- 1Applied Parasitology Research Laboratory, Department of Biology, Faculty of Science, Srinakharinwirot University, Bangkok, Thailand
- 2School of Biodiversity, One Health, and Veterinary Medicine, University of Glasgow, Glasgow, United Kingdom
- 3James Watt School of Engineering, University of Glasgow, Glasgow, United Kingdom
- 4Food Control Department, Faculty of Veterinary Medicine, Kafrelsheikh University, Kafr-El-Sheikh, Egypt
- 5Kreavet, Kruibeke, Belgium
- 6Research and Innovation Unit for Diagnosis of Medical and Veterinary Important Parasites, Faculty of Science, Srinakharinwirot University, Bangkok, Thailand
The bovine lungworm, Dictyocaulus viviparus (Bloch, 1782), is highly pathogenic and disease outbreaks can be difficult to predict and manage. Rapid and accurate diagnosis is vital, but without a sensitive diagnostic test this remains challenging in clinical practice. High performance molecular detection tools are therefore required to improve the diagnosis of this parasite and promote the implementation of strategic control measures. Loop-mediated isothermal amplification (LAMP), a rapid DNA assay, offers potential for field-based detection. Here we report a novel LAMP assay (DviLAMP), that was designed to target the D. viviparus internal transcribed spacer 2 (ITS2) ribosomal DNA region. Firstly, genomic DNA was extracted from a single D. viviparus L1 larva to amplify and clone the ITS2 into the recombinant plasmid (DviITS2). The DviLAMP successfully detected the target, with results shown by gel electrophoresis and real-time analysis, in addition to point-of-care amenable end-point detection: colorimetry and lateral flow dipstick (LFD). Analytical sensitivity can detect 0.5 ng DviITS2 following 45 min of incubation at 64°C, increasing to just 1 pg following 90 min of incubation. Using the same primers, other nematodes of cattle, Ostertagia ostertagi and Cooperia oncophora, were also detectable both by gel electrophoresis and real-time. However, when FITC and biotin tagged primers were incorporated to adapt the DviLAMP to LFD end-point detection, the LFD showed specific detection of D. viviparus. Further development of DviLAMP as a point-of-care test could significantly improve the sensitivity of lungworm diagnosis in the field.
Highlights
• A LAMP primer set was designed to detect Dictyocaulus viviparus ITS2 DNA.
• Conventional and colorimetric LAMP assays were tested for specificity and sensitivity.
• DviLAMP specifically detects Dictyocaulus viviparus on the lateral flow dipstick.
1 Introduction
Dictyocaulus viviparus (Bloch, 1782), the bovine lungworm, is the cause of parasitic bronchitis (dictyocaulosis) or “husk” and is a recognized cause of high morbidity and mortality in cattle. The disease occurs most frequently in calves in their first grazing season but can occasionally affect adult cattle. Dictyocaulus viviparus is found globally but is most prevalent in Europe, North and South America, and Australia (1). The economic cost of D. viviparus in the UK and Europe has recently been estimated at € 16 million and € 139 million, respectively (2).
Outbreaks of lungworm are sporadic and unpredictable but have been increasing in the UK (3). Due to the lack of a sensitive test, diagnosis relies on clinical signs, grazing history, and time of year. Coughing while at pasture is the major clinical sign but is not specific to lungworm (1). The classical diagnosis of D. viviparus infection is based on morphological identification of L1 larvae in the feces following Baermannisation (4, 5). However, false negatives are a problem with this approach due to the significant impact of storage temperature and time on larval recovery (6). Alternative molecular detection methods such as protein-based ELISA assays were developed to detect anti-lungworm antibodies (7) and antigen (8). The antibody level in bulk tank samples from dairy herds has been investigated as diagnostic strategy, but despite high specificity and a correlation between positive samples and disease outbreaks, this approach lacks sensitivity (4, 9). Techniques based on DNA detection by polymerase chain reaction (PCR) have also been developed to detect and identify D. viviparus from various ruminant hosts in a research setting (10).
The internal transcribed spacer 2 (ITS2) ribosomal DNA locus can be used for the identification of multiple strongyle species including D. viviparus by PCR (11–13). However, PCR-based approaches are time consuming and rely on specialized equipment, such as the thermocycler, that limits application in veterinary clinics or on farm. Loop-mediated isothermal amplification (LAMP) is an interesting technique that has been attracting growing attention for pathogen diagnostics (14–16). Several studies have been successful in developing LAMP assays for helminth parasites, with versatile and accessible options for read out such as colorimetry and lateral flow dipstick (LFD) (17–22).
Therefore, our study aimed to develop a new diagnostic tool for D. viviparus based on LAMP coupled with simple and point-of-care amenable end-point detection, including colorimetry and lateral flow. Real-time LAMP was used to confirm positive and negative results, in addition to determining the limit of detection in sensitivity analyses. Our proof-of-concept DviLAMP is the first step towards a sensitive and specific diagnostic tool for rapid and convenient detection of the bovine lungworm.
2 Materials and methods
2.1 DNA extraction
Dictyocaulus viviparus L1 larvae were recovered from cattle feces from a Scottish farm (23). Crude gDNA was extracted by lysis from individual larvae. Lysis buffer included 10 μL of Direct PCR Lysis Reagent (Cell, Viagen Biotech), 0.5 μL of 1 M DTT, and 0.1 μL of Proteinase K (100 mg/mL). The L1 was then incubated at 60°C for 120 min, followed by 85°C for 45 min to denature the Proteinase K. Individual Ostertagia ostertagi (Stiles, 1892) and Cooperia oncophora (Railliet, 1898) L3 from cattle feces from Scottish farms were processed in the same manner to obtain gDNA. Extracted DNA was diluted 20-fold and stored at −20°C.
2.2 Plasmid preparations
To amplify the ITS2 region, a PCR reaction (final volume 20 μL) was set up, including 4 μL of 5X Phusion Green GC Buffer, 0.4 μL of 10 mM dNTPs solution (N0447S, NEB), 0.4 μL of 10 μM of each generic forward and reverse primer [ITS2GF and ITS2GR, respectively, from a previous publication (24)] (Eurofins Genomics), 0.2 μL of 2 U/μL Phusion High-Fidelity DNA Polymerase (F-534S, Thermo Scientific), 1 μL of DNA template, and DEPC H2O (AM9906, Ambion, Invitrogen). A thermocycler (Biometra TAdvanced, Analytik Jena) with the following thermal conditions was used: denature at 98°C for 30 s, 40 cycles of 98°C for 20 s, annealing temperatures (61°C for first 15 cycles, and 58°C for later 25 cycles) for 20 s, and 72°C for 20 s, followed by a final extension at 72°C for 5 min. The PCR product was visualized by 2% agarose gel (NBS-AG500, NBS Biologicals) electrophoresis under UV (FireReader, UVITEC) to verify the PCR amplicon size (~593–596 bp for D. viviparus; ~378–381 bp for O. ostertagi and C. oncophora). Next, the remaining volume (15 μL) was purified using the Monarch PCR and DNA Cleanup Kit (T1030S, NEB). Purified PCR products from five individual L1 samples were pooled. For A tailing, 2 μL of the purified amplicon (~10 ng/μL) was mixed with 2 μL of 5X GoTaq Flexi Reaction buffer, 2 μL of GoTaq Flexi DNA Polymerase (Promega), 2 μL of 1 mM dATP (10216018, Invitrogen), 0.6 μL of 25 mM MgCl2, and 1.4 μL of DEPC H2O, and then incubated at 72°C for 20 min. The PCR product was ligated into the plasmid (pCR™ TOPO™ vector) and the recombinant plasmid was transformed into competent cells according to the TA Cloning Kit with One Shot™ TOP10 E. coli (Invitrogen) manufacturer’s instructions. After incubation on LB plates at 37°C overnight, the recombinant plasmid containing colonies were identified by blue-white colony selection, followed by PCR to check the plasmids contained the desired ITS2 product. The recombinant plasmid (DviITS2) was extracted using a Monarch Plasmid Miniprep Kit (T1010S, NEB) and the DNA concentration was evaluated by Qubit (high sensitivity dsDNA quantification assay).
