- 1Microbiology Lab, Department of Zoology, Government College University, Lahore, Punjab, Pakistan
- 2Department of Zoology, Abdul Wali Khan University Mardan, Mardan, Khyber Pakhtunkhwa, Pakistan
- 3King Abdulaziz City for Science and Technology, Riyadh, Saudi Arabia
- 4Department of Pharmacology and Toxicology, College of Pharmacy, King Saud University, Riyadh, Saudi Arabia
- 5United States National Tick Collection, The James H. Oliver Jr. Institute for Coastal Plain Science, Georgia Southern University, Statesboro, GA, United States
- 6Department of Biology, Georgia Southern University, Statesboro, GA, United States
- 7Laboratory of Animal Microbiology, Graduate School of Agricultural Science/Faculty of Agriculture: Tohoku University, Aoba-ku, Sendai, Japan
There is limited information on the occurrence of Hyalomma turanicum and Hyalomma asiaticum ticks, as well as associated Rickettsia, Anaplasma, and Ehrlichia species in Pakistan. Addressing this knowledge gap, the current study aimed at morphomolecular confirmation of these ticks and molecular assessment of associated Rickettsiales bacteria (Rickettsia, Anaplasma, and Ehrlichia spp.) in Balochistan, Pakistan. A total of 314 ticks were collected from 74 of 117 (63.2%) hosts, including 41 of 74 (55.4%) goats and 33 of 74 (44.5%) sheep. Subsequently, a subset of microscopically identified ticks was subjected to DNA extraction and PCR to amplify 16S rDNA and cox1 fragments. Additionally, gltA, ompA, and ompB fragments were targeted for Rickettsia spp. and 16S rDNA fragments for both Anaplasma and Ehrlichia spp. The 16S rDNA and cox1 sequences of Hy. turanicum demonstrated 100% identity with those of the same species previously reported from Pakistan. The 16S rDNA and cox1 sequences of Hy. asiaticum exhibited 99.52 and 100% identities, respectively, with corresponding species reported from China, Kazakhstan, and Turkey. The gltA, ompA, and ompB fragments associated with Hy. turanicum showed 100% identities with Rickettsia aeschlimannii reported from Egypt, Italy, Kazakhstan, Kenya, Pakistan, and Senegal. The 16S rDNA sequences of Anaplasma sp. and Ehrlichia sp. associated with both Hy. asiaticum and Hy. turanicum exhibited 99.67 and 100% identities with unknown Anaplasma sp. and Ehrlichia sp. reported from Morocco and Pakistan, respectively. In the 16S rDNA and cox1 phylogenetic trees of ticks, Hy. turanicum and Hy. asiaticum from the current study clustered with their respective species. Similarly, in gltA, ompA, and ompB phylogenetic trees of Rickettsia, R. aeschlimannii of the present study clustered with the same species, whereas Anaplasma sp. and Ehrlichia sp. of this study clustered with undetermined Anaplasma spp. and Ehrlichia spp. in the 16S rDNA phylogenetic tree of Anaplasmataceae. Among the DNA samples from the screened ticks, a coinfection rate of R. aeschlimannii, Anaplasma sp., and Ehrlichia sp. (2 of 80, 2.5%) was observed in Hy. turanicum, whereas individual infection rates were noted as follows: R. aeschlimannii (8 of 80, 10%), Anaplasma sp. (5 of 80, 6.3%), and Ehrlichia sp. (5 of 80, 6.3%). This study marks the first record of molecular characterization of Hy. turanicum and Hy. asiaticum as well as the detection of associated R. aeschlimannii, Anaplasma sp., and Ehrlichia sp. in Balochistan, Pakistan.
1 Introduction
Ticks are hematophagous ectoparasites and leading vectors of important tickborne pathogens that affect all classes of terrestrial and semi-terrestrial animals, including livestock. Losses due to tick infestation could be either direct or indirect and are affected by certain climatic factors, including fluctuation in temperature, humidity, photoperiod, and vegetation, as well as host availability (1–3). The Hyalomma genus is among the prominent genus of ticks found in Asia, Africa, and Southern Europe with 27 reported species (4), while in Pakistan, 14 species of this genus have been reported (5–7). The several species complexes in this genus share many morphological traits, making accurate identification challenging. Within Hyalomma (Euhyalomma) marginatum complex, Hyalomma glabrum Delpy 1949, Hyalomma isaaci Sharif 1928, Hy. marginatum Koch 1844, Hyalomma rufipes Koch 1844, and Hyalomma turanicum Pomerantzev 1946 are closely related species (8). Of these species, Hy. turanicum is found in Africa and Asia (Afghanistan, Bahrain, China, Egypt, India, Iran, Iraq, Israel, Jordan, Kazakhstan, Kyrgyzstan, Nepal, Oman, Pakistan, Qatar, Saudi Arabia, Syria, Tajikistan, Turkmenistan, Uzbekistan, and Yemen) (9, 10). Within Hyalomma (Euhyalomma) asiaticum complex, Hyalomma asiaticum Schulze & Schlottke 1929, Hyalomma dromedarii Koch 1844, Hyalomma impeltatum Schulze & Schlottke 1929, Hyalomma schulzei Olenev 1931, and Hyalomma somalicum Tonelli Rondelli 1935 are closely related (8, 11). Among these species, Hy. asiaticum is limited to Asia (Afghanistan, Armenia, Azerbaijan, China, Georgia, Iran, Iraq, Kazakhstan, Kyrgyzstan, Mongolia, Pakistan, Russia, Syria, Tajikistan, Turkey, Turkmenistan, and Uzbekistan) (9, 10). Key morphological features of these species are adanal plates, spiracular plates, setae, genital aperture, and scutum/conscutum (12, 13).
Morphological-based identification of Hyalomma species creates more challenges, which resulted in the re-description of its several species (14, 15). Recent studies of molecular identification have improved our understanding of the inter- and intra-specific diversity of Hyalomma ticks, which may help in understanding its taxonomy and epidemiology (9, 16, 17).
Within the order Rickettsiales, the genus Rickettsia consists of obligate intracellular Gram-negative bacteria that affect veterinary and human health by causing various diseases such as Rocky Mountain spotted fever (18, 19). In this order, Ehrlichia species have unique cell tropisms within the vertebrate hosts and are causative agents of tickborne fever in ruminants, while Anaplasma species cause human granulocytic anaplasmosis (20, 21). Several Rickettsia, Anaplasma, and Ehrlichia species of medical importance are vectored by Hyalomma ticks, including Hy. turanicum and Hy. asiaticum (22, 23). Moreover, these tick species are also competent vectors of various viruses, including the Crimean–Congo hemorrhagic fever (CCHF) viruses (24). In Pakistan, Hyalomma ticks are expected to be associated with tickborne diseases such as babesiosis, theileriosis, rickettsiosis, and CCHF (7, 25–27). Pakistan’s economy is mostly based on agriculture, which contributes ≈ 24% to its gross domestic product and employs ≈ 40% of its labor force (53). Consequently, these TBDs can cause significant damage to the country’s economy.
Though Hyalomma ticks are highly important in both veterinary and medical aspects, morphomolecular studies of Hy. turanicum and Hy. asiaticum in Pakistan in general and in Balochistan province in particular are limited. Additionally, Hy. turanicum and Hy. asiaticum ticks from Pakistan have been largely overlooked in terms of pathogen research, including Rickettsia, Anaplasma, and Ehrlichia spp. Therefore, the study represents the first attempt to investigate the occurrence of Hy. turanicum and Hy. asiaticum ticks and Rickettsiales bacteria associated with these tick species. This study specifically aims to determine whether Hy. turanicum and Hy. asiaticum are associated with Rickettsia, Anaplasma, and Ehrlichia spp. in the study region.
2 Materials and methods
2.1 Ethical consideration
Prior to performing this study, approval was given by the Advanced Studies and Research Board (REG-ACAD-ASRB-55/23/035) of the Government College University, Lahore, Pakistan. All animal or herd owners were verbally informed, and permission was taken from them.
