- 1Avian Disease Laboratory, College of Veterinary Medicine, Konkuk University, Seoul, Republic of Korea
- 2Wildlife Disease Research Team, National Institute of Wildlife Disease Control and Prevention, Gwangju, Republic of Korea
- 3Zoonotic Disease Research Center, Konkuk University, Seoul, Republic of Korea
- 4Birdflu Veterinarian LLC, Watkinsville, GA, United States
- 5Wildlife Health Laboratory, College of Veterinary Medicine, Konkuk University, Seoul, Republic of Korea
1 Introduction
Highly pathogenic avian influenza (HPAI) subtype H5Nx viruses of the A/Goose/Guangdong/1/1996 (Gs/Gd) lineage have caused substantial economic losses in the poultry industry and represent a significant public health concern (1). Since its first detection in 1996, it has diverged into 10 genetically distinct hemagglutinin (HA) clades (0–9) and subclades (2). Of these, the clade 2.3.4.4b HPAI H5N1 viruses have caused outbreaks in wild birds, poultry, and mammals in broad geographical regions including Asia, Europe, Africa, North America, South America, and Antarctica since 2020 (3–5). In particular, wild aquatic birds have played a key role in the maintenance and global spread of clade 2.3.4.4b HPAI viruses (6–8).
In South Korea, the clade 2.3.4.4b HPAI H5N1 viruses caused multiple outbreaks in wild birds, including 67 reported cases from October 2021 to March 2022, and 174 reported cases from October 2022 to March 2023 (9, 10). During October 2022-March 2023, the HPAI H5N1 virus was detected in 22 wild bird species (11). Particularly, there was a mass die-off event of 221 hooded cranes (Grus monacha) in Suncheon Bay, South Korea during November–December 2022. We conducted whole genome sequencing and comparative phylogenetic analysis of the isolates from hooded cranes. To elucidate the histopathological changes induced by clade 2.3.4.4b H5N1 HPAI viruses in hooded cranes, histopathological evaluation and immunohistochemistry were conducted on a hooded crane (sample no. 22WC-042) found dead in Suncheon Bay during November 2022 (Supplementary Figure S1).
2 Methods
2.1 Virus isolation and genome sequencing
Unprecedentedly, 221 hooded cranes (Grus monacha) were found dead in Suncheon Bay, South Korea during November–December 2022. The National Institute of Wildlife Disease Control and Prevention (NIWDC) of South Korea conducted virus isolation and collected internal organs from 211 carcasses to investigate the viral distribution. Oral and cloacal swab samples obtained from birds were placed in phosphate-buffered saline (PBS) containing 400 mg/mL antibiotic-antimycotic (Gibco, Grand Island, NY, USA) and thoroughly homogenized by vortexing for 1 min. For the isolation and identification of the virus, supernatant of swab samples was filtered using a 0.45 μm Syringe Filter. Filtered samples were inoculated into the allantoic cavities of 9–11-day-old specific pathogen-free embryonated eggs. After incubation for 72 h at 37°C, allantoic fluid was collected, and RNA extraction was performed using the Maxwell® RSC simply RNA Tissue Kit (Promega, Madison, WI, USA) following the manufacturer's instructions. Subsequently, the samples were screened with the matrix (M) and H5 genes using real-time reverse transcription-PCR (rRT-PCR) (12). Complementary DNA synthesis was carried out using the SuperScript III First-Strand Synthesis system (Invitrogen, Carlsbad, CA, USA), followed by PCR targeting the hemagglutinin (HA) and neuraminidase (NA) genes to determine pathogenicity and subtypes (13). Among the 194 H5N1 HPAI virus-positive samples, 15 samples representing each outbreak week underwent next-generation sequencing (NGS) as previously described (14).
2.2 Phylogenetic analysis
The viral strains with the highest similarity to isolated H5N1 HPAIVs were identified using Basic Local Alignment Search Tool (BLAST) of the Global Initiative on Sharing all Avian Influenza Data (GISAID) database. For phylogenetic analysis, reference sequences were obtained from GISAID database. To select reference sequences, representative sequences were chosen by extracting clusters containing genome sequences of viruses from hooded cranes, and the ElimDupes program was used to exclude sequences showing more than 99.8% similarity (https://www.hiv.lanl.gov/content/sequence/elimdupesv2/elimdupes.html). The reference sequences and sequences isolated from the hooded crane were aligned using MAFFT (https://mafft.cbrc.jp/alignment/software/). Maximum-likelihood (ML) trees for 8 genes were constructed using RAxML v8.0 (15) with general time-reversible + Gamma model, 1,000 bootstrap replicates. Genotypes were determined based on criteria from a previous study (11).
