- 1Key Laboratory of Animal Disease and Human Health of Sichuan Province, College of Veterinary Medicine, Sichuan Agricultural University, Chengdu, China
- 2Key Laboratory of Qinghai-Tibetan Plateau Animal Genetic Resource Reservation and Utilization, Ministry of Education, Southwest Minzu University, Chengdu, Sichuan, China
Introduction: Lactic acid bacteria (LAB) are Gram-positive bacteria that produce lactic acid during fermentation, with some strains enhancing host health by modulating the gut microbiota, boosting immune responses, and reducing inflammation.
Methods: In this study, 6 LAB strains were isolated from two dog milk samples, and their probiotic properties were comprehensively evaluated. The evaluation included growth properties, stress resistance, antipathogen activity, adhesion activity, safety assessment, antioxidant capacity, and prebiotic metabolites assessment.
Results: In comparison to the control strain Lactobacillus rhamnosus LGG, all 6 LAB isolates exhibited favorable probiotic properties. Additionally, the results of the antioxidant tests indicated that these strains demonstrated high tolerance to 0.5 mmol/L H2O2 and exhibited significant scavenging abilities for the free radicals 1,1-diphenyl-2-trinitrophenylhydrazine (DPPH) and hydroxyl (OH−). Furthermore, the 6 LAB isolates were found to produce elevated concentrations of prebiotic metabolites, including exopolysaccharides (EPS), γ-aminobutyric acid (GABA), and bile salt hydrolase (BSH).
Discussion: This study presents a comprehensive analysis of LAB isolates derived from canine milk. These isolates exhibited multifunctional properties, with strain L221 performing the best overall, making it a promising candidate for probiotic use in dogs.
Introduction
The definition of probiotics, according to the World Health Organization, is that they are live microorganisms that provide health benefits to the host when administered in sufficient quantities (1). Lactic acid bacteria (LAB), including Lactobacillus, Lactococcus, Pediococcus, Enterococcus, and Streptococcus, are commonly found in milk, feces, fermented dairy products, and the gastrointestinal tract, with certain strains exhibiting probiotic potential (2–6). Milk and its products are generally considered the main source of LAB and have always been widely used to isolate probiotics (7, 8). Numerous studies have shown that certain LAB can improve digestion, maintain intestinal microbiota balance, preventing infections, and boost the immune system (3–6, 9, 10). Since the discovery of the multifunctional properties of certain LAB strains, the incorporation of LAB into food, cosmetics, and medicine has become increasingly popular (11–16).
Dog is a popular choice for household pets worldwide. Changes in diet, environment, and weakened immune systems can make them vulnerable to digestive disorders, such as gastroenteritis, pancreatitis, and inflammatory bowel disease. These conditions can disrupt the body's intestinal microbiota, leading to symptoms such as vomiting, diarrhea, allergies, and obesity (17, 18). As the number of pet dogs increases and human life quality improves, pet owners are increasingly considering scientific feeding methods and functional pet products to meet their dogs' health and nutritional needs (19). LAB is commonly used as a food additive or nutritional supplement by pet owners to improve the health of dogs due to its beneficial functions (10).
Furthermore, while existing studies have primarily focused on the basic properties of LAB strains, such as their stress resistance, antibacterial activity, and adhesion capabilities, there remains a significant gap in the evaluation of their antioxidant properties and production of beneficial metabolites, particularly in the context of canine health (20–23). The current research aims to bridge this gap by not only screening for LAB strains with multifunctional potential derived from canine milk but also by extensively assessing their antioxidant capacity and the production of probiotic metabolites, which are crucial for their therapeutic efficacy. By focusing on these underexplored properties, this study aims to identify LAB strains that possess enhanced safety profiles and broader probiotic properties, tailored specifically for canine health.
Materials and methods
Sample collection
Milk samples, ~1 mL each, were obtained from two healthy domestic Beagle dogs that had not been administered antibiotics or probiotics for at least 1 month prior to sample collection. The samples were immediately transported to the laboratory under cold chain conditions and stored at 4°C until further analysis.
Isolation and purification and species identification
The Isolation, Purification and Species Identification was conducted based on Zhang's report with minor adjustments (24). To isolate LAB strains, 0.1 mL of canine milk was inoculated onto de Man, Rogosa, and Sharpe (MRS) agar plates (Hopebio, Qingdao, China) and incubated aerobically at 37°C for 24–72 h until distinct colonies appeared. Isolated colonies were subjected to three rounds of purification by re-culturing on MRS agar plates, followed by enrichment in MRS broth. All subsequent bacterial counting work is completed by Halo counter HD-4 Cell Counter (Hiscore Inc., China). The 16S rRNA genes of all isolates were amplified using universal primers 27F (5′-AGGTTTTTGATCCTGGCCAG-3′) and 1492R (5′-TACGACTTAACCCCAATCGC-3′). The resulting amplicons were sequenced by Sangon Bioengineering (Chengdu) Co., Ltd., and the sequences were aligned using BLAST against GenBank bacterial nucleic acid databases. Species identification was performed using MEGA6 software (Mega Limited, Auckland, New Zealand), employing the neighbor-joining method and the Kimura 2-parameter model to construct a phylogenetic tree. A sequence similarity threshold of >99% was used to define species.
Growth performance evaluation
Growth performance was evaluated by measuring the growth curve of LAB isolates as previously described by Liu et al. (25). A 50 μL aliquot (1%) of LAB culture in the mid-exponential phase was inoculated into 50 mL of fresh MRS broth and incubated at 37°C for 48 h. Optical density (OD600) measurements were taken every 2 h during the first 12 h and subsequently every 4 h up to 48 h.
Tolerance for simulating the GIT environment
Tolerance of the LAB isolates to simulated gastrointestinal conditions was tested according to the method of Zhang et al. (24), with slight adjustments to simulate the specific gastrointestinal fluid conditions in dogs. The LAB isolates were incubated in MRS broth for 12 h. Then, 10 ml of the fresh cultures were centrifuged at 8,000 × g for 10 min at 25°C to obtain bacterial cells. Bacterial cells were suspended in 10 mL of simulated gastric juice and incubated at 37°C for 3 h. Cells were then harvested by centrifugation and transferred into 10 mL of simulated intestinal juice for incubation at 37°C for an additional 4 h. Bacterial survival at 0, 3, and 7 h was assessed using colony-forming units (CFU) determined by plate counting. Survival rates were calculated as the ratio of surviving CFUs at each time point to the previous time point count.
Antipathogenic activity detection
Antibacterial activity
The Antibacterial activity detection was conducted based on Zhang's report with minor adjustments (24). The antibacterial potential of LAB isolates was tested using the Oxford cup method against four common enteropathogens: Staphylococcus aureus ATCC 25923, Escherichia coli ATCC 25922, Salmonella enterica H9812, and Pseudomonas aeruginosa PAO1. Cell-free supernatants (CFS) were obtained by centrifuging LAB cultures at 4,500 × g for 10 min and filtering the supernatant through a 0.22 μm sterile filter. The pathogenic bacteria were cultured in LB broth, diluted to about 107 CFU/mL, and 100 μL of each suspension was spread onto LB agar plates. Three sterile Oxford cups were placed on each plate and filled with 200 μL of CFS. Plates were incubated at 37°C for 24 h, and inhibition zones were measured using a caliper to determine antibacterial activity.
Co-aggregative ability with pathogens
Co-aggregation of LAB with the four pathogens was assessed by mixing equal volumes (2 mL) of LAB and pathogen suspensions, followed by incubation at 37°C for 2 h. Absorbance (A600) was measured before and after incubation for both individual and mixed cultures. The co-aggregation rate was calculated using the formula: Co-aggregation rate (%) = 1 – Amix / [ (ALAB + Apathogen) / 2] × 100.
Adhesion activity detection
Auto-aggregation activity
According to the report by Zhang et al. (24). Auto-aggregation activity was measured by adjusting the LAB culture to 1 × 108 CFU/mL in phosphate-buffered saline (PBS). After incubation at 37°C for 6 h, the OD600 of the upper layer of the bacterial suspension was measured, and auto-aggregation was calculated as: Auto-aggregation rate (%) = 1 – (A1/A0) × 100, where A0 is the initial OD and A1 is the OD at 6 h.
Cell surface hydrophobicity
The hydrophobicity of LAB strains in various organic solvents was determined using a modified version of the method reported by Kos (26). The LAB strains that were activated were introduced into MRS liquid medium at a concentration of 1% (v/v) and incubated overnight. Afterward, they were centrifuged at 8,000 × g for 10 min at 4°C to collect the microorganisms. The microorganisms were then washed three times with sterile phosphate-buffered saline (PBS, pH = 7.4) and resuspended in PBS. The absorbance of the lactobacilli suspension was adjusted to OD600 = 0.60 ± 0.05 (A0). LAB strains suspension (3 mL) was mixed with 1 mL of different organic solvents (ethyl acetate, xylol, and trichloromethane), vortexed and shaken for 2 min, and then allowed to stand for 20 min. The OD value of the aqueous phase was measured at 600 nm using a UV spectrophotometer (A1). The experiment was repeated three times. The hydrophobicity of LAB was calculated using the following formula: Hydrophobicity rate (%) = (1-A1/A0) × 100.
Adhesion to Caco-2 cells
The Adhesion to Caco-2 cells was conducted based on Wang's report with minor adjustments (27). Caco-2 cells were cultured in a flask until they reached a sub-confluent state of 80–90%. They were then digested with 0.25% trypsin and counted using a hemocytometer. The concentration of viable cells was adjusted to 1 × 105 cells/mL (VC) using DMEM medium. The cell suspension was added to a 12-well cell culture plate at a volume of 1 mL per well. The plate was then incubated at 37°C and 5% CO2 until the cells formed a confluent monolayer, typically within 24–48 h. Prior to the adhesion assay, the cells were cultured for 1 day, and the medium was replaced with high-glucose DMEM (without antibiotics) to avoid any influence of antibiotic resistance on the adhesion assay. At the beginning of the assay, the cells were washed three times with sterile PBS and 1 mL of a LAB suspension at a concentration of 1 × 108 CFU/mL (V0) was added to each well. The cells were then incubated at 37°C and 5% CO2 in a constant-temperature cell culture incubator for 2 h. After the incubation period, the cells were washed three times with sterile PBS to remove any unattached LAB. The cells were digested using 0.25% trypsin, which should be handled with care due to its potential hazards. After complete digestion, the cells were collected and subjected to 10-fold gradient dilution. The number of viable adherent LAB after dilution on MRS solid media was determined using plate colony counting (V1) after incubating the plates at 37°C for 48 h in an anaerobic jar. The experiment was conducted three times. The adhesion rate and adhesion index of LAB to Caco-2 cells were calculated using the following formula: Adhesion rate (%) = (V1 / V0) × 100; Adhesion index (CFU/cell) = V1 / VC.
