- 1Department of Parasitology, Faculty of Veterinary Medicine, Assiut University, Assiut, Egypt
- 2Department of Zoology, Faculty of Science, Assiut University, Assiut, Egypt
- 3Department of Parasitology, Faculty of Veterinary Medicine, Aswan University, Aswan, Egypt
- 4Department of Medical Parasitology, Faculty of Medicine, Kafrelsheikh University, Kafr El Sheikh, Egypt
- 5Departamento de Sanidad Animal, Grupo de Investigación en Sanidad Animal y Zoonosis (GISAZ), Universidad de Córdoba, Córdoba, Spain
- 6Department of Parasitology, Faculty of Veterinary Medicine, Zagazig University, Zagazig, Egypt
- 7Department of Biology, College of Science, University of Hafr Al Batin, Hafr Al Batin, Saudi Arabia
- 8Department of Biology, College of Science, Princess Nourah bint Abdulrahman University, Riyadh, Saudi Arabia
- 9CIBERINFEC, ISCIII CIBER de Enfermedades Infecciosas, Instituto de Salud Carlos III, Madrid, Spain
- 10Department of Zoonoses, Faculty of Veterinary Medicine, Sohag University, Sohag, Egypt
Introduction: Rodents, thriving in human-altered environments, pose significant public health risks due to their role as reservoirs for numerous zoonotic parasites. Among these, Cryptosporidium spp. are recognized globally as leading causes of waterborne and foodborne diarrheal illnesses in humans. The specific role of fat sand rats (Psammomys obesus) in the transmission of Cryptosporidium spp. in Egypt and the genotypic characteristics of the circulating species in these animals remain poorly understood.
Methods: In this study, a total of 150 individual fat sand rat stool samples were collected from the saline marsh periurban areas of Abu-Rawash, Giza, Egypt. The samples were initially screened for the presence of Cryptosporidium spp. using light and scanning electron microscopy to characterize the parasite’s oocysts. Furthermore, molecular identification and characterization of the parasite were carried out on selected microscopy-positive samples (n = 30) using conventional polymerase chain reaction (PCR) targeting the Cryptosporidium oocyst wall protein (COWP) gene. A subset of these positive samples by PCR was subjected to sequencing, with the resulting sequences deposited in GenBank™ and analyzed through phylogenetic methods.
Results: Conventional microscopy revealed that 46.7% (70/150; 95% CI: 38.7–54.6) of the analyzed stool samples contained structures consistent with Cryptosporidium oocysts. Moreover, the molecular analysis confirmed Cryptosporidium species in DNA from all 30 stool samples previously identified as heavily infected through microscopy. Notably, the phylogenetic analysis identified Cryptosporidium parvum (C. parvum) in the sequenced samples, likely originating from the rats’ native habitats. These identified species have been deposited in GenBank™ under the accession numbers OM817461 (C. parvum FSA-1), OM817462 (C. parvum FSA-2), and OM817463 (C. parvum FSA-3) and revealed closed genetic identity with those species reported from human and other animal species in the same geographic location.
Conclusion: Overall, this study represents the first morphological and genetic identification of C. parvum isolated from fecal samples of fat sand rats trapped from periurban areas in Egypt. These findings provide valuable insights into the potential zoonotic implications of rodents in disease transmission at the national level, offering crucial information for public health awareness campaigns and informing local authorities.
1 Introduction
Cryptosporidium spp., protozoan parasites from the phylum Apicomplexa, are significant contributors to the global incidence of diarrheal diseases in humans and a wide range of animal species (1, 2). These organisms are responsible for cryptosporidiosis, a disease with considerable zoonotic potential. The epidemiological profile of these foodborne and waterborne parasites encompasses a wide range of domestic and wild animals, in addition to humans. Transmission of the infection occurs through consumption of contaminated water, food, or vegetables, as well as through direct contact with infected animals or humans (3, 4). While some infections may be asymptomatic, symptomatic cases often experience prolonged symptoms such as moderate-to-severe diarrhoea, lose or watery stools, stomach cramps, gastrointestinal discomfort, and mild fever (5, 6). It should be stressed that the genus Cryptosporidium comprises multiple pathogenic species, among them C. parvum, which is predominantly associated with human infections. In 2016 alone, acute infections caused by C. parvum led to over 48,000 deaths globally, especially affecting children with diarrhoea (7).
Despite the urgent need, there are currently no commercially available vaccines to protect humans or animals against cryptosporidiosis (8). Effective control strategies require integrated approaches and a One Health perspective to address the interconnected health of humans, animals, and the environment. The conventional method for diagnosing the infection involves microscopically examining stained fecal samples. This approach is widely used due to its simplicity and cost-effectiveness (9). On the other hand, molecular methods, such as genotyping and subtyping, have higher sensitivity and aid in tracing infection sources, understanding pathogen circulation in specific populations and regions, and assessing potential zoonotic spillover (10–12).
Importantly, rodent-borne diseases pose significant global health challenges, often categorized as neglected tropical diseases (13, 14). The transmission of pathogens between rodents and humans is more likely in areas where they cohabit, such as agricultural regions, peri-urban zones, and densely populated urban environments, especially when rodent populations are abundant (14, 15). Urbanization and climate change further exacerbate the spread of diseases carried by rodents, including bacteria, viruses, and parasites. Cryptosporidium spp. are particularly prevalent among rodents, with studies documenting a diverse array of species and genotypes, totalling 14 species and over 57 genotypes (1, 14). Among these, C. parvum is well-known for infecting humans and other mammals, as well as for its extensive genetic diversity within rodent populations (16).
Psammomys obesus (P. obesus), also referred as the fat sand rat, is a diurnal rodent belonging to the family Muridae (17, 18). It is found in North Africa, ranging from Mauritania to Egypt and Sudan, and extends into the Arabian Peninsula (19). This species is commonly predominantly residing in sandy desert habitats, although it can also inhabit rocky areas or saline marshlands. It exhibits a highly selective diet, consuming only the stems and leaves of plants within the Amaranthaceae family (20). It is important to highlight that P. obesus is recognized as can act as reservoir host for numerous zoonotic diseases (14). However, based on existing literature, there is currently no available information regarding the potential role of sand rats in transmitting Cryptosporidium spp. in Egypt. Therefore, our aim was to investigate and identify Cryptosporidium spp. in stool samples from fat sand rats collected from saline marsh areas (Navigations) of the Abu-Rawash, Egypt.
