA3-BHD6 and pfosA3-HN68 plasmids. Furthermore, sequence analysis results show that both pfosA3-BHD6 and pfosA3-HN68 plasmids contain multiple essential modules required for conjugative transfer, such as conjugative transfer protein TrbD, Type IV secretion system genes (including tra series and tab series), plasmid replication origin oriT, and relaxase enzyme. Plasmid conjugation experiments further confirm the horizontal transmission capability of these two plasmids. Although there are currently no specific research reports on fosA3-positive IncX1-type plasmids, several heterozygous plasmids carrying the fosA3 classified as IncX1 type have been identified in the NCBI database. For example, the heterozygous plasmid pXH992_2 (accession number CP019395.1) carrying the IncR-IncX1 type from urine samples of patients in Zhejiang, the plasmid pSXC4-2 carrying IncFIN/IncHI1B/IncR/IncX1 types from E. coli strains isolated from chicken sources in Jiangsu, and the plasmid pHNMCC14 carrying IncFII/IncN/IncN/IncX1 types from chicken-derived E. coli strains in Guangzhou. Additionally, it is worth noting that elements related to conjugative transfer such as oriT, T4SS, T4CP and relaxase were not found in the plasmid pfosA3-LA3. Therefore, this may be a reason for the inability of fosA3-positive plasmids in strain LA3 to undergo conjugative transfer.
The dissemination of ARGs is not confined to plasmids, it also includes other mobile genetic structures, such as transposons and insertion elements. IS26 plays a crucial role in the spread and mobilization of fosA3 (63). It can facilitate the transfer of ARGs by forming a circular intermediate structure, thereby promoting their dissemination through replicative transposition (71). In this study, it was found that there is an insertion element IS26 both upstream and downstream of the fosA3 in three fosA3-positive plasmids, forming a specific genetic structure of IS26-hp-hp-hp-fosA3-IS26. This is a common genetic structure for fosA3, which has been reported in multiple studies (9). Previous research has shown that fosA3 often co-disseminates with blaCTX-M (7, 55). In the plasmids pfosA3-BHD6 and pfosA3-HN68, it was found that both blaTEM and blaCTX-M are present upstream of fosA3. Additionally, there is an IS26 sequence upstream of blaCTX-M, along with a truncated 131 bp IS1 family mobile element. Six genetic structures of fosA3 were identified in animal-derived E. coli, with three core structures similar to those in this study. Additionally, three unique genetic structures were found: IS26-CTX-M-15-IS5/IS1182-fosA3-orf1-orf2-orf3-IS26; IS26-fosA3-orf1-IS26; and IS26-CTX-M-55-TEM-1-IS26-fosA3-orf1-IS26 (10). Two new genetic environments of fosA3 were first discovered in ESBL-producing E. coli from chicken meat in China (ISEcp1-blaCTX-M-65-ΔIS903D-IS26-fosA3-orf1-orf2-Δorf3-IS26 and IS26- ISEcp1-blaCTX-M-3-orf477-blaTEM-1-IS26-fosA3-orf1-orf2-Δorf3-IS26) (32). These findings provide critical insights into understanding of the transmission mechanism and genetic background of fosA3.
Our findings revealed that FOS resistance E. coli in waterfowl filed in selected regions is mainly due to the plasmid-mediated fosA3. The presence of both fosA3 and blaCTX-M may have contributed to the rapid spread of MDR bacteria. The spread of fosA3 is primarily attributed to the horizontal transfer of plasmids with high conjugation ability, along with clonal dissemination. It is significant to note that the fosA3 is often flanked by IS26, which could be crucial for its transfer between plasmids. Considering the ecological role of waterfowl and the clinical importance of FOS, the emergence of FOS resistance could potentially spread through the food chain and the environment, posing a risk to public health. Therefore, reducing the use of related antibiotics in Chinese poultry farming may be effective in curbing the spread of the fosA3 in E. coli.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary material.
The study was approved by the Experimental Operation Guidelines and Animal Welfare Committee of Sichuan Agricultural University. The study was conducted in accordance with the local legislation and institutional requirements.