2.2 Study area
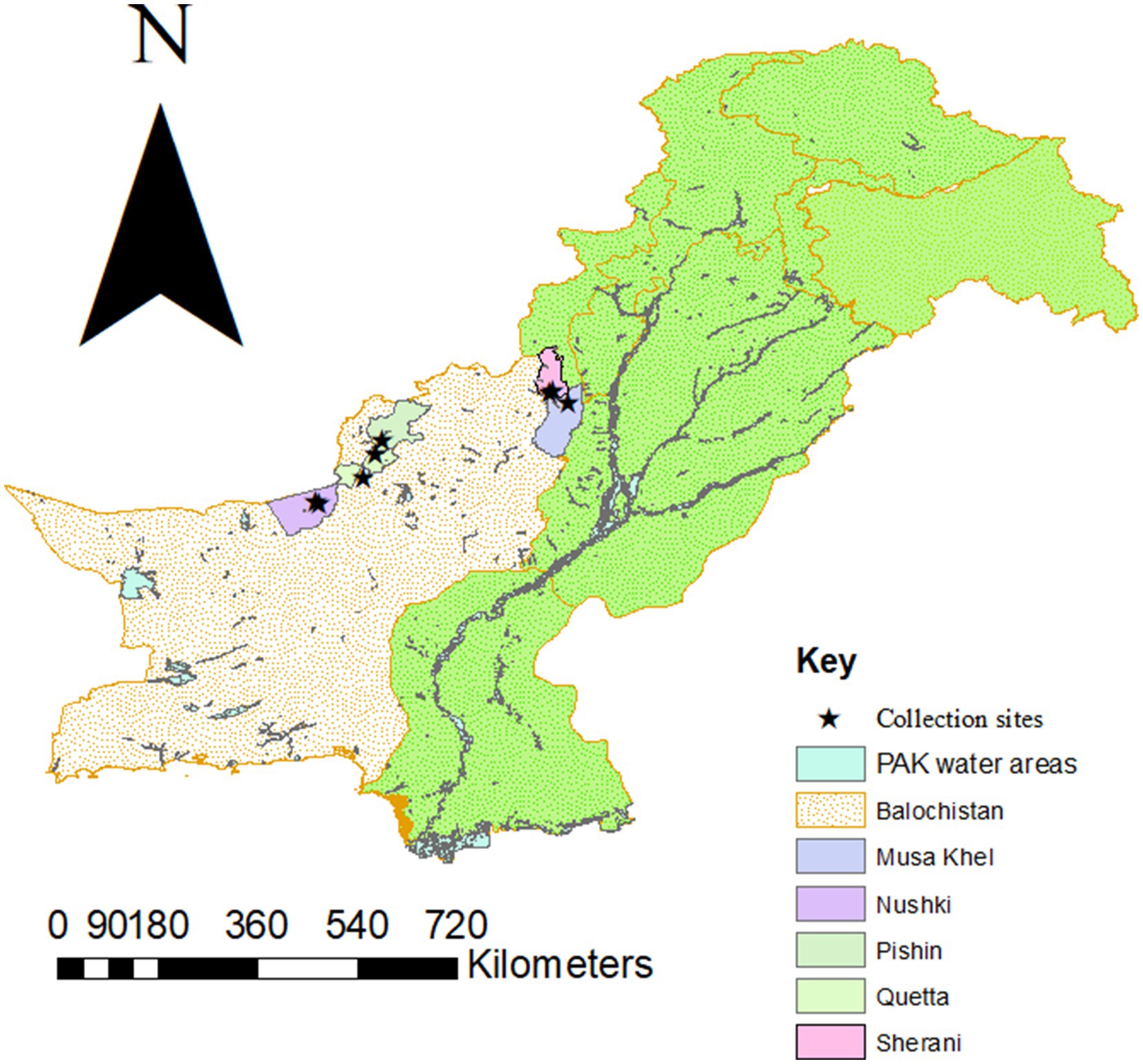
The present study was executed in five districts of the Balochistan province, Pakistan: Musakhail (30°50′52.1″N 69°57′18.0″E), Nushki (29°28′27.1″N 65°39′57.2″E), Pishin (30°44′17.9″N 67°17′05.3″E), Quetta (30°10′47.6″N 67°05′38.8″E), and Sherani (29°46′60″ N 68°31′0″ E). For creating a map (Figure 1) using ArcGIS v. 10.3.1, the coordinates of the above-collected sites were documented using the Global Positioning System. The choice of these districts was strategic because they share borders with neighboring countries (Afghanistan and Iran), making them key areas for cross-border animal movement and trade. Additionally, Quetta, as the provisional capital, serves as a major hub for the transportation of livestock, further increasing the importance of these districts. Livestock, particularly sheep and goats, are more prevalent in these areas, forming the backbone of the local economy.
2.3 Collection and preservation of ticks
Ticks were collected from goats and sheep from April 2023 to April 2024 in the specified districts of Balochistan. To minimize potential damage to the specimens, a meticulous approach was employed, delicately detaching ticks from the skin of hosts using forceps. The collected specimens underwent a thorough cleaning process with distilled water prior to washing with 70% absolute ethanol. For preservation and further analysis, the ticks were stored in a solution made of 95% ethanol, 4% distilled water, and 1% glycerol.
2.4 Morphological identification
Tick specimens were meticulously examined under a stereo-zoom microscope (Luxeo 6Z, LABOMED, USA). Morphological identification was carried out at both the life stage and species level, with the help of established morphological identification keys specified to Hyalomma species (12, 15).
2.5 DNA extraction and PCR
A total of 80 morphologically identified ticks were randomly selected for molecular analysis, including four males and four females of each species per host per district. Subsequently, the homogenized specimens were incubated until dry, involving dissection and grinding with sterile scissors. DNA extraction followed the standard phenol–chloroform method with minor adjustments (28). After extraction, DNA was quantified using a NanoDrop spectrophotometer (NanoQ, Optizen, Daejeon, South Korea) and stored at −20°C for further analysis.
Conventional PCRs (GE-96G, BIOER, Hangzhou, China) were performed to amplify partial fragments of 16S ribosomal DNA and cox1 genes of ticks from the extracted DNA (Table 1). Additionally, the extracted genomic DNA was molecularly assessed for rickettsial citrate synthase (gltA), outer membrane protein A and B (ompa and ompB), and Anaplasmataceae 16S rDNA (Table 1).
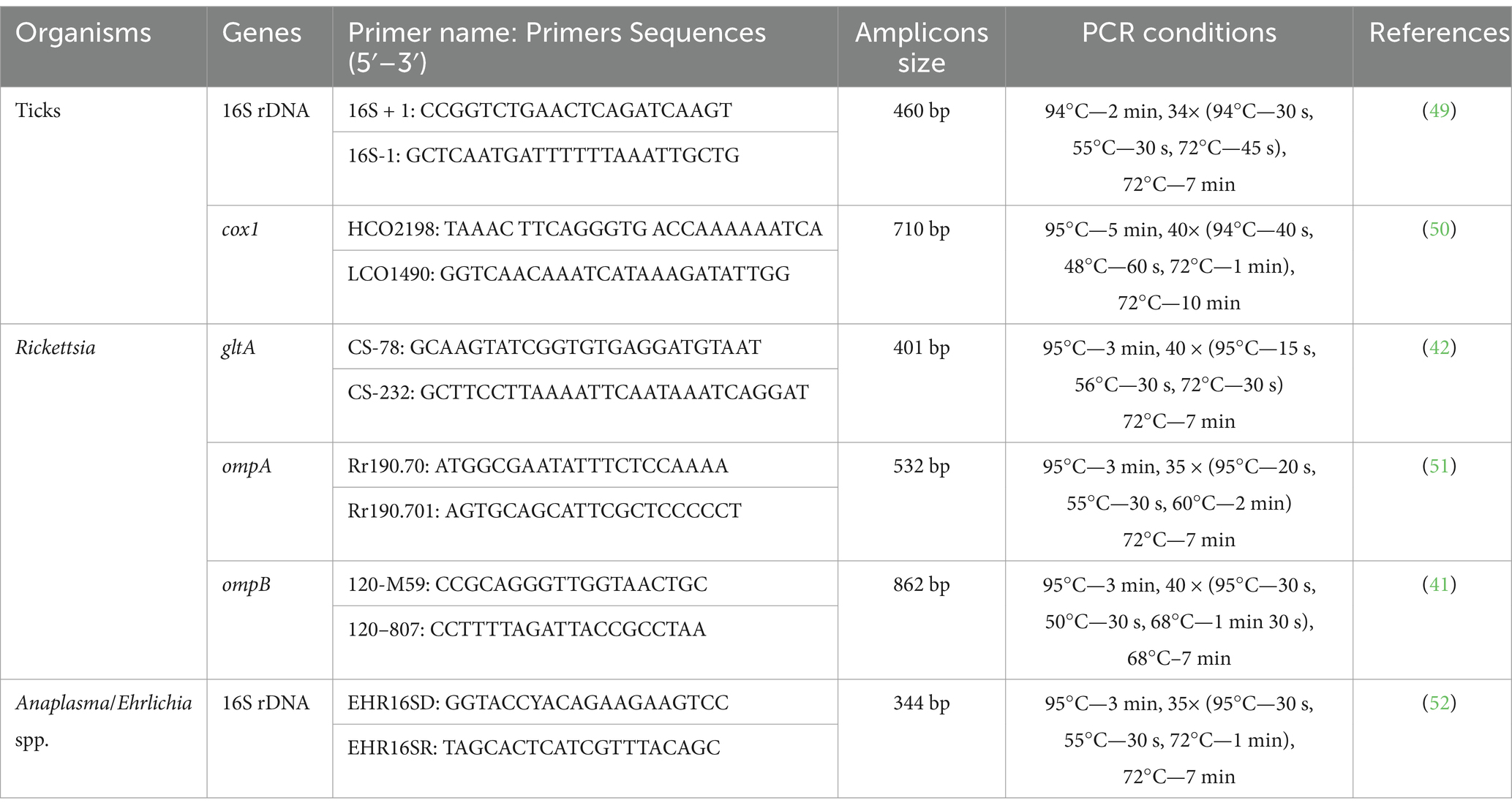
Table 1. List of primers and PCR conditions that were used for the molecular identification of ticks and pathogens.