2 Materials and methods
2.1 Ethics statement
The ethical approval was obtained from the Committee of the Faculty of Veterinary Medicine, Assiut University, Egypt (No. 06/ 2042/ 0160).
2.2 Rodent collections
From December 2022 to November 2023, a total of 150 fecal samples were collected from fat sand rats in multiple saline marsh areas (Navigations) across 10 locations in the Abu Rawash area of Giza. Abu Rawash, situated in Giza Governorate, Egypt. This area is part of Egypt’s Western Desert, on the periphery of Greater Cairo. The area features a semi-arid climate with hot summers and mild winters. The map of the sampling studied area is shown in Figure 1. Sand rats were captured using 30 cage traps placed near burrow entrances as described elsewhere (19). These traps were baited with fresh sunflower seeds and either peanut or sesame butter, positioned at sunset, and retrieved early the following morning before sunrise. The captured rats were euthanized using CO2 inhalation as described elsewhere (21), and identified based on established criteria (22, 23), and then transported to the Parasitology Laboratory at Faculty of Veterinary Medicine, Assiut University (Egypt).
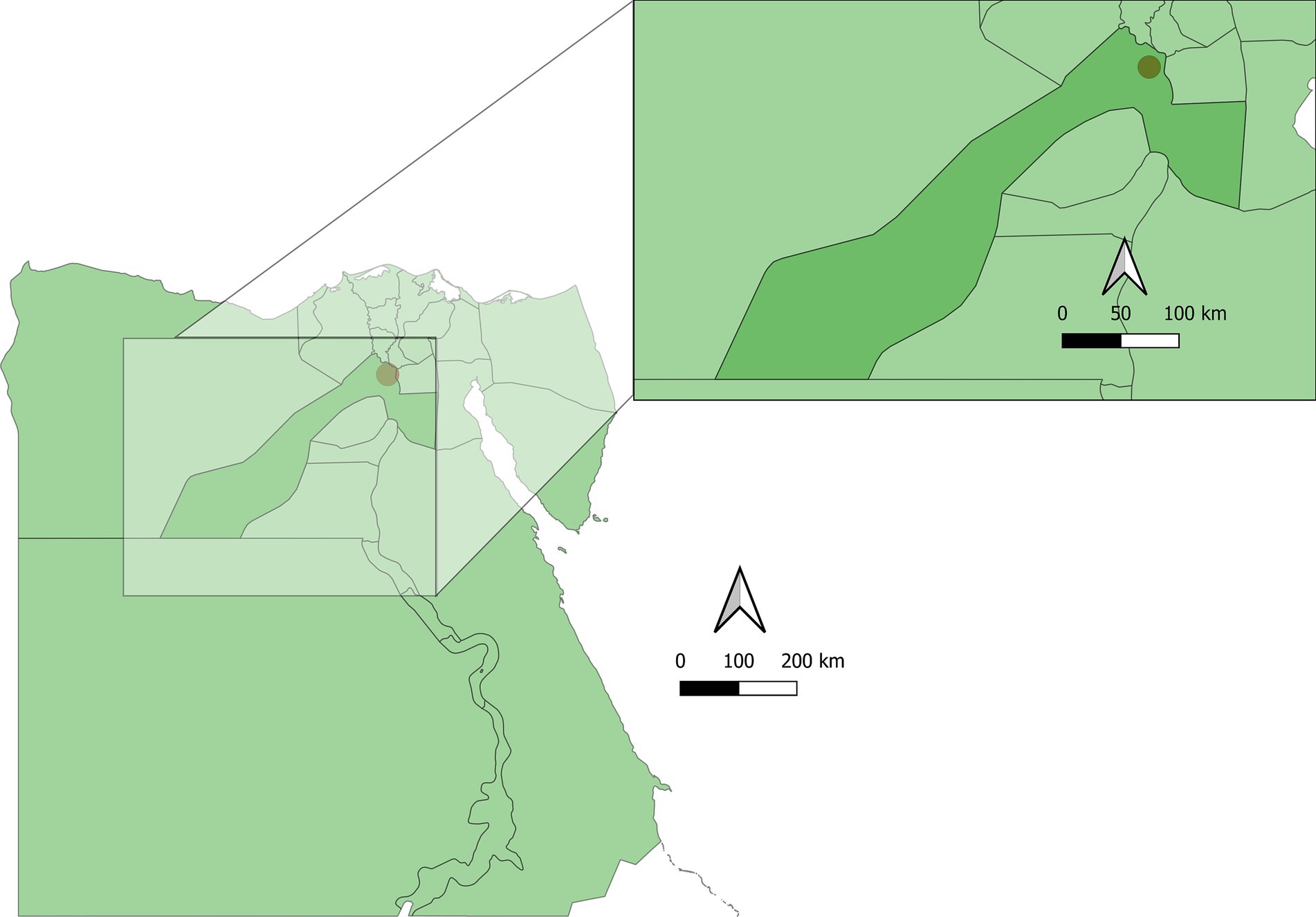
Figure 1. Map of the sampling sites and studied area. GIS files Copyright 2024 Pareto Software, LLC released under the creative commons by 4.0 license, https://simplemaps.com.
2.3 Parasitological investigation
For parasitological examination, fecal samples (approximately 500 mg) were collected directly from the intestine of each rat. Direct examination and sedimentation methods were employed, as previously described (24). Briefly, each sample was promptly processed and examined using routine direct fecal smear techniques with normal saline and Lugol’s iodine preparations. Additionally, fresh or preserved stool specimens (in 10% formalin) underwent concentration via centrifugal sedimentation in formalin-ether and formalin-ethyl acetate solutions, as well as centrifugal flotation in saturated Sheather’s sugar solution. Both the supernatant and sediment were fixed with methanol for 5 min before being smeared onto glass slides and stained using the modified Ziehl Neelsen technique. The samples stained with Ziehl Neelsen were then examined under light microscopy (1,000X magnification).