Determination of biofilm forming ability
Research indicates that LAB with a strong ability to form biofilm exhibit better resistance to heat and freezing (28). The biofilm-forming ability of LAB strains was determined using crystal violet staining (29). The concentration of LAB was adjusted to 1 × 107 CFU/mL. The LAB suspension was inoculated into 96-well cell culture plates at a volume of 200 μL per well. The plates were then incubated in a 37°C incubator for 24 h to form a stable biofilm. Blank MRS liquid medium was used as a control. The bacteria were washed three times with sterile PBS to remove the planktonic bacteria, and then dried at room temperature for 15 min. Next, they were fixed in 200 μL of methanol solution for 15 min and dried again at room temperature for 10 min. The sample was immersed in a 1% crystal violet solution (200 μL) for 20 min, rinsed three times with distilled water, and air-dried for 10 min. It was then eluted in a 33% acetic acid solution (200 μL) for 10 min, and the resulting decolorized solution was analyzed at a wavelength of 595 nm using an enzyme marker. The OD value of the decolorized solution at 595 nm was measured using an enzyme counter (the control was recorded as A0 and the LAB were recorded as A). The strength of LAB biofilm formation ability was evaluated based on the following criteria: no biofilm formation ability (–): A <A0; weak biofilm forming ability (+): A0 <A ≤ 2A0; moderate biofilm forming ability (++): 2A0 <A ≤ 4A0; and strong biofilm forming ability (+++): A > 4A0. The experiments were repeated three times.
Safety assessment
Hemolytic activity
The Safety assessment test was conducted based on Zhang's report with minor adjustments (24). LAB isolates were streaked onto blood agar plates and incubated at 37°C for 48 h. Staphylococcus aureus ATCC 25923 was used as a positive control. Hemolysis was assessed by the presence of clear zones (β-hemolysis), green zones (α-hemolysis), or no zones (γ-hemolysis).
Antibiotic susceptibility
The susceptibility of the selected LAB strains to antibiotics was assessed using the disc-diffusion test. A total of 14 antimicrobials (Shunyoubio, Shanghai, China) were tested, including penicillin G (P, 10 μg), ampicillin (AMP, 10 μg), amoxicillin (AML, 25 μg), erythromycin (E, 15 μg), Cefuroxim (CXM, 30 μg), and cefotaxime (CTX). Oxacillin (OX, 5 μg), Cefazolin (KZ, 30 μg), Norfloxacin (NOR, 5 μg), Rifampicin (RD, 5 μg), clindamycin (DA, 10 μg), chloramphenicol (C, 30 μg), tetracycline (TE, 30 μg), and vancomycin (VA, 30 μg). Fresh overnight cultures of each LAB strain were diluted to a concentration of 108 CFU/ml. Subsequently, 100 μl of the diluted cultures were spread on MRS agar plates and dried. Three uniform antibiotic discs were manually placed on the surface of the dried MRS plates. The plates were then inverted and incubated for 48 h under anaerobic conditions at 37°C. Antibiotic susceptibility was classified as resistant (R), moderately susceptible (M), or sensitive (S) based on the diameter of the zone of inhibition (mm) according to the parameters of the Clinical and Laboratory Standards Institute (65).
Antioxidant capacity assessment
Inoculate 2% (v/v) of the activated LAB culture into the MRS liquid medium. After an overnight culture, centrifuge the mixture at 4°C at 8,000 × g for 10 min. Collect the supernatant to obtain a cell-free supernatant. Then, resuspend the pellet in PBS and adjust the LAB concentration to 1 × 109 CFU/mL to obtain a bacterial suspension.
Tolerance to H2O2
The tolerance of LAB to H2O2 was measured using the method reported by Xiong et al. (30). A liquid culture of LAB with a concentration of 1 × 108 CFU/mL was inoculated into MRS liquid culture medium containing 0, 0.5, 1.0, 1.5, and 2.0 mmol/L H2O2 at an inoculum volume of 2% (v/v). The mixture was incubated for 8 h at 37°C in a constant-temperature incubator, and the OD value of the culture medium was measured at a wavelength of 600 nm using a UV spectrophotometer. The experiment was repeated three times.
DPPH radical scavenging ability
The ability of LAB to scavenge the 1,1-diphenyl-2-picrylhydrazyl (DPPH) free radical was determined using the method described in the literature (31). Two milliliter of 0.2 mmol/L DPPH absolute ethanol solution was added to a centrifuge tube containing 1 mL of cell-free supernatant or bacterial suspension of lactic acid bacteria. The mixture was vortexed and left to react for 30 min at room temperature in the dark at 4°C. The supernatant was collected by centrifuging at 8,000 × g for 10 min. The OD value of the supernatant was measured at a wavelength of 517 nm using a UV spectrophotometer (ODsample). Anhydrous ethanol was used as the blank group instead of DPPH absolute ethanol solution (ODblank). Distilled water was used as the control group instead of the reaction sample (ODcontrol). The experiment was conducted three times. Afterward, we calculated the DPPH free radical scavenging rate of LAB using the following formula: DPPH free radical scavenging rate (%) = [1– (ODsample – ODblank) / ODcontrol] × 100.
Determination of OH− free radical scavenging ability
The OH− scavenging capability was determined following the protocol outlined by Alam et al. with certain adjustments (32). Five hundred microliters of LAB cell-free supernatant or suspension were added to a centrifuge tube, followed by 1 mL of 0.1% 1,10-phenanthroline, 1 mL of PBS, 1 mL of 2.5 mmol/L FeSO4, and 1 mL of 20 mmol/L H2O2. After incubating for 1.5 h in a water bath set at 37°C, we measured the OD536 of the reaction mixture (ODsample). In the blank group, we substituted a consistent volume of absolute ethanol for H2O2 (ODblank). Similarly, in the control group, we replaced the sample solution with an equivalent volume of distilled water (ODcontrol). We determined the OH− radical scavenging rate using the following formula: OH− free radical scavenging rate (%) = [(ODsample − ODcontrol)/ (ODblank− ODcontrol)] × 100.
Determination of O2− free radical scavenging ability
The ability of LAB to scavenge O2− free radicals was evaluated using the method described by Liu et al. (33). LAB cell-free supernatant or bacterial suspension (100 μL) was mixed with 2.8 mL of 0.05 mol/L Tris-HCl (pH 8.2) and 100 μL of 0.05 mol/L pyrogallol. The mixture was vortexed and incubated at 25°C in the dark. After 4 min, the reaction was stopped by adding 1 mL of 8 mol/L HCl. To measure the OD value of the reaction solution at a wavelength of 320 nm (ODsample), use a UV spectrophotometer. Adjust to zero with distilled water, which replaces the sample for reaction as a control group (ODcontrol). The experiment was repeated three times. Then, calculate the O2− free radical scavenging rate of LAB using the following formula: O2− free radical scavenging rate (%) = [1 − ODsample / ODcontrol] × 100.
Metabolite determination
Determination of exopolysaccharides production capacity
The ability of LAB to produce EPS was determined using the method described by Ren et al. (34). To prepare the cell-free LAB supernatant, follow the same procedure as in the antioxidant test, as previously described. Mix the supernatant with 800 mg/mL trichloroacetic acid to obtain a final concentration of 40 mg/mL. Incubate the mixture at 4°C in a temperature-controlled incubator overnight. Centrifuge the mixture at 4°C at 8,000 × g for 10 min to collect the supernatant. Add 250 μL of 6% phenol solution and 1 mL of concentrated sulfuric acid to 250 μL of supernatant. Mix well in an ice-water bath and cool to room temperature. Next, add 200 μL of the reaction solution to a 96-well cell culture plate and measure the OD value of the reaction solution at a wavelength of 490 nm using a microplate reader, with the blank set to the reagent mixture without the supernatant. To measure the concentration of EPS produced by LAB, prepare a standard curve using glucose solutions with concentrations of 3.125, 6.25, 12.5, 25, 50, and 100 mg/L in the presence of trichloroacetic acid, phenol, and sulfuric acid. Repeat the experiment three times to ensure accuracy.
Determination of gamma-aminobutyric acid production ability
The ability of LAB to produce GABA was determined using the Berthelot colorimetric method as reported by Zhang et al. (35). The LAB strains were activated and then introduced into a fermentation medium containing glucose, yeast extract, and peptone (GYP), with an inoculation volume of 2% (v/v). After being cultured overnight, the LAB were centrifuged at 8,000 × g for 10 min at 4°C. The supernatant should be collected and mixed with 200 μL of 0.2 mol/L borate buffer (pH 9.0), 1 mL of 6% phenol, and 0.4 mL of sodium hypochlorite solution with an available chlorine content of 5.5%. After adding these components, vortex the mixture. Place the mixture in a boiling water bath for 10 min, followed by an ice water bath for 20 min. Then, add 2 mL of a 60% ethanol solution and mix thoroughly using a vortex. Measure the OD value of the reaction solution at a wavelength of 645 nm using a UV spectrophotometer and adjust to zero with distilled water. To calculate the concentration of GABA produced by LAB, draw a standard curve using GABA standards with concentrations of 0, 0.2, 0.4, 0.6, 0.8, and 1.0 g/L. Repeat the experiment three times.