2.3 DNA sequencing and species confirmation
Plasmids were sequenced by TubeSeq Supreme (Eurofins Genomics), using both forward and reverse primers. Sequences for each clone were aligned and assembled using MEGA 11 software and the species confirmed using BLASTn (National Center for Biotechnology Information (NCBI), https://blast.ncbi.nlm.nih.gov/Blast.cgi). The 5.8S and 28S rDNA regions were annotated based on a previous study (24) to identify the precise ITS2 region. Then, the assembled DNA sequences were aligned by MEGA 11 software, exported to FASTA format, and uploaded into MultAlin online software [(25), http://multalin.toulouse.inra.fr/multalin/multalin] to generate a plain text DNA sequence alignment. DNA sequences from this study were published under accession numbers: PP970511-PP970515 for D. viviparus; PP968975-PP968976 for O. ostertagi; and PP968977 for C. oncophora.
2.4 LAMP primer design
A consensus sequence of the D. viviparus ITS2 region with partial 5.8S rDNA sequence was derived from the five picked clones (Figure 1). Primer design was carried out with Geneious Prime software (Version 2023.2.1, Dotmatics). The forward outer primer was designed manually, with a fixed start at the 5′ end of the 5.8S rDNA sequence, then imported into PrimerExplorer V5.1 The loop primers were also designed manually using Geneious Prime (Version 2023.2.1, Dotmatics). General properties of the primers were investigated in-silico using the OligoAnalyzer Tool [Integrated DNA Technologies (IDT), https://sg.idtdna.com/calc/analyzer] to assess the following criteria: length, Tm, ΔG, hairpin, homodimer, and heterodimer formation. The selected DviLAMP primers for this study (Table 1) were ordered from IDT and Eurofins Genomics in lyophilized form.
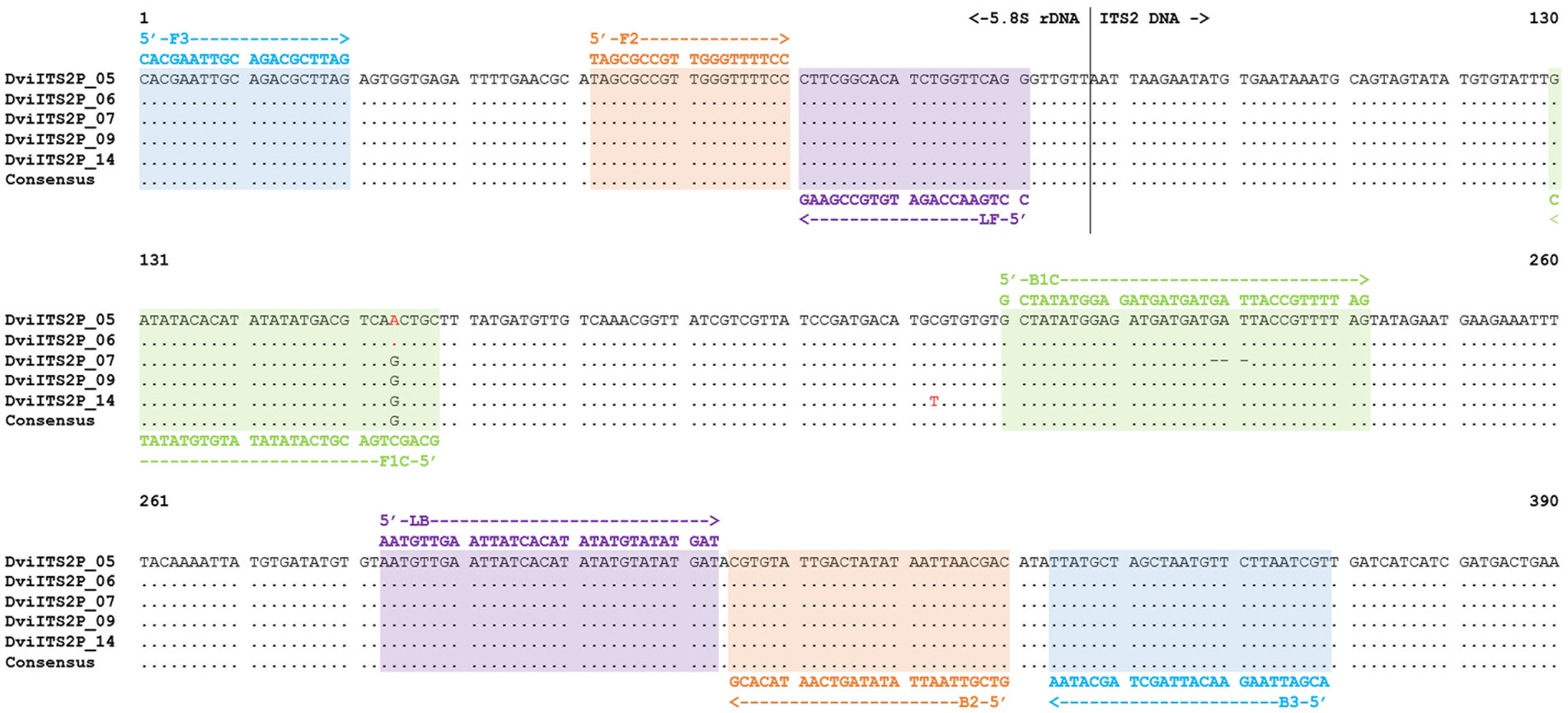
Figure 1. ITS2 with partial 5.8S rDNA sequence alignment of Dictyocaulus viviparus (clones 5, 6, 7, 9, and 14) highlighted with different DviLAMP primer DNA sequences; variable sites in the DNA sequence are presented as red letters.

Table 1. DviLAMP primers for Dictyocaulus viviparus lungworm detection: [BIO], biotin; [FITC], fluorescein-5-isothiocyanate; and bp, base pair.
2.5 Conventional and colorimetric LAMP
The DviLAMP primer mix was prepared in a 10X mixture solution, including 16 μM each of forward inner primer (FIP) and backward inner primer (BIP), 8 μM each of forward loop primer (LF) and backward loop primer (LB), and 4 μM each of forward outer primer (F3) and backward outer primer (B3), prior to using in colorimetric and real-time LAMP. For colorimetric LAMP, a 1X LAMP-reaction mixture (12.5 μL final volume) containing 6.25 μL of WarmStart Colorimetric LAMP 2X Master Mix (M1800S, NEB), 2.5 μL of 5 M betaine (Sigma-Aldrich), 1.25 μL of 10X DviLAMP primer mix, 1.5 μL of DEPC H2O, and 1 μL of the DNA template (20 ng/μL–1 pg/μL DviITS2 plasmid) was used. For each batch of experiments, a negative control was run using 1 μL DEPC H2O in place of the DNA template. The LAMP assay was performed in a thermocycler (Biometra TAdvanced, Analytik Jena) at 64°C for three different reaction durations: 45, 60, and 90 min, with a final 80°C incubation for 10 min to terminate the reaction. The post-reaction tubes were left until they reached room temperature (~20–25°C), then were mixed well and spun down. The solution color (yellow as positive; or pink as negative) was assessed in good light on a white paper background and recorded by taking a photo with an iPad camera. After recording the color change, 5 μL of LAMP product was then visualized using 2% agarose gel electrophoresis.
2.5.1 Real-time LAMP
For real time LAMP, the 12.5 μL reaction mixture (1X) contained 1.25 μL of 10X isothermal amplification buffer, 1.25 μL of 10X DviLAMP primer mix, 1.75 μL of 10 mM/base dNTPs, 0.75 μL of 100 mM MgSO4, 2.5 μL of 5 M Betaine, 0.5 μL of 8 U/μL Bst 2.0 WarmStart DNA Polymerase (M0538S, NEB), 0.25 μL of 50X LAMP Fluorescent Dye (B1700S, NEB), 0.375 μL of 1 μM passive reference dye (600530, Agilent), 2.875 μL of DEPC H2O, and 1 μL of the DNA template (20 ng/μL–1 pg/μL DviITS2 plasmid or H2O). The thermal profile was controlled under AriaMx Real-time PCR System (G8830A, Agilent Technologies), at 64°C for 60 min for specificity validation and 90 min for sensitivity determination, then 80°C for 10 min. High resolution melt (HRM) analysis of the LAMP product was then performed at 95°C for 30 s, 65°C for 30 s, and 95°C for 30 s, with 0.5°C resolution and 5 s for soak time. FAM and ROX channels were selected for fluorescence detection and as the reference dye, respectively. The real-time result with three technical replicates for each DNA template was recorded and analyzed using AriaMx software (Agilent). Normalized fluorescence with the reference dye (Rn) and the first derivative of the normalized fluorescence multiplied by −1 [−Rn´(T)] values were measured to investigate the fluorescence signal data (as Y-axis) of the amplification plot and melt curve, respectively.