SZ: Conceptualization, Funding acquisition, Investigation, Methodology, Writing – original draft, Writing – review & editing. JY: Investigation, Methodology, Visualization, Writing – original draft. QianY: Data curation, Writing – original draft. QL: Data curation, Writing – review & editing. ZZ: Data curation, Writing – review & editing. MW: Writing – review & editing, Data curation, Funding acquisition. RJ: Writing – review & editing, Formal analysis. SC: Validation, Writing – review & editing. ML: Writing – review & editing, Validation. DZ: Writing – review & editing, Project administration. XZ: Data curation, Writing – review & editing. YW: Data curation, Writing – review & editing. QiaoY: Writing – review & editing, Data curation. JH: Writing – review & editing, Project administration. XO: Writing – review & editing, Data curation. DS: Conceptualization, Writing – review & editing. BT: Conceptualization, Writing – review & editing. ZW: Conceptualization, Writing – review & editing. YH: Writing – review & editing, Conceptualization, Software. AC: Writing – review & editing, Funding acquisition, Supervision.
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This work was supported by the National Key Research and Development Program of China (2023YFD1800200), the earmarked fund for China Agriculture Research System (CARS-42-17), Sichuan Veterinary Medicine and Drug Innovation Group of China Agricultural Research System (SCCXTD-2025-18).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fvets.2025.1481822/full#supplementary-material
1. Tolley, A, Bansal, A, Murerwa, R, and Howard, DJ. Cost-effectiveness of point-of-care diagnostics for AMR: a systematic review. J Antimicrob Chemother. (2024) 79:dkae067. doi: 10.1093/jac/dkae067
2. Hou, J, Long, X, Wang, X, Li, L, Mao, D, Luo, Y, et al. Global trend of antimicrobial resistance in common bacterial pathogens in response to antibiotic consumption. J Hazard Mater. (2023) 442:130042. doi: 10.1016/j.jhazmat.2022.130042
3. Bellucci, MC, Romani, C, Sani, M, and Volonterio, A. Dual antibiotic approach: synthesis and antibacterial activity of antibiotic–antimicrobial peptide conjugates. Antibiotics. (2024) 13:783. doi: 10.3390/antibiotics13080783
4. Hendlin, D, Stapley, EO, Jackson, M, Wallick, H, Miller, AK, Wolf, FJ, et al. Phosphonomycin, a new antibiotic produced by strains of streptomyces. Science. (1969) 166:122–3. doi: 10.1126/science.166.3901.122
5. Falagas, ME, Vouloumanou, EK, Samonis, G, and Vardakas, KZ. Fosfomycin. Clin Microbiol Rev. (2016) 29:321–47. doi: 10.1128/CMR.00068-15
6. Putensen, C, Ellger, B, Sakka, SG, Weyland, A, Schmidt, K, Zoller, M, et al. Current clinical use of intravenous fosfomycin in ICU patients in two European countries. Infection. (2019) 47:827–36. doi: 10.1007/s15010-019-01323-4
7. Cheng, K, Fang, L-X, Ge, Q-W, Wang, D, He, B, Lu, J-Q, et al. Emergence of fosA3 and blaCTX–M–14 in multidrug-resistant Citrobacter freundii isolates from flowers and the retail environment in China. Front Microbiol. (2021) 12:586504. doi: 10.3389/fmicb.2021.586504
8. Hu, J, Chen, L, Li, G, Pan, Y, Lu, Y, Chen, J, et al. Prevalence and genetic characteristics of fosB-positive staphylococcus aureus in duck farms in Guangdong, China in 2020. J Antimicrob Chemother. (2023) 78:802–9. doi: 10.1093/jac/dkad014
9. Mattioni Marchetti, V, Hrabak, J, and Bitar, I. Fosfomycin resistance mechanisms in enterobacterales: an increasing threat. Front Cell Infect Microbiol. (2023) 13:1178547. doi: 10.3389/fcimb.2023.1178547