2.3 Histopathology and immunohistochemistry
Among the specimens confirmed to be infected with clade 2.3.4.4b H5N1 HPAI virus, necropsies were conducted on a hooded crane (sample no. 22WC-042) collected on 17 November 2022 (Supplementary Figure S1). Organs including trachea, liver, spleen, kidney, pancreas, and cecal tonsils were fixed in 4% paraformaldehyde, embedded in paraffin, and sectioned into 5 μm-thick slices. These sections were subsequently stained with hematoxylin and eosin (H&E) for histopathological analysis and evaluated according to Landmann et al. (16). Immunohistochemical staining was carried out to detect the presence of the influenza virus antigen. The tissue sections were first incubated overnight at 4°C with goat anti-influenza A virus antibody (25 μg/ml, Invitrogen). After the overnight incubation, the sections were then incubated for 2 h at room temperature with biotinylated horse anti-goat IgG antibody (Vector Laboratories, Inc., USA). This was followed by incubation with horseradish peroxidase-conjugated streptavidin (Vector Laboratories, Inc.) for 1 h at room temperature. The positive signal indicating the presence of the virus was visualized using either diaminobenzidine (DAB) or Vector Red1 (Vector Laboratories, Inc.) as a substrate, and the sections were counterstained with methyl green for better contrast.
3 Descriptive results
Out of 221 dead hooded cranes, 194 tested positive for clade 2.3.4.4b H5N1 HPAI, and NGS was performed on 15 of the isolates. ML phylogenies analysis of the eight genes showed that all genes of H5N1 HPAI viruses isolated from hooded cranes in Korea and Japan clustered together and showed high sequence identities, indicating their close genetic relatedness (Supplementary Figure S2). Genotype of H5N1 HPAI viruses were analyzed according to the genotypic criteria established in a previous study (11). All viruses were classified as Kor22-23C genotype, except the A/hooded crane/Korea/22WC046-2/2022 virus which was Kor22-23B genotype, indicating that Kor22-23C was the dominant genotype among the hooded cranes (Table 1). The A/Hooded crane/Korea/WC042/2022 (H5N1) virus [22WC-042], isolated from the necropsied individuals, also showed 100% nucleotide sequence identity with the virus isolated from hooded cranes in Kagoshima, Japan, according to BLAST results (Supplementary Table S1), suggesting virus transmission between Korea and Japan.
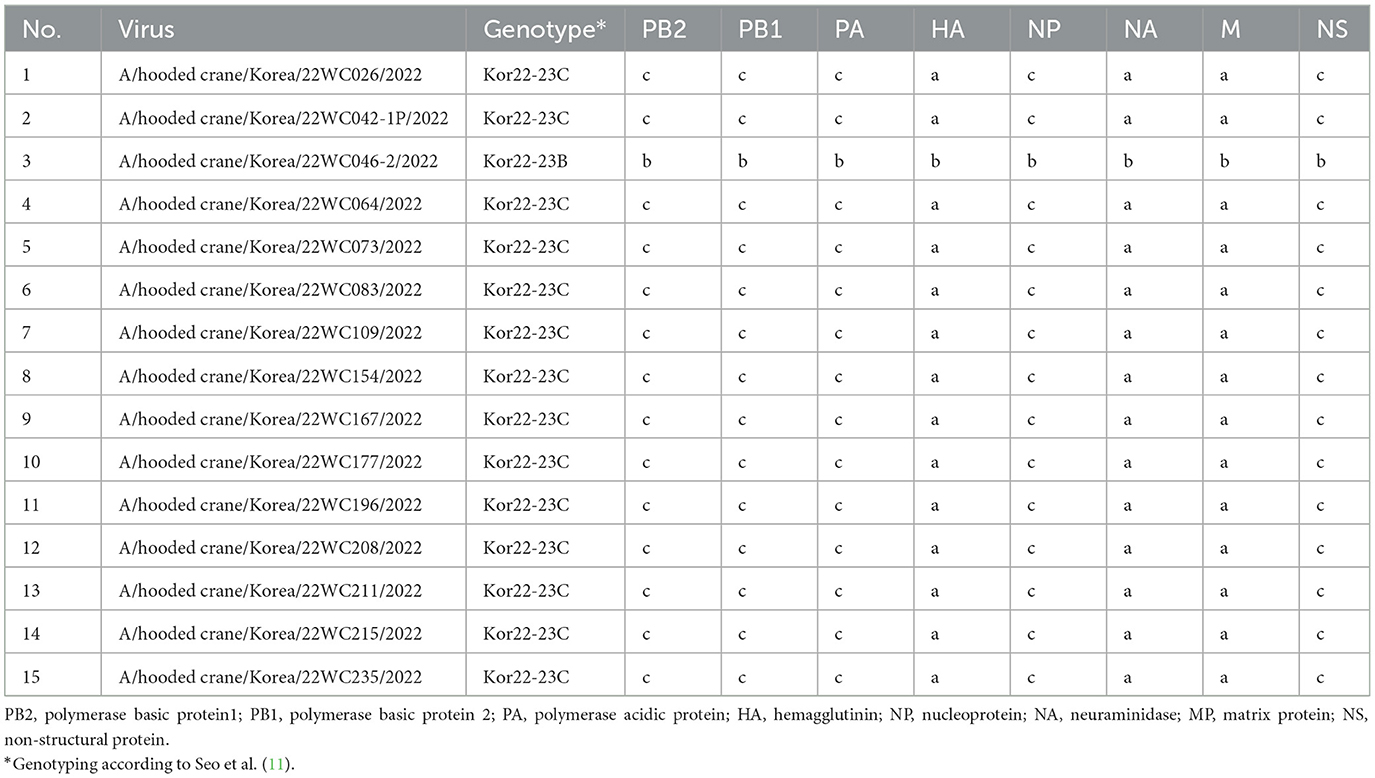
