
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Vet. Sci., 22 January 2025
Sec. Oncology in Veterinary Medicine
Volume 11 - 2024 | https://doi.org/10.3389/fvets.2024.1473421
Introduction: As the lifespan of companion animals extends, the incidence of tumor also increases. Among these tumors, lymphoma is reported as the most prevalent hematopoietic tumor with a 80-90% prevalence rate. Ongoing research spans multiple domains, aiming to uncover novel therapeutic targets, including small molecular weight inhibitors, antibody treatments, and subtype-specific selective agents.
Methods: Transcriptional profiling was performed on canine lymphoma samples to identify genes and functional pathways associated with pathogenesis, treatment response, and prognosis. Additionally, genes with potential relevance to the clinical characteristics of T-cell lymphoma (TCL), which is characterized by a low treatment response and poor prognosis, were identified through a comparative analysis of different lymphoma subtypes.
Results: Within the canine lymphoma group, HERC5 showed consistent upregulation, a gene similarly implicated in human acute myeloid leukemia but previously no reports exist. Additionally, noteworthy genes, including IKZF2, CCL4, SAA1, and CD40, exhibited differential expression in the TCL group compared to the B-cell lymphoma (BCL) group.
Discussion: The upregulation of HERC5 may impact on canine lymphoma pathogenicity. Furthermore, the upregulation of IKZF2, CCL4, and SAA1, along with the downregulation of CD40, may contribute to adverse clinical characteristics of TCL in dogs.
Lymphoma is a prevalent hematopoietic tumor that results from the malignant transformation of lymphocytes (1). It exhibits diverse subtypes, classified based on morphology, immunophenotype, and molecular variations (2, 3). To differentiate between T-cell and B-cell types, complementary diagnostic tests, such as flow cytometry and immunohistochemistry, are conducted (2, 3). Notably, B-cell lymphoma (BCL) generally demonstrates a more favorable response to treatment and prognosis compared to T-cell lymphoma (TCL) in dogs (4), with previous reports indicating lower complete remission rates and shorter survival times, approximately 40%, in dogs with TCL compared to in those with BCL (5, 6). Because the prognosis varies depending on the type of lymphoma, it is important to differentiate between BCL and TCL (5, 6).
Recently, numerous studies have been conducted in the field of genetics to characterize subtypes, diagnose, and understand the prognosis of lymphoma (7–9). “Transcriptome” encompasses all transcripts, including mRNAs, non-coding RNAs, and small RNAs, present within a cell at a given developmental stage or physiological state (7). Analyzing splicing patterns, post-transcriptional modifications, and expression levels of the transcriptome in a particular environment is crucial for understanding genome functionality and gaining insights into tumor biology (7–9). A protocol has been established to classify melanoma, osteosarcoma, lung cancer, B-cell lymphoma, and T-cell lymphoma using canine transcriptome data (8). The analysis identified a total of 625 genes exhibiting significant differences, and Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis indicated 11 pathways associated with the progression of lymphoma in dogs (9). Further research is necessary to accurately diagnose canine lymphoma and predict prognosis. While numerous studies have focused on the transcriptome of B-cell lymphoma (BCL) compared to normal samples in dogs, there is a paucity of research examining the differences between T-cell lymphoma (TCL) and B-cell lymphoma (BCL) (9–11).
The current study aims to compare transcriptomic profiles between normal samples and lymphoma, including each lymphoma subtype, to analyze gene expression differences. Furthermore, it seeks to explore associated biochemical pathways for lymphoma diagnosis and prognosis.
The study included a population of 13 client-owned dogs diagnosed with multicentric lymphoma and 7 dogs without lymphoma. The clinical characteristics of each participant are detailed in Table 1. The 13 dogs in the lymphoma group were referred for internal medicine evaluation to Chonnam National University Teaching Hospital in 2020–2022 (Identification code No. CNU IACUC-YB-2021-166, No. CNU IACUC-YB-2022-121).
The staging of lymphoma cases was performed in accordance with the World Health Organization TNM classification system for canine lymphoma (12). Among the dogs diagnosed with lymphoma, 10% (1/10) were classified as stage III, 70% (7/10) as stage IV, and 20% (2/10) as stage V. Additionally, 60% (6/10) of dogs were classified as TCL, while 40% (4/10) were identified as BCL (Table 2).
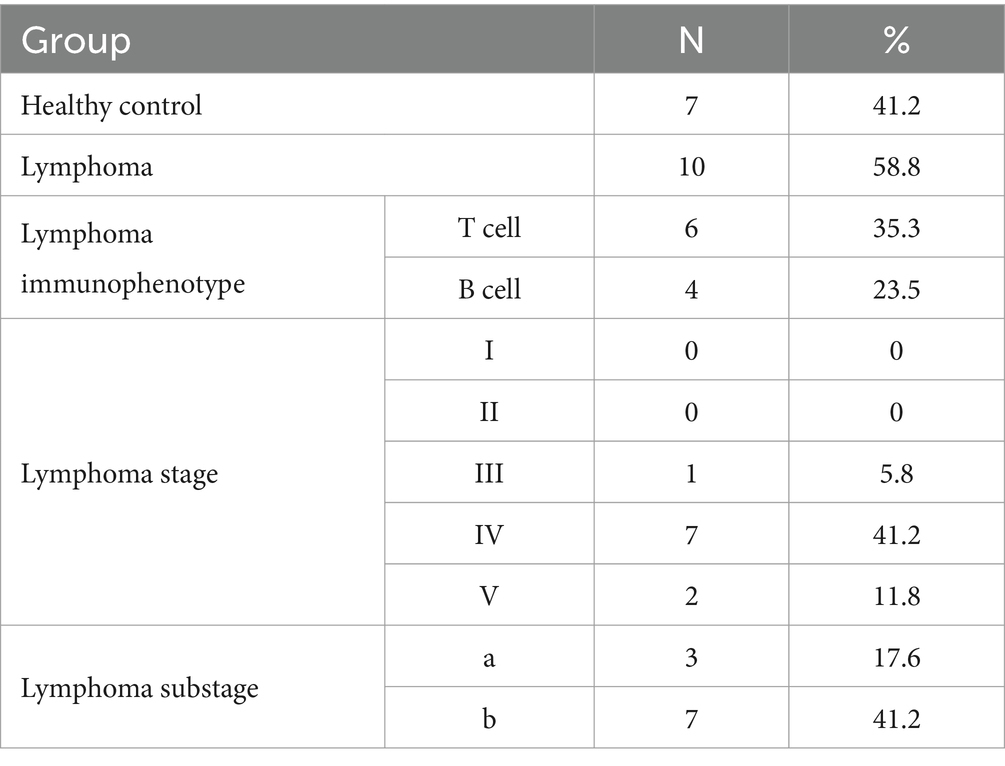
Table 2. Classification of healthy, lymphoma and the World Health Organization’s clinical staging system.