2.4 Scanning electron microscopy
For scanning electron microscopy, fecal samples are thoroughly mixed in a 10-mL saturated sugar solution (specific gravity 1.2) and then centrifuged at 1050 × g for 2 min (25). After centrifugation, 2 mL of the fluid from the meniscus, which contains the oocysts, is preserved in 70% alcohol until further use (26). Cryptosporidium sp. oocysts isolated from stool samples were then fixed in 5% glutaraldehyde in sodium cacodylate buffer for 1.5 h following the protocol described elsewhere (25, 26). They were then rinsed in distilled water and dehydrated in ethanol. After critical point drying, the samples were affixed onto stubs, coated with either carbon, then subsequently examined using a Joel JSM 35 Scanning Electron Microscope at 20 kV at Assiut University (Egypt).
2.5 Molecular detection
2.5.1 DNA extraction and PCR amplification
Genomic DNA was extracted from 30 heavily infected stool samples, which had tested positive for the parasite via microscopic examination, using the QIAamp DNA Mini Kit (Qiagen, Valencia, CA, USA), according to the manufacturer’s instructions. A nested PCR technique was employed to amplify a specific fragment of the COWP gene following the protocols detailed in previous studies (27, 28). The initial PCR reaction targeted a 769-bp fragment of the COWP gene, using the following primer set: BCOWPF (5’-ACCGCTTCTCAACAACCATCTTGTCCTC-3′) and BCOWPR (5’-CGCACCTGTTCCCACTCAATGTAAACCC-3′). In the secondary reaction, a 553-bp fragment of the COWP gene was amplified using the primers COWP-Forward (5’-GGACTGAAATA CAGGCATTATCTTG-3′) and COWP-Reverse (5′-GTAGATAATGG AAGAGATTGTG-3′), as previously described (29). PCR amplification was carried out in a total volume of 25 μL and the reaction mixture comprised 2.5 μL of template DNA, 1× PCR buffer (Promega, Madison, WI, USA), 250 μM of each dNTP, 1.5 mM MgCl2, 10 pmoles of each primer, and 1.25 U of Taq DNA polymerase (28). The cycling conditions employed for the molecular identification of Cryptosporidium species in this study are provided in Supplementary Table 1. Subsequently, the resulting amplicons were electrophoresed on 1.5% agarose gels stained with RedSafe™ Nucleic Acid Staining solution and images were captured using a gel documentation system.
2.5.2 Sequencing and phylogenetic analyses
PCR products from five positive samples, which displayed distinct and sharp bands, were purified using the QIAquick PCR Product Extraction Kit (Qiagen, Valencia, CA, USA). Following this, the sequence reaction was carried out using the Bigdye Terminator V3.1 cycle sequencing kit (Perkin-Elmer, Waltham, MA, USA), and subsequent purification was performed using Centrisep spin columns.
DNA sequences were then acquired using the Applied Biosystems 3,130 genetic analyzer manufactured (manufactured by Hitachi, Tokyo, Japan) and a Basic Local Alignment Search Tool (BLAST® analysis) (30) was conducted to determine sequence identity with GenBankRM accessions. The final C. parvum sequences were named based on the primary subtype families identified through BLASTn searches, ensuring they exhibited >99% identity and adhered to the nomenclature method described elsewhere (31). These selected sequences of the COWP gene were deposited in GenBank via the National Center for Biotechnology Information (NCBI) under accession numbers OM817461 for C. parvum FSA-1, OM817462 for C. parvum FSA-2, and OM817463 for C. parvum FSA-3. To ensure a thorough comparison, sequences from various host species, including human samples from Egypt and other countries, were carefully selected. Alignment of these sequences was performed using ClustalW (32). A phylogenetic tree was constructed using the maximum likelihood method and the Tamura 3-parameter model (33), incorporating the bootstrap method with 1,000 replicates and MEGA Software (Version 11) was utilized for this analysis, selecting models based on the lowest Bayesian Information Criterion (BIC) (34).
3 Results
3.1 Frequency and morphological characteristics of Cryptosporidium spp. detected by microscopic methods
In this study, microscopic examination of fecal samples collected from sand rats using centrifugal flotation method revealed that 46.7% (70/150, 95% CI: 38.68–54.65) of the samples had compatible forms with Cryptosporidium spp. Using direct methods and centrifugal sedimentation, Cryptosporidium spp. was detected in 20% (30/150, 95% CI: 13.60–26.40) and 26.7% (40/150, 95% CI: 19.59–33.74) of the tested samples, respectively. The oocysts detected exhibited uniform morphological features. As depicted in Figures 2A,B, microscopic analysis of fecal samples obtained from sand rats, stained with Ziehl-Neelsen dye, indicated the presence of Cryptosporidium spp. oocysts, which appeared spherical in shape and ranged in size from 4 to 6 μm. They also exhibited a characteristic red to pink coloration and are surrounded by a clear halo. The oocyst wall is notably thick, smooth, and colourless, lacking structures such as sporocysts, micropyles, and polar granules. Inner structures are minimally visible under a light microscope. When employing direct smears and the flotation method with Sheather’s solution, the oocysts of the parasite were observed as transparent circular structures with discernible walls (Figure 2). On the other hand, electron microscopy revealed that these oocysts are spherical cysts with thin, rough membranes of varying sizes (Figures 2C,D).