Table 1. Genotypes of clade 2.3.4.4b H5N1 highly pathogenic avian influenza viruses isolated from the hooded crane carcasses in Suncheon Bay, Korea.
On November 1, 2022, the first death of a hooded crane due to clade 2.3.4.4b H5N1 HPAI virus was reported in the Izumi Plain in Japan. Subsequently, on November 13, 2022, the first death of a hooded crane caused by the same virus strain was reported in Suncheon Bay, South Korea. Both Suncheon Bay and Izumi Plain, where the deaths were reported, are known as major wintering sites for hooded cranes in East Asia and lie along the same migratory route (Supplementary Figure S1) (17, 18). Given that at least 3,000 hooded cranes were confirmed to have migrated between Japan and Korea during November 2022 (19), it is highly likely that the virus was introduced during migration of hooded crane population. Taking into account the mass mortality of 1,476 hooded cranes in the Izumi Plain during the same period, it represents the death of 10% of the total hooded crane population (20).
The histopathologic and immunohistochemical findings were consistent with previously reported HPAI virus infections in aquatic birds including multiple organs exhibiting necrotic and inflammatory lesions with corresponding positive immunostaining for influenza A virus within these areas, which indicates systemic infection (Figure 1) (21, 22). The trachea showed degeneration and necrosis of epithelial cells, accompanied by the infiltration of inflammatory cells such as lymphocytes and macrophages. Immunostaining was positive in some epithelial cells, including the brush border. In the pancreas, localized inflammatory cell infiltration, and vacuolar degeneration of acinar glandular epithelium was observed with immunostaining positivity acinar epithelium. The spleen showed focal hemorrhage, lymphoid depletion, and multifocal necrotic foci. Nuclear fragmentation and ghost nuclei were observed within the necrotic foci, with strong immunostaining. In the kidney, necrosis of tubules was observed with immunostaining of epithelial cells of the affected tubules. In the cecal tonsil, lymphoid depletion was observed in the region of the germinal center and immunostaining was strongly positive in the affected area. No lesions or immunostaining were identified in the liver. A limitation of our histopathology data is the exclusion of brain tissue, which precludes the evidence of potential neurotropism.