For dogs with lymphoma, lymph node tissues were collected through either ultrasound-guided fine needle aspiration or biopsy procedures on enlarged peripheral or abdominal lymph nodes at initial diagnosis. The collected tissues were preserved in RNAprotect Tissue Reagent (Qiagen, Hilden, Germany) and stored at −80°C until further analysis.
In the control group, one dog underwent anesthesia, and tissue samples were collected from six dogs post-euthanasia. Following a thorough disinfection procedure, an incision was made in the left hind leg muscle to access and isolate the left popliteal lymph node. The lymph node and adjacent fat tissue were separated, and then preserved in a similar manner to the lymphoma samples.
All dogs undergoing biopsy for the diagnosis of lymphoma, as well as one dog in the control group, were anesthetized. For pre-medication, which included sedation and induction, glycopyrrolate (5 μg/kg, SC; Mobinul® Injection, Myungmoon, South Korea), medetomidine (5–10 μg/kg, IM; Tomidin® Injection, Provet Veterinary Products, Turkey), and alfaxalone (1.5–2 mg/kg, IV; Alfaxan® Injection, JUROX, Australia) were administered. During the surgical procedure, respiratory anesthesia was maintained with isoflurane (2–3%, inhalation; Terrel™ Solution, Piramal Critical Care, USA).
Furthermore, for the sedation and induction of six dogs in the control group, medetomidine (5–10 μg/kg, IM; Tomidin® Injection, Provet Veterinary Products, Turkey) and alfaxalone (1.5–2 mg/kg, IV; Alfaxan® Injection, JUROX, Australia) were utilized. Subsequently, 20% potassium chloride (20 mL/body, IV; Potassium Chloride-40® Injection, DaeHan Pharm, South Korea) was employed as part of the euthanasia protocol.
A total of 13 dogs diagnosed with lymphoma and 7 dogs without lymphoma were included in the transcriptome analysis. Three of the lymphoma samples did not provide sufficient RNA for analysis and therefore excluded from the study. The isolation of total RNA from each tissue was performed using the Transzol-based RNA extraction protocol.
The concentration and purity of total RNA were assessed utilizing the ND-2000 Spectrophotometer (NanoDrop, Wilmington, USA), while the integrity was evaluated with the Agilent RNA 6000 Nano Kit on a 2,100 Bioanalyzer (Agilent Technologies, Palo Alto, USA). For RNA labeling and hybridization, the Agilent One-Color Microarray-Based Gene Expression Analysis protocol (Agilent Technology, Version 6.5, 2010) was utilized. The labeled complementary RNAs (cRNAs) were purified using the RNAeasy Mini Kit (Qiagen). The hybridization solution was dispensed onto the gasket slide and then assembled onto the Agilent Unrestricted AMADID Release GE 4x44K (Canine V2). The hybridized array was then scanned using the Agilent Microarray Scanner D (Agilent Technologies). Data extraction from the microarray was conducted using the Agilent Feature Extraction software version 11.0 (Agilent Technologies).
Subsequently, the raw data for each gene were automatically summarized according to the Agilent feature extraction protocol, resulting in the generation of a raw data text file that contained expression data for each gene probed on the array. A comparative analysis of expression profiles among analogous samples was conducted utilizing the Agilent Canine GE 4X44k v2 data.
Differential expression analysis between the control and lymphoma groups was conducted using R version 3.3.2. p-values underwent Benjamini and Hochberg methods to account for multiple testing. Significance was defined as an FDR-adjusted p-value (P) <0.05 and a log2 fold change ≥2 for the selection of differentially expressed genes (DEGs). Subsequently, DEGs were analyzed for gene enrichment and functional annotation utilizing the KEGG. Data analysis and visualization of DEGs were also performed using R version 3.3.2.
To evaluate the normality of the variables, the Shapiro–Wilk test was used, and the data were expressed as mean ± standard error of the mean. The statistical significance of the expression data was assessed through fold change analysis and independent t-tests, with the null hypothesis positing no difference between groups. The false discovery rate (FDR) control was achieved by adjusting p-values using the Benjamini–Hochberg methods. Hierarchical cluster analysis, based on complete linkage and Euclidean distances, was conducted for DEGs. Significance was set at p < 0.05.
A total of 43,603 genes were identified through transcriptomic analysis. Based on the set criteria, a total of 282 DEGs were identified in lymphoma vs. control groups, which included 22 upregulated genes and 260 downregulated genes. To visually depict the distinct gene expression patterns, a hierarchical clustering heatmap was constructed, illustrating the divergence between the lymphoma and control groups (Figure 1A).