2.5.2 Lateral flow LAMP
Both conventional and colorimetric LAMP reactions gave the same result on lateral flow dipstick (LFD) so were used interchangeably in this study. For colorimetric LAMP, the reaction mixture for LFD was the same as section 2.5 except in this case, tagged primers were used, where biotin and fluorescein-5-isothiocyanate (FITC) tagged primers (FIPL, BIPL, LFL, and LBL) replaced untagged primers (FIP, BIP, LF, and LB, respectively), as presented in Table 1. For conventional LAMP with LFD (without colorimetry), the 12.5 μL final volume contained 1.25 μL of 10X isothermal amplification buffer, 1.25 μL of 10X DviLAMP primer mix (a pair of biotin and FITC tagged primers), 1.75 μL of 10 mM/base dNTPs (N0447S, NEB), 0.75 μL of 100 mM MgSO4, 2.5 μL of 5 M Betaine, 0.5 μL of 8 U/μL Bst 2.0 WarmStart DNA Polymerase (NEB), 3.5 μL of DEPC H2O, and 1 μL of the DNA template (20 ng/μL–10 pg/μL DviITS2 plasmid) or H2O (for negative control). For both LAMP assays the thermal profile was the same as described in section 2.5. After incubation, 5 μL of the LAMP product was mixed with 45 μL of a commercial lateral flow buffer (from the LFD kit, Milenia Genline HybriDetect, Germany) in a 1.5 mL microcentrifuge tube. The LFD was submerged in the mixture for 5 min and then removed to record the result. To select the optimum pair of biotin/FITC tagged primers, four schemes trialing different positions of biotin and FITC tagging, including (I) [BIO]-FIPL and [FITC]-BIPL; (II) [FITC]-LFL and [BIO]-LBL; (III) [BIO]-FIPL and [FITC]-LFL; and (IV) [BIO]-LBL and [FITC]-BIPL, with LAMP incubation at 64°C for 60 min, were tested with LFD (see results section 3.2).
2.6 Analytical sensitivity and specificity of the LAMP assay
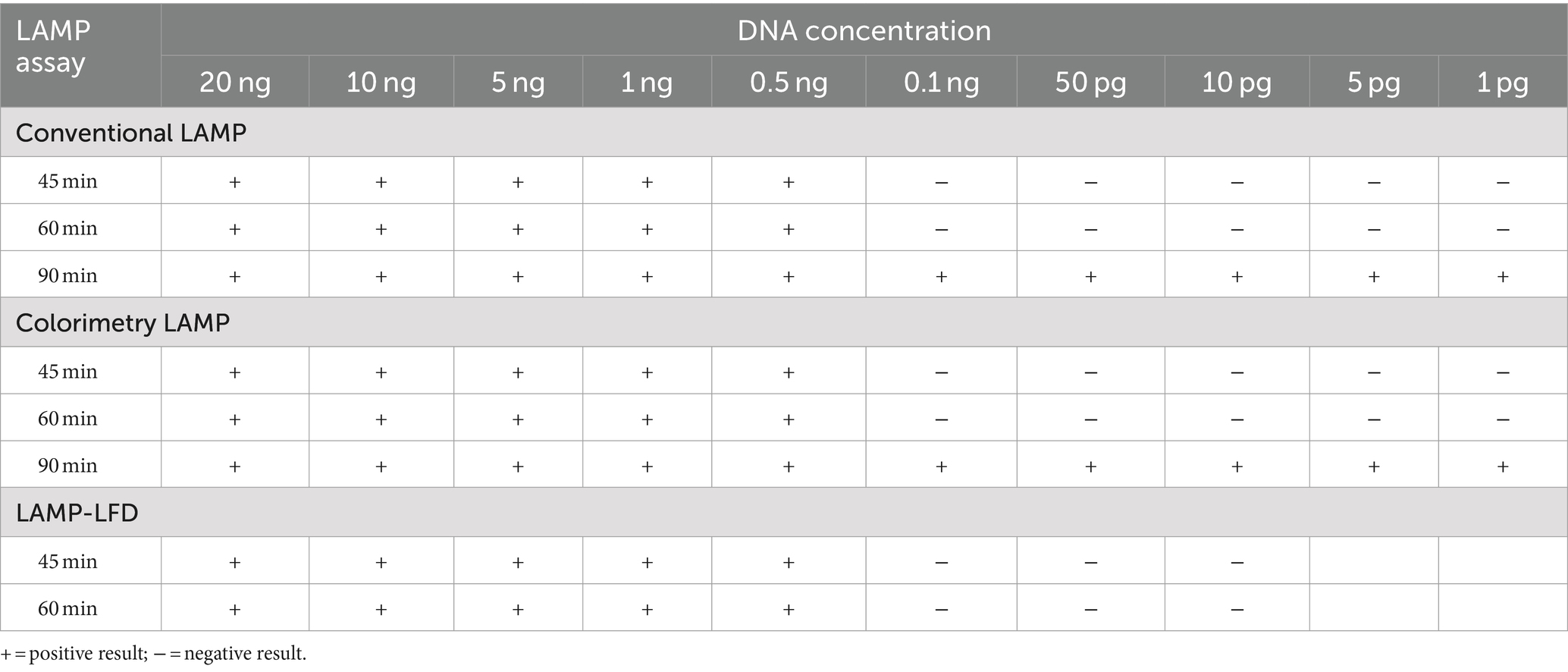
To test the analytical sensitivity, DviLAMP with 45, 60, and 90 min of incubation at 64°C was applied to various DviITS2 plasmid DNA concentrations from 20 ng to 1 pg. To test the analytical specificity, O. ostertagi and C. oncophora ITS2 plasmids were incorporated separately, and together, with the DviITS2 plasmid and tested by the DviLAMP with 60 min of incubation at 64°C. The results were analyzed by colorimetry, gel electrophoresis, real-time, and LFD.
3 Results
3.1 DNA sequences
In this study, we made use of seven ITS2 sequences with flanking 5.8S and 28S rDNA regions from three species; five D. viviparus clones (593–596 bp with 97.22–100% identity to other D. viviparus DNA sequences in the NCBI database) and one sequence each for O. ostertagi (378 bp with 98.14–100% identity to other O. ostertagi DNA sequences in the NCBI database) and C. oncophora (381 bp with 99.21–99.74% identity to other C. oncophora DNA sequences in the NCBI database). Two DNA sequences from D. viviparus (clone 5 and 6) were 100% identical so only four variants were found. The predicted primer hybridization sites on the DNA alignments are shown in Figure 1 (D. viviparus) and Supplementary Figure S1 (O. ostertagi and C. oncophora). BLASTn results and DNA sequence annotations are shown in Supplementary Table S1.
3.2 Analytical sensitivity
The analytical sensitivity results in this study are summarized in Table 2. After 45 min incubation, colorimetric DviLAMP detected down to 0.5 ng of DviITS2 plasmid, turning from pink to pale yellow/orange (Figure 2A). The color change to a brighter yellow continued until 60 min (Figure 2B). Extending incubation of the DviLAMP to 90 min increased sensitivity, allowing detection of 1 pg of DviLAMP plasmid, which was the lowest DNA concentration tested in this study (Figure 2C). All positive colorimetry results were supported by the appearance of a smear-like ladder on a gel, typical of a LAMP reaction, and there were no products observed for any negative (pink color mixture) result (Figure 2). The real-time amplification plot also supported the sensitivity test (Figure 3). Florescence signals for 20 ng to 0.5 ng began to increase in exponential phase from ~35–45 min, with the late linear or plateau phase at 60 min or more. Increasing florescence signal was detected for all DNA concentrations and the Ct value or the time of the highest change in signal (exponential phase) (Figure 3A), could be calculated for all concentrations except for 5 pg and 1 pg. However, the florescence signals for these two lowest DNA concentrations were detected by 90 min. HRM analysis (Figure 3B) confirmed the varied sizes of LAMP products from different DNA concentrations.
