- Key Laboratory of Animal Production, Product Quality and Security, Ministry of Education, Jilin Province Key Laboratory of Animal Nutrition and Feed Science, College of Animal Science and Technology, Jilin Agricultural University, Changchun, China
Introduction: The study evaluated the enzymatic hydrolysis processing on physicochemical properties and protein digestive dynamics of soybean meal (SBM), as well as the relationship between protein secondary structure and digestive parameters was established.
Methods: Scanning Electron Microscopy (SEM) and Fourier Transform Infrared Spectroscopy (FTIR) were employed to analyze the microstructure and protein structure of the SBM and enzymatic hydrolysis processed soybean meal (ESBM). SBM and ESBM were incubated with pepsin at pH 3.5 and 39°C for 30 min, then with pancreatin at pH 6.8 for 0–240 min. The in vitro protein digestive dynamics were described as the release dynamics of amino acids and low molecular weight peptides (AA_LMW).
Results: The results showed that enzymatic hydrolysis processing did not alter the chemical composition of SBM, but changed its microstructure and protein structure. After enzymatic hydrolysis processing, the size of blocky structures of SBM decreased, exhibiting a fibrous surface and a relatively loose internal structure. The β-sheet content of ESBM was lower than that of SBM (p < 0.05), while the α-helix, β-turn, and α-helix/β-sheet content was higher than that of SBM (p < 0.05). The release rates (k) of AA_LMW in SBM and ESBM were 0.0123 min−1 and 0.0733 min−1, respectively. Enzymatic hydrolysis processing increased the CPfast content of SBM (p < 0.05) and decreased the CPslow and CPresistant contents (p < 0.05). α-helix, β-turn, and the ratio of α-helix to β-sheet were positively correlated with CPfast and k (p < 0.05) and were negatively correlated with CPslow and CPresistant (p < 0.05). β-sheet was negatively correlated with CPfast and k (p < 0.05) and was positively correlated with CPslow and CPresistant (p < 0.05).
Discussion: Enzymatic hydrolysis processing altered the digestive dynamics of SBM, increased the CPfast content and the release rate of AA_LMW, which might be attributed to the structure changes of SBM.
1 Introduction
Dietary protein plays a critical role in animal production as a major feed component. In current feed evaluation systems, the nutritional value of protein ingredients in diets for pigs is typically evaluated based on the content of essential amino acids and their ileal digestibility (1, 2). Considering factors such as convenience and cost, researchers have also explored in vitro digestion methods to simulate and predict the ileal digestibility of amino acids and proteins in pigs, providing a rapid evaluation of the value of protein sources (3). However, these data do not explain the dynamic process of protein digestion along the gastrointestinal tract.
Protein sources with similar digestibility may exhibit distinct protein digestive dynamics, thereby affecting the postprandial absorption and metabolism of the end products of protein digestion. For instance, although both casein and whey proteins are considered highly digestible in humans (3), the rate and extent of the increase in amino acids and peptides in plasma exhibited significant differences (4). The casein group showed a slow and sustained postprandial increase of amino acids and peptides. In contrast, the whey protein group showed a rapid but transient postprandial increase of amino acids and peptides in plasma. Based on the rate and the extent of the postprandial increase of plasma AAs and peptides, Boirie et al. (4) categorized protein sources into fast protein and slow protein. Recently, Mai Anh Ton Nu et al. (5) further categorized protein sources into fast, slow, and resistant proteins based on the extent of in vitro protein digestion over different periods. The rate of protein digestion affects the deposition of body protein (4, 6). The slow protein can reduce body weight loss and conserve mobilization of body protein in sows (7), increase litter weight gain in piglets, and improve protein efficiency during lactation (8). Additionally, the synchrony of starch and protein digestion also affects energy and protein utilization efficiency in pigs (9). The protein digestive dynamics can be affected by the chemical composition, the protein classification, and the physicochemical properties of the protein sources (10). Different physical, chemical, and enzymatic methods can be used to modify the structure and physicochemical properties of protein sources.
Soybean meal (SBM) is the most common protein source in pig diets (11). However, SBM, a byproduct of oil extraction from soybean, also contains residual anti-nutritional factors that limit its use in piglet diets (12–15). To improve the utilization efficiency of SBM in pigs and reduce the use limitations, various processing methods such as extrusion (16), enzymatic hydrolysis (17), and fermentation (18) have been studied. These processing methods alter the physical structure of SBM, reduce their anti-nutritional factor content, and increase protein utilization efficiency in pigs. Additionally, the reduction of undigested protein entering the hindgut helps regulate hindgut pH and improve gut health (19, 20). The structure of protein influences the accessibility of enzymes for protein digestion. Some commonly used feed ingredients in pig diets exhibit structural constraints in their natural protein composition (21). Some studies have found that the secondary structure of proteins can influence both the rate and extent of protein digestion. Protein sources with a high proportion of α-helix or a high ratio of α-helix to β-sheets structures tend to have faster digestion rates and higher digestibility within the same timeframe (22–24).
Understanding the changes in structure and protein digestive dynamics of protein sources after processing can guide the optimal processing of protein sources, thereby further improving the utilization efficiency of protein by pigs. These could provide important references for evaluating the nutritional value of ingredients and their application in pig production. Therefore, we selected SBM as the research subjects in the present study, which aimed to investigate how enzymatic hydrolysis processing affects its physicochemical properties and the protein digestive dynamics represented by the release dynamics of amino acids and low molecular weight peptides (AA_LMW). Furthermore, a correlation between dynamic parameters and the protein secondary structure was established. We hypothesized that enzymatic hydrolysis processing alters the microstructure and molecular structure of the SBM, thereby affecting the protein digestive dynamics of SBM.
2 Materials and methods
2.1 Materials
SBM and enzymatic hydrolysis processed soybean meal (ESBM; K-Protein) were provided by Liaoning Complete Biotechnology Co. (Liaoning, China). The ESBM was produced using a combination of asynchronous enzymatic hydrolysis methods. Briefly, enzymatic hydrolysis processing steps were as follows: (1) SBM was ground and mixed using water to 50–60% moisture content, including an enzyme mixture (Proteases, Liaoning Province Feed Pre-digested Technical Innovation Center, Liaoning, China) as a processing aid; (2) SBM underwent enzymatic hydrolysis in the enzymatic hydrolysis tank at approximately 50°C for 6 h. No other method was used to control the pH, and the pH value range remained within 5.5–6.5 during the processing; (3) then the enzyme mixture was inactivated at temperature ≥95°C for 10–15 min; (4) drying and cooling to stabilize the product with moisture ≤12%. Three samples were randomly collected from SBM and ESBM and ground by a hammer mill through a 1 mm sieve for subsequent analysis with three replicates per analysis. Sub-samples were collected and stored at 4°C. To eliminate the interference of sample sources on the test results, the SBM before and after the enzymatic hydrolysis treatment were the same source and the same batch. Meanwhile, to improve the representativeness of the samples, the newly produced SBM, purchased in bulk by typical enzymatic hydrolysis processing enterprises, was chosen as the research object.
2.2 Scanning electron microscopy (SEM)
A SEM (ZEISS GeminiSEM500, Oberkochen, Germany) was employed to observe the microstructure of SBM and ESBM samples at an acceleration voltage of 2 kV. Before SEM, the samples were glued to the sample loading table, and gold was sprayed on the surface. The software Image J (Image J 1.53e, National Institutes of Health, USA) was employed to analyze the particle area of blocky structures.