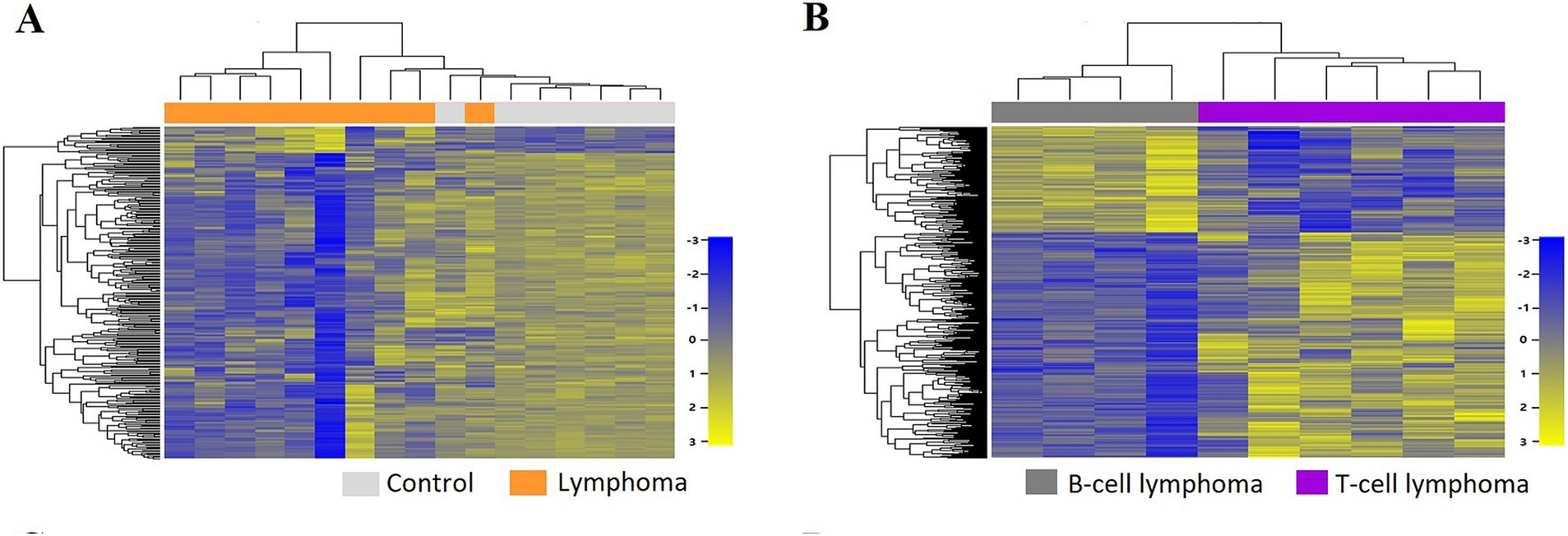
Figure 1. Two-way hierarchical clustering heatmap of differentially expressed genes (DEGs) identified in the lymphoma group compared with the control group (A) and in the T-cell lymphoma (TCL) group compared with the B-cell lymphoma (BCL) group (B).
Among the DEGs, five most highly expressed DEGs were MX1 (MX dynamin-like GTPase 1), HERC5 (HECT and RLD domain-containing E3 ubiquitin protein ligase 5), OAS1 (2′-5′-oligoadenylate synthetase 1), and DDX58 (DEAD box polypeptide 58). In contrast, the five DEGs with the lowest expression levels were CAMP (Cathelicidin antimicrobial peptide), NMB (Neuromedin B), JAM2 (junctional adhesion molecule 2), IL7R (interleukin 7 receptor), and GRAP2 (GRB2-related adaptor protein 2). Detailed information is available in Table 3, and a visual representation in Supplementary Figure S1A.
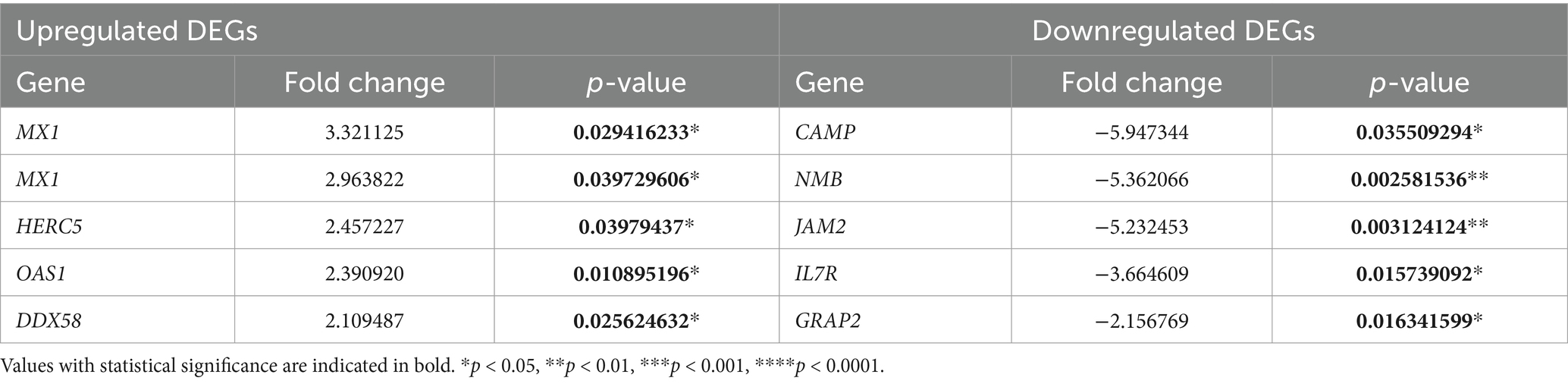
Table 3. The top five upregulated and downregulated DEGs in the lymphoma group vs. the control group.