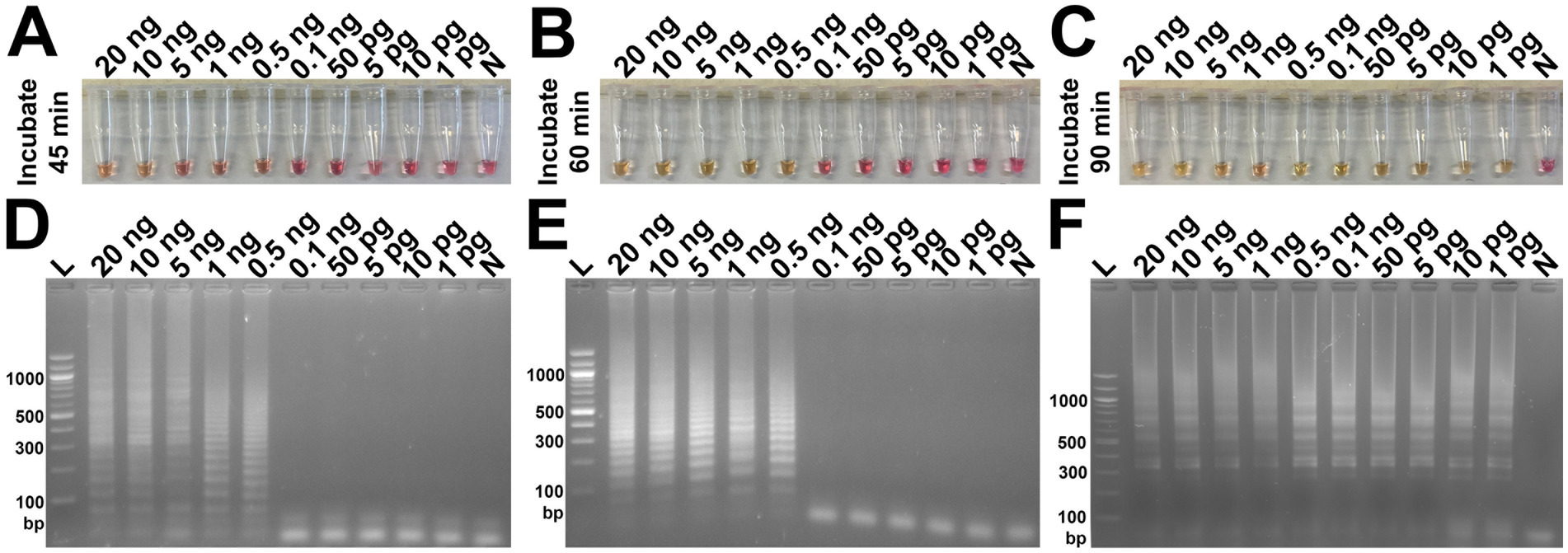
Figure 2. Analytical sensitivity test of the DviLAMP based on colorimetry with naked eye observation (A–C) and gel electrophoresis (D–F) with different incubation periods, including 45 min (A,D), 60 min (B,E), and 90 min (C,F); and various DNA concentrations (20 ng to 1 pg): L, DNA ladder; N, negative control.
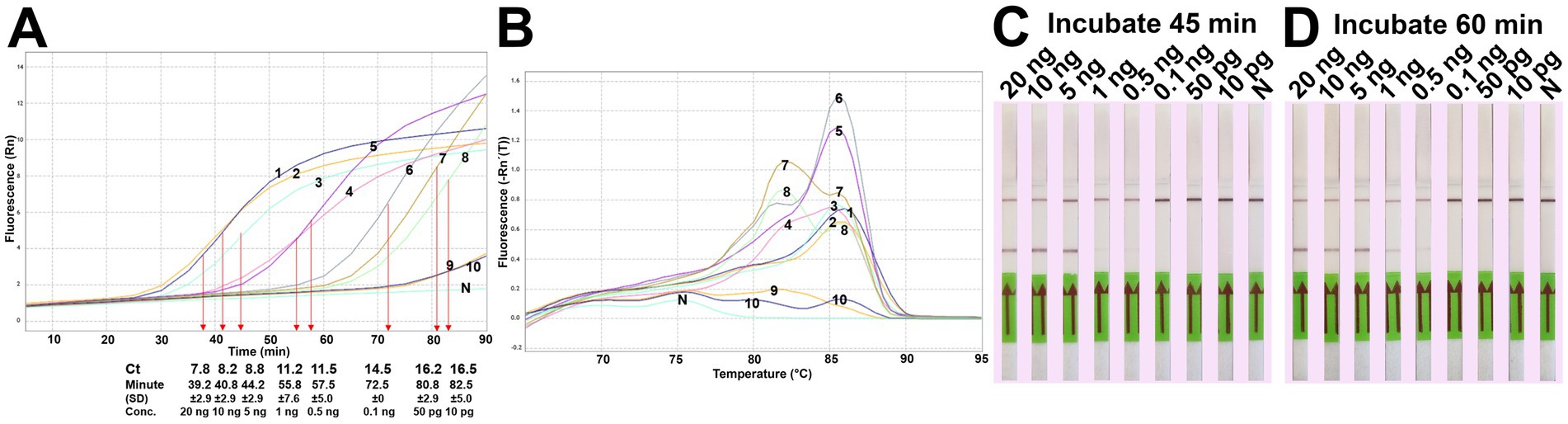
Figure 3. DviLAMP with real-time detection (A), melting curve plot (B) and lateral flow (C,D). The real-time fluorescence signal was calculated from three replicates. Ct values and time of the highest change in the fluorescence were calculated and indicated in the amplification plot. Numbering in the amplification plot indicates the DviITS2 plasmid DNA concentrations as follows: 1 = 20 ng; 2 = 10 ng; 3 = 5 ng; 4 = 1 ng; 5 = 0.5 ng; 6 = 0.1 ng; 7 = 50 pg; 8 = 10 pg; 9 = 5 pg; and 10 = 1 pg.
For the LFD assay, we tested four pairs of biotin and FITC labelled primers (Figure 4A). Test line appearance could be observed clearly from two schemes (Figure 4B), which were [FITC]-LFL and [BIO]-LBL (scheme II); and [BIO]-FIPL and [FITC]-LFL (scheme III). For scheme III, the test line was stronger than scheme II but the control line was faint. Therefore, we decided to choose the biotin-FITC tagged primers from scheme II to test the sensitivity of DviLAMP-LFD for 45 min and 60 min reaction time (Figures 3C,D). The test line was visible from 20 ng to 0.5 ng of DNA for both incubation times, indicating the limit of detection for the DviLAMP was 0.5 ng. For the lowest amounts of input DNA, 1 ng and 0.5 ng, lines were visible but faint after 45 min of incubation but were clearly visible after 60 min of incubation. All four variants of the D. viviparus ITS2 DNA in this study were detected by DviLAMP (Supplementary Figure S2).
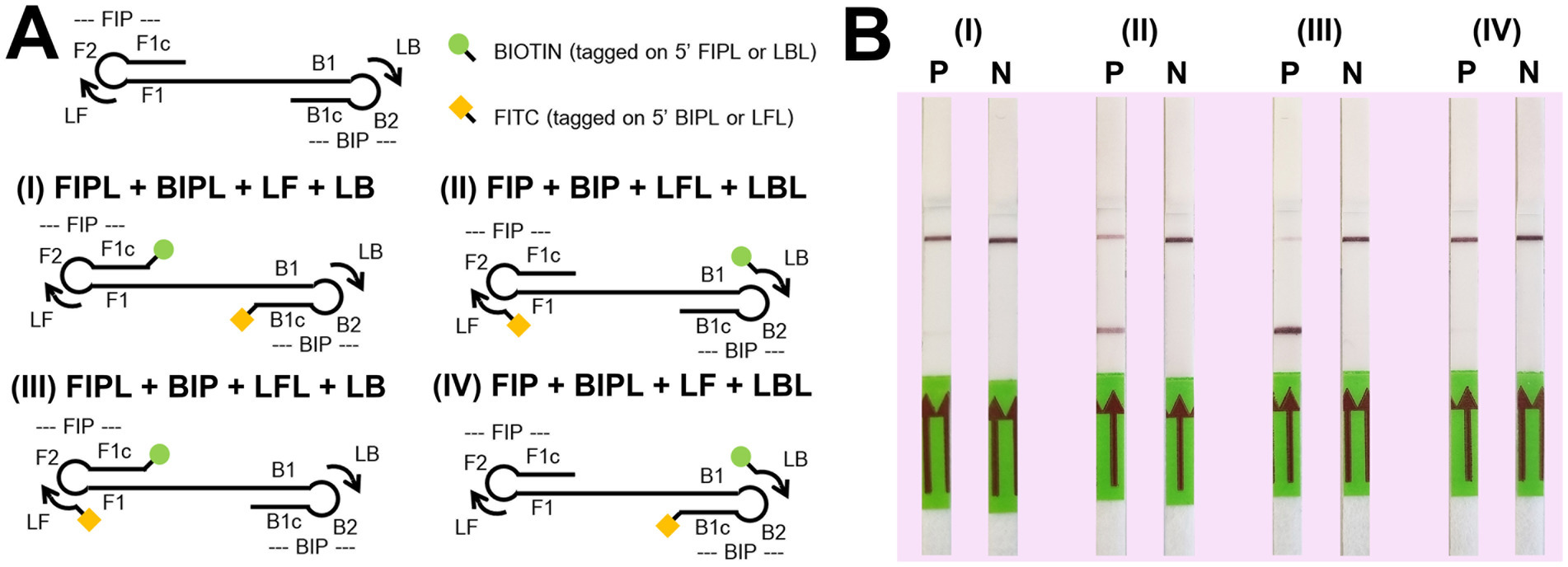
Figure 4. Biotin (BIO) and fluorescein-5-isothiocyanate (FITC) position tagging test. Graphics for four tagged position schemes (A) including (I) [BIO]-FIPL and [FITC]-BIPL; (II) [FITC]-LFL and [BIO]-LBL; (III) [BIO]-FIPL and [FITC]-LFL; and (IV) [BIO]-LBL and [FITC]-BIPL. The P (positive, 10 ng of Dictyocaulus viviparus as DNA template) and N (negative, H2O) results on LFD for each scheme after amplification for 60 min (B).
