
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Vet. Sci. , 21 October 2024
Sec. Livestock Genomics
Volume 11 - 2024 | https://doi.org/10.3389/fvets.2024.1465389
This article is part of the Research Topic Insights in Livestock Genomics View all 6 articles
Meat and its derivatives serve as crucial sources of protein, vitamins, minerals, and other essential nutrients for humans. Pork stands as China’s primary animal-derived food product consumed widely across diverse dietary structures; evaluating intramuscular fat content becomes pivotal in assessing its quality standards. Nonetheless, the intricate molecular mechanisms governing intramuscular fat deposition remain elusive. Our study utilized sequencing technology to scrutinize longitudinal development stages within Ningxiang pig’s longest dorsal muscles aiming to unravel these underlying mechanisms. In three distinct comparisons (30d vs. 90d, 90d vs. 150d and 150d vs. 210d) there were 578, 1,000 and 3,238 differentially expressed mRNA, along with 16, 158 and 85 lncRNAs were identified. STEM analysis unveiled six enriched model profiles for lncRNAs while seven such profiles emerged for mRNAs; notably, multiple shared model profiles existed between both RNA types. Enriched analysis highlighted numerous genes from mRNA profile8 and lncRNA profile7 significantly associated with pathways linked to fat deposition. Weight Gene Co-Expression Network Analysis (WGCNA) revealed that differential expression modules (DMEs) & differential expression lncRNAs primarily clustered within cyan, dark slate blue and pale turquoise modules. Furthermore, target genes PKD2 (MSTRG21592.MTRSG8859 and MTRSG18175), COL5A1 (MTRSG9969 and MTRSG180) and SOX13 (MTRSG21592 and MTRSG9088) as core components all intricately tied into processes related to fat deposition. This study lays the groundwork for deeper exploration into the molecular mechanisms underlying LDM fat deposition traits, and it also presents candidate genes for future molecular marker-assisted breeding.
As the primary source of human meat on Earth, domestic pigs contribute 40% of global meat consumption (1). Geographical analysis indicates that pig farming was widespread across most regions of the world, with Asia leading with a 60% share (2). According to statistics from FAO Statistical Databases (FAOSTAT: FAO Statistical Databases (FAO, 2023).), in 2023, China’s pig inventory accounted for 58.16% of global pig inventory. As a major producer and consumer of pigs, the quality of pork in China is crucial. The Ningxiang (NX) pig, a renowned local breed, is cherished by consumers for its exceptional meat quality and flavor. Research has shown that the intramuscular fat (IMF) content in NX pigs stands at 5%, significantly higher than the 2% found in commercial pigs (3). Furthermore, indicators such as tenderness and water holding capacity of NX pigs are notably superior to those of commercial pigs (4). However, compared to breeds like Duroc, the growth cycle of NX pigs is longer, leading to higher feeding costs. The skeletal muscles of pigs are primarily characterized by hyperplasia and hypertrophy. Following birth, muscle fiber cells are fixed in structure allowing only for the hypertrophy of these cells to increase muscle mass as is also the case with adipocytes (5). Therefore an in-depth exploration into skeletal muscles is crucial for understanding reasons behind high IMF content in NX pigs.
Long non-coding RNAs (lncRNAs) are prevalent in animals and lacks the capacity for coding. Initially, they were thought to disrupt translation, but recent advances in transcription and molecular biology have revealed their crucial role in various biological processes, including gene imprinting (6), chromatin remodeling (7), splicing regulation (8), mRNA degradation and translation (9). Research indicates that the lncRNA Has2os consistently increases during skeletal muscle differentiation. However, its knockout inhibits differentiation. Experiments further revealed that Has2os influences muscle cell differentiation by modulating the JNK/MAPK pathway (10). Some lncRNAs regulate muscle development by collaborating with miRNAs and exerting a competitive inhibitory effect. For example, miR-101 and lncRNA MALAT1 regulate muscle development via the Mef2A-p38/MAPK signaling pathway (11). Furthermore, lncRNAs not only play a regulatory role in the growth and differentiation of muscle cells but also partially relieve the binding of miRNA to mRNA through competing endogenous RNA (ceRNA), thereby regulating adipocyte differentiation. AI504432 can reduce the inhibitory effect of miRNA on the Fasn gene by competitively binding to miR-1a-3p, thereby promoting adipocyte differentiation (12). Therefore, lncRNAs have regulatory effects on fat and muscle cells.
Drawing from our prior research, it has been established that the types of muscle fibers and the growth and development of NX pigs’ muscles are regulated by various non-coding RNAs (13), including lncRNA, across different developmental stages (14). Additionally, lncRNAs in the liver and subcutaneous adipose tissue at various stages were also investigated (14). Consequently, this study primarily analyzes the IMF in the LDM of NX pigs at different developmental stages, laying a foundation for understanding the mechanisms underlying IMF variations.
This experiment utilized half-sib animals, born to the same male parent and four female parents under identical rearing conditions. Males were castrated at ages 30, 90, 150, and 210 day and slaughtered at these specific ages to ensure that all pigs slaughtered at a given time point were full siblings. Each time point included three slaughtered animals, totaling twelve in the study. After slaughter, samples of the dorsal muscle from the same region were frozen in liquid nitrogen and stored at −80°C. This study was approved by Use Committee of Hunan Agricultural University, Changsha city, Hunan Province, China and Institutional Animal Care under approval number 2021–13.
The total RNA from tissue samples was then checked by the Nanodrop2000 (NanoDrop Technologies, Wilmington, DE, United States) for extracted RNA concentration and purity, followed by RNA integrity detection using agarose gel electrophoresis and RIN values determined using Agilent2100 (Agilent Technologies, Santa Clara, CA, United States). The requirements for a single library construction are total RNA of 5 ug, concentration of 250 ng/uL, OD 260/280 between 1.8 and 2.2. The Ribo Zero Magnetic kit (Epicentre, Madison, WI, United States) is employed to eliminate ribosomal RNA. Subsequently, the TruSeqTM Stranded Total RNA Library Prep Kit is utilized to construct strand-specific libraries for the detection of lncRNA and mRNA. Sequencing of strand-specific libraries and small RNA libraries was conducted on the HiSeq 4,000 platform using PE150 and SE50, respectively. The deep sequencing was executed by Majorbio Biopharmaceutical Technology Co., Ltd. (Shanghai, China).
Due to the presence of many interfering sequences and low-quality reads in the original sequencing data, we used Sickle1 and SeqPrep2 to filter out the original sequence information for sequencing, removing uninserted fragments and fragments carrying connecting sub sequences, and screening out reads with N values greater than 10%. Align and locate the obtained clean reads with the reference genome of NX pigs, while cutting the large reads into smaller segments aligned with the reference genome.
Select RNA with a length greater than 200 bp that does not overlap with known mRNA, and then score the selected RNA using CNCI 2.0 (15). RNA with a score of<0 will be excluded, and the remaining RNA will be scored using CPC 0.9-r2 (16). RNA with a score of<1 will be excluded, and the remaining RNA will be considered lncRNA.
StringTie was used to display the expression levels of mRNA and lncRN by calculating the FPKM of each sample. Then use R package edge R to calculate the mRNA and lncRNA differentially expressed with log2 (fold change) <−1 or log2 (fold change) >1 and statistical significance (p < 0.005). To investigate the enrichment of differentially expressed genes in various closed groups, we employed Gene Ontology (GO) and the Kyoto Encyclopedia of Genes and Genomes (KEGG).