Each PCR was performed in a 20 μL volume, including 1 μL forward and reverse primers each (concentration of 10 μM), 4 μL of “nuclease-free” PCR water, 2 μL of genomic DNA (100 ng/μL), and 12 μL of DreamTaq MasterMix (2X) (Thermo Fisher Scientific, Inc., Waltham, MA, USA). The primers and thermocycling conditions used in this study are listed in Table 1. To ensure the reliability of the PCRs, positive control samples containing DNA of Hyalomma anatolicum (for ticks) and DNA of Rickettsia massiliae (for bacterial species), and a negative control sample consisting of PCR water (nuclease-free), were part of each PCR. The amplified products of the tick and its pathogen from the PCRs were run on a gel made of 2% agarose. After PCR, the obtained bands were made visible under through UV from a Gel Documentation system (BioDoc-It™ Imaging Systems, UVP, LLC).
2.6 DNA sequencing and phylogenetic analyses
PCR-positive amplified products of the expected size were purified using the DNA Clean and Concentrator Kit (Zymo Research, Irvine, CA, USA) and submitted for bidirectional sequencing (Macrogen Inc., Seoul, South Korea) using the Sanger sequencing method. The resulting sequences were proceeded and purified through SeqMan v. 5 (DNASTAR, Inc., Madison, WI, USA) to remove low-quality and contaminated regions. Trimmed and consensus sequences were then analyzed using the Basic Local Alignment Search Tool (BLAST) at the National Center for Biotechnology Information (NCBI)1 (29). Sequences with high homology were downloaded in FASTA format. The downloaded sequences, along with an outgroup, were aligned using ClustalW (30) in the BioEdit alignment editor V.7.0.5 tool (Raleigh, NC, USA) (31).
Phylogenetic trees were constructed using MEGA-X (32) with the following specifications: Neighbor-Joining method, Tamura–Nei model, and 1,000 bootstrap replicates. Additionally, coding sequences were aligned using the MUSCLE alignment method (33).
3 Results
3.1 Morphological identification
Morphologically, Hy. turanicum was identified based on different features. The conscutum exhibited a yellow–brown color, with large punctations being rare, while medium- and small-sized punctations were relatively dense and covered the entire conscutum. The perforated portion of dorsal prolongation of the spiracular plate is relatively narrow; circumspiracular setae moderately dense; proximally moderately sized dorsal ivory-colored enamel spot (Figures 2C, D). While the scutum of the female was dark, ranging from reddish brown to nearly black, the genital aperture was wide, deep, and rounded (U-shaped), with the vestibular portion of the vagina bulging to some extent.
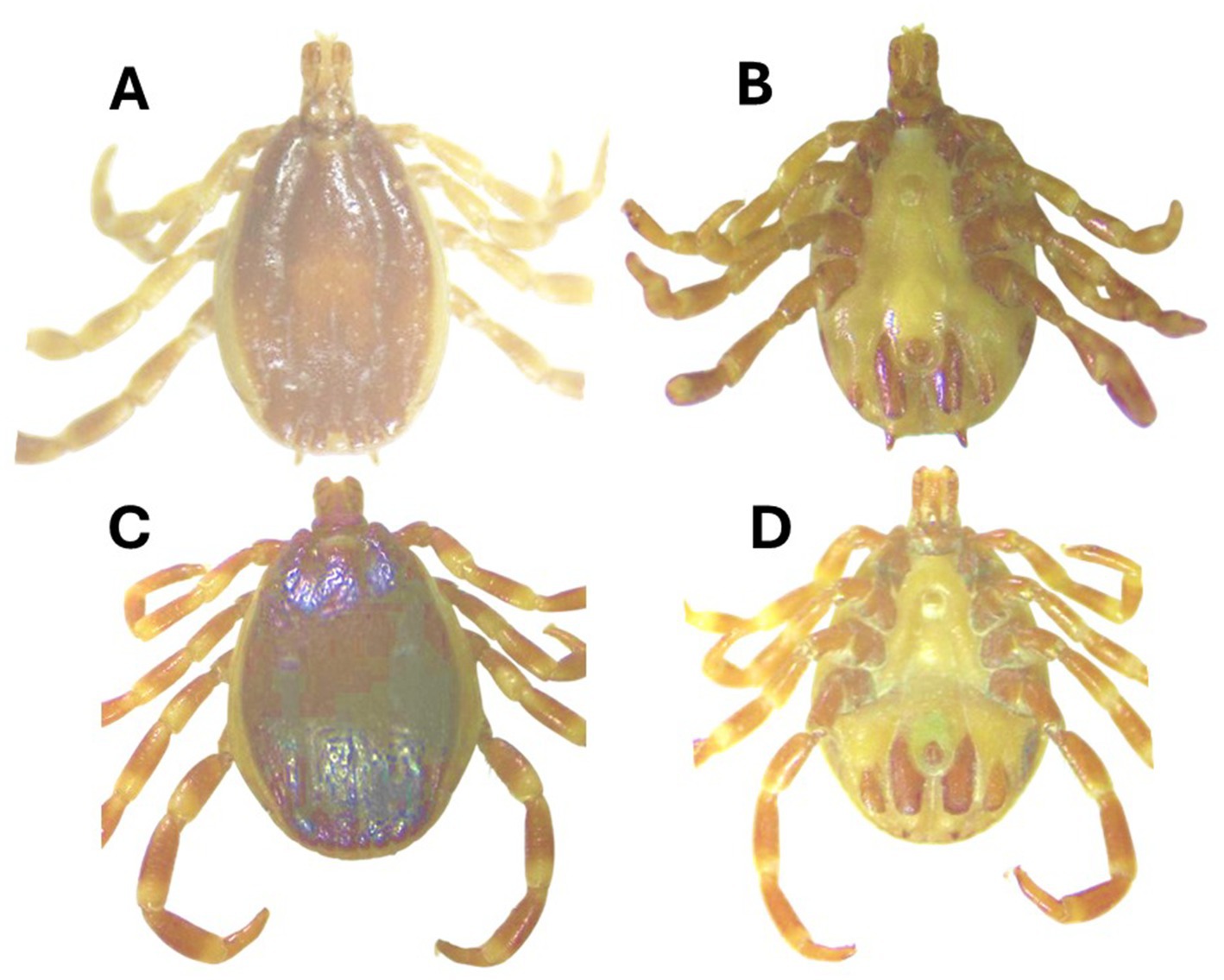
Figure 2. Hyalomma asiaticum male [(A) dorsal, (B) ventral] and Hyalomma turanicum male [(C) dorsal and (D) ventral] were collected in this study.
The Hy. asiaticum tick was morphologically identified based on different features. The conscutum exhibited a yellow to red–brown coloration. The cervical grooves were remarkably deep, while the marginal grooves were short. The adanal plates were long, narrow, and nearly straight, tapering slightly posteriorly from the median projection. (Figures 2A, B). While the scutum of the female was yellow– to red–brown, the genital aperture was narrow, U-shaped with the vestibular portion of the vagina distinctly bulging.
3.2 Hosts and ticks
Altogether, 314 ticks were collected from five districts of Balochistan; Musakhail, Nushki, Pishin, Quetta, and Sherani. These ticks included 165 (56.18%) Hy. asiaticum, comprising 104 (53.07%) females and 61 (32.03%) males, as well as 149 (43.81%) Hy. turanicum, including 96 (54.77%) females and 53 (31.12%) males. All these ticks were collected from 74 of the 117 examined hosts, demonstrating an overall prevalence of infestation of 63.2%. Moreover, goats exhibited a high prevalence of infestation (41 of 74, 55.4%) compared to sheep (33 of 74, 44.5%). Detailed results about ticks, small ruminants, and Rickettsiales bacteria are summarized in Table 2.