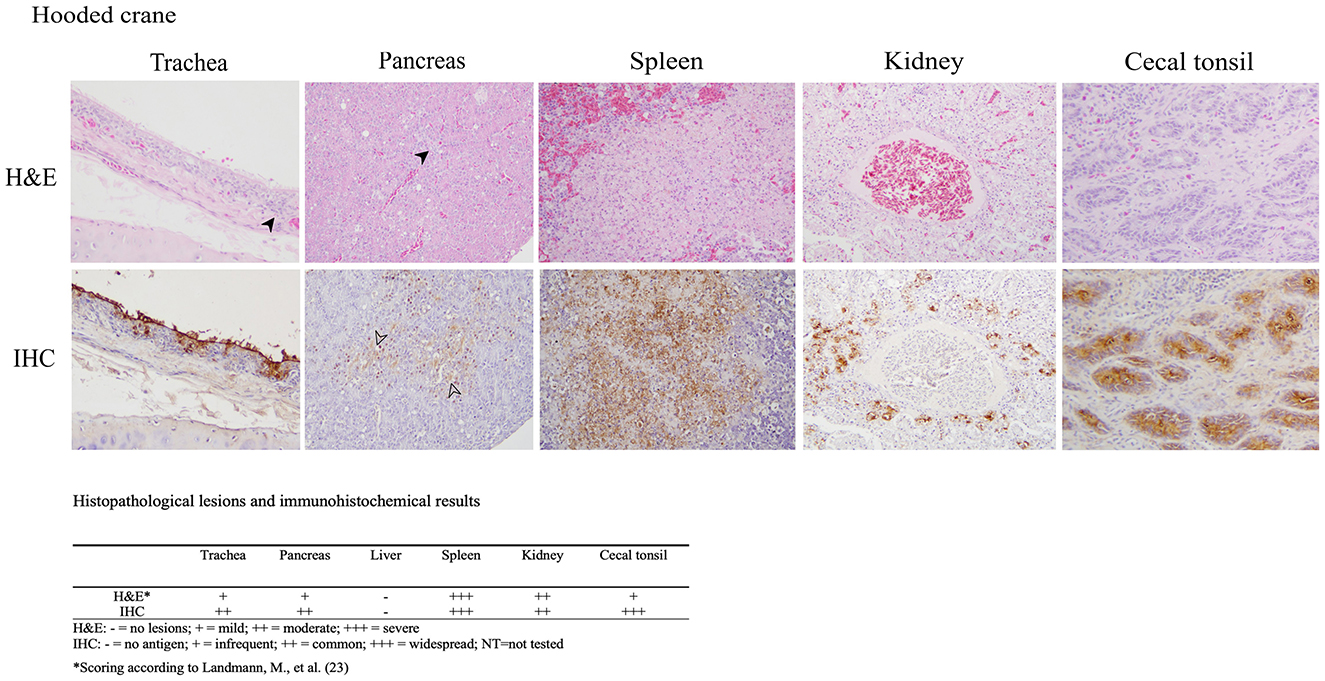
Figure 1. Results of H&E staining and immunohistochemical (IHC) testing in tissues of hooded crane (Grus monacha). Virus staining is the brown coloration. Trachea (magnification x200); infiltration of inflammatory cells (arrowhead), viral antigen in epithelial cells. Pancreas (magnification x200); inflammation, vacuolar degeneration (arrowhead). Spleen (magnification x400); necrotic foci with strong immunostaining positivity. Kidney (magnification x200); nuclear fragmentation and condensed nuclei in necrostic tubular epithelium with viral antigen in necrotic epithelium. Cecal tonsil (magnification x200); lymphoid depletion and viral antigen in the affected area. The lesion and immunostaining positivity scores are summarized under the images.
4 Conclusion
As the evolution and spread of clade 2.3.4.4b H5Nx HPAI virus continues, the host range and virulence are continuously changing. Unlike Anseriformes and Charadriiforms, hooded crane is not a previously reported natural host of influenza A virus (23), but we found mass die-off of this species and evidence of systemic infection. The genetic and pathological data established in this study would be useful as reference data for genomic surveillance and pathobiological study of HPAI viruses. In addition, hooded cranes are designated as a vulnerable species by the International Union for the Conservation of Nature (IUCN), raising concern not only for wide spread of HPAI but also for wildlife conservation. There is a need for a cross-border cooperation effort on active surveillance of HPAI in wild birds to monitor the evolution and spread of HPAI.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary material.
Author contributions
Y-RS: Writing – original draft. S-HL: Writing – review & editing. SJ: Writing – review & editing. HC: Writing – review & editing. DK: Writing – review & editing. D-JK: Writing – review & editing. Y-JS: Writing – review & editing. HJ: Writing – review & editing. SL: Writing – review & editing. C-SS: Writing – review & editing. DS: Writing – review & editing. D-HL: Writing – original draft, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This research was financially supported by a grant from the National Institute of Wildlife Disease Control and Prevention (NIWDC) (grant number 2023-016) and the Ministry of Environment, Republic of Korea.