2.3 Fourier transform infrared spectroscopy (FTIR)
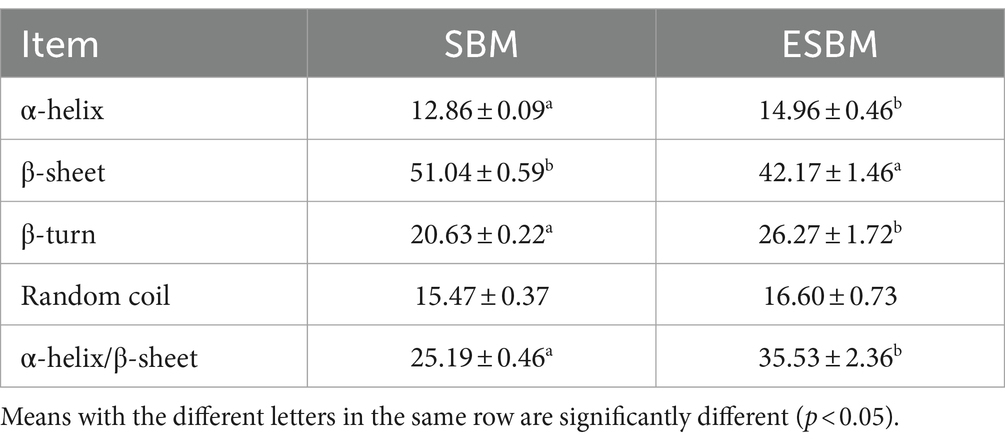
The spectral data of samples were obtained using the FTIR spectroscopy (ALPHA II, Bruker, Germany). Raw spectra were recorded in the 4,000–400 cm−1 spectral range with 32 co-added scans at a resolution of 4 cm−1. Each sample was analyzed in triplicate. The software OMNIC (OMNIC 8.2, Thermo Nicolet Corp., Madison, USA) was employed to analyze the spectral data. The software Peakfit (Systat PeakFit 4.12, SeaSolve Software Inc., CA, USA) was utilized to correct and analyze each spectrum and to determine the relative content of the protein secondary structures as indicated by the following bands: α-helix (1,650–1,660 cm−1), β-sheet (1,600–1,640 cm−1), β-turn (1,660–1,670 cm−1), and random coil (1,640–1,650 cm−1) (25).
2.4 In vitro protein digestive dynamics
The in vitro protein digestion method was modified based on the method of Boisen and Fernandez (3, 10). For the simulation of protein digestion in the stomach, 1.0 g samples and five 6 mm diameter glass beads were placed into a 50 mL plastic centrifuge tube, with three replicates for each sample at each time point. Then, 10 mL of porcine pepsin solution (1 mg/mL P7000 Sigma) was added. The pH was adjusted to 3.5 with 1 M HCl or 1 M NaOH, and each centrifuge tube was placed in the Electric Water Bath Shaker at 39°C under continuous stirring. The incubation time with pepsin was 30 min. After incubating with pepsin for 30 min, adjust the solution to pH 6.8 using 0.2 M NaOH. The protein digestion in the small intestine was simulated by adding 10 mL of porcine pancreatin phosphate buffer solution (pH 6.8, 5 mg/mL, Sigma P7545). The incubation with pancreatin was continued in the Electric Water Bath Shaker at 39°C under continuous stirring. The incubation times with pancreatin were 0, 15, 30, 60, 90, 120, 180, and 240 min, respectively.
At each time point, the centrifuge tubes with samples were taken out and immediately placed in a −20°C refrigerator for 20 min to cool, and then centrifuged (10 min, 8,000 rpm, 4°C) to separate the insoluble protein fraction (IPF) and the soluble protein fraction (SPF). After centrifugation, the soluble fraction (supernatant) was transferred to a 50 mL volumetric flask and diluted with de-mineralized water to 50 mL. 10 mL of the diluted soluble fraction were retained for nitrogen (N) content analysis to calculate the N solubility at each digestion time point. 8 mL of the diluted soluble fraction was taken and mixed with 2 mL of 20% sulfosalicylic acid. Invert to mix thoroughly, then centrifuge (10 min, 8,000 rpm, 4°C) to separate amino acids and soluble low molecular weight peptides (AA_LMW) and soluble high molecular weight peptides (HMW). The supernatant was retained for N content analysis to calculate the AA_LMW.
2.5 Chemical analysis
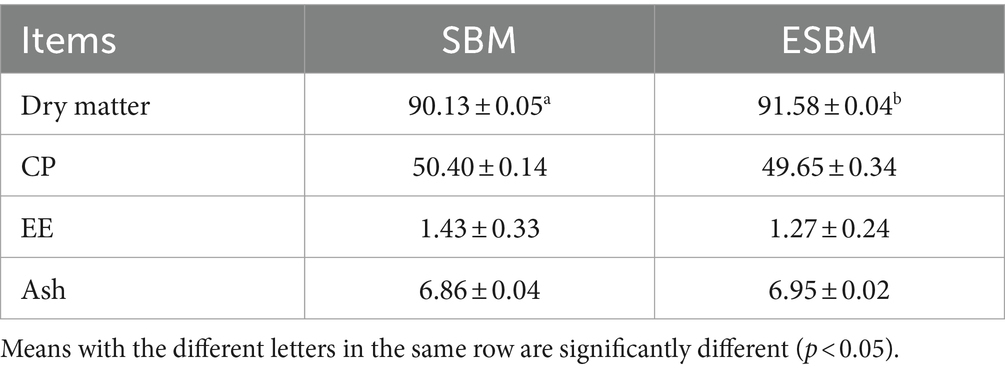
All chemical analyses were conducted following standard laboratory methods. Determinations included analysis of dry matter (DM) (26), crude protein (CP) (27), ether extract (EE) (28), and ash (29).
2.6 Calculation
The N Solubility was calculated by equation 1:
Where Nsample (mg) is the amount of N in 1 g of sample, and NSPF (mg) is the amount of N in the SPF during the in vitro digestion with pepsin and pancreatin.
The N content in AA_LMW was calculated by equation 2:
Where Nsample (mg) is the amount of N in 1 g of sample, and NAA_LMW is the amount of N in the AA_LMW during the in vitro digestion with pepsin and pancreatin.
The N content in HMW was calculated by equation 3:
Where Nsample (mg) is the amount of N in 1 g of sample, NSPF (mg) is the amount of N in the SPF during the in vitro digestion with pepsin and pancreatin, and NAA_LMW is the amount of N in the AA_LMW during the in vitro digestion with pepsin and pancreatin.
The N content in IPF was calculated by equation 4:
Where Nsample (mg) is the amount of N in 1 g of sample, and NSPF (mg) is the amount of N in the SPF during the in vitro digestion with pepsin and pancreatin.
The dynamics of AA_LMW for SBM and ESBM during the incubations were described by an exponential equation 5 (30):
Where Dt (%) is the N content in AA_LMW at incubation time t (min), D0 (%) is the N content in the AA_LMW at 0 min after adding pancreatin, ΔD (%) is the maximum N content in the AA_LMW (asymptotic line), and k is the rate constant.