10. Zhang, X, Ma, M, Cheng, Y, Huang, Y, Tan, Y, Yang, Y, et al. Spread and molecular characteristics of enterobacteriaceae carrying fosA-like genes from farms in China. Microbiol Spectr. (2022) 10:e0054522. doi: 10.1128/spectrum.00545-22
11. Han, L, Lu, X-Q, Liu, X-W, Liao, M-N, Sun, R-Y, Xie, Y, et al. Molecular epidemiology of fosfomycin resistant E. coli from a pigeon farm in China. Antibiotics. (2021) 10:777. doi: 10.3390/antibiotics10070777
12. Wang, X-R, Lian, X-L, Su, T-T, Long, T-F, Li, M-Y, Feng, X-Y, et al. Duck wastes as a potential reservoir of novel antibiotic resistance genes. Sci Total Environ. (2021) 771:144828. doi: 10.1016/j.scitotenv.2020.144828
13. Li, Q, Yang, J, Wang, M, Jia, R, Chen, S, Liu, M, et al.Global distribution and genomic characteristics analysis of avian-derived mcr-1-positive Escherichia coli. Ecotoxicol Environ Saf. (2024) 285:117109. doi: 10.1016/j.ecoenv.2024.117109
14. Zhang, S, Wen, J, Wang, Y, Zhong, Z, Wang, M, Jia, R, et al. Decoding the enigma: unveiling the molecular transmission of avian-associated tet(X4)-positive E. coli’ in Sichuan Province, China. Poult Sci. (2023) 102:103142. doi: 10.1016/j.psj.2023.103142
15. Zhang, S, Shu, Y, Wang, Y, Zhong, Z, Wang, M, Jia, R, et al. High rate of multidrug resistance and integrons in Escherichia coli isolates from diseased ducks in select regions of China. Poult Sci. (2023) 102:102956. doi: 10.1016/j.psj.2023.102956
16. Zhang, S, Chen, S, Abbas, M, Wang, M, Jia, R, Chen, S, et al. High incidence of multi-drug resistance and heterogeneity of mobile genetic elements in Escherichia coli isolates from diseased ducks in Sichuan province of China. Ecotoxicol Environ Saf. (2021) 222:112475. doi: 10.1016/j.ecoenv.2021.112475
17. Zurfluh, K, Treier, A, Schmitt, K, and Stephan, R. Mobile fosfomycin resistance genes in Enterobacteriaceae—an increasing threat. MicrobiologyOpen. (2020) 9:e1135. doi: 10.1002/mbo3.1135
18. Lee, S-Y, Park, Y-J, Yu, JK, Jung, S, Kim, Y, Jeong, SH, et al. Prevalence of acquired fosfomycin resistance among extended-spectrum β-lactamase-producing Escherichia coli and klebsiella pneumoniae clinical isolates in Korea and IS26-composite transposon surrounding fosA3. J Antimicrob Chemother. (2012) 67:2843–7. doi: 10.1093/jac/dks319
19. Zhang, S, Yang, H, Rehman, MU, Yang, K, Dong, M, Yang, J, et al. Class 1 integrons as predominant carriers in Escherichia coli isolates from waterfowls in Hainan, China. Ecotoxicol Environ Saf. (2019) 183:109514. doi: 10.1016/j.ecoenv.2019.109514
20. Zhang, S, Abbas, M, Rehman, MU, Wang, M, Jia, R, Chen, S, et al. Updates on the global dissemination of colistin-resistant Escherichia coli: an emerging threat to public health. Sci Total Environ. (2021) 799:149280. doi: 10.1016/j.scitotenv.2021.149280
21. Zhang, S, Wen, J, Wang, Y, Wang, M, Jia, R, Chen, S, et al. Dissemination and prevalence of plasmid-mediated high-level tigecycline resistance gene tet (X4). Front Microbiol. (2022) 13:969769. doi: 10.3389/fmicb.2022.969769
22. Rehman, MU, Yang, H, Zhang, S, Huang, Y, Zhou, R, Gong, S, et al. Emergence of Escherichia coli isolates producing NDM-1 carbapenemase from waterfowls in Hainan island, China. Acta Trop. (2020) 207:105485. doi: 10.1016/j.actatropica.2020.105485