Determination of bile salt hydrolase producing ability
The production of BSH by LAB was determined following the method described in Wang et al. (36). The resulting supernatant was collected to obtain a cell-free extract. To prepare the antioxidant test samples, the bacterial suspension was mixed with 10 mmol/L dithiothreitol, sonicated for 10 min in an ice bath, and then centrifuged at 4°C and 8,000 × g for 10 min. To 10 μL of LAB cell-free supernatant or cell-free extract, add 180 μL of 0.1 mol/L PBS (pH 6.0) and 10 μL of 0.1 mol/L sodium taurocholate solution. Then, add 200 μL of 15% trichloroacetic acid and react for 1 min. Centrifuge the mixture at 4°C, 8,000 × g for 10 min and collect the supernatant. Add 100 μL of the supernatant to 1.9 mL of ninhydrin chromogenic solution and vortex. Vortex the mixture and place it in a 37°C water bath for 30 min. Put the mixture in a boiling water bath for 15 min and then in an ice water bath for 3 min. The OD value of the reaction solution was measured at a wavelength of 570 nm using a UV spectrophotometer. Trichloroacetic acid was added to the sample, followed by the addition of sodium taurocholate solution as a control group for the reaction. A standard curve was constructed using glycine standards at concentrations of 0, 0.1, 0.2, 0.3, 0.4, and 0.5 μmol/L. Enzyme activity was defined as the production of 1 μg of glycine per minute at 37°C, with 1 unit of enzyme activity being equivalent to this amount. The experiment was repeated three times.
Results
In this study, six LAB strains were isolated from the milk of two dogs. These isolates were phenotypically characterized and the results are summarized in Supplementary Table 1. These strains were identified as Gram-positive, rod-shaped or Cocci-shaped, and catalase-negative bacteria. To identify potential probiotic LAB candidates, using the well-characterized reference strain Lactobacillus rhamnosus (LGG) as a control, the following assessments were performed on these six strains: species identification, growth performance, stress resistance, antipathogenic activity, adhesion activity, safety assessment, antioxidant capacity, and metabolite determination.
Species identification
The 16S rRNA sequences of the 6 LAB isolates obtained were compared with the sequences in GenBank. The strains were identified as members of the Lactobacillus and Enterococcus genera, which are commonly recognized as lactic acid bacteria. Specifically, the strains were related to Lactobacillus johnsonii (L218), Lactobacillus reuteri (L219), Enterococcus faecium (L220), Lactobacillus acidophilus (L221), Lactobacillus animalis (L222), and Enterococcus faecalis (L223), with sequence similarity exceeding 98%. The phylogenetic tree, constructed based on the 16S rRNA gene sequences and shown in Figure 1, provides a visual representation of the genetic relatedness among these isolates.
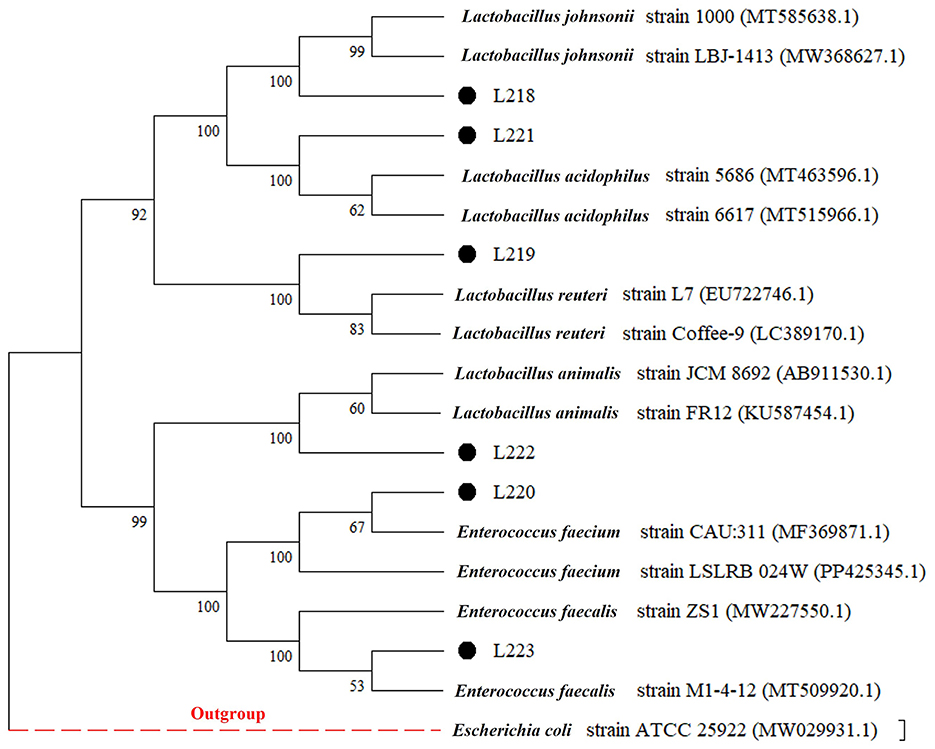
Figure 1. Phylogenetic tree (based on neighbor-joining method) of six LAB strains created with the data from 16S rRNA gene analysis results (Bootstrap value was 1,000 repeats. Escherichia coli ATCC 25922 was used as outgroup).
Growth performance
The growth conditions of the six lactic acid bacteria strains and the control strain LGG are shown in Figure 2a. It was obvious that the growth cycles of all strains were relatively similar, entering the exponential growth phase after about 4 h, and the OD600 value increased exponentially. After 24 h of culture, the growth rate decreased and entered a stable phase. Among them, Lactobacillus rhamnosus LGG showed a significantly stronger growth speed and performance compared to the other strains.
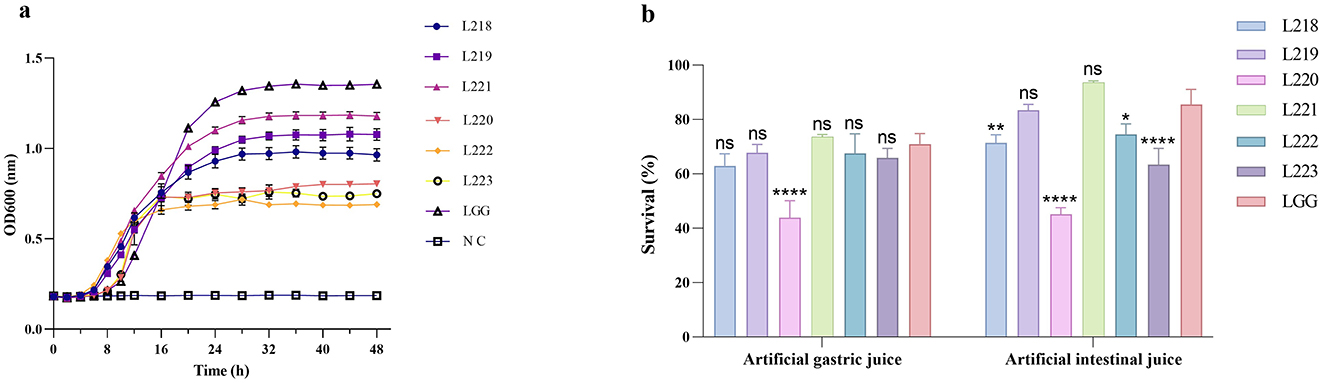
Figure 2. (a) Growth curve; (b) survival of the potential probiotic isolates in the artificial gastric and intestinal juices. *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001; ns, not significant (p ≥ 0.05). These comparisons are specifically made relative to the control group (LGG).
Resistant capacity
Figure 2b shows the survival rate results of LAB strains under simulated gastrointestinal tract (GIT) conditions. After being treated with artificial gastric juice for 3 h, except for Enterococcus faecium L220, other isolated strains showed higher resistance to simulated gastric juice, with survival rates exceeding 60%. The specific average survival rates for these strains ranged from 43.91 to 73.65%, and there was no significant difference compared with the control group, which was treated with the same artificial gastric juice (P > 0.05). Seven hours after incubating the surviving cells in artificial intestinal fluid, strains L221 (93.59%) and L219 (83.39%) showed better resistance to artificial intestinal fluid, with no significant difference compared with the control group (P > 0.05).
Antipathogenic activity detection
Antibacterial activity
Table 1 shows that the six strains exhibit varying levels of antagonistic activity against four common intestinal pathogenic bacteria. The control strain LGG exhibited the strongest antagonistic activity against Escherichia coli and Staphylococcus aureus. Its antagonistic activity against Escherichia coli was significantly better than that of the other isolated strains (P < 0.05). L221 and L222 were the isolates with the strongest antagonistic activity against Salmonella and Pseudomonas aeruginosa, respectively. Both were superior to the control strain LGG (P < 0.05).
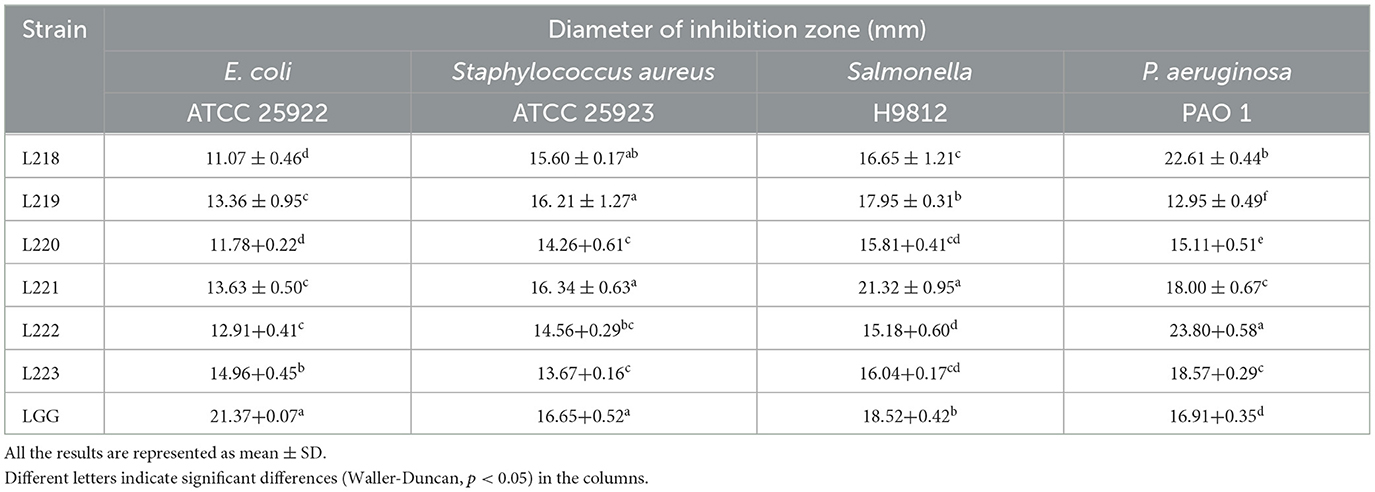
Table 1. Antagonistic activity of potential probiotic strains from canine milk samples against pathogenic bacteria by the Oxford cup method.