Acknowledgments
We gratefully acknowledge all data contributors, i.e., the authors and their originating laboratories responsible for obtaining the specimens, and their submitting laboratories for generating the genetic sequence and metadata and sharing via the GISAID Initiative, on which this research is based.
Conflict of interest
DS was employed by Birdflu Veterinarian LLC, a private poultry veterinary practice.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fvets.2024.1499440/full#supplementary-material
Supplementary Figure 1 | The geographical coordinates (latitude and longitude) of the sample collection sites. Suncheon Bay and Izumi Plain are wintering sites for hooded cranes and are located on the same migration route.
Supplementary Figure 2 | Maximumlikelihood tree of (a) PB2, (b) PB1, (c) PA, (d) HA, (e) NP, (f) NA, (g) M, and (h) NS genes. Each color range represents different internal gene types. Viruses from hooded cranes in Korea are marked with blue dots, while those from hooded cranes in Japan are marked with red dots.
References
1. Kwon JH, Bertran K, Lee DH, Criado MF, Killmaster L, Pantin-Jackwood MJ, et al. Diverse infectivity, transmissibility, and pathobiology of clade 2.344 H5Nx highly pathogenic avian influenza viruses in chickens. Emerg Microbes Infect. (2023) 12:2218945. doi: 10.1080/22221751.2023.2218945
2. Health/Food WHOWOfA Group AOHNEW. Revised and updated nomenclature for highly pathogenic avian influenza A (H5N1) viruses. Influenza Other Respirat Virus. (2014) 8:384–8. doi: 10.1111/irv.12230
3. Adlhoch C, Fusaro A, Gonzales JL, Kuiken T, Melidou A, Mirinavičiute G, et al. Avian influenza overview April - June 2023. EFSA J. (2023) 21:e08191. doi: 10.2903/j.efsa.2023.8191
4. Venkatesan P. Avian influenza spillover into mammals. Lancet Microbe. (2023) 4:e492. doi: 10.1016/S2666-5247(23)00173-8
5. Bennison A, Byrne AM, Reid SM, Lynton-Jenkins JG, Mollett B, Silva DD, et al. Detection and spread of high pathogenicity avian influenza virus H5N1 in the Antarctic Region. bioRxiv [preprint] bioRxiv.2023:2023.11. 23.568045. doi: 10.1101/2023.11.23.568045
6. Gaidet N, Cappelle J, Takekawa JY, Prosser DJ, Iverson SA, Douglas DC, et al. Potential spread of highly pathogenic avian influenza H5N1 by wildfowl: dispersal ranges and rates determined from large-scale satellite telemetry. J Appl Ecol. (2010) 47:1147–57. doi: 10.1111/j.1365-2664.2010.01845.x
7. Bevins SN, Shriner SA, Cumbee JC Jr, Dilione KE, Douglass KE, Ellis JW, et al. Intercontinental movement of highly pathogenic avian influenza A(H5N1) clade 2.3.4.4 virus to the United States, 2021. Emerg Infect Dis. (2022) 28:1006–11. doi: 10.3201/eid2805.220318
8. Claes F, Morzaria SP, Donis RO. Emergence and dissemination of clade 2.344 H5Nx influenza viruses-how is the Asian HPAI H5 lineage maintained. Curr Opin Virol. (2016) 16:158–63. doi: 10.1016/j.coviro.2016.02.005
9. National Institute of Wildlife Disease Control and Prevention RoK. Highly Pathogenic Avian Influenza Outbreak Status in Wild Birds during the Winter Season of '22-'23 2023. Available at: https://www.me.go.kr/niwdc/web/board/read.do?menuId=50&boardMasterId=780&boardCategoryId=&boardId=1593630 (accessed April 28, 2024).
10. National Institute of Wildlife Disease Control and Prevention RoK. Current Situation of Domestic Highly Pathogenic Avian Influenza (HPAI) Outbreaks. (2022). Available at: https://www.me.go.kr/niwdc/web/board/read.do?pagerOffset=20&maxPageItems=10&maxIndexPages=10&searchKey=&searchValue=&menuId=50&orgCd=&boardId=1526750&boardMasterId=780&boardCategoryId=&decorator= (accessed May 20th, 2022).