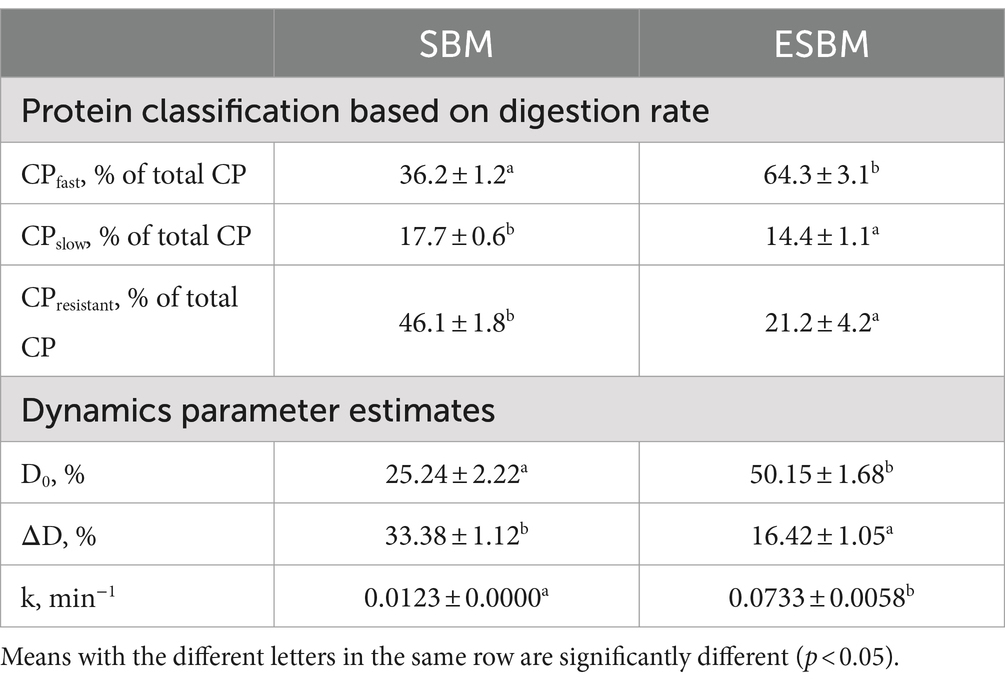
Based on the N content of AA_LMW during in vitro digestion, the protein fractions of ingredients were classified into fast protein (CPfast), slow protein (CPslow), and resistant protein (CPresistance), where CPfast and CPslow correspond to the amount of CP digested within the first 30 min and between 30 and 240 min, respectively, and CPresistance = 100 − CPfast − CPslow.
2.7 Statistical analysis
Statistical analysis was performed using SPSS statistical software (SPSS 26.0, IBM Corp., Armonk, NY, USA), with the Independent-Samples T-test used to analyze the data presented as mean ± standard deviation. Correlation analysis was used to analyze the relationship between protein secondary structure, protein classification, and in vitro protein digestive dynamics parameters. Heat graphs were generated using Origin software (Origin 2022, OriginLab Corp., Northampton, MA, USA). The level of significance was determined as p < 0.05.
3 Results
3.1 Chemical composition
The chemical composition of SBM and ESBM are presented in Table 1. The DM content of ESBM was significantly higher than that of SBM (p < 0.05), while the CP, EE, and ash contents (DM basis) did not exhibit significant differences between SBM and ESBM (p > 0.05).
3.2 Microstructural characterization
The SEM images of SBM and ESBM are presented in Figure 1. The internal structure of SBM was relatively compact, primarily comprising spherical, large blocky, and flaky structures. Following enzymatic hydrolysis processing, the morphological structure of SBM changed, and the blocky structures decreased in size (Particle area μm2: SBM 236.0 ± 137.9b; ESBM 103.6 ± 81.0a; p < 0.05), exhibiting a fibrous surface and a relatively loose internal structure.
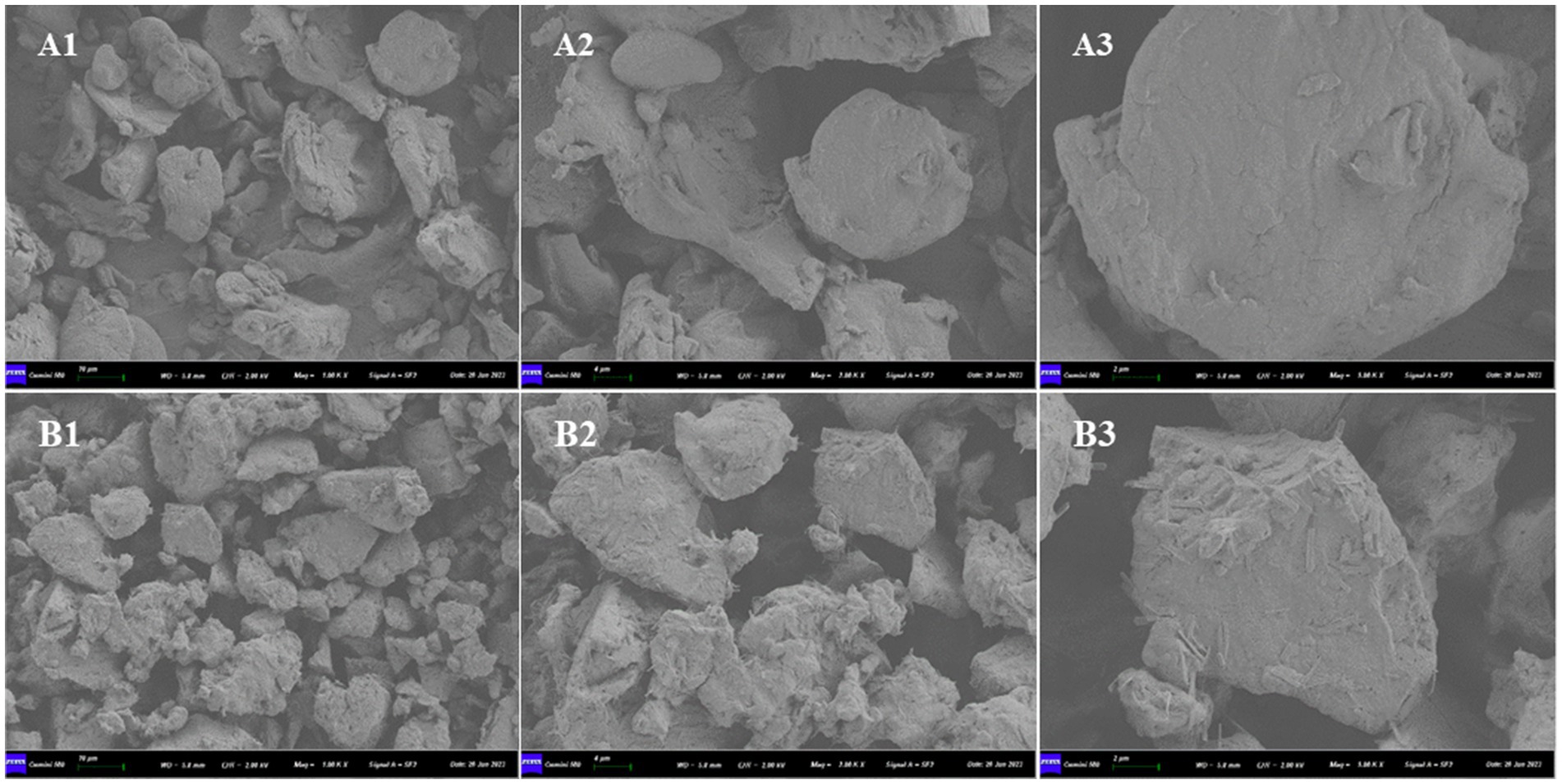
Figure 1. Scanning electron microscopy: (A) SBM and (B) ESBM, (1) magnification: 1,000×, (2) magnification: 2,000×, (3) magnification: 5,000×.