3.3 Analytical specificity
Our results showed that changing the position of the biotin and FITC labels impacted the ability of DviLAMP with LFD to detect the target (D. viviparus) and non-target (O. ostertagi and C. oncophora) ITS2 plasmids (Figure 5). There was a notable difference in the specificity of the assay dependent on which primers were labelled. While all three parasites were detected by gel electrophoresis in both schemes II and III, and by LFD in scheme III, only D. viviparus was detected by the LFD in scheme II. These findings support the choice of biotin and FITC tagged primers in scheme II for specific detection of D. viviparus.
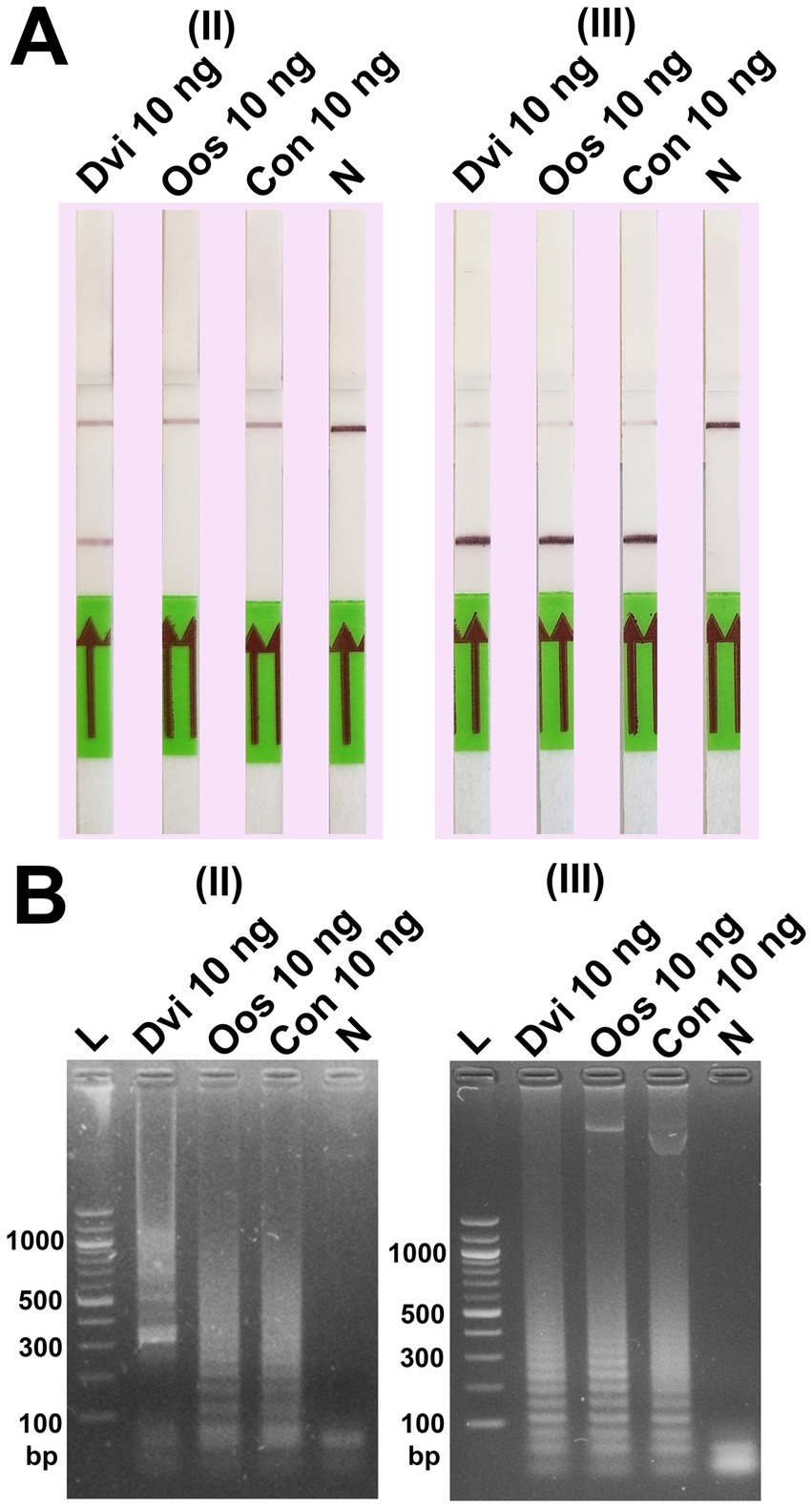
Figure 5. Specificity test of the DviLAMP with two biotin-FITC tagged position schemes, (II) [FITC]-LFL and [BIO]-LBL; and (III) [BIO]-FIPL and [FITC]-LFL, showing the results by LFD (A) and gel electrophoresis (B) for Dictyocaulus viviparus (Dvi), Ostertagia ostertagi (Oos), and Cooperia oncophora (Con) after amplification for 60 min.
Finally, ITS2 plasmids from all three species were combined and used as template for DviLAMP (Figure 6). Whether mixed in a 1:1 ratio with either O. ostertagi or C. oncophora plasmids, or in a 1:1:1 reaction mixture with both, D. viviparus DNA could still be specifically detected with LFD and colorimetry (LFD test line appearance and yellow color change, Figures 6A,C, respectively). While gel electrophoresis and real time LAMP amplification showed amplified LAMP product for all three species when tested individually (Figures 6B,D), this was not reflected in the LFD or colorimetry results. HRM peaks for these species presented no obvious difference (Figure 6E). No LFD test line was visible for O. ostertagi or C. oncophora and colorimetry showed no change for O. ostertagi and a slight change for C. oncophora to an orange shade of pink (Figure 6C). Therefore, only D. viviparus can be detected and differentiated from a negative result by colorimetry and LFD, even in the presence of DNA of other species.