Co-aggregative ability with pathogens
Table 2 shows that the co-aggregation rates of the six LAB strains against four common pathogens. The strains with the highest co-aggregation ability against all four pathogens was Lactobacillus reuteri strain L219, including Escherichia coli (68.03%), Staphylococcus aureus (69.70%), Salmonella (68.10%), and Pseudomonas aeruginosa (69.60%).
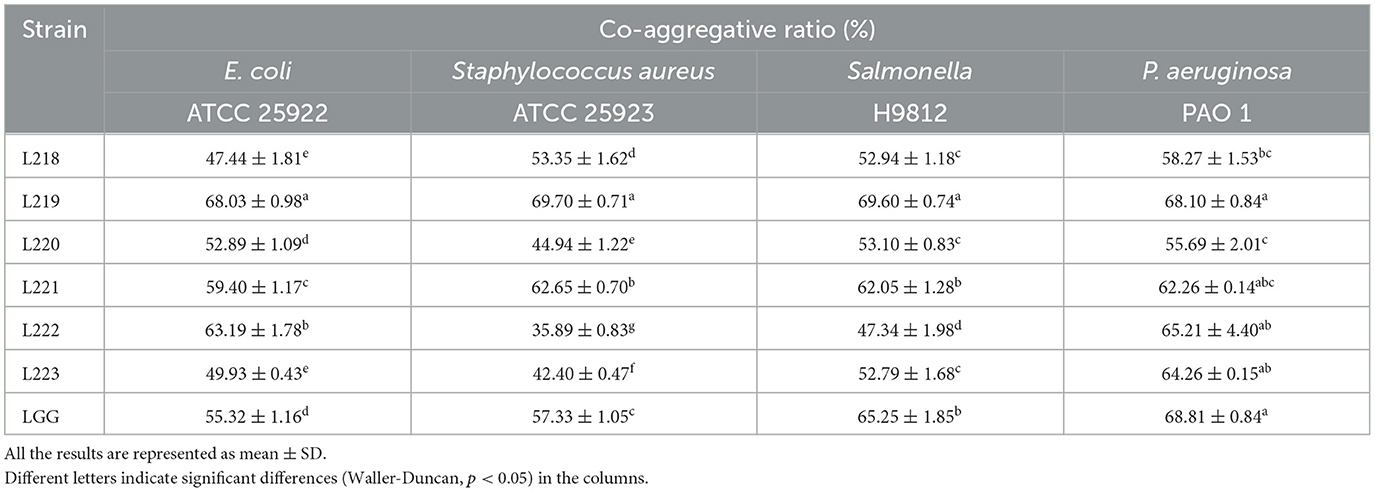
Table 2. Co-aggregative activity of potential probiotic strains from canine milk samples against pathogenic bacteria.
Adhesion activity detection
To evaluate the adhesion ability of the six isolates, we performed the following experiments: auto-aggregation assay, adhesion ability assay of Caco-2 cell line, cell surface hydrophobicity assay, and biofilm formation ability assay.
Auto-aggregation activity
The auto-aggregation ability is shown in Figure 3a. L221 (88.93%) has the highest self-agglutination rate, followed by LGG (78.31%) and L219 (74.40%).
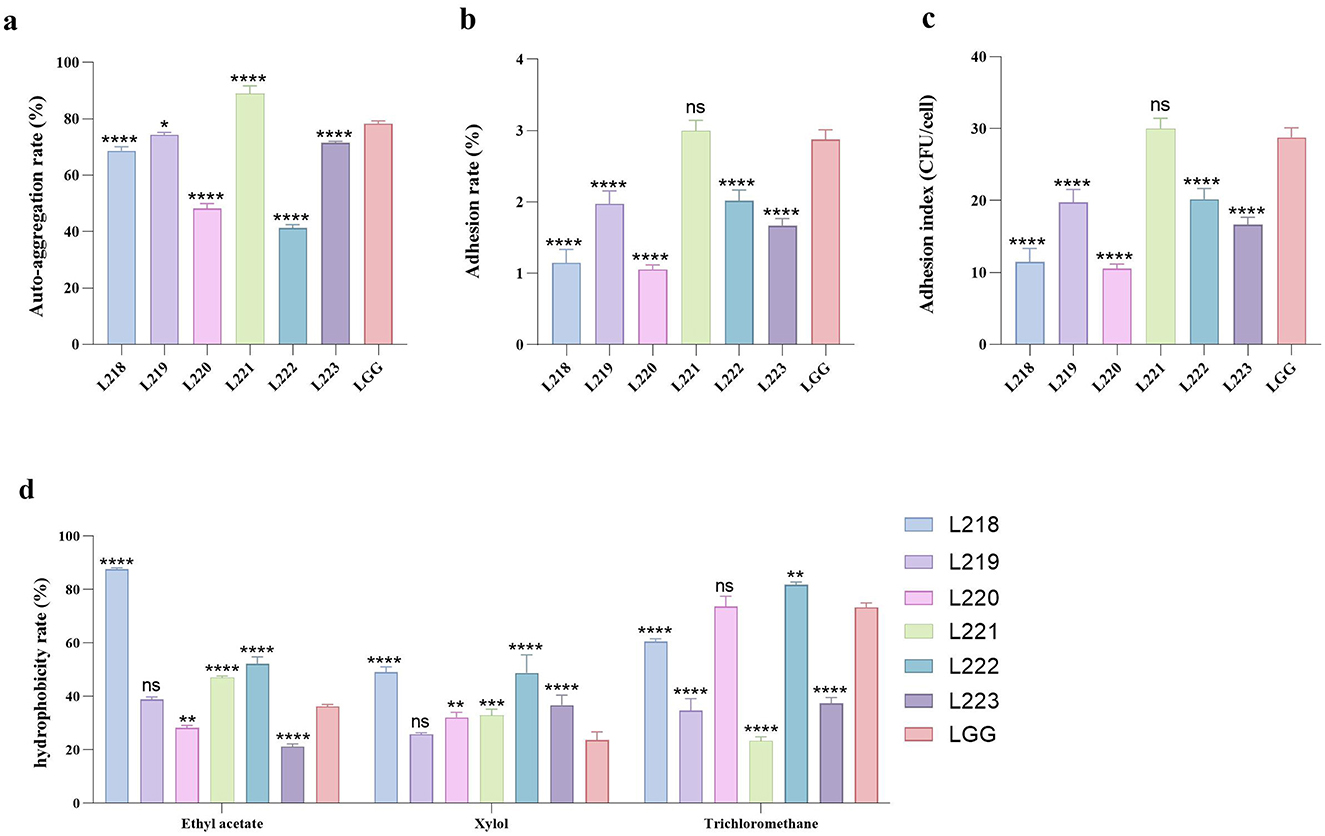
Figure 3. All the results are represented as mean ± SD. (a) Auto-aggregation rate of LAB strains. (b) Adhesion rate of LAB strains to Caco-2 cell lines. (c) Adhesion index of LAB strains to Caco-2 cell lines. (d) The cell surface hydrophobicity of LAB strains. *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001; ns, not significant (p ≥ 0.05). These comparisons are specifically made relative to the control group (LGG).
Adhesion to Caco-2 cells
Figures 3b, c show the adhesion ability of the six LAB strains to the Caco-2 cell line. Among them, Strain L221 showed the strongest adhesion ability (adhesion rate 3.00%, 30.00 CFU/cell), followed by control strain LGG (adhesion rate 2.88%, 28.77 CFU/cell).
Cell surface hydrophobicity
Figure 3d displays the results of cell surface hydrophobicity. Significant differences were observed in the hydrophobicity of various LAB strains when exposed to different solutions. L218 exhibited the highest hydrophobicity against two organic solvents: Ethyl acetate (87.63%) and Xylol (49.03%). L222 exhibited the highest hydrophobicity toward the organic solvent Trichloromethane (81.76%).
Determination of biofilm forming ability
Supplementary Table 2 displays the biofilm formation ability of the six isolates. Strains L218, L219, L220, and L223 exhibited moderate biofilm formation ability (++), while strain L221, L222 and LGG demonstrated strong biofilm formation ability (+++).
Safety assessment
Hemolytic activity
The hemolytic activity test results showed that these six isolates were non-hemolytic (Supplementary Figure 1).
Antibiotic susceptibility
Table 3 shows the susceptibility results of six isolates to 14 antibiotics. The resistance rates (includes resistance and intermediate) were 0% (0/6) to penicillin G, 0% (0/6) to amoxicillin, 0% (0/6) to amoxicillin, 16.67% (1/6) to erythromycin, 0% (0/6) to Cefuroxim, 0% (0/6) to cefotaxime, 83.33% (5/6) to Oxacillin, 0% (0/6) to Cefazolin, 100% (6/6) to Norfloxacin, 33.33% (2/6) to Rifampicin, 16.67% (1/6) to clindamycin, 0% (0/6) to chloramphenicol, 33.33% (2/6) to tetracycline, 100% (6/6) to vancomycin. Isolates L219, L221, and L222 had the highest sensitivity to 14 antibiotics (78.57%), followed by L218 (71.43%), L220 (64.29%), and L223 (64.29%).