3.3 Infrared spectra of protein sources
The FTIR absorption spectra of the SBM and ESBM are presented in Figure 2A. After the enzymatic hydrolysis processing, the position of the Amide I band peaks (1,700–1,600 cm−1) in ESBM shifted toward shorter wavenumbers, and the intensity of the absorption peaks of the Amide I and II bands in ESBM decreased. The analysis results are shown in Table 2 and Figures 2B,C. The β-sheet content of ESBM was significantly lower than that of SBM (p < 0.05), whereas the α-helix, β-turn, or the ratio of α-helix to β-sheet content of ESBM was significantly higher than that of SBM (p < 0.05).
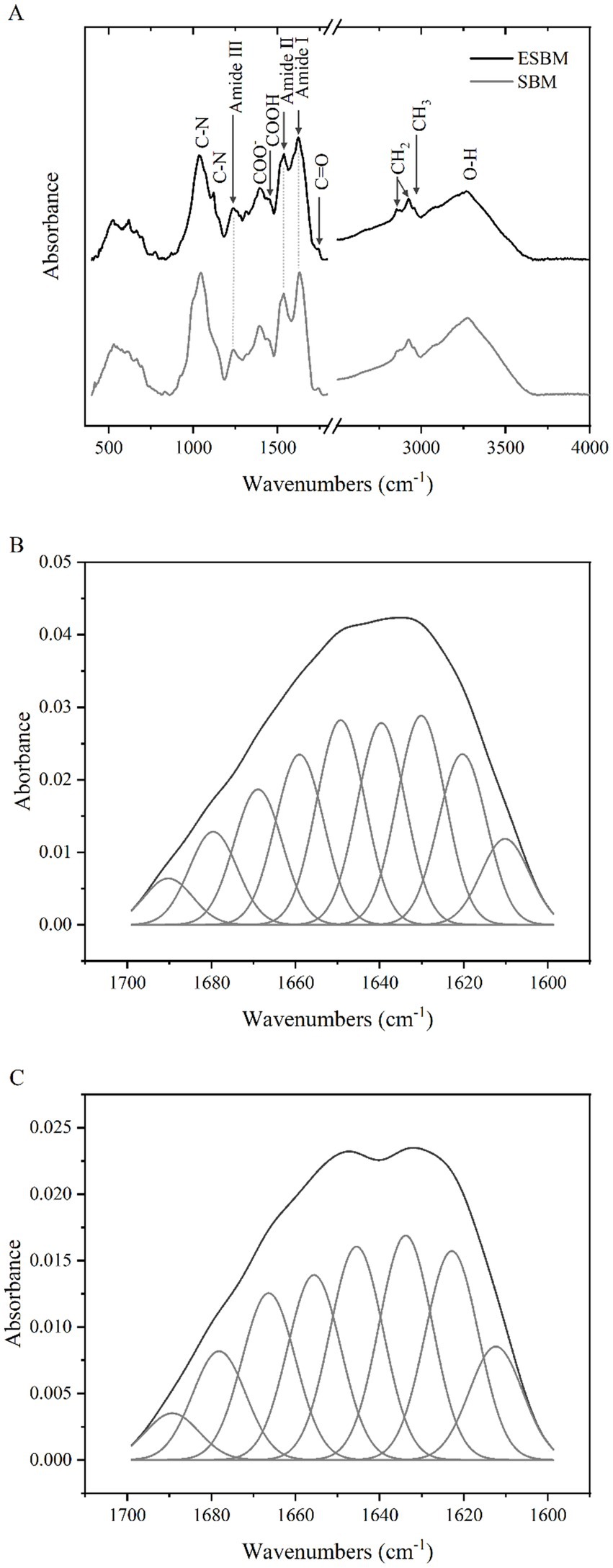
Figure 2. FTIR absorption spectra of SBM and ESBM (A), Curve fitting of amide I band spectra of SBM (B), Curve fitting of amide I band spectra of ESBM (C).
3.4 Separation of soluble N into different molecular weight fractions
Table 3 presents the distribution of N into different molecular weight fractions. During in vitro digestion at 0, 15, 30, 120, 180, and 240 min, no significant differences were observed in N solubility between SBM and ESBM (p > 0.05). At 60 and 90 min of in vitro digestion, the N solubility of ESBM was, respectively, 9.8 and 8.1% higher than that of SBM (p < 0.05). Notably throughout all stages of in vitro protein digestion from 0 to 240 min, the N present in the AA_LMW of ESBM was significantly higher than that in SBM (p < 0.05), while the N present in the HMW was significantly lower compared to SBM (p < 0.05). During this process, the N solubility of SBM increased from an average of approximately 47% to about 80% within the first 30 min. At the end of pancreatin incubation (240 min), the N solubility of SBM reached about 82%, with about 54% attributed to AA_LMW and about 28% to HMW fractions, respectively. Similarly, for ESBM during in vitro digestion, N solubility rose from an average initial value of approximately 52% to about 81% within the first 30 min. At the end of pancreatin incubation (240 min), the ESBM showed approximately 90% N solubility, of which about 79% was present in AA_LMW and about 11% in HMW. Both SBM and ESBM exhibited a higher proportion of N present in the AA_LMW compared to the HMW fraction at the end of pancreatin incubation.