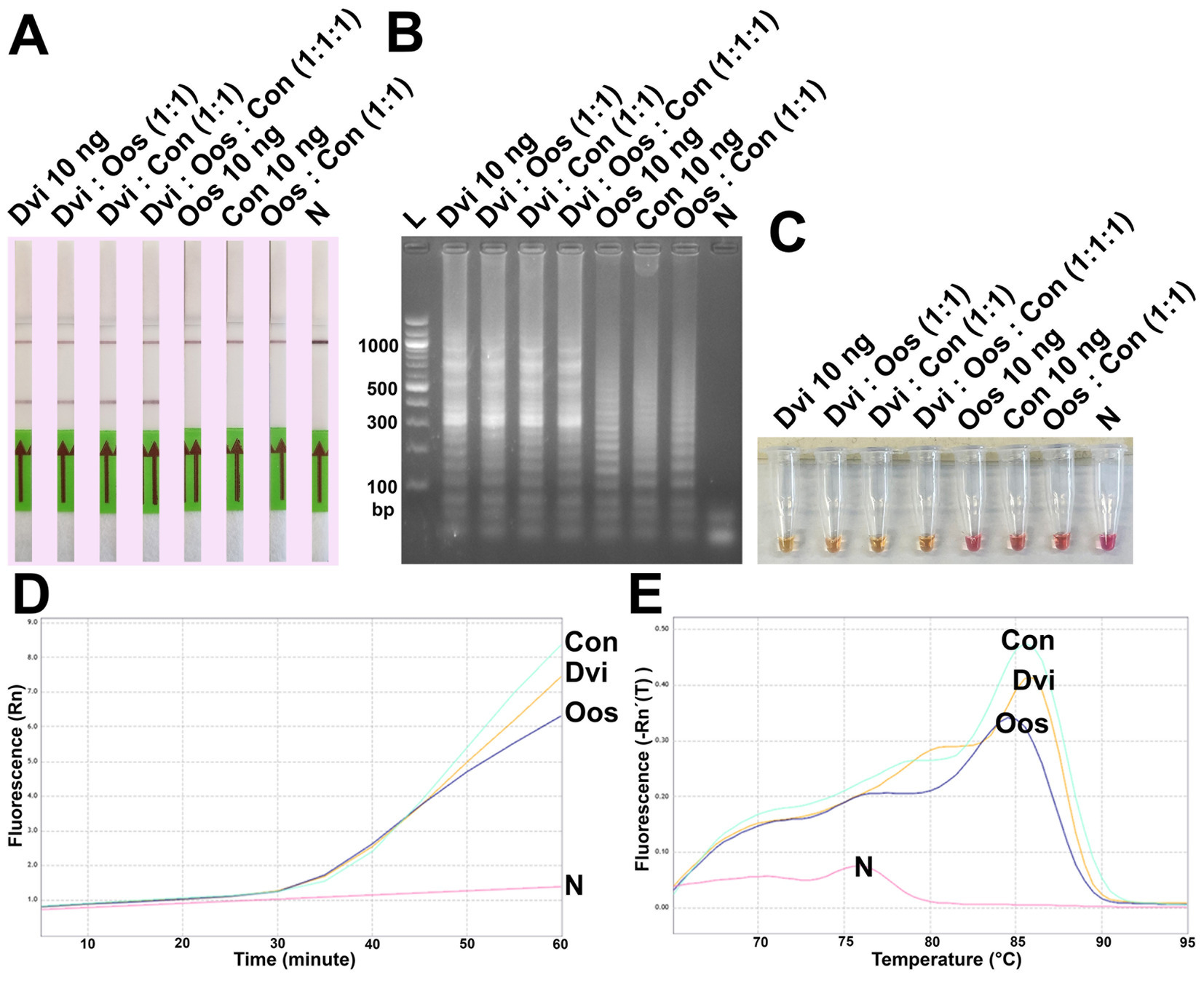
Figure 6. Analytical specificity test of the DviLAMP by LFD (A), gel electrophoresis (B), and colorimetry (C); the amplification plot (D) and the melting plot (E) showing the fluorescence increasing for Dictyocaulus viviparus (Dvi), Ostertagia ostertagi (Oos), and Cooperia oncophora (Con) for 60 min of the incubation. The amplification plot and melting plot show the results for each species individually from three technical replicates.
4 Discussion
A commercial vaccine using radiation-attenuated D. viviparus larvae (Bovilis® Huskvac, Intervet UK Ltd.) has been used successfully to control this parasite in the UK for many years (26). However, uptake of vaccination has declined with the increasing use of long-acting macrocyclic lactones for simultaneous control of lungworm and gastrointestinal nematodes. Suppressive drug treatment is not sustainable due to the strong selection for anthelmintic resistant individuals, with macrocyclic lactone resistant D. viviparus recently detected on a dairy farm in Scotland (23). The use of targeted anthelmintic treatment could improve the sustainability of parasite control, however lungworm outbreaks are severe and difficult to control, so strategic treatment approaches require sensitive and rapid diagnostic tests. A rapid detection tool using LAMP with a quantitative analysis (22) could be used to evaluate parasite burden and inform rational control, for example early treatment of at risk calves or post-treatment testing for drug efficacy. In addition to helping famers control lungworm in their herds, improving the accuracy of D. viviparus diagnosis in the field would inform epidemiological studies to better understand, predict, and control this highly pathogenic and economically important disease.
As a first step in developing a LAMP assay we investigated sequence variation at the conserved ITS2 locus in D. viviparus larvae from a farm in Scotland. Four sequence variations of the D. viviparus ITS2 DNA region (453–456 bp) were found in this study, from pooled PCR products amplified from five L1, all of which were detected by DviLAMP. Published ITS2 sequences from geographically separated D. viviparus (GenBank database, NCBI) showed additional genetic diversity, but the lack of full length 5.8S and ITS2 DNA sequence from these populations prevented assessment of primer hybridization in this study. Future work to examine a wider range of D. viviparus populations from different farms should be implemented to assess the sensitivity of the test in a wider context. In this study we found that the limit of detection was 0.5 ng with a 45 min reaction time. It may be possible to lower the limit of detection by increasing the reaction time to 90 min. In the real-time LAMP, we found that 1 pg of template could be detected after an 85–90 min reaction time. However, although increasing the reaction time can also increase the sensitivity of the reaction, this increase in sensitivity may not be practical for a rapid on-site test. Therefore, a balance between a user-friendly reaction time and detectable sensitivity, should be considered. A single L1 could be expected to have much less than 1 ng of DNA, however an active infection would likely have DNA from adults, multiple L1 and also recently ingested L3 from pasture. Therefore, a sensitivity of 0.5 ng or 1 pg is theoretically suitable for a field based diagnostic test.
We tested the ability of DviLAMP to differentiate between D. viviparus and other nematodes commonly found in bovine feces. The DviLAMP primers can amplify D. viviparus, O. ostertagi and C. oncophora ITS2 plasmids as observed by both gel electrophoresis and real-time amplification plots. However, the reactions with O. ostertagi and C. oncophora template produced a lower quantity of LAMP products when compared to the bright LAMP ladder band above 300 bp visible on gel electrophoresis for D. viviparus. The result indicates that a high quantity of longer LAMP products are produced only from D. viviparus template. However, the quantity of LAMP product alone is not sufficient evidence for differentiation between species, with amplification for all three species shown by real-time analyses. HRM analysis can be used to discriminate rumen fluke species (27), but although there is a peak of fluorescence signal at ~80°C for D. viviparus, further work is required to clearly differentiate non-target species in our study.
The amplification results for O. ostertagi and C. oncophora are consistent with the DNA alignment (Supplementary Figure S1) which showed the potential hybridized regions for the DviLAMP primers in forward sites, with a 100% match for F3 and F2, one mismatch in LF, and seven bases matching at the 3′ site of F1C. This reflects the high conservation of the DNA sequence in the 5.8S rDNA region. However, the sequence divergence in the remaining primers sited in the ITS2 region are expected to provide species specificity: there are no hits on either O. ostertagi or C. oncophora ITS2 sequences for the backward sites of DviLAMP primer sequences, B1C and LB, or for the 3′ nine bases of the B3 primer. We hypothesize the amplified LAMP product from non-target species showed a lesser effect on the colorimetry result (Figure 6C), due to an uncertain amount of H+ (LAMP by-product) produced by the less efficient reaction with incomplete loop structure formation from the primer hybridization on the forward site only. Consistent with this expectation, we show that the biotin-FITC tagging position on the primer or DNA probe strongly impacts the results, and correct positioning is essential for accurate diagnosis. The biotin tagged primer in scheme II specifically targets the D. viviparus ITS2 region only, so this is the key to the specificity of DviLAMP-LFD. This assumption is based on in-silico DNA sequence investigation. This could be explored further by the previously demonstrated LEC-LAMP assay design (28) to add further species specific discrimination of the LAMP products. However, the inclusion of this technology lay outside the current study’s scope, the focus of which was developing a species-specific assay for detecting bovine lungworm. This aim has been successfully demonstrated using LAMP-LFD. Nonetheless, it will be of great interest to explore this in future work. The detection of both C. oncophora and O. ostertagia ITS2 sequence by the lungworm primers developed herein opens the possibility to develop a triplex LEC-LAMP assay, using a similar methodology to that described previously for levamisole resistance (17) and bacterial species differentiation and antimicrobial resistance gene variants (28, 29). The LEC-LAMP assay design offers added advantage that it is also amenable to LFD end-point detection, although further work is necessary to optimize this (17).