In the lymphoma group, 21 significant KEGG pathways were identified. The ten most prominent pathways included hematopoietic cell lineage, measles, T-cell receptor signaling, cancer pathways, cytokine–receptor interaction, primary immunodeficiency, viral myocarditis, PI3K–Akt signaling, arrhythmogenic right ventricular cardiomyopathy, and coronavirus disease, as presented in Table 4.
In the comparative analysis of the control group and the lymphoma groups, as well as between the control group and each lymphoma subtype, a greater number of DEGs exhibited downregulation rather than upregulation. Conversely, when comparing the comparison between the TCL group and the BCL group, there was higher incidence of upregulated genes (n = 378) compared to downregulated genes (n = 177), resulting in a total of 555 DEGs. The expression patterns of these DEGs in the TCL group compared to the BCL group are illustrated in a hierarchical clustering heatmap (Figure 1B).
Among the DEGs identified, the five most highly expressed DEGs were IKZF2 (Ikaros family zinc finger 2, Helios), LOC102155886 (serum amyloid A protein-like), CCL4 (C-C motif chemokine ligand 4), IL1R2 (interleukin 1 receptor, type II), and SAA1 (serum amyloid A1). In contrast, the five DEGs with the lowest expression levels included LOC484343 (sialic acid-binding immunoglobulin-like lectin 10), PLEKHA5 (pleckstrin homology domain-containing family A member 5), GRK4 (G protein-coupled receptor kinase 4), MYRIP (myosin VIIA and Rab-interacting protein), and CD40 (cluster of differentiation 40 molecule). Further details are available in Table 5, and a visual representation is provided in Supplementary Figure S1B.
The KEGG pathway analysis revealed a total of 85 significant pathways within the TCL group. The ten most prominent pathways identified include: human T-cell leukemia virus 1 infection, focal adhesion, lipid and atherosclerosis, pathways in cancer, axon guidance, transcriptional misregulation in cancer, fluid shear stress and atherosclerosis, cell adhesion molecules, Yersinia infection, and lysosome (Table 6).
The current study conducted a comparative analysis between lymphoma and control subjects, and sequential transcriptome analysis of TCL and BCL in dogs. In the comparison between lymphoma dogs and controls, we found that the upregulation of HERC5 may be associated with the pathogenic mechanisms underlying canine lymphoma. Furthermore, we identified the upregulation of IKZF2, CCL4, and SAA1, as well as the downregulation of CD40 in dogs with TCL compared to those with BCL, all of which were reported to be associated with shorter survival times in humans.
Among the DEGs that exhibited significant expression alternations in the lymphoma group compared with the control group, it has been reported that elevated expression of MX1 is significantly associated with higher tumor grade and lymphovascular invasion (13), elevated expression of OAS1 correlates with higher tumor grade and reduced overall survival (14, 15), and elevated expression of DDX58 is associated with increased microsatellite instability and tumor mutation burden, which are related to decreased overall survival and progression-free survival (16), indicating a poor prognosis in human cancers. This study also confirmed high expression of these genes in the lymphoma group.
A recent study has indicated a downregulation of HERC5 expression in human acute myeloid leukemia; however, there are no related reports within the field of veterinary medicine (17). Conversely, IL7R has been indicated as a favorable prognostic maker and a potential target for immunotherapy in human lung adenocarcinoma (18). This protein encoded by IL7R plays a critical role in inhibiting tumor growth by modulating the proportion of immune infiltrating cells within the tumor’ s immune microenvironment. Notably, low expression of IL7R has been correlated with increased tumor growth and lower survival rates (18). In our study, we confirmed lower expression of IL7R in the lymphoma group compared to the control group.
It has been reported that increased expression of CAMP in human breast cancer (19). Additionally, NMB exhibited increased expression in various human tumors, including breast (20, 21), lung (22, 23), colon (24), and ovary (25). In the present study, a decreased expression of both genes was identified, and there are no confirmed reports related to expression in lymphoma or veterinary science. This discrepancy may be attributed to species- or tumor-specific differences, and further research is needed.
Regarding the enriched pathways in the lymphoma group, it is noteworthy that the T-cell receptor signaling pathway (26, 27) and the PI3K-Akt signaling pathway (28, 29) have been associated with lymphoma in previous literature. Among the DEGs, IKZF2 identified as one of the most prominently upregulated genes. This gene has been reported to play a critical role in T-cell development, differentiation, and function as a transcription inhibitor (30). A recent study highlighted the upregulation of IKZF2 in human cutaneous TCL and proposed its potential as a target gene for treatment (Figure 2B) (30), as well as in T-cell acute lymphoblastic leukemia in humans (31). Furthermore, in this study identified that IKZF2 was also upregulated in the canine TCL group when compared to the canine BCL group. This finding suggests that IKZF2 may serve as a relevant gene associated with poorer clinical outcomes in TCL rather than BCL in dogs.