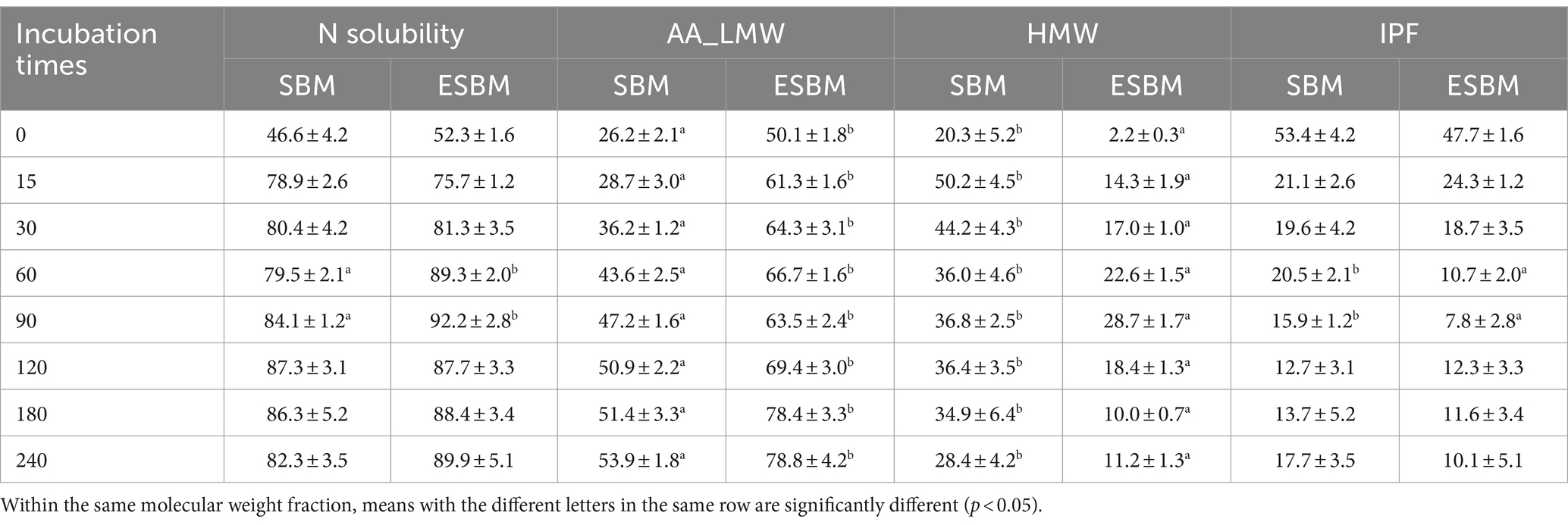
Table 3. N solubility and N present in the AA_LMW, HMW, and IPF as a proportion (%) of total N of SBM and ESBM at different time points during the sequential incubation with pancreatin.
Enzymatic hydrolysis processing correspondingly resulted in a significant increase in the CPfast content of SBM (p < 0.05), while concurrently leading to a significant reduction in both the contents of CPslow and CPresistant (p < 0.05) (Table 4).
3.5 Release dynamics of the N present in the AA_LMW
The N content in the AA_LMW fraction increased rapidly during the initial 120 min of in vitro digestion. Subsequently, the release rate relatively stabilized. Therefore, the N release dynamics parameters of AA_LMW were calculated based on data from the first 120 min. The initial N present in the AA_LMW D0 and the release rate k were significantly higher in ESBM compared to SBM (p < 0.05) (Table 4). During the incubation with pancreatin, SBM and ESBM had the mean N release rates k in AA_LMW of 0.0123 min−1 and 0.0733 min−1, with D0 of 25.24 and 50.15%, and ΔD of 33.38 and 16.42%, respectively (Table 4).
3.6 Correlation between protein secondary structures and protein classification as well as in vitro protein digestive dynamics
The results of correlation analysis between the content of protein secondary structure, protein classification, and in vitro protein digestive dynamics were presented in Figure 3. The content of α-helix was positively correlated with CPfast, D0, and k (p < 0.05), while negatively correlated with CPslow, CPresistant, and ΔD (p < 0.05). Conversely, the β-sheet showed a significant negative correlation with CPfast, D0, and k (p < 0.05) but a positive correlation with CPslow, CPresistant, and ΔD (p < 0.05). The β-turn exhibited a significant positive correlation with CPfast, D0, and k (p < 0.05) while exhibiting a negative correlation with CPslow, CPresistant, and ΔD (p < 0.05). Random coil was significantly negatively correlated with ΔD (p < 0.05), while correlations with other components were not significant. Additionally, the ratio of α-helix to β-sheet was significantly positively correlated with CPfast, D0, and k (p < 0.05), and negatively correlated with CPslow, CPresistant, and ΔD (p < 0.05).
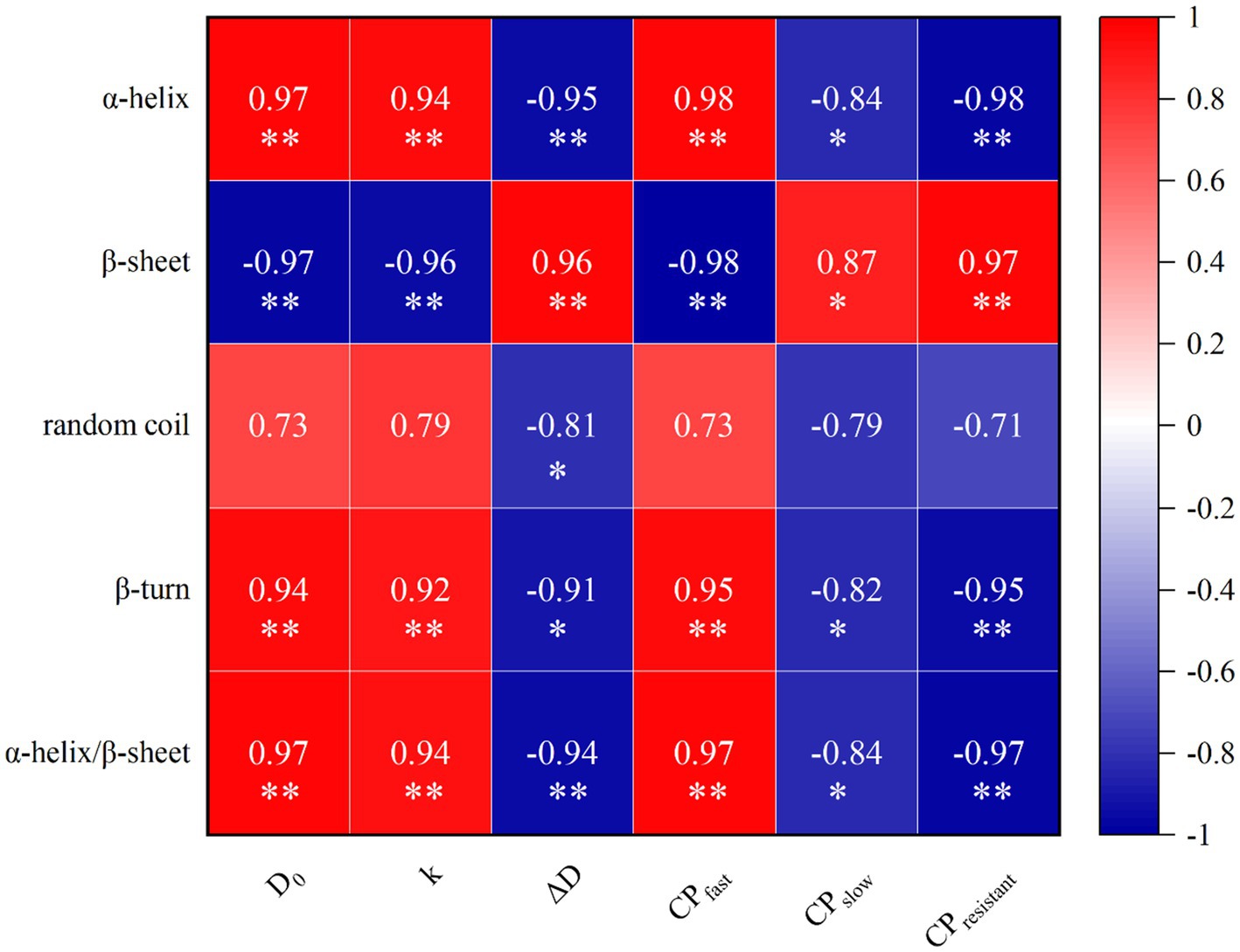
Figure 3. Correlation of protein secondary structure, protein classification and dynamic parameters of protein digestion in vitro. Heatmaps depict Pearson’s correlation coefficients. Deep red and dark blue represent stronger correlation coefficients and p values. Light red and light blue represent weaker correlation coefficients and p values. *p < 0.05; **p < 0.01.