23. Zhang, S, Guo, X, Wang, Y, Zhong, Z, Wang, M, Jia, R, et al. Implications of different waterfowl farming on cephalosporin resistance: investigating the role of blaCTX-M-55. Poult Sci. (2023) 102:102929. doi: 10.1016/j.psj.2023.102929
24. Yang, H, Rehman, MU, Zhang, S, Yang, J, Li, Y, Gao, J, et al. High prevalence of CTX-M belonging to ST410 and ST889 among ESBL producing E. coli isolates from waterfowl birds in china’s tropical island, Hainan. Acta Trop. (2019) 194:30–5. doi: 10.1016/j.actatropica.2019.03.008
25. Clinical and Laboratory Standards Institute. Performance Standards for Antimicrobial Susceptibility Testing 2022: M100-ED31. Wayne, PA: Clinical and Laboratory Standards Institute (2022). https://clsi.org/standards-development/resources/
26. Kinsella, PM, Smibert, OC, Whitlam, JB, Steven, M, Masia, R, and Gandhi, RG. Successful use of azithromycin for Escherichia coli–associated renal allograft malakoplakia: a report of two cases. Eur J Clin Microbiol Infect Dis. (2021) 40:2627–31. doi: 10.1007/s10096-021-04270-x
27. Mohammed, EJ, Hasan, KC, and Allami, M. Phylogenetic groups, serogroups and virulence factors of uropathogenic Escherichia coli isolated from patients with urinary tract infection in Baghdad, Iraq. Iran J Microbiol. (2022) 14:445–57. doi: 10.18502/ijm.v14i4.10230
28. Jolley, KA, Bray, JE, and Maiden, MCJ. Open-access bacterial population genomics: BIGSdb software, the PubMLST.org website and their applications. Wellcome Open Res. (2018) 3:124. doi: 10.12688/wellcomeopenres.14826.1
29. Tatusova, T, DiCuccio, M, Badretdin, A, Chetvernin, V, Nawrocki, EP, Zaslavsky, L, et al. NCBI prokaryotic genome annotation pipeline. Nucleic Acids Res. (2016) 44:6614–24. doi: 10.1093/nar/gkw569
30. Yang, T-Y, Lu, P-L, and Tseng, S-P. Update on fosfomycin-modified genes in enterobacteriaceae. J Microbiol Immunol Infect. (2019) 52:9–21. doi: 10.1016/j.jmii.2017.10.006
31. Liu, F, Tian, A, Wang, J, Zhu, Y, Xie, Z, Zhang, R, et al. Occurrence and molecular epidemiology of fosA3-bearing Escherichia coli from ducks in Shandong province of China. Poult Sci. (2022) 101:101620. doi: 10.1016/j.psj.2021.101620
32. Jiang, W, Men, S, Kong, L, Ma, S, Yang, Y, Wang, Y, et al. Prevalence of plasmid-mediated fosfomycin resistance gene fosA3 among CTX-M-producing Escherichia coli isolates from chickens in China. Foodborne Pathog Dis. (2017) 14:210–8. doi: 10.1089/fpd.2016.2230
33. Khoshnood, S, Shahi, F, Jomehzadeh, N, Montazeri, EA, Saki, M, Mortazavi, SM, et al. Distribution of genes encoding resistance to macrolides, lincosamides, and streptogramins among methicillin-resistant Staphylococcus aureus strains isolated from burn patients. Acta Microbiol Immunol Hung. (2019) 66:387–98. doi: 10.1556/030.66.2019.015
34. Garbacz, K, Wierzbowska, M, Kwapisz, E, Kosecka-Strojek, M, Bronk, M, Saki, M, et al. Distribution and antibiotic-resistance of different Staphylococcus species identified by matrix assisted laser desorption ionization-time of flight mass spectrometry (MALDI-TOF MS) isolated from the oral cavity. J Oral Microbiol. (2021) 13:1983322. doi: 10.1080/20002297.2021.1983322
35. Li, S, Zhu, Y, Zhong, G, Huang, Y, and Jones, KC. Comprehensive assessment of environmental emissions, fate, and risks of veterinary antibiotics in China: an environmental fate modeling approach. Environ Sci Technol. (2024) 58:5534–47. doi: 10.1021/acs.est.4c00993