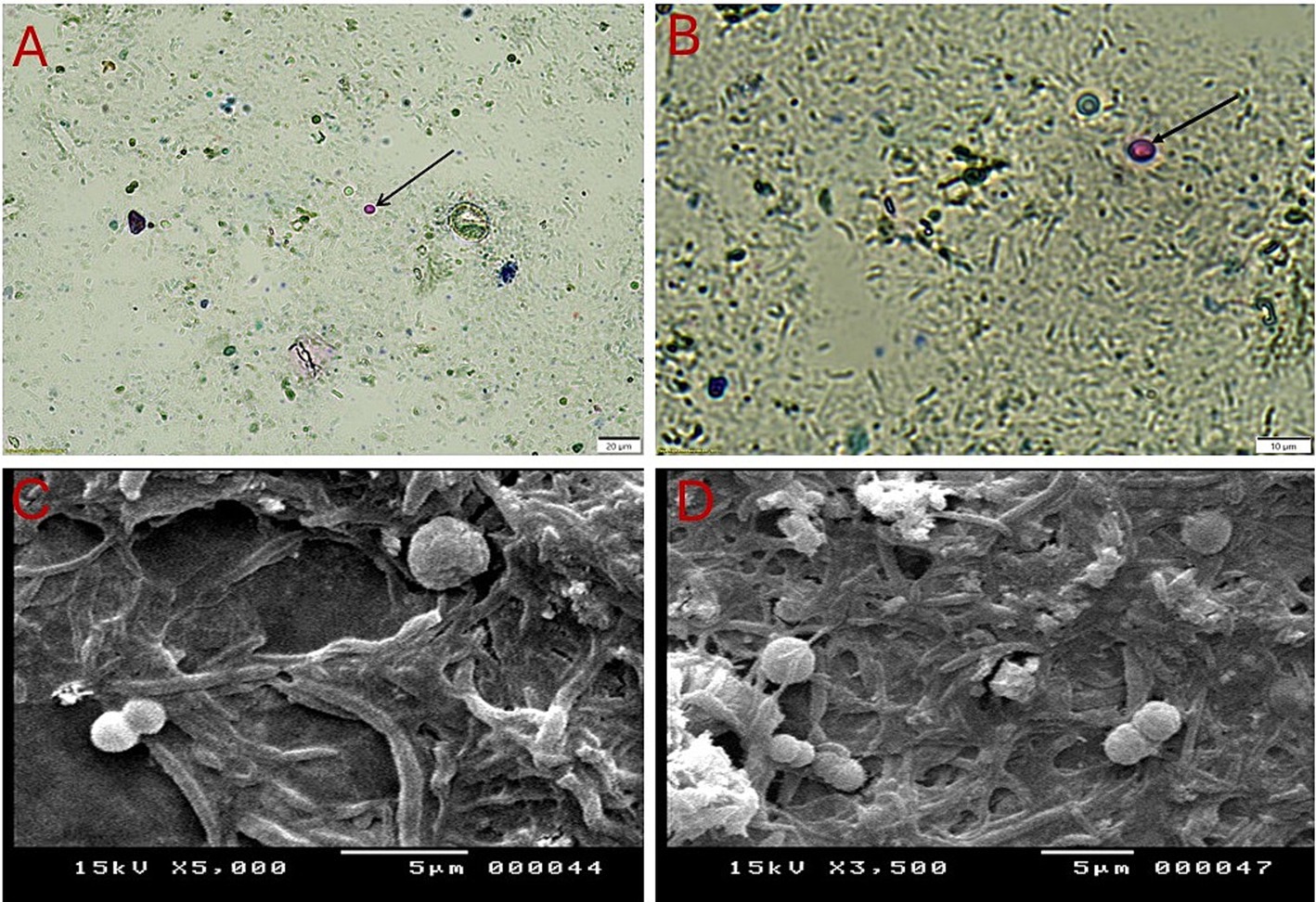
Figure 2. Cryptosporidium oocysts collected from the feces of sand rats (Psammomys obesus) captured using light and scanning electron microscopy. (A,B) Cryptosporidium parvum oocysts stained with Ziehl-Neelsen at different magnifications: (A) to be x100 and (B) to be x400. The oocysts appear spherical, stained red to pink, and are surrounded by a transparent halo. (C,D) Scanning electron micrographs (SEM) showing rounded oocysts.
3.2 Genetic characteristics of Cryptosporidium species/subtypes
Molecular analysis verified the presence of Cryptosporidium species in DNA extracted from 30 stool samples that had been identified as heavily infected based on microscopic examination. In this study, a total of five PCR-positive samples were subjected for sequencing. However, due to insufficient DNA, two Cryptosporidium spp.-positive samples were excluded from the genetic analyses. The phylogenetic analysis confirmed the presence of C. parvum (n = 3) among the studied samples (Figure 3). The sequences generated in this research have been submitted to the GenBank database under the accession numbers OM817461 for C. parvum FSA-1, OM817462 for C. parvum FSA-2, and OM817463 for C. parvum FSA-3. Figure 3 presents the Neighbor-Joining analysis comparing various Cryptosporidium species/genotypes alongside the isolated from sand rats in Egypt.
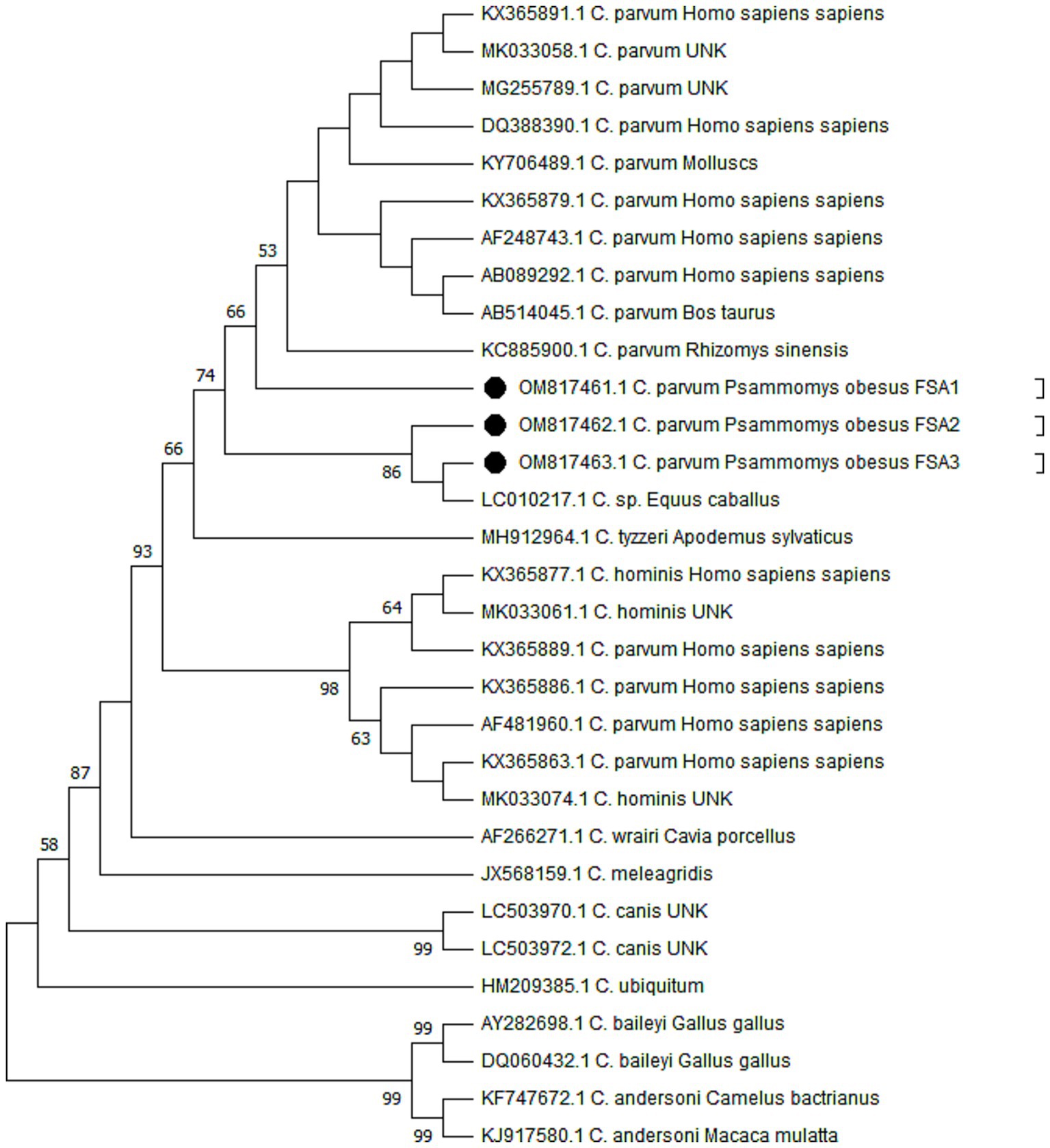
Figure 3. Phylogenetic trees constructed by the Maximum Likelihood method and Tamura 3-parameter model. The bootstrap consensus tree was inferred from 1,000 replicates and used to represent the evolutionary history of the taxa analyzed. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1,000 replicates) is shown next to the branches when values were larger than 50. The tree with the highest log likelihood (−1825.56) is shown. Initial tree(s) for the heuristic search were obtained automatically by applying Neighbor-Join and BioNJ algorithms to a matrix of pairwise distances estimated using the Tamura 3 parameter model, and then selecting the topology with superior log likelihood value. A discrete Gamma distribution was used to model evolutionary rate differences among sites (5 categories) (+G, parameter = 0.5552). This analysis involved 31 nucleotide sequences with a total of 478 positions in the final dataset. Evolutionary analyses were conducted in MEGA11. The sequence retrieved from the positive sand rats is given in black circles (accession numbers OM817461, OM817462, and OM817463).
4 Discussion
Wild rodents play a crucial role in transmitting zoonotic pathogens within their ecosystems, highlighting their significant public health implications (35, 36). Their prolific reproductive capacity and frequent interaction with humans facilitate the spread of various pathogens through faecal shedding. More importantly, their close association with human habitats in urban, rural, and natural environments increases the risk of zoonotic pathogen transmission through contaminated food, water, and environmental surfaces (37). Therefore, understanding these dynamics is essential for implementing targeted disease surveillance and management measures to mitigate the risks associated with zoonotic diseases transmitted by wild rodents.