4 Discussion
Enzymatic hydrolysis processing did not alter the CP, EE, and Ash content of SBM. The variation in the DM content of SBM before and after enzymatic hydrolysis processing could probably be attributed to the drying process that follows wet enzymatic hydrolysis. The microstructure of ingredients significantly impacts their digestibility. Therefore, SEM was employed to examine how the microstructure of SBM changed after enzymatic hydrolysis. Compared to SBM, ESBM exhibited a fibrous surface and a relatively loose internal structure. This might increase the effective contact surface area of ESBM with enzymes during digestion, thereby accelerating hydrolysis. Studies have shown that the extrusion-enzyme hydrolysis treatment significantly altered the conformational and functional properties of soybean protein (31).
The present study investigated the molecular spectral band characteristics of proteins. As shown in Figure 2A, the peaks observed at 3,266–3,282 cm−1 correspond to the stretching vibrations of water molecules (O–H) in the SBM and ESBM, indicating a reduction in moisture content in SBM after the enzymatic hydrolysis processing. Amide I, II, and III bands are typically assigned to peaks located at 1,700–1,610 cm−1, 1,600–1,500 cm−1, and 1,330–1,220 cm−1, respectively (32). The position of the Amide I band peaks (1,700–1,600 cm−1) changed, indicating changes in the protein molecular structure of SBM after the enzymatic hydrolysis processing. The peaks observed at 1,623.05–1,631.20 cm−1 are mainly attributed to the stretching vibrations of the C=O group in the Amide I band. The peaks observed at 1,537.20–1,538.25 cm−1 are mainly attributed to the stretching vibrations of the C–N group in the Amide II band. The peaks observed at 1,236.39–1,238.26 cm−1 are mainly attributed to the stretching vibrations of the C–N group in the Amide III band. We focused on the protein regions to evaluate the effect of the enzymatic hydrolysis processing on the proteins in SBM. The amide I band is most frequently used for conformational analysis (33). In the present study, after enzymatic hydrolysis of SBM, the α-helix, β-turn, and the ratio of α-helix to β-sheet content were significantly higher and β-sheet content was significantly lower than before. This indicates that the enzymatic processing results in conversion between different protein secondary structures in SBM. It has been reported that fermentation treatment can also cause changes in the protein secondary structure of SBM (34). Different fermentation methods have different effects on the protein secondary structure in corn-soybean meal diets (25).
The in vitro method is an effective approach to evaluate the protein quality of ingredients. It has broad application prospects in evaluating the nutritional value of feed. However, there are few studies on the dynamics of in vitro protein digestion in pigs and poultry (5, 10, 22, 24, 35, 36). Typically, in vitro protein digestion methods were according to a two-step method described by Boisen and Fernández (3) or with modification (3, 10, 36). Nevertheless, there is still no standardized and unified in vitro protein digestion method. For example, the incubation time and pH value in the gastric phase vary significantly in different trials (3, 10). It has been reported that low pH (pH 2.0) during in vitro gastric digestion may lead to overestimation of the protein digestibility (37), and that pH 3.5 is more likely to mimic the internal environment of the porcine stomach than pH 2.0 (38). Therefore, we used a pH of 3.5 in this trial. Enzymatic hydrolysis processing changed SBM’s protein digestibility and affected N distribution in the different fractions during digestion. Chen et al. (10) observed that N solubility rapidly increased from 47–52% to 80–85% in the first 30 min of the small intestine phase during in vitro protein digestion of SBM, and eventually (210 min) reached 91–93%, which is similar to the results in the present study (47, 80, and 82%, respectively). The N present in the AA_LMW of SBM increased from 9–11% to 22–30%, and eventually (210 min) reached 32–44% (10), it was much lower than in vitro results in the present study (26, 36, and 54%, respectively). The gaps may be related to the different sources and processing of SBM. In the present study, the changes in N solubility during in vitro protein digestion of ESBM were not significant with SBM (except for 60 and 90 min), which is different from the report by Mai Anh Ton et al. (5), probably due to differences in the source and production process of SBM. However, the N present in the AA_LMW of ESBM during in vitro digestion was higher than that of SBM at all time points (17–32% higher). This indicates that the increase in the N present in the AA_LMW of ESBM during in vitro digestion was mainly due to the hydrolysis of the HMW fraction and not related to changes in the N solubility rate.
During in vitro protein digestion, the N content in the HMW fraction of ESBM was lower than that of SBM at all time points. It may be attributed to the internal hydrogen bonds of the protein being disrupted after SBM processing, and the protein was depolymerized to low molecular weight peptides (17, 31). The sequential hydrolysis of intermediate peptides is considered to be the rate-limiting step in protein digestion and absorption during protein digestion in animals (39). We may be able to consider the N present in the HMW as intermediate peptides produced during protein digestion in vitro to guide animal production. Studies have shown that free amino acid and small peptide content of SBM increased, antigen degraded, and protein digestibility increased after treatments such as extrusion, enzymatic hydrolysis, and fermentation (16–18, 40).
Enzymatic hydrolysis processing also led to changes in the contents of CPfast, CPslow, and CPresistant in SBM and ESBM. Some studies have found that slow protein can reduce body weight loss and conserve mobilization of body protein in sows (7), increase litter weight gain in piglets, and improve protein efficiency during lactation (8). Combined with the results from SEM and FTIR, the internal hydrogen bonds of proteins in SBM might be disrupted and the structure altered after enzymatic hydrolysis processing. The increased N release rate and content of AA_LMW during digestion may be related to the structural changes. In animals, proteins need to be hydrolyzed into free amino acids or di-and tri-peptides before they can be transported into enterocytes in the small intestine mucosa for subsequent utilization (41). Therefore, compared with N solubility, the release dynamics of N in the AA_LMW can better reflect the in vivo release dynamics of amino acids, di-and tri-peptides from ingredients (42). The dynamic pattern of dietary glucose release could affect the contents and patterns of portal amino acids, thereby enhancing the utilization efficiency of dietary nitrogen (43). Therefore, the release dynamics of N in the AA_LMW can be used to further develop the concept of synchronization release among glucose and AA_LMW, which is likely to improve the utilization efficiency of protein in animals.
The protein secondary structure can partly predict changes in protein classification and in vitro protein digestive dynamics. In this study, the content of α-helix or β-turn or the ratio of α-helix to β-sheet was positively correlated with CPfast, D0, and k. The content of β-sheet was positively correlated with CPslow, CPresistant, and ΔD. The nutritional value of proteins is closely related to their secondary structure (44). Some studies similar to current results that the content of α-helix or the ratio of α-helix to β-sheet were positively correlated with protein digestibility and amino acid release coefficients, while β-sheets showed a negative correlation with these factors (22, 24). Possibly because β-sheet is rich in hydrogen bonds, which can prevent the digestive enzymatic activities of proteins (22).