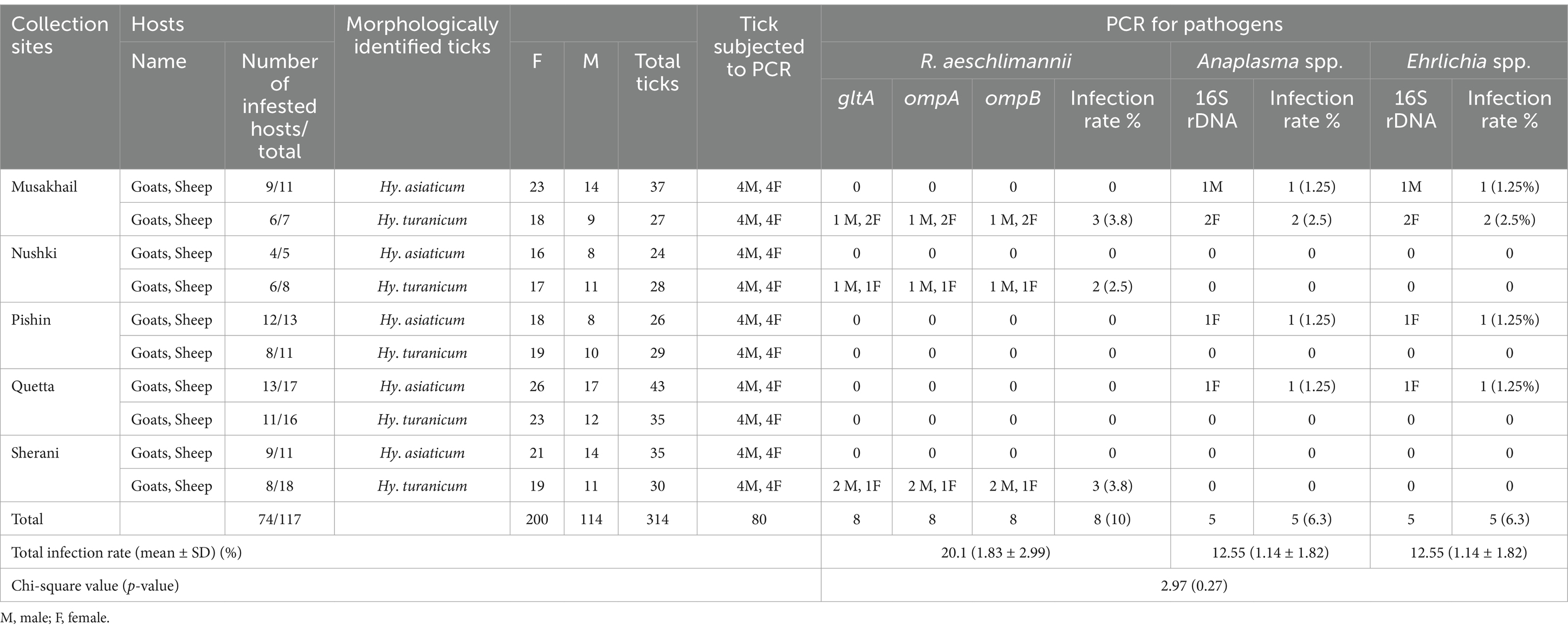
Table 2. Collection sites of collected specimens and their molecular characterization with infection rate.
3.3 Molecular outcomes
DNA was extracted from 80 ticks, including 40 Hy. turanicum and 40 Hy. asiaticum. Altogether, 53 bidirectional sequences were obtained, including at least one 16S rDNA and one cox1 for each female and male of each tick species per district. Consensus sequences of ticks were obtained as follows: 16S rDNA for Hy. turanicum (410 bp), 16S rDNA for Hy. asiaticum (420 bp), cox1 for Hy. turanicum (531 bp), and cox1 for Hy. asiaticum (611 bp). Considering the closest identity, 16S rDNA and cox1 sequences for Hy. turanicum were 100% identical with the corresponding species reported from Pakistan. Similarly, the 16S rDNA sequences for Hy. asiaticum showed a maximum identity of 99.52%, while its cox1 sequences revealed a maximum identity of 100% with conspecific from China, Kazakhstan, and Turkey.
For each amplified fragment of Rickettsia spp., bidirectional sequences were obtained, which gave consensus sequences as follows: gltA (369 bp), ompA (471 bp), and ompB (790 bp) genes. The obtained sequences showed 100% identity with R. aeschlimannii reported from Egypt, Italy, Kazakhstan, Kenya, Pakistan, and Senegal.
Among the Anaplasmataceae, 8 of 80 (9.52%) samples were positive for 16S rDNA, including four Hy. turanicum and four Hy. asiaticum ticks. From the amplified fragments of Anaplasmataceae, bidirectional sequences were obtained, which resulted in the following consensus sequences: 16S rDNA (301 bp) for Anaplasma sp. and 16S rDNA sequence (333 bp) for Ehrlichia sp. The former sequence showed 99.67% identity with unknown Anaplasma sp. reported from Morocco, while the latter sequence revealed 100% identity with an undetermined Ehrlichia sp. reported from Pakistan.
The overall detection rates were as follows: 10% (8 of 80) for R. aeschlimannii associated with Hy. turanicum, 6.3% (5 of 80) for Anaplasma sp., and 6.3% (5 of 80) for Ehrlichia sp. associated with both Hy. turanicum and Hy. asiaticum. Additionally, coinfection with R. aeschlimannii, Anaplasma sp., and Ehrlichia sp. was found in 2 of 80 (2.5%) Hy. turanicum ticks. Detailed information about the infection rates of R. aeschlimannii, Anaplasma spp., and Ehrlichia spp. is shown in Table 2.
Following are the GenBank accession numbers of all consensus sequences of this study: 16S rDNA of Hy. turanicum (PQ218624) and Hy. asiaticum (PQ218629), cox1 of Hy. turanicum (PQ218744), and Hy. asiaticum (PQ218788). R. aeschlimannii, gltA (PQ221477), ompA (PQ227817), and ompB (PQ227818), 16S rDNA for Anaplasma sp. (PQ213859), and Ehrlichia sp. (PQ214321).
3.4 Phylogenetic outcomes
The phylogenetic tree constructed 16S rDNA sequences revealed that Hy. turanicum clustered with the same species recorded from Iraq and Pakistan (KU130480, KU130482, and KU130483), while Hy. asiaticum grouped with the corresponding species reported from China and Kazakhstan (JX051085 MK530106, OR486026, and OR486010) (Figure 3A). The phylogenetic tree constructed for the cox1 sequences showed that Hy. turanicum clustered with the related species from Saudi Arabia, Pakistan, and Iraq (OQ799122, PP716472, KU130649, KU130646, and MH094478), while Hy. asiaticum grouped with the related species described from Kazakhstan, China, and Turkey (OM368315, KU364332, and MW546281) (Figure 3B).
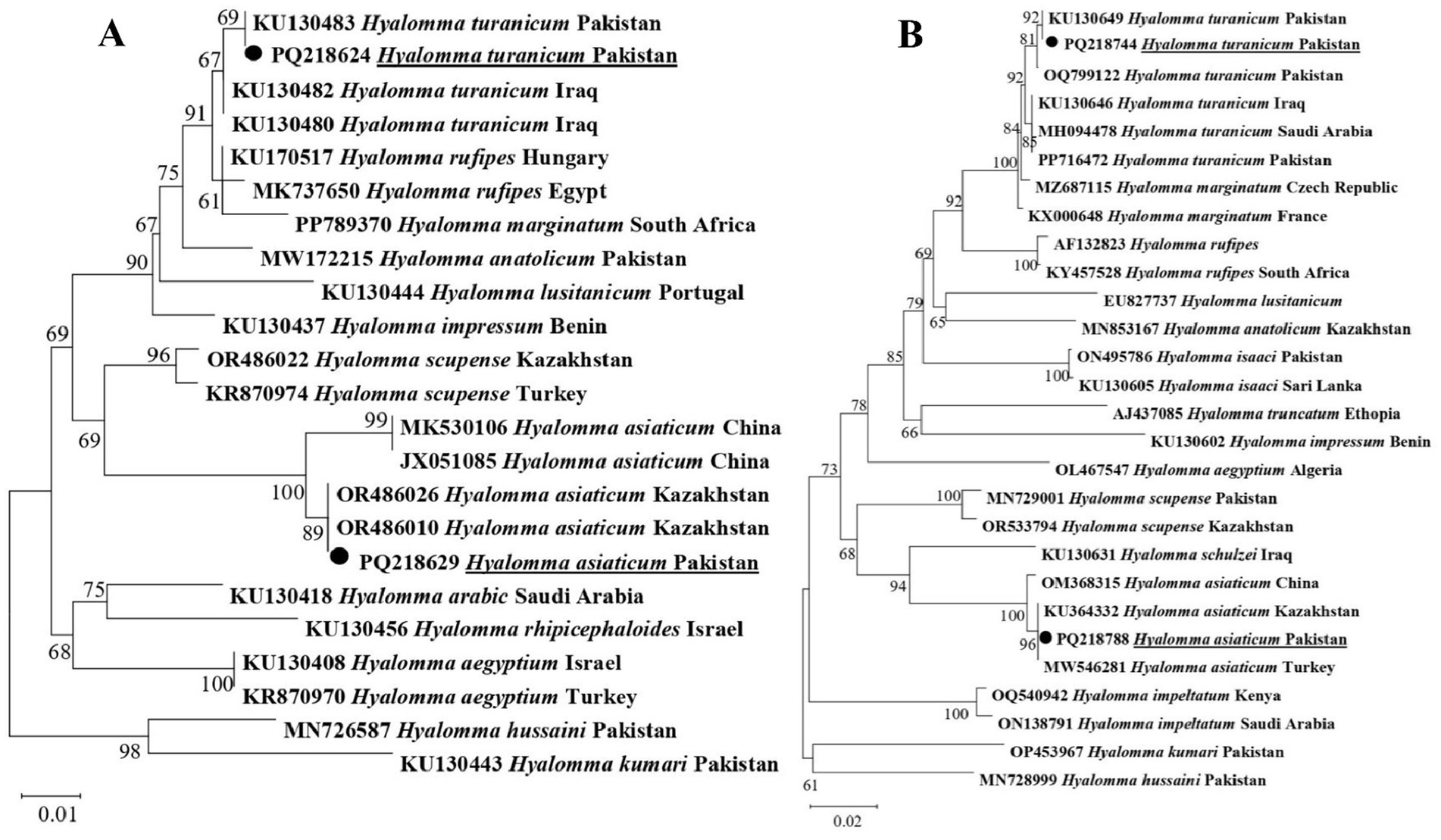
Figure 3. Phylogenetic tree constructed for Hyalomma spp. based on 16S rDNA (A) and cox1 (B) sequences. Each sequence was identified by accession number, species name, and origin country. Sequences of Hyalomma hussaini and Hyalomma kumari were used as an outgroup. The acquired sequences have been highlighted.