This present study provides novel insights into the role of sand rats in transmitting Cryptosporidium spp. and underscores their potential zoonotic implications in saline marsh and periurban areas in Giza, Egypt. Using a combination of microscopic techniques, our study revealed that 46.7% of the analysed fecal samples from rodents tested positive for Cryptosporidium spp., which represents a significantly higher detection rates compared to previous global studies. For instance, Zhao et al. (38) reported a lower prevalence of 28.6% in brown rats in Hainan Province (China) using molecular techniques, while Li et al. (39) found a prevalence of 12.2% in bamboo rats in another study in Guangdong Province (China) using PCR and sequence analyses of the small subunit rRNA gene. In another study in the Canary Islands (Spain), García-Livia et al. (40) documented prevalence rates of 13.9 and 10.3% among Rattus rattus and Mus musculus domesticus, respectively, using molecular methods. On the other hand, close results were recorded in a previous study in El Hierro, Canary Islands (Spain), with an overall prevalence of 48.6% for C. parvum of among murine species studied, Rattus rattus and Mus musculus domesticus (41). Given that the current data were derived from direct observation of oocysts in stool specimens using microscopic methods, which are routine but known for their low sensitivity, specificity and reliance on expertise (9), this fluctuation across studies can also be attributed to several factors that include the diversity of host species, abundance of rodents in studied area, sample sizes, geographical variations, climatic conditions and environmental influences, effectiveness of rodent control measures, hygiene practices, rodents’ access to human habitats and other animals, as well as variations in the diagnostic methodologies employed (39–43).
It should be noted that PCR is an increasingly preferred molecular method for detecting Cryptosporidium species in fecal samples due to its superior sensitivity and specificity over traditional microscopy (44, 45). It is therefore evident that PCR-based techniques with high sensitivity, specificity, and rapidly features can differentiate among species and genotypes in different specimens, including water, stool, and animal or human tissues (43). Adopting these molecular methods is essential not only to validate or confirm microscopic findings but also to identify circulating genotypes and species of the parasite. This approach might be crucial for gaining insights into the potential zoonotic transmission of the disease from rodents to humans. As depicted, the molecular analysis of examined stool samples revealed C. parvum based on the amplification of Cryptosporidium COWP gene. The BLAST analysis deposited the identified genotype under accession numbers: (OM817461 C. parvum FSA-1, OM817462 C. parvum FSA-2, andOM 817,463 C. parvum FSA-3). Similarly, a previous investigation (39) in China reported a Cryptosporidium bamboo rat genotype I, C. parvum, besides Cryptosporidium bamboo rat genotype III, C. occultus, and C. muris, whereas another study (42) documented C. ditrichi in Apodemus flavicollis (yellow-necked mouse) and C. andersoni in Craseomys rufocanus (grey-sided vole). Nonetheless, another study (38) in China recognized Cryptosporidium rat genotype I and IV, Cryptosporidium suis-like genotype and C. ubiquitum. In a prior study (40) conducted in the Canary Islands (Spain), C. tyzzeri, C. meleagridis, C. muris and Cryptosporidium sp. rat genotype I and II/III were identified among the examined rodents. More interestingly, the present study revealed a close genetic resemblance between the identified C. parvum parasites in rodents from this research and those previously recorded in GenBank, showing over 98% identity with human isolates from Egypt and other countries (46–49). This finding suggests that rodents may play a role in perpetuating this pathogen among humans.