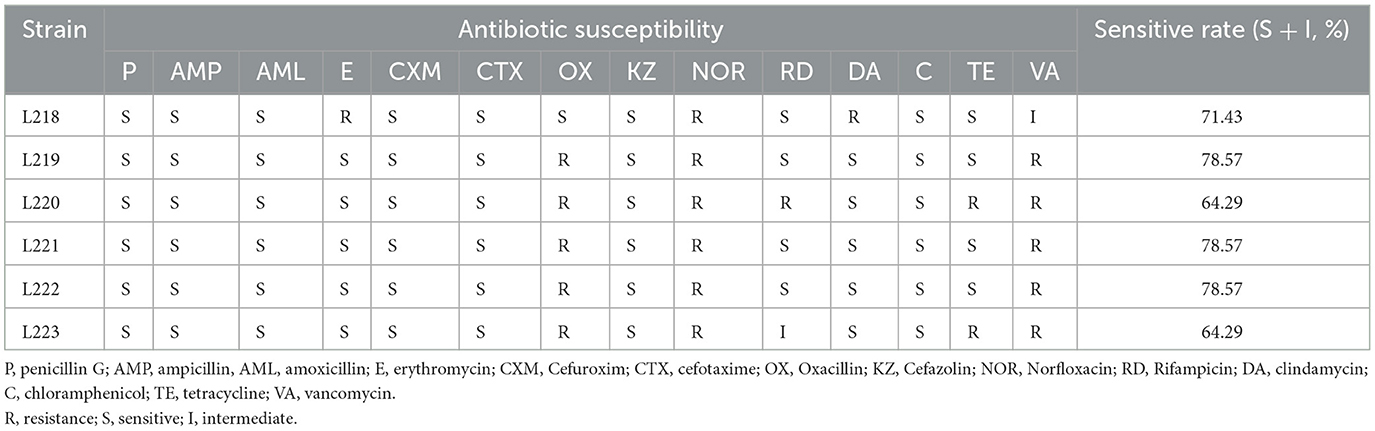
Table 3. Antibiotic susceptibility of 6 LAB strains isolated from canine milk to different antibiotics.
Antioxidant capacity assessment
Tolerance to H2O2
Table 4 displays the tolerance of 6 LAB strains to varying concentrations of H2O2. The results showed that all six isolates could survive well in 0.5 mmol/L H2O2, with survival rates ranging from 86.01 to 98.96%. Except for isolates L220 and L221, the survival rates of the other strains were found to be statistically significantly higher than the control strain LGG (P < 0.05). When the concentration increased to 1 mmol/L H2O2, the survival rate of strains L220, L221, and LGG exceeded 28%. When the concentration increased to 1.5 and 2 mmol/L H2O2, the overall survival rate was lower than 10%.
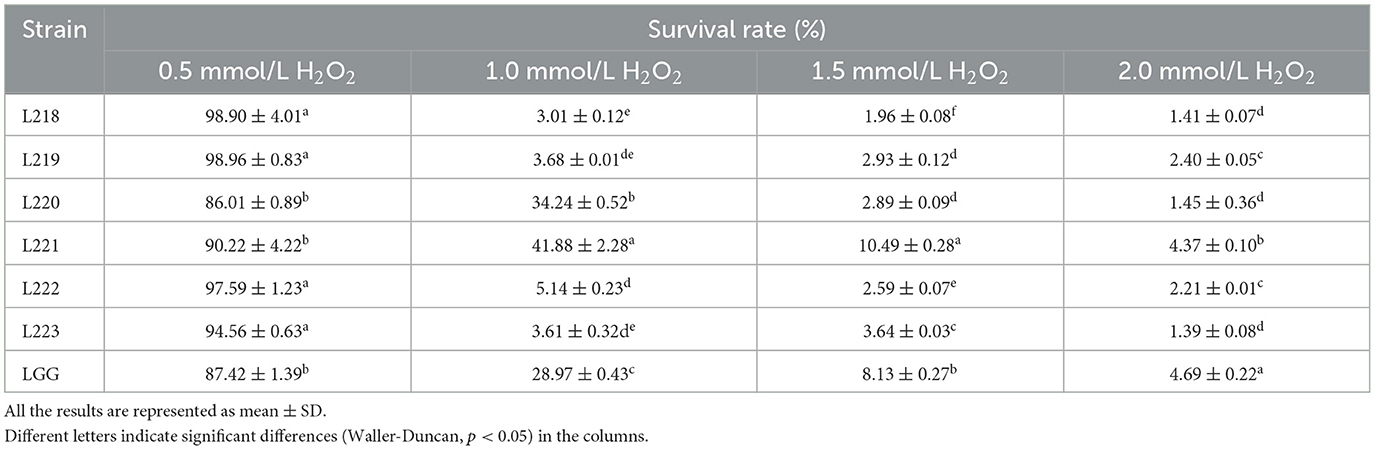
Table 4. Livability of potential probiotic strains from canine milk samples in different concentrations of H2O2 environment.
Free radical scavenging ability
The results of the free radical scavenging experiment are shown in Table 5. The cell-free supernatant of the six LAB isolates was always better than the bacterial suspension. The clearance rate of DPPH by bacterial suspension is 18.05% ± 0.15 to 24.40% ± 0.35, all were higher than the control strain LGG (17.65 ± 0.36). The highest clearance rate (24.40 ± 0.35) was observed in strain L219. Interestingly, the overall DPPH clearance rate of the supernatant for the isolated strains remained within the range of 77.91% ± 1.09 to 88.44% ± 0.99, all lower than the control strain LGG (93.43 ± 1.16). The clearance rate of OH− in bacterial suspension is 11.27% ± 1.05 to 22.38% ± 3.73, all lower than the control strain LGG (39.67 ± 1.35). The removal rate of OH- in the supernatant liquid is 58.00–71.40%. Strain L223 showed the highest removal rate (71.40 ± 1.57). The isolated strains showed no ability to clear O2− from bacterial suspensions or cell-free supernatants, which is consistent with the control strain LGG.
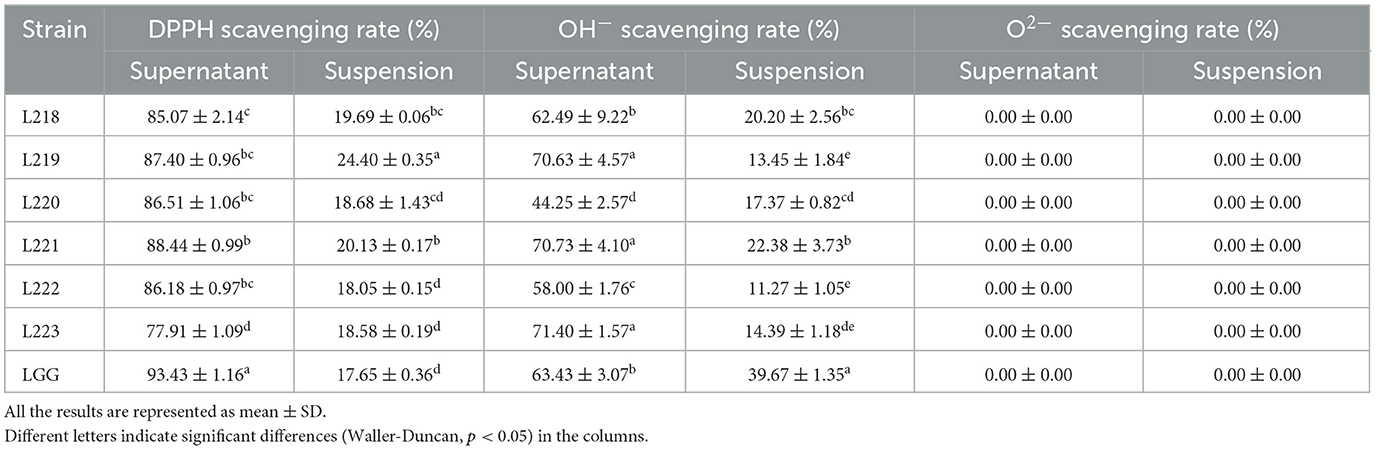
Table 5. DPPH, OH−, and O2− radical scavenging activity of 6 LAB strains isolated from canine milk samples.
Metabolite determination
Figure 4 shows the results of metabolite assessment for six isolates. All strains showed good EPS production capacity, with values exceeding 548.52 mg/L (Figure 3a). Strain L221 has the highest EPS production capacity of 635.81 mg/L, significantly higher than the control strain LGG (597.09 mg/L).
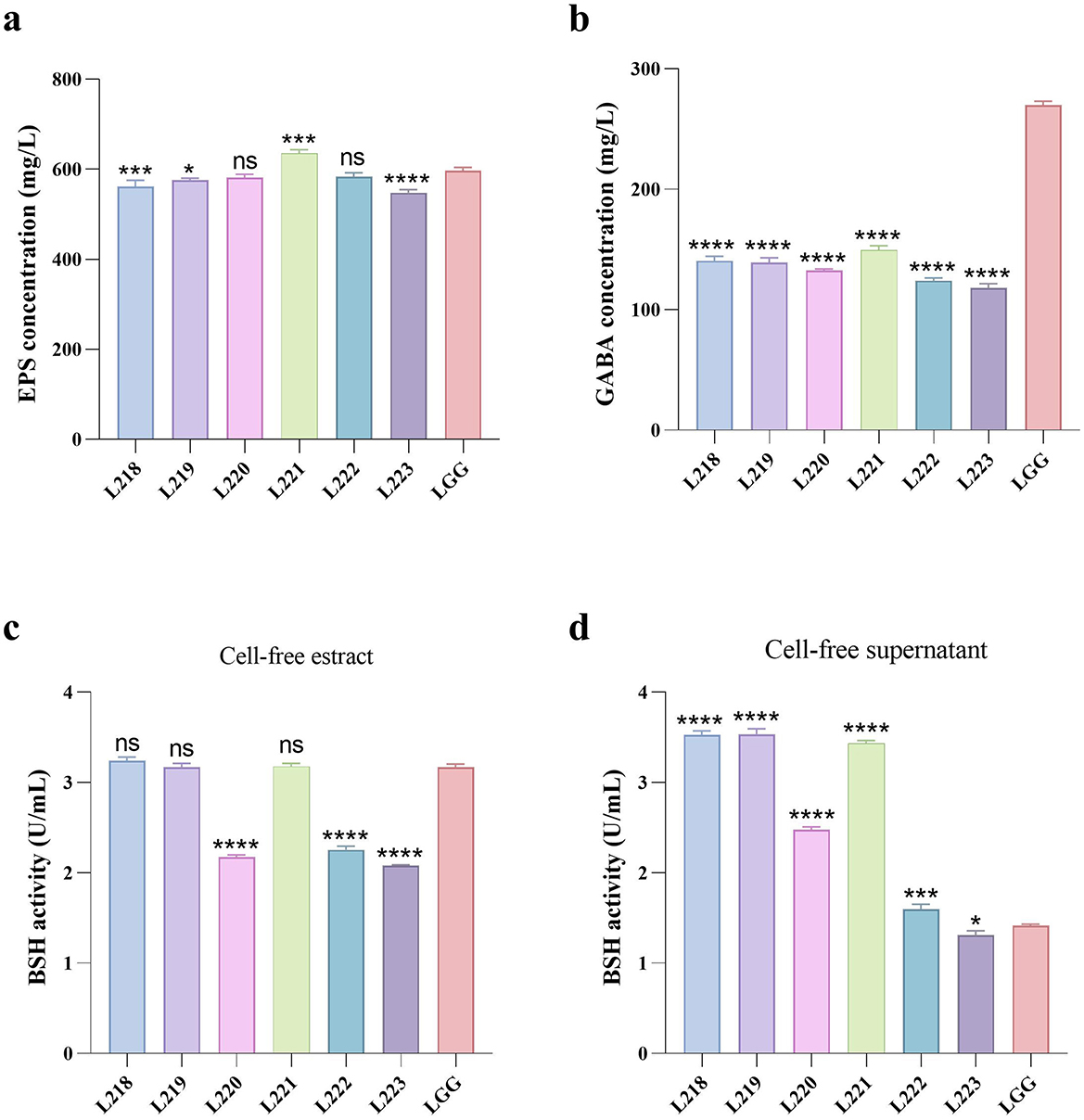
Figure 4. Metabolites production abilities of seven LAB strains. All the results are represented as mean ± SD. (a) EPS production ability. (b) GABA production ability. (c) BSH production ability of cell-free extract. (d) BSH production ability of cell-free supernatant. *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001; ns, not significant (p ≥ 0.05). These comparisons are specifically made relative to the control group (LGG).