LncRNA may bind to neighboring target genes through cis interactions, thereby regulating gene expression. Therefore, we used a Python script to select the target genes of lncRNA, and functional analysis was performed on the upstream and downstream coding genes of lncRNA using BLAST2GSO (17). The significantly expressed p < 0.005. TargetScan and miRanda were used to predict the association between lncRNA and mRNA, and Pearson correlation coefficients were used to evaluate the association between lncRNA and mRNA. Finally, Cyto-scape software was used to visualize the regulatory network.
STEM was utilized to categorize the genes and lncRNA expression patterns. Following the importation of mRNA and lncRNA data from four distinct time periods, they will be organized into clusters based on genes exhibiting similar expression trends, subsequently being assigned to the same module. The colored background indicates a significant trend (adjusted p ≤ 0.05 by Bonferroni correction), while the gray one indicates no significant trend. Modules of the same color demonstrate similar trends in gene expression.
Initially, a gene co-expression network is established. The Pearson correlation coefficient is then calculated for the expression patterns of pairs of genes. Using this correlation coefficient, a gene network is constructed. Subsequently, genes with closely related expression patterns are grouped into modules based on a defined threshold (β value). Feature values are assigned to these modules, and GO enrichment analysis is conducted on the individual genes within them.
Using the Animal Total RNA Kit (Tiangen, China) to extract total RNA from muscle of NX pigs, Prime-script RT Master Kit (Thermo Scientific, United States) and random primers were used to reverse the total RNA into cDNA. Then, Primer 5.0 was used to design primers, and all reactions were carried out three times for each sample. The relative expression levels of lncRNAs and mRNAs were was calculated via the comparative CT method (2–△△CT). Please refer to previous papers for details (18).
The unpaired two-tailed T-test was utilized to assess the differential expression of mRNA and lncRNA at 2 distinct time points; data were showed as ± SEM.
The LDM of NX pigs at four different developmental stages was detected to be 128,342,221, 97,849,492, 105,862,852, and 11,0513,258 clean reads with Q30 exceeding 95.24% (Table 1). In total, 3,170 novel RNAs were identified, which comprised of 1,443 intergenic, 47 sense intro overlapping, 828 antisense, 817 sense exons and 35 bidirectional (Supplementary material S2). The majority of lncRNAs consist of 2 or 3 exons (Figure 1A). Additionally, lncRNAs are generally shorter in length and exhibit lower expression levels compared to mRNA (Figures 1B,C). Furthermore, data pertaining to open reading frames reveals that lncRNAs typically possess shorter open reading frames than mRNA (Figure 1D). Analysis of the mRNA expression profiles reveals a similarity in expression patterns at 30d and 90d, with a majority of mRNA exhibiting low levels of expression. However, there is a notable increase in high-expression genes during the middle and later stages (Figure 1E). In contrast, lncRNA demonstrates high expression at 30d and 90d but relatively weak expression at 210d, suggesting a potential attenuation of regulatory control in the later stages of development (Figure 1F).
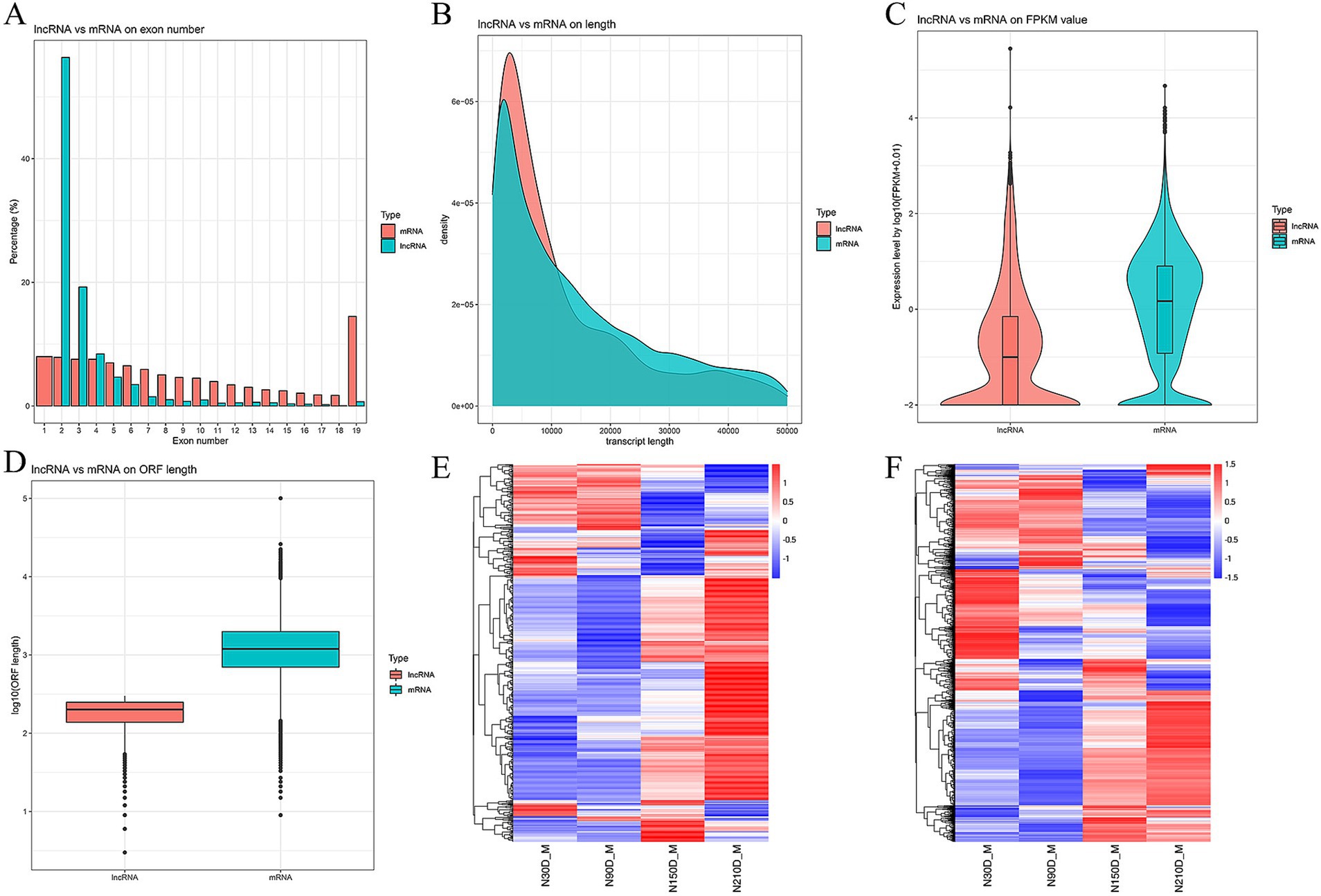
Figure 1. Comparison between lncRNA and mRNA on exon number (A), length (B), fragments per kilobase of exon per million mapped reads (FRKM) value (C), and open reading frame (ORF) length (D). Heat maps depicting differentially expressed mRNAs (E) and lncRNAs (F) along with their expression modes.
To further explore the expression differences of mRNA and lncRNA across various time periods, we conducted pairwise comparisons among four time points, resulting in three closed groups: 30d vs. 90d, 90d vs. 150d, and 150d vs. 210d. Within these three closed groups 578, 1,000 and 3,238 differentially expressed mRNA, along with 16, 158 and 85 lncRNAs were identified (Figures 2A,B).
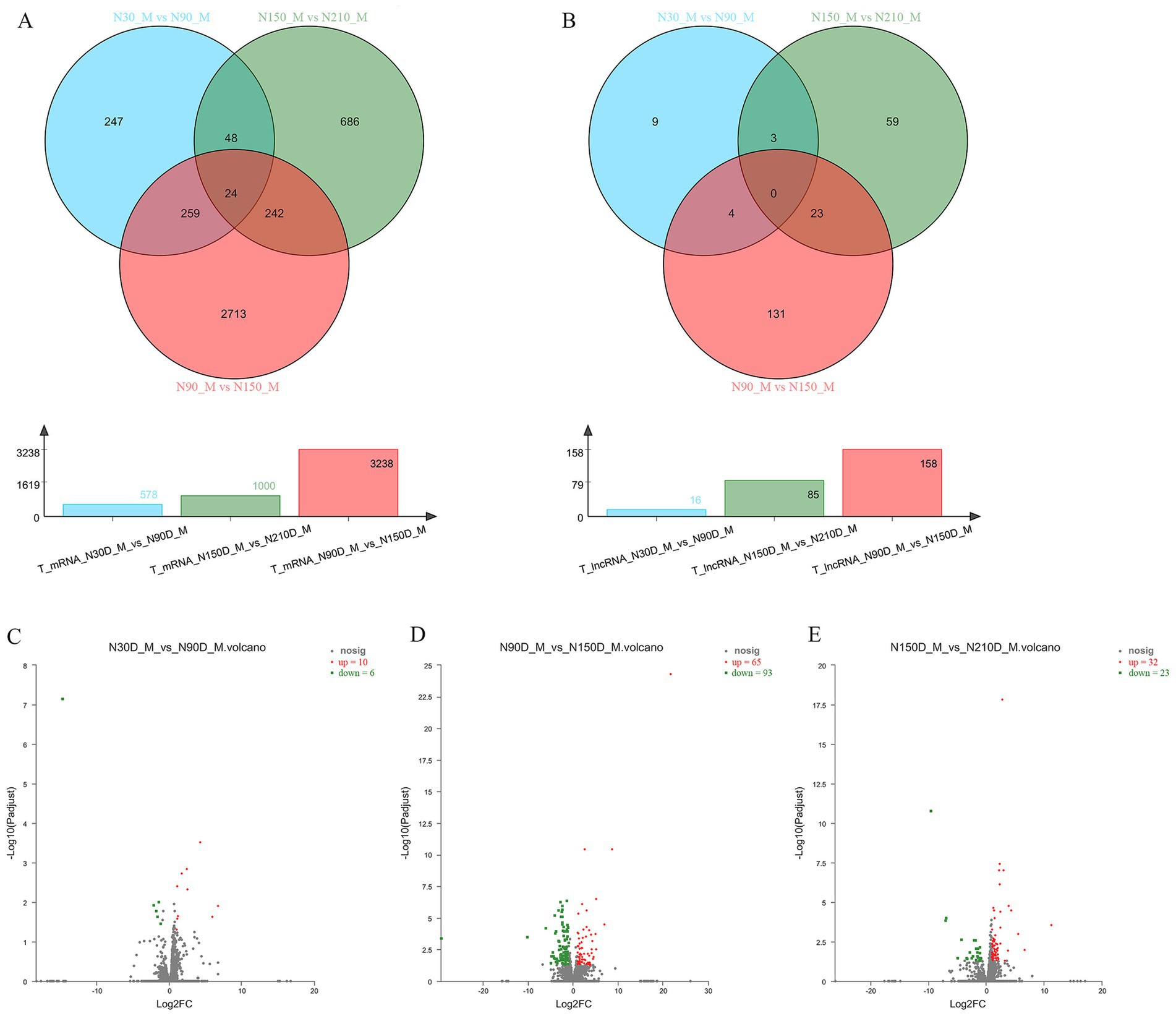
Figure 2. The Venn diagram illustrates the co-expression of mRNA and lncRNA across three distinct groups at four developmental stages, while the bar chart displays the number of co-expressed mRNA and lncRNA in each group (A,B). Volcano plot demonstrating significant up- and down-regulation of genes within three distinct groups (C–E).
By comparing the differentially expressed genes within each group, there were 10 upregulated genes and 6 downregulated genes in the group of 30d vs. 90d, 65 upregulated genes and 93 downregulated genes in the group of 90d vs. 150d, and 32 upregulated genes and 23 downregulated genes in the group of 150d vs. 210d (Figures 2C–D; Supplementary materials S2–S4).
Through STEM analysis of four developmental stages, all lncRNA in the LDM were categorized into 3 cluster profiles, including 6 enriched model profiles. The expression trends for profiles 16, 15, 25 and 13 generally increased, whereas those for profiles 8 and 7 initially decreased before increasing (Figure 3A). The mRNA in the LDM was divided into 3 cluster profiles, with a total of 7 enriched model profiles. Specifically, the trends for profiles 16 and 15 are upward, whereas for profiles 3, 9 and 0, the trends are gradually decreasing (Figure 3B).
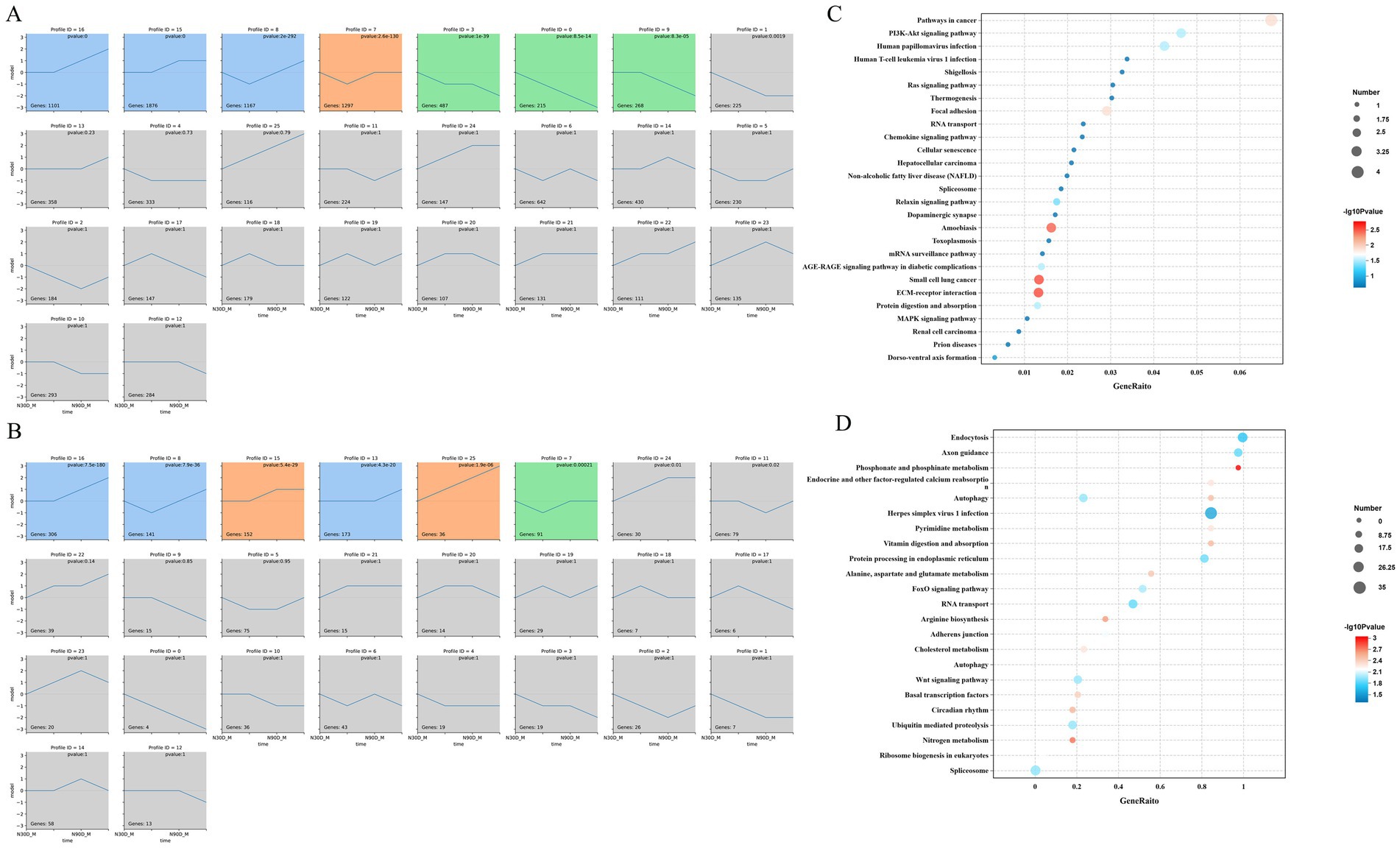
Figure 3. STEM analysis of the mRNA (A) and lncRNA (B). Number on the top means module number. The p-value in the module indicated the statistically significant p-value. Number in the lower-left corner indicate the numbers of mRNAs and lncRNAs in each module. The same color means there belong to same cluster. The enrichment pathway of the mRNA profile 8 (C) and lncRNA profile 7 (D) were show in the picture.
Through enrichment analysis of mRNA from different modules, it was found that mRNA in profile 8 is significantly enriched in pathways such as phosphonate and phosphinate metabolism, Nitrogen metabolism, Wnt signaling pathway, and arginine biosynthesis (Figure 3C). The lncRNA in profile 7 is mainly enriched in signaling pathways such as ECM-receptor interaction, Focal adhesion and PI3K-Akt signaling pathway (Figure 3D).
To explore the potential functional regulatory relationship and mechanism of lncRNA regulatory protein coding genes in the expression process, WGCNA was employed to construct the co-expression network of protein coding genes and lncRNA in the development of muscle tissue in NX pigs. The aim was to investigate the hub gene in different modules and their functions. All samples were available for subsequent analysis under cluster classification. A total of 19,978 mRNA and 2,683 lncRNA obtained by transcription were included in the analysis, while 1,682 lncRNA and 487 mRNA did not meet the requirements and were eliminated. Ultimately, 18,296 mRNA and 2,196 lncRNA were analyzed. These lncRNAs and mRNAs were divided into 13 modules (Figures 4A–C), with a focus on the top three modules (cyan, darkslateblue and paleturquoise) for subsequent analysis (Figure 4D). In the cyan module, genes PAK2, LARS and BIRC6 are positioned centrally and considered as hub genes. Within the darkslateblue module, genes PKD2, SOX13 and PITPNM2 also hold central positions and are regarded as hub genes. Similarly, in the paleturquoise module, genes COL5A1, OLFML2A and P3H3 occupy central positions and are recognized as hub genes (Figures 4E–G and Table 2).
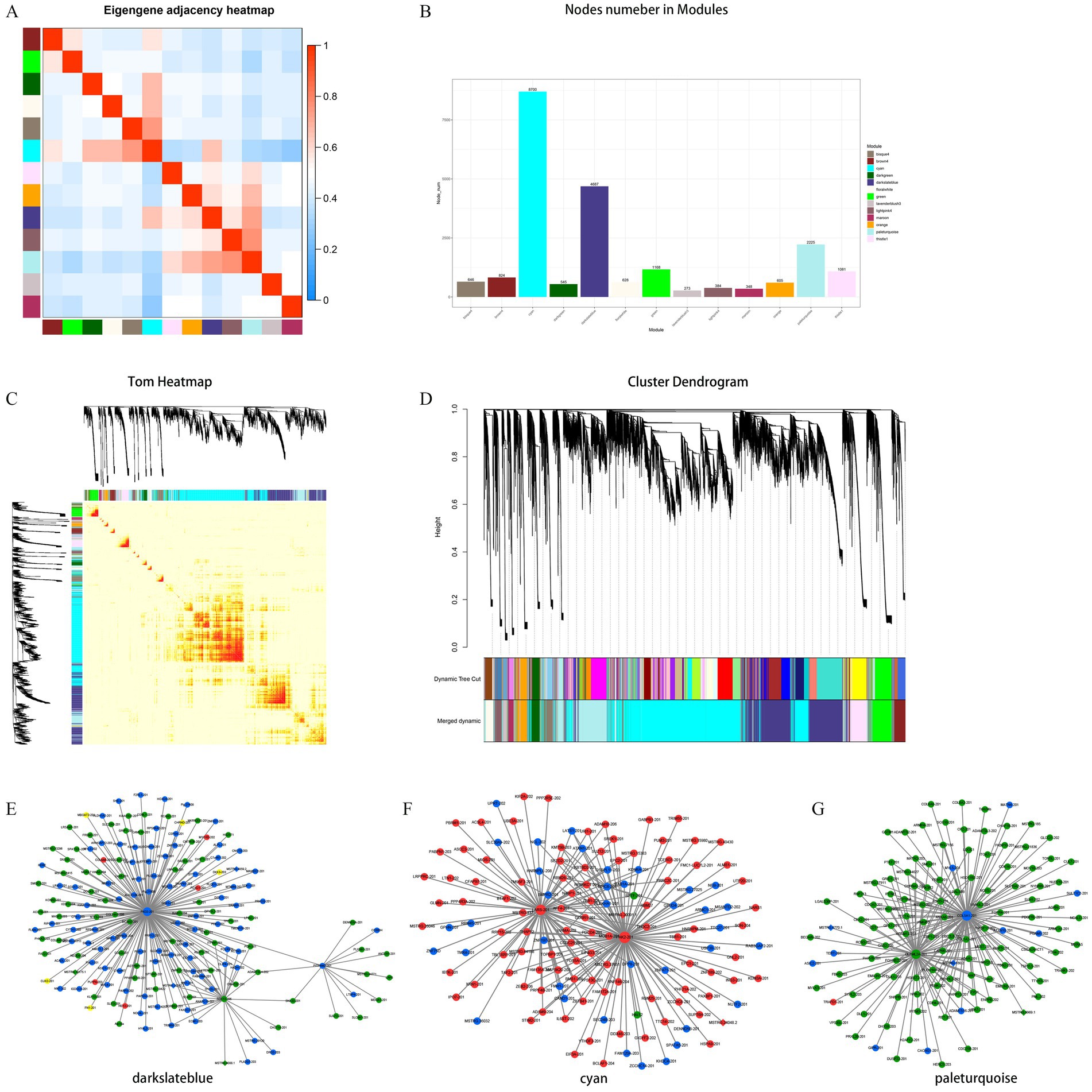
Figure 4. Hierarchical clustering dendrogram of module eigengenes and heatmap of adjacencies (A,C). Number of the lncRNA or mRNA in the 13 modules, the different color means different cluster (B). Hierarchical clustering dendrogram of lncRNA–mRNA co-expression modules. Each branches represents a cluster of lncRNAs or mRNAs. Dynamic tree cut represents original split module, and merged dynamic represents final merge module (D). The darkslateblue represent cellular component module (E), the cyan means biological process module (F), the paleturquoise means molecular function module (G). The up-regulated types are represented in red, while the down-regulated types are depicted in green. Types exhibiting both up- and down-regulation are considered different and are indicated in yellow. Diamonds represent mRNA genes, while rounds represent lncRNA genes.
Six lncRNAs exhibiting high expression levels were randomly selected at four developmental stages of NX pig muscle tissue (30, 90, 150, 210 days after birth), and quantitative analysis was conducted using RT-qPCR with the primers listed in Table 3. Additionally, three genes including ELOVL5 associated with fatty acid metabolism were chosen for quantitative analysis using RT-qPCR with the primers specified in Table 4. As depicted in Figure 5, the RNA-seq data and RT-qPCR results showed a high degree of consistency, validating the reliability of the sequencing outcomes.
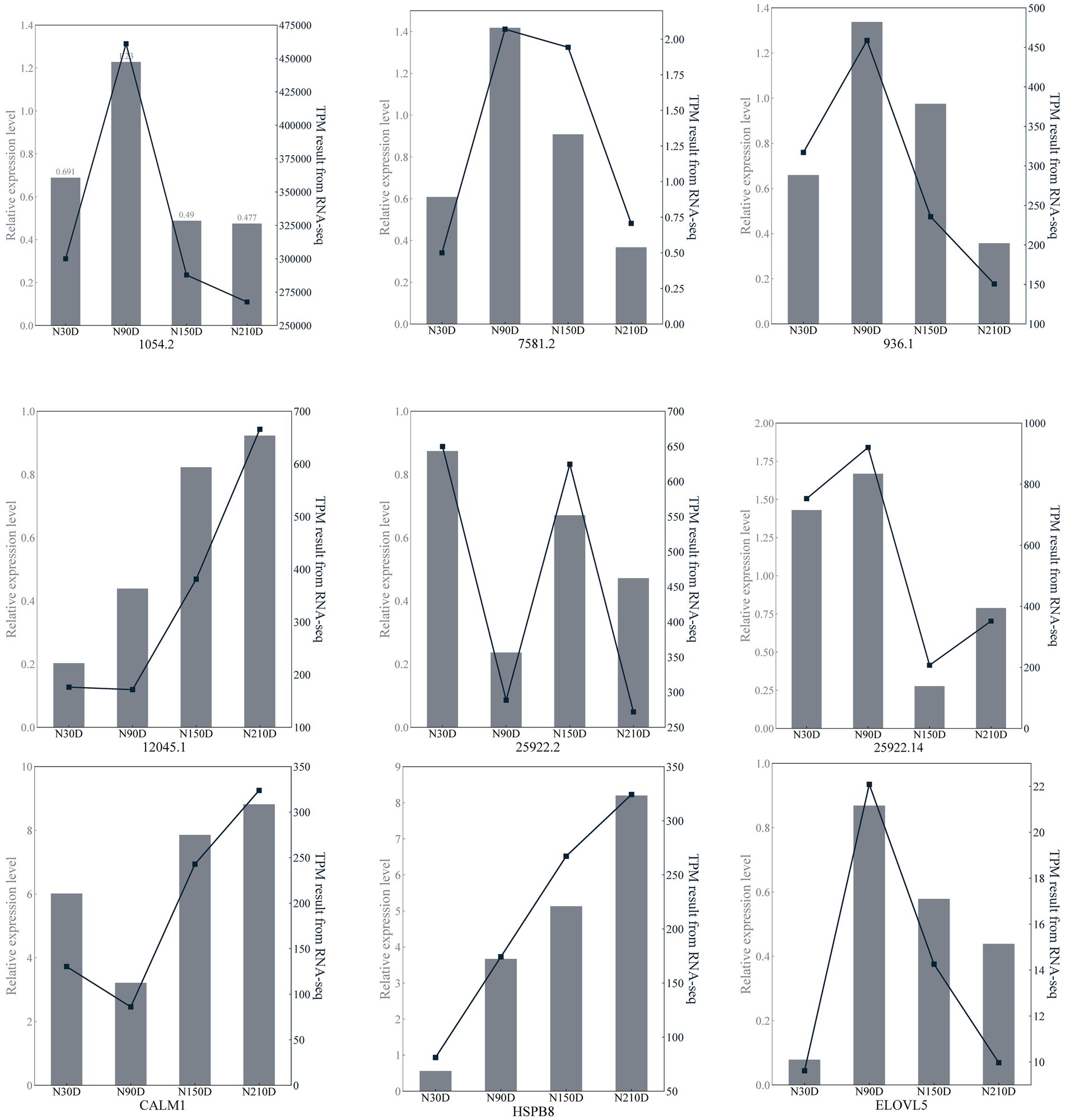
Figure 5. Transcription patterns of MSTRG.1054.2, MSTRG.7581.2, MSTRG.963.1, MSTRG.12045.1, MSTRG.25922.2, MSTRG.25922.14, CLAM1, HSPB8, ELOVL5 compared to expression patterns in the RNA-seq.
Muscle growth and development primarily manifest as the growth and hypertrophy of muscle cells, with minimal changes in the number of muscle fibers. Currently, pork serves as the primary source of animal protein for human consumption. The longissimus dorsi muscle is a key component in evaluating pork quality. Studies on pork quality predominantly focus on the characteristics of muscle fiber and IMF, with IMF content showing a positive correlation with meat tenderness, flavor, and juiciness. NX pigs are renowned for their high-quality pork. An increasing body of research suggests that lncRNA plays a significant role in muscle growth and development, IMF deposition, and fatty acid composition. In this study, a substantial number of differentially expressed mRNAs (DEGs) and lncRNAs (DELs) were identified in 4 stages of NX pig muscle tissues: 4816 DEG and 259 DEL (p < 0.05). Among the DEGs and DELs, the greatest difference was observed between 150 days vs. 210 days that indicating significant differences in muscle development at 210 days compared to other time points.
KEGG enrichment analysis of DEGs at four time points revealed that numerous upregulated DEGs were associated with immune pathways during the early growth phase of NX pigs. Specifically, upregulated DEGs at 30 vs. 90 days were primarily enriched in immune signaling pathways, including the T cell receptor signaling pathway, Natural killer cell mediated cytotoxicity and Primary immunodeficiency. In the 90 vs. 150 days comparison, upregulated genes were primarily enriched in pathways such as MAPK signaling pathway, IL-17 signaling pathway, NOD-like receptor signaling pathway and TNF signaling pathway (Supplementary materials S5, S6). In the later growth phase, most upregulated DEGs were associated with pathways related to fat accumulation, including the Wnt signaling pathway, cGMP−PKG signaling pathway and MAPK signaling pathway in the 150 vs. 210 days comparison (Supplementary material S7). 10 lncRNAs expressions were found to be involved in the regulation related to lipid metabolism in 30vs90 days. This indicates that 90 day is an important stage for lipid metabolism during the development of NX pigs. Additionally, when compared with 150 days, it was observed that 23 DELs were downregulated at 210 days. Their target genes were enriched in phospholipase activity regulation and lipase activity regulation, suggesting a reduction in the expression level of lncRNAs involved in lipid regulation function at 210 days. Lipase present in the blood promotes triglyceride decomposition to provide fatty acids for triglyceride synthesis and fat deposition promotion. However, as lipase mainly participates in body fat decomposition, a decline in its regulatory function can lead to obesity and further promote fat deposition. At 210 days, reduced lncRNA expression levels regulating lipase activity accelerated fat deposition within muscle tissue.
This study employed STEM analysis to uncover the dynamic expression patterns of lncRNA and mRNA during the development of LDM in NX pigs. The analysis revealed that profiles 16, 8, 15 and 7 exhibited similar expression trends across both omics, suggesting a high degree of correlation between these modules during pig development. Predictions of lncRNA target genes revealed that certain protein-coding genes within profiles 15 and 16 of the mRNA module were also targeted by lncRNA profiles 15 and 16. The target gene of MSTRG.19683.1, MSTRG.20242.2 and MSTRG.860.1 in module 15 is LEPR, while the target gene of MSTRG.19683.1, MSTRG.20242.2 and MSTRG.860.1 is KLF7; additionally, the gene of MSTRG1058 0.1 in module 16 is SMGS1.
The LEPR gene was categorized as a type I cytokine receptor, capable of binding with leptin to regulate lipid metabolism in the body (19). Previous research indicates that this gene serves as a genetic marker linked to growth speed and fattening (20). In Suzhong pigs, which are hybridized with foreign breeds in China, high levels of LEPR expression were detected in both back fat and the LDM, with significantly elevated expression in high-fat pigs compared to low-fat ones (21); KLF7 gene is widely present in animals and is a type of regulatory factor related to transcription and metabolism. It is a member of the SP/KLF transcription factor family and its gene coding sequence exhibits high homology among mammals (22). Induced differentiation of preadipocytes reveals that KLF7 exerts a negative regulatory effect on these cells. Overexpression of KLF7 hinders the differentiation of preadipocytes, leading to reduced expression of PPARγ and A-FABP, whereas overexpression in mature adipocytes has no impact on the expression of PPARγ and A-FABP (23). In broiler chickens, this gene has also been linked to fat traits such as very low density lipoprotein levels and abdominal fat weight (24). In mice fed a high-fat diet, the gene shows significantly elevated expression. Overexpression of this gene can lead to reduced expression of adiponectin and leptin, as well as hexokinase in skeletal muscle, accelerating energy metabolism and enhancing muscle fat deposition (25);SMGS1 gene encodes sphingomyelin synthase, a protein composed of 413 amino acids, which shares up to 97% sequence similarity with human proteins and is extensively expressed in the body (26). Sphingomyelin synthase catalyzes the synthesis of sphingomyelin and diglycerides from lecithin and ceramide. Sphingomyelin synthase modulates lipid raft receptors by regulating sphingomyelin synthesis, thereby influencing fatty acid metabolism. Overexpression of SMGS1 at the cellular level has been observed to enrich the production of triglycerides with polyunsaturated fatty acids (27). Currently, sphingomyelin synthase has emerged as a new target for research in the treatment of arteriosclerosis (28). Based on the STEM decomposition results, the IMF content of LDM in NX pigs is modulated by a variety of lncRNAs during the developmental process and lncRNAs can impact fat deposition and metabolism of NX pigs through diverse metabolic pathways.
To investigate the potential functional regulatory relationship between lncRNA and mRNA during body development, a co-expression network of lncRNA and lncRNA in NX pig muscle tissue development was established to identify the core genes in different modules. The co-expression network analysis revealed PAK2, LARS and BIRC6 as the core genes in the cyan module. PKD2, SOX13 and PITPNM2 were identified as core genes in the darkslateblue module. In the paleturquoise module, the core genes are COL5A1, OLFML2A and P3H3 (Figure 4).
PKD2 (protein kinase D2) serves as an effector for diacylglycerol and protein kinase C (PKC), and also acts as a central regulator of nutritional homeostasis (29). It facilitates lipid uptake in the intestine by directly interacting with intestinal cells. Research indicates that PKD is activated by free fatty acids (FFAs) and diacylglycerol (DAG) (30) and inhibiting PKD pathways results in heightened energy expenditure in adipocytes (31). Within the mRNA-lncRNA co-expression network of NX pig muscle tissue, this gene was found to be associated with three lncRNAs (MSTRG.21592.2, MSTRG.8859.2 and MSTRG.18175.1) and two protein-encoding genes ACADL and FADS3 which are related to fatty acid metabolism. ACADL encodes a long-chain-specific acyl-CoA dehydrogenase, serving as the initial catalyst for releasing energy through mitochondrial fatty acid oxidation. It exhibits activity toward unsaturated acyl-CoA ranging from 6 to 24 carbons in length, with a preference for chains containing 8 to 18 carbons (22). Linkage disequilibrium analysis revealed a significant correlation between porcine ACADL polymorphism and IMF content in the LDM (32). The FADS3 gene facilitates conversion of n-6 polyunsaturated fatty acids (PUFAs) to n-3 PUFAs, thereby enhancing levels of n-3 PUFAs within the body (33).
COL5A1 (Collagen Type V Alpha 1 Chain) encodes a low-abundance fibrous collagen alpha chain. These fibrous collagen molecules form trimers and can be composed of one or more types of alpha chains. Research has showed that the deletion of the PLXND1 gene in zebrafish can lead to increased expression of COL5A1, which in turn promotes the proliferation and differentiation of adipocytes, resulting in morphological hyperplasia of visceral adipose tissue (34). Additionally, Sheng et al. identified COL5A1 as a core gene through WGCNA analysis of adipose tissue from cow (35), while Chen et al. also found COL5A1 to be a core gene through analysis of LDM in pigs, findings consistent with this research (36). MSTRG.9969.1 and MSTRG.180.2 were associated with this gene by co-expression network suggested they will regulate the expression of COL5A1. SOX13 (SRY-Box Transcription Factor 13) is a member of the SOX protein family, extensively expressed in body tissues. The protein, containing an HMG domain, can specifically bind to the DNA sequence AACAAT, endowing it with transcription factor activity. Previous research indicates that islet cell antibody 12 (ICA12) corresponds to the SOX13 protein (37). The growth rate of SOX13 deficient mice is faster than that of normal mice, while inhibiting the differentiation of αβ T cells and the Wnt signaling pathway (38). By WGCNA analysis there were MSTRG.21592.2 and MSTRG.9088.1 are associated with SOX13. The aforementioned studies indicate that, in the developmental process of NX pigs, lncRNA may influence the IMF deposition in LDM by regulating core genes.
In this study unveiled the expression profiles of mRNA and lncRNA at four time points during the development of LDM in NX pigs. STEM analysis revealed multiple expression modules with similar trends between DEGs and DELs at the four developmental time points, while enrichment analysis indicated that numerous mRNAs in profile8 were enriched in pathways associated with fat development, such as the Wnt signaling pathway. Additionally, several lncRNAs were also found to be enriched in PI3K-Atk and other signaling pathways related to fat development. WGCNA analysis demonstrated that DEGs and DELs are primarily divided into three main modules (cyan module, darkslateblue module and paleturquoise module). Core gene analysis within these modules identified PKD2, SOX13 and COL5A1 are implicated in adipose tissue development or adipocyte differentiation, and all three genes are target genes of lncRNA within the network. However, further research is needed to explore the impact of genes including PAK2, LARS, BIRC6, PKD2, SOX13, PITPNM2, COL5A1, OLFML2A and P3H3 on IMF in porcine LDM, as well as the effects of candidate lncRNAs MSTRG.21592.2, MSTRG.8859.2, MSTRG.18175.1, MSTRG.9969.1 and MSTRG.180.2 on meat quality via molecular regulation.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found at: https://www.ncbi.nlm.nih.gov/bioproject/, PRJNA721288.
The animal study was approved by Animal Science and Technology College of Hunan Agriculture University (No. 2021-13). The study was conducted in accordance with the local legislation and institutional requirements.
WC: Writing – original draft, Writing – review & editing. FY: Methodology, Writing – review & editing. SL: Formal analysis, Writing – review & editing. ZL: Software, Writing – review & editing. YG: Data curation, Writing – review & editing. HM: Writing – review & editing.
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This research was funded by the design and cultivation of the new breed in pigs with high-quality meat (2023ZD04046-05), Major Science and Technology Special Plan of Yunnan Province (202202AE090032 and 202102AE090046-3), Postgraduate Scientific Research Innovation Project of Hunan Province (CX20230713), Postgraduate Scientific Research Innovation Project of Hunan Agriculture University (2023XC006), and the Hunan Provincial Department of Education Scientific Research Project (23B1097).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fvets.2024.1465389/full#supplementary-material
1. Underwood, WJ, McGlone, JJ, Swanson, J, Anderson, KA, and Anthony, R. Agricultural Animal Welfare In: K Bayne and PV Turner, editors. Laboratory Animal Welfare. American College of Laboratory Animal Medicine. Boston: Academic Press (2014). 233–78.
2. Ganges, L, Crooke, HR, Bohórquez, JA, Postel, A, Sakoda, Y, Becher, P, et al. Classical swine fever virus: the past, present and future. Virus Res. (2020) 289:198151. doi: 10.1016/j.virusres.2020.198151
3. He, J, Wu, X-L, Zeng, Q, Li, H, Ma, H, Jiang, J, et al. Genomic mating as sustainable breeding for Chinese indigenous Ningxiang pigs. PLoS One. (2020) 15:e0236629. doi: 10.1371/journal.pone.0236629
4. Feng, Z., Guo, J., Kong, X., Wang, W., Li, F., Nyachoti, M., et al. (2012). Molecular cloning and expression profiling of G protein coupled receptor 120 in landrace pig and different Chinese indigenous pig breeds. J. Food Agric. Environ. (2012).
5. Ashmore, CR, Addis, PB, and Doerr, L. Development of muscle fibers in the fetal pig. J Anim Sci. (1973) 36:1088–93. doi: 10.2527/jas1973.3661088x
6. Farhadova, S, Ghousein, A, Charon, F, Surcis, C, Gomez-Velazques, M, Roidor, C, et al. The long non-coding RNA Meg3 mediates imprinted gene expression during Stem cell differentiation. Nucleic Acids Res. (2024) 52:6183–200. doi: 10.1093/nar/gkae247
7. Yu, J, Wang, W, Yang, J, Zhang, Y, Gong, X, Luo, H, et al. LncRNA PSR regulates vascular remodeling through encoding a novel protein Arteridin. Circ Res. (2022) 131:768–87. doi: 10.1161/CIRCRESAHA.122.321080
8. Zhang, F, Wang, H, Yu, J, Yao, X, Yang, S, Li, W, et al. LncRNA CRNDE attenuates Chemoresistance in gastric Cancer via SRSF6-regulated alternative splicing of PICALM. Mol Cancer. (2021) 20:6. doi: 10.1186/s12943-020-01299-y
9. Zhu, Y, Zhou, B, Hu, X, Ying, S, Zhou, Q, Xu, W, et al. LncRNA LINC00942 promotes Chemoresistance in gastric Cancer by suppressing MSI2 degradation to enhance C-Myc mRNA stability. Clin Transl Med. (2022) 12:e703. doi: 10.1002/ctm2.703
10. Chen, W, Chen, W, Liu, P, Qian, S, Tao, S, Huang, M, et al. Role of lncRNA Has2os in skeletal muscle differentiation and regeneration. Cells. (2022) 11:3497. doi: 10.3390/cells11213497
11. Liu, S, Xie, S, Chen, H, Li, B, Chen, Z, Tan, Y, et al. The functional analysis of transiently upregulated miR-101 suggests a “braking” regulatory mechanism during Myogenesis. Sci China Life Sci. (2021) 64:1612–23. doi: 10.1007/s11427-020-1856-5
12. Wang, S, Wang, R, Hu, Y, Zhang, Y, Yuan, Q, Luo, Y, et al. Long noncoding RNA AI504432 upregulates FASN expression by sponging miR-1a-3p to promote lipogenesis in senescent adipocytes. Cell Signal. (2024) 120:111232. doi: 10.1016/j.cellsig.2024.111232
13. Yu, Z, Xu, X, Ai, N, Wang, K, Zhang, P, Li, X, et al. Integrated analysis of circRNA, lncRNA, miRNA and mRNA to reveal the ceRNA regulatory network of postnatal skeletal muscle development in Ningxiang pig. Front Cell Dev Biol. (2023) 11:11. doi: 10.3389/fcell.2023.1185823
14. Gong, Y, Zhang, Y, Li, B, Xiao, Y, Zeng, Q, Xu, K, et al. Insight into liver lncRNA and mRNA profiling at four developmental stages in Ningxiang pig. Biology. (2021) 10:310. doi: 10.3390/biology10040310
15. Sun, L, Luo, H, Bu, D, Zhao, G, Yu, K, Zhang, C, et al. Utilizing sequence intrinsic composition to classify protein-coding and long non-coding transcripts. Nucleic Acids Res. (2013) 41:e166–6. doi: 10.1093/nar/gkt646
16. Kong, L, Zhang, Y, Ye, Z-Q, Liu, X-Q, Zhao, S-Q, Wei, L, et al. CPC: assess the protein-coding potential of transcripts using sequence features and support vector machine. Nucleic Acids Res. (2007) 35:W345–9. doi: 10.1093/nar/gkm391
17. Conesa, A, Götz, S, García-Gómez, JM, Terol, J, Talón, M, and Robles, M. Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics. (2005) 21:3674–6. doi: 10.1093/bioinformatics/bti610
18. Gong, Y, He, J, Li, B, Xiao, Y, Zeng, Q, Xu, K, et al. Integrated analysis of lncRNA and mRNA in subcutaneous adipose tissue of Ningxiang pig. Biology. (2021) 10:726. doi: 10.3390/biology10080726
19. Ruiz-Cortés, ZT, Men, T, Palin, MF, Downey, BR, Lacroix, DA, and Murphy, BD. Porcine leptin receptor: molecular structure and expression in the ovary. Mol Reprod Dev. (2000) 56:465–74. doi: 10.1002/1098-2795(200008)56:4<465::AID-MRD4>3.0.CO;2-Q
20. Pérez-Montarelo, D, Fernández, A, Folch, JM, Pena, RN, Ovilo, C, Rodríguez, C, et al. Joint effects of porcine leptin and leptin receptor polymorphisms on productivity and quality traits. Anim Genet. (2012) 43:805–9. doi: 10.1111/j.1365-2052.2012.02338.x
21. Georgescu, SE, Manea, MA, Dinescu, S, and Costache, M. Comparative study of leptin and leptin receptor gene expression in different swine breeds. Genet Mol Res GMR. (2014) 13:7140–8. doi: 10.4238/2014.February.14.15
22. Yang, X, Liang, M, Tang, Y, Ma, D, Li, M, Yuan, C, et al. KLF7 promotes adipocyte inflammation and glucose metabolism disorder by activating the PKCζ/NF-κB pathway. FASEB J. (2023) 37:e23033. doi: 10.1096/fj.202300005R
23. Matsuo, H, Kondo, Y, Kawasaki, T, Tokuyama, S, and Imamura, N. Borrelidin isolated from Streptomyces Sp. inhibited adipocyte differentiation in 3T3-L1 cells via several factors including GATA-binding protein 3. Biol Pharm Bull. (2015) 38:1504–11. doi: 10.1248/bpb.b15-00257
24. Sun, Y, Xu, H, Li, J, Peng, M, Jia, Z, Kong, L, et al. Genome-wide survey identifies TNNI2 as a target of KLF7 that inhibits chicken Adipogenesis via downregulating FABP4. Biochim Biophys Acta Gene Regul Mech. (2023) 1866:194899. doi: 10.1016/j.bbagrm.2022.194899
25. Wang, C, Qiao, S, Zhao, Y, Tian, H, Yan, W, Hou, X, et al. The KLF7/PFKL/ACADL Axis modulates cardiac metabolic Remodelling during cardiac hypertrophy in male mice. Nat Commun. (2023) 14:959. doi: 10.1038/s41467-023-36712-9
26. Guillén, N, Navarro, MA, Surra, JC, Arnal, C, Fernández-Juan, M, Cebrián-Pérez, JA, et al. Cloning, characterization, expression and comparative analysis of pig Golgi membrane sphingomyelin synthase 1. Gene. (2007) 388:117–24. doi: 10.1016/j.gene.2006.10.013
27. Deevska, GM, Dotson, PP II, Karakashian, AA, Isaac, G, Wrona, M, Kelly, SB, et al. Novel interconnections in lipid metabolism revealed by overexpression of sphingomyelin Synthase-1. J Biol Chem. (2017) 292:5110–22. doi: 10.1074/jbc.M116.751602
28. Li, Z, Fan, Y, Liu, J, Li, Y, Huan, C, Bui, HH, et al. Impact of sphingomyelin synthase 1 deficiency on sphingolipid metabolism and atherosclerosis in mice. Arterioscler Thromb Vasc Biol. (2012) 32:1577–84. doi: 10.1161/ATVBAHA.112.251538
29. Sumara, G, Formentini, I, Collins, S, Sumara, I, Windak, R, Bodenmiller, B, et al. Regulation of PKD by the MAPK P38delta in insulin secretion and glucose homeostasis. Cell. (2009) 136:235–48. doi: 10.1016/j.cell.2008.11.018
30. Mayer, AE, Löffler, MC, Loza Valdés, AE, Schmitz, W, El-Merahbi, R, Viera, JT, et al. The kinase PKD3 provides negative feedback on cholesterol and triglyceride synthesis by suppressing insulin signaling. Sci Signal. (2019) 12:eaav9150. doi: 10.1126/scisignal.aav9150
31. Löffler, MC, Mayer, AE, Trujillo Viera, J, Loza Valdes, A, El-Merahbi, R, Ade, CP, et al. Protein kinase D1 deletion in adipocytes enhances energy dissipation and protects against adiposity. EMBO J. (2018) 37:e99182. doi: 10.15252/embj.201899182
32. Kojima, M, and Degawa, M. Causes of sex differences in serum cholesterol and triglyceride levels in Meishan pigs. Biol Pharm Bull. (2024) 47:606–10. doi: 10.1248/bpb.b23-00895
33. Brenna, JT, and Kothapalli, KSD. New understandings of the pathway of long-chain polyunsaturated fatty acid biosynthesis. Curr Opin Clin Nutr Metab Care. (2022) 25:60–6. doi: 10.1097/MCO.0000000000000810
34. Minchin, JEN, Dahlman, I, Harvey, CJ, Mejhert, N, Singh, MK, Epstein, JA, et al. Plexin D1 determines body fat distribution by regulating the type V collagen microenvironment in visceral adipose tissue. Proc Natl Acad Sci USA. (2015) 112:4363–8. doi: 10.1073/pnas.1416412112
35. Sheng, H, Pan, C, Wang, S, Yang, C, Zhang, J, Hu, C, et al. Weighted gene co-expression network analysis identifies key modules and central genes associated with bovine subcutaneous adipose tissue. Front Vet Sci. (2022) 9:914848. doi: 10.3389/fvets.2022.914848
36. Chen, C, Ren, H, Li, H, Deng, Y, Cui, Q, Zhu, J, et al. Identification of crucial modules and genes associated with Backfat tissue development by WGCNA in Ningxiang pigs. Front Genet. (2023) 14:1234757. doi: 10.3389/fgene.2023.1234757
37. Park, M, Kim, KH, Jaiswal, V, Choi, J, Chun, JL, Seo, KM, et al. Effect of black ginseng and silkworm supplementation on obesity, the transcriptome, and the gut microbiome of diet-induced overweight dogs. Sci Rep. (2021) 11:16334. doi: 10.1038/s41598-021-95789-8
Keywords: Ningxiang pig, muscle, transcriptome, lncRNA, mRNA
Citation: Chen W, Yang F, Liufu S, Li Z, Gong Y and Ma H (2024) Integrated analysis of muscle lncRNA and mRNA of Chinese indigenous breed Ningxiang pig in four developmental stages. Front. Vet. Sci. 11:1465389. doi: 10.3389/fvets.2024.1465389
Received: 16 July 2024; Accepted: 08 October 2024;
Published: 21 October 2024.
Edited by:
Johann Sölkner, University of Natural Resources and Life Sciences Vienna, AustriaReviewed by:
Yu Bai, Tianjin University of Science and Technology, ChinaCopyright © 2024 Chen, Yang, Liufu, Li, Gong and Ma. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yan Gong, MTM5MTAwMDgxNzVAMTM5LmNvbQ==; Haiming Ma, bWFoYWltaW5nMjAwMEAxNjMuY29t
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.