36. Torres, RT, Cunha, MV, Araujo, D, Ferreira, H, Fonseca, C, and Palmeira, JD. A walk on the wild side: wild ungulates as potential reservoirs of multi-drug resistant bacteria and genes, including Escherichia coli harbouring CTX-M beta-lactamases. Environ Pollut. (2022) 306:119367. doi: 10.1016/j.envpol.2022.119367
37. Zhu, Z, Jiang, S, Qi, M, Liu, H, Zhang, S, Liu, H, et al. Prevalence and characterization of antibiotic resistance genes and integrons in Escherichia coli isolates from captive non-human primates of 13 zoos in China. Sci Total Environ. (2021) 798:149268. doi: 10.1016/j.scitotenv.2021.149268
38. Tian, X, Zhang, L, Li, C, Xia, D, and Ying, J. Genome sequence of a sequence type 1 NDM-5-producing carbapenem-resistant Klebsiella pneumoniae in China. J Glob Antimicrob Resist. (2024) 38:271–4. doi: 10.1016/j.jgar.2024.05.001
39. Wang, D, Fang, L-X, Jiang, Y-W, Wu, D-S, Jiang, Q, Sun, R-Y, et al. Comparison of the prevalence and molecular characteristics of fosA3 and fosA7 among salmonella isolates from food animals in China. J Antimicrob Chemother. (2022) 77:1286–95. doi: 10.1093/jac/dkac061
40. Yang, X, Liu, W, Liu, Y, Wang, J, Lv, L, Chen, X, et al. F33: a-: B-, IncHI2/ST3, and IncI1/ST71 plasmids drive the dissemination of fosA3 and blaCTX−M−55/−14/−65 in Escherichia coli from chickens in China. Front Microbiol. (2014) 5:688. doi: 10.3389/fmicb.2014.00688
41. Zhang, L-J, Yang, J-T, Chen, H-X, Liu, W-Z, Ding, Y-L, Chen, R-A, et al. F18:a-:b1 plasmids carrying blaCTX-M-55 are prevalent among Escherichia coli isolated from duck–fish polyculture farms. Antibiotics. (2023) 12:961. doi: 10.3390/antibiotics12060961
42. Wu, J, Huang, Y, Rao, D, Zhang, Y, and Yang, K. Evidence for environmental dissemination of antibiotic resistance mediated by wild birds. Front Microbiol. (2018) 9:745. doi: 10.3389/fmicb.2018.00745
43. Wang, Y, Zhang, R, Li, J, Wu, Z, Yin, W, Schwarz, S, et al. Comprehensive resistome analysis reveals the prevalence of NDM and MCR-1 in chinese poultry production. Nat Microbiol. (2017) 2:1–7. doi: 10.1038/nmicrobiol.2016.260
44. Saki, M, Amin, M, Savari, M, Hashemzadeh, M, and Seyedian, SS. Beta-lactamase determinants and molecular typing of carbapenem-resistant classic and hypervirulent Klebsiella pneumoniae clinical isolates from southwest of Iran. Front Microbiol. (2022) 13:1029686. doi: 10.3389/fmicb.2022.1029686
45. Cen, D-J, Sun, R-Y, Mai, J-L, Jiang, Y-W, Wang, D, Guo, W-Y, et al. Occurrence and transmission of blaNDM-carrying enterobacteriaceae from geese and the surrounding environment on a commercial goose farm. Appl Environ Microbiol. (2021) 87:e00087–21. doi: 10.1128/AEM.00087-21
46. Pérez-Etayo, L, González, D, and Vitas, AI. Clonal complexes 23, 10, 131 and 38 as genetic markers of the environmental spread of extended-Spectrum β-lactamase (ESBL)-Producing E. coli. Antibiot Basel Switz. (2022) 11:1465. doi: 10.3390/antibiotics11111465
47. García-Meniño, I, García, V, Mora, A, Díaz-Jiménez, D, Flament-Simon, SC, Alonso, MP, et al. Swine enteric colibacillosis in Spain: pathogenic potential of mcr-1 ST10 and ST131 E. coli isolates. Front Microbiol. (2018) 9:2659. doi: 10.3389/fmicb.2018.02659
48. Freitag, C, Michael, GB, Li, J, Kadlec, K, Wang, Y, Hassel, M, et al. Occurrence and characterisation of ESBL-encoding plasmids among Escherichia coli isolates from fresh vegetables. Vet Microbiol. (2018) 219:63–9. doi: 10.1016/j.vetmic.2018.03.028