5 Conclusion
SBM underwent enzymatic hydrolysis processing, resulting in a decrease in the content of β-sheet and an increase in the content of α-helix or β-turn or the ratio of α-helix to β-sheet. Simultaneously, the size of the blocky structure of ESBM became smaller than that of SBM, exhibiting a fibrous surface and a relatively loose internal structure. These alterations might enhance the effective contact surface area of ESBM with enzymes during digestion, thereby accelerating hydrolysis and resulting in a higher CPfast content and release rate of AA_LMW in ESBM compared with SBM. These results provide valuable insights for optimizing the processing methods of protein sources to improve the utilization efficiency of protein in pig feed.
Data availability statement
The original contributions presented in the study are included in the article/supplementary material, further inquiries can be directed to the corresponding authors.
Author contributions
DW: Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Writing – original draft. HD: Conceptualization, Writing – review & editing. XD: Conceptualization, Writing – review & editing. YZ: Data curation, Methodology, Writing – review & editing. JZ: Conceptualization, Investigation, Writing – review & editing. RL: Conceptualization, Investigation, Writing – review & editing. ZG: Conceptualization, Investigation, Writing – review & editing. QZ: Conceptualization, Formal analysis, Supervision, Writing – review & editing. ZS: Conceptualization, Funding acquisition, Resources, Supervision, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This research was supported by the National Natural Science Foundation of China (Grant No. 32394043), the National Key Research and Development Program of China (2021YFD1300201) and Jilin Province Key Research and Development Program of China (20220202044NC).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The authors declare that no Gen AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. NRC. Nutrient requirements of swine. Eleventh Revised ed. Washington, DC, USA: National Academy Press (2012).
2. CVB. Chemical composition and nutritional values of feedstuffs. Wageningen, The Netherlands: CVB (2021).
3. Boisen, S, and Fernandez, JA. Prediction of the apparent ileal digestibility of protein and amino acids in feedstuffs and feed mixtures for pigs by in vitro analyses. Anim Feed Sci Tech. (1995) 51:29–43. doi: 10.1016/0377-8401(94)00686-4
4. Boirie, Y, Dangin, M, Gachon, P, Vasson, MP, Maubois, JL, and Beaufrère, B. Slow and fast dietary proteins differently modulate postprandial protein accretion. P Natl Acad Sci USA. (1997) 94:14930–5. doi: 10.1073/pnas.94.26.14930
5. Anh, TNM, Ingrid, L, Lupatsch, I, Zannatta, JS, Schulze, H, and Zijlstra, RT. Thermo-mechanical and enzyme-facilitated processing of soybean meal enhanced in vitro kinetics of protein digestion and protein and amino acid digestibility in weaned pigs. J Anim Sci. (2020) 98:skaa224. doi: 10.1093/jas/skaa224
6. Dangin, M, Boirie, Y, Garcia-Rodenas, C, Gachon, P, Fauquant, J, Callier, P, et al. The digestion rate of protein is an independent regulating factor of postprandial protein retention. Am J Physiol Endoc M. (2001) 280:E340–8. doi: 10.1152/ajpendo.2001.280.2.E340
7. Ye, H, Langendijk, P, Jaworski, NW, Wu, Y, Bai, Y, Lu, D, et al. Protein digestion kinetics influence maternal protein loss, litter growth, and nitrogen utilization in lactating sows. Front Nutr. (2022) 9:9. doi: 10.3389/fnut.2022.862823
8. Ye, H, Soede, NM, Kemp, B, Wang, J, Jaworski, NW, and Langendijk, P. Dietary Cp and digestion kinetics influence Bw loss, litter weight gain, and reproduction by affecting postprandial amino acid metabolism in lactating sows. Animal. (2024) 18:101184. doi: 10.1016/j.animal.2024.101184
9. van den Borne, JJ, Schrama, JW, Heetkamp, MJ, Verstegen, MW, and Gerrits, WJ. Synchronising the availability of amino acids and glucose increases protein retention in pigs. Animal. (2007) 1:666–74. doi: 10.1017/s1751731107736741
10. Chen, H, Wierenga, PA, Hendriks, WH, and Jansman, AJM. In vitro protein digestion kinetics of protein sources for pigs. Animal. (2019) 13:1154–64. doi: 10.1017/S1751731118002811
11. Jezierny, D, Mosenthin, R, and Bauer, E. The use of grain legumes as a protein source in pig nutrition: a review. Anim Feed Sci Tech. (2010) 157:111–28. doi: 10.1016/j.anifeedsci.2010.03.001
12. Mukherjee, R, Chakraborty, R, and Dutta, A. Role of fermentation in improving nutritional quality of soybean meal — a review. Asian Austral J Anim. (2016) 29:1523–9. doi: 10.5713/ajas.15.0627
13. Zheng, L, Li, D, Li, ZL, Kang, LN, Jiang, YY, Liu, XY, et al. Effects of Bacillus fermentation on the protein microstructure and anti-nutritional factors of soybean meal. Lett Appl Microbiol. (2017) 65:520–6. doi: 10.1111/lam.12806
14. Yuan, L, Chang, J, Yin, Q, Lu, M, Di, Y, Wang, P, et al. Fermented soybean meal improves the growth performance, nutrient digestibility, and microbial flora in piglets. Anim Nutr. (2017) 3:19–24. doi: 10.1016/j.aninu.2016.11.003
15. Garcia, RM, Arruda, G, Chaves, R, Ribeiro, C, Mendes, M, and Cantarelli, VS. Psiv-2 digestibility evaluation of fermented soybean for nursery piglets. J Anim Sci. (2019) 97:184. doi: 10.1093/jas/skz122.324
16. Kong, F, Zeng, Q, Li, Y, and Guo, X. Effect of steam explosion on structural characteristics of Β−Conglycinin and morphology, chemical compositions of soybean meal. Front Nutr. (2022) 9:9. doi: 10.3389/fnut.2022.896664
17. Wang, Z, Li, L, Yuan, D, Zhao, X, Cui, S, Hu, J, et al. Reduction of the allergenic protein in soybean meal by enzymatic hydrolysis. Food Agr Immunol. (2014) 25:301–10. doi: 10.1080/09540105.2013.782268
18. Yang, A, Zuo, L, Cheng, Y, Wu, Z, Li, X, Tong, P, et al. Degradation of major allergens and allergenicity reduction of soybean meal through solid-state fermentation with microorganisms. Food Funct. (2018) 9:1899–909. doi: 10.1039/C7FO01824J
19. Rist, VTS, Weiss, E, Eklund, M, and Mosenthin, R. Impact of dietary protein on microbiota composition and activity in the gastrointestinal tract of piglets in relation to gut health: a review. Animal. (2013) 7:1067–78. doi: 10.1017/s1751731113000062
20. Xia, J, Fan, H, Yang, J, Song, T, Pang, L, Deng, H, et al. Research progress on diarrhoea and its mechanism in weaned piglets fed a high-protein diet. J Anim Physiol An N. (2022) 106:1277–87. doi: 10.1111/jpn.13654
21. Salazar-Villanea, S, Hendriks, WH, Bruininx, EM, Gruppen, H, and van der Poel, AF. Protein structural changes during processing of vegetable feed ingredients used in swine diets: implications for nutritional value. Nutr Res Rev. (2016) 29:126–41. doi: 10.1017/s0954422416000056
22. Bai, M, Qin, G, Sun, Z, and Long, G. Relationship between molecular structure characteristics of feed proteins and protein in vitro digestibility and solubility. Asian Austral J Anim. (2016) 29:1159–65. doi: 10.5713/ajas.15.0701
23. Carbonaro, M, Maselli, P, and Nucara, A. Relationship between digestibility and secondary structure of raw and thermally treated legume proteins: a Fourier transform infrared (Ft-Ir) spectroscopic study. Amino Acids. (2012) 43:911–21. doi: 10.1007/s00726-011-1151-4
24. Zhao, Y, Zhu, Y, Qin, G, Pan, L, Sun, H, Bao, N, et al. Physicochemical properties of dietary protein as predictors for digestibility or releasing percentage of amino acids in monogastrics under in-vitro conditions. Ital J Anim Sci. (2022) 21:507–21. doi: 10.1080/1828051X.2022.2048975
25. Li, L, Zhang, Q, Yuan, X, Yang, H, Qin, S, Hong, L, et al. Study of the molecular structure of proteins in fermented maize-soybean meal-based rations based on Ftir spectroscopy. Food Chem. (2024) 441:138310. doi: 10.1016/j.foodchem.2023.138310
26. ISO. Animal feeding stuffs. Determination of moisture and other volatile matter content. Geneva, Switzerland: International Organization for Standardization (1999).