In a phylogenetic tree developed for rickettsial gltA, R. aeschlimannii was detected in Hy. turanicum grouped with the sequences of the corresponding species described from Egypt, Kenya, and Senegal (HQ335148, KX227772, KX227768, and HM050285) (Figure 4A). In a phylogenetic developed for rickettsial ompA, R. aeschlimannii clustered with the sequences of the same species recorded from Italy and Pakistan (JN944634 and OQ632790) (Figure 4B). Additionally, in the phylogenetic tree obtained for rickettsial ompB, R. aeschlimannii grouped with the corresponding species reported from Italy, Kazakhstan, and Pakistan (MH532261, MW430414, and OR351961) (Figure 4C).
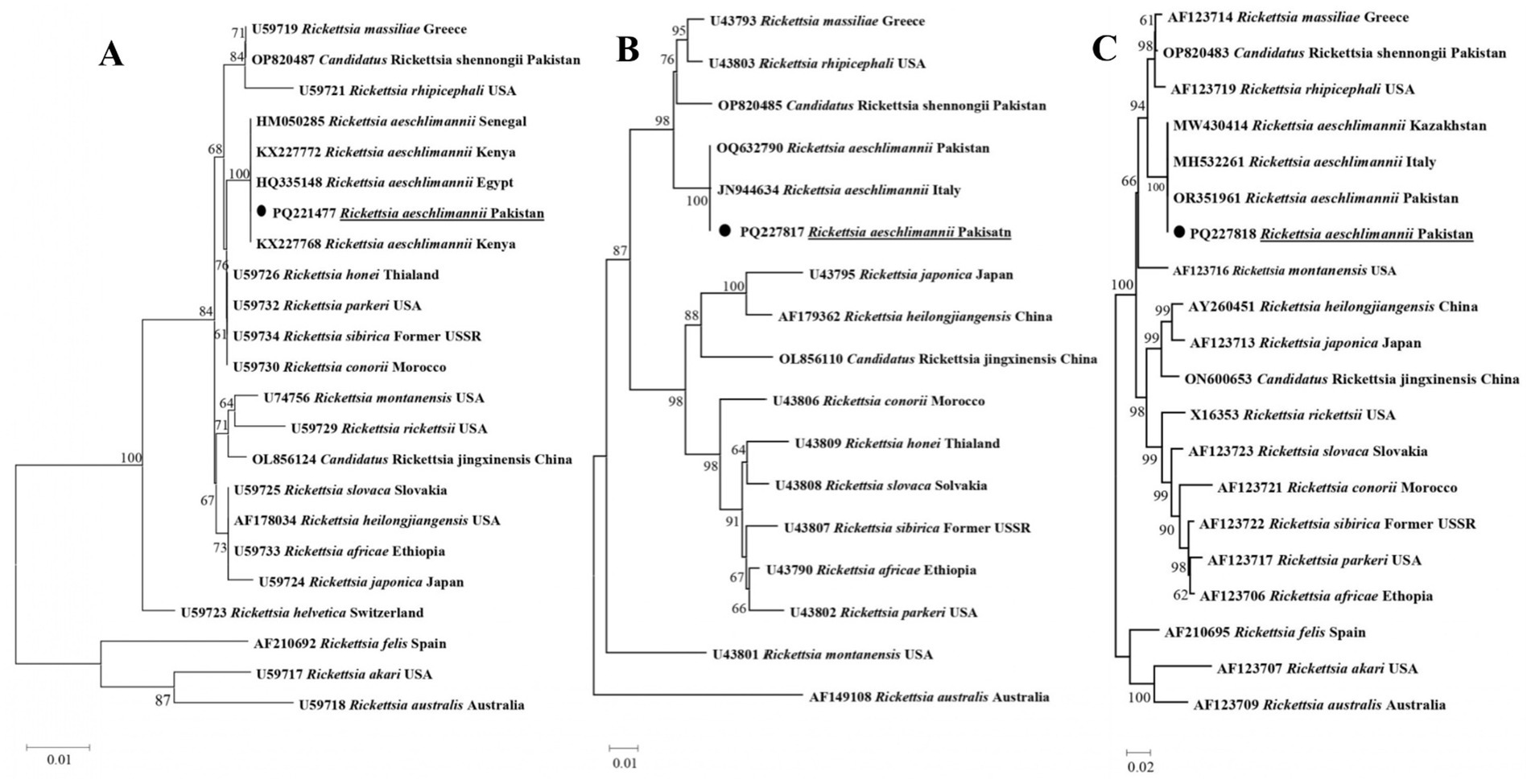
Figure 4. Phylogenetic tree constructed for Rickettsia spp. based on gltA (A), ompA (B), and ompB (C). Each sequence was identified by accession number, species name, and origin country. Rickettsia akari and Rickettsia australis were jointly used as outgroups, and the acquired sequence has been highlighted.
In the phylogenetic tree for Anaplasmataceae 16S rDNA sequences, Anaplasma sp. associated with Hy. asiaticum and Hy. turanicum was clustered with an undetermined Anaplasma sp. described from Morocco (OK606072) (Figure 5). In the same phylogenetic tree, Ehrlichia sp. recorded in the same ticks was grouped with an undermined Ehrlichia sp. previously reported from Pakistan (MH250197) (Figure 5).
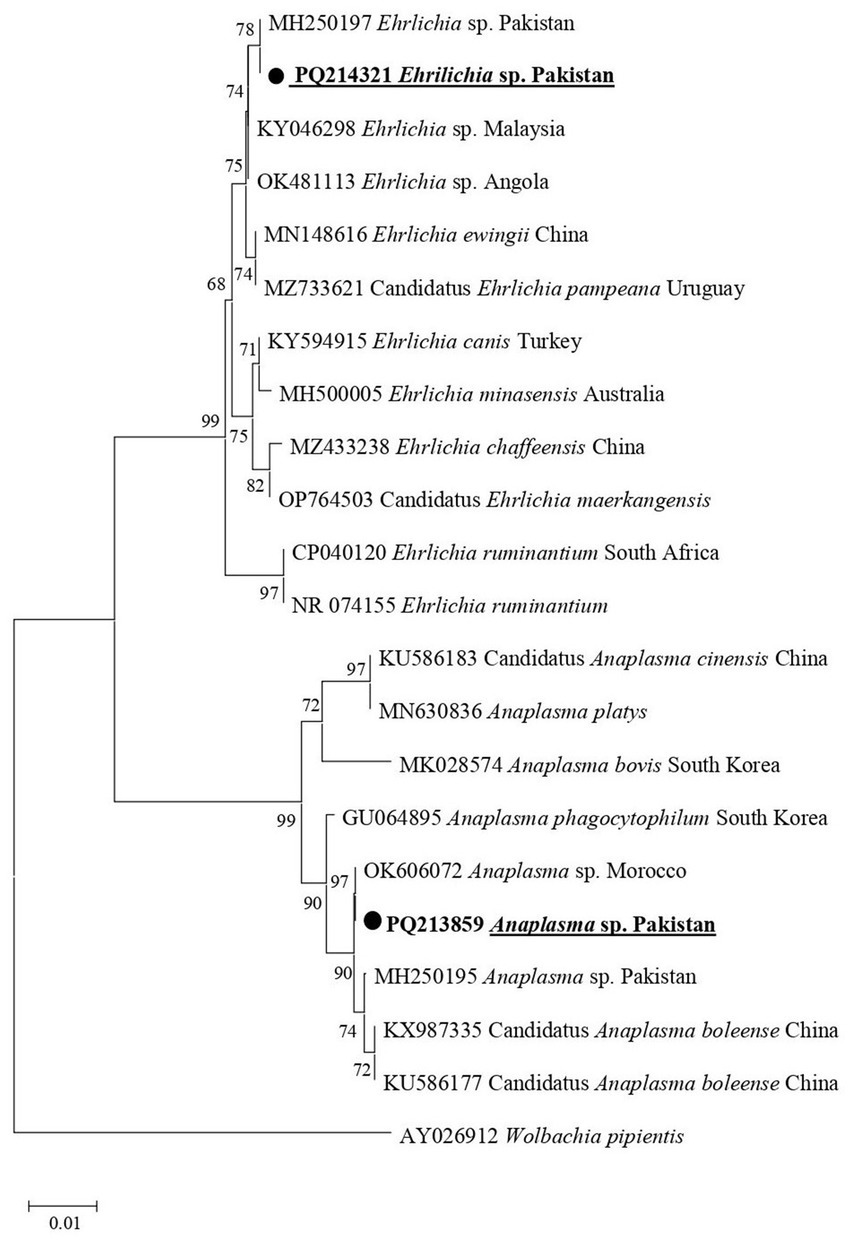
Figure 5. Phylogenetic tree constructed for 16S rDNA sequences of Anaplasma spp. and Ehrlichia spp. Each sequence was identified by providing the accession number, species name, and origin country. The sequence of Wolbachia pipientis was used as an outgroup, and the acquired sequences have been highlighted.