The GABA production capacity of the six isolates ranged from 118.19 to 149.51 mg/L (Figure 3b), Significantly lower than the control strain LGG (269.81 mg/L). The BSH production capacity of cell-free extracts of the isolates ranged from 2.08 to 3.24 U/mL (Figure 3c), while the cell-free supernatants of the isolates ranged from 1.31 to 3.54 U/mL (Figure 3d).
Discussion
In this study, we isolated six LAB strains from canine milk samples via 16S rRNA molecular identification. The results of the basic characteristics test showed that all six strains exhibited robust growth and demonstrated resistance to the artificial gastrointestinal tract environment. Among them, Lactobacillus acidophilus L221 performed best, possibly due to its unique cell wall structure and acid and bile resistance mechanisms (37, 38). The growth curve reflects the growth and metabolic capabilities of bacteria, suggesting their potential for rapid activation upon reaching the intestinal colon and their ability to establish and maintain a large number of colonies for a long time, laying the foundation for their probiotic effects in animal intestines (39). The strains' good tolerance to the gastrointestinal tract environment, as assessed by their ability to survive in artificial gastric and intestinal fluids, suggests that they can reach the intestine intact and undergo further reproductive growth (40).
Anti-pathogenic activity and safety properties are considered the most important properties for probiotics. In dogs, prevalent bacterial pathogenic microorganisms such as Escherichia coli, Staphylococcus aureus, Salmonella, and Pseudomonas aeruginosa are responsible for a spectrum of diseases including ulcerative keratitis, otitis media, pyoderma, urinary tract infections, skin and wound infections, as well as various respiratory tract infections (41–46). In this study, we found that the cell-free supernatants derived from six isolates exhibited robust antagonistic activity against these four common pathogens. This suggests that our LAB candidate strains hold promise for potential applications in the treatment and prevention of these diseases in dogs in the future.
The functionality of LAB in resisting pathogenic bacterial invasion, maintaining intestinal flora balance, and modulating immune responses is contingent upon their adherence to host intestinal epithelial cells (47). This adhesion is related to cell surface hydrophobicity and autoaggregation activity. Enhanced cell surface hydrophobicity facilitates interactions between LAB strains and intestinal epithelial cells, while auto-aggregation activity enables LAB strains to achieve high cell densities in the intestine (48). In this study, six canine-derived LAB strains were found to be hydrophobic to organic solvents (ethyl acetate, xylene, and chloroform). In addition, these strains showed better self-agglutination ability and adhesion to Caco-2 cells. Notably, Lactobacillus acidophilus (L221) exhibited the most robust properties, which may be attributed to the strain's ability to produce a large amount of EPS. Similar results were reported by Kos et al. for Lactobacillus acidophilus M92 (26). Compared to probiotics in the free state, probiotics in the periplasmic state have been shown to have excellent gastrointestinal tolerance and adhesion capabilities (49). In this study, all six potential LAB strains showed strong biofilm forming ability, suggesting their capacity to adhere, colonize, and replicate in the gastrointestinal tract.
When the body undergoes oxidative stress, it produces large amounts of reactive oxygen species (ROS), including H2O2, DPPH, OH−, and O2− radicals. Excess ROS attack proteins, lipids, nucleic acids, and other biomolecules, further exacerbating oxidative stress. Oxidative damage to these biomolecules can trigger apoptosis and contributing to various diseases such as inflammation, cancer, atherosclerosis, aging, and degenerative diseases (50). Numerous studies have demonstrated that LAB possess a potent antioxidant capacity, capable of inhibiting oxidative stress and mitigating the damage caused by associated diseases (51). Lactobacilli have been shown to exert their antioxidant capacity through both ROS scavenging and redox systems (52–54). It is noteworthy that the antioxidant capacity and mechanisms may vary among different LAB species. In this study, the six canine-derived LAB strains demonstrated high tolerance to 0.5 mmol/L H2O2, as well as high scavenging capacity for DPPH and OH− radicals. However, they did not show any O2− radical scavenging capacity, which is consistent with the findings of Kuda et al. (55). In addition, Lactobacillus acidophilus L221 showed the ability to tolerate 1.0 and 1.5 mmol/L H2O2 and the strongest scavenging ability against DPPH and OH− radicals. Lactobacillus acidophilus L221 exhibited the best comprehensive in vitro antioxidant effect, likely due to its production of antioxidative metabolites, such as γ-aminobutyric acid (GABA) and exopolysaccharides, which enhance its ability to scavenge ROS (56).
Beneficial metabolite production is a crucial factor in evaluating functional probiotics. EPS, produced by Lactobacillus during reproduction and metabolism, is an important metabolite that promotes animal health. Studies have shown that EPS can have beneficial effects on the organism through antibacterial, antiviral, antioxidant, antitumor, and immunomodulatory effects (9). Therefore, the screening for LAB strains that produce EPS and the quantitative analysis of EPS have garnered considerable attention. Hamet et al. screened 28 strains of Lactobacillus spp with EPS production capacities ranging from 20 to 370 mg/L (57). However, the six strains of LAB strains evaluated in this study demonstrated a strong EPS production capacity ranging from 548.52–635.81 mg/L. The highest strain was Lactobacillus acidophilus L221, suggesting potential multifunctional effects. This study is one of the few to evaluate the EPS production capacity of LAB canis. LAB produces GABA, an inhibitory neurotransmitter in the mammalian central nervous system. GABA has been investigated for its physiological roles, such as stimulating appetite, aiding digestion, managing epilepsy, suppressing cancer cell growth, and boosting immune function (58, 59). Therefore, the screening of GABA-producing LAB is a current research focus. However, there are few reports on GABA-producing LAB of canine origin. In this experiment, we discovered that 6 strains of LAB strains produced 118.18–149.51 g/L of GABA, which is comparable to the 0.16 g/L produced by Lactobacillus plantarum 8014 as reported by Li et al. (60). This suggests that the six LAB strains have the potential to be probiotics. Bile salt hydrolase (BSH) is an intracellular enzyme produced by intestinal flora during growth and reproduction. It regulates the balance of bile acids in the host, affects lipid metabolism, and controls cholesterol, as well as regulates intestinal diseases (61). Therefore, it is important to screen for LAB that produce BSH. Pinto et al. found that BSH activity was absent in all seven Lactobacillus isolates examined (62). Tsai et al. screened 800 strains of Lactobacillus and found only 22 with BSH activity (63). In the present study, six strains of LAB were found to have BSH activity, ranged from 1.31 to 3.54 u/ml, suggesting potential probiotic functions.
Compared to other well-known probiotic strains, L221 demonstrated a distinct advantage, particularly in its strong gastrointestinal tolerance, superior adhesion ability, robust anti-pathogenic activity against common bacterial pathogens, and potent antioxidant effects (8, 27, 64). These exceptional attributes not only highlight L221's potential in enhancing gut health and modulating immune responses, but also position it as a versatile probiotic with applications in the treatment and prevention of various canine diseases (10). Given the similarities in gastrointestinal microbiota across species, the strain's ability to improve gut health and support immune function may extend beyond dogs, offering potential benefits for other companion animals, livestock, and even human health applications. However, some limitations should be acknowledged, such as the lack of in vivo validation of L221's efficacy and safety in dogs, as well as the need for long-term studies to assess its potential side effects. Future research could focus on evaluating the long-term therapeutic effects of L221 in canine health and exploring its interactions with other gut microbiota.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary material, further inquiries can be directed to the corresponding author.
Ethics statement
The animal studies were approved by Sichuan Agricultural University Animal Ethical and Welfare Committee. The studies were conducted in accordance with the local legislation and institutional requirements. Written informed consent was obtained from the owners for the participation of their animals in this study.
Author contributions
YL: Writing – original draft, Writing – review & editing. YZ: Writing – original draft. LC: Writing – original draft. JX: Writing – review & editing. ZZhon: Writing – review & editing. HL: Writing – review & editing. HF: Writing – review & editing. ZZhou: Writing – review & editing. GP: Writing – original draft, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This study was financially supported by the National Science and Technology Department's “13th Five-Year” Special Subproject of China (No. 2016YFD0501009).