49. Umair, M, Mohsin, M, Ali, Q, Qamar, MU, Raza, S, Ali, A, et al. Prevalence and genetic relatedness of extended spectrum-β-lactamase-producing Escherichia coli among humans, cattle, and poultry in Pakistan. Microb Drug Resist. (2019) 25:1374–81. doi: 10.1089/mdr.2018.0450
50. Roer, L, Overballe-Petersen, S, Hansen, F, Schønning, K, Wang, M, Røder, BL, et al. Escherichia coli sequence type 410 is causing new international high-risk clones. mSphere. (2018) 3:e00337-18. doi: 10.1128/msphere.00337-18
51. Nandanwar, N, Janssen, T, Kühl, M, Ahmed, N, Ewers, C, and Wieler, LH. Extraintestinal pathogenic Escherichia coli (ExPEC) of human and avian origin belonging to sequence type complex 95 (STC95) portray indistinguishable virulence features. Int J Med Microbiol. (2014) 304:835–42. doi: 10.1016/j.ijmm.2014.06.009
52. Castañeda-Barba, S, Top, EM, and Stalder, T. Plasmids, a molecular cornerstone of antimicrobial resistance in the one health era. Nat Rev Microbiol. (2024) 22:18–32. doi: 10.1038/s41579-023-00926-x
53. Wachino, J, Yamane, K, Suzuki, S, Kimura, K, and Arakawa, Y. Prevalence of fosfomycin resistance among CTX-M-producing Escherichia coli clinical isolates in Japan and identification of novel plasmid-mediated Fosfomycin-modifying enzymes. Antimicrob Agents Chemother. (2010) 54:3061–4. doi: 10.1128/aac.01834-09
54. Farajzadeh Sheikh, A, Moradi Bandbal, M, and Saki, M. Emergence of multidrug-resistant Shigella species harboring extended-spectrum beta-lactamase genes in pediatric patients with diarrhea from southwest of Iran. Mol Biol Rep. (2020) 47:7097–106. doi: 10.1007/s11033-020-05776-x
55. Li, X, Hu, H, Zhu, Y, Wang, T, Lu, Y, Wang, X, et al. Population structure and antibiotic resistance of swine extraintestinal pathogenic Escherichia coli from China. Nat Commun. (2024) 15:5811. doi: 10.1038/s41467-024-50268-2
56. Menck-Costa, MF, Baptista, AAS, Justino, L, Sanches, MS, De Souza, M, Nishio, EK, et al. High-frequency detection of fosA3 and blaCTX–M–55 genes in Escherichia coli from longitudinal monitoring in broiler chicken farms. Front Microbiol. (2022) 13:846116. doi: 10.3389/fmicb.2022.846116
57. Hou, J, Huang, X, Deng, Y, and He, L. Dissemination of the fosfomycin resistance gene fosA3 with CTX-M β-lactamase genes and rmtB carried on IncFII plasmids among Escherichia coli isolates from pets in China | antimicrobial agents and chemotherapy. Antimicrob Agents Chemother. (2012) 56:2135–8. doi: 10.1128/AAC.05104-11
58. Ito, R, Mustapha, MM, Tomich, AD, Callaghan, JD, McElheny, CL, Mettus, RT, et al. Widespread Fosfomycin resistance in gram-negative Bacteria attributable to the chromosomal fosA gene. MBio. (2017) 8:e00749-17. doi: 10.1128/mBio.00749-17
59. Wang, J, Wang, Y, Wang, Z-Y, Wu, H, Mei, C-Y, Shen, P-C, et al. Chromosomally located fosA7 in salmonella isolates from China. Front Microbiol. (2021) 12:781306. doi: 10.3389/fmicb.2021.781306
60. Güneri, CÖ, Stingl, K, Grobbel, M, Hammerl, JA, and Kürekci, C. Different fosA genes were found on mobile genetic elements in Escherichia coli from wastewaters of hospitals and municipals in Turkey. Sci Total Environ. (2022) 824:153928. doi: 10.1016/j.scitotenv.2022.153928
61. Zhang, L-J, Gu, X-X, Zhang, J, Yang, L, Lu, Y-W, Fang, L-X, et al. Characterization of a fosA3 carrying IncC–IncN plasmid from a multidrug-resistant ST17 salmonella Indiana isolate. Front Microbiol. (2020) 11:1582. doi: 10.3389/fmicb.2020.01582