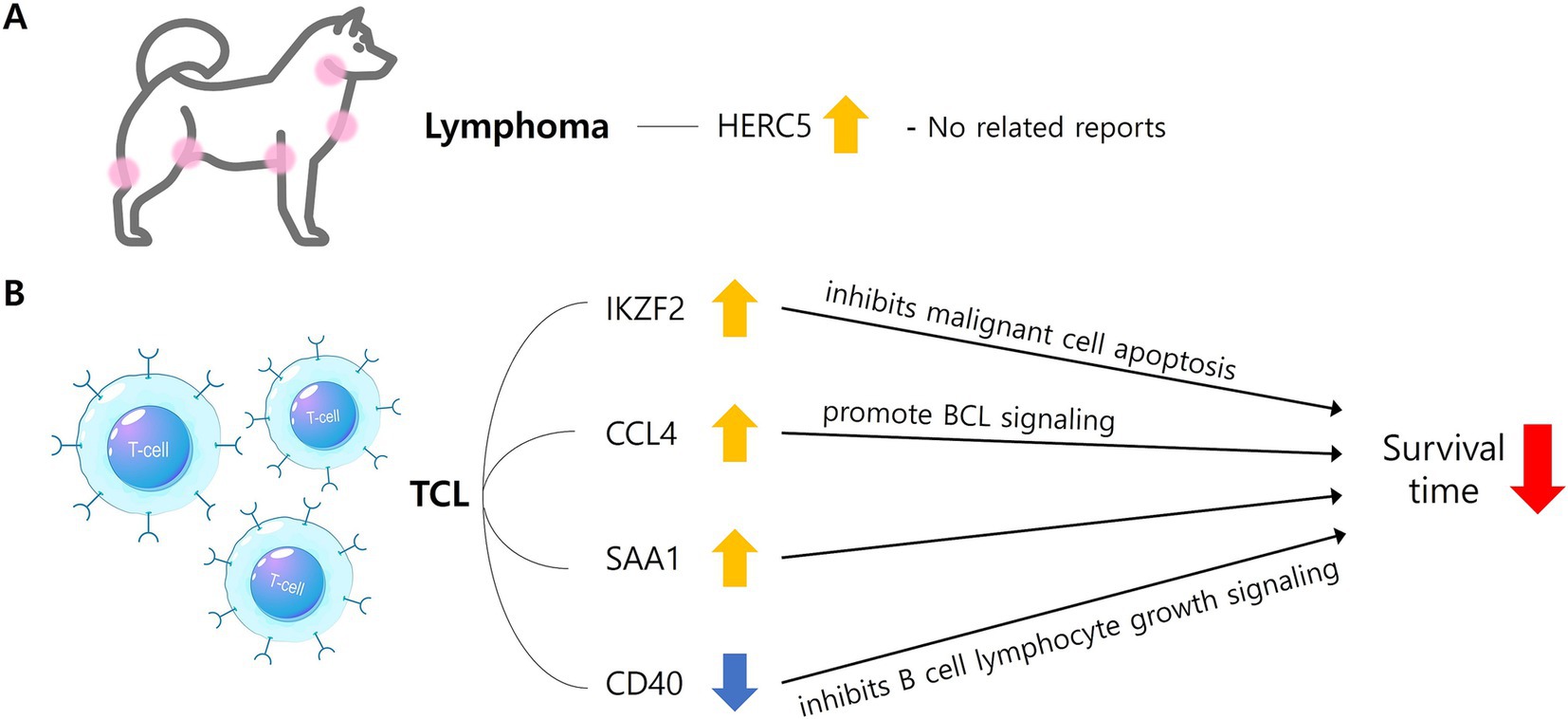
Figure 2. The mechanism by which significant DEGs identified in the lymphoma (A) and TCL group (B) are involved in characteristics. In lymphoma group, HERC5 gene was upregulated (yellow arrow), but there were no related reports with lymphoma in dogs (A). In TCL group, IKZF2, CCL4, and SAA were upregulated (yellow arrow) and CD40 was downregulated (blue arrow), and each was reported to be involved in a shorter survival time through the above mechanisms (B). DEGs, Differentially expressed genes; TCL, T-cell lymphoma; BCL, B-cell lymphoma; HERC5, HECT and RLD domain containing E3 ubiquitin protein ligase 5; IKZF2, IKAROS family zinc finger 2; CCL4, C-C motif chemokine ligand 4; SAA1, Serum amyloid A1; CD40, Cluster of differentiation 40 molecule.
There are reports indicating high concentrations of CCL4 are associated with reduced overall survival and progression-free survival (32, 33). High expression of SAA is significantly related to extranodal lesions, elevated LDH (Lactate Dehydrogenase) levels, and high NCCN-IPI (National Comprehensive Cancer Network-International Prognostic Index) scores, with shorter survival times compared to the control group (34) in human diffuse large B-cell lymphoma (DLBCL), indicating a poor prognosis (Figure 2B). Furthermore, it has been reported that IL1R2 is upregulated in breast cancer, and higher expression is correlated with lower survival rates in humans (35). These findings suggest that these three genes may contribute to the characteristics of canine TCL.
Among the downregulated DEGs, PLEKHA5, GRK4, MYRIP, and CD40 have established associations with various human cancers (36–39). PLEKHA5 (39) has been related with melanoma, while GRK4 (36) and MYRIP (38) have been associated with hepatocellular carcinoma in humans. These genes exhibit to have low expression levels in human cancers, and their downregulation has been correlated with the promotion of tumor metastasis and poorer clinical outcomes. Furthermore, CD40 is downregulated in human DLBCL, and lower expression linked to decreased survival rates (37). This suggests that CD40 may be a gene of significance concerning poorer clinical outcomes in TCL as opposed to BCL in dogs (Figure 2B). Among the enriched pathways identified in the canine TCL group compared to the canine BCL group, human T-cell leukemia virus 1 infection (40–42) and transcriptional misregulation in cancer (43) have previously been associated with lymphoma. The roles of the confirmed DEGs identified in this study are summarized in Table 7 (44–53).
This study faced several limitations including small sample sizes and discrepancies in group numbers, hindering comprehensive variance analysis of the entire cohort. The exclusive focus on one species (dog) and a single breed (Beagle), within the control group restricted genetic diversity, contrasting with the actual clinical data involving client-owned dogs. Furthermore, the lack of longitudinal follow-up data for the study subjects prevented establishing prognostic evaluations based on differences in gene expression. Despite these limitations, this pioneering study is the first to conduct transcriptome profiling of both normal and lymphoma tissues in dogs, including a comparative analysis between TCL and BCL.
In conclusion, the current study demonstrates that the regulation of specific genes, which has not previously been reported in canine TCL, may function as a prognostic indicator. A comprehensive understanding of these regulatory mechanisms may facilitate the quality of care provided to dogs affected by lymphoma, thereby potentially extending the average lifespan of companion animals. Consequently, this study is considerable significance for both pet owners and their canine companions.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/geo/, GSE285369.