For a user-friendly diagnostic test, the results should be obtained in the shortest time possible. Three different incubation times from 45 min to 90 min were tested, and the result indicated improving sensitivity for LAMP, colorimetry, and LFD by longer incubation. Incubation times for other published LAMP assays vary from about 15–60 min (15, 16) and the differing incubation times could be due to several factors in addition to DNA concentration, for example, DNA integrity and the presence of inhibitors from different extracted specimen types. This highlights that sample preparation/extraction before DNA amplification needs to be considered. To develop DviLAMP as a point-of-care diagnostic test additional steps relating to sample preparation need to be undertaken. The assay could be used to detect larvae in feces, which would require a simple method to isolate D. viviparus DNA and remove potential inhibitors. DviLAMP could potentially be developed to detect eggs and/or larvae directly in bovine nasal mucus, which would potentially allow earlier detection of infection than using feces. However, the amount of free D. viviparus DNA in mucus of infected calves is currently unknown so this may also require a DNA extraction step. Farmer preference and practicalities of handling youngstock will also dictate the most appropriate sampling approach. In addition, LAMP could be further developed to detect anthelmintic resistance mutations, as has been shown for levamisole resistant Haemonchus contortus (Rudolphi, 1803) Cobb, 1898 using colorimetric, SNP specific enzymatic cleavage, and restriction analysis (17, 28). A future application of LAMP in point-of-care and field-based detection could combine a microfluidic chip (for mixing reagents and incubation) with a convenient measurement (e.g., colorimetry, lateral flow, electrochemistry) and digital data processing, to present, analyze, and store results (30–33). In addition, there is growing interest in the potential for combining LAMP with portable sequencing technologies such as Oxford Nanopore (34). Thus, our newly designed and validated LAMP primer set for the detection of D. viviparus ITS2 could be integrated into a future farm-side sequencing assay.
In conclusion, we have developed a novel proof of concept LAMP assay for D. viviparus with various methods of end point detection, including colorimetry, gel electrophoresis, real-time with HRM, and LFD. Its application with colorimetry and LFD can detect target DNA specifically and sensitively down to a concentration of 0.5 ng within 45–60 min of incubation at 64°C. Although non-target nematodes, O. ostertagi and C. oncophora, can be detected by gel electrophoresis and real-time, they were effectively discriminated from the positive D. viviparus result in colorimetry and LFD assays. Therefore, the DviLAMP represents a major step in the development of a field-based diagnostic tool for D. viviparus to improve parasite control and livestock health.
Data availability statement
The data presented in the study are deposited in the Nucleotide database (https://www.ncbi.nlm.nih.gov/nuccore/), accession numbers: PP970511-PP970515, PP968975-PP968977.
Ethics statement
The animal studies were approved by University of Glasgow MVLS College Ethics Committee (Project No: 200210097). The studies were conducted in accordance with the local legislation and institutional requirements. Written informed consent was obtained from the owners for the participation of their animals in this study.
Author contributions
SN: Conceptualization, Formal analysis, Investigation, Methodology, Visualization, Writing – original draft, Writing – review & editing. PC: Data curation, Validation, Writing – review & editing, Supervision. MS: Resources, Supervision, Writing – review & editing, Data curation, Methodology. JM: Supervision, Writing – review & editing, Conceptualization, Project administration, Resources. AA: Data curation, Methodology, Supervision, Writing – review & editing, Software. TC: Funding acquisition, Project administration, Software, Supervision, Writing – review & editing. RL: Conceptualization, Funding acquisition, Project administration, Resources, Supervision, Writing – review & editing, Writing – original draft.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. We acknowledge funding by a Lord Kelvin Adam Smith (LKAS) Fellowship awarded to RL. SN is funded by the Royal Golden Jubilee Ph.D. Program [N41A650118] under the National Research Council of Thailand and the Government of Thailand. RL and JM are funded by a Wellcome Clinical Research Career Development Fellowship awarded to RL [216614/Z/19/Z]. PC is funded by a James Herriot Scholarship from the University of Glasgow.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fvets.2024.1454065/full#supplementary-material
SUPPLEMENTARY FIGURE S1 | ITS2 with flanking 5.8S and 28S rDNA sequences alignment of Ostertagia ostertagi and Cooperia oncophora highlighted with different primers DNA sequences for the DviLAMP; variable sites in the DNA sequence are presented as red letters; gray letters indicating non-hybridized primer region.
SUPPLEMENTARY FIGURE S2 | Detection of various DviITS2 plasmid variations (clone 5, 7, 9, and 14) at 10 ng using the DviLAMP (60 min reaction time) with lateral flow dipstick (LFD) (A), colorimetry (B), and gel electrophoresis (C). L, DNA ladder; and N, negative control.
Footnotes
References
1. Forbes, A . Lungworm in cattle: epidemiology, pathology and immunobiology. Livestock. (2018) 23:59–66. doi: 10.12968/live.2018.23.2.59
2. Charlier, J, Rinaldi, L, Musella, V, Ploeger, HW, Chartier, C, Vineer, HR, et al. Initial assessment of the economic burden of major parasitic helminth infections to the ruminant livestock industry in Europe. Prev Vet Med. (2020) 182:105103. doi: 10.1016/j.prevetmed.2020.105103
3. McCarthy, C, and van Dijk, J. Spatiotemporal trends in cattle lungworm disease (Dictyocaulus viviparus) in Great Britain from 1975 to 2014. Vet Rec. (2020) 186:642–2. doi: 10.1136/vr.105509
4. Charlier, J, Williams, DJ, Ravinet, N, and Claerebout, E. To treat or not to treat: diagnostic thresholds in subclinical helminth infections of cattle. Trends Parasitol. (2023) 39:139–51. doi: 10.1016/j.pt.2022.11.014
5. van Wyk, JA, and Mayhew, E. Morphological identification of parasitic nematode infective larvae of small ruminants and cattle: a practical lab guide. Onderstepoort J Vet Res. (2013) 80:539. doi: 10.4102/ojvr.v80i1.539
6. Rode, B, and Jørgensen, RJ. Baermannization of Dictyocaulus spp. from faeces of cattle, sheep and donkeys. Vet Parasitol. (1989) 30:205–11. doi: 10.1016/0304-4017(89)90016-2
7. Fiedor, C, Strube, C, Forbes, A, Buschbaum, S, Klewer, AM, von Samson-Himmelstjerna, G, et al. Evaluation of a milk ELISA for the serodiagnosis of Dictyocaulus viviparus in dairy cows. Vet Parasitol. (2009) 166:255–61. doi: 10.1016/j.vetpar.2009.09.002
8. Goździk, K, Engström, A, and Höglund, J. Optimization of in-house ELISA based on recombinant major sperm protein (rMSP) of Dictyocaulus viviparus for the detection of lungworm infection in cattle. Res Vet Sci. (2012) 93:813–8. doi: 10.1016/j.rvsc.2011.09.016
9. Vanhecke, M, Charlier, J, Strube, C, and Claerebout, E. Association between Dictyocaulus viviparus bulk tank milk antibody levels and farmer-reported lungworm outbreaks. Vet Parasitol. (2020) 288:109280. doi: 10.1016/j.vetpar.2020.109280
10. Pyziel, AM, Laskowski, Z, and Höglund, J. Development of a multiplex PCR for identification of Dictyocaulus lungworms in domestic and wild ruminants. Parasitol Res. (2015) 114:3923–6. doi: 10.1007/s00436-015-4657-y