4 Discussion
Sheep and goats are commonly raised animals in Pakistan, particularly in rural areas where these domestic animals serve as a significant source of income (34, 35). Despite the diverse range of ticks and TBDs, which cause significant economic losses in the livestock sector in rural areas of Pakistan, there is limited knowledge about the occurrence of Hy. turanicum and Hy. asiaticum ticks, as well as the pathogens associated with them. Few studies have been conducted to characterize Hyalomma ticks and/or associated pathogens from different regions of Pakistan (5–7, 9, 17). Herein, we morphologically and molecularly characterized two Hyalomma species, Hy. turanicum and Hy. asiaticum, for the first time from Balochistan, Pakistan. Additionally, this research also detected R. aeschlimannii in Hy. turanicum and undetermined Anaplasma sp. and Ehrlichia sp. in both Hy. turanicum and Hy. asiaticum.
Morphological identification of Hyalomma ticks, including Hy. turanicum and Hy. asiaticum, presents challenges (12, 15). In addition to morphological comparisons within their species, Hy. turanicum and Hy. asiaticum were also compared to their closest relatives within their respective complexes. Hyalomma turanicum was distinguished from Hy. marginatum by the presence of dense punctations on the conscutum or scutum, moderately narrow dorsal prolongation of the spiracular plate, and denser circumspiracular setae (12). Similarly, Hy. asiaticum was differentiated from Hy. dromedarii by characteristics such as: in males, a posteromedian groove that does not reach the parma and straighter adanal plates; and in females, a more U-shaped genital operculum and bulging preatrial fold (15).
To validate the morphological identification of ticks and ascertain the presence of a diverse array of pathogens, molecular confirmation holds significance (36, 37). Comprehensive molecular studies on the Hyalomma ticks have been conducted using the genetic markers; 16S rDNA and coxI (6, 7). In addition to clustering with the same species in the monophyletic clade, Hy. turanicum in the current study formed a sister clade with Hy. marginatum and Hy. rufipus within the subgenus Euhyalomma (38), corroborating previous studies that categorize them as members of the Hy. marginatum complex based on both morphological and genetic data (12, 38). Similarly, Hy. asiaticum clustered with the same species in the monophyletic clade and formed a sister clade with Hyalomma schulzei within the subgenus Euhyalomma, consistent with earlier studies that identify them as members of the Hy. asiaticum group, supported by morphological and genetic data (15, 38).
Rickettsia spp., including R. aeschlimannii, are zoonotic pathogens carried by different arthropod vectors, including Hyalomma ticks (18, 26, 39). The detection of Rickettsia spp. in ticks is significant not only for recognizing infected ticks but also for assessing the risk of transmission to humans (40). Considering suitability (41, 42), genetic markers such as gltA, ompA, and ompB were employed in this study to detect and phylogenetically analyze Rickettsia spp. in Hyalomma ticks. Previously, R. aeschlimannii has been detected in Hy. turanicum, Ha. bispinosa, and Ha. montgomeryi in Pakistan (43). This study further expanded the range of competent tick hosts and the geographical distribution of R. aeschlimannii in Pakistan by detecting it in Hy. turanicum in the Balochistan province. Transmission from animals to humans occurs through the bite of an infected tick, resulting in Mediterranean spotted fever (39). Additionally, Rickettsia amblyommii, R. massiliae, Rickettsia conorii, and Rickettsia hoogstraalii have been detected in other Hyalomma ticks, including Hy. hussaini, Hy. anatolicum, Hy. dromedarii, Hy. Turanicum, and Hy. kumari (7, 25, 44, 45). By clustering R. aeschlimannii with identical species from various regions within the spotted fever group, all three phylogenetic trees mutually validated each other. Anaplasma and Ehrlichia spp. are primarily transmitted by ticks and can cause diseases in humans and animals (46, 47). Partial fragments of 16S rDNA, widely acknowledged as a reliable molecular marker for characterizing tick-associated Anaplasma and Ehrlichia spp. (20), were utilized in this study. The detection of Anaplasma and Ehrlichia spp. in Hyalomma ticks in the present study, as well as Anaplasma marginale, Anaplasma ovis, and Anaplasma centrale, in other Hyalomma ticks, including Hy. anatolicum and Hyalomma scupense, and undetermined Ehrlichia spp. in Hyalomma ticks, including Hy. dromedarii and Hy. anatolicum, suggests a greater diversity of these tickborne microorganisms in Pakistan. Herein, Anaplasma sp. shared phylogenetic similarities with Candidatus Anaplasma boleense, and Ehrlichia sp. with Ehrlichia ewingii, indicating potential zoonotic implications. The detection of R. aeschlimannii, Anaplasma sp., and Ehrlichia sp. in Hyalomma ticks in the study region indicates risks to livestock workers and farmers, especially in rural areas. In addition, it may be associated with economic losses and food security by affecting livestock. Therefore, understanding various aspects of these bacteria, such as their ecology, genetic diversity, and pathogenicity, is necessary to mitigate the associated risks in Pakistan.
The presence of Hy. turanicum and Hy. asiaticum, along with their notable abundance in the Balochistan province of Pakistan, may be attributed to nomadic migration, resulting in the uncontrolled movement of domestic animals between Balochistan, Afghanistan, and Iran (48). Other factors contributing to the prevalence of these tick species could include the presence of suitable hosts and the arid or semi-arid climate of the study area (6, 26). Alternative approaches implemented in the study area, such as cohabiting with diverse hosts in shared habitats, managing densely populated herds, and employing mixed grazing methods, may positively impact the presence of these ticks. Additionally, these conditions may influence the association between Hyalomma ticks and bacterial species. As the present study is based on relatively a small size sample size and is restricted to the Balochistan province of Pakistan, future studies should aim to increase the sample size, expand the geographic coverage, and investigate the vector competence of Hyalomma species for associated bacteria.
5 Conclusion
This study addresses a significant knowledge gap concerning Hy. turanicum and Hy. asiaticum, which were morphologically and genetically characterized for the first time in Balochistan, Pakistan. Additionally, the study molecularly assessed R. aeschlimannii in Hy. turanicum, as well as Anaplasma sp. and Ehrlichia sp. in both Hy. turanicum and Hy. asiaticum. Notably, it records the coinfection of all these three bacterial species in Hy. turanicum. This study may help to understand the identity, molecular epidemiology, and geographic distribution of Hyalomma ticks and associated pathogens. The findings of the present study indicate that public awareness and surveillance of tick-borne diseases in the region are needed, which may mitigate the risks to public health and livestock.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/supplementary material.
Ethics statement
The animal studies were approved by the Advanced Studies and Research Board (REG-ACAD-ASRB-55/23/035) of the Government College University, Lahore, Pakistan. All animal or herds owners were verbally informed, and permission was taken from them. The studies were conducted in accordance with the local legislation and institutional requirements. Written informed consent was obtained from the owners for the participation of their animals in this study.