The animal studies were approved by The Institutional Animal Care and Use Committee at Chonnam National University (CNU IACUC-YB-2021-166, CNU IACUC-YB-2022-121 and BMC-IACUC-R-2022-13). The studies were conducted in accordance with the local legislation and institutional requirements. Written informed consent was obtained from the owners for the participation of their animals in this study.
YK: Conceptualization, Formal analysis, Investigation, Writing – original draft. JK: Investigation, Writing – review & editing. YS: Investigation, Writing – review & editing. KJ: Investigation, Writing – review & editing. SK: Writing – review & editing. H-JK: Conceptualization, Funding acquisition, Supervision, Writing – review & editing, Writing – original draft.
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This study was supported by the Basic Science Research Program through the National Research Foundation of Korea (NRF), funded by the Ministry of Education (NRF-2023R1A2C1005348), and conducted with research funds from the Advanced Technology Center (grant no. 20014237) dedicated to the Korea Evaluation Institute of Industrial Technology (KEIT), an affiliate of the Ministry of Trade, Industry, and Energy.
The authors are grateful for all dogs and to the dog owners for participating in our investigations.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fvets.2024.1473421/full#supplementary-material
1. Valli, VE, Myint, MS, Barthel, A, Bienzle, D, Caswell, J, Colbatzky, F, et al. Classification of canine malignant lymphomas according to the World Health Organization criteria. Vet Pathol. (2011) 48:198–211. doi: 10.1177/0300985810379428
2. Valli, VE, Bienzle, D, and Meuten, DJ. Tumors of the hemolymphatic system In: DJ Meuten, editor. Tumors in domestic animals. 5th ed. Ames, Iowa: Wiley (2016). 203–321.
3. Thalheim, L, Williams, LE, Borst, LB, Fogle, JE, and Suter, SE. Lymphoma immunophenotype of dogs determined by immunohistochemistry, flow cytometry, and polymerase chain reaction for antigen receptor rearrangements. J Vet Intern Med. (2013) 27:1509–16. doi: 10.1111/jvim.12185
4. Starrak, G, Berry, CR, Page, RL, Johnson, JL, and Thrall, DE. Correlation between thoracic radiographic changes and remission/survival duration in 270 dogs with lymphosarcoma. Vet Radiol Ultrasound. (1997) 38:411–8. doi: 10.1111/j.1740-8261.1997.tb00863.x
5. Limmer, S, Nerschbach, V, Eberle, N, Teske, E, and Betz, DS. Efficacy and tolerability of a 12-week combination chemotherapy followed by lomustine consolidation treatment in canine B- and T-cell lymphoma. Acta Vet Scand. (2022) 64:36. doi: 10.1186/s13028-022-00660-z
7. Zhong, W, Mark, G, and Michael, S. RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet. (2009) 10:57–63. doi: 10.1038/nrg2484
8. Gregory, J, John, B, David, G, Gurmit, G, Christina, M, Matthew, B, et al. Transcriptomic profiling in canines and humans reveals cancer specific gene modules and biological mechanisms common to both species. PLoS Comput Biol. (2021) 17:e1009450. doi: 10.1371/journal.pcbi.1009450
9. Sheet, S, Oh, YI, Arora, D, Choi, BH, Ko, M, Nam, Y, et al. Insight into the potential candidate genes and signaling pathways involved in lymphoma disease in dogs using a comprehensive whole blood transcriptome analysis. Gene Combis. (2022) 838:146735. doi: 10.1016/j.gene.2022.146735
10. Cascione, L, Giudice, L, Ferraresso, S, Marconato, L, Giannuzzi, D, Napoli, S, et al. Long non-coding RNAs as molecular signatures for canine B-cell lymphoma characterization. Noncoding RNA. (2019) 5:47. doi: 10.3390/ncrna5030047
11. Giannuzzi, D, Marconato, L, Cascione, L, Comazzi, S, Elgendy, R, Pegolo, S, et al. Mutational landscape of canine B-cell lymphoma profiled at single nucleotide resolution by RNA-seq. PLoS One. (2019) 14:e0215154–4. doi: 10.1371/journal.pone.0215154
12. Owen, LN. World health organization. TNM classification of tumors in domestic animals. 1st ed. Geneva: World Health Organization (1980).
13. Aljohani, AI, Joseph, C, Kurozumi, S, Mohammed, OJ, Miligy, IM, Green, AR, et al. Myxovirus resistance 1 (MX1) is an independent predictor of poor outcome in invasive breast cancer. Breast Cancer Res Treat. (2020) 181:541–51. doi: 10.1007/s10549-020-05646-x
14. Lu, L, Wang, H, Fang, J, Zheng, J, Liu, B, Xia, L, et al. Overexpression of OAS1 is correlated with poor prognosis in pancreatic cancer. Front Oncol. (2022) 12:944194. doi: 10.3389/fonc.2022.944194
15. Zhang, Y, and Yu, C. Prognostic characterization of OAS1/OAS2/OAS3/OASL in breast cancer. BMC Cancer. (2020) 20:575. doi: 10.1186/s12885-020-07034-6
16. Huang, Z, Yi, L, Jin, L, Chen, J, Han, Y, Zhang, Y, et al. Systematic analysis of virus nucleic acid sensor DDX58 in malignant tumor. Front Microbiol. (2022) 13:1085086. doi: 10.3389/fmicb.2022.1085086
17. Niu, P, Yao, B, Wei, L, Zhu, H, Fang, C, and Zhao, Y. Construction of prognostic risk prediction model based on high-throughput sequencing expression profile data in childhood acute myeloid leukemia. Blood Cells Mol Dis. (2019) 77:43–50. doi: 10.1016/j.bcmd.2019.03.008
18. Wang, X, Chang, S, Wang, T, Wu, R, Huang, Z, Sun, J, et al. IL7R is correlated with immune cell infiltration in the tumor microenvironment of lung adenocarcinoma. Front Pharmacol. (2022) 13:857289. doi: 10.3389/fphar.2022.857289
19. Chen, J, Shin, VY, Ho, JCW, Siu, MT, Cheuk, IWY, and Kwong, A. Functional implications of cathelicidin antimicrobial protein in breast cancer and tumor-associated macrophage microenvironment. Biomol Ther. (2020) 10:688. doi: 10.3390/biom10050688
20. Giacchetti, S, Gauvillé, C, Crémoux, PD, Bertin, L, Berthon, P, Abita, JP, et al. Characterization, in some human breast cancer cell lines, of gastrin-releasing peptide-like receptors which are absent in normal breast epithelial cells. Int J Cancer. (1990) 46:293–8. doi: 10.1002/ijc.2910460226
21. Gugger, M, and Reubi, JC. Gastrin-releasing peptide receptors in non-neoplastic and neoplastic human breast. Am J Pathol. (1999) 155:2067–76. doi: 10.1016/S0002-9440(10)65525-3
22. Giaccone, G, Battey, JF, Gazdar, AF, Oie, HK, Draoui, M, and Moody, TW. Neuromedin B is present in lung cancer cell lines. Cancer Res. (1992) 52:2732s–6s.