Acknowledgments
We would like to thank Jinpeng Yang, Ruihu Wu, Wancheng Li, and other laboratory partners for their help in our experiments.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The authors declare that no Generative AI was used in the creation of this manuscript.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fvets.2024.1505854/full#supplementary-material
References
1. FAO and WHO. Probiotics in Food: Health and Nutritional Properties and Guidelines for Evaluation. Rome: Food and Agriculture Organization of the United Nations; World Health Organization (2006).
2. Agagündüz D, Yilmaz B, Sahin TÖ, Güneşliol BE, Ayten S, Russo P, et al. Dairy lactic acid bacteria and their potential function in dietetics: the food–gut-health axis. Foods. (2021) 10:3099. doi: 10.3390/foods10123099
3. Bender MJ, McPherson AC, Phelps CM, Pandey SP, Laughlin CR, Shapira JH, et al. Dietary tryptophan metabolite released by intratumoral Lactobacillus reuteri facilitates immune checkpoint inhibitor treatment. Cell. (2023) 186:1846–62.e26. doi: 10.1016/j.cell.2023.03.011
4. Fan X, Mai C, Zuo L, Huang J, Xie C, Jiang Z, et al. Herbal formula BaWeiBaiDuSan alleviates polymicrobial sepsis-induced liver injury via increasing the gut microbiota Lactobacillus johnsonii and regulating macrophage anti-inflammatory activity in mice. Acta Pharm Sin B. (2023) 13:1164–79. doi: 10.1016/j.apsb.2022.10.016
5. Heumel S, de Rezende Rodovalho V, Urien C, Specque F, Brito Rodrigues P, Robil C, et al. Shotgun metagenomics and systemic targeted metabolomics highlight indole-3-propionic acid as a protective gut microbial metabolite against influenza infection. Gut Microbes. (2024) 16:2325067. doi: 10.1080/19490976.2024.2325067
6. Lau HC-H, Zhang X, Ji F, Lin Y, Liang W, Li Q, et al. Lactobacillus acidophilus suppresses non-alcoholic fatty liver disease-associated hepatocellular carcinoma through producing valeric acid. EBioMedicine. (2024) 100:104952. doi: 10.1016/j.ebiom.2023.104952
7. Plessas S, Nouska C, Karapetsas A, Kazakos S, Alexopoulos A, Mantzourani I, et al. Isolation, characterization and evaluation of the probiotic potential of a novel Lactobacillus strain isolated from Feta-type cheese. Food Chem. (2017) 226:102–8. doi: 10.1016/j.foodchem.2017.01.052
8. Reuben RC, Roy PC, Sarkar SL, Rubayet Ul Alam ASM, Jahid IK. Characterization and evaluation of lactic acid bacteria from indigenous raw milk for potential probiotic properties. J Dairy Sci. (2020) 103:1223–37. doi: 10.3168/jds.2019-17092
9. Werning ML, Hernández-Alcántara AM, Ruiz MJ, Soto LP, Dueñas MT, López P, et al. Biological functions of exopolysaccharides from lactic acid bacteria and their potential benefits for humans and farmed animals. Foods. (2022) 11:1284. doi: 10.3390/foods11091284
10. Yang Q, Wu Z. Gut probiotics and health of dogs and cats: benefits, applications, and underlying mechanisms. Microorganisms. (2023) 11:2452. doi: 10.3390/microorganisms11102452
11. Abedi E, Hashemi SMB. Lactic acid production – producing microorganisms and substrates sources-state of art. Heliyon. (2020) 6:e04974. doi: 10.1016/j.heliyon.2020.e04974
12. Mejía-Avellaneda LF, Suárez H, Jiménez H, Mesa L. Challenges and opportunities for the production of lactic acid bacteria inoculants aimed for ensiling processes. Crit Rev Biotechnol. (2022) 42:1028–44. doi: 10.1080/07388551.2021.1988508
13. de Deus C, Eduardo de Souza Brener C, Marques da Silva T, Somacal S, Queiroz Zepka L, Jacob Lopes E, et al. Co-encapsulation of Lactobacillus plantarum and bioactive compounds extracted from red beet stem (Beta vulgaris L.) by spray dryer. Food Res Int. (2023) 167:112607. doi: 10.1016/j.foodres.2023.112607
14. Yin Y, Liao Y, Li J, Pei Z, Wang L, Shi Y, et al. Lactobacillus plantarum GX17 benefits growth performance and improves functions of intestinal barrier/intestinal flora among yellow-feathered broilers. Front Immunol. (2023) 14:1195382. doi: 10.3389/fimmu.2023.1195382
15. Rao W, Wu J, Fang Z, Chen Z, Wu J, Fang X. Antibacterial mechanism of metabolites of Lactobacillus plantarum against Pseudomonas lundensis and their application in dry-aged beef preservation. Food Chem. (2024) 460:140463. doi: 10.1016/j.foodchem.2024.140463
16. Sabahi S, Noori SMA, Ekrami A, Hosseini SA, Seyedtabib M, Akrami S. Application of Lactobacillus casei and Lactobacillus plantarum to develop dried functional apple and banana. Microsc Res Tech. (2024) 87:2636–42. doi: 10.1002/jemt.24639
17. Rhimi S, Kriaa A, Mariaule V, Saidi A, Drut A, Jablaoui A, et al. The nexus of diet, gut microbiota and inflammatory bowel diseases in dogs. Metabolites. (2022) 12:1176. doi: 10.3390/metabo12121176
18. Fan Z, Bian Z, Huang H, Liu T, Ren R, Chen X, et al. Dietary strategies for relieving stress in pet dogs and cats. Antioxidants. (2023) 12:545. doi: 10.3390/antiox12030545
19. White BL. Insights-driven development of humanized foods for pets. Meat Muscle Biol. (2023) 6:14397. doi: 10.22175/mmb.14397
20. Jang H-J, Son S, Kim J-A, Jung MY, Choi Y-J, Kim D-H, et al. Characterization and functional test of canine probiotics. Front Microbiol. (2021) 12:625562. doi: 10.3389/fmicb.2021.625562
21. Morales B, Spadetto L, Calvo MÀ, Yeste M, Arosemena L, Rigau T, et al. Evaluation of the probiotic in vitro potential of lactic acid-producing bacteria from canine vagina: possible role in vaginal health. Animals. (2022) 12:796. doi: 10.3390/ani12060796
22. Quilodrán-Vega SR, Muñoz-Flores C, Pino A, Buldres P, Sandoval F, Aguirre A, et al. Isolation, characterization, and immunomodulatory activity evaluation of probiotic strains from colostrum and canine milk. Front Vet Sci. (2023) 10:1266064. doi: 10.3389/fvets.2023.1266064
23. Zhang Y, Zhao M, Li Y, Liang S, Li X, Wu Y, et al. Potential probiotic properties and complete genome analysis of Limosilactobacillus reuteri LRA7 from dogs. Microorganisms. (2024) 12:1811. doi: 10.3390/microorganisms12091811
24. Zhang W, Lai S, Zhou Z, Yang J, Liu H, Zhong Z, et al. Screening and evaluation of lactic acid bacteria with probiotic potential from local Holstein raw milk. Front Microbiol. (2022) 13:918774. doi: 10.3389/fmicb.2022.918774
25. Liu W, Chen M, Duo L, Wang J, Guo S, Sun H, et al. Characterization of potentially probiotic lactic acid bacteria and bifidobacteria isolated from human colostrum. J Dairy Sci. (2020) 103:4013–25. doi: 10.3168/jds.2019-17602
26. Kos B, Susković J, Vuković S, Simpraga M, Frece J, Matosić S. Adhesion and aggregation ability of probiotic strain Lactobacillus acidophilus M92. J Appl Microbiol. (2003) 94:981–7. doi: 10.1046/j.1365-2672.2003.01915.x
27. Wang J, Pu Y, Zeng Y, Chen Y, Zhao W, Niu L, et al. Multi-functional potential of five lactic acid bacteria strains derived from giant panda (Ailuropoda melanoleuca). Probiotics Antimicrob Proteins. (2023) 15:668–81. doi: 10.1007/s12602-021-09881-6
28. Cheow WS, Hadinoto K. Biofilm-like Lactobacillus rhamnosus probiotics encapsulated in alginate and carrageenan microcapsules exhibiting enhanced thermotolerance and freeze-drying resistance. Biomacromolecules. (2013) 14:3214–22. doi: 10.1021/bm400853d
29. Stepanovic S, Vukovic D, Dakic I, Savic B, Svabic-Vlahovic M. A modified microtiter-plate test for quantification of staphylococcal biofilm formation. J Microbiol Methods. (2000) 40:175–9. doi: 10.1016/S0167-7012(00)00122-6
30. Xiong L, Ni X, Niu L, Zhou Y, Wang Q, Khalique A, et al. Isolation and preliminary screening of a Weissella confusa Strain from Giant Panda (Ailuropoda melanoleuca). Probiotics Antimicrob Proteins. (2019) 11:535–44. doi: 10.1007/s12602-018-9402-2
31. Lin MY, Chang FJ. Antioxidative effect of intestinal bacteria Bifidobacterium longum ATCC 15708 and Lactobacillus acidophilus ATCC 4356. Dig Dis Sci. (2000) 45:1617–22. doi: 10.1023/A:1005577330695
32. Alam M.d.N, Bristi NJ, Rafiquzzaman M. Review on in vivo and in vitro methods evaluation of antioxidant activity. Saudi Pharm J. (2013) 21:143–152. doi: 10.1016/j.jsps.2012.05.002
33. Liu W, Wang H, Pang X, Yao W, Gao X. Characterization and antioxidant activity of two low-molecular-weight polysaccharides purified from the fruiting bodies of Ganoderma lucidum. Int J Biol Macromol. (2010) 46:451–7. doi: 10.1016/j.ijbiomac.2010.02.006
34. Ren Z, Cui T, Zeng J, Chen L, Zhang W, Xu X, et al. Molecule targeting glucosyltransferase inhibits Streptococcus mutans biofilm formation and virulence. Antimicrob Agents Chemother. (2016) 60:126–35. doi: 10.1128/AAC.00919-15