11. Höglund, J, Wilhelmsson, E, Christensson, D, Mörner, T, Waller, P, and Mattsson, JG. ITS2 sequences of Dictyocaulus species from cattle, roe deer and moose in Sweden: molecular evidence for a new species. Int J Parasitol. (1999) 29:607–11. doi: 10.1016/S0020-7519(98)00229-X
12. Pyziel, AM, Laskowski, Z, Demiaszkiewicz, AW, and Höglund, J. Interrelationships of Dictyocaulus spp. in wild ruminants with morphological description of Dictyocaulus cervi n. sp. (Nematoda: Trichostrongyloidea) from Red Deer Cervus elaphus. J Parasitol. (2017) 103:506–18. doi: 10.1645/16-75
13. Schnieder, T, Epe, C, and von Samson-Himmelstjerna, G. Species differentiation of lungworms (Dictyocaulidae) by polymerase chain reaction/restriction-fragment-length polymorphism of second internal transcribed spacers of ribosomal DNA. Parasitol Res. (1996) 82:392–4. doi: 10.1007/s004360050134
14. Deng, MH, Zhong, LY, Kamolnetr, O, Limpanont, Y, and Lv, ZY. Detection of helminths by loop-mediated isothermal amplification assay: a review of updated technology and future outlook. Infect Dis Poverty. (2019) 8:20. doi: 10.1186/s40249-019-0530-z
15. García-Bernalt Diego, J, Fernández-Soto, P, and Muro, A. LAMP in neglected tropical diseases: a focus on parasites. Diagnostics. (2021) 11:521. doi: 10.3390/diagnostics11030521
16. Garg, N, Ahmad, FJ, and Kar, S. Recent advances in loop-mediated isothermal amplification (LAMP) for rapid and efficient detection of pathogens. Curr Res Microb Sci. (2022) 3:100120. doi: 10.1016/j.crmicr.2022.100120
17. Antonopoulos, A, Higgins, O, Doyle, SR, Bartley, D, Morrison, A, Shalaby, MM, et al. Real-time single-base specific detection of the Haemonchus contortus S168T variant associated with levamisole resistance using loop-primer endonuclease cleavage loop-mediated isothermal amplification. Mol Cell Probes. (2024) 73:101946. doi: 10.1016/j.mcp.2023.101946
18. Khangembam, R, Tóth, M, Vass, N, Várady, M, Czeglédi, L, Farkas, R, et al. Point of care colourimetric and lateral flow LAMP assay for the detection of Haemonchus contortus in ruminant faecal samples. Parasite. (2021) 28:82. doi: 10.1051/parasite/2021078
19. Melville, L, Kenyon, F, Javed, S, McElarney, I, Demeler, J, and Skuce, P. Development of a loop-mediated isothermal amplification (LAMP) assay for the sensitive detection of Haemonchus contortus eggs in ovine faecal samples. Vet Parasitol. (2014) 206:308–12. doi: 10.1016/j.vetpar.2014.10.022
20. Nak-on, S, Sabaijai, M, Raksaman, A, Panich, W, Tejangkura, T, and Chontananarth, T. Visualization and development of colorimetric loop-mediated isothermal amplification for the rapid detection and diagnosis of paramphistome infection: colorimetric PAR-LAMP. Parasitol Res. (2024) 123:126. doi: 10.1007/s00436-024-08150-z
21. Nak-on, S, Tejangkura, T, and Chontananarth, T. Multi-detection for paramphistomes using novel manually designed PAR-LAMP primers and its application in a lateral flow dipstick (LAMP-LFD) system. Vet Parasitol. (2023) 317:109905. doi: 10.1016/j.vetpar.2023.109905
22. Panich, W, Nak-on, S, Sabaijai, M, Raksaman, A, Puttharugsa, C, Tejangkura, T, et al. Evaluation of semi-quantitative colorimetric assays based on loop-mediated isothermal amplification indicators by using image analysis. Anal Biochem. (2024) 688:115481. doi: 10.1016/j.ab.2024.115481
23. Campbell, P, Forbes, A, McIntyre, J, Bartoschek, T, Devine, K, O'Neill, K, et al. Inefficacy of ivermectin and moxidectin treatments against Dictyocaulus viviparus in dairy calves. Vet Rec. (2024) 195:e4265. doi: 10.1002/vetr.4265
24. Bisset, SA, Knight, JS, and Bouchet, CLG. A multiplex PCR-based method to identify strongylid parasite larvae recovered from ovine faecal cultures and/or pasture samples. Vet Parasitol. (2014) 200:117–27. doi: 10.1016/j.vetpar.2013.12.002
25. Corpet, F . Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res. (1988) 16:10881–90. doi: 10.1093/nar/16.22.10881
26. McKeand, JB . Vaccine development and diagnostics of Dictyocaulus viviparus. Parasitology. (2000) 120:17–23. doi: 10.1017/S0031182099005727
27. Buddhachat, K, Sriuan, S, Nak-On, S, and Chontananarth, T. Differentiating paramphistome species in cattle using DNA barcoding coupled with high-resolution melting analysis (Bar-HRM). Parasitol Res. (2023) 122:769–79. doi: 10.1007/s00436-022-07769-0
28. Higgins, O, and Smith, TJ. Loop-primer endonuclease cleavage-loop-mediated isothermal amplification technology for multiplex pathogen detection and single-nucleotide polymorphism identification. J Mol Diagn. (2020) 22:640–51. doi: 10.1016/j.jmoldx.2020.02.002
29. Higgins, O, Chueiri, A, O’Connor, L, Lahiff, S, Burke, L, Morris, D, et al. Portable differential detection of ctx-m esbl gene variants, blactx-m-1 and blactx-m-15, from escherichia coli isolates and animal fecal samples using loop-primer endonuclease cleavage loop-mediated isothermal amplification. Microbiol. Spectr. (2023) 11:e0331622. doi: 10.1128/spectrum.03316-22
30. Bai, H, Wang, Y, Li, X, and Guo, J. Electrochemical nucleic acid sensors: competent pathways for mobile molecular diagnostics. Biosens Bioelectron. (2023) 237:115407. doi: 10.1016/j.bios.2023.115407
31. Sharma, AS, and Lee, NY. Advancements in visualizing loop-mediated isothermal amplification (LAMP) reactions: a comprehensive review of colorimetric and fluorometric detection strategies for precise diagnosis of infectious diseases. Coord Chem Rev. (2024) 154:112069. doi: 10.1016/j.ccr.2024.215769
32. Trotter, M, Borst, N, Thewes, R, and von Stetten, F. Review: electrochemical DNA sensing—principles, commercial systems, and applications. Biosens Bioelectron. (2020) 154:112069. doi: 10.1016/j.bios.2020.112069
33. Zhang, H, Xu, Y, Fohlerova, Z, Chang, H, Iliescu, C, and Neuzil, P. LAMP-on-a-chip revising microfluidic platforms for loop-mediated DNA amplification. TrAC Trends Anal Chem. (2019) 113:44–53. doi: 10.1016/j.trac.2019.01.015
Keywords: nematode, lungworm, diagnosis, loop-mediated isothermal amplification, colorimetry, lateral flow system
Citation: Nak-on S, Campbell P, Shalaby MM, McIntyre J, Antonopoulos A, Chontananarth T and Laing R (2024) Development of a loop-mediated isothermal amplification detection assay for Dictyocaulus viviparus (Bloch, 1782) lungworm: DviLAMP. Front. Vet. Sci. 11:1454065. doi: 10.3389/fvets.2024.1454065
Edited by:
Nicola Pugliese, University of Bari Aldo Moro, ItalyReviewed by:
Rayan De Paula, Federal University of Minas Gerais, BrazilLynsey Melville, Moredun Research Institute, United Kingdom
Copyright © 2024 Nak-on, Campbell, Shalaby, McIntyre, Antonopoulos, Chontananarth and Laing. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sirapat Nak-on, Mjk0MjAxNW5Ac3R1ZGVudC5nbGEuYWMudWs=; bWF4bW9zaXJhcGF0QGdtYWlsLmNvbQ==; Roz Laing, cm9zYWxpbmQubGFpbmdAZ2xhc2dvdy5hYy51aw==
†ORCID: Sirapat Nak-on, https://orcid.org/0000-0001-8726-5973
Paul Campbell, https://orcid.org/0000-0001-5418-6566
Maha Mansour Shalaby, https://orcid.org/0000-0002-8153-0278
Jennifer McIntyre, https://orcid.org/0000-0003-2579-6693
Alistair Antonopoulos, https://orcid.org/0000-0002-9232-9246
Thapana Chontananarth, https://orcid.org/0000-0002-4241-6966
Roz Laing, https://orcid.org/0000-0002-6191-0759
 Sirapat Nak-on
Sirapat Nak-on Paul Campbell
Paul Campbell Maha Mansour Shalaby
Maha Mansour Shalaby Jennifer McIntyre
Jennifer McIntyre Alistair Antonopoulos
Alistair Antonopoulos Thapana Chontananarth
Thapana Chontananarth Roz Laing
Roz Laing