23. Siegfried, JM, Krishnamachary, N, Gaither Davis, A, Gubish, C, Hunt, JD, and Shriver, SP. Evidence for autocrine actions of neuromedin B and gastrin-releasing peptide in non-small cell lung cancer. Pulm Pharmacol Ther. (1999) 12:291–302. doi: 10.1006/pupt.1999.0210
24. Chave, HS, Gough, AC, Palmer, K, Preston, SR, and Primrose, JN. Bombesin family receptor and ligand gene expression in human colorectal cancer and normal mucosa. Br J Cancer. (2000) 82:124–30. doi: 10.1054/bjoc.1998.0888
25. Sun, B, Schally, AV, and Halmos, G. The presence of receptors for bombesin/GRP and mRNA for three receptor subtypes in human ovarian epithelial cancers. Regul Pept. (2000) 90:77–84. doi: 10.1016/S0167-0115(00)00114-2
26. Agostinelli, C, Rizvi, H, Paterson, J, Shende, V, Akarca, AU, Agostini, E, et al. Intracellular TCR-signaling pathway. Am J Surg Pathol. (2014) 38:1349–59. doi: 10.1097/PAS.0000000000000309
27. Serwold, T, Hochedlinger, K, Swindle, J, Hedgpeth, J, Jaenisch, R, and Weissman, IL. T-cell receptor-driven lymphomagenesis in mice derived from a reprogrammed T cell. Proc Natl Acad Sci USA. (2010) 107:18939–43. doi: 10.1073/pnas.1013230107
28. Aresu, L, Ferraresso, S, Marconato, L, Cascione, L, Napoli, S, Gaudio, E, et al. New molecular and therapeutic insights into canine diffuse large B-cell lymphoma elucidates the role of the dog as a model for human disease. Haematologica. (2019) 104:e256–9. doi: 10.3324/haematol.2018.207027
29. Majchrzak, A, Witkowska, M, and Smolewski, P. Inhibition of the PI3K/Akt/mTOR signaling pathway in diffuse large B-cell lymphoma: current knowledge and clinical significance. Molecules. (2014) 19:14304–15. doi: 10.3390/molecules190914304
30. Xu, B, Liu, F, Gao, Y, Sun, J, Li, Y, Lin, Y, et al. High expression of IKZF2 in malignant T cells promotes disease progression in cutaneous T cell lymphoma. Acta Derm Venereol. (2021) 101:adv00613–3. doi: 10.2340/actadv.v101.570
31. Nakase, K, Ishimaru, F, Fujii, K, Tabayashi, T, Kozuka, T, Sezaki, N, et al. Overexpression of novel short isoforms of Helios in a patient with T-cell acute lymphoblastic leukemia. Exp Hematol. (2002) 30:313–7. doi: 10.1016/S0301-472X(01)00796-2
32. Ek, S, Björck, E, Högerkorp, C-M, Nordenskjöld, M, Porwit-MacDonald, A, and Borrebaeck, C. Mantle cell lymphomas acquire increased expression of CCL4, CCL5 and 4-1BB-L implicated in cell survival. Int J Cancer. (2006) 118:2092–7. doi: 10.1002/ijc.21579
33. Takahashi, K, Sivina, M, Hoellenriegel, J, Oki, Y, Hagemeister, FB, Fayad, L, et al. CCL3 and CCL4 are biomarkers for B cell receptor pathway activation and prognostic serum markers in diffuse large B cell lymphoma. Br J Haematol. (2015) 171:726–35. doi: 10.1111/bjh.13659
34. Hu, L, Hou, SL, Liu, S, Li, X, Li, L, Cui, JY, et al. Clinical value of serum amyloid a and misfolded transthyretin for relapsed/refractory diffuse large B-cell lymphoma patients. Zhongguo Shi Yan Xue Ye Xue Za Zhi. (2020) 28:1923–32. doi: 10.19746/j.cnki.issn.1009-2137.2020.06.022
35. Zhang, L, Qiang, J, Yang, X, Wang, D, Rehman, A, He, X, et al. IL1R2 blockade suppresses breast tumorigenesis and progression by impairing USP15-dependent BMI1 stability. Adv Sci. (2020) 7:1901728–8. doi: 10.1002/advs.201901728
36. Luo, Y, Wang, Z, Xiao, S, Li, R, and Jiang, X. G protein-coupled receptor kinase 4 is a novel prognostic factor in hepatocellular carcinoma. Dis Markers. (2022) 2022:1–2628879. doi: 10.1155/2022/2628879
37. Song, G, Ni, H, Zou, L, Wang, S, Tian, F, Liu, H, et al. Expression of CD40 is a positive prognostic factor of diffuse large B-cell lymphoma treated with R-CHOP (rituximab, cyclophosphamide, doxorubicin, vincristine, and prednisone). Onco Targets Ther. (2016) 9:3799–805. doi: 10.2147/OTT.S96992
38. Yang, JD, Seol, SY, Leem, SH, Kim, YH, Sun, Z, Lee, JS, et al. Genes associated with recurrence of hepatocellular carcinoma: integrated analysis by gene expression and methylation profiling. J Korean Med Sci. (2011) 26:1428. doi: 10.3346/jkms.2011.26.11.1428