27. ISO. Animal feeding stuffs. Determination of N content and calculation of crude protein content-part 1: Kjeldahl method. Geneva, Switzerland: International Organization for Standardization (2005).
28. ISO. Animal feeding stuffs. Determination of fat content. Geneva, Switzerland: International Organization for Standardization (1999).
29. ISO. Animal feeding stuffs. Determination of crude ash content. Geneva, Switzerland: International Organization for Standardization (2002).
30. Ørskov, ER, and McDonald, I. The estimation of protein degradability in the rumen from incubation measurements weighted according to rate of passage. J Agric Sci. (1979) 92:499–503. doi: 10.1017/S0021859600063048
31. Ma, W, Qi, B, Sami, R, Jiang, L, Li, Y, and Wang, H. Conformational and functional properties of soybean proteins produced by extrusion-hydrolysis approach. Int J Anal Chem. (2018) 2018:9182508–11. doi: 10.1155/2018/9182508
32. Zhang, QH, Huang, XQ, Li, MY, Liu, YX, Zhang, JW, Gao, XP, et al. Study on secondary structure of meat protein by Ftir. Food Ferment Ind. (2015) 41:247–51. doi: 10.13995/j.cnki.11-1802/ts.201510045
33. Long, G, Ji, Y, Pan, H, Sun, Z, Li, Y, and Qin, G. Characterization of thermal denaturation structure and morphology of soy Glycinin by Ftir and Sem. Int J Food Prop. (2015) 18:763–74. doi: 10.1080/10942912.2014.908206
34. Miao, X, Niu, H, Sun, M, Li, D, Hua, M, Wang, J, et al. Structural characterization and properties of modified soybean meal protein via solid-state fermentation by Bacillus Subtilis. Molecules. (2023) 28:8015. doi: 10.3390/molecules28248015
35. Bryan, D, Abbott, DA, and Classen, HL. Digestion kinetics of protein sources determined using an in vitro chicken model. Anim Feed Sci Tech. (2019) 248:106–13. doi: 10.1016/j.anifeedsci.2019.01.002
36. Bryan, DD, Abbott, DA, and Classen, HL. Development of an in vitro protein digestibility assay mimicking the chicken digestive tract. Anim Nutr. (2018) 4:401–9. doi: 10.1016/j.aninu.2018.04.007
37. Wilfart, A, Jaguelin-Peyraud, Y, Simmins, H, Noblet, J, van Milgen, J, and Montagne, L. Kinetics of enzymatic digestion of feeds as estimated by a stepwise in vitro method. Anim Feed Sci Tech. (2008) 141:171–83. doi: 10.1016/j.anifeedsci.2007.05.021
38. Merchant, HA, McConnell, EL, Liu, F, Ramaswamy, C, Kulkarni, RP, Basit, AW, et al. Assessment of gastrointestinal Ph, fluid and lymphoid tissue in the Guinea pig, rabbit and pig, and implications for their use in drug development. Eur J Pharm Sci. (2011) 42:3–10. doi: 10.1016/j.ejps.2010.09.019
39. Sklan, D, and Hurwitz, S. Protein digestion and absorption in young chicks and turkeys. J Nutr. (1980) 110:139–44. doi: 10.1093/jn/110.1.139
40. Yáñez, JL, Woyengo, TA, Jha, R, Van Kempen, TATG, and Zijlstra, RT. Nutrient digestibility of soybean products in grower-finisher Pigs1. J Anim Sci. (2019) 97:4598–607. doi: 10.1093/jas/skz290
41. Loveday, SM. Protein digestion and absorption: the influence of food processing. Nutr Res Rev. (2023) 36:544–59. doi: 10.1017/S0954422422000245
42. Greenberg, NA, and Shipe, WF. Comparison of the abilities of trichloroacetic, picric, sulfosalicylic, and tungstic acids to precipitate protein hydrolysates and proteins. J Food Sci. (1979) 44:735–7. doi: 10.1111/j.1365-2621.1979.tb08487.x
43. Li, Z, Li, Y, Zhao, Y, Wang, G, Liu, R, Li, Y, et al. Effects of the kinetic pattern of dietary glucose release on nitrogen utilization, the portal amino acid profile, and nutrient transporter expression in intestinal enterocytes in piglets. J Anim Sci Biotechno. (2024) 15:49. doi: 10.1186/s40104-024-01000-z
44. Theodoridou, K, and Yu, P. Application potential of Atr-Ft/Ir molecular spectroscopy in animal nutrition: revelation of protein molecular structures of canola meal and Presscake, as affected by heat-processing methods, in relationship with their protein digestive behavior and utilization for dairy cattle. J Agr Food Chem. (2013) 61:5449–58. doi: 10.1021/jf400301y
Keywords: enzymatic hydrolysis, soybean meal, protein, structure, digestive dynamics
Citation: Wang D, Du H, Dang X, Zhao Y, Zhang J, Liu R, Ge Z, Zhong Q and Sun Z (2024) Enzymatic hydrolysis processing of soybean meal altered its structure and in vitro protein digestive dynamics in pigs. Front. Vet. Sci. 11:1503817. doi: 10.3389/fvets.2024.1503817
Edited by:
Yun Ji, China Agricultural University, ChinaReviewed by:
Hongbin Pan, Yunnan Agricultural University, ChinaBolin Zhang, Zunyi Normal University College, China
Zaisen Wang, Langston University, United States
Copyright © 2024 Wang, Du, Dang, Zhao, Zhang, Liu, Ge, Zhong and Sun. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Qingzhen Zhong, cWluZ3poZW56aG9uZ0AxNjMuY29t; Zewei Sun, c3VuemV3ZWlAamxhdS5lZHUuY24=
 Dapeng Wang
Dapeng Wang Honglin Du
Honglin Du Jiaxuan Zhang
Jiaxuan Zhang Rujie Liu
Rujie Liu Qingzhen Zhong
Qingzhen Zhong Zewei Sun
Zewei Sun