The current study encounters some methodological limitations that might require consideration when interpreting the results. Primarily, due to financial constraints, the initial screening was based on light microscopy, and we were unable to conduct molecular analysis on all retrieved samples. Furthermore, COWP-based tools have demonstrated limited effectiveness in genotyping some Cryptosporidium spp. in animals due to their narrow specificity (50). Despite these limitations, our study represents the first molecular confirmation of C. parvum in sand rats in Egypt. These findings reflect that sand rats might act as reservoirs for C. parvum in this country. Given the zoonotic significance of C. parvum and the close genetic similarity between the sequences obtained and those found in humans in Egypt, the presence of rodents in human-inhabited areas—be they urban, rural, or natural—heightens the risk of pathogen transmission through contaminated food, water, and environmental surfaces. Further investigations are necessary to accurately ascertain the potential circulation of other Cryptosporidium spp. among rodents and other animals’ reservoirs cross the country.
5 Conclusion
This study provides novel preliminary insights into the potential role of sand rats in the transmission of Cryptosporidium spp. using a set of microscopic and molecular methods. Notably, it is the first molecular study to characterize C. parvum in sand rats (Psammomys obesus), which enhances our understanding for the potential zoonotic importance played by rodents in the transmission of cryptosporidiosis. Our findings emphasize the critical importance for public health education on the role of rodents in maintaining epidemiological foci of the pathogen and the necessity for enforcing strict hygienic measures by local authorities to eradicate rodents and prevent their access to human habitats. Conducting larger-scale, molecular-based epidemiological surveys will allow us to examine the prevalence and genetic characteristics of Cryptosporidium across diverse animal species and environmental samples. These studies will be vital in identifying infection sources, elucidating transmission pathways, and reducing the risk of disease spread from sand rats to humans.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary material, further inquiries can be directed to the corresponding author.
Ethics statement
The animal study was approved by the Ethics Committee of the Faculty of Veterinary Medicine, Assiut University, Egypt (No. 06/2024/0160). The study was conducted in accordance with the local legislation and institutional requirements.
Author contributions
SM: Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Project administration, Software, Supervision, Validation, Visualization, Writing – original draft, Writing – review & editing. FAn: Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Project administration, Resources, Software, Supervision, Validation, Visualization, Writing – original draft, Writing – review & editing. AGa: Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Project administration, Software, Supervision, Validation, Visualization, Writing – original draft, Writing – review & editing. MG: Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Project administration, Software, Supervision, Validation, Writing – review & editing. RM: Data curation, Formal analysis, Investigation, Software, Validation, Visualization, Writing – review & editing. AGo: Conceptualization, Data curation, Formal analysis, Investigation, Project administration, Resources, Software, Supervision, Validation, Visualization, Writing – review & editing. FAl: Data curation, Formal analysis, Funding acquisition, Resources, Software, Validation, Visualization, Writing – review & editing. HA: Data curation, Formal analysis, Funding acquisition, Resources, Software, Validation, Visualization, Writing – review & editing. IG-B: Data curation, Formal analysis, Validation, Writing – review & editing. EE: Conceptualization, Data curation, Formal analysis, Funding acquisition, Investigation, Methodology, Resources, Software, Supervision, Validation, Visualization, Writing – original draft, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. EE received support through a postdoctoral fellowship from the María Zambrano Program at the University of Córdoba, funded by the Program of Requalification of the Spanish University System, sponsored by the Spanish Ministry of Universities and financed by the European Union-NextGenerationEU.
Acknowledgments
This study was supported by Princess Nourah bint Abdulrahman University Researchers Supporting Project No. (PNURSP2025R401), Princess Nourah bint Abdulrahman University, Riyadh, Saudi Arabia. RMP was supported by a postdoctoral contract (POSTDOC2100041) at the University of Córdoba (ROR code 05yc77b46) from the Consejería de Transformación Económica, Industria, Conocimiento y Universidades of the Junta de Andalucía (ROR code 01jem9c82) Regional Government (Andalucía, Spain).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fvets.2024.1488508/full#supplementary-material
References
1. Ryan, U, Zahedi, A, Feng, Y, and Xiao, L. An update on zoonotic Cryptosporidium species and genotypes in humans. Animals. (2021) 11, 1–22. doi: 10.3390/ANI11113307
2. Kotloff, KL. The burden and etiology of diarrheal illness in developing countries. Pediatr Clin N Am. (2017) 64:799–814. doi: 10.1016/J.PCL.2017.03.006
3. Strand, TM, and Lundkvist, Å. Rat-borne diseases at the horizon. A systematic review on infectious agents carried by rats in Europe 1995-2016. Infect Ecol Epidemiol. (2019) 9:1553461. doi: 10.1080/20008686.2018.1553461
4. Tallant, C, Huddleston, P, Alshanberri, A, and Misra, S. Acute, severe cryptosporidiosis in an immunocompetent pediatric patient. Clin Pract. (2016) 6:837. doi: 10.4081/cp.2016.837
5. Fayer, R, Trout, JM, and Jenkins, MC. Infectivity of Cryptosporidium parvum oocysts stored in water at environmental temperatures. J Parasitol. (1998) 84:1165–9. doi: 10.2307/3284666