62. Hao, J, Zeng, Z, Xiao, X, Ding, Y, Deng, J, Wei, Y, et al. Genomic and phenotypic characterization of a colistin-resistant Escherichia coli isolate co-harboring blaNDM-5, blaOXA-1, and blaCTX-M-55 isolated from urine. Infect Drug Resist. (2022) 15:1329–43. doi: 10.2147/IDR.S355010
63. Lv, L, Huang, X, Wang, J, Huang, Y, Gao, X, Liu, Y, et al. Multiple plasmid vectors mediate the spread of fosA3 in extended-spectrum-β-lactamase-producing enterobacterales isolates from retail vegetables in China. mSphere. (2020) 5:e00507-20. doi: 10.1128/mSphere.00507-20
64. Xiaomin, S, Yiming, L, Yuying, Y, Zhangqi, S, Yongning, W, and Shaolin, W. Global impact of mcr-1-positive enterobacteriaceae bacteria on “one health.”. Crit Rev Microbiol. (2020) 46:565–77. doi: 10.1080/1040841X.2020.1812510
65. Sadek, M, Ortiz de la Rosa, JM, Ramadan, M, Nordmann, P, and Poirel, L. Molecular characterization of extended-spectrum ß-lactamase producers, carbapenemase producers, polymyxin-resistant, and fosfomycin-resistant enterobacterales among pigs from Egypt. J Glob Antimicrob Resist. (2022) 30:81–7. doi: 10.1016/j.jgar.2022.05.022
66. Zhao, W, Li, W, Du, X-D, and Yao, H. Hybrid IncFIA/FIB/FIC(FII) plasmid co-carrying blaNDM-5 and fosA3 from an Escherichia coli ST117 strain of retail chicken. Int J Food Microbiol. (2022) 382:109914. doi: 10.1016/j.ijfoodmicro.2022.109914
67. Tian, X, Fang, R, Wu, Q, Zheng, X, Zhao, Y, Dong, G, et al. Emergence of a multidrug-resistant ST 27 Escherichia coli co-harboring blaNDM-1, mcr-1, and fosA3 from a patient in China. J Antibiot. (2020) 73:636–41. doi: 10.1038/s41429-020-0306-5
68. Peng, Z, Li, X, Hu, Z, Li, Z, Lv, Y, Lei, M, et al. Characteristics of carbapenem-resistant and colistin-resistant Escherichia coli co-producing NDM-1 and MCR-1 from pig farms in China. Microorganisms. (2019) 7:482. doi: 10.3390/microorganisms7110482
69. Lupo, A, Saras, E, Madec, J-Y, and Haenni, M. Emergence of blaCTX-M-55 associated with fosA, rmtB and mcr gene variants in Escherichia coli from various animal species in France. J Antimicrob Chemother. (2018) 73:867–72. doi: 10.1093/jac/dkx489
70. Wang, J, Zeng, Z-L, Huang, X-Y, Ma, Z-B, Guo, Z-W, Lv, L-C, et al. Evolution and comparative genomics of f33:a−:B− plasmids carrying blaCTX-M-55 or blaCTX-M-65 in escherichia coli and Klebsiella pneumoniae isolated from animals, food products, and humans in China. mSphere. (2018) 3:e00137–18. doi: 10.1128/msphere.00137-18
Keywords: waterfowl, Escherichia coli, Fosfomycin resistance, fosA3, plasmid -mediated
Citation: Zhang S, Yang J, Yang Q, Li Q, Zhong Z, Wang M, Jia R, Chen S, Liu M, Zhu D, Zhao X, Wu Y, Yang Q, Huang J, Ou X, Sun D, Tian B, Wu Z, He Y and Cheng A (2025) High prevalence of plasmid-mediated Fosfomycin resistance in waterfowl-derived Escherichia coli strains: insights into genetic context and transmission dynamics in China. Front. Vet. Sci. 12:1481822. doi: 10.3389/fvets.2025.1481822
Received: 16 August 2024; Accepted: 31 January 2025;
Published: 21 March 2025.
Edited by:
Benti Deresa Gelalcha, The University of Tennessee, Knoxville, United StatesReviewed by:
Morteza Saki, Ahvaz Jundishapur University of Medical Sciences, IranCopyright © 2025 Zhang, Yang, Yang, Li, Zhong, Wang, Jia, Chen, Liu, Zhu, Zhao, Wu, Yang, Huang, Ou, Sun, Tian, Wu, He and Cheng. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shaqiu Zhang, c2hhcWl1ODZAaG90bWFpbC5jb20=; Anchun Cheng, Y2hlbmdhbmNodW5AdmlwLjE2My5jb20=
†These authors have contributed equally to this work and share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.