11. Seo Y-R, Cho AY, Si Y-J, Lee S-I, Kim D-J, Jeong H, et al. Evolution and spread of highly pathogenic avian influenza A (H5N1) clade 2.3. 4.4 b virus in wild birds, South Korea, 2022–2023. Emerg Infect Dis. (2024) 30:299. doi: 10.3201/eid3002.231274
12. Spackman E, Senne D, Bulaga L, Myers T, Perdue M, Garber L, et al. Development of real-time RT-PCR for the detection of avian influenza virus. Avian Dis. (2003) 47:1079–82. doi: 10.1637/0005-2086-47.s3.1079
13. Hoffmann E, Stech J, Guan Y, Webster R, Perez D. Universal primer set for the full-length amplification of all influenza A viruses. Arch Virol. (2001) 146:2275–89. doi: 10.1007/s007050170002
14. Lee D-H. Complete genome sequencing of influenza A viruses using next-generation sequencing. Anim Influenza Virus. (2020) 2123:69–79. doi: 10.1007/978-1-0716-0346-8_6
15. Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. (2014) 30:1312–3. doi: 10.1093/bioinformatics/btu033
16. Landmann M, Scheibner D, Graaf A, Gischke M, Koethe S, Fatola OI, et al. A semiquantitative scoring system for histopathological and immunohistochemical assessment of lesions and tissue tropism in avian influenza. Viruses. (2021) 13:868. doi: 10.3390/v13050868
17. Mi C, Møller AP, Guo Y. Annual spatio-temporal migration patterns of Hooded Cranes wintering in Izumi based on satellite tracking and their implications for conservation. Avian Res. (2018) 9:1–9. doi: 10.1186/s40657-018-0114-9
18. Jinming L, Yongjie W, Fan Y, Zhijun L. Effects of human disturbance on the Hooded Crane (Grus monacha) at stopover sites in northeastern China. Avian Research. (2012) 3:206–16. doi: 10.5122/cbirds.2012.0024
19. Ministry of Environment RoK. Confirmation of 156,000 Birds in 200 Major Wintering Sites for Migratory Birds in December 2022. Available at: https://www.me.go.kr/home/web/board/read.do?menuId=10525&boardMasterId=1&boardCategoryId=39&boardId=1568845 (accessed April 28, 2024).
20. Ministry of Environment RoK. National Institute of Wildlife Disease Control and Prevention Collaborates with Specialized Agencies and Local Governments in Japan for Avian Influenza Response. (2023). Available at: https://www.me.go.kr/home/web/board/read.do?menuId=10525&boardMasterId=1&boardCategoryId=39&boardId=1607490 (accessed April 28, 2024).
21. Spackman E, Pantin-Jackwood MJ, Lee SA, Prosser D. The pathogenesis of a 2022 North American highly pathogenic clade 2.3 44 b H5N1 avian influenza virus in mallards (Anas platyrhynchos). Avian Pathology. (2023) 52:219–28. doi: 10.1080/03079457.2023.2196258
22. Kim H-R, Kwon Y-K, Jang I, Lee Y-J, Kang H-M, Lee E-K, et al. Pathologic changes in wild birds infected with highly pathogenic avian influenza A (H5N8) viruses, South Korea, 2014. Emerg Infect Dis. (2015) 21:775. doi: 10.3201/eid2105.141967
Keywords: highly pathogenic avian influenza virus, H5N1, clade 2.3.4.4b, South Korea, wild bird, hooded crane
Citation: Seo Y-R, Lee S-H, Jeong S, Cho H, Kim D, Kim D-J, Si Y-J, Jeong H, Lee S, Song C-S, Swayne DE and Lee D-H (2024) Genetic and pathological analysis of hooded cranes (Grus monacha) naturally infected with clade 2.3.4.4b highly pathogenic avian influenza H5N1 virus in South Korea in the winter of 2022. Front. Vet. Sci. 11:1499440. doi: 10.3389/fvets.2024.1499440
Received: 20 September 2024; Accepted: 16 October 2024;
Published: 06 November 2024.
Edited by:
Iryna Goraichuk, Agricultural Research Service (USDA), United StatesReviewed by:
Laura Roberts, Western Cape Department of Agriculture, South AfricaValerie Shearn-Bochsler, United States Department of the Interior, United States
Copyright © 2024 Seo, Lee, Jeong, Cho, Kim, Kim, Si, Jeong, Lee, Song, Swayne and Lee. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Dong-Hun Lee, ZG9uZ2h1bmxlZUBrb25rdWsuYWMua3I=
 Ye-Ram Seo
Ye-Ram Seo Sun-Hak Lee
Sun-Hak Lee Sol Jeong2
Sol Jeong2 Chang-Seon Song
Chang-Seon Song Dong-Hun Lee
Dong-Hun Lee