6. Fayer, R, Graczyk, TK, Lewis, EJ, Trout, JM, and Farley, CA. Survival of infectious Cryptosporidium parvum oocysts in seawater and eastern oysters (Crassostrea virginica) in the Chesapeake Bay. Appl Environ Microbiol. (1998) 64:1070–4. doi: 10.1128/AEM.64.3.1070-1074.1998
7. Khalil, IA, Troeger, C, Rao, PC, Blacker, BF, Brown, A, Brewer, TG, et al. Morbidity, mortality, and long-term consequences associated with diarrhoea from Cryptosporidium infection in children younger than 5 years: a meta-analyses study. Lancet Glob Health. (2018) 6:e758–68. doi: 10.1016/S2214-109X(18)30283-3
8. da Silva, DRR, Oliveira, T, Marta, B, Baptista, C, Bottaro, M, and Bresciani, K. Vaccine development for cryptosporidiosis: systematic review. Res. Soc. Develop. (2021) 10:e18910615540–08. doi: 10.33448/rsd-v10i6.15540
9. McHardy, IH, Wu, M, Shimizu-Cohen, R, Couturier, MR, and Humphries, RM. Detection of intestinal protozoa in the clinical laboratory. J Clin Microbiol. (2014) 52:712–20. doi: 10.1128/JCM.02877-13
10. Vioque, F, Dashti, A, Santín, M, Ruiz-Fons, F, Köster, PC, Hernández-Castro, C, et al. Wild micromammal host spectrum of zoonotic eukaryotic parasites in Spain. Occurrence and genetic characterisation. Transbound Emerg Dis. (2022) 69:e2926–42. doi: 10.1111/tbed.14643
11. Robertson, LJ, Clark, CG, Debenham, JJ, Dubey, JP, Kváč, M, Li, J, et al. Are molecular tools clarifying or confusing our understanding of the public health threat from zoonotic enteric protozoa in wildlife? Int J Parasitol Parasites Wildl. (2019) 9:323–41. doi: 10.1016/j.ijppaw.2019.01.010
12. Zhang, L, Guo, W, and Lv, C. Modern technologies and solutions to enhance surveillance and response systems for emerging zoonotic diseases. Science in One Health. (2024) 3:100061. doi: 10.1016/j.soh.2023.100061
13. Tomassone, L, Berriatua, E, De Sousa, R, Duscher, GG, Mihalca, AD, Silaghi, C, et al. Neglected vector-borne zoonoses in Europe: into the wild. Vet Parasitol. (2018) 251:17–26. doi: 10.1016/j.vetpar.2017.12.018
14. Zhang, K, Fu, Y, Li, J, and Zhang, L. Public health and ecological significance of rodents in Cryptosporidium infections. One Health. (2022) 14:100364. doi: 10.1016/j.onehlt.2021.100364
15. Bryja, J, Nesvadbová, J, Heroldová, M, Jánová, E, Losík, J, Trebatická, L, et al. Common vole (Microtus arvalis) population sex ratio: biases and process variation. Can J Zool. (2005) 83:1391–9. doi: 10.1139/z05-133
16. Xiao, L, Morgan, UM, Limor, J, Escalante, A, Arrowood, M, Shulaw, W, et al. Genetic diversity within Cryptosporidium parvum and related Cryptosporidium species. Appl Environ Microbiol. (1999) 65:3386–91. doi: 10.1128/AEM.65.8.3386-3391.1999
17. El-Salkh, BA, Khidr, HA, Zaki, ZT, and Basuony, MI. Anatomical, histological and histochemical studies on some organs of True Desert rodents in the Egyptian habitats. Egypt J Hosp Med. (2008) 33:578–306. doi: 10.21608/ejhm.2008.17622
18. Musser, GG, and Carleton, MD. Super-family Muroidea In: DE Wilson and DM Reeder, editors. Mammal species of the world: A taxonomic and geographic reference. 3rd ed: Baltimore, Maryland, USA: Johns Hopkins University Press (2005). 894–1531.
19. Fichet-Calvet, E, Jomâa, I, Ben, IR, and Ashford, RW. Reproduction and abundance of the fat sand rat (Psammomys obesus) in relation to weather conditions in Tunisia. J Zool. (1999) 248:15–26. doi: 10.1111/J.1469-7998.1999.TB01018.X
20. Daly, M, and Daly, S. Spatial distribution of leaf-eatingsaharan gerbil (Psammomys obesus) in relation to its food. Mammalia. (1974) 38:591–603. doi: 10.1515/mamm.1974.38.4.591
21. National Research Council. AVMA guidelines for the euthanasia of animals; guide for the care and use of laboratory animals. 8th ed. Schaumburg, IL: American Veterinary Medical Association (2020).
22. Berengere, B. Taxonomie et identification des Gerbillusde l’Afrique de l’Ouest. Ed. Université Pierre et MarieCurie, Paris VI, 2003.
23. Silberberg, R. morphometric observations on vertebral bone of aging sand rats. Spine (Phila Pa 1976). 1988 Feb;13(2):202-8. doi: 10.1097/00007632-198802000-00013
24. Garcia, LS. Diagnostic medical parasitology. 5th ed. Washington DC, USA: American Society for Microbiology Press. (2006). 57–37.
25. Thompson, A. Review of “Cryptosporidium and cryptosporidiosis” by Ronald Fayer and Lihua Xiao Parasit vectors (2008) 1:47. doi: 10.1186/1756-3305-1-47
26. Reduker, DW, Speer, CA, and Blixt, JA. Ultrastructure of Cryptosporidium parvum oocysts and excysting sporozoites as revealed by high resolution scanning electron microscopy. J Protozool. (1985) 32:708–11. doi: 10.1111/j.1550-7408.1985.tb03106.x
27. Spano, F, Putignani, L, McLauchlin, J, Casemore, DP, and Crisanti, A. PCR-RFLP analysis of the Cryptosporidium oocyst wall protein (COWP) gene discriminates between C. Wrairi and C. Parvum, and between C. parvum isolates of human and animal origin. FEMS Microbiol Lett. (1997) 150:209–17. doi: 10.1016/s0378-1097(97)00115-8
28. Pedraza-Díaz, S, Amar, C, Nichols, GL, and McLauchlin, J. Nested polymerase chain reaction for amplification of the Cryptosporidium oocyst wall protein gene. Emerg Infect Dis. (2001) 7:49–56. doi: 10.3201/eid0701.700049
29. Feltus, DC, Giddings, CW, Schneck, BL, Monson, T, Warshauer, D, and McEvoy, JM. Evidence supporting zoonotic transmission of Cryptosporidium spp. in Wisconsin. J Clin Microbial. (2006) 44:4303–8. doi: 10.1128/JCM.01067-06
30. Altschul, SF, Gish, W, Miller, W, Myers, EW, and Lipman, DJ. Basic local alignment search tool. J Mol Biol. (1990) 215:403–10. doi: 10.1016/S0022-2836(05)80360-2
31. Sulaiman, IM, Hira, PR, Zhou, L, Al-Ali, FM, Al-Shelahi, FA, Shweiki, HM, et al. Unique endemicity of cryptosporidiosis in children in Kuwait. J Clin Microbiol. (2005) 43:2805–9. doi: 10.1128/JCM.43.6.2805-2809.2005
32. Clustal, W: A multiple sequence alignment program for DNA or proteins. Available at: http://www.ebi.ac.uk/Tools/clustalw/.