35. Zhang Q, Xiang J, Zhang L, Zhu X, Evers J, van der Werf W, et al. Optimizing soaking and germination conditions to improve gamma-aminobutyric acid content in japonica and indica germinated brown rice. J Funct Foods. (2014) 10:283–91. doi: 10.1016/j.jff.2014.06.009
36. Wang Z, Zeng X, Mo Y, Smith K, Guo Y, Lin J. Identification and characterization of a bile salt hydrolase from Lactobacillus salivarius for development of novel alternatives to antibiotic growth promoters. Appl Environ Microbiol. (2012) 78:8795–802. doi: 10.1128/AEM.02519-12
37. Azcarate-Peril MA, Altermann E, Hoover-Fitzula RL, Cano RJ, Klaenhammer TR. Identification and inactivation of genetic loci involved with Lactobacillus acidophilus acid tolerance. Appl Environ Microbiol. (2004) 70:5315–22. doi: 10.1128/AEM.70.9.5315-5322.2004
38. Altermann E, Russell WM, Azcarate-Peril MA, Barrangou R, Buck BL, McAuliffe O, et al. Complete genome sequence of the probiotic lactic acid bacterium Lactobacillus acidophilus NCFM. Proc Natl Acad Sci USA. (2005) 102:3906–12. doi: 10.1073/pnas.0409188102
39. Navarro-Pérez ML, Fernández-Calderón MC, Vadillo-Rodríguez V. Decomposition of growth curves into growth rate and acceleration: a novel procedure to monitor bacterial growth and the time-dependent effect of antimicrobials. Appl Environ Microbiol. (2022) 88:e0184921. doi: 10.1128/aem.01849-21
40. Liu C, Han F, Cong L, Sun T, Menghe B, Liu W. Evaluation of tolerance to artificial gastroenteric juice and fermentation characteristics of Lactobacillus strains isolated from human. Food Sci Nutr. (2021) 10:227. doi: 10.1002/fsn3.2662
41. Cengiz S, Okur S, Oz C, Turgut F, Gumurcinler B, Sevuk NS, et al. Prevalence and clonal diversity of methicillin-resistant Staphylococcus aureus and methicillin-resistant Staphylococcus pseudintermedius isolated from dogs and cats with eye discharge. Acta Microbiol Immunol Hung. (2023) 70:134–41. doi: 10.1556/030.2023.01899
42. Faccin M, Wiener DJ, Rech RR, Santoro D, Rodrigues Hoffmann A. Common superficial and deep cutaneous bacterial infections in domestic animals: a review. Vet Pathol. (2023) 60:796–811. doi: 10.1177/03009858231176558
43. Viitanen SJ, Tuomisto L, Salonen N, Eskola K, Kegler K. Escherichia coli-associated follicular cystitis in dogs: clinical and pathologic characterization. J Vet Intern Med. (2023) 37:1059–66. doi: 10.1111/jvim.16719
44. Jangsangthong A, Lugsomya K, Apiratwarrasakul S, Phumthanakorn N. Distribution of sequence types and antimicrobial resistance of clinical Pseudomonas aeruginosa isolates from dogs and cats visiting a veterinary teaching hospital in Thailand. BMC Vet Res. (2024) 20:234. doi: 10.1186/s12917-024-04098-5
45. LeCuyer TE, Sellon RK, Byrne BA, Daniels JB, Diaz-Campos DV, Hendrix GK, et al. Multicenter molecular investigation of recurrent Escherichia coli bacteriuria in dogs. Vet Microbiol. (2024) 288:109914. doi: 10.1016/j.vetmic.2023.109914
46. Yildiz M, Demirbilek SK. Investigation of prevalence and antimicrobial resistance of Salmonella in pet dogs and cats in Turkey. Vet Med Sci. (2024) 10:e1513. doi: 10.1002/vms3.1513
47. Poimenidou SV, Skarveli A, Saxami G, Mitsou EK, Kotsou M, Kyriacou A. Inhibition of Listeria monocytogenes growth, adherence and invasion in Caco-2 cells by potential probiotic lactic acid bacteria isolated from fecal samples of healthy neonates. Microorganisms. (2023) 11:363. doi: 10.3390/microorganisms11020363
48. Rajab S, Tabandeh F, Shahraky MK, Alahyaribeik S. The effect of lactobacillus cell size on its probiotic characteristics. Anaerobe. (2020) 62:102103. doi: 10.1016/j.anaerobe.2019.102103
49. Wang X, Cao Z, Zhang M, Meng L, Ming Z, Liu J. Bioinspired oral delivery of gut microbiota by self-coating with biofilms. Sci Adv. (2020) 6:eabb. (1952). doi: 10.1126/sciadv.abb1952
50. Chaudhary P, Janmeda P, Docea AO, Yeskaliyeva B, Abdull Razis AF, Modu B, et al. Oxidative stress, free radicals and antioxidants: potential crosstalk in the pathophysiology of human diseases. Front Chem. (2023) 11:1158198. doi: 10.3389/fchem.2023.1158198
51. Nakagawa H, Miyazaki T. Beneficial effects of antioxidative lactic acid bacteria. AIMS Microbiol. (2017) 3:1–7. doi: 10.3934/microbiol.2017.1.1
52. Feng T, Wang J. Oxidative stress tolerance and antioxidant capacity of lactic acid bacteria as probiotic: a systematic review. Gut Microbes. (2020) 12:1801944. doi: 10.1080/19490976.2020.1801944
53. Nejati R, Nematollahi A, Doraghi HK, Sayadi M, Alipanah H. Probiotic bacteria alleviate chlorpyrifos-induced rat testicular and renal toxicity: a possible mechanism based on antioxidant and anti-inflammatory activity. Basic Clin Pharmacol Toxicol. (2023) 133:743–56. doi: 10.1111/bcpt.13945
54. Zhao T, Wang H, Liu Z, Liu Y, DeJi Li B, et al. Recent perspective of lactobacillus in reducing oxidative stress to prevent disease. Antioxidants. (2023) 12:769. doi: 10.3390/antiox12030769
55. Kuda T, Kawahara M, Nemoto M, Takahashi H, Kimura B. In vitro antioxidant and anti-inflammation properties of lactic acid bacteria isolated from fish intestines and fermented fish from the Sanriku Satoumi region in Japan. Food Res Int. (2014) 64:248–55. doi: 10.1016/j.foodres.2014.06.028
56. Vinothkanna A, Sathiyanarayanan G, Rai AK, Mathivanan K, Saravanan K, Sudharsan K, et al. Exopolysaccharide produced by probiotic Bacillus albus DM-15 isolated from ayurvedic fermented dasamoolarishta: characterization, antioxidant, and anticancer activities. Front Microbiol. (2022) 13:832109. doi: 10.3389/fmicb.2022.832109
57. Hamet MF, Piermaria JA, Abraham AG. Selection of EPS-producing Lactobacillus strains isolated from kefir grains and rheological characterization of the fermented milks. LWT Food Sci Technol. (2015) 63:129–35. doi: 10.1016/j.lwt.2015.03.097
58. Dicks LMT. Gut bacteria and neurotransmitters. Microorganisms. (2022) 10:1838. doi: 10.3390/microorganisms10091838
59. Bravo JA, Forsythe P, Chew MV, Escaravage E, Savignac HM, Dinan TG, et al. Ingestion of Lactobacillus strain regulates emotional behavior and central GABA receptor expression in a mouse via the vagus nerve. Proc Natl Acad Sci USA. (2011) 108:16050–5. doi: 10.1073/pnas.1102999108
60. Li Y, Chen X, Shu G, Ma W. Screening of gamma-aminobutyric acid-producing lactic acid bacteria and its application in Monascus-fermented rice production. Acta Sci Pol Technol Aliment. (2020) 19:387–94. doi: 10.17306/J.AFS.2020.0868
61. Bourgin M, Kriaa A, Mkaouar H, Mariaule V, Jablaoui A, Maguin E, et al. Bile salt hydrolases: at the crossroads of microbiota and human health. Microorganisms. (2021) 9:1122. doi: 10.3390/microorganisms9061122
62. Pinto A, Barbosa J, Albano H, Isidro J, Teixeira P. Screening of bacteriocinogenic lactic acid bacteria and their characterization as potential probiotics. Microorganisms. (2020) 8:393. doi: 10.3390/microorganisms8030393
63. Tsai C-C, Lin P-P, Hsieh Y-M, Zhang Z, Wu H-C, Huang C-C. Cholesterol-lowering potentials of lactic acid bacteria based on bile-salt hydrolase activity and effect of potent strains on cholesterol metabolism in vitro and in vivo. ScientificWorldJournal. (2014) 2014:690752. doi: 10.1155/2014/690752
64. Kim K-T, Kim J-W, Kim S-I, Kim S, Nguyen TH, Kang C-H. Antioxidant and anti-inflammatory effect and probiotic properties of lactic acid bacteria isolated from canine and feline feces. Microorganisms. (2021) 9:1971. doi: 10.3390/microorganisms9091971
65. Biosafe. Antimicrobial Susceptibility Testing. MIC Testing (n.d.). Available at: https://www.biosafe.fi/antimicrobial-susceptibility-testing-mic-testing (accessed January 6, 2024).
Keywords: lactic acid bacteria, canine milk, adhesion, antioxidant capacity, metabolite
Citation: Liu Y, Zeng Y, Chen L, Xin J, Zhong Z, Liu H, Fu H, Zhou Z and Peng G (2024) Isolation and evaluation of multi-functional properties of lactic acid bacteria strains derived from canine milk. Front. Vet. Sci. 11:1505854. doi: 10.3389/fvets.2024.1505854
Received: 03 October 2024; Accepted: 08 November 2024;
Published: 28 November 2024.
Edited by:
Gulbeena Saleem, University of Veterinary and Animal Sciences, PakistanReviewed by:
Sadia Chaman, University of Veterinary and Animal Sciences, PakistanBushra Nisar Khan, University of the Punjab, Pakistan
Copyright © 2024 Liu, Zeng, Chen, Xin, Zhong, Liu, Fu, Zhou and Peng. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Guangneng Peng, cGduLnNpY2F1QDE2My5jb20=
†These authors have contributed equally to this work and share first authorship
 Yunjiang Liu
Yunjiang Liu Yueyan Zeng1†
Yueyan Zeng1† Zhijun Zhong
Zhijun Zhong Ziyao Zhou
Ziyao Zhou Guangneng Peng
Guangneng Peng