Author contributions
ZU: Formal analysis, Investigation, Methodology, Writing – original draft, Writing – review & editing. IL: Formal analysis, Investigation, Methodology, Software, Writing – original draft, Writing – review & editing. MK: Investigation, Methodology, Validation, Visualization, Writing – original draft, Writing – review & editing. AbdA: Funding acquisition, Investigation, Methodology, Project administration, Writing – original draft, Writing – review & editing. MA: Funding acquisition, Investigation, Methodology, Project administration, Resources, Writing – original draft, Writing – review & editing. DA: Formal analysis, Investigation, Methodology, Writing – original draft, Writing – review & editing. TT: Funding acquisition, Investigation, Methodology, Resources, Writing – original draft, Writing – review & editing. AbiA: Conceptualization, Funding acquisition, Investigation, Methodology, Project administration, Resources, Supervision, Writing – original draft, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. The authors acknowledge the financial support provided by the Higher Education Commission (HEC), Pakistan, Pakistan Science Foundation (PSF). The researchers supporting project number (RSP2024R494), King Saud University, Riyadh, Saudi Arabia. This work was supported by JSPS KAKENHI Grant Numbers JP20KK0154, JP22H02522, JP23K23787, JSPS Bilateral Program Grant Number JPJSBP120239937, SHONOGI INFECTIOUS DISEASE RESEARCH PROMOTION FOUNDATION, Japan Association for Livestock New Technology Research Foundation, and Kieikai Research Foundation.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Footnotes
References
1. Ali, A, Zahid, H, Zeb, I, Tufail, M, Khan, S, Haroon, M, et al. Risk factors associated with tick infestations on equids in Khyber Pakhtunkhwa, Pakistan, with notes on Rickettsia massiliae detection. Parasit Vectors. (2021) 14:363–12. doi: 10.1186/s13071-021-04836-w
2. Cvek, M, Broznić, D, Puškadija, D, Blagonić, B, Kirin, I, Pustijanac, E, et al. Investigation and spatial distribution of hard ticks by geographical information system (gis) in the region of Istria, Croatia. J Appl Sci. (2023) 13:9483. doi: 10.3390/app13169483
3. Younas, M, Ashraf, K, Rashid, MI, Ijaz, M, Suleman, M, and Chohan, TA. Expression and purification of recombinant multi-epitope protein of Rhipicephalus microplus tick and its antigenicity in the rabbit model. Pak Vet J. (2023):43. doi: 10.29261/pakvetj/2023.086
4. Hekimoglu, O, and Ozer, AN. Distribution and phylogeny of Hyalomma ticks (Acari: Ixodidae) in Turkey. Exp Appl Acarol. (2017) 73:501–19. doi: 10.1007/s10493-017-0192-0
5. Ali, A, Khan, MA, Zahid, H, Yaseen, PM, Qayash Khan, M, Nawab, J, et al. Seasonal dynamics, record of ticks infesting humans, wild and domestic animals and molecular phylogeny of Rhipicephalus microplus in Khyber Pakhtunkhwa Pakistan. Front Physiol. (2019) 10:793. doi: 10.3389/fphys.2019.00793
6. Alam, S, Khan, M, Alouffi, A, Almutairi, MM, Ullah, S, Numan, M, et al. Spatio-temporal patterns of ticks and molecular survey of Anaplasma marginale, with notes on their phylogeny. Microorganisms. (2022) 10:1663. doi: 10.3390/microorganisms10081663
7. Ullah, S, Alouffi, A, Almutairi, MM, Islam, N, Rehman, G, Ul Islam, Z, et al. First report of Rickettsia conorii in Hyalomma kumari ticks. Animals. (2023) 13:1488. doi: 10.3390/ani13091488
8. Apanaskevich, DA, Schuster, AL, and Horak, IG. The genus Hyalomma: VII. Redescription of all parasitic stages of H.(Euhyalomma) dromedarii and H.(E.) schulzei (Acari: Ixodidae). J Med Entomol. (2008) 45:817–31. doi: 10.1093/jmedent/45.5.817
9. Sands, AF, Apanaskevich, DA, Matthee, S, Horak, IG, Harrison, A, Karim, S, et al. Effects of tectonics and large scale climatic changes on the evolutionary history of Hyalomma ticks. Mol Phyl Evol. (2017) 114:153–65. doi: 10.1016/j.ympev.2017.06.002
10. Guglielmone, AA, Nava, S, and Robbins, RG. Geographic distribution of the hard ticks (Acari: Ixodida: Ixodidae) of the world by countries and territories. Zootaxa. (2023) 5251:1–274. doi: 10.11646/zootaxa.5251.1.1
11. Apanaskevich, DA, and Horak, IG. The genus Hyalomma Koch, 1844. IX. Redescription of all parasitic stages of H.(Euhyalomma) impeltatum Schulze & Schlottke, 1930 and H.(E.) somalicum Tonelli Rondelli, 1935 (Acari: Ixodidae). Syst Parasitol. (2009) 73:199–218. doi: 10.1007/s11230-009-9190-x
12. Apanaskevich, DA, and Horak, IG. The genus Hyalomma Koch, 1844: V. Re-evaluation of the taxonomic rank of taxa comprising the H.(Euhyalomma) marginatum Koch complex of species (Acari: Ixodidae) with redescription of all parasitic stages and notes on biology. Int J Acarol. (2008) 34:13–42. doi: 10.1080/01647950808683704
13. Apanaskevich, DA, Filippova, NA, and Horak, IG. The genus Hyalomma Koch, 1844. X. Redescription of all parasitic stages of H.(Euhyalomma) scupense Schulze, 1919 (= H. Detritum Schulze)(Acari: Ixodidae) and notes on its biology. Folia Parasitol (Praha). (2010) 57:69–78. doi: 10.14411/fp.2010.009
14. Apanaskevich, D, and Horak, I. The genus Hyalomma Koch, 1844. I. Reinstatement of Hyalomma (Euhyalomma) glabrum Delpy, 1949 (Acari, Ixodidae) as a valid species with a redescription of the adults, the first description of its immature stages and notes on its biology. Onderstepoort J Ve. 73:1–12. doi: 10.4102/ojvr.v73i1.164
15. Apanaskevich, DA, and Horak, IG. The genus Hyalomma. XI. Redescription of all parasitic stages of H.(Euhyalomma) asiaticum (Acari: Ixodidae) and notes on its biology. Exp Appl Acarol. (2010) 52:207–20. doi: 10.1007/s10493-010-9361-0
16. Khan, M, Islam, N, Khan, A, Islam, ZU, Muñoz-Leal, S, Labruna, MB, et al. New records of Amblyomma gervaisi from Pakistan, with detection of a reptile-associated borrelia sp. Ticks Tick-Borne Dis. (2022) 13:102047. doi: 10.1016/j.ttbdis.2022.102047
17. Jabeen, F, Mushtaq, M, Qayyum, M, Hasan, M, Zafar, MA, Riaz, A, et al. Tick taxonomy and nucleotide sequence analysis by internal transcribed spacer 2 (ITS 2) in large ruminants of Pothohar, Pakistan. Pak Vet J. (2022) 42:554–60. doi: 10.29261/pakvetj/2022.063
18. Davison, HR, Pilgrim, J, Wybouw, N, Parker, J, Pirro, S, Hunter-Barnett, S, et al. Genomic diversity across the rickettsia and ‘Candidatus Megaira’genera and proposal of genus status for the Torix group. Nat Commun. (2022) 13:2630. doi: 10.1038/s41467-022-30385-6
19. Hussain, S, Saqib, M, Ashfaq, K, and Sindhu, ZUD. First molecular evidence of Coxiella burnetii in ticks collected from dromedary camels in Punjab, Pakistan. Pak Vet J. (2021):10. doi: 10.29261/pakvetj/2021.073
20. Battilani, M, De Arcangeli, S, Balboni, A, and Dondi, F. Genetic diversity and molecular epidemiology of Anaplasma. Infect Genet Evol. (2017) 49:195–211. doi: 10.1016/j.meegid.2017.01.021
21. Barradas, PF, Mesquita, JR, Ferreira, P, Gärtner, F, Carvalho, M, Inácio, E, et al. Molecular identification and characterization of rickettsia spp. and other tick-borne pathogens in cattle and their ticks from Huambo, Angola. Ticks Tick-Borne dis. (2021) 12:101583. doi: 10.1016/j.ttbdis.2020.101583
22. Parola, P, Paddock, CD, and Raoult, D. Tick-borne rickettsioses around the world: emerging diseases challenging old concepts. Clin Microbiol Rev. (2005) 18:719–56. doi: 10.1128/CMR.18.4.719-756.2005
23. Yu, Z, Wang, H, Wang, T, Sun, W, Yang, X, and Liu, J. Tick-borne pathogens and the vector potential of ticks in China. Parasites Vectors. (2015) 8:24–8. doi: 10.1186/s13071-014-0628-x
24. de la Fuente, J, Ghosh, S, Lempereur, L, Garrison, A, Sprong, H, Lopez-Camacho, C, et al. Interventions for the control of Crimean-Congo hemorrhagic fever and tick vectors. npj Vaccines. (2024) 9:181. doi: 10.1038/s41541-024-00970-5
25. Karim, S, Budachetri, K, Mukherjee, N, Williams, J, Kausar, A, Hassan, MJ, et al. A study of ticks and tick-borne livestock pathogens in Pakistan. PLoS Negl Trop Dis. (2017) 11:e0005681. doi: 10.1371/journal.pntd.0005681
26. Khan, M, Almutairi, MM, Alouffi, A, Tanaka, T, Chang, S-C, Chen, C-C, et al. Molecular evidence of borrelia theileri and closely related borrelia spp. in hard ticks infesting domestic animals. Front Vet Sci. (2023) 10:1297928. doi: 10.3389/fvets.2023.1297928
27. Ullah, Z, Khan, M, Liaqat, I, Kamran, K, Alouffi, A, Almutairi, MM, et al. Unveiling misconceptions among small-scale farmers regarding ticks and tick-borne diseases in Balochistan, Pakistan. J Vet Sci. (2024) 11:497. doi: 10.3390/vetsci11100497
28. Sambrook, J. Molecular cloning: a laboratory manual Cold Spring Habor Laboratory USA, New York. (1989).
29. Altschul, SF, Gish, W, Miller, W, Myers, EW, and Lipman, DJ. Basic local alignment search tool. J Mol Biol. (1990) 215:403–10. doi: 10.1016/S0022-2836(05)80360-2
30. Thompson, JD, Higgins, DG, and Gibson, TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. (1994) 22:4673–80. doi: 10.1093/nar/22.22.4673
31. Hall, T, Biosciences, I, and Carlsbad, C. BioEdit: an important software for molecular biology. GERF Bull Biosci. (2011) 2:60–1.