39. Zhang, H, Zhu, H, Deng, G, Zito, CR, Oria, VO, Rane, CK, et al. PLEKHA5 regulates tumor growth in metastatic melanoma. Cancer. (2020) 126:1016–30. doi: 10.1002/cncr.32611
40. Bangham, CRM. HTLV-1 persistence and the oncogenesis of adult T cell leukemia/lymphoma. Blood. (2023) 141:2299–306. doi: 10.1182/blood.2022019332
41. Tan, BJY, Sugata, K, Reda, O, Matsuo, M, Uchiyama, K, Miyazato, P, et al. HTLV-1 infection promotes excessive T cell activation and transformation into adult T cell leukemia/lymphoma. J Clin Invest. (2021) 131:e150472. doi: 10.1172/JCI150472
42. Zhang, L, Wei, J, Wang, L, Huang, S, and Chen, J. Human T-cell lymphotropic virus type 1 and its oncogenesis. Acta Pharmacol Sin. (2017) 38:1093–103. doi: 10.1038/aps.2017.17
43. Kuang, Z, Guo, L, and Li, X. Identification of key genes and pathways associated with classical Hodgkin lymphoma by bioinformatics analysis. Mol Med Rep. (2017) 16:4685–93. doi: 10.3892/mmr.2017.7158
44. Boerner, JL, Nechiporchik, N, Mueller, KL, Polin, L, Heilbrun, L, Boerner, SA, et al. Protein expression of DNA damage repair proteins dictates response to topoisomerase and PARP inhibitors in triple-negative breast Cancer. K Borgmann, editor. PLoS One. (2015); 10::e0119614. doi: 10.1371/journal.pone.0119614
45. Wong, J, Pung, YF, Sze, NSK, and Chin, KC. HERC5 is an IFN-induced HECT-type E3 protein ligase that mediates type I IFN-induced ISGylation of protein targets. Proc Natl Acad Sci USA. (2006) 103:10735–40. doi: 10.1073/pnas.0600397103
46. Justesen, J, Hartmann, R, and Kjeldgaard, NO. Gene structure and function of the 2′-5′-oligoadenylate synthetase family. Cell Mol Life Sci. (2000) 57:1593–612. doi: 10.1007/pl00000644
47. Rebouillat, D, and Hovanessian, AG. The human 2′, 5’-Oligoadenylate Synthetase family: interferon-induced proteins with unique enzymatic properties. J Interf Cytokine Res. (1999) 19:295–308. doi: 10.1089/107999099313992
48. Coffelt, SB, Waterman, RS, Florez, L, Höner zu Bentrup, K, Zwezdaryk, KJ, Tomchuck, SL, et al. Ovarian cancers overexpress the antimicrobial protein hCAP-18 and its derivative LL-37 increases ovarian cancer cell proliferation and invasion. Int J Cancer. (2008) 122:1030–9. doi: 10.1002/ijc.23186
49. Srivastava, S, Mohanty, A, Nam, A, Singhal, S, Salgia, R, Margarita, M, et al. Chemokines and NSCLC: emerging role in prognosis, heterogeneity, and therapeutics. Semin Cancer Biol. (2022) 86:233–46. doi: 10.1016/j.semcancer.2022.06.010
50. Coffelt, SB, Marini, FC, Watson, K, Zwezdaryk, KJ, Dembinski, JL, LaMarca, HL, et al. The pro-inflammatory peptide LL-37 promotes ovarian tumor progression through recruitment of multipotent mesenchymal stromal cells. Proc Natl Acad Sci. (2009) 106:3806–11. doi: 10.1073/pnas.0900244106
51. Miyazaki, M, Lamharzi, N, Schally, AV, Halmos, G, Szepeshazi, K, Groot, K, et al. Inhibition of growth of MDA-MB-231 human breast cancer xenografts in nude mice by bombesin/gastrin-releasing peptide (GRP) antagonists RC-3940-II and RC-3095. Eur J Cancer. (1998) 34:710–7. doi: 10.1016/S0959-8049(97)10123-X
52. Gabay, C, and Kushner, I. Acute-phase proteins and other systemic responses to inflammation. N Engl J Med. (1999) 340:448–54. doi: 10.1056/NEJM199902113400607
Keywords: lymphoma, T-cell, B-cell, transcriptome, microarray, heterogeneity, dog
Citation: Kim Y, Kim J, Song Y, Jang K, Kim SE and Kim H-J (2025) Sequential transcriptome profiling: comparative analysis of normal and canine lymphoma preceding detailed T-cell and B-cell subtype comparison. Front. Vet. Sci. 11:1473421. doi: 10.3389/fvets.2024.1473421
Received: 31 July 2024; Accepted: 05 December 2024;
Published: 22 January 2025.
Edited by:
Maria Elena Turba, Genefast srl, ItalyReviewed by:
Byeongteck Kang, Chungbuk National University, Republic of KoreaCopyright © 2025 Kim, Kim, Song, Jang, Kim and Kim. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ha-Jung Kim, a2ltaGo2MTRAam51LmFjLmty
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.