33. Tamura, K. Estimation of the number of nucleotide substitutions when there are strong transition-transversion and G+C-content biases. Mol Biol Evol. (1992) 9:678–87. doi: 10.1093/oxfordjournals.molbev.a040752
34. Tamura, K, Stecher, G, and Kumar, S. MEGA11: molecular evolutionary genetics analysis version 11. Mol Biol Evol. (2021) 38:3022–7. doi: 10.1093/molbev/msab120
35. Meerburg, BG, Singleton, GR, and Kijlstra, A. Rodent-borne diseases and their risks for public health. Crit Rev Microbiol. (2009) 35:221–70. doi: 10.1080/10408410902989837
36. Dahmana, H, Granjon, L, Diagne, C, Davoust, B, Fenollar, F, and Mediannikov, O. Rodents as hosts of pathogens and related zoonotic disease risk. Pathogens. (2020) 9, 1–21. doi: 10.3390/pathogens9030202
37. Bradley, CA, and Altizer, S. Urbanization and the ecology of wildlife diseases. Trends Ecol Evol. (2007) 22:95–102. doi: 10.1016/j.tree.2006.11.001
38. Zhao, W, Zhou, H, Huang, Y, Xu, L, Rao, L, Wang, S, et al. Cryptosporidium spp. in wild rats (Rattus spp.) from the Hainan Province, China: molecular detection, species/genotype identification and implications for public health. Int J Parasitol Parasites Wildl. (2019) 9:317–21. doi: 10.1016/j.ijppaw.2019.03.017
39. Li, F, Zhao, W, Zhang, C, Guo, Y, Li, N, Xiao, L, et al. Cryptosporidium species and C. parvum subtypes in farmed bamboo rats. Pathogens. (2020) 9, 1–11. doi: 10.3390/pathogens9121018
40. García-Livia, K, Martín-Alonso, A, and Foronda, P. Diversity of Cryptosporidium spp. in wild rodents from the Canary Islands, Spain. Parasit Vectors. (2020) 13:445. doi: 10.1186/s13071-020-04330-9
41. Feliu, C, López, M, Gómez, MS, Torres, J, Sánchez, S, Miquel, J, et al. Parasite fauna of rodents (Murinae) from El Hierro (Canary Islands, Spain): a multidisciplinary approach. Acta Parasitol. (2012) 57:171–8. doi: 10.2478/s11686-012-0016-7
42. Kivistö, R, Kämäräinen, S, Huitu, O, Niemimaa, J, and Henttonen, H. Zoonotic Cryptosporidium spp. in wild rodents and shrews. Microorganisms. (2021) 9, 1–13. doi: 10.3390/microorganisms9112242
43. Saki, J, Foroutan-Rad, M, and Asadpouri, R. Molecular characterization of Cryptosporidium spp. in wild rodents of southwestern Iran using 18s rRNA gene nested-PCR-RFLP and sequencing techniques. J Trop Med. (2016) 2016:1–6. doi: 10.1155/2016/6834206
44. Templeton, TJ, Lancto, CA, Vigdorovich, V, Liu, C, London, NR, Hadsall, KZ, et al. The Cryptosporidium oocyst wall protein is a member of a multigene family and has a homolog in toxoplasma. Infect Immun. (2004) 72:980–7. doi: 10.1128/IAI.72.2.980-987.2004
45. Fayer, R. General biology In: R Fayer and L Xiao, editors. Cryptosporidium and 394 cryptosporidiosis. CRC Press: Boca Raton (2008). 1–42.
46. Ghallab, MMI, Khater, MM, Tyler, KM, Bouzid, M, and El-Badry, AA., Genotypic variance analysis of Cryptosporidium isolates from stool of children in Egypt. Cairo: Cairo University. (2016). 1–164 p.
47. Wielinga, PR, de Vries, A, van der Goot, TH, Mank, T, Mars, MH, Kortbeek, LM, et al. Molecular epidemiology of Cryptosporidium in humans and cattle in the Netherlands. Int J Parasitol. (2008) 38:809–17. doi: 10.1016/j.ijpara.2007.10.014
48. Pedraza-Díaz, S, Amar, C, and McLauchlin, J. The identification and characterisation of an unusual genotype of Cryptosporidium from human faeces as Cryptosporidium meleagridis. FEMS Microbiol Lett. (2000) 189:189–94. doi: 10.1016/S0378-1097(00)00278-0
49. Akiyoshi, DE, Feng, X, Buckholt, MA, Widmer, G, and Tzipori, S. Genetic analysis of a Cryptosporidium parvum human genotype 1 isolate passaged through different host species. Infect Immun. (2002) 70:5670–5. doi: 10.1128/IAI.70.10.5670-5675.2002
Keywords: Cryptosporidium parvum, morphology, molecular identification, sand rat, Egypt
Citation: Mohamed SA-A, Anwar FAS, Gareh A, Ghallab MMI, Martínez R, Gouda AA, Alajmi FE, Alzaylaee H, García-Bocanegra I and Elmahallawy EK (2025) First microscopic and molecular identification of Cryptosporidium spp. in fat sand rats (Psammomys obesus) in Egypt and their potential zoonotic implications. Front. Vet. Sci. 11:1488508. doi: 10.3389/fvets.2024.1488508
Edited by:
Rudi Cassini, University of Padua, ItalyReviewed by:
Lisa Guardone, University of Pisa, ItalyMosaab Omar Mohamed, Qassim University, Saudi Arabia
Copyright © 2025 Mohamed, Anwar, Gareh, Ghallab, Martínez, Gouda, Alajmi, Alzaylaee, García-Bocanegra and Elmahallawy. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ehab Kotb Elmahallawy, c2EyZWxlbGVAdWNvLmVz
 Sara Abdel-Aal Mohamed1
Sara Abdel-Aal Mohamed1 Ahmed Gareh
Ahmed Gareh Remigio Martínez
Remigio Martínez Asmaa Aboelabbas Gouda
Asmaa Aboelabbas Gouda Ehab Kotb Elmahallawy
Ehab Kotb Elmahallawy