32. Kumar, S, Stecher, G, Li, M, Knyaz, C, and Tamura, K. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. (2018) 35:1547–9. doi: 10.1093/molbev/msy096
33. Edgar, RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. (2004) 32:1792–7. doi: 10.1093/nar/gkh340
34. Fao, F. Food and agriculture organization of the United Nations. Rome (2018):403. Available at: http://faostat.fao.org (Accessed July 11 2024).
35. Tila, H, Khan, M, Almutairi, MM, Alouffi, A, Ahmed, H, Tanaka, T, et al. First report on detection of Hepatozoon ayorgbor in Rhipicephalus haemaphysaloides and Hepatozoon colubri in Haemaphysalis sulcata and Hyalomma anatolicum: risks of spillover of Hepatozoon spp. from wildlife to domestic animals. Front Vet Sci. (2023) 10:1255482. doi: 10.3389/fvets.2023.1255482
36. Lv, J, Wu, S, Zhang, Y, Chen, Y, Feng, C, Yuan, X, et al. Assessment of four DNA fragments (COI, 16S rDNA, ITS2, 12S rDNA) for species identification of the Ixodida (Acari: Ixodida). Parasit Vectors. (2014) 7:1–11. doi: 10.1186/1756-3305-7-93
37. Mans, BJ, Chitimia-Dobler, L, Pienaar, R, de Castro, M, Khan, M, Almutairi, MM, et al. Mitochondrial genome and nuclear ribosomal RNA analysis place Alveonasus lahorensis within the Argasinae and suggest that the genus Alveonasus is paraphyletic. Parasitology. (2024):1–10. doi: 10.1017/S0031182024000441
38. Uiterwijk, M, Ibáñez-Justicia, A, van de Vossenberg, B, Jacobs, F, Overgaauw, P, Nijsse, R, et al. Imported Hyalomma ticks in the Netherlands 2018–2020. Parasit Vectors. (2021) 14:244. doi: 10.1186/s13071-021-04738-x
39. Parola, P, Paddock, CD, Socolovschi, C, Labruna, MB, Mediannikov, O, Kernif, T, et al. Update on tick-borne rickettsioses around the world: a geographic approach. Clin Microbiol Rev. (2013) 26:657–702. doi: 10.1128/CMR.00032-13
40. Parola, P, and Raoult, D. Ticks and tickborne bacterial diseases in humans: an emerging infectious threat. Clin Infect Dis. (2001) 32:897–928. doi: 10.1086/319347
41. Roux, V, and Raoult, D. Phylogenetic analysis of members of the genus rickettsia using the gene encoding the outer-membrane protein rOmpB (ompB). Int J Syst Evol Microbiol. (2000) 50:1449–55. doi: 10.1099/00207713-50-4-1449
42. Labruna, MB, Whitworth, T, Horta, MC, Bouyer, DH, McBride, JW, Pinter, A, et al. Rickettsia species infecting Amblyomma cooperi ticks from an area in the state of Sao Paulo, Brazil, where Brazilian spotted fever is endemic. J Clin Microbiol. (2004) 42:90–8. doi: 10.1128/JCM.42.1.90-98.2004
43. Majid, A, Almutairi, MM, Alouffi, A, Tanaka, T, Yen, T-Y, Tsai, K-H, et al. First report of spotted fever group Rickettsia aeschlimannii in Hyalomma turanicum, Haemaphysalis bispinosa, and Haemaphysalis montgomeryi infesting domestic animals: updates on the epidemiology of tick-borne Rickettsia aeschlimannii. Front Microbiol. (2023) 14:1283814. doi: 10.3389/fmicb.2023.1283814
44. Ghafar, A, Cabezas-Cruz, A, Galon, C, Obregon, D, Gasser, RB, Moutailler, S, et al. Bovine ticks harbour a diverse array of microorganisms in Pakistan. Parasit Vectors. (2020) 13:1–15. doi: 10.1186/s13071-019-3862-4
45. Aneela, A, Almutairi, MM, Alouffi, A, Ahmed, H, Tanaka, T, da Silva, VI, et al. Molecular detection of Rickettsia hoogstraalii in Hyalomma anatolicum and Haemaphysalis sulcata: updated knowledge on the epidemiology of tick-borne Rickettsia hoogstraalii. J Vet Sci. (2023) 10:605. doi: 10.3390/vetsci10100605
46. Dahlgren, FS, Heitman, KN, Drexler, NA, Massung, RF, and Behravesh, CB. Human granulocytic anaplasmosis in the United States from 2008 to 2012: a summary of national surveillance data. Am J Trop Med Hyg. (2015) 93:66–72. doi: 10.4269/ajtmh.15-0122
47. Abbas, SN, Ijaz, M, Abbas, RZ, Saleem, MH, and Mahmood, AK. Molecular characterization, risk factor analysis and hematological alterations associated with Anaplasma phagocytophilum in domestic cats of Pakistan. Pak Vet J. (2023) 43:493–9. doi: 10.29261/pakvetj/2023.082
48. Pourhossein, B, Irani, AD, and Mostafavi, E. Major infectious diseases affecting the Afghan immigrant population of Iran: a systematic review and meta-analysis. Epidemiol Health. (2015):37. doi: 10.4178/epih/e2015002
49. Mangold, A, Bargues, M, and Mas-Coma, S. Mitochondrial 16S rDNA sequences and phylogenetic relationships of species of Rhipicephalus and other tick genera among Metastriata (Acari: Ixodidae). J Parasitol Res. (1998) 84:478–84. doi: 10.1007/s004360050433
50. Folmer, R, Nilges, M, Folkers, P, Konings, R, and Hilbers, C. A model of the complex between single-stranded DNA and the single-stranded DNA binding protein encoded by gene V of filamentous bacteriophage M13. J Mol Biol. (1994) 240:341–57. doi: 10.1006/jmbi.1994.1449
51. Regnery, RL, Spruill, CL, and Plikaytis, B. Genotypic identification of rickettsiae and estimation of intraspecies sequence divergence for portions of two rickettsial genes. J Bacteriol. (1991) 173:1576–89. doi: 10.1128/jb.173.5.1576-1589.1991
52. Inokuma, H, Raoult, D, and Brouqui, P. Detection of Ehrlichia platys DNA in brown dog ticks (Rhipicephalus sanguineus) in Okinawa Island, Japan. J Clin Microbiol. (2000) 38:4219–21. doi: 10.1128/JCM.38.11.4219-4221.2000
53. PES. (2024). Available at: https://finance.gov.pk/survey_2024.html (Accessed 16 July 2024).
Keywords: Hyalomma turanicum, Hyalomma asiaticum, Rickettsia aeschlimannii, Anaplasma sp., Ehrlichia sp., coinfection, phylogeny
Citation: Ullah Z, Liaqat I, Khan M, Alouffi A, Almutairi MM, Apanaskevich DA, Tanaka T and Ali A (2024) Investigation of Hyalomma turanicum and Hyalomma asiaticum in Pakistan, with notes on the detection of tickborne Rickettsiales. Front. Vet. Sci. 11:1500930. doi: 10.3389/fvets.2024.1500930
Edited by:
Mughees Aizaz Alvi, University of Agriculture, Faisalabad, PakistanReviewed by:
Hafiz Ishfaq Ahmad, University of Veterinary and Animal Sciences, PakistanJose Reck, Desidério Finamor Veterinary Research Institute (IPVDF), Brazil
Copyright © 2024 Ullah, Liaqat, Khan, Alouffi, Almutairi, Apanaskevich, Tanaka and Ali. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Abid Ali, uop_ali@yahoo.com; Tetsuya Tanaka tetsuya.tanaka.a3@tohoku.ac.jp
 Zafar Ullah
